Abstract
Androgen receptor splice variants are alternatively spliced variants of androgen receptor that are C-terminally truncated and lack the canonical ligand-binding domain. Accumulating evidence has indicated a significant role of androgen receptor splice variants in mediating resistance of castration-resistant prostate cancer to current therapies and in predicting therapeutic responses. As such, there is an urgent need to target androgen receptor splicing variants for more effective treatment of castration-resistant prostate cancer. Identification of precise and critical targeting points to deactivate androgen receptor splicing variants relies on a deep understanding of how they are generated and the mechanisms of their action. In this review, we will focus on the emerging data on their generation, clinical significance, and mechanisms of action as well as the therapeutic impact of these findings.
Keywords: androgen receptor, splice variant, prostate cancer, dimerization, cofactors
Introduction
The androgen receptor (AR) signaling pathway remains active in castration-resistant prostate cancer (CRPC) (reviewed in (Egan, et al. 2014; Kahn, et al. 2014; Knudsen and Scher 2009)). Mechanisms leading to AR reactivation include intra-tumoral androgen production, alterations in AR expression level and structure, AR gene mutations, and transcriptional activation of AR by non-androgen ligands and AR cofactors as well as by the crosstalk with growth factor signaling pathways (reviewed in (Egan et al. 2014; Kahn et al. 2014; Knudsen and Scher 2009)). Abiraterone and enzalutamide were developed to target intra-tumoral androgen production and AR overexpression, respectively, but resistance inevitably occurs during treatment (Attard, et al. 2008; de Bono, et al. 2011; Fizazi, et al. 2012; Ryan, et al. 2013; Scher, et al. 2012; Tran, et al. 2009). Recent studies have implicated constitutively-active androgen receptor splice variants (AR-Vs) as a potential driver of resistance to these treatments (Antonarakis, et al. 2014; Cao, et al. 2014; Efstathiou, et al. 2012; Li, et al. 2013; Mostaghel, et al. 2011; Nadiminty, et al. 2013; Yamamoto, et al. 2015).
As a steroid receptor, AR activity is not only regulated by ligand binding but also affected by protein-protein interactions, including homodimerization and interactions with cofactors (reviewed in (Chan and Dehm 2014; Gelmann 2002)). The recruitment and formation of multiple protein complexes are required to activate or repress downstream gene expression. The same rules can be applied to AR-Vs. Several studies indicated that AR-Vs may activate different gene profiles in a cell-context-specific manner (reviewed in (Lu, et al. 2015)). However, the factors that determine the specificity of AR-V-regulated gene sets are currently unknown. One appealing mechanism is that selective gene targeting is achieved by different protein complexes formed between AR-Vs and their partners and/or cofactors, which may induce the formation of novel interfaces to allow the installment of the general transcriptional machinery. In this review, we summarize the recent discoveries about the mechanisms of AR-V generation, their clinical values, and the modulation of their transcriptional activity by their dimerization partners and cofactors in CRPC.
AR-V structure
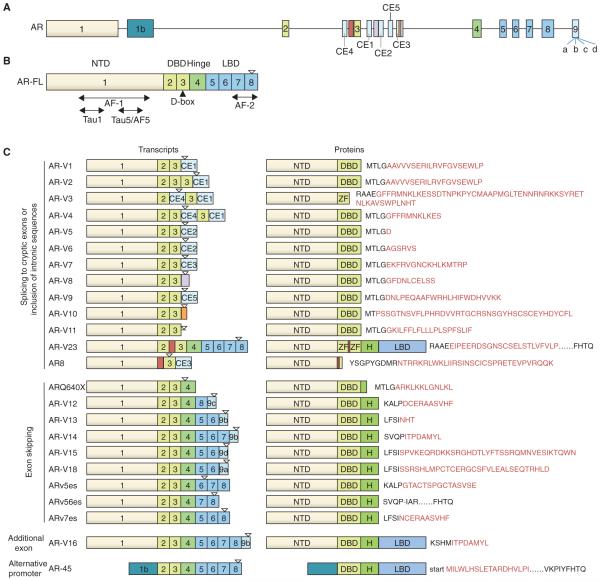
To date, over 20 AR variants have been identified in human prostate cancer cell models and clinical specimens (Figure 1). Some of these variants are constitutively active, such as AR-V7 and ARv567es (aka AR-V12), while some others are conditionally active, depending on the cellular context, such as AR-V1 and AR-V9 (Hu, et al. 2011). Except AR45, which is truncated in the N-terminal domain, all the other variants contain an intact N-terminal domain but lack portions of the ligand-binding domain (LBD). Since the N-terminal domain harbors the two trans-activating regions (Tau1 and Tau5/AF5), AR45 loses trans-activating ability and acts as a dominant-negative variant to inhibit the function of the full-length AR (AR-FL) by forming a heterodimer with AR-FL (Ahrens-Fath, et al. 2005).
Figure 1. Schematic representation of the structure of AR-FL and AR-V transcripts and proteins.
A. AR gene structure with canonical exons and the cryptic exons (CE). B. AR-FL mRNA structure showing exons encoding the N-terminal domain (NTD; exon 1), DNA-binding domain (DBD; exons 2 and 3), hinge region (part of exons 3 and 4), and ligand-binding domain (LBD; exons 5-8). AF-1, Tau1, Tau5/AF-5, and AF-2 are activation function domains. Filled triangle depicts the D-box, which mediates AR-V/AR-V, AR-V/AR-FL, and AR-FL/AR-FL dimerization. C. mRNA and protein structures of AR-Vs. AR-V-specific peptide sequences are indicated in red, and the “-“ in ARv56es indicates a unique junction. Inverted open triangle depicts translation stop. Drawings are not to scale. Exon 9 harbors four cryptic 3’ splicing sites, and the corresponding cryptic exons are indicated as 9a, 9b, 9c, and 9d.
Most AR-Vs contain an intact DNA-binding domain (DBD), however, AR8 does not have a functional DBD, and AR-V3 (aka AR6) lacks the second zinc finger of the DBD. As a result of missing DBD, AR8 cannot function as a transcription factor (Yang, et al. 2011). Instead, it primarily locates on the plasma membrane and promotes EGF-induced Src activation and AR-FL phosphorylation and transactivation (Yang et al. 2011). In contrast, AR-V3, which still contains the AR DNA-binding interface residing in the first zinc finger of DBD, can constitutively activate AR-responsive promoters in prostate cancer cells (Dehm, et al. 2008). Another variant, AR23, has in-frame insertions between the two zinc fingers by retaining part of the intronic sequence. AR23 was shown to exhibit exclusively cytoplasmic activities, promoting the transcriptional activity of nuclear factor-κB while decreasing the activity of activator protein-1 (Jagla, et al. 2007). It is likely that the insertion of the intronic sequence has scrambled the DBD.
Following the DBD is the hinge region, encoded by part of exon 3 and exon 4, which has been shown to harbor the canonical nuclear localization signal. The nuclear localization property is important for the variants to perform their trans-activating function. Although some of the variants do not have the nuclear localization signal, they are still primarily located in the nucleus or have a basal level sufficient for ligand-independent transcriptional activity (Chan, et al. 2012). Some mechanisms were proposed for their nuclear localization, such as the existence of a nuclear localization signal-like sequence, lack of nuclear export signal (Chan et al. 2012; Saporita, et al. 2003), or tyrosine phosphorylation in the N-terminal domain (Karaca, et al. 2015). However, these mechanisms do not appear to satisfactorily cover all the variants with predominant nuclear localization. Characterization of the involved amino acid sequences and understanding the mechanisms that govern their nuclear localization may offer the potential to block AR-V nuclear localization.
AR-V production
AR-Vs may arise from multiple mechanisms. Genomic rearrangement of the AR gene has been associated with AR-V generation (Li, et al. 2011; Li, et al. 2012b; Nyquist, et al. 2013). Modeling gene rearrangement in prostate cancer cells showed expression of ARv567es without AR-FL in clonally selected cells (Nyquist et al. 2013). While AR gene rearrangement could contribute to AR-V production in the subset of prostate cancers with AR-Vs being the predominant form of AR expressed, other mechanisms, such as the involvement of specific splicing factors, may underlie the co-expression of AR-FL and AR-Vs observed in many prostate cancer specimens (Miyamoto, et al. 2015). Nadiminty et al. showed that the splicing factor hnRNPA1 is upregulated and correlated with AR-V7 expression level in 22Rv1 cells with acquired enzalutamide resistance (Nadiminty, et al. 2015). The recruitment of hnRNPA1 to the AR-V7 and AR-V3 (aka AR-1/2/2b) splicing sites in AR pre-mRNA is increased, but no significant change is observed in the recruitment of hnRNPA1 to the AR-FL splicing sites, suggesting hnRNPA1 can selectively regulate the generation of AR-Vs (Nadiminty et al. 2015). In addition, Liu et al. reported that splicing factors U2AF65 and ASF/SF2 can recognize the binding sites near AR exon 3B and facilitate the recruitment of RNA spliceosome to the AR-V7 3’ splicing site in VCaP and LNCaP95 cells after androgen deprivation (Liu, et al. 2014b). Unlike the hnRNPA1 in the aforementioned study, the expression of U2AF65 and ASF/SF2 is not changed after androgen deprivation. Instead, increased AR-V7 production is due to elevated AR pre-mRNA substrates for those splicing factors (Liu et al. 2014b). Furthermore, the study of Ferraldeschi et al. implicated a role for the molecular chaperone, HSP90, in AR-V7 splicing (Ferraldeschi, et al. 2016). HSP90 inhibition leads to the disruption of AR-V7 splicing and reduction of AR-V7 level (Ferraldeschi et al. 2016). Finally, another study highlighted the interplay between the noncoding RNA PCGEM1 and splicing factors in contributing to AR splicing (Zhang, et al. 2016). Androgen deprivation was found to induce PCGEM1 redistribution into nuclear speckles, and the interaction between PCGEM1 and U2AF65 promotes AR-V7 splicing (Zhang et al. 2016). Thus, mechanisms governing AR-V production could be cell-context specific and may involve more layers of regulation beyond splicing factors.
Clinical relevance
Accumulating clinical studies showed an association between AR-V expression and prostate cancer progression, therapy resistance, and poor clinical outcome. AR-Vs can be detected in benign prostate tissues, hormone-naïve prostate cancers, and CRPC samples, with the most frequent and highest expression detected in CRPC samples (Abeshouse, et al. 2015; Antonarakis et al. 2014; Guo, et al. 2009; Hornberg, et al. 2011; Hu, et al. 2009; Miyamoto et al. 2015; Qu, et al. 2015; Robinson, et al. 2015; Sun, et al. 2010; Welti, et al. ; Zhang, et al. 2011). Higher expression of AR-V7 in hormone-naïve prostate tumors has been shown to correlate with increased risk of biochemical recurrence following radical prostatectomy (Guo et al. 2009; Hu et al. 2009) and more rapid progression to CRPC (Qu et al. 2015). Moreover, high levels of AR-V7 mRNA or nuclear AR-V7 protein or detectable expression of ARv567es mRNA in CRPCs are associated with a shorter survival of the patients (Hornberg et al. 2011; Qu et al. 2015; Welti et al.). Thus, AR-V expression appears to be associated with a more lethal form of the disease. In support of these clinical evidences, preclinical studies showed that expression of AR-V7 or ARv567es in prostate cancer cell lines or in prostate epithelium of transgenic mice can induce epithelial-to-mesenchymal transition markers (Cottard, et al. 2013; Liu, et al. 2013; Sun, et al. 2014) and castration-resistant growth of prostate cancer cells (Guo et al. 2009; Sun et al. 2010; Watson, et al. 2010).
AR-Vs have also been shown to confer both primary and acquired resistance to abiraterone and enzalutamide in preclinical models (Cao et al. 2014; Li et al. 2013; Mostaghel et al. 2011; Nadiminty et al. 2013; Yamamoto et al. 2015). Significantly, AR-V7 has been indicated to have prognostic value in CRPC patients treated with abiraterone or enzalutamide. In a prospective phase 2 study, Efstathiou et al. assessed the AR-V7 protein levels in bone marrow biopsies from 60 patients with bone metastatic CRPC before and after enzalutamide treatment (Efstathiou, et al. 2015). The presence of AR-V7 is associated with primary resistance to enzalutamide (Efstathiou et al. 2015). In another study, Antonarakis et al. evaluated the AR-V7 mRNA level in circulating tumor cells from metastatic CRPC patients before the initiation of enzalutamide or abiraterone treatment (Antonarakis et al. 2014). None of the patients with AR-V7-positive circulating tumor cells showed a response of prostate-specific antigen (PSA) to enzalutamide or abiraterone, and AR-V7 positivity is associated with a shorter progression-free survival of the patients (Antonarakis et al. 2014). Together, these studies suggested the potential of using AR-Vs as a predictive marker of response to enzalutamide and abiraterone.
The role of AR-Vs in therapy resistance may not be limited to androgen-directed therapies. Preclinical studies indicated that AR-Vs might also contribute to taxane resistance. Paclitaxel, docetaxel, and cabazitaxel are members of the taxane family of chemotherapeutic agents. They bind to and stabilize microtubules to suppress microtubule dynamics (Martin and Kyprianou 2015; Mellado, et al. 2016). Recent literature suggested that androgen-induced AR-FL nuclear import, which is facilitated by microtubule, can be inhibited by paclitaxel and docetaxel (Darshan, et al. 2011; van Soest, et al. 2013; Zhang, et al. 2015; Zhu, et al. 2010) but may or may not by cabazitaxel (de Leeuw, et al. 2015; Martin, et al. 2016; van Soest et al. 2013; Zhang et al. 2015). In contrast to AR-FL, AR-V7 is not only resistant to docetaxel and paclitaxel inhibition of nuclear translocation (Martin, et al. 2015; Thadani-Mulero, et al. 2014; Zhang et al. 2015) but also attenuates the ability of docetaxel and paclitaxel to retain AR-FL in the cytoplasm (Zhang et al. 2015). The role of AR-V7 in contributing to taxane resistance is further supported by the ability of the AR N-terminal domain antagonist EPI-001/002 to enhance the response of AR-V7-expressing CRPC cells to docetaxel treatment in vitro and in vivo (Martin et al. 2015). However, such role of AR-V7 does not appear to be supported by clinical evidences. Two groups examined AR-V7 mRNA levels in circulating tumor cells from CRPC patients before the initiation of taxane chemotherapy. Both showed that the response to taxanes seems to be independent of the AR-V7 status of circulating tumor cells (Antonarakis, et al. 2015; Onstenk, et al. 2015). In these studies, the methodologies for isolating circulating tumor cells heavily depend on the expression of an epithelial cell marker on circulating tumor cells (Antonarakis et al. 2015; Onstenk et al. 2015). Whether the exclusion of circulating tumor cells that have undergone epithelial-to-mesenchymal transition in these studies contributes to the conflicting clinical and preclinical findings awaits to be determined.
Homodimerization and heterodimerization of AR-Vs.
Homodimerization is an essential step for AR-FL to activate target gene expression. The consecutive steps leading to AR-FL homodimerization has been well elucidated (van Royen, et al. 2012). Upon ligand binding in the cytoplasm, AR-FL forms intramolecular N-terminal and C-terminal (N/C) interactions, which facilitate nuclear translocation of AR-FL (van Royen et al. 2012). In the nucleus, the intramolecular interactions are followed by a D-box-dimerization-dependent transition to intermolecular N/C interaction (van Royen et al. 2012). Both the intra- and inter-molecular N/C interactions and D-box/D-box interactions are required for AR-FL dimerization (van Royen et al. 2012). Mutations in the N-terminal domain or the D-box can both cause the loss of AR-FL transcriptional ability (van Royen et al. 2012). The knowledge on AR-FL transactivation paved the path for the understanding of AR-V transactivation and identification of critical trans-activating steps for therapeutic targeting. We recently showed that, like liganded AR-FL, dimerization is also required for AR-Vs to transactivate target genes. AR-V7 and ARv567es can not only homodimerize but also heterodimerize with each other, and the dimerization is mediated by D-box-D-box interactions (Xu, et al. 2015). We further showed that the D-box mutants of AR-V7 and ARv567es lose the ability to transactivate target genes and to induce castration-resistant cell growth (Xu et al. 2015). These findings highlight the potential of targeting AR D-box to inhibit the activity of both AR-FL and AR-Vs for CRPC treatment.
Interplays between AR-Vs and AR-FL
The function of AR-Vs to regulate gene expression has mainly been investigated independent of AR-FL. However, since AR-Vs are often co-expressed with AR-FL in clinical specimens (Miyamoto et al. 2015), the interplays between AR-Vs and AR-FL may be an important mechanism of their actions. Coimmunoprecipitation of ARv567es and AR-FL (Sun et al. 2010) as well as co-occupancy of AR-V7 and AR-FL on the PSA promoter (Cao et al. 2014) indicate that there may be direct interactions between AR-FL and AR-Vs. By using two different assays to detect protein dimerization, we recently showed that both AR-V7 and ARv567es can heterodimerize with AR-FL (Xu et al. 2015). The heterodimerization induces androgen-independent AR-FL nuclear localization and transcriptional activity (Cao et al. 2014). Interestingly, we found that the promoter of the canonical AR target PSA is co-occupied by AR-V7 and AR-FL whereas the promoter of the UBE2C gene is bound by AR-V7 only (Cao et al. 2014), suggesting that the AR-V/AR-FL dimers and the AR-V/AR-V dimers may regulate different sets of target genes. This is supported by transcriptome and metabolome data showing that AR-Vs can regulate some canonical AR targets as well as a distinct set of genes/pathways (Chan, et al. 2015; Guo et al. 2009; Hu, et al. 2012; Li et al. 2013; Lu, et al. 2014; Shafi, et al. 2015). Identifying the respective binding sites for AR-V/AR-FL dimers and AR-V/AR-V dimers across the genome and elucidating whether the genes and pathways regulated by the AR-V/AR-FL dimers fully overlap with those regulated by AR-FL homodimer could be vital in understanding how AR-Vs contribute to castration resistance.
AR-V cofactors
It has long been appreciated that AR cofactors play critical roles in modulating AR activity. However, cofactors for AR-Vs have been scarcely addressed. Although the vast majority of AR-Vs have a truncated C-terminal domain, cofactors that interact with the N-terminal domain of AR-FL may still be involved in modulating the activity of AR-Vs. Several groups have profiled the AR-V transcriptome (Chan et al. 2015; Guo et al. 2009; Hu et al. 2012; Li et al. 2013; Lu et al. 2014), however, AR-V gene expression profiles lack consistency in different prostate cancer models (reviewed in (Lu et al. 2015)). It is possible that AR-Vs can recruit distinct cofactors that may confer target specificity in different cellular context. Since AR-Vs can evade androgen-directed therapies, investigation on the cofactors shared by AR-FL and AR-Vs may provide more potent therapeutic targets.
Theoretically, cofactors bound to the N-terminal domain of AR-FL should interact with AR-Vs as well. Gli2, which binds to the Tau5/AF5 region in the N-terminal domain of AR to enhance AR-FL activity (Chen, et al. 2010), can also coactivate AR-V7 and ARv567es (Li, et al. 2014b). However, there are also cofactors that interact with AR-FL and AR-Vs through different interfaces and with differing affinity. For example, the transcriptional coactivator FHL2 can increase AR-FL transcriptional activity in an agonist- and AF-2-dependent manner (Muller, et al. 2000). Since most AR-Vs do not contain the AF-2 domain, it is expected that FHL2 would not influence AR-V activity. However, a recent study showed that FHL2 can also act as an AR-V7 coactivator (McGrath, et al. 2013). The authors showed that FHL2 accumulates aberrantly in the nucleus in CRPC cells and that nuclear-accumulated FHL2 directly binds to AR-V7 and enhances its transcriptional activity (McGrath et al. 2013). Although the interacting domain on AR-V7 has not been mapped out, it is clear that FHL2 interacts with AR-FL and AR-V7 through binding to different domains. This case also indicates that AR reactivation in CRPC can be contributed by not only altered expression of cofactors but also deregulated subcellular localization of cofactors. Another example is the cofactor MED1. In the presence of androgen, MED1 has been shown to interact with the LBD of AR-FL and enhance androgen-dependent AR-FL activity (Wang, et al. 2002). In androgen-deprived condition, PI3K/AKT phosphorylated MED1 (p-MED1) can enhance both AR-FL- and ARv567es-mediated UBE2C transcription through an enhancer-promoter chromatin-looping mechanism (Chen, et al. 2011; Liu, et al. 2015b; Wang, et al. 2009). However, ARv567es, with the assistance of the pioneer factor FOXA1, has been shown to be more potent than AR-FL to recruit p-MED1 to the promoter and enhancer regions of UBE2C (Liu et al. 2015b). This may underlie the preferential regulation of UBE2C by AR-Vs over AR-FL in cells co-expressing AR-FL and AR-Vs (Cao et al. 2014; Hu et al. 2012). Together, these findings suggest that MED1 could not only regulate AR-FL activity through different mechanisms when androgen is present versus absent but also contribute to a switch from AR-FL signaling to AR-V signaling.
AR-FL and AR-Vs may compete for binding to certain cofactors in different biological conditions, leading to the activation of different sets of genes. The RhoGTPase guanine nucleotide exchange factor, Vav3, found to be an AR-FL coactivator (Rao, et al. 2012), was shown to also enhance AR-V7 and ARv567es transactivation and increase AR-V7 nuclear localization by directly binding to AR-V7 (Peacock, et al. 2012). Immunoprecipitation assay showed that AR-V7 may compete with AR-FL for interacting with Vav3 (Peacock et al. 2012).
Some cofactors may regulate not only the activity but also the abundance of AR-FL and AR-Vs. The pioneer factor GATA-binding protein 2 (GATA2) is reported to facilitate AR-FL genomic binding by colocalizing with FOXA1 at the enhancer regions of target genes to produce an accessible chromatin environment for AR-FL prior to androgen stimulation and by recruiting MED1 to sustain basal enhancer-promoter chromatin looping in the absence of androgen (He, et al. 2014; Wang, et al. 2007; Wu, et al. 2014). It has also been shown to interact with AR-V7 and colocalize with AR-Vs on chromatin (He et al. 2014). At the same time, GATA2 induces the transcription of the AR gene and thereby the levels of both AR-FL and AR-Vs through binding to an extended promoter region of the AR gene (Bohm, et al. 2009; He et al. 2014; Wang et al. 2007; Wu et al. 2014). Interestingly, androgen-bound AR represses GATA2 expression (He et al. 2014). Under androgen-deprived condition, with the disruption of this negative-feedback regulatory loop, GATA2 contributes to both overexpression and increased activity of AR-FL and AR-Vs (He et al. 2014). As a result, inhibiting GATA2 has dual efficacy to inactivate AR signaling by ablating both the expression and transcriptional activities of AR-FL and AR-Vs (He et al. 2014). Preclinical data showed that the small molecule inhibitor of GATA2, K7174, significantly decreased the viability of various GATA2+/AR+ prostate cancer cell lines in vitro and inhibited the growth of LNCaP-abl xenograft tumors (He et al. 2014). All these data suggested a promising role for GATA2 inhibitors in CRPC treatment.
When coexisting in the same cells, corepressors and coactivators may compete for AR-V binding. FOXO1 is a well-known corepressor of AR (Attard, et al. 2009; Dong, et al. 2006; Fan, et al. 2007; Liu, et al. 2008). It can inhibit androgen-independent activation of AR-FL (Liu et al. 2008) and the constitutive activity of different AR-Vs (Bohrer, et al. 2013; Mediwala, et al. 2013). In addition, FOXO1 represses SRC-1-enhanced transcriptional activity of AR-V5 through competing with SRC-1 for binding to the Tau5/AF5 region of AR-V5. Although the Tau5/AF5 region is present in almost all AR-Vs, SRC-1 can only selectively enhance the transcriptional activity of AR-V5 but not the other variants (Bohrer et al. 2013), indicating that the transactivation of different AR-Vs may involve distinct cofactors and mechanisms. It is also possible that different AR-V dimers may recruit specific cofactors to activate gene expression. Further mechanistic study of how these cofactors contribute to CRPC and their interplay with AR-Vs would drive the development of strategies that effectively target the cofactors.
AR-V7 degradation
In addition to blocking the production and activity of AR-Vs, inducing AR-V protein degradation is another attractive approach to suppress AR-V signaling. Li et al. recently showed that, similar to AR-FL, the AR-V7 protein can be degraded through an Mdm2-mediated ubiquitin-proteasome degradation process and that this process can be accelerated by Akt signaling but repressed by protein phosphatase-1 (PP-1) (Li, et al. 2015b). Co-targeting the AR and Akt signaling pathways is being tested for CRPC treatment. Would co-suppressing the two pathways lead to a more robust induction of AR-Vs and AR-V-driven tumor progression? If so, would co-targeting the Akt pathway with an agent that can induce AR-V degradation (Cao, et al. 2013; Kwegyir-Afful, et al. 2015; Li, et al. 2012a; Liu, et al. 2014a; Sun, et al. 2015; Yamashita, et al. 2012; Yu, et al. 2014; Zengerling, et al. 2012) alleviate this problem? These questions need to be carefully addressed.
Therapeutic targeting of AR-Vs
Various approaches have been explored to disrupt AR-V signaling, such as targeting AR N-terminal domain (Myung, et al. 2013) or DNA-binding interface (Dalal, et al. 2014; Li, et al. 2014a), inducing AR-V protein degradation (Cao et al. 2013; Kwegyir-Afful et al. 2015; Li et al. 2012a; Liu et al. 2014a; Sun et al. 2015; Yamashita et al. 2012; Yu et al. 2014; Zengerling et al. 2012), reducing AR-V expression (Mashima, et al. 2010; Zhan, et al. 2013), inhibiting AR-V chromatin binding (Chan et al. 2015; Li, et al. 2015a), disrupting AR-FL and AR-V dimerization (Streicher, et al. 2014), or using antisense oligonucleotides against exons shared by AR-FL and AR-Vs (Yamamoto et al. 2015). Here, we describe several AR-V-targeting agents that have entered clinical trials for cancer treatment. EPI-001/002 is a small-molecule, non-steroidal AR antagonist that covalently binds to AR N-terminal domain to inhibit the transcriptional activities of AR-FL and AR-Vs (Andersen, et al. 2010; Kato, et al. 2016; Myung et al. 2013; Sadar 2011). It has also been found to act as a selective PPARγ modulator to inhibit AR expression (Brand, et al. 2015). EPI-001/002 showed excellent anti-tumor efficacy in various preclinical models of CRPC, and its successor, EPI-506, is being investigated in a phase I/II study in metastatic CRPC patients (Andersen et al. 2010; Brand et al. 2015; Martin and Kyprianou 2015; Myung et al. 2013; Sadar 2011).
Galeterone is another small molecule that can inhibit AR-V activity. It was shown to target AR signaling at three levels, blocking androgen synthesis by inhibiting CYP17, inducing proteasomal degradation of AR-FL and AR-V7, and preventing the binding of androgen to the AR (Handratta, et al. 2005; Kwegyir-Afful et al. 2015; Njar and Brodie 2015; Purushottamachar, et al. 2013; Yu et al. 2014). A Phase III trial comparing galeterone to enzalutamide in treatment-naïve metastatic CRPC patients with AR-V7-positive prostate tumors was launched in 2015. Unfortunately, the trial was terminated in July of 2016 due to the unlikelihood of galeterone to show improved radiographic progression-free survival in these patients than enzalutamide. Further clinical testing is being planned in metastatic CRPC patients who rapidly progress on enzalutamide or abiraterone or have developed acquired resistance to enzalutamide.
Niclosamide is an FDA-approved anti-helminthic drug that was found to inhibit AR-V7 expression and activity (Liu et al. 2014a). It also targets the IL6-Stat3-AR axis (Liu, et al. 2015a). Niclosamide inhibits prostate cancer cell growth and overcomes enzalutamide and abiraterone resistance in preclinical models (Liu, et al. 2016; Liu et al. 2015a; Liu et al. 2014a). These promising preclinical data led to the Phase I trial of niclosamide in combination with enzalutamide in AR-V-positive metastatic CRPC patients.
Several second-generation HSP90 inhibitors are in active clinical trials for cancer treatment. HSP90 interacts with the LBD of AR-FL as a molecular chaperone, which is critical for proper folding, hormone binding, and transcriptional activity of AR-FL (Ai, et al. 2009; Vanaja, et al. 2002). It does not affect the activities of AR-Vs (Shafi, et al. 2013; Vanaja et al. 2002). However, it has been reported to play a role in AR-V7 splicing (Ferraldeschi et al. 2016). The second generation HSP90 Inhibitor Onalespib was shown to block AR-V7 mRNA splicing and reduce AR-V7 level, supporting further clinical investigation of HSP90 inhibitors against AR-V7-expressing CRPC (Ferraldeschi et al. 2016).
Several bromodomain and extra-terminal domain family (BET) inhibitors have shown promising efficacies in preclinical studies and are currently under clinical trials in CRPC patients (Asangani, et al. 2014). Many AR cofactors contain a bromodomain(s), which recognizes acetylated lysine residues and is essential for chromatin binding and remodeling as well as coactivation. A BET inhibitor, JQ1, was shown to disrupt the recruitment of both AR-FL and AR-Vs to target gene loci and block their transactivation (Asangani et al. 2014; Chan et al. 2015). While it is likely that not all of these agents can be translated into the clinic, these emerging drugs provided promising and innovative approaches to effectively target AR-Vs for treating CRPC.
Conclusion
The seminar discovery of AR-Vs has revolutionized the field of prostate cancer. It not only broadened our view of how to tackle the AR signaling axis but also provided a most promising prognostic marker for predicting responses of CRPC to current androgen-directed therapies. Various approaches have been explored to disrupt both AR-FL and AR-V signaling. Several emerging drugs, such as EPI-001/002, galaterone, niclosamide, HSP90 inhibitors, and BET inhibitors, have shown excellent efficacies to overcome resistance of CRPC to current therapies in preclinical models and are currently under clinical investigations. These innovative approaches of AR targeting have marked the beginning of a new era in prostate cancer treatment. Further investigations of the mechanisms of how AR-Vs are activated to regulate downstream genes and crosstalk with other signaling pathways will continue to benefit the search for more effective therapeutic approaches and targets. Finally, the prognostic impact of AR-Vs may allow the implementation of a tailored therapeutic strategy in selecting patients who may benefit from specific treatment at specific point of disease progression. This will be a major step forward in precision medicine for advanced prostate cancer.
Acknowledgments
Funding
This work was supported by the following grants: NIH/NCI R01CA188609; DOD W81XWH-15-1-0439, W81XWH-16-1-0317, and W81XWH-14-1-0485; National Natural Science Foundation of China Projects 81272851 and 81430087.
Footnotes
Declaration of interest
The authors declare no conflict of interest that could be perceived as prejudicing the impartiality of this review.
Reference
- Abeshouse A, Ahn J, Akbani R, Ally A, Amin S, Andry Christopher D, Annala M, Aprikian A, Armenia J, Arora A, et al. The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrens-Fath I, Politz O, Geserick C, Haendler B. Androgen receptor function is modulated by the tissue-specific AR45 variant. FEBS J. 2005;272:74–84. doi: 10.1111/j.1742-4658.2004.04395.x. [DOI] [PubMed] [Google Scholar]
- Ai J, Wang Y, Dar JA, Liu J, Liu L, Nelson JB, Wang Z. HDAC6 regulates androgen receptor hypersensitivity and nuclear localization via modulating Hsp90 acetylation in castration-resistant prostate cancer. Mol Endocrinol. 2009;23:1963–1972. doi: 10.1210/me.2009-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen RJ, Mawji NR, Wang J, Wang G, Haile S, Myung JK, Watt K, Tam T, Yang YC, Banuelos CA, et al. Regression of Castrate-Recurrent Prostate Cancer by a Small-Molecule Inhibitor of the Amino-Terminus Domain of the Androgen Receptor. Cancer Cell. 2010;17:535–546. doi: 10.1016/j.ccr.2010.04.027. [DOI] [PubMed] [Google Scholar]
- Antonarakis ES, Lu C, Luber B, Wang H, Chen Y, Nakazawa M, Nadal R, Paller CJ, Denmeade SR, Carducci MA, et al. Androgen Receptor Splice Variant 7 and Efficacy of Taxane Chemotherapy in Patients With Metastatic Castration-Resistant Prostate Cancer. JAMA Oncol. 2015;1:582–591. doi: 10.1001/jamaoncol.2015.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, Chen Y, Mohammad TA, Chen Y, Fedor HL, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N.Engl.J Med. 2014;371:1028–1038. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, Escara-Wilke J, Wilder-Romans K, Dhanireddy S, Engelke C, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510:278–282. doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attard G, Reid AH, Yap TA, Raynaud F, Dowsett M, Settatree S, Barrett M, Parker C, Martins V, Folkerd E, et al. Phase I clinical trial of a selective inhibitor of CYP17, abiraterone acetate, confirms that castration-resistant prostate cancer commonly remains hormone driven. J.Clin.Oncol. 2008;26:4563–4571. doi: 10.1200/JCO.2007.15.9749. [DOI] [PubMed] [Google Scholar]
- Attard G, Swennenhuis JF, Olmos D, Reid AH, Vickers E, A'Hern R, Levink R, Coumans F, Moreira J, Riisnaes R, et al. Characterization of ERG, AR and PTEN gene status in circulating tumor cells from patients with castration-resistant prostate cancer. Cancer Res. 2009;69:2912–2918. doi: 10.1158/0008-5472.CAN-08-3667. [DOI] [PubMed] [Google Scholar]
- Bohm M, Locke WJ, Sutherland RL, Kench JG, Henshall SM. A role for GATA-2 in transition to an aggressive phenotype in prostate cancer through modulation of key androgen-regulated genes. Oncogene. 2009;28:3847–3856. doi: 10.1038/onc.2009.243. [DOI] [PubMed] [Google Scholar]
- Bohrer LR, Liu P, Zhong J, Pan Y, Angstman J, Brand LJ, Dehm SM, Huang H. FOXO1 binds to the TAU5 motif and inhibits constitutively active androgen receptor splice variants. Prostate. 2013;73:1017–1027. doi: 10.1002/pros.22649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand LJ, Olson ME, Ravindranathan P, Guo H, Kempema AM, Andrews TE, Chen X, Raj GV, Harki DA, Dehm SM. EPI-001 is a selective peroxisome proliferator-activated receptor-gamma modulator with inhibitory effects on androgen receptor expression and activity in prostate cancer. Oncotarget. 2015;6:3811–3824. doi: 10.18632/oncotarget.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao B, Liu X, Li J, Liu S, Qi Y, Xiong Z, Zhang A, Wiese T, Fu X, Gu J, et al. 20(S)-protopanaxadiol-aglycone downregulation of the full-length and splice variants of androgen receptor. International Journal of Cancer. 2013;132:1277–1287. doi: 10.1002/ijc.27754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao B, Qi Y, Zhang G, Xu D, Zhan Y, Alvarez X, Guo Z, Fu X, Plymate SR, Sartor O, et al. Androgen receptor splice variants activating the full-length receptor in mediating resistance to androgen-directed therapy. Oncotarget. 2014;5:1646–1656. doi: 10.18632/oncotarget.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SC, Dehm SM. Constitutive activity of the androgen receptor. Adv Pharmacol. 2014;70:327–366. doi: 10.1016/B978-0-12-417197-8.00011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SC, Li Y, Dehm SM. Androgen receptor splice variants activate androgen receptor target genes and support aberrant prostate cancer cell growth independent of canonical androgen receptor nuclear localization signal. J Biol Chem. 2012;287:19736–19749. doi: 10.1074/jbc.M112.352930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SC, Selth LA, Li Y, Nyquist MD, Miao L, Bradner JE, Raj GV, Tilley WD, Dehm SM. Targeting chromatin binding regulation of constitutively active AR variants to overcome prostate cancer resistance to endocrine-based therapies. Nucleic Acids Res. 2015;43:5880–5897. doi: 10.1093/nar/gkv262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Feuerstein MA, Levina E, Baghel PS, Carkner RD, Tanner MJ, Shtutman M, Vacherot F, Terry S, de la Taille A, et al. Hedgehog/Gli supports androgen signaling in androgen deprived and androgen independent prostate cancer cells. Mol Cancer. 2010;9:89. doi: 10.1186/1476-4598-9-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Zhang C, Wu D, Chen H, Rorick A, Zhang X, Wang Q. Phospho-MED1-enhanced UBE2C locus looping drives castration-resistant prostate cancer growth. EMBO J. 2011;30:2405–2419. doi: 10.1038/emboj.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottard F, Asmane I, Erdmann E, Bergerat JP, Kurtz JE, Ceraline J. Constitutively active androgen receptor variants upregulate expression of mesenchymal markers in prostate cancer cells. PLoS One. 2013;8:e63466. doi: 10.1371/journal.pone.0063466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal K, Roshan-Moniri M, Sharma A, Li H, Ban F, Hassona MD, Hsing M, Singh K, LeBlanc E, Dehm S, et al. Selectively targeting the DNA-binding domain of the androgen receptor as a prospective therapy for prostate cancer. J Biol Chem. 2014;289:26417–26429. doi: 10.1074/jbc.M114.553818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darshan MS, Loftus MS, Thadani-Mulero M, Levy BP, Escuin D, Zhou XK, Gjyrezi A, Chanel-Vos C, Shen R, Tagawa ST, et al. Taxane-induced blockade to nuclear accumulation of the androgen receptor predicts clinical responses in metastatic prostate cancer. Cancer Res. 2011;71:6019–6029. doi: 10.1158/0008-5472.CAN-11-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, Chi KN, Jones RJ, Goodman OB, Saad F, et al. Abiraterone and Increased Survival in Metastatic Prostate Cancer. New England Journal of Medicine. 2011;364:1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw R, Berman-Booty LD, Schiewer MJ, Ciment SJ, Den RB, Dicker AP, Kelly WK, Trabulsi EJ, Lallas CD, Gomella LG, et al. Novel actions of next-generation taxanes benefit advanced stages of prostate cancer. Clin Cancer Res. 2015;21:795–807. doi: 10.1158/1078-0432.CCR-14-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehm SM, Schmidt LJ, Heemers HV, Vessella RL, Tindall DJ. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res. 2008;68:5469–5477. doi: 10.1158/0008-5472.CAN-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong XY, Chen C, Sun X, Guo P, Vessella RL, Wang RX, Chung LW, Zhou W, Dong JT. FOXO1A is a candidate for the 13q14 tumor suppressor gene inhibiting androgen receptor signaling in prostate cancer. Cancer Res. 2006;66:6998–7006. doi: 10.1158/0008-5472.CAN-06-0411. [DOI] [PubMed] [Google Scholar]
- Efstathiou E, Titus M, Tsavachidou D, Tzelepi V, Wen S, Hoang A, Molina A, Chieffo N, Smith LA, Karlou M, et al. Effects of abiraterone acetate on androgen signaling in castrate-resistant prostate cancer in bone. J.Clin.Oncol. 2012;30:637–643. doi: 10.1200/JCO.2010.33.7675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efstathiou E, Titus M, Wen S, Hoang A, Karlou M, Ashe R, Tu SM, Aparicio A, Troncoso P, Mohler J, et al. Molecular characterization of enzalutamide-treated bone metastatic castration-resistant prostate cancer. Eur Urol. 2015;67:53–60. doi: 10.1016/j.eururo.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan A, Dong Y, Zhang H, Qi Y, Balk SP, Sartor O. Castration-resistant prostate cancer: Adaptive responses in the androgen axis. Cancer Treat.Rev. 2014;40:426–433. doi: 10.1016/j.ctrv.2013.09.011. [DOI] [PubMed] [Google Scholar]
- Fan W, Yanase T, Morinaga H, Okabe T, Nomura M, Daitoku H, Fukamizu A, Kato S, Takayanagi R, Nawata H. Insulin-like growth factor 1/insulin signaling activates androgen signaling through direct interactions of Foxo1 with androgen receptor. J Biol Chem. 2007;282:7329–7338. doi: 10.1074/jbc.M610447200. [DOI] [PubMed] [Google Scholar]
- Ferraldeschi R, Welti J, Powers MV, Yuan W, Smyth T, Seed G, Riisnaes R, Hedayat S, Wang H, Crespo M, et al. Second-Generation HSP90 Inhibitor Onalespib Blocks mRNA Splicing of Androgen Receptor Variant, 7 in Prostate Cancer Cells. Cancer Res. 2016;76:2731–2742. doi: 10.1158/0008-5472.CAN-15-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fizazi K, Scher HI, Molina A, Logothetis CJ, Chi KN, Jones RJ, Staffurth JN, North S, Vogelzang NJ, Saad F, et al. Abiraterone acetate for treatment of metastatic castration-resistant prostate cancer: final overall survival analysis of the COU-AA-301 randomised, double-blind, placebo-controlled phase 3 study. Lancet Oncol. 2012;13:983–992. doi: 10.1016/S1470-2045(12)70379-0. [DOI] [PubMed] [Google Scholar]
- Gelmann EP. Molecular biology of the androgen receptor. J.Clin.Oncol. 2002;20:3001–3015. doi: 10.1200/JCO.2002.10.018. [DOI] [PubMed] [Google Scholar]
- Guo Z, Yang X, Sun F, Jiang R, Linn DE, Chen H, Chen H, Kong X, Melamed J, Tepper CG, et al. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 2009;69:2305–2313. doi: 10.1158/0008-5472.CAN-08-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handratta VD, Vasaitis TS, Njar VC, Gediya LK, Kataria R, Chopra P, Newman D, Jr., Farquhar R, Guo Z, Qiu Y, et al. Novel C-17-heteroaryl steroidal CYP17 inhibitors/antiandrogens: synthesis, in vitro biological activity, pharmacokinetics, and antitumor activity in the LAPC4 human prostate cancer xenograft model. J Med Chem. 2005;48:2972–2984. doi: 10.1021/jm040202w. [DOI] [PubMed] [Google Scholar]
- He B, Lanz RB, Fiskus W, Geng C, Yi P, Hartig SM, Rajapakshe K, Shou J, Wei L, Shah SS, et al. GATA2 facilitates steroid receptor coactivator recruitment to the androgen receptor complex. Proc Natl Acad Sci U S A. 2014;111:18261–18266. doi: 10.1073/pnas.1421415111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornberg E, Ylitalo EB, Crnalic S, Antti H, Stattin P, Widmark A, Bergh A, Wikstrom P. Expression of androgen receptor splice variants in prostate cancer bone metastases is associated with castration-resistance and short survival. PLoS.One. 2011;6:e19059. doi: 10.1371/journal.pone.0019059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R, Dunn TA, Wei S, Isharwal S, Veltri RW, Humphreys E, Han M, Partin AW, Vessella RL, Isaacs WB, et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 2009;69:16–22. doi: 10.1158/0008-5472.CAN-08-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R, Isaacs WB, Luo J. A snapshot of the expression signature of androgen receptor splicing variants and their distinctive transcriptional activities. Prostate. 2011;71:1656–1667. doi: 10.1002/pros.21382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R, Lu C, Mostaghel EA, Yegnasubramanian S, Gurel M, Tannahill C, Edwards J, Isaacs WB, Nelson PS, Bluemn E, et al. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res. 2012;72:3457–3462. doi: 10.1158/0008-5472.CAN-11-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagla M, Feve M, Kessler P, Lapouge G, Erdmann E, Serra S, Bergerat JP, Ceraline J. A splicing variant of the androgen receptor detected in a metastatic prostate cancer exhibits exclusively cytoplasmic actions. Endocrinology. 2007;148:4334–4343. doi: 10.1210/en.2007-0446. [DOI] [PubMed] [Google Scholar]
- Kahn B, Collazo J, Kyprianou N. Androgen receptor as a driver of therapeutic resistance in advanced prostate cancer. Int J Biol Sci. 2014;10:588–595. doi: 10.7150/ijbs.8671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaca M, Liu Y, Zhang Z, De Silva D, Parker JS, Earp HS, Whang YE. Mutation of androgen receptor N-terminal phosphorylation site Tyr-267 leads to inhibition of nuclear translocation and DNA binding. PLoS One. 2015;10:e0126270. doi: 10.1371/journal.pone.0126270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Banuelos CA, Imamura Y, Leung JK, Caley DP, Wang J, Mawji NR, Sadar MD. Cotargeting Androgen Receptor Splice Variants and mTOR Signaling Pathway for the Treatment of Castration-Resistant Prostate Cancer. Clin Cancer Res. 2016;22:2744–2754. doi: 10.1158/1078-0432.CCR-15-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen KE, Scher HI. Starving the addiction: new opportunities for durable suppression of AR signaling in prostate cancer. Clin.Cancer Res. 2009;15:4792–4798. doi: 10.1158/1078-0432.CCR-08-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwegyir-Afful AK, Ramalingam S, Purushottamachar P, Ramamurthy VP, Njar VC. Galeterone and VNPT55 induce proteasomal degradation of AR/AR-V7, induce significant apoptosis via cytochrome c release and suppress growth of castration resistant prostate cancer xenografts in vivo. Oncotarget. 2015;6:27440–27460. doi: 10.18632/oncotarget.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ban F, Dalal K, Leblanc E, Frewin K, Ma D, Adomat H, Rennie PS, Cherkasov A. Discovery of small-molecule inhibitors selectively targeting the DNA-binding domain of the human androgen receptor. J Med Chem. 2014a;57:6458–6467. doi: 10.1021/jm500802j. [DOI] [PubMed] [Google Scholar]
- Li H, Xie N, Gleave ME, Dong X. Catalytic inhibitors of DNA topoisomerase II suppress the androgen receptor signaling and prostate cancer progression. Oncotarget. 2015a;6:20474–20484. doi: 10.18632/oncotarget.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Chen M, Truong S, Yan C, Buttyan R. Determinants of Gli2 co-activation of wildtype and naturally truncated androgen receptors. Prostate. 2014b;74:1400–1410. doi: 10.1002/pros.22855. [DOI] [PubMed] [Google Scholar]
- Li X, Liu Z, Xu X, Blair CA, Sun Z, Xie J, Lilly MB, Zi X. Kava components down-regulate expression of AR and AR splice variants and reduce growth in patient-derived prostate cancer xenografts in mice. PLoS.One. 2012a;7:e31213. doi: 10.1371/journal.pone.0031213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Alsagabi M, Fan D, Bova GS, Tewfik AH, Dehm SM. Intragenic rearrangement and altered RNA splicing of the androgen receptor in a cell-based model of prostate cancer progression. Cancer Res. 2011;71:2108–2117. doi: 10.1158/0008-5472.CAN-10-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Chan SC, Brand LJ, Hwang TH, Silverstein KA, Dehm SM. Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer Res. 2013;73:483–489. doi: 10.1158/0008-5472.CAN-12-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Hwang TH, Oseth LA, Hauge A, Vessella RL, Schmechel SC, Hirsch B, Beckman KB, Silverstein KA, Dehm SM. AR intragenic deletions linked to androgen receptor splice variant expression and activity in models of prostate cancer progression. Oncogene. 2012b;31:4759–4767. doi: 10.1038/onc.2011.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Xie N, Gleave ME, Rennie PS, Dong X. AR-v7 protein expression is regulated by protein kinase and phosphatase. Oncotarget. 2015b;6:33743–33754. doi: 10.18632/oncotarget.5608. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Liu C, Armstrong C, Zhu Y, Lou W, Gao AC. Niclosamide enhances abiraterone treatment via inhibition of androgen receptor variants in castration resistant prostate cancer. Oncotarget. 2016;7:32210–32220. doi: 10.18632/oncotarget.8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Lou W, Armstrong C, Zhu Y, Evans CP, Gao AC. Niclosamide suppresses cell migration and invasion in enzalutamide resistant prostate cancer cells via Stat3-AR axis inhibition. Prostate. 2015a;75:1341–1353. doi: 10.1002/pros.23015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Lou W, Zhu Y, Nadiminty N, Schwartz CT, Evans CP, Gao AC. Niclosamide inhibits androgen receptor variants expression and overcomes enzalutamide resistance in castration-resistant prostate cancer. Clin Cancer Res. 2014a;20:3198–3210. doi: 10.1158/1078-0432.CCR-13-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Sprenger C, Sun S, Epilepsia KS, Haugk K, Zhang X, Coleman I, Nelson PS, Plymate S. AR variant ARv567es induces carcinogenesis in a novel transgenic mouse model of prostate cancer. Neoplasia. 2013;15:1009–1017. doi: 10.1593/neo.13784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Sprenger C, Wu PJ, Sun S, Uo T, Haugk K, Epilepsia KS, Plymate S. MED1 mediates androgen receptor splice variant induced gene expression in the absence of ligand. Oncotarget. 2015b;6:288–304. doi: 10.18632/oncotarget.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LL, Xie N, Sun S, Plymate S, Mostaghel E, Dong X. Mechanisms of the androgen receptor splicing in prostate cancer cells. Oncogene. 2014b;33:3140–3150. doi: 10.1038/onc.2013.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Li S, Gan L, Kao TP, Huang H. A transcription-independent function of FOXO1 in inhibition of androgen-independent activation of the androgen receptor in prostate cancer cells. Cancer Res. 2008;68:10290–10299. doi: 10.1158/0008-5472.CAN-08-2038. [DOI] [PubMed] [Google Scholar]
- Lu J, Lonergan PE, Nacusi LP, Wang L, Schmidt LJ, Sun Z, van der Steen T, Boorjian SA, Kosari F, Vasmatzis G, et al. The cistrome and gene signature of androgen receptor splice variants in castration-resistant prostate cancer cells. J Urol. 2014 doi: 10.1016/j.juro.2014.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Van der Steen T, Tindall DJ. Are androgen receptor variants a substitute for the full-length receptor? Nat Rev Urol. 2015;12:137–144. doi: 10.1038/nrurol.2015.13. [DOI] [PubMed] [Google Scholar]
- Martin SK, Banuelos CA, Sadar MD, Kyprianou N. N-terminal targeting of androgen receptor variant enhances response of castration resistant prostate cancer to taxane chemotherapy. Molecular Oncology. 2015;9:628–639. doi: 10.1016/j.molonc.2014.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SK, Kyprianou N. Exploitation of the Androgen Receptor to Overcome Taxane Resistance in Advanced Prostate Cancer. Adv Cancer Res. 2015;127:123–158. doi: 10.1016/bs.acr.2015.03.001. [DOI] [PubMed] [Google Scholar]
- Martin SK, Pu H, Penticuff JC, Cao Z, Horbinski C, Kyprianou N. Multinucleation and Mesenchymal-to-Epithelial Transition Alleviate Resistance to Combined Cabazitaxel and Antiandrogen Therapy in Advanced Prostate Cancer. Cancer Res. 2016;76:912–926. doi: 10.1158/0008-5472.CAN-15-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashima T, Okabe S, Seimiya H. Pharmacological targeting of constitutively active truncated androgen receptor by nigericin and suppression of hormone-refractory prostate cancer cell growth. Mol.Pharmacol. 2010;78:846–854. doi: 10.1124/mol.110.064790. [DOI] [PubMed] [Google Scholar]
- McGrath MJ, Binge LC, Sriratana A, Wang H, Robinson PA, Pook D, Fedele CG, Brown S, Dyson JM, Cottle DL, et al. Regulation of the transcriptional coactivator FHL2 licenses activation of the androgen receptor in castrate-resistant prostate cancer. Cancer Res. 2013;73:5066–5079. doi: 10.1158/0008-5472.CAN-12-4520. [DOI] [PubMed] [Google Scholar]
- Mediwala SN, Sun H, Szafran AT, Hartig SM, Sonpavde G, Hayes TG, Thiagarajan P, Mancini MA, Marcelli M. The activity of the androgen receptor variant AR-V7 is regulated by FOXO1 in a PTEN-PI3K-AKT-dependent way. Prostate. 2013;73:267–277. doi: 10.1002/pros.22566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellado B, Jimenez N, Marin-Aguilera M, Reig O. Diving Into Cabazitaxel's Mode of Action: More Than a Taxane for the Treatment of Castration-Resistant Prostate Cancer Patients. Clin Genitourin Cancer. 2016;14:265–270. doi: 10.1016/j.clgc.2015.12.030. [DOI] [PubMed] [Google Scholar]
- Miyamoto DT, Zheng Y, Wittner BS, Lee RJ, Zhu H, Broderick KT, Desai R, Fox DB, Brannigan BW, Trautwein J, et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science. 2015;349:1351–1356. doi: 10.1126/science.aab0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostaghel EA, Marck BT, Plymate SR, Vessella RL, Balk S, Matsumoto AM, Nelson PS, Montgomery RB. Resistance to CYP17A1 Inhibition with Abiraterone in Castration-Resistant Prostate Cancer: Induction of Steroidogenesis and Androgen Receptor Splice Variants. Clinical Cancer Research. 2011;17:5913–5925. doi: 10.1158/1078-0432.CCR-11-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller JM, Isele U, Metzger E, Rempel A, Moser M, Pscherer A, Breyer T, Holubarsch C, Buettner R, Schule R. FHL2, a novel tissue-specific coactivator of the androgen receptor. EMBO J. 2000;19:359–369. doi: 10.1093/emboj/19.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung JK, Banuelos CA, Fernandez JG, Mawji NR, Wang J, Tien AH, Yang YC, Tavakoli I, Haile S, Watt K, et al. An androgen receptor N-terminal domain antagonist for treating prostate cancer. J.Clin.Invest. 2013;123:2948–2960. doi: 10.1172/JCI66398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadiminty N, Tummala R, Liu C, Lou W, Evans CP, Gao AC. NF-kappaB2/p52:c-Myc:hnRNPA1 Pathway Regulates Expression of Androgen Receptor Splice Variants and Enzalutamide Sensitivity in Prostate Cancer. Mol Cancer Ther. 2015;14:1884–1895. doi: 10.1158/1535-7163.MCT-14-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadiminty N, Tummala R, Liu C, Yang J, Lou W, Evans CP, Gao AC. NF-kappaB2/p52 Induces Resistance to Enzalutamide in Prostate Cancer: Role of Androgen Receptor and Its Variants. Mol.Cancer Ther. 2013;12:1629–1637. doi: 10.1158/1535-7163.MCT-13-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njar VC, Brodie AM. Discovery and development of Galeterone (TOK-001 or VN/124-1) for the treatment of all stages of prostate cancer. J Med Chem. 2015;58:2077–2087. doi: 10.1021/jm501239f. [DOI] [PubMed] [Google Scholar]
- Nyquist MD, Li Y, Hwang TH, Manlove LS, Vessella RL, Silverstein KA, Voytas DF, Dehm SM. TALEN-engineered AR gene rearrangements reveal endocrine uncoupling of androgen receptor in prostate cancer. Proc Natl Acad Sci U S A. 2013;110:17492–17497. doi: 10.1073/pnas.1308587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onstenk W, Sieuwerts AM, Kraan J, Van M, Nieuweboer AJ, Mathijssen RH, Hamberg P, Meulenbeld HJ, De Laere B, Dirix LY, et al. Efficacy of Cabazitaxel in Castration-resistant Prostate Cancer Is Independent of the Presence of AR-V7 in Circulating Tumor Cells. Eur Urol. 2015;68:939–945. doi: 10.1016/j.eururo.2015.07.007. [DOI] [PubMed] [Google Scholar]
- Peacock SO, Fahrenholtz CD, Burnstein KL. Vav3 enhances androgen receptor splice variant activity and is critical for castration-resistant prostate cancer growth and survival. Mol Endocrinol. 2012;26:1967–1979. doi: 10.1210/me.2012-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purushottamachar P, Godbole AM, Gediya LK, Martin MS, Vasaitis TS, Kwegyir-Afful AK, Ramalingam S, Ates-Alagoz Z, Njar VC. Systematic structure modifications of multitarget prostate cancer drug candidate galeterone to produce novel androgen receptor down-regulating agents as an approach to treatment of advanced prostate cancer. J Med Chem. 2013;56:4880–4898. doi: 10.1021/jm400048v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y, Dai B, Ye D, Kong Y, Chang K, Jia Z, Yang X, Zhang H, Zhu Y, Shi G. Constitutively active AR-V7 plays an essential role in the development and progression of castration-resistant prostate cancer. Sci Rep. 2015;5:7654. doi: 10.1038/srep07654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S, Lyons LS, Fahrenholtz CD, Wu F, Farooq A, Balkan W, Burnstein KL. A novel nuclear role for the Vav3 nucleotide exchange factor in androgen receptor coactivation in prostate cancer. Oncogene. 2012;31:716–727. doi: 10.1038/onc.2011.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, Montgomery B, Taplin ME, Pritchard CC, Attard G, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan CJ, Smith MR, de Bono JS, Molina A, Logothetis CJ, de Souza P, Fizazi K, Mainwaring P, Piulats JM, Ng S, et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. N Engl J Med. 2013;368:138–148. doi: 10.1056/NEJMoa1209096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadar MD. Small molecule inhibitors targeting the "achilles' heel" of androgen receptor activity. Cancer Res. 2011;71:1208–1213. doi: 10.1158/0008-5472.CAN_10-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saporita AJ, Zhang Q, Navai N, Dincer Z, Hahn J, Cai X, Wang Z. Identification and characterization of a ligand-regulated nuclear export signal in androgen receptor. J Biol Chem. 2003;278:41998–42005. doi: 10.1074/jbc.M302460200. [DOI] [PubMed] [Google Scholar]
- Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, de WR, Mulders P, Chi KN, Shore ND, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N.Engl.J.Med. 2012;367:1187–1197. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- Shafi AA, Cox MB, Weigel NL. Androgen receptor splice variants are resistant to inhibitors of Hsp90 and FKBP52, which alter androgen receptor activity and expression. Steroids. 2013;78:548–554. doi: 10.1016/j.steroids.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafi AA, Putluri V, Arnold JM, Tsouko E, Maity S, Roberts JM, Coarfa C, Frigo DE, Putluri N, Sreekumar A, et al. Differential regulation of metabolic pathways by androgen receptor (AR) and its constitutively active splice variant, AR-V7, in prostate cancer cells. Oncotarget. 2015;6:31997–32012. doi: 10.18632/oncotarget.5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streicher W, Luedeke M, Azoitei A, Zengerling F, Herweg A, Genze F, Schrader MG, Schrader AJ, Cronauer MV. Stilbene induced inhibition of androgen receptor dimerization: implications for AR and ARDeltaLBD-signalling in human prostate cancer cells. PLoS ONE. 2014;9:e98566. doi: 10.1371/journal.pone.0098566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Chen HG, Li W, Yang X, Wang X, Jiang R, Guo Z, Chen H, Huang J, Borowsky AD, et al. Androgen receptor splice variant AR3 promotes prostate cancer via modulating expression of autocrine/paracrine factors. J Biol Chem. 2014;289:1529–1539. doi: 10.1074/jbc.M113.492140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Indran IR, Zhang ZW, Tan MH, Li Y, Lim ZL, Hua R, Yang C, Soon FF, Li J, et al. A novel prostate cancer therapeutic strategy using icaritin-activated arylhydrocarbon-receptor to co-target androgen receptor and its splice variants. Carcinogenesis. 2015;36:757–768. doi: 10.1093/carcin/bgv040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Sprenger CC, Vessella RL, Haugk K, Soriano K, Mostaghel EA, Page ST, Coleman IM, Nguyen HM, Sun H, et al. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. J.Clin.Invest. 2010;120:2715–2730. doi: 10.1172/JCI41824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thadani-Mulero M, Portella L, Sun S, Sung M, Matov A, Vessella RL, Corey E, Nanus DM, Plymate SR, Giannakakou P. Androgen receptor splice variants determine taxane sensitivity in prostate cancer. Cancer Res. 2014;74:2270–2282. doi: 10.1158/0008-5472.CAN-13-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V, Wongvipat J, Smith-Jones PM, Yoo D, Kwon A, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science. 2009;324:787–790. doi: 10.1126/science.1168175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Royen ME, van Cappellen WA, de VC, Houtsmuller AB, Trapman J. Stepwise androgen receptor dimerization. J.Cell Sci. 2012;125:1970–1979. doi: 10.1242/jcs.096792. [DOI] [PubMed] [Google Scholar]
- van Soest RJ, van Royen ME, de Morree ES, Moll JM, Teubel W, Wiemer EA, Mathijssen RH, de Wit R, van Weerden WM. Cross-resistance between taxanes and new hormonal agents abiraterone and enzalutamide may affect drug sequence choices in metastatic castration-resistant prostate cancer. Eur J Cancer. 2013;49:3821–3830. doi: 10.1016/j.ejca.2013.09.026. [DOI] [PubMed] [Google Scholar]
- Vanaja DK, Mitchell SH, Toft DO, Young CY. Effect of geldanamycin on androgen receptor function and stability. Cell Stress Chaperones. 2002;7:55–64. doi: 10.1379/1466-1268(2002)007<0055:eogoar>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Li W, Liu XS, Carroll JS, Janne OA, Keeton EK, Chinnaiyan AM, Pienta KJ, Brown M. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol.Cell. 2007;27:380–392. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Li W, Zhang Y, Yuan X, Xu K, Yu J, Chen Z, Beroukhim R, Wang H, Lupien M, et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138:245–256. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Sharma D, Ren Y, Fondell JD. A coregulatory role for the TRAP-mediator complex in androgen receptor-mediated gene expression. J Biol Chem. 2002;277:42852–42858. doi: 10.1074/jbc.M206061200. [DOI] [PubMed] [Google Scholar]
- Watson PA, Chen YF, Balbas MD, Wongvipat J, Socci ND, Viale A, Kim K, Sawyers CL. Constitutively active androgen receptor splice variants expressed in castration-resistant prostate cancer require full-length androgen receptor. Proc.Natl.Acad.Sci.U.S.A. 2010;107:16759–16765. doi: 10.1073/pnas.1012443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welti J, Rodrigues DN, Sharp A, Sun S, Lorente D, Riisnaes R, Figueiredo I, Zafeiriou Z, Rescigno P, de Bono JS, et al. Analytical Validation and Clinical Qualification of a New Immunohistochemical Assay for Androgen Receptor Splice Variant-7 Protein Expression in Metastatic Castration-resistant Prostate Cancer. European Urology. doi: 10.1016/j.eururo.2016.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Sunkel B, Chen Z, Liu X, Ye Z, Li Q, Grenade C, Ke J, Zhang C, Chen H, et al. Three-tiered role of the pioneer factor GATA2 in promoting androgen-dependent gene expression in prostate cancer. Nucleic Acids Res. 2014;42:3607–3622. doi: 10.1093/nar/gkt1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Zhan Y, Qi Y, Cao B, Bai S, Xu W, Gambhir SS, Lee P, Sartor O, Flemington EK, et al. Androgen Receptor Splice Variants Dimerize to Transactivate Target Genes. Cancer Res. 2015;75:3663–3671. doi: 10.1158/0008-5472.CAN-15-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Loriot Y, Beraldi E, Zhang F, Wyatt AW, Nakouzi NA, Mo F, Zhou T, Kim Y, Monia BP, et al. Generation 2.5 antisense oligonucleotides targeting the androgen receptor and its splice variants suppress enzalutamide-resistant prostate cancer cell growth. Clin Cancer Res. 2015;21:1675–1687. doi: 10.1158/1078-0432.CCR-14-1108. [DOI] [PubMed] [Google Scholar]
- Yamashita S, Lai KP, Chuang KL, Xu D, Miyamoto H, Tochigi T, Pang ST, Li L, Arai Y, Kung HJ, et al. ASC-J9 suppresses castration-resistant prostate cancer growth through degradation of full-length and splice variant androgen receptors. Neoplasia. 2012;14:74–83. doi: 10.1593/neo.111436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Guo Z, Sun F, Li W, Alfano A, Shimelis H, Chen M, Brodie AM, Chen H, Xiao Z, et al. Novel membrane-associated androgen receptor splice variant potentiates proliferative and survival responses in prostate cancer cells. J Biol Chem. 2011;286:36152–36160. doi: 10.1074/jbc.M111.265124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Cai C, Gao S, Simon NI, Shen HC, Balk SP. Galeterone prevents androgen receptor binding to chromatin and enhances degradation of mutant androgen receptor. Clin Cancer Res. 2014;20:4075–4085. doi: 10.1158/1078-0432.CCR-14-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zengerling F, Streicher W, Schrader AJ, Schrader M, Nitzsche B, Cronauer MV, Hopfner M. Effects of sorafenib on C-terminally truncated androgen receptor variants in human prostate cancer cells. Int.J.Mol.Sci. 2012;13:11530–11542. doi: 10.3390/ijms130911530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Y, Cao B, Qi Y, Liu S, Zhang Q, Zhou W, Xu D, Lu H, Sartor O, Kong W, et al. Methylselenol prodrug enhances MDV3100 efficacy for treatment of castration-resistant prostate cancer. International Journal of Cancer. 2013;133:2225–2233. doi: 10.1002/ijc.28202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Liu X, Li J, Ledet E, Alvarez X, Qi Y, Fu X, Sartor O, Dong Y, Zhang H. Androgen receptor splice variants circumvent AR blockade by microtubule-targeting agents. Oncotarget. 2015;6:23358–23371. doi: 10.18632/oncotarget.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Morrissey C, Sun S, Ketchandji M, Nelson PS, True LD, Vakar-Lopez F, Vessella RL, Plymate SR. Androgen receptor variants occur frequently in castration resistant prostate cancer metastases. PLoS.One. 2011;6:e27970. doi: 10.1371/journal.pone.0027970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhou N, Huang J, Ho TT, Zhu Z, Qiu Z, Zhou X, Bai C, Wu F, Xu M, et al. Regulation of androgen receptor splice variant AR3 by PCGEM1. Oncotarget. 2016;7:15481–15491. doi: 10.18632/oncotarget.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu ML, Horbinski CM, Garzotto M, Qian DZ, Beer TM, Kyprianou N. Tubulin-targeting chemotherapy impairs androgen receptor activity in prostate cancer. Cancer Res. 2010;70:7992–8002. doi: 10.1158/0008-5472.CAN-10-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]