Abstract
Engineered microbial biosynthesis of plant natural products can support manufacturing of complex bioactive molecules and enable discovery of non-naturally occurring derivatives. Purine alkaloids, including caffeine (coffee), theophylline (antiasthma drug), theobromine (chocolate), and other methylxanthines, play a significant role in pharmacology and food chemistry. Here, we engineered the eukaryotic microbial host Saccharomyces cerevisiae for the de novo biosynthesis of methylxanthines. We constructed a xanthine-to-xanthosine conversion pathway in native yeast central metabolism to increase endogenous purine flux for the production of 7-methylxanthine, a key intermediate in caffeine biosynthesis. Yeast strains were further engineered to produce caffeine through expression of several enzymes from the coffee plant. By expressing combinations of different N-methyltransferases, we were able to demonstrate re-direction of flux to an alternate pathway and develop strains that support the production of diverse methylxanthines. We achieved production of 270 μg/L, 61 μg/L, and 3700 μg/L of caffeine, theophylline, and 3-methylxanthine, respectively, in 0.3-L bench-scale batch fermentations. The constructed strains provide an early platform for de novo production of methylxanthines and with further development will advance the discovery and synthesis of xanthine derivatives.
Keywords: plant natural products, purine alkaloids, methylxanthines, synthetic biology, yeast
1. Introduction
Plant natural products exhibit a diverse range of therapeutic activities, ranging from anti-microbial, analgesic, to anti-cancer activities (Cragg and Newman, 2013). Unfortunately, the production process of these important compounds often represents a significant barrier toward inexpensive compound discovery for pharmaceutical development and commercial use (Leonard et al., 2009). Traditional production of many natural products relies on laborious and resource intensive cultivation and extraction of the desired compounds from plant biomass. These strategies face long production times and the intensive resource requirements associated with growing the natural plant hosts, which can result in a low or fluctuating supply of essential medicines (Leonard et al., 2009). Synthetic chemistry can provide an alternative approach to synthesizing natural products; however, toxic chemicals and petroleum-based sources are often required and many natural products are too complex for conventional synthesis. The desire to implement scalable, greener synthesis strategies has led researchers to engineer microorganisms, such as yeast or bacteria, to produce valuable plant-derived compounds (Sun et al., 2015).
Several recent examples have highlighted the exciting potential of engineered biosynthetic pathways in microbial hosts for the production of natural products (Brown et al., 2015; Galanie et al., 2015; Paddon et al., 2013). In particular, heterologous expression of entire biosynthetic pathways in yeast enables fermentative production of complex molecules in a single vessel, leading to increased efficiency and yield by eliminating intermediate isolation steps. Other benefits of heterologous expression in microbes include flexibility in engineering alternate pathways, and the potential for producing intermediates that do not naturally accumulate in the native plant host (Thodey et al., 2014). In addition, our ability to reconstruct biosynthetic pathways in microbial hosts can facilitate the synthesis of semisynthetic derivatives, which can further clinical application of natural products (Thodey et al., 2014).
Alkaloids are an important class of structurally-diverse, nitrogen-containing plant natural products. Purine alkaloids are essential for life - two of the five bases in nucleic acids, adenine and guanine are purine alkaloids. Organisms can make purines, but can also scavenge from the environment, or salvage endogenous molecules (Peifer et al., 2012). In plants, purines are utilized in secondary metabolism resulting in the synthesis of purine alkaloids commonly found as xanthine-based compounds (e.g., caffeine, theophylline, and 3-methylxanthine) that have a significant role in pharmacology and food chemistry. In particular, caffeine represents one of the most consumed psychoactive substances in the world, and also helps reverse the effects of sleep deprivation on alertness and cognition (Nall et al., 2016). Theophylline, another xanthine derivative, was first prescribed for the treatment of asthma in 1937 and has since been widely used as an oral treatment of both asthma and chronic obstructive pulmonary disease (COPD) (Page, 2010). Other xanthine derivatives have also been demonstrated to have clinical effects on various biomedical targets, including adenosine receptors (Muller and Jacobson, 2011), acetylcholinesterase (Mohamed et al., 2013), phosphodiesterases (Fisone et al., 2004), ryanodine receptors (Guerreiro et al., 2011), monoamine oxidase (Carradori and Silvestri, 2015), and GABA A receptors (Daly, 2007). Although derivatives can be produced through chemical synthesis, the process is difficult due to the challenges in achieving selective alkylation or modification of each of the nitrogen atoms (Summers et al., 2015).
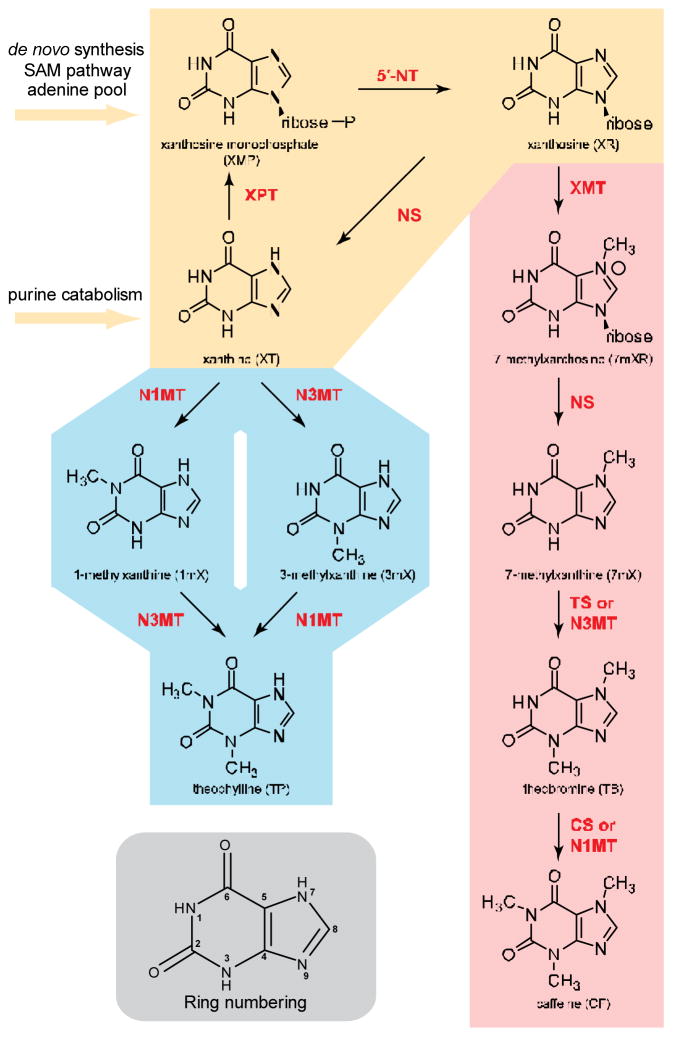
Engineering a caffeine biosynthesis pathway in microbial hosts can provide a promising route to the production of methylxanthines and their derivatives. Three N-methylation reactions and a nucleosidation reaction are involved in the pathway to convert endogenous xanthine to caffeine (Fig. 1). The first enzyme in the pathway (xanthosine N-methyltransferase, XMT) methylates xanthosine at the N7 position to yield 7-methylxanthosine (Misako and Kouichi, 2004). Subsequent removal of ribose from 7-methylxanthosine by endogenous yeast nucleosidases produces 7-methylxanthine, which serves as a substrate for the next N-methyltransferase (theobromine synthase, TS) to produce theobromine. The last step occurs at the N1 position and is catalyzed by caffeine synthase (CS) to yield caffeine as the final product (Ishida et al., 2009). Multiple variants have been identified for the different pathway enzymes from the plants Coffea arabica (coffee) and Camellia sinensis (tea) (Ashihara et al., 2008; Kato et al., 2010). In vitro characterization studies have demonstrated that these enzyme variants exhibit a range of catalytic activities and substrate specificities (Uefuji et al., 2003; Yoneyama et al., 2006).
Fig. 1.
(1.5 column) Engineered pathway for de novo production of diverse methylxanthines. Yellow: changes to central metabolism. Pink: major caffeine biosynthetic pathway from plants. Blue: minor pathway engineered in S. cerevisiae. Enzymes are abbreviated in red: 5′-NT, 5′-nucleotidase; XPT, xanthine phosphoribosyltransferase; NS, nucleosidase; XMT, xanthosine N-methyltransferase; TS, theobromine synthase; N3MT, N3-methyltransferase; CS, caffeine synthase; N1MT, N1-methyltransferase. Gray: Xanthine ring numbering.
Several of the enzymes involved in caffeine biosynthesis have demonstrated activity when expressed in microbial hosts. However, full reconstruction of the pathway and de novo production of xanthine-based alkaloids has not been previously achieved. Earlier work demonstrated the partial reconstruction of this pathway in two separately engineered hosts (Escherichia coli or Saccharomyces cerevisiae) via the heterologous expression of two enzymes: an XMT from C. arabica (CaXMT1) and a CS from C. sinensis (CsTCS) (Jin et al., 2014). The biosynthesis of 7-methylxanthine was demonstrated in the bacterial system, whereas the biosynthesis of caffeine from fed xanthosine was demonstrated in the yeast system. In a more recent study, researchers produced methylxanthines in E. coli strains via the heterologous expression of an enzyme that converts fed theophylline to 3-methylxanthine (Algharrawi et al., 2015). A reliable route to de novo synthesis of plant purine alkaloids from basic nutrient sources in a microbial host has remained unachieved to date. The challenge to total biosynthesis of purine alkaloids may lie in the need for re-directing metabolic flux from endogenous nucleotide biosynthesis, which is one of the most energy-consuming yet critical segments of central metabolism. In contrast, most other classes of alkaloids have precursors from amino acids (Trenchard et al., 2015) or fatty acids (Brown et al., 2015).
In this work, we describe the de novo biosynthesis of plant purine alkaloids and diverse methylxanthines using the microbial host S. cerevisiae. We began with rational modifications to the native yeast central metabolic pathway to improve endogenous purine flux, enabling the total synthesis of a key intermediate 7-methylxanthine from simple nutrient sources (yellow, Fig. 1). We then completed the caffeine pathway (pink, Fig. 1) by heterologously expressing a variety of enzymes from C. arabica, and identified enzyme variants that exhibit the desired activity in the context of the microbial host for caffeine production. Finally, by combining different N-methyltransferases, we were able to re-direct flux to an alternate pathway (blue, Fig. 1) and further engineer selective production of diverse methylxanthines. A bench scale 0.3-L bioreactor experiment resulted in titers as high as 274 μg/L, 61.4 μg/L and 3700 μg/L of caffeine, theophylline, and 3-methylxanthine, respectively.
2. Materials and Methods
2.1 Methylxanthines
The following molecules were purchased from Sigma-Aldrich (St. Louis, MO): xanthine (LOT# 103k5326), xanthosine dehydrate (LOT# SLBG2848V), 1-methylxanthine (LOT# BCBH6759V), 7-methylxanthine (LOT# BCBM7725V), paraxanthine (LOT# 118K1448), theobromine (LOT# 094K2500), theophylline (LOT# MKBS5105V) and caffeine (LOT# MKBQ2780V). 3-methylxanthine (LOT# 1733J) was obtained from MP Biomedicals (Burlingame, CA). Abbreviations of chemicals used in this study: xanthine (XT), xanthosine (XR), xanthosine monophosphate (XMT), 1-methylxanthine (1mX), 3-methylxanthine (3mX), 7-methylxanthine (7mX), 7-methylxanthosine (7mXR), theophylline (TP), theobromine (TB), paraxanthine (PX), and caffeine (CF).
2.2 Yeast strain and plasmid construction
Oligonucleotides used in this work were synthesized by either Stanford Protein and Nucleic Acid Facility (Stanford, CA) or Integrated DNA Technologies (IDT; Coralville, CA). Gene sequences for enzymes yCCS1 and yXMT1 were codon-optimized to increase expression in S. cerevisiae using GeneArt GeneOptimizer program (Thermo Fisher Scientific), and synthesized as standard gBlock fragments for assembly (IDT). Each genetic fragment was subsequently PCR amplified, and assembled into a full size coding sequence using the Gibson method (Gibson et al., 2009). DNA polymerase Pfu Hotstart or PfuUltra II (Agilent Technologies) was used to amplify DNA fragments shorter or longer than 2 kb, respectively. PCR products were cleaned up using QIAquick PCR purification kit (QIAGEN) and eluted in water or 10 mM Tris-HCl, pH 8.5.
The engineered S. cerevisiae (budding yeast) strains in this work were derived from the strain CSY893, commonly known as CEN.PK2-1D (MATα ura3-52; trp1-289; leu2-3, 112; trp1-1; his3Δ1; MAL2-8L; SUC2) (Entian and Kötter, 2007). Yeast strains were grown non-selectively in either yeast extract peptone dextrose (YPD) media, or yeast nitrogen base (YNB) defined media (Becton, Dickinson and Company (BD)) supplemented with synthetic complete amino acid mixture (YNB-SC; Clontech). Yeast strains with auxotrophic selection markers (either URA, LEU, URA/TRP, or URA/HIS) were grown selectively in standard yeast nitrogen base (YNB) defined media (BD) and supplemented with the proper amino acid drop out solution (Clontech) to maintain selection (YNB-DO). The YNB media was supplied with 2% dextrose as the carbon source and its pH was adjusted to 5.8 using 5 M sodium hydroxide. Yeast YPD or YNB agar plates were also made for each selection condition.
To integrate the upstream pathway genes CaXMT1 (or yXMT1), PA0065, and XPT1 into the genome of CSY893 (CEN.PK2-1D), individual holding vectors were constructed, which comprised the gene flanked by a unique promoter and terminator pair (Table S1). Holding vectors were constructed by amplifying the backbone, including the promoter and terminator, and assembling with the gene insert via Gibson cloning (Gibson et al., 2009) and sequence verified. Yeast chromosomal modifications were made using the CRISPRm method (Ryan et al., 2014) and yeast DNA assembler (Shao et al., 2009). Expression cassettes were amplified with Expand High Fidelity Polymerase (Roche) using primers with 20–40 bp overlap for gap repair in yeast.
Each assembly was flanked by an integration microhomology region of 30–40 base pairs. 50–300 ng of each PCR product was co-transformed with a CRISPR-Cas9/gRNA expression vector targeted to Lyp1 (Addgene plasmid # 60847) using standard yeast electroporation. Successful integrants were isolated by plating transformations onto leucine-deficient selection plates and chromosomal modifications were verified by yeast colony PCR (Kwiatkowski et al., 1990). Engineered yeast strains used in this work are listed in Table 1.
Table 1.
Yeast strains used in this study.
| Name | Description | References |
|---|---|---|
| CSY893 | CEN.PK2-1Dα (ura3-52; trp1-289; leu2-3, 112; his3Δ1; MAL2-8C; SUC2) | (Entian and Kötter, 2007) |
| CSY1121 | CSY893 + pCS3523 | This study |
| CSY1122 | CSY893 with lyp1::LEU2, PGPD-PA0065-TADH1, PTEF1-CaXMT1-TCYC1 | This study |
| CSY1123 | CSY893 with lyp1::PTPI1-XPT1-TSTE2, LEU2, PTEF1-CaXMT1-TCYC1 | This study |
| CSY1124 | CSY893 with lyp1::PTPI1-XPT1-TSTE2, LEU2, PGPD-PA0065-TADH1 | This study |
| CSY1125 | CSY893 with lyp1::PTPI1-XPT1-TSTE2, LEU2, PGPD-PA0065-TADH1, PTEF1-CaXMT1-TCYC1 | This study |
| CSY1126 | CSY893 with lyp1::PTPI1-XPT1-TSTE2, LEU2, PGPD-PA0065-TADH1, PTEF1-yXMT1-TCYC1 | This study |
| CSY1127 | CSY1126 + pCS3530 | This study |
| CSY1128 | CSY1126 + pCS3531 | This study |
| CSY1129 | CSY1125 + pCS3530 | This study |
| CSY1130 | CSY1125 + pCS3531 | This study |
| CSY1131 | CSY1125 + pCS3523 + pCS3531 | This study |
| CSY1132 | CSY1125 + pCS3524 + pCS3531 | This study |
| CSY1133 | CSY1125 + pCS3523 + pCS3532 | This study |
| CSY1134 | CSY1125 + pCS3523 + pCS3533 | This study |
| CSY1135 | CSY1125 + pCS3526 | This study |
| CSY1136 | CSY1124 + pCS3526 | This study |
| CSY1137 | CSY1124 + pCS3527 | This study |
| CSY1138 | CSY1124 + pCS3531 | This study |
| CSY1139 | CSY1124 + pCS3531 + pCS3525 | This study |
| CSY1140 | CSY1124 + pCS3531 + pCS3528 | This study |
| CSY1149 | CSY1125 + pCS1 | This study |
| CSY1150 | CSY1125 + pCS3618 | This study |
| CSY1151 | CSY1133 + pCS1 | This study |
| CSY1152 | CSY1133 + pCS3618 | This study |
The yeast plasmid-based genetic expression cassettes in this work (Table 2) were constructed using Gateway Cloning Technology (Life Technologies). Each enzyme coding sequence was amplified by PCR and recombined into pDONR211 (Life Technologies) with BP clonase II to construct its corresponding pENTR vector. The expression plasmid for each enzyme was subsequently built by recombining its pENTR vector with a pAG expression destination vector (Alberti et al., 2007) using LR clonase II (Life Technologies). All yeast expression plasmids were sequence-verified (Elim Biopharmaceuticals Inc) and transformed into a TOP10 E. coli strain (Life Technologies) for propagation and storage. Plasmids were prepared from overnight selective LB liquid media (EMD Millipore) under either ampicillin (50 mg/L) or kanamycin (50 mg/L) selection, using Econospin columns (Epoch Life Science) according to the manufacturer’s instructions. To build yeast strains with plasmid-based expression cassettes, one or two plasmids were directly transformed into yeast cells using the standard lithium acetate protocol (Gietz and Schiestl, 2007). Yeast cells harboring plasmids were grown under the appropriate selection conditions for plasmid maintenance.
Table 2.
Plasmids used in this study.
| Name | Description | References |
|---|---|---|
| Gateway destination vectors | ||
| pAG414GPD-ccdB | Centromeric TRP PGPD-ccdB-TCYC1 | (Alberti et al., 2007) |
| pAG416ADH1-ccdB | Centromeric URA PADH1-ccdB-TCYC1 | This study |
| pAG416GPD-ccdB | Centromeric URA PGPD-ccdB-TCYC1 | (Alberti et al., 2007) |
| pAG416HXT7-ccdB | Centromeric URA PHXT7-ccdB-TCYC1 | This study |
| pAG423GPD-ccdB | 2μ HIS PGPD-ccdB-TCYC1 | (Alberti et al., 2007) |
| Holding vectors | ||
| pCS2656 | PGPD-T6ODM-TADH1 | (Thodey et al., 2014) |
| pCS2657 | PTEF1-CODM-TCYC1 | (Thodey et al., 2014) |
| pCS2661 | PTPI1-COR1.3-TSTE2 | (Thodey et al., 2014) |
| pCS3428 | PTEF1-yXMT1-TCYC1 | This study |
| pCS3429 | PTEF1-CaXMT1-TCYC1 | This study |
| pCS3430 | PGPD-PA0065-TADH1 | This study |
| pCS3431 | PTPI1-XPT1-TSTE2 | This study |
| Gateway product vectors | ||
| pCS3523 | Centromeric TRP PGPD-CaXMT1-TCYC1 | This study |
| pCS3524 | 2μ HIS PGPD-CaXMT1-TCYC1 | This study |
| pCS3525 | Centromeric TRP PGPD-CaMXMT1-TCYC1 | This study |
| pCS3526 | Centromeric URA PGPD-CaMXMT1-TCYC1 | This study |
| pCS3527 | Centromeric URA PGPD-CaMXMT2-TCYC1 | This study |
| pCS3528 | Centromeric TRP PGPD-CaDXMT1-TCYC1 | This study |
| pCS3529 | Centromeric URA PGPD-CaDXMT1-TCYC1 | This study |
| pCS3530 | Centromeric URA PGPD-CaCCS1-TCYC1 | This study |
| pCS3531 | Centromeric URA PGPD-yCCS1-TCYC1 | This study |
| pCS3532 | Centromeric URA PHXT7-yCCS1-TCYC1 | This study |
| pCS3533 | Centromeric URA PADH1-yCCS1-TCYC1 | This study |
| pCS3618 | Centromeric HIS PGPD-SAM2-TCYC1 | This study |
| Control vectors | ||
| pCS1 | Centromeric HIS empty control | This study |
2.3 Culture conditions for assays
Yeast cultures were grown in two different scales for metabolite production: 1) milliliter lab-scale 96-well plates and 2) liter-scale bioreactor. In the lab-scale closed-batch condition (200 to 300 μL) cultures were grown in 2 mL deep-well 96-well plates (USA Scientific) covered by an AeraSeal film (Excel Scientific). An approximate 1:10 ratio was kept between the culture and container volumes to maintain good aeration of the cultures. The plate-based cultures were grown in an LT-X plate shaker (Adolf Kühner), shaking at 480 rpm with 80% humidity. Single colonies of each strain were inoculated in triplicate into YNB media under appropriate dropout selection conditions, and grown overnight at 30°C. The next day, the stationary phase culture was back-diluted and grown for an additional 3 or 6 days before harvesting the media for metabolite characterization.
For enhanced closed-batch culture conditions, strains were cultured in a Biostat Q-plus bioreactor with 0.5 L univessel size (Sartorius Stedim Biotech). Initial media volume was 0.3 L of 5χ YNB-DO at pH 5.5 with 10% glucose. Single colonies of freshly transformed yeast strains were streaked on YNB-DO agar, grown for 24 hours, and then inoculated into 3 mL of the appropriate YNB-DO media and grown in 16 × 150 mm glass culture tubes for 24 hours at 30 °C in a shaking incubator at 260 rpm. Cultures were back-diluted 1:10 in 50 mL YNB-DO in 250-mL shake flasks and grown to OD600 4 to 6, concentrated, and used to inoculate the bioreactor vessels. Dissolved oxygen was calibrated at 100%. Process parameters were held constant during the fermentation at 30°C, with controlled pH at 5.5, stirring at 300 rpm and 0.2 lpm air flow. Samples of 1 to 2 mL were taken to measure OD600 and methylxanthine concentrations throughout the course of the fermentation.
2.4 Analysis of metabolite production
Aliquots of yeast culture samples were centrifuged at 15,000 rpm (10 min, 4°C) to remove cells and debris from the media. The supernatant was carefully sampled without disturbing the cell pellet and analyzed by LC–MS/MS using an Agilent 1260 infinity binary pump HPLC and Agilent 6420 triple quadrupole mass spectrometer with an electrospray ionization source. Chromatography was performed with a Zorbax Eclipse Plus C18 column (2.1 × 50 mm, 1.8 μm; Agilent Technologies). Mobile phase A was water with 0.1% formic acid, phase B was acetonitrile with 0.1% formic acid, and the flow rate was 0.4 mL/min at 40 °C. 2 μL samples were injected and separated with the following method: 2% B for 0.5 min, 2–10% B gradient for 4.5 min, 95% B for 2.5 min, and re-equilibrate for 2 min. The eluent was directed to the MS for 0.8–7 min with ESI source gas temperature at 350°C, gas flow of 11 L/min, nebulizer pressure of 40 PSI. The quantification of small molecules was based on the integrated peak area of the MRM chromatograms. The MRM transitions used are listed in Table 3, each of which was determined using the MassHunter Optimizer software (Agilent) with appropriate standards. MRM peak areas were compared to a calibration curve of external standard peak areas to determine concentration.
Table 3.
MRM transition for purine alkaloid compound identification and quantification.
| Compound | Quantifier | Qualifier | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| MRM Transition | Fragmentor | CE | MRM Transition | Fragmentor | CE | |
| 1-methylxanthine | 167→110 | 124 | 21 | 167→55 | 124 | 33 |
| 3-methylxanthine | 167→124 | 118 | 17 | 167→42 | 113 | 33 |
| 7-methylxanthine | 167→124 | 113 | 17 | 167→42 | 113 | 33 |
| Theobromine | 181→138 | 108 | 17 | 181→110 | 108 | 21 |
| Theophylline | 181→124 | 108 | 17 | 181→96 | 108 | 25 |
| Paraxanthine | 181→124 | 108 | 17 | 181→96 | 108 | 25 |
| Caffeine | 195→138 | 122 | 17 | 195→110 | 122 | 25 |
3. Results
3.1 Engineering a methylxanthine scaffold-producing microbial strain for de novo biosynthesis
E. coli and S. cerevisiae are commonly used microbial hosts for heterologous metabolite production because of their rapid growth, ability to stably maintain plasmids, and abundant genetic tools supporting pathway manipulation (Pourmir and Johannes, 2012). We selected S. cerevisiae over E. coli as our host for plant natural product production due to a closer genealogical distance between plants and fungi, leading to potentially better functional heterologous expression of plant enzymes (Ostergaard et al., 2000). In addition, the end-product of this pathway (caffeine) is reported to exhibit antibacterial activity, which can inhibit growth (Almeida et al., 2006; Sandlie et al., 1980) and therefore limit final production levels (Jin et al., 2014).
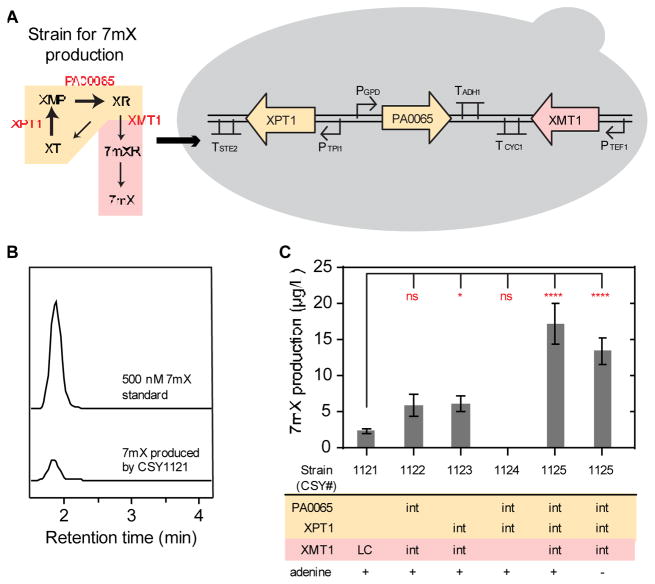
Our initial efforts focused on re-directing metabolic flux from central metabolism into our target caffeine pathway, specifically converting endogenous xanthosine into 7-methylxanthosine and then to 7-methylxanthine. Production levels of 7-methylxanthine in yeast culture medium were used to quantify redirection of flux into the heterologous pathway. In our early experiments we observed that intracellular concentrations of xanthosine were not detected in cell lysate, which is supported by other reports in the literature (Peifer et al., 2012). Moreover, 7-methylxanthosine is not available as a standard, making it difficult to quantify this intermediate. Thus, we set up the early pathway to produce 7-methylxanthine, which is the next pathway product after nucleosidation of 7-methylxanthosine by endogenous yeast enzymes. We selected an XMT variant (C. arabica XMT1; CaXMT1 [Q9AVK0]) that had been previously validated for activity in yeast (Jin et al., 2014), and expressed its cDNA under a strong constitutive yeast promoter (PGPD) in a low-copy plasmid. We transformed the CaXMT1-harboring plasmid into our host strain CSY893, and grew the engineered strain (CSY1121) at 30°C for 6 days. The production of 7-methylxanthine was confirmed through comparison of the retention time and fragmentation to a 7-methylxanthine standard (Fig. 2B). We were able to detect the production of 7-methylxanthine in the culture supernatant (2.3 ± 0.3 μg/L, Fig. 2C), indicating that the heterologous pathway was able to interface with yeast central metabolism. However, the accumulation of 7-methylxanthine was low, and thus we examined modifications to increase flux through this heterologous pathway.
Fig. 2.
(1.5 column) De novo synthesis of 7-methylxanthine. (A) The pathway (left) represents the engineered xanthine-to-xanthosine conversion in S. cerevisiae strain for the production of the intermediate 7-methylxanthine. Two enzymes, XPT1 (yeast endogenous, with one additional copy) and PA0065 (a 5′-NT from P. aeruginosa), are used for xanthine-to-xanthosine conversion, while XMT1 (from C. arabica) supports the production of 7-methylxanthine. The schematic yeast cell (right) illustrates the associated genetic promoters and terminators for enzyme expression when integrated in the genome. (B) LC–MS/MS analysis of 7-methylxanthine produced in the media of strain CSY1121 (bottom) and a 7-methylxanthine standard for comparison (top, + ESI MRM 167 → 124). (C) Effect of different genetic modifications and adenine supply on 7-methylxanthine production after 3 days of culture (YNB-DO with 2% dextrose). Relevant genetic contents for each strain are displayed below the horizontal bar under the plot (see Table 1 for complete strain information). Production of 7-methylxanthine was analyzed by LC–MS/MS and data are reported as the mean and standard deviation of biological triplicates. Abbreviations: int: integrated gene copy, LC: low-copy plasmid. Notations: significance summary: p > 0.05 (ns), p ≤ 0.05 (*), p ≤ 0.01 (**), p ≤ 0.001 (***), and p ≤ 0.0001 (****).
We hypothesized that the pathway bottleneck was due to limited availability of endogenous xanthosine (Peifer et al., 2012), as its ribose group can be actively removed to produce xanthine by the same group of endogenous nucleosidases. However, direct knockout of these nucleosidases is not feasible as they are required for ribose salvage and nucleotide degradation in yeast (Xu et al., 2013). In addition, de novo synthesis of purines, including xanthosine, is energetically expensive. Thus, we focused on tuning the salvage pathway for xanthosine, i.e., making xanthosine from xanthine to increase the available flux to 7-methylxanthine. This salvage pathway occurs in two steps, first converting xanthine to xanthosine monophosphate (XMP) and then to xanthosine. We targeted two enzymes to engineer this xanthosine-xanthine flux reversion - xanthine phosphoribosyltransferase (XPT) and 5′-nucleotidase (5′-NT). We identified an endogenous XPT (XPT1 [P47165]) with confirmed activity to convert xanthine to XMP; however, the reported endogenous 5′-NT exhibits low activity on purine backbones other than inosine monophosphate (IMP) (Guetsova et al., 1999). Thus, we identified a 5′-NT from Pseudomonas aeruginosa (PA0065 [Q9I767]), which had recently been characterized as a 5′-NT with significant activity against XMP (Kuznetsova et al., 2005), to complete the proposed xanthosine-xanthine flux reversion pathway. The overexpression of XPT1 and the heterologous expression of PA0065, together with the endogenous nucleosidases, completed a cyclic pathway for xanthine-to-xanthosine conversion in yeast (yellow, Fig. 1).
We integrated a copy of XPT1 and PA0065 into the yeast genome using strong constitutive promoters (PTPI1 and PGPD, respectively) (Fig. 2A). Individual modifications to purine central metabolism, i.e., overexpressing XPT1 or only expressing PA0065, resulted in approximately 2.5-fold improvement in 7-methylxanthine production compared to strain CSY1121, which relies on natural production of xanthosine (Fig. 2C). This result indicates that the overexpression of XPT1 is able to exploit xanthine as an endogenous source with the yeast 5′-NT catalyzing the phosphate removal step. This result also demonstrates that the bacterial 5′-NT (PA0065) is sufficient at pulling flux from XMP to increase production of xanthosine available for the heterologous pathway. The combination of both modifications resulted in a nearly 8-fold improvement in the production of 7-methylxanthine after 3 days of growth (Fig. 2C). Control experiments confirmed that the yeast strain could not produce 7-methylxanthine when expressing only XPT1 and PA0065 without CaXMT1 (Fig. 2C).
Endogenous xanthosine can be synthesized through three major routes in yeast: adenosine released from the SAM pathway, IMP originating from de novo purine synthesis, and the cellular adenine nucleotide pool (Ashihara et al., 2008). To ensure that our strains are capable of de novo production of xanthosine and were not relying on exogenously fed adenine, we tested the strain CSY1121 grown in the absence of adenine in the culture media. Removing adenine resulted in no significant reduction in production of 7-methylxanthine (p > 0.1), suggesting that the purine alkaloids of the heterologous pathway can be synthesized from an endogenous metabolite in S. cerevisiae grown on a simple carbon source with no additional specific substrate feeding (Fig. 2C).
3.2 Constructing the de novo biosynthetic caffeine pathway and improving its production through copy number and enzyme variant engineering
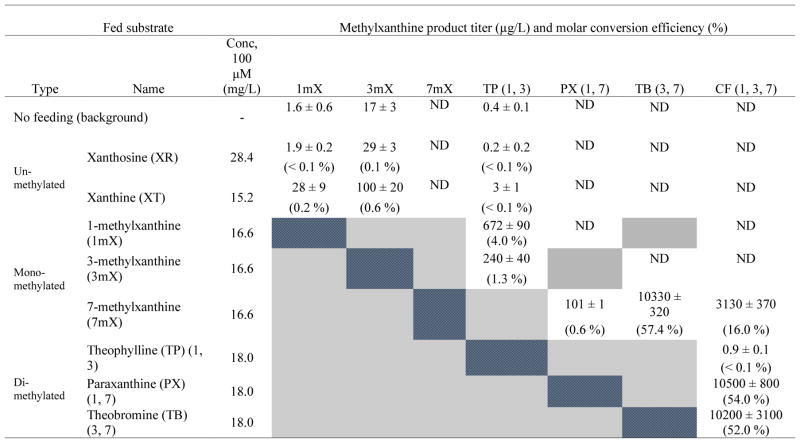
The next two steps in the caffeine biosynthesis pathway are the conversion of 7-methylxanthine to theobromine by methylating the N3 position, followed by methylation at N1 to yield caffeine. We investigated four different N-methyltransferases native to the coffee plant C. arabica (CaMXMT1, CaMXMT2, CaDXMT1, and CCS1) for their functional activities in S. cerevisiae. CaMXMT1 and CaMXMT2 are TS variants responsible for the second methylation step in the pathway while CaDXMT1 is a CS that acts on the last methylation step (Uefuji et al., 2003). CCS1, on the other hand, is known to exhibit dual-substrate activities that can carry out both methylation steps to convert 7-methylxanthine and theobromine to caffeine (Mizuno et al., 2003). To evaluate the functional activities of these N-methyltransferases in yeast, we expressed individual enzymes under the control of a constitutive promoter (PGPD) on a low-copy plasmid, and transformed each of them into our background strain (CSY893). Cells harboring each N-methyltransferase were grown with 100 μM (approximately 15.2 to 28.4 mg/L) of their reported substrates and analyzed after a three-day growth period for the production of respective methylxanthines. Commercially-available methylxanthines were used as standards to validate and quantify their production in yeast (Fig. S1). The data indicate that both TS variants are functional in yeast although CaMXMT1 was observed to be more active than CaMXMT2 in terms of producing theobromine (column “TB” in Tables S2A and S2B). This result is consistent with the KM values of these enzymes previously characterized in vitro (Uefuji et al., 2003). Production of caffeine was detected from both cell cultures harboring either CaDXMT1 or CCS1, but to different extents (column “CF” in Tables 4 and S2C). CCS1 was observed to be bi-functional and significantly more active than CaDXMT1 in terms of caffeine biosynthesis, which is also in agreement with their previously reported KM values (Mizuno et al., 2003; Uefuji et al., 2003). CCS1 had the highest activity, resulting in 57.4% conversion of 100 μM (16.6 mg/L) fed 7-methylxanthine to theobromine (compared to 40.8% and 27% for CaMXMT1 and CaMXMT2) and 52% conversion of 100 μM (18.0 mg/L) fed theobromine to caffeine (compared to 2.4% for CaDXMT1; row “7mX” in Tables 4 and S2). No methylxanthine production was detected from our background strain when feeding the substrates, supporting the activities of the N-methyltransferases specific to their heterologous expression and the corresponding substrates (Fig. S2). Taken together, these results demonstrate that all of the N-methyltransferases of C. arabica are functionally active in S. cerevisiae.
Table 4.
Comparison of methylxanthine production using yCCS1-only strain (CSY893 + pCS3531) under different fed (100 μM) intermediate substrates.
Note: The grayed-out regions are methylxanthine products that are not feasible in the reactions when feeding the indicated substrates and the blue-meshed regions represent the fed substrates themselves. Product titer is presented in the upper part of a cell and the molar conversion efficiency is in the bottom part of a cell. Molar conversion efficiency is calculated by the molarity ratio between the product and the fed substrate.
Abbreviations: ND: not detectable.
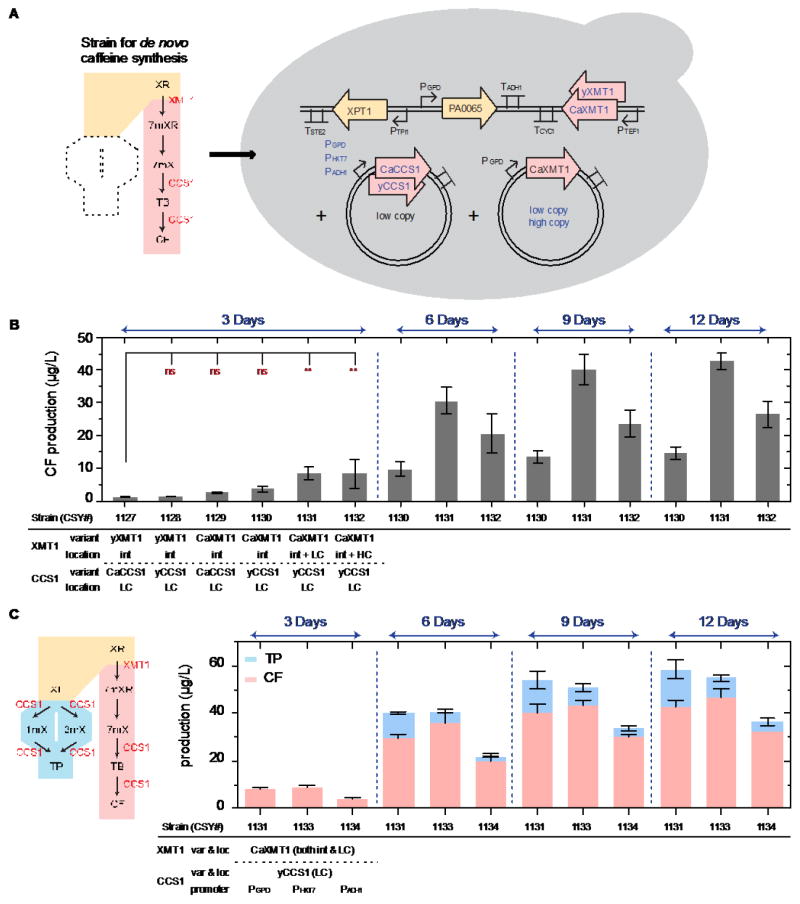
Based on these results, CCS1 was used to develop yeast strains capable of de novo caffeine biosynthesis. We added a low-copy plasmid expressing CCS1 into our optimized 7-methylxanthine producing strains (Fig. 3A) and observed a final de novo caffeine titer of 2.6 ± 0.2 μg/L after 3 days of growth (Fig. 3B). To further improve the caffeine titer, we first evaluated the effect of codon optimization and generated DNA sequence variants for each enzyme used in the biosynthesis pathway (i.e., CaXMT1 and CaCCS1 were codon-optimized to increase expression in S. cerevisiae to generate yXMT1 and yCCS1, respectively). Combinations of yeast codon-optimized and C. arabica native variants were tested, and we observed that the best combination (CaXMT1 with yCCS1; CSY1130) resulted in slightly improved caffeine titers (3.2 ± 0.2 μg/L; Fig. 3B). We examined the yXMT1 variant by expressing the enzyme alone, and observed a reduction in 7-methylxanthine levels (Fig. S3), which is likely to be the cause of the observed reduction in caffeine titers when expressing yXMT1 with CCS1 variants. Upon examining accumulation of pathway intermediates, we identified that the relatively low activity of CaXMT1 (0.5% conversion of xanthosine to 7-methylxanthine; Table S2D) was the primary bottleneck in the pathway. Therefore, we tested the effect of CaXMT1 copy number on production. An additional copy of CaXMT1 expressed on a low-copy plasmid (CSY1131) resulted in 3-fold improvement in caffeine titer, whereas expression from a high-copy plasmid (CSY1132) did not result in any significant change in caffeine production. It has been shown that artificial 2-micron plasmids are associated with instability in yeast (Ludwig and Bruschi, 1991), which may result in the observed little-to-no change in production when CaXMT1 is expressed from a high-copy plasmid. Furthermore, viability experiments (Fig. S4B) indicate that cell viability and growth is reduced in cells harboring the high-copy plasmid, which may indicate a higher metabolic burden on the cell for maintaining the plasmid and expressing the enzyme at higher levels. By increasing the culture time of CSY1131 to 6 days, we observed a final caffeine titer of 31 ± 4 μg/L, which is a 12-fold increase compared to our original de novo strain (Fig. 3B). Titers continued to modestly increase after 9 and 12 days of growth (Fig. 3B).
Fig. 3.
(1.5 column) Caffeine production and genetic optimization strategies. (A) The pathway (left) represents the de novo caffeine synthesis route (pink), with an additional enzyme CCS1 (from C. arabica) to convert 7-methylxanthine into caffeine. The schematic yeast cell (right) illustrates the optimization strategies used to improve caffeine titer, including: copy number, yeast-codon optimization, and expression level tuning (in blue font color). (B) DNA copy number and enzyme codon optimization for increased caffeine titer after 3, 6, 9, and 12 days of culture (YNB-DO with 2% dextrose). (C) An alternate route (blue, left) when expressing CCS1 in the engineered S. cerevisiae strains. The plot shows the production of both caffeine and theophylline of each strain with different CCS1 expression levels under the control of three promoters (PGPD, PHXT7, and PADH1) after 3, 6, 9, and 12 days of culture (YNB-DO with 2% dextrose). Production of caffeine and theophylline was analyzed by LC–MS/MS, and data are reported as the mean and standard deviation of biological triplicates. Relevant genetic contents for each strain are displayed below the horizontal bar under each plot (see Table 1 for complete strain information). Abbreviations: int: integrated gene copy, LC: low-copy plasmid. Notations: significance summary: p > 0.05 (ns), p ≤ 0.05 (*), p ≤ 0.01 (**), p ≤ 0.001 (***), and p ≤ 0.0001 (****).
Although we achieved improvements in caffeine titers, we noticed an accumulation of a dimethylated alternative product. This compound was confirmed to be theophylline by analyzing the compound with a chromatography method optimized for separation of paraxanthine and theophylline (see Fig. S5 and Supporting Information). We hypothesized that yCCS1 was able to methylate xanthine at the N1 and N3 positions to produce theophylline. We validated this activity by expressing yCCS1 alone in the background strain, and observed small but measureable levels of these compounds after 3 days (1.6 ± 0.6, 17 ± 3, and 0.4 ± 0.1 μg/L of 1-, 3-methylxanthine, and theophylline, respectively; Table 4). Moreover, we further confirmed the substrate branch point of this alternate pathway by feeding a yCCS1-harboring strain either xanthine or xanthosine. Feeding of xanthine to the yCCS1-harboring strain resulted in at least a 6-fold increase in production of 1-, 3-methylxanthine, and theophylline, while feeding xanthosine led to no significant increase in the level of these methylxanthines (Table 4). Overall, this suggests that yCCS1 is capable of directly methylating xanthine at the N1 and N3 positions in our S. cerevisiae host.
To mitigate the production of this alternative product, we examined the impact of promoter strength and timing on production titer and specificity. In particular, yCCS1 was expressed from a low-copy plasmid under the control of three different promoters: PGPD (constitutively strong), PHXT7 (late-stage strong), and PADH1 (constitutively weak) (Partow et al., 2010). After 6 days of culture, cells harboring either PHXT7- or PADH1-regulated yCCS1 resulted in more specific production of caffeine (88 and 91%, respectively) compared to those with the original PGPD (74%). However, the strong promoter PHXT7 maintained the high titers of total caffeine (36 ± 4 μg/L), whereas the weak PADH1 resulted in 45% reduction in final caffeine titer (Fig. 3C).
3.3. Leveraging enzyme specificity to engineer alternate methylxanthine production platforms
Due to the broad substrate specificity of the methyltransferases, alternate pathways to caffeine have been proposed (Ashihara, 2006). These alternate routes are understudied; thus, we first investigated the specificities of each of the methyltransferases in the context of the heterologous host. Xanthine, xanthosine, various monomethylated xanthines (e.g. 1-, 3-, and 7-methylxanthine) and dimethylated xanthines (e.g., theophylline, theobromine, and paraxanthine) were fed at 100 μM concentrations (approximately 16 to 18 mg/L) to yeast strains harboring each individual methyltransferase (Tables 4 and S2) in a low-copy plasmid. The data is consistent with previous in vitro studies (Misako and Kouichi, 2004), indicating that each enzyme is capable of accepting several xanthine-based substrates. However, we found one exception compared with previous specificity data: strains harboring yCCS1, CaMXMT1, or CaMXMT2, which are capable of methylating N1 and N3 (Fig. 4), resulted in relatively high conversion to 1- and 3-methylxanthine when fed xanthine (Tables 4, S2A, and S2B).
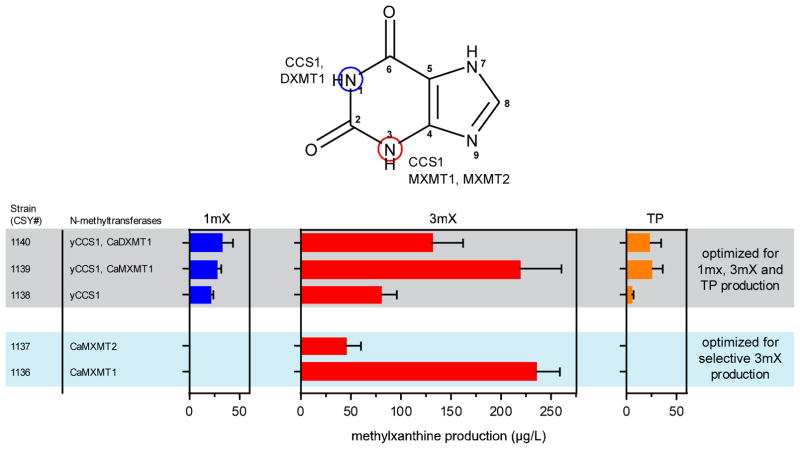
Fig. 4.
(two columns) Engineering methylxanthine production using an alternate pathway. The top molecular structure illustrates methyltransferases used in the study and their corresponding N-methylation position on the xanthine backbone. The plot below represents the titer of four different methylxanthines produced in the media of the engineered strains, optimized for 1-, 3-methylxanthine and theophylline production (grey) and for 3-methylxanthine production (light blue). Production of methylxanthines was analyzed by LC–MS/MS and data are reported as the mean and standard deviation of biological triplicates. Relevant genetic contents for each strain are displayed next to the vertical bar on the left of the plot (see Table 1 for complete strain information).
Given our early observation of theophylline accumulation in our caffeine strains (Fig. 3C), we first sought to engineer strains capable of selective production of theophylline. We hypothesized that adding either N1-methyltransferase (N1MT) or N3-methyltransferase (N3MT) to our engineered strains would help increase the flux to theophylline, which is methylated at N1 and N3. We removed CaXMT1 from our initial caffeine producing strain (CSY1130), and observed that the production of 7-methylxanthine and its downstream compounds was completely abolished. Although a higher accumulation level of 3-methylxanthine was achieved, no significant improvement in theophylline titer was observed (CSY1138 in Fig. 4). Addition of a N1MT (CaDXMT1) to this strain (CSY1138) resulted in a 5-fold increase in theophylline production (23.1 ± 11.5 μg/L; CSY1140 in Fig. 4), and a similar increase in theophylline titer was observed when adding a N3MT (CaMXMT1; 25.4 ± 10.5 μg/L, CSY1139 in Fig. 4). In both strains, we also found small increases in 1- and 3-methylxanthine, whereas 7-methylxanthine and its downstream compounds were not observed, indicating a successful diversion of the biosynthesis from the original caffeine route to our targeted theophylline pathway.
We next examined the feasibility of using our strains to selectively produce a monomethylated xanthine (i.e., 3-methylxanthine) at a higher titer, as the reactions required for this product tend to be challenging in synthetic chemistry. Selective production of 3-methylxanthine was observed in small amounts when expressing CaMXMT1 or CaMXMT2 on a low-copy plasmid in the background strain (row “No feeding” in Tables S2A and S2B). The promiscuous yCCS1was substituted with either CaMXMT1 or CaMXMT2 (as an N3MT) in our 7-methylxanthine producing strain (CSY1125) to increase selectivity in methylating at the N3 position. We tested CaMXMT1 by building strain CSY1136 and observed selective production of 3-methylxanthine (156 ± 5.6 μg/L; Fig. 4), which is 3-fold higher than that observed when using yCCS1 as an N3MT (CSY1130 in Fig. 4). We hypothesized that by completely abolishing flux towards the caffeine route (i.e., removing CaXMT1), it is possible to further increase the production of 3-methylxanthine. We expressed the CaMXMT1 in the CaXMT1-knockout strain (CSY1124) and obtained another 1.5-fold increase in 3-methylxanthine titer (235 ± 23 μg/L; CSY1136 in Fig. 4). Lower production titer was observed when using CaMXMT2 as the N3MT (44 ± 15 μg/L; CSY1137 in Fig. 4); nevertheless, both strains produced no detectable amount of other methylxanthines in the culture medium.
3.4. Demonstrating bench scale batch fermentation of optimized yeast strains for methylxanthine production
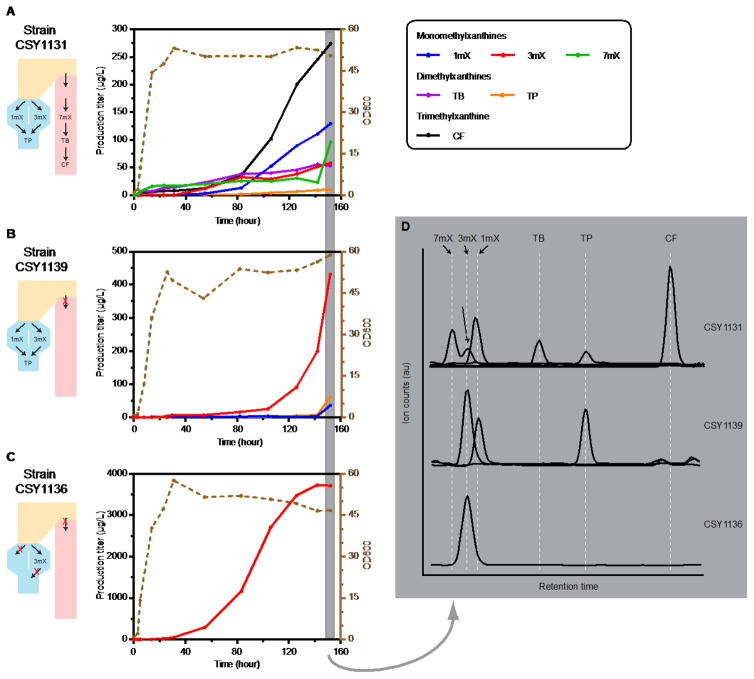
To further evaluate the engineered strains for de novo methylxanthine production, 0.3-L bench scale batch fermentations were performed in a bioreactor in parallel. 5X media concentration was used to boost cell density to an OD600 of 50. We monitored the accumulation of methylxanthine products over the course of the fermentation. Target products were detected in the culture medium after 14 hours, increased in concentration with increasing cell density, and significantly accumulating in the stationary phase over the 6-day period (Fig. 5). Strains CSY1131, CSY1139, and CSY1136 accumulated 274 μg/L, 61.4 μg/L, and 3710 μg/L of caffeine, theophylline, and 3-methylxanthine target molecules, respectively, in the culture medium during fermentation. These titers were 9-fold, ~3-fold, and 16-fold higher, respectively, than those achieved from the same strains in the plate-based assays. In particular, the production of 3-methylxanthine by strain CSY1136 was highly specific, resulting in no alternative products in the culture medium and plateaued after a six-day culture period (CSY1136 in Fig. 5D). In addition, strains CSY1131 and CSY1139 exhibited a continuing accumulation of methylxanthine products over the course of the fermentation; however, longer fermentation times did not result in increases in titers (Fig. S6).
Fig. 5.
(2 column) Bench scale batch fermentation for the production of diverse methylxanthines. Cell density and concentrations of indicated metabolites as a function of time for the fermentation of S. cerevisiae strains: (A) CSY1131, (B) CSY1139, and (C) CSY1136. Corresponding engineered pathways for each strain are displayed below its name. At indicated time points, samples were taken, diluted, and analyzed for cell density (OD600, brown y-axis on the right) through spectrometry and methylxanthine production through LC-MS/MS (black y-axis on the left). (D) LC-MS/MS chromatograms showing accumulation of diverse methylxanthines. The analysis was performed on media supernatant collected at 152 hour of culture. LC traces shown are the quantifier +ESI MRM of each compound (see details in Table 3).
4. Discussion
We demonstrated the reconstruction of a heterologous purine biosynthesis pathway in S. cerevisiae, and the de novo production of caffeine and other methylxanthines from simple carbon and nitrogen sources (i.e., sugar, ammonia). We successfully re-directed the endogenous metabolic flux to produce methylxanthines, while maintaining the ability of the engineered strains to grow normally (up to an observed OD600 ~50). The production of all monomethylated (1-, 3-, or 7-methylxanthine), dimethylated (theophylline, paraxanthine, and theobromine), and trimethylated (caffeine) xanthine products was achieved in the engineered strains. We further increased the yield and selectivity of methylxanthine production through optimizing the enzyme copy number, engineering temporal expression profiles, and optimizing the fermentation process. The three final engineered strains synthesized the following compounds de novo: 270 μg/L of caffeine (44% of total 620 μg/L produced methylxanthines; CSY1131), 61 μg/L of theophylline (11% of total 530 μg/L produced methylxanthines; CSY1139), and 3700 μg/L of 3-methyxanthine (~100% of total produced methylxanthines; CSY1136) in sub-liter scale batch fermenters.
Although our reconstructed methylxanthine pathway is of intermediate length (five steps from the central precursor XMP), this pathway interfaces significantly with the endogenous purine biosynthesis pathway. Specifically, the metabolites are at the intersection of central pathway routes that are critical for nucleotide de novo synthesis (Rolfes, 2006), co-substrate production (Zrenner et al., 2006), and cellular waste degradation (Vogels and Van der Drift, 1976), which causes engineering complexity in implementing the heterologous pathway in the yeast hosts. We addressed these challenges by using an engineered cyclic pathway (i.e., the conversion among XMP, xanthosine, and xanthine) that enables a dynamic allocation of central metabolic resources to achieve a fine balance between cellular growth and heterologous production. Toxicity assays (see Supplemental Results) of the end-products performed at concentrations up to 150 fold of current titers demonstrated no significant impact on growth or viability of the cultures (Fig. S7). Furthermore, in the fermentation experiments, we observed a growth profile that is comparable to previously reported examples (Galanie and Smolke, 2015; Thodey et al., 2014), suggesting the genetic modifications in central metabolism of our engineered strains, and the production of end-products have no or undetectable impacts on cellular growth. Our work therefore successfully implements the re-direction of endogenous purine alkaloid flux from central metabolism to a heterologous biosynthesis pathway in yeast hosts.
Previous studies described engineered yeast strains that are able to synthesize caffeine (380 μg/L) from 100 μM fed xanthosine (28 mg/L) (Jin et al., 2014), suggesting that the endogenous level of xanthosine in yeast is insufficient to support this heterologous biosynthesis pathway. Our work describes the redirection of xanthosine metabolic flux into the caffeine pathway, allowing the de novo synthesis of caffeine in yeast hosts (8.5 μg/L after 3 days). Furthermore, we observed a ~30-fold increase in caffeine titers (280 μg/L after 6 days) through optimizing growth conditions in sub-liter scale batch fermenters. Methylxanthines have also been produced by feeding a methylated substrate (theophylline) to bacteria expressing a bacterial demethylase (Algharrawi et al., 2015). This approach requires feeding high levels of methylated substrates to the engineered cells and may face limitations on the type and quantity of substrates that can be fed to or accessed by the host.
Our engineered yeast strains can serve as a microbial platform to optimize the production of intermediates in natural biosynthetic pathways. For example, monomethylated xanthines are non-accumulating metabolites in tea and coffee plants, and are also challenging to obtain through traditional chemical synthesis, since N-methylation reactions are typically non-selective (Minami et al., 2008). We demonstrated the use of a microbial platform to construct a metabolic route that selectively produces monomethylated xanthines (3-methylxanthine from CSY1136, and 7-methylxanthine from CSY1125). The current pharmaceutical applications of monomethylated xanthines and their derivatives remain largely unexploited. Further optimizing these microbial production platforms and incorporating other enzymes to synthesize different derivatives can potentially serve to provide a cheap and easy-to-access supply of these molecules to enable future studies on their therapeutic potential (Li and Vederas, 2009).
In our substrate cross-feeding experiments, we observed that the pathway enzymes exhibit a range of substrate specificities (e.g., only methylating particular substrates) and selectivities (e.g., only methylating at a particular position). For example, XMT1 exhibits both high substrate specificity (xanthosine) and strong methylation selectivity (N7 methylation; Table S2D), whereas CCS1 can act on various xanthine derivatives and perform both N1 and N3 methylations (Table 4). In some cases, increased enzyme promiscuity, as a result of losing the native regulation in heterologous expression systems, can lead to undesirable complexity of pathway reconstruction (Galanie et al., 2015). Here, we leveraged the non-specific characteristic of “jack-of-all-trades” enzymes as a substitute for a missing step to complete an alternate pathway (blue, Fig. 1) to enable the production of diverse methylxanthines that are not part of the original biosynthetic pathway. However, the compounds in the alternate pathway accumulate to low levels and are present as a minor portion of the overall methylxanthine pool (e.g., theophylline, ~ 20 μg/L and < 10% of the pool). The current market price of methylxanthines is estimated to be between $10–200 per kg (see Supplemental Information). At this price range it is estimated that titers greater than ~25 g/L would be required to approach commercialization of production of these particular methylxanthines through microbial fermentation. The quantity and specificity of target compound production can be improved with further engineering of the described pathways, including spatial engineering to localize enzymatic reactions (Thodey et al., 2014), and feedback regulation to redirect metabolic fluxes into target pathways (Zhang et al., 2012). While intracellular levels of SAM (see Supplemental Results; Fig. S8) do not appear to be limiting, future strain optimization for routing flux from central metabolism, including increasing endogenous levels of XMP from the SAM production, may improve production titers. As another example, transporter engineering may be a useful strategy to increase the quantity of our end-products; in particular intermediates in each of the pathways accumulated at high levels in the media compared to the intracellular concentrations (see Supplemental Results; Fig. S9. Finally, the activity of many of the heterologous enzymes in this pathway, mainly XMT1 and CCS1, have high Km values (78 μM and 75.1–125.6 μM, respectively) (Mizuno et al., 2003; Uefuji et al., 2003; Yoneyama et al., 2006). Thus, directed evolution of pathway enzymes can also be leveraged to improve the activity of each methyltransferase on its specific substrate (Michener and Smolke, 2012).
The engineered yeast strains and reconstructed pathways described in this work provide a foundation for biosynthesis of other natural purine alkaloids and non-naturally occurring derivatives. In addition, the described microbial platforms may be used for discovering and characterizing novel enzymes for bioremediation of xanthine-derived compounds, such as degrading caffeine in wastewater (Gokulakrishnan et al., 2005). Together with the recent advances in the total biosynthesis of other plant alkaloids (Brown et al., 2015; DeLoache et al., 2015; Galanie et al., 2015; Trenchard et al., 2015), this work provides another example of microbial biosynthesis as a flexible technology to address challenges in producing valuable plant-derived compounds.
Supplementary Material
Highlights.
Diverse methylxanthines are de novo produced from sugar in engineered yeast in a 0.3-L batch scale bioreactor.
Modifications to central metabolism allow for heterologous reconstruction of methylxanthine biosynthesis pathway.
Promoter swapping enables redirecting metabolic flux between pathways to increase product type and specificity.
Acknowledgments
We thank Dr. Alan Crozier and Dr. Hiroshi Sano for their generous donations of the plasmids harboring the enzymes CaXMT1, CaMXMT1, CaMXMT2, CaDXMT1, and CCS1 used in this work. We thank S. Galanie, Y. Li, C. Schmidt, M. Siddiqui, K. Thodey, B. Townshend, and I. Trenchard for discussions and feedback in the preparation of this manuscript. We also thank K. Thodey for help with setting up the bioreactor experiments; and Y. Li for Gateway plasmids. This work was supported by funds from the National Institutes of Health (grant to C.D.S, AT007886), Natural Sciences and Engineering Research Council of Canada (postdoctoral fellowship to M.M.), Stanford Bio-X Institute, Ministry of Education of Taiwan, Siebel Scholars Foundation (graduate student fellowships to Y.-H.W).
Appendix A. Supporting Information
Supplementary data associated with this article can be found in the online version.
Footnotes
Author contributions
M.M., Y.-H.W., M.N.W., and C.D.S. conceived of this project. M.M., Y.-H.W., and C.D.S. designed the experiments. M.M, Y.-H.W, A.C conducted the experiments. M.M., Y.-H.W., and C.D.S. analyzed the data and discussed the results. All authors wrote and edited the manuscript.
Competing interests statement
The authors declare competing financial interests in the form of a pending patent application.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alberti S, Gitler AD, Lindquist S. A suite of Gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast. 2007;24:913–919. doi: 10.1002/yea.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Algharrawi KH, Summers RM, Gopishetty S, Subramanian M. Direct conversion of theophylline to 3-methylxanthine by metabolically engineered E. coli. Microbial cell factories. 2015;14:203. doi: 10.1186/s12934-015-0395-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida AA, Farah A, Silva DA, Nunan EA, Gloria MB. Antibacterial activity of coffee extracts and selected coffee chemical compounds against enterobacteria. Journal of agricultural and food chemistry. 2006;54:8738–8743. doi: 10.1021/jf0617317. [DOI] [PubMed] [Google Scholar]
- Ashihara H. Metabolism of alkaloids in coffee plants. Brazilian Journal of Plant Physiology. 2006;18:1–8. [Google Scholar]
- Ashihara H, Sano H, Crozier A. Caffeine and related purine alkaloids: biosynthesis, catabolism, function and genetic engineering. Phytochemistry. 2008;69:841–856. doi: 10.1016/j.phytochem.2007.10.029. [DOI] [PubMed] [Google Scholar]
- Brown S, Clastre M, Courdavault V, O’Connor SE. De novo production of the plant-derived alkaloid strictosidine in yeast. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:3205–3210. doi: 10.1073/pnas.1423555112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carradori S, Silvestri R. New Frontiers in Selective Human MAO-B Inhibitors. Journal of medicinal chemistry. 2015;58:6717–6732. doi: 10.1021/jm501690r. [DOI] [PubMed] [Google Scholar]
- Cragg GM, Newman DJ. Natural products: a continuing source of novel drug leads. Biochimica et biophysica acta. 2013;1830:3670–3695. doi: 10.1016/j.bbagen.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly JW. Caffeine analogs: biomedical impact. Cellular and molecular life sciences : CMLS. 2007;64:2153–2169. doi: 10.1007/s00018-007-7051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLoache WC, Russ ZN, Narcross L, Gonzales AM, Martin VJ, Dueber JE. An enzyme-coupled biosensor enables (S)-reticuline production in yeast from glucose. Nature chemical biology. 2015;11:465–471. doi: 10.1038/nchembio.1816. [DOI] [PubMed] [Google Scholar]
- Entian K-D, Kötter P. 25 Yeast Genetic Strain and Plasmid Collections. In: Ian S, Michael JRS, editors. Methods in Microbiology. Academic Press; 2007. pp. 629–666. [Google Scholar]
- Fisone G, Borgkvist A, Usiello A. Caffeine as a psychomotor stimulant: mechanism of action. Cellular and molecular life sciences : CMLS. 2004;61:857–872. doi: 10.1007/s00018-003-3269-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanie S, Smolke CD. Optimization of yeast-based production of medicinal protoberberine alkaloids. Microbial cell factories. 2015;14:144. doi: 10.1186/s12934-015-0332-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanie S, Thodey K, Trenchard IJ, Filsinger Interrante M, Smolke CD. Complete biosynthesis of opioids in yeast. Science. 2015;349:1095–1100. doi: 10.1126/science.aac9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nature protocols. 2007;2:31–34. doi: 10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
- Gokulakrishnan S, Chandraraj K, Gummadi SN. Microbial and enzymatic methods for the removal of caffeine. Enzyme Microb Tech. 2005;37:225–232. [Google Scholar]
- Guerreiro S, Marien M, Michel PP. Methylxanthines and ryanodine receptor channels. Handbook of experimental pharmacology. 2011:135–150. doi: 10.1007/978-3-642-13443-2_5. [DOI] [PubMed] [Google Scholar]
- Guetsova ML, Crother TR, Taylor MW, Daignan-Fornier B. Isolation and characterization of the Saccharomyces cerevisiae XPT1 gene encoding xanthine phosphoribosyl transferase. Journal of bacteriology. 1999;181:2984–2986. doi: 10.1128/jb.181.9.2984-2986.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida M, Kitao N, Mizuno K, Tanikawa N, Kato M. Occurrence of theobromine synthase genes in purine alkaloid-free species of Camellia plants. Planta. 2009;229:559–568. doi: 10.1007/s00425-008-0847-5. [DOI] [PubMed] [Google Scholar]
- Jin L, Bhuiya MW, Li M, Liu X, Han J, Deng W, Wang M, Yu O, Zhang Z. Metabolic engineering of Saccharomyces cerevisiae for caffeine and theobromine production. PloS one. 2014;9:e105368. doi: 10.1371/journal.pone.0105368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Kitao N, Ishida M, Morimoto H, Irino F, Mizuno K. Expression for caffeine biosynthesis and related enzymes in Camellia sinensis. Zeitschrift fur Naturforschung C, Journal of biosciences. 2010;65:245–256. doi: 10.1515/znc-2010-3-413. [DOI] [PubMed] [Google Scholar]
- Kuznetsova E, Proudfoot M, Sanders SA, Reinking J, Savchenko A, Arrowsmith CH, Edwards AM, Yakunin AF. Enzyme genomics: Application of general enzymatic screens to discover new enzymes. FEMS microbiology reviews. 2005;29:263–279. doi: 10.1016/j.femsre.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski TJ, Jr, Zoghbi HY, Ledbetter SA, Ellison KA, Chinault AC. Rapid identification of yeast artificial chromosome clones by matrix pooling and crude lysate PCR. Nucleic acids research. 1990;18:7191–7192. doi: 10.1093/nar/18.23.7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard E, Runguphan W, O’Connor S, Prather KJ. Opportunities in metabolic engineering to facilitate scalable alkaloid production. Nature chemical biology. 2009;5:292–300. doi: 10.1038/nchembio.160. [DOI] [PubMed] [Google Scholar]
- Li JW, Vederas JC. Drug discovery and natural products: end of an era or an endless frontier? Science. 2009;325:161–165. doi: 10.1126/science.1168243. [DOI] [PubMed] [Google Scholar]
- Ludwig DL, Bruschi CV. The 2-micron plasmid as a nonselectable, stable, high copy number yeast vector. Plasmid. 1991;25:81–95. doi: 10.1016/0147-619x(91)90019-s. [DOI] [PubMed] [Google Scholar]
- Michener JK, Smolke CD. High-throughput enzyme evolution in Saccharomyces cerevisiae using a synthetic RNA switch. Metabolic engineering. 2012;14:306–316. doi: 10.1016/j.ymben.2012.04.004. [DOI] [PubMed] [Google Scholar]
- Minami H, Kim JS, Ikezawa N, Takemura T, Katayama T, Kumagai H, Sato F. Microbial production of plant benzylisoquinoline alkaloids. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:7393–7398. doi: 10.1073/pnas.0802981105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misako K, Kouichi M. Caffeine synthase and related methyltransferases in plants. Frontiers in bioscience : a journal and virtual library. 2004;9:1833–1842. doi: 10.2741/1364. [DOI] [PubMed] [Google Scholar]
- Mizuno K, Okuda A, Kato M, Yoneyama N, Tanaka H, Ashihara H, Fujimura T. Isolation of a new dual-functional caffeine synthase gene encoding an enzyme for the conversion of 7-methylxanthine to caffeine from coffee (Coffea arabica L.) FEBS letters. 2003;534:75–81. doi: 10.1016/s0014-5793(02)03781-x. [DOI] [PubMed] [Google Scholar]
- Mohamed T, Osman W, Tin G, Rao PP. Selective inhibition of human acetylcholinesterase by xanthine derivatives: in vitro inhibition and molecular modeling investigations. Bioorganic & medicinal chemistry letters. 2013;23:4336–4341. doi: 10.1016/j.bmcl.2013.05.092. [DOI] [PubMed] [Google Scholar]
- Muller CE, Jacobson KA. Xanthines as adenosine receptor antagonists. Handbook of experimental pharmacology. 2011:151–199. doi: 10.1007/978-3-642-13443-2_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nall AH, Shakhmantsir I, Cichewicz K, Birman S, Hirsh J, Sehgal A. Caffeine promotes wakefulness via dopamine signaling in Drosophila. Scientific reports. 2016;6:20938. doi: 10.1038/srep20938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostergaard S, Olsson L, Nielsen J. Metabolic engineering of Saccharomyces cerevisiae. Microbiology and molecular biology reviews : MMBR. 2000;64:34–50. doi: 10.1128/mmbr.64.1.34-50.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddon CJ, Westfall PJ, Pitera DJ, Benjamin K, Fisher K, McPhee D, Leavell MD, Tai A, Main A, Eng D, et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 2013;496:528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- Page CP. Doxofylline: a “novofylline”. Pulmonary pharmacology & therapeutics. 2010;23:231–234. doi: 10.1016/j.pupt.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Partow S, Siewers V, Bjorn S, Nielsen J, Maury J. Characterization of different promoters for designing a new expression vector in Saccharomyces cerevisiae. Yeast. 2010;27:955–964. doi: 10.1002/yea.1806. [DOI] [PubMed] [Google Scholar]
- Peifer S, Barduhn T, Zimmet S, Volmer DA, Heinzle E, Schneider K. Metabolic engineering of the purine biosynthetic pathway in Corynebacterium glutamicum results in increased intracellular pool sizes of IMP and hypoxanthine. Microbial cell factories. 2012;11:138. doi: 10.1186/1475-2859-11-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourmir A, Johannes TW. Directed evolution: selection of the host organism. Computational and structural biotechnology journal. 2012;2:e201209012. doi: 10.5936/csbj.201209012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfes RJ. Regulation of purine nucleotide biosynthesis: in yeast and beyond. Biochemical Society transactions. 2006;34:786–790. doi: 10.1042/BST0340786. [DOI] [PubMed] [Google Scholar]
- Ryan OW, Skerker JM, Maurer MJ, Li X, Tsai JC, Poddar S, Lee ME, DeLoache W, Dueber JE, Arkin AP, et al. Selection of chromosomal DNA libraries using a multiplex CRISPR system. eLife. 2014;3 doi: 10.7554/eLife.03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandlie I, Solberg K, Kleppe K. The effect of caffeine on cell growth and metabolism of thymidine in Escherichia coli. Mutation research. 1980;73:29–41. doi: 10.1016/0027-5107(80)90133-5. [DOI] [PubMed] [Google Scholar]
- Shao Z, Zhao H, Zhao H. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic acids research. 2009;37:e16. doi: 10.1093/nar/gkn991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers RM, Mohanty SK, Gopishetty S, Subramanian M. Genetic characterization of caffeine degradation by bacteria and its potential applications. Microbial biotechnology. 2015;8:369–378. doi: 10.1111/1751-7915.12262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Shen X, Jain R, Lin Y, Wang J, Sun J, Wang J, Yan Y, Yuan Q. Synthesis of chemicals by metabolic engineering of microbes. Chemical Society reviews. 2015;44:3760–3785. doi: 10.1039/c5cs00159e. [DOI] [PubMed] [Google Scholar]
- Thodey K, Galanie S, Smolke CD. A microbial biomanufacturing platform for natural and semisynthetic opioids. Nature chemical biology. 2014;10:837–844. doi: 10.1038/nchembio.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trenchard IJ, Siddiqui MS, Thodey K, Smolke CD. De novo production of the key branch point benzylisoquinoline alkaloid reticuline in yeast. Metabolic engineering. 2015;31:74–83. doi: 10.1016/j.ymben.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uefuji H, Ogita S, Yamaguchi Y, Koizumi N, Sano H. Molecular cloning and functional characterization of three distinct N-methyltransferases involved in the caffeine biosynthetic pathway in coffee plants. Plant physiology. 2003;132:372–380. doi: 10.1104/pp.102.019679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogels GD, Van der Drift C. Degradation of purines and pyrimidines by microorganisms. Bacteriological reviews. 1976;40:403–468. doi: 10.1128/br.40.2.403-468.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YF, Letisse F, Absalan F, Lu W, Kuznetsova E, Brown G, Caudy AA, Yakunin AF, Broach JR, Rabinowitz JD. Nucleotide degradation and ribose salvage in yeast. Molecular systems biology. 2013;9:665. doi: 10.1038/msb.2013.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama N, Morimoto H, Ye CX, Ashihara H, Mizuno K, Kato M. Substrate specificity of N-methyltransferase involved in purine alkaloids synthesis is dependent upon one amino acid residue of the enzyme. Molecular genetics and genomics : MGG. 2006;275:125–135. doi: 10.1007/s00438-005-0070-z. [DOI] [PubMed] [Google Scholar]
- Zhang F, Carothers JM, Keasling JD. Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat Biotechnol. 2012;30:354–359. doi: 10.1038/nbt.2149. [DOI] [PubMed] [Google Scholar]
- Zrenner R, Stitt M, Sonnewald U, Boldt R. Pyrimidine and purine biosynthesis and degradation in plants. Annual review of plant biology. 2006;57:805–836. doi: 10.1146/annurev.arplant.57.032905.105421. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.