Abstract
Peroxy-caged luciferin (PCL-1) probe was first used to image hydrogen peroxide in living systems (Van de Bittner GC et al. Proc Natl Acad Sci U S A. 2010; 107:21316–21). Recently this probe was shown to react with peroxynitrite more potently than with hydrogen peroxide (Sieracki et al. Free Radic Biol Med. 2013; 61:40–50) and was suggested to be a more suitable probe for detecting peroxynitrite under in vivo conditions. In this work, we investigated in detail the products formed from the reaction between PCL-1 and hydrogen peroxide, hypochlorite, and peroxynitrite. HPLC analysis showed that hydrogen peroxide reacts slowly with PCL-1, forming luciferin as the only product. Hypochlorite reaction with PCL-1 yielded significantly less luciferin, as hypochlorite oxidized luciferin to form a chlorinated luciferin. Reaction between PCL-1 and peroxynitrite consists of a major and minor pathway. The major pathway results in luciferin and the minor pathway produces a radical-mediated nitrated luciferin. Radical intermediate was characterized by spin trapping. We conclude that monitoring of chlorinated and nitrated products in addition to bioluminescence in vivo will help identify the nature of oxidant responsible for bioluminescence derived from PCL-1.
Keywords: boronate probes, luciferin, bioluminescence, spin traps
Graphical Abstract
1. Introduction
Reactive oxygen species (ROS) and reactive nitrogen species (RNS) have emerged as important mediators of cellular signaling and damage [1–3]. ROS and RNS comprise of different species of very diverse chemical reactivity, lifetime and target specificity in extracellular and intracellular milieu [4,5]. The term ‘ROS’ typically refers to superoxide radical anion (O2•−), hydrogen peroxide (H2O2), hydroxyl radical (•OH), lipid peroxy radicals (LOO•), lipid hydroperoxides (LOOH), and singlet oxygen (1O2) and ‘RNS’ refers to peroxynitrite (ONOO−) and nitrogen dioxide radical (•NO2). Rigorous identification of those species is crucial for understanding their role in cellular signaling and pathology.
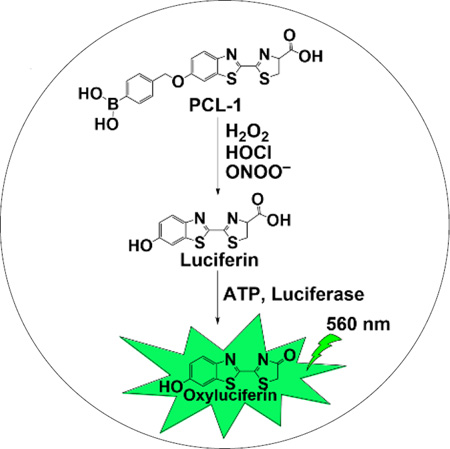
Molecular imaging of ROS/RNS is an emerging area of research in redox and free radical biology [6–8]. Bioluminescence or fluorescence modalities are typically used. Peroxy-caged luciferin (PCL-1) (Fig. 1) is one of the first cell-permeable small molecular weight probes used to image ROS in living systems [9,10]. H2O2 slowly reacts with PCL-1 probe (k = 1.2 M−1s−1 [11]) to form luciferin in situ that is oxidized by the luciferase enzyme (using ATP as a co-factor) emitting a green bioluminescent signal [9,10] (Fig. 1). Upon oxidation, PCL-1 probe eliminates the para-quinone methide (QM), with the formation of luciferin which gets oxidized to oxyluciferin in luciferase-transfected cells generating bioluminescence (Fig. 1). It was shown that administration of bolus H2O2 to mice overexpressing luciferase increased the bioluminescent signal from PCL-1 [9,10]. Subsequently, PCL-1 was shown to be oxidized to luciferin in the presence of hypochlorite (HOCl) and ONOO− [11]. Importantly, it was shown that in the presence of plasma, the probability of oxidation of PCL-1 probe by H2O2 is very low; however, under this condition, the probe was still oxidized by ONOO−. This is consistent with our previous reports demonstrating that ONOO− reacts directly and rapidly with boronate probes forming corresponding phenols as the major products. This brings into question the identity of the oxidant(s) responsible for in vivo bioluminescence measurements using PCL-1 probe.
Figure 1.
A hypothetical scheme showing the application of PCL-1 probe for bioluminescent detection of H2O2, HOCl or ONOO− in luciferase-transfected mice tumor xenografts.
We have demonstrated that in addition to H2O2, other biologically-relevant oxidants, including HOCl and ONOO−, are able to oxidize aromatic boronates to the corresponding phenolic products [12–15]. More recently, we have shown that selected amino acid hydroperoxides (AA-OOH) and protein hydroperoxides (Pr-OOH) also oxidize boronic compounds [16]. With all of the oxidants tested so far, the mechanism appears to be similar; a 1:1 stoichiometry between oxidant and boronate probes was observed, resulting in the same major product. However, the main difference is the rate constant of the reaction between different oxidants and the boronate probe. The rate constants varied from 100, 101, 104 to 106 M−1s−1 for H2O2, AA-OOH, HOCl and ONOO−, respectively [13,16]. Therefore, depending on the experimental settings, boronates may be used to detect different oxidants by monitoring the reaction products. Unlike other listed oxidants, ONOO− oxidizes boronic compounds in two pathways: major (~90%), non-radical pathway, leading to the corresponding phenol; and minor (~10%), radical pathway, forming a phenyl-type radical, nitrogen dioxide (•NO2) and stable products formed from them [12,13,17]. We propose that these ONOO−-specific products may serve as specific markers for ONOO−. By determining the ONOO−-specific products, we recently confirmed the formation of ONOO− from nitroxyl (HNO) reaction with O2 [18] and tested the effect of inhibition of NADPH oxidase on the production of ONOO− by activated macrophages [19].
Here we investigate in detail the products formed from the oxidative and nitrative chemistry of PCL-1 that will help to better interpret in vivo bioluminescence results. We compared the products formed during the oxidation of PCL-1 by H2O2, HOCl and ONOO−, the likely in vivo inflammatory oxidants. As several oxidant species react with PCL-1 to generate bioluminescence, the oxidant-specific minor product(s) may be used to confirm the identity of ROS/RNS species.
2. Materials and methods
2.1. Chemicals, preparation of solutions
PCL-1 probe was synthesized as described below. D-Luciferin (potassium salt) was purchased from Gold Biotechnology. H2O2 and HOCl were from Sigma-Aldrich. ONOO− was synthesized as described elsewhere [12] and stored at −80 °C. L-NAME and DPI were from Cayman. All other chemicals were from Sigma-Aldrich and were of highest purity available. The stock solutions of ONOO−, HOCl and H2O2 were prepared freshly each day and the concentration was determined by spectrophotometry, using the extinction coefficients values of 1.7×103 M−1cm−1 (at 302 nm, in 0.1 M NaOH), 350 M−1cm−1 (at 292 nm, in 0.1 M NaOH) and 39.4 M−1cm−1 (at 240 nm, in water), respectively. PCL-1 stock solution (1 mM) was typically prepared using ethanol (EtOH) as a solvent to minimize scavenging of HOCl by DMSO, a solvent typically used for boronate probes. Of the four organic solvent (EtOH, acetonitrile, DMSO and DMF) tested for interference with HOCl-induced oxidation of coumarin boronic acid (CBA) to hydroxycoumarin (COH), EtOH exhibited the smallest inhibitory effect (Suppl. Fig. 1). For the spin trapping of phenyl radical, DMSO was used to prepare the stock solution of PCL-1 to avoid scavenging of phenyl radical by EtOH. 5-Diisopropoxyphosphoryl-5-methyl-1-pyrroline N-oxide (DIPPMPO) was synthesized according to the published procedure [20].
2.2. HPLC analyses
HPLC analyses of PCL-1 and its oxidation products were performed using Agilent 1100 HPLC equipped with UV-vis absorption and fluorescence detectors. The compounds were loaded onto Kinetex C18 column (Phenomenex, 100 mm × 4.6 mm, 2.6 µm) equilibrated with 10% of acetonitrile in water, containing 0.1% trifluoroacetic acid. The products were eluted by an increase of the acetonitrile concentration from 10–100% over 7 min. The flow rate was kept at 1.5 mL/min. PCL-1 and the products containing luciferin moiety were detected by monitoring absorbance at 330 nm, and the product of water added to QM was detected at 220 nm. Additionally, luciferin was also monitored using the fluorescence detector with the excitation set at 330 nm and emission set at 520 nm.
2.3. LC-MS analyses
LC-MS analyses of PCL-1, its oxidation products and spin adducts were performed using Shimadzu LC-MS 8030 triple quadrupole mass detector coupled to Shimadzu Nexera 2 UHPLC system. The reaction mixture was injected on Cortecs C18 column (Waters, 50 mm × 2 mm, 1.6 µm) equilibrated with 10% of acetonitrile in water containing 0.1% of formic acid. The compound was eluted by increasing the acetonitrile concentration in the mobile phase from 10–80% over 4 min. The flow rate was set at 0.5 mL/min, and the flow was diverted to waste during the first minute and after 4 min, counting from the time of injection. PCL-1, luciferin, Luc-Bz-NO2, Luc-Bz-H and Luc-Cl were detected as positive ions using multiple reaction monitoring (MRM) mode, using the primary/fragment ion pairs of 415>135, 281>235, 416>234, 371>91 and 315>269, respectively. Luc-Bz-OH was detected in positive mode using single ion monitoring (SIM), set at the m/z value of 387.
2.4. EPR spin-trapping
EPR spin trapping experiments were performed using Bruker EMX EPR spectrometer, as reported previously [17]. The instrument parameters were as follows: scan range, 150 G; time constant, 1.28 ms; scan time, 84 s; modulation amplitude, 1 G; modulation frequency, 100 kHz; receiver gain, 1×105; and microwave power, 20 mW. The spectra shown are the averages of 5 scans.
2.5. NMR analyses
NMR analyses for determination of the structure of synthesized standards of Luc-Bz-NO2 and Luc-Cl were performed at the Aix-Marseille Université (Spectropole). 1H NMR and 13C NMR spectra were recorded with a Bruker DPX 600 spectrometer at 400.13 or 600.13 MHz and 75.54 MHz, respectively. Solutions were prepared in CDCl3 as a solvent, using TMS or CDCl3 as internal reference for 1H NMR and 13C NMR spectra respectively. Chemical shifts (δ) are reported in ppm, and coupling constant J values in hertz (Hz). NMR peak multiplicities are described as follows: s, singlet; d, doublet; dd, doublet of doublets; brdd, broad doublet of doublets; and m, multiplets.
2.6. Synthesis of PCL-1 and standards of the products
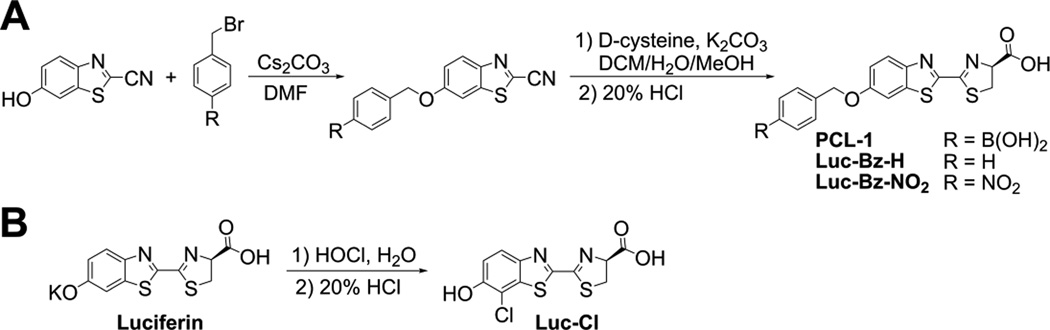
PCL-1, Luc-Bz-H and Luc-Bz-NO2 (Fig. 2A) were synthesized by modifying the published protocol [9] using 4-(bromomethyl)benzeneboronic acid pinacol ester, benzyl bromide, and 4-nitrobenzyl bromide, respectively, as the starting materials. The synthetic protocol was modified to include the use of free amino acid, D-cysteine, rather than hydrochloride form. The crude products were purified on a silica column (hexane:ethyl acetate, 9:1). This synthetic method allowed us to obtain the compound of sufficient purity, without the need for HPLC-based purification. Therefore, relatively large amounts of the probe can be conveniently prepared using this protocol. Nitrobenzylated luciferin (Luc-Bz-NO2) structure was confirmed by NMR analysis: 1H NMR, (400.13 MHz): δ 8.29 (2H, d, J = 8.4), 8.10 (1H, dd, J = 8.7, 18.6), 7.87 (1H, d, J = 7.9), 7.77 (2H, d, J = 7.5), 7.38-7.29 (1H, m), 5.40 (2H, s), 5.2 (1H, m), 3.81-3.61 (2H, m).
Figure 2.
Synthetic pathways used to obtain (A) PCL-1 probe, Luc-Bz-H and Luc-Bz-NO2 and (B) Luc-Cl standards.
Chloroluciferin (Luc-Cl) was synthesized by reacting D-luciferin with HOCl, as shown in Figure 2B. D-Luciferin potassium salt (100 mg, 0.314 mmol) was dissolved in deionized water (10 mL). The solution was stirred in the dark, and HOCl (208 µL, 10 mM) was added. The reaction’s progress was monitored by HPLC. After depletion of luciferin, hydrochloric acid (2 M, 20 mL) was added to the mixture. The precipitate was filtered off, washed with water until washings were neutral, and dried under reduced pressure. Crude product was purified using preparative HPLC. The position of chlorination was determined based on NMR analyses: 1H NMR, (600.13 MHz): δ 7.94 (1H, d, J = 9.0), 7.30 (1H, d, J = 9.0), 5.35 (1H, br.dd, J = 10.4, 8.4), 3.75 (1H, dd, J = 10.9, 10.4), 3.71 (1H, dd, J = 10.9, 8.4); 13C NMR (75.47 MHz): δ 171.3 (1C, s), 163.3, (1C, s), 157.4 (1C, s), 153.4 (1C), 145.7 (1C), 137.1 (1C), 123.5 (1C), 117.6 (1C), 110.5 (1C), 78.9 (1C), 35.2 (1C). The lack of a signal of proton at the carbon atom C-2, in the 1H NMR spectrum of chlorinated luciferin (Suppl. Fig. 2A), points to its replacement by the chlorine atom. The assignment has been confirmed by 13C NMR (Suppl. Fig. 2B) and 2D-NMR analyses (Suppl. Fig. 3). The 13C chemical shift of the carbon atom C-2 at 110.5 ppm combined with the 3J correlation between proton atom Hb (7.3) and the carbon atom C2 prove that the chlorine atom is located in the position C-2 The NMR spectra of chloroluciferin, including two-dimensional sequences (heteronuclear multiple bond coherence, HMBC and heteronuclear single quantum coherence, HSQC) are shown in Supplementary Figures 2–3.
2.7. Cell culture and extraction of PCL-1 oxidation products
RAW 264.7 cells were cultured in DMEM medium (Invitrogen) supplemented with 10% FBS, 2 mM L-glutamine, 100 units/mL penicillin and 100 µg/mL streptomycin. For stimulation of the cells to produce nitric oxide, cells were incubated overnight (12–16 h) with LPS (0.5 µg/mL) and IFNγ (50 units/mL). To stimulate NADPH oxidase-dependent superoxide production, the cells were washed and treated with phorbol 12-myristate 13-acetate (PMA, 1 µM) in DPBS supplemented with pyruvic acid and glucose (DPBS-GP). At the time of addition of PMA, PCL-1 probe (100 µM) was also added and the cells were incubated for 1 h at 37 °C in a CO2-free incubator. Where indicated, L-NAME (1 mM) or diphenyleneiodonium (DPI, 0.1–10 µM) was added 30 min before PMA. After 1 h, an aliquot of the medium was collected and frozen in liquid nitrogen. Cells were washed twice with ice-cold PBS, harvested and centrifuged (1 min, 1000g). The supernatant was discarded and the cell pellet was frozen in liquid nitrogen.
For LC-MS analyses, cells were lysed in ice-cold DPBS containing 0.1% Triton X100. The cell lysates and cell media were mixed (1:1) with ice-cold acetonitrile and left for 30 min on ice, followed by centrifugation (30 min, 20,000g, 4 °C). Clear supernatants were subsequently mixed (1:1) with ice-cold 0.1% formic acid in water and centrifugated (15 min, 20,000g, 4 °C). The supernatants were analyzed by LC-MS, as described above.
3. Results
3.1. Identification of the primary product formed during the oxidation of PCL-1
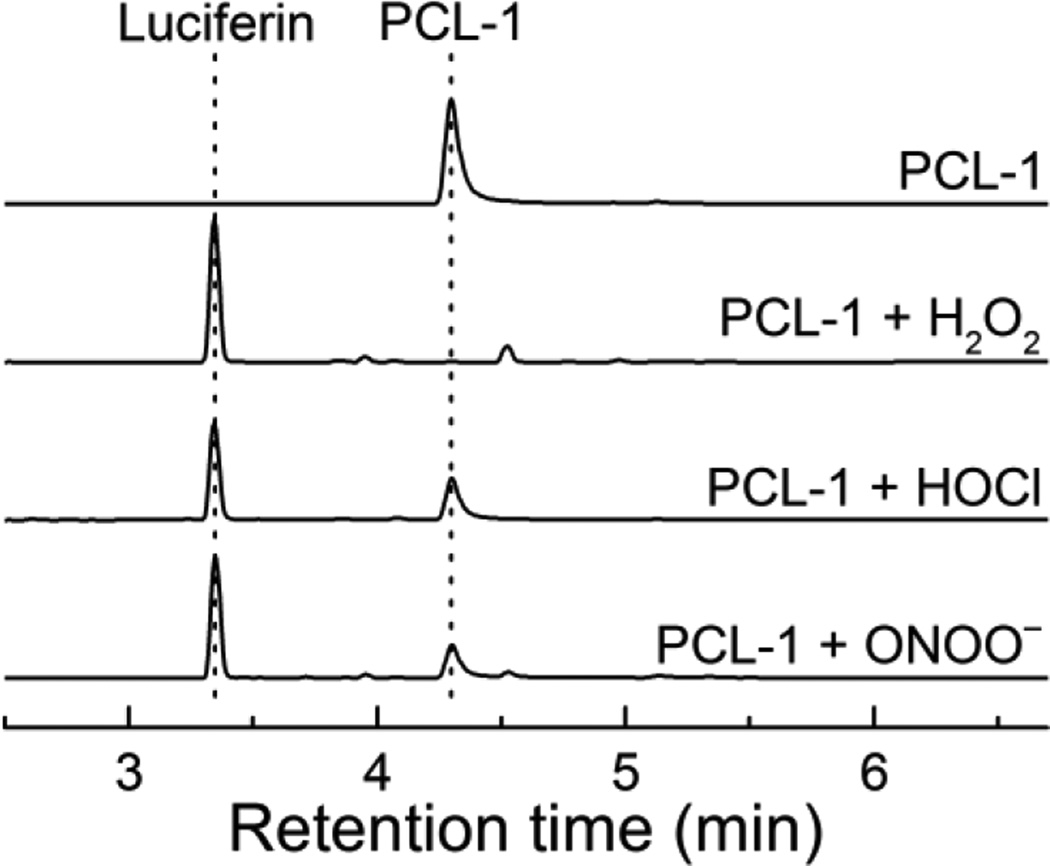
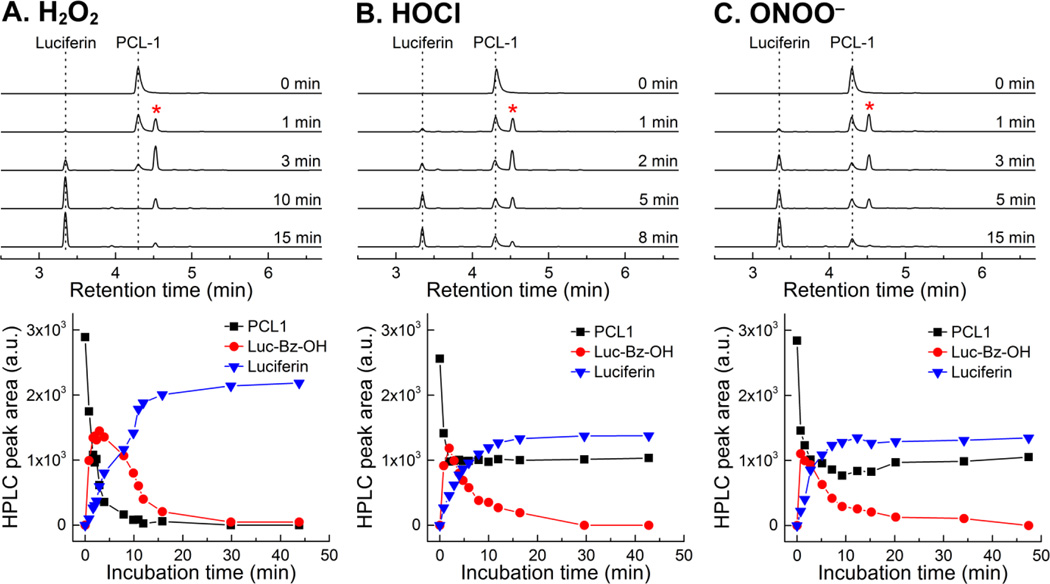
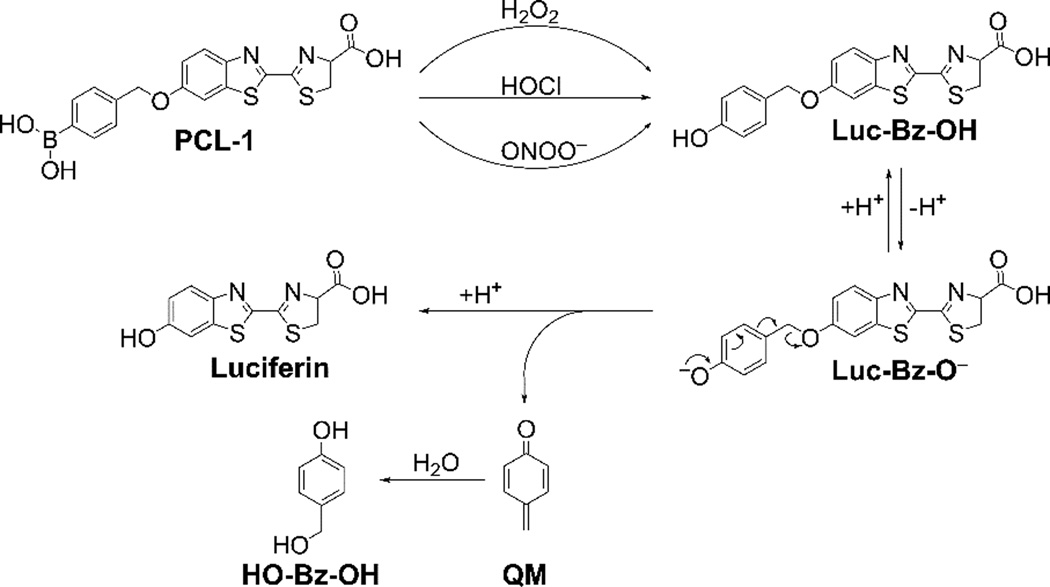
To understand the mechanism of oxidation of PCL-1 probe to luciferin, we detected and quantitated the products formed upon PCL-1 reaction with H2O2, HOCl and ONOO−. In agreement with previous reports [9–11], luciferin was detected as the major stable product in all cases (Fig. 3). However, HPLC-based monitoring of the product formation over time indicates that before luciferin is formed, elution of a peak (marked by an asterisk next to the PCL-1 probe peak) corresponding to a different species, was observed (Fig. 4A–C, top). As this species is formed from PCL-1 reaction with all three oxidants and its decomposition is accompanied by luciferin formation, we attributed this species to the primary phenolic product (Luc-Bz-OH in Fig. 5) formed prior to the elimination of the QM (Fig. 5). Oxidation of PCL-1 by ONOO− or HOCl resulted in the formation of the primary product formed immediately after mixing (< 1 min), as compared to the time resolution of the HPLC analyses (Fig. 4B and C, bottom). This is consistent with relatively high rate constants of the reaction of boronates with HOCl and ONOO− (104 and 106 M−1s−1, respectively) [12,13]. Under the conditions used, the reaction of PCL-1 with ONOO− was completed within 1 s, and with HOCl within 1 min, after mixing. In contrast, the reported rate constant of the reaction of boronates with H2O2 is ~1 M−1s−1 [12]. Thus, the reaction half-life when using 10 mM H2O2 can be calculated to be 70 s. This is consistent with the observed time course of PCL-1 consumption (Fig. 4A, bottom) and slower formation of the intermediate Luc-Bz-OH (Fig. 4, bottom), as compared to the other oxidants tested. When Luc-Bz-OH was generated rapidly from HOCl and ONOO− reaction with PCL-1, its decomposition kinetics could be easily monitored. The elimination of the QM (Fig. 5) moiety follows the first-order kinetics, with the rate constant of ~2×10−3 s−1 determined at pH 7.4 from the time-dependent decay of Luc-Bz-OH (Fig. 4B–C, bottom).
Figure 3.
HPLC chromatograms obtained after a 30-min incubation of PCL-1 probe (100 µM) alone or in the presence of H2O2 (10 mM), HOCl (80 µM) or ONOO− (80 µM) in aqueous solutions containing phosphate buffer (0.1 M, pH 7.4) and dtpa (10 µM). The traces were collected using the absorption detector set at 330 nm.
Figure 4.
Time course of the conversion of PCL-1 probe (0.1 mM) into luciferin product in the presence of (A) H2O2 (10 mM), (B) HOCl (80 µM) and (C) ONOO− (80 µM) in aqueous solutions containing phosphate buffer (0.1 M, pH 7.4) and dtpa (10 µM). The traces were collected using the absorption detector set at 330 nm. The asterisk indicates the position of a Luc-Bz-OH.
Figure 5.
Scheme showing the oxidative conversion of PCL-1 probe into luciferin, with the elimination of para-quinone methide (QM).
Next we confirmed the identity of the primary product detected under acidic conditions, using HPLC and mass spectrometry. The mass of the detected species (m/z = 385) is consistent with the deprotonated phenol Luc-Bz-O− (Fig. 5). The decomposition of Luc-Bz-OH into luciferin is accompanied by elimination of QM through a self-immolative reaction mechanism [21]. We reasoned that in aqueous solution, QM would quickly react with water to form 4-hydroxybenzyl alcohol (HO-Bz-OH, Fig. 5 [22]). The decomposition of Luc-Bz-OH intermediate produced two HPLC peaks: one corresponding to luciferin, and another one at a shorter retention time. This peak detected at a shorter retention time co-eluted with the authentic standard of HO-Bz-OH, but not of 4-methylresorcinol. Thus we conclude that in the absence of other nucleophiles, QM exclusively produces HO-Bz-OH in aqueous solutions.
3.2. Oxidation of PCL-1 by H2O2
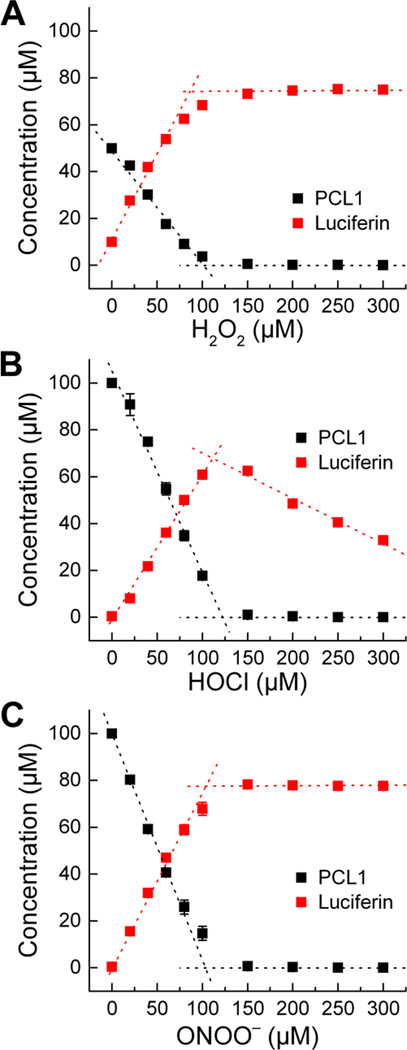
As shown in Figure 6, oxidation of PCL-1 probe by H2O2 (10 mM) yields the highest level of luciferin as compared to the other two oxidants tested. This is consistent with our previous studies using other boronates, demonstrating a 1:1 stoichiometry for the boronate:H2O2 reaction, with the corresponding phenol as the sole product. Because of the slow reaction kinetics, a 24-h incubation was needed to ensure completion of the reaction between PCL-1 and H2O2. As shown in Figure 6A, the PCL-1 probe was completely consumed, and the yield of luciferin reached maximum when the probe was reacted with equimolar amount of H2O2, confirming a 1:1 reaction stoichiometry. Of note, the yield of the product was only ca. 75%. This could be explained by the low stability of PCL-1 and/or luciferin over a 24-h incubation period in aqueous solutions. Indeed, the PCL-1 concentration determined after 24-h incubation was only 50 µM, indicating a significant decomposition of the probe during the prolonged incubation period. Under those conditions luciferin was one of the products formed in the absence of H2O2, but only accounted for 20% of the amount of PCL-1 decomposed.
Figure 6.
HPLC-based titration of PCL-1 (100 µM) with (A) H2O2 (24 h), (B) HOCl (30 min) and (C) ONOO− (30 min) in aqueous solutions containing phosphate buffer (0.1 M, pH 7.4) and dtpa (10 µM).
3.3. Oxidation of PCL-1 by HOCl
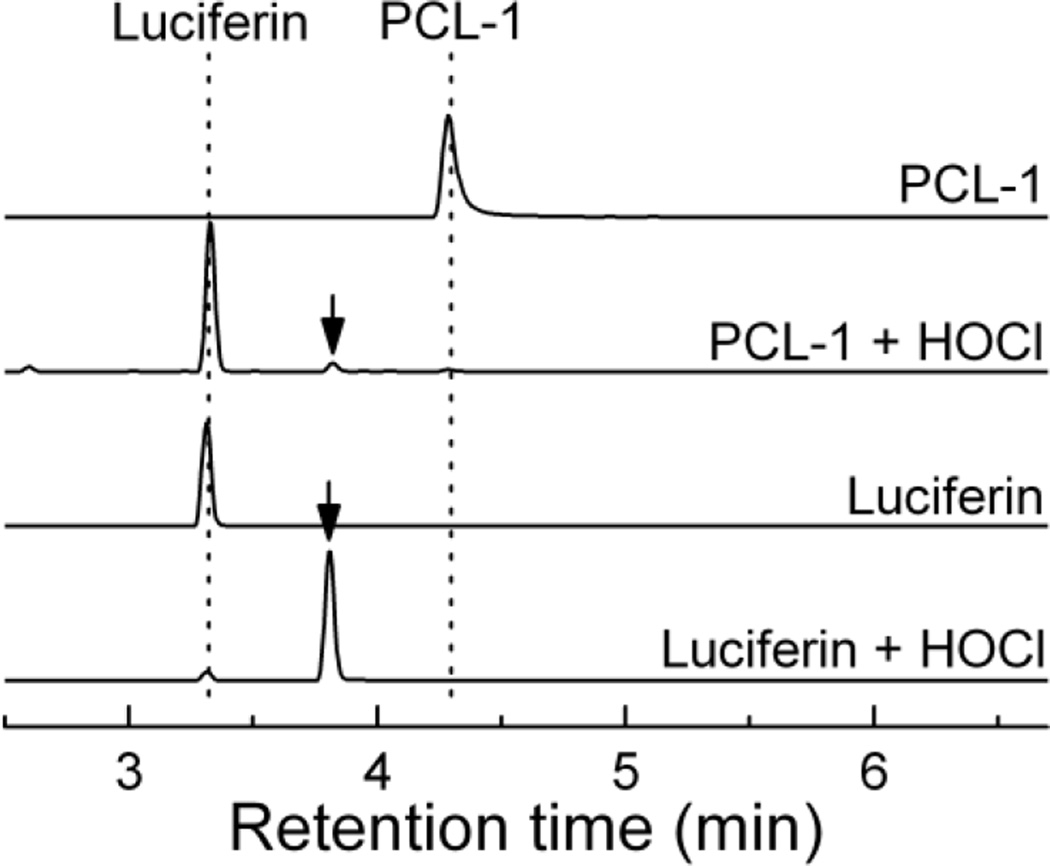
The stoichiometric analysis of the reaction between PCL-1 probe and HOCl is shown in Figure 6B. Unlike reaction with H2O2, a slight excess of HOCl was required for complete consumption of PCL-1. Again, the yield of luciferin was significantly less than 100%, despite the fact that the probe was stable enough over the incubation period (30 min). Based on the effect of ethanol on the extent of oxidation of CBA to COH (Suppl. Fig. 1), interference by EtOH (10%) used as a solvent for PCL-1 cannot account for more than 10% decrease in the yield of luciferin. We tentatively attribute the lower yield of luciferin to formation of additional, minor product(s) of the reaction of PCL-1 with HOCl (not detected). With excess HOCl, the product luciferin undergoes further reaction with HOCl, leading to decreased yield of luciferin. This is supported by the detection of a new HPLC peak assigned to chlorinated luciferin under the conditions of excess of HOCl (Fig. 7). The assignment of this HPLC peak to chlorinated luciferin was supported by the observation that this product was also formed when authentic luciferin was reacted with HOCl (Fig. 7). The structure was further confirmed by mass spectrometry (m/z = 315), and the position of chlorination was established using NMR analyses (see Materials and methods section).
Figure 7.
HPLC chromatograms of the reaction mixtures of PCL-1 and luciferin with HOCl. PCL-1 (0.1 mM) was mixed with HOCl (175 µM), and luciferin (90 µM) was reacted with 80 µM HOCl. The arrow indicates the position of a new peak attributed to chloroluciferin (Luc-Cl). Reactions were carried out in aqueous solutions containing phosphate buffer (0.1 M, pH 7.4) in the absence of dtpa, with 10% EtOH in case of PCL-1 reaction.
3.4. Oxidation of PCL-1 by ONOO−
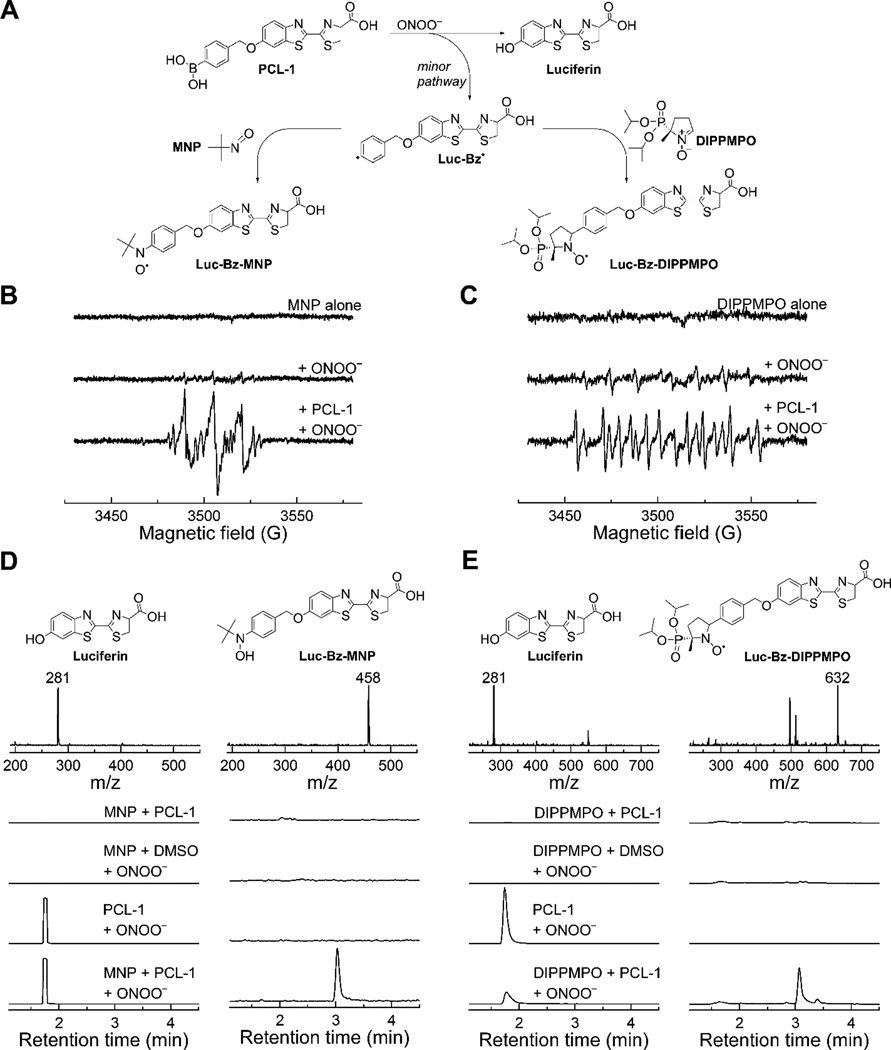
Similar to H2O2 and HOCl, ONOO− also oxidized PCL-1 probe, forming a maximal yield of luciferin at an equimolar ratio of the probe and ONOO− (Fig. 6C). Although the yield of luciferin was significantly lower than 100%, this was expected for ONOO− reaction with PCL-1 based on two pathways of oxidation: major (non-radical) pathway yielding luciferin, and minor (radical-mediated) pathway yielding products derived from the phenyl radical formed. To demonstrate the occurrence of the free radical pathway and formation of the corresponding phenyl radical, we performed EPR spin trapping experiments, using 2-methyl-2-nitroso propane (MNP) and DIPPMPO spin traps (Fig. 8). The basis of the formation and trapping of the PCL-1-derived phenyl radical is shown in Figure 8A. With both spin traps (MNP and DIPPMPO) in the reaction mixtures of PCL-1 with ONOO−, we recorded the EPR spectra that were qualitatively different than observed in reaction mixtures of spin traps with ONOO−, but without the PCL-1 probe (Fig. 8B,C). Although the signal intensity was significantly lower than detected for simple 4-acetylphenylboronic acid, we were able to attribute the spectra to the spin adducts of the PCL-1-derived phenyl radical. The spectrum obtained with MNP spin trap consisted of three major lines (due to the hyperfine splitting from the nitrogen atom), with an additional structure (due to the hyperfine splitting caused by the phenyl ring hydrogen atoms) (Fig. 8B). With DIPPMPO cyclic nitrone spin trap, the spectrum (Fig. 8C) corresponds to a mixture of spin adducts. To confirm the formation of the phenyl radical adduct, we performed LC-MS analysis of the reaction mixture containing both MNP (Fig. 8D) and DIPPMPO (Fig. 8E) spin traps. When PCL-1 was reacted with ONOO− the peaks of luciferin were detected with both spin traps, regardless of the presence of the spin trap. In the presence of DIPPMPO, we observed a decrease in peak intensity due to luciferin, which we attribute to ion suppression by excess of DIPPMPO eluting over the whole LC-MS analysis time. In the presence of PCL-1, ONOO− and MNP, the spin adduct of MNP and Luc-Bz• radical was detected (Fig. 8D, m/z = 458, peak detected at 3.03 min). Similarly, only in the presence of PCL-1, ONOO− and DIPPMPO did we detect the spin adduct of Luc-Bz• to DIPPMPO (Fig. 8E, m/z = 632, peak detected at 3.07 min). This peak can be assigned to the spin adduct present in the form of nitroxide and/or protonated nitrone. The spin trapping results confirm the formation of the phenyl radical during the oxidation of PCL-1 by ONOO−. Notably, under the conditions used, no elimination of the QM occurred from PCL-1-derived phenyl radical. To detect the stable end-products derived from the phenyl radical, we synthesized the authentic standards of the expected compounds, including Luc-Bz-NO2 (the product of recombination of Luc-Bz• phenyl and •NO2 radicals) and Luc-Bz-H (Luc-Bz• reduction product) and performed LC-MS analyses of the reaction mixtures.
Figure 8.
Spin trapping of the phenyl radical formed during the reaction of PCL-1 with ONOO−. (A) Scheme of the formation and trapping of Luc-Bz• radicals; (B) EPR spectra registered using MNP spin trap; (C) EPR spectra registered with the use of DIPPMPO spin trap; (D) LC-MS analyses of luciferin and Luc-Bz-MNP spin adduct; (E) LC-MS analyses of luciferin and Luc-Bz-DIPPMPO spin adduct. Incubation mixture contained the following compounds: PCL-1 (250 µM), DIPPMPO (10 mM) or MNP (40 mM), in Tris-HCl buffer (100 mM, pH 9.5) containing dtpa (100 µM), catalase (100 U/ml), and DMSO (0.25 %). The reaction mixture was transferred to an EPR capillary immediately after bolus addition of ONOO− (resulting in the 200 µM ONOO− concentration in the sample), and the spectra were recorded at room temperature.
3.5. LC-MS analyses
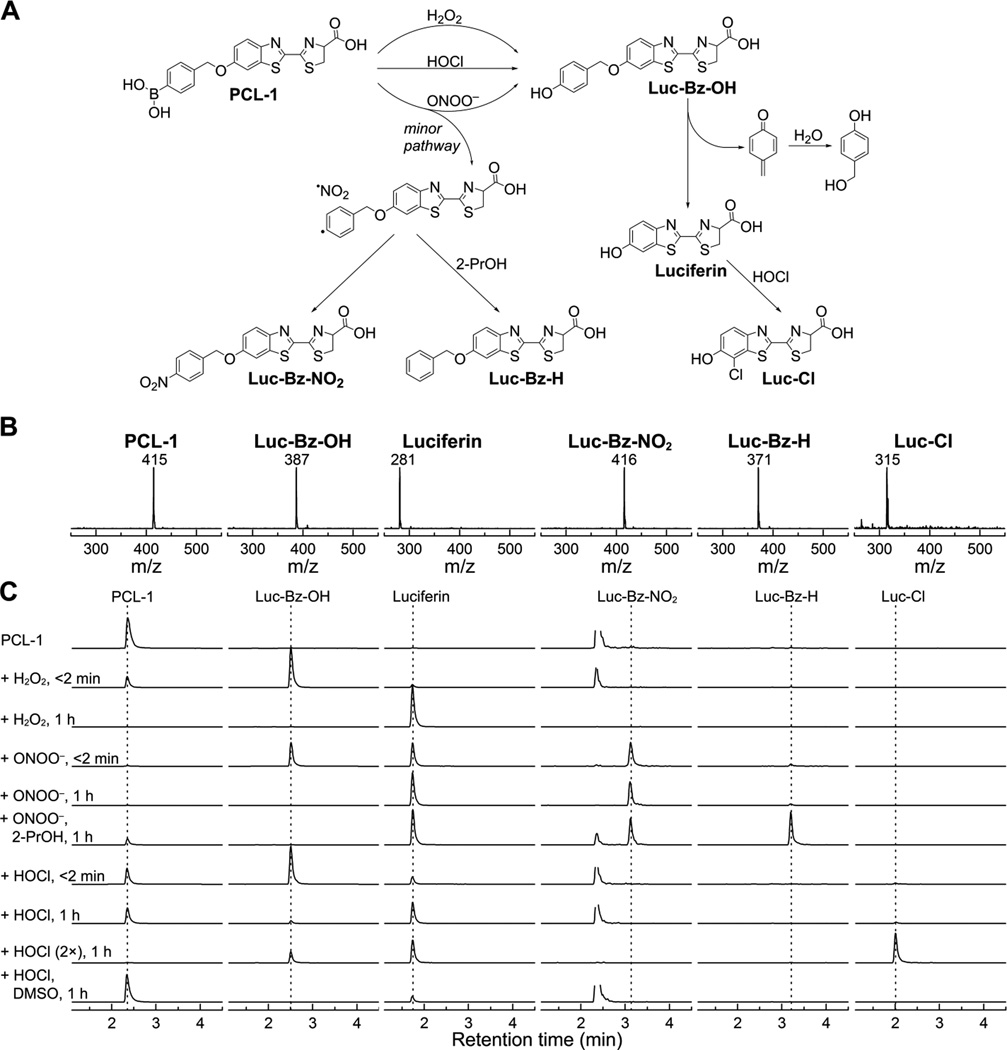
Figure 9 shows the product analyses of the reaction of oxidation of PCL-1 by three oxidants: H2O2, HOCl, and ONOO−. The proposed reaction mechanism and the structures of compounds detected are shown in Figure 9A and the mass spectra of every analyte are presented in Figure 9B. With all three oxidants, the peak detected immediately (injected in less than 2 min) after mixing corresponds to Luc-Bz-OH, which upon further incubation (1 h) decomposes to form luciferin (Figure 9C). No other products were detected in the presence of H2O2. The LC-MS peaks observed in the Luc-Bz-NO2 channel matched the retention time of PCL-1 and were attributed to relatively low resolution of the mass detector and only one unit difference (415 vs. 416) of the m/z values for PCL-1 and Luc-Bz-NO2 (Fig. 9B). The nitrated product (Luc-Bz-NO2) was observed with ONOO− immediately after mixing, and its yield did not change upon further incubation. This is consistent with a rapid recombination of Luc-Bz• and •NO2 radicals. Small amounts of the Luc-Bz• reduction product (Luc-Bz-H, Fig. 9) was also detected. The yield of Luc-Bz-H was significantly increased when the reaction between PCL-1 and ONOO− was performed in the presence of 2-propanol (2-PrOH), a known scavenger of phenyl radicals. During the reaction between PCL-1 and HOCl, an additional product, Luc-Cl (Fig. 9) detected under the conditions of excess HOCl was attributed to the chlorinated product from luciferin. Addition of dimethyl sulfoxide (DMSO), a known HOCl scavenger (Suppl. Fig. 1), inhibited oxidation of PCL-1 and formation of the Luc-Cl product.
Figure 9.
LC-MS analyses of the products of PCL-1 oxidation. (A) Scheme of the transformation of PCL-1, leading to luciferin and oxidant-specific minor products, (B) online mass spectra recorded for each product and (C) LC-MS traces of the reaction mixtures of PCL-1 (100 µM) alone or after addition of H2O2 (10 mM), ONOO− (80 µM) or HOCl (90 µM or 200 µM).
3.6. Oxidation of PCL-1 by activated RAW 264.7 cells
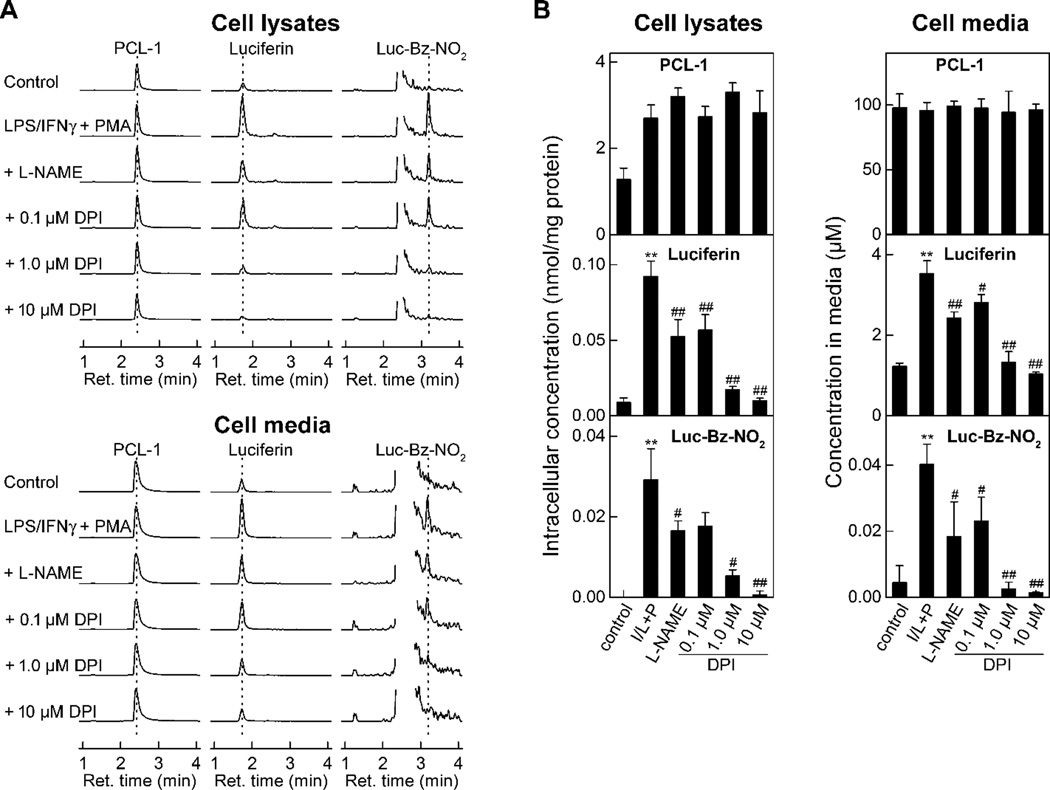
To test the feasibility of formation and detection of the peroxynitrite-specific product, Luc-Bz-NO2 in biological systems, we utilized RAW 264.7 cells activated to produce ONOO− (Fig. 10). Stimulation of RAW 264.7 cells with lipopolysaccharide (LPS), interefon γ (IFNγ) and phorbol myristate acetate (PMA) leads to the formation of ONOO− [14,19] and induces oxidation of PCL-1 to luciferin and nitration to Luc-Bz-NO2. These products were detected both intracellularily (Fig. 10A) and in the cell media (Fig. 10B) and were inhibitable by preincubation of the cells with L-NAME or DPI in a concentration-dependent manner. We have recently demonstrated that DPI blocks the formation of ONOO− in activated macrophages [19].
Figure 10.
Profiles of PCL-1 oxidation products formed in RAW 264.7 macrophages activated to produce ONOO−. RAW 264.7 cells were activated to produce ONOO− by overnight incubation of LPS (0.5 µg/ml) and IFNγ (50 units/mL) followed by addition of 1 µM PMA (I/L+P). At the time of addition of PMA, PCL-1 (100 µM) was also added and the cells were incubated for 1 h before harvesting. L-NAME (1 mM) or DPI (0.1–10 µM) were added 30 min before addition of PMA. (A) LC-MS/MS traces of PCL-1, luciferin and Luc-Bz-NO2; (B) Results of quantitative analyses of the LC-MS/MS data. ** - p<0.01 vs. control; #and ##- p<0.05 and p<0.01, respectively vs. I/L+P.
4. Discussion
4.1. Rigorous identification of ROS and RNS
The use of molecular probes for detection and quantification of reactive cellular oxidizing and nitrating species requires detailed knowledge of the probes’ chemistry, reaction kinetics, and possibly the identification of the oxidant-specific product(s), as well. In many cases, ROS/RNS-specific marker products have been characterized [12–14,18,19,20,22–24]. Because of overlapping fluorescence characteristics of products formed from the reaction between fluorophores and oxidants, it is nearly impossible to categorically identify specific oxidants formed in cells using confocal fluorescence technique [23,25]. For example, both 2-hydroxyethidium (specific marker product of hydroethidine and superoxide) and ethidium (non-specific, two electron oxidation product of hydroethidine and various oxidants) have overlapping fluorescence spectral characteristics. HPLC-based methods are clearly more suitable for separating, identifying and quantifying them by comparing with appropriate standards [26]. The most significant progress in ROS detection over the last decade was the development of a new class of boronate-based probes, initially proposed for specific detection of H2O2. H2O2 slowly oxidizes boronates to the corresponding hydroxyl derivatives (phenolic products in case of aromatic boronates) [12,13]. The design of boronate probes is typically based on the substitution of the hydroxyl group of the fluorescent compound with the boronate moiety. When direct substitution of the hydroxyl group leads to undesired intramolecular interactions or is synthetically challenging, a simpler boronobenzylation is used (e.g., PCL-1 probe), leading to the corresponding boronobenzyl derivatives.
In a series of papers we have characterized the mechanism of the reaction of boronates with peroxynitrite and identified minor, ONOO−-specific products, in which the boronate moiety is replaced by the nitro group [12,13,17,19,27,28]. While the nitration of the phenolic products is a common feature of the chemical reactivity of peroxynitrite and peroxidase/H2O2/nitrite systems, replacement of the boronate moiety by the nitro group is specific for ONOO− and results in nitrobenzene-like product [13,17]. Incubation of boronates with myeloperoxidase(MPO)/H2O2/nitrite systems does not produce the nitrobenzene-like product, but leads to nitration of the phenolic product (formed during the reaction of boronates with H2O2), leading to nitrophenols (in contrast to nitrobenzenes) [27,28].
One of the advantages of boronate probes is that the reaction chemistry and kinetics remain unchanged, independent of the actual scaffold used in the probe design, with a few exceptions. The advantage of in vivo probes based on boronates’ chemistry is the relatively low probability for interference of the heme proteins, and biological reductants, which limit the in vivo application of other probes, including hydroethidine and cyclic nitrone spin traps. Recently, a boronate-caged positron emission tomography (PET) tracer was used to image H2O2 in renal carcinoma cells [29]. As PET imaging can be readily translated to the clinical setting, boronatecaged PET tracers may find wider applications in oxidative stress/nitrative stress imaging in vivo.
4.2. Potential applications
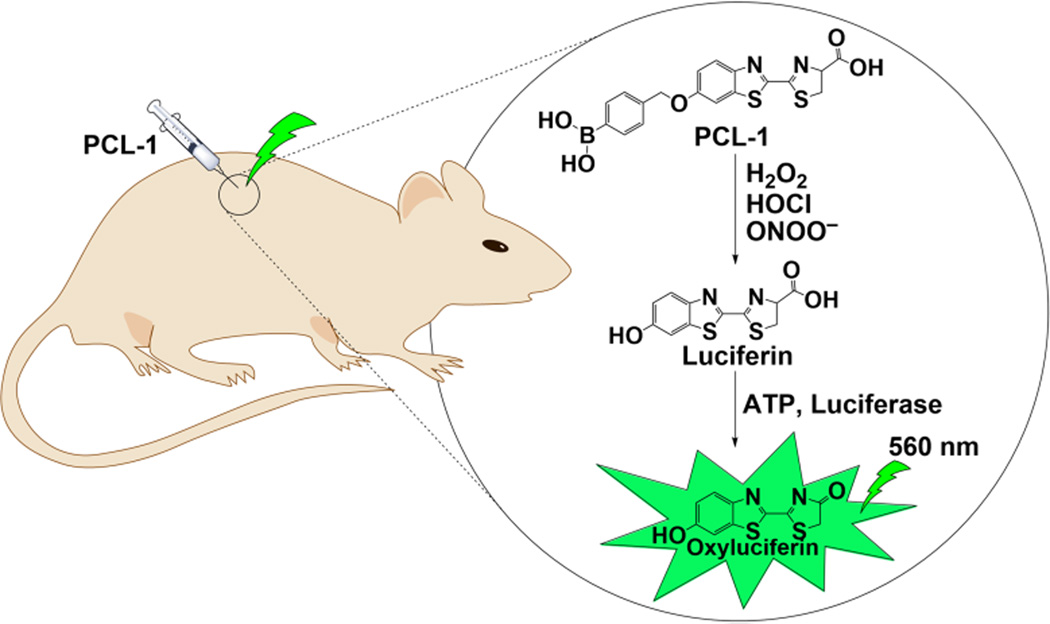
The application of PCL-1 probe to monitor production of oxidants in tumor and/or tumor environment by monitoring bioluminescence in rodent models may enable better understanding of the role of ROS/RNS in tumor growth and immunosuppressive effects of tumor microenvironment. This in turn may provide a better approach to increase the efficiency of cancer immunotherapy and/or combination of redox modulators with the standard-of-care drugs. Typically tumor growth is assessed by measuring the intensity of bioluminescence signal (light intensity) in luciferase-transfected cancer cell mice xenografts [30]. The substrate, luciferin, is injected as needed and the green bioluminescent signal intensity from the luciferase-transfected tumor cells is measured [30]. Using the PCL-1 probe, one can monitor in vivo bioluminescence imaging of tumor-derived ROS/RNS. Mice bearing luciferase-transfected cancer cells are administered with PCL-1 probe on different days after tumor implantation. This approach enables selective monitoring of ROS/RNS in tumor cells due to selective localization of luciferase in tumor cells. Upon reaction with H2O2 or ONOO− or HOCl, luciferin is formed in situ (from PCL-1) which is oxidized by the luciferase enzyme (using ATP as a co-factor) to generate green bioluminescence (Fig. 1). As bioluminescence will depend not only on ROS/RNS but also on tumor size, the number of tumor cells, and intracellular ATP, parallel analysis should be performed with luciferin as the substrate. To distinguish between H2O2 and ONOO−, appropriate antioxidant enzymes (e.g., PEG catalase), nitric oxide synthase inhibitors or superoxide dismutase mimetics inhibitors may be used in addition to measuring the specific nitrated product derived from PCL-1 in tumor tissues.
5. Conclusions
5.1
In this work we have identified the primary product of oxidation of the PCL-1 probe, Luc-Bz-OH (Fig. 5). Decomposition of Luc-Bz-OH (via a self-immolative reaction) leads to the formation of luciferin with the elimination of QM. In the absence of other nucleophiles, QM reacts with water to form HO-Bz-OH (Fig. 5). However, in a cellular environment, other nucleophiles, including thiols, will likely react with QM.
5.2
The major product identified by HPLC with all three oxidants tested was luciferin. However, in the case of ONOO−, the reaction proceeds via two pathways, with the minor pathway leading to the formation of ONOO−-specific minor product, Luc-Bz-NO2, via intermediate phenyl radical Luc-Bz• (Fig. 9A). This radical has been detected using the spin trapping technique, and the nitrone adduct identified by LC-MS.
5.3
The minor, ONOO−-specific product, Luc-Bz-NO2 is formed by activated macrophages incubated in the presence of the PCL-1 probe, and can be detected and quantified by LC-MS analyses.
5.4
Although, reaction with HOCl seems to proceed via a single, non-radical pathway, the product formed, luciferin, undergoes further reaction with HOCl, leading to the formation of Luc-Cl, a product specific for HOCl.
5.5
Here we propose the combination of non-invasive bioluminescence monitoring of oxidant production in vivo in luciferase-expressing cells, with HPLC or LC-MS analyses of tissues to detect oxidant-specific minor products. This will provide more detailed information on the identity(ies) of the species detected. Identification of the oxidant produced under pathophysiological conditions will allow for more precise interventions to inhibit their formation.
Supplementary Material
Highlights.
PCL-1 reacts with H2O2, HOCl, and ONOO− to form luciferin as the major product.
ONOO− oxidizes PCL-1 nearly a million times more rapidly than does H2O2.
Nitrobenzylated luciferin is a minor, ONOO−-specific product of PCL-1.
PCL-1 reaction with excess HOCl generates a chlorinated luciferin.
Minor products of PCL-1 may serve as specific markers for ONOO− and HOCl.
Acknowledgments
This work was supported by grants from NIH (R01 HL073056) to B.K. The LC-MS analyses were performed in Medical College of Wisconsin Cancer Center Redox and Bioenergetics Shared Resource.
Abbreviations
- AA-OOH
amino acid hydroperoxide
- DIPPMPO
diisopropoxyphosphoryl-5-methyl-1-pyrroline N-oxide
- DPI
diphenyleneiodonium
- HO-Bz-OH
4-hydroxybenzyl alcohol
- L-NAME
L-NG-Nitroarginine methyl ester
- LOO•
lipid peroxy radicals
- LOOH
lipid hydroperoxide
- Luc-Cl
chloroluciferin
- MNP
2-methyl-2-nitroso propane
- MRM
multiple reaction monitoring
- PCL-1
peroxy-caged luciferin
- Pr-OOH
protein hydroperoxide
- QM
para-quinone methide
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
- SIM
single ion monitoring
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Jacek Zielonka, Email: jzielonk@mcw.edu.
Radosław Podsiadły, Email: radoslaw.podsiadly@p.lodz.pl.
Monika Zielonka, Email: mzielonka@mcw.edu.
Micael Hardy, Email: micael.hardy@univ-provence.fr.
Kalyanaraman Balaraman, Email: balarama@mcw.edu.
References
- 1.Finkel T. Signal transduction by reactive oxygen species. J Cell Biol. 2011;194:7–15. doi: 10.1083/jcb.201102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Janssen-Heininger YM, Mossman BT, Heintz NH, Forman HJ, Kalyanaraman B, Finkel T, Stamler JS, Rhee SG, van der Vliet A. Redox-based regulation of signal transduction: principles, pitfalls, and promises. Free Radic Biol Med. 2008;45:1–17. doi: 10.1016/j.freeradbiomed.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winterbourn CC. Reconciling the chemistry and biology of reactive oxygen species. Nat Chem Biol. 2008;4:278–286. doi: 10.1038/nchembio.85. [DOI] [PubMed] [Google Scholar]
- 4.Murphy MP, Holmgren A, Larsson NG, Halliwell B, Chang CJ, Kalyanaraman B, Rhee SG, Thornalley PJ, Partridge L, Gems D, Nyström T, Belousov V, Schumacker PT, Winterbourn CC. Unraveling the biological roles of reactive oxygen species. Cell Metab. 2011;13:361–366. doi: 10.1016/j.cmet.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forman HJ, Fukuto JM, Torres M. Redox signaling: thiol chemistry defines which reactive oxygen and nitrogen species can act as second messengers. Am J Physiol Cell Physiol. 2004;287:C246–C256. doi: 10.1152/ajpcell.00516.2003. [DOI] [PubMed] [Google Scholar]
- 6.Abe K, Takai N, Fukumoto K, Imamoto N, Tonomura M, Ito M, Kanegawa N, Sakai K, Morimoto K, Todoroki K, Inoue O. In vivo imaging of reactive oxygen species in mouse brain by using [3H]hydromethidine as a potential radical trapping radiotracer. J Cereb Blood Flow Metab. 2014;34:1907–1913. doi: 10.1038/jcbfm.2014.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Y, Ding J1, Liang T, Abdel-Halim ES, Jiang L, Zhu JJ. FITC doped rattle-type silica colloidal particle-based ratiometric fluorescent sensor for biosensing and imaging of superoxide anion. ACS Appl Mater Interfaces. 2016;8:6423–6430. doi: 10.1021/acsami.6b01031. [DOI] [PubMed] [Google Scholar]
- 8.Carroll VN1, Truillet C, Shen B, Flavell RR, Shao X, Evans MJ, VanBrocklin HF, Scott PJ, Chin FT, Wilson DM. [(11)C]Ascorbic and [(11)C]dehydroascorbic acid, an endogenous redox pair for sensing reactive oxygen species using positron emission tomography. Chem Commun (Camb) 2016;52:4888–4890. doi: 10.1039/c6cc00895j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van de Bittner GC, Dubikovskaya EA, Bertozzi CR, Chang CJ. In vivo imaging of hydrogen peroxide production in a murine tumor model with a chemoselective bioluminescent reporter. Proc Natl Acad Sci U S A. 2010;107:21316–21321. doi: 10.1073/pnas.1012864107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van de Bittner GC, Bertozzi CR, Chang CJ. Strategy for dual-analyte luciferin imaging: in vivo bioluminescence detection of hydrogen peroxide and caspase activity in a murine model of acute inflammation. J Am Chem Soc. 2013;135:1783–1795. doi: 10.1021/ja309078t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sieracki NA, Gantner BN, Mao M, Horner JH, Ye RD, Malik AB, Newcomb ME, Bonini MG. Bioluminescent detection of peroxynitrite with a boronic acid-caged luciferin. Free Radic Biol Med. 2013;61:40–50. doi: 10.1016/j.freeradbiomed.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sikora A, Zielonka J, Lopez M, Joseph J, Kalyanaraman B. Direct oxidation of boronates by peroxynitrite: mechanism and implications in fluorescence imaging of peroxynitrite. Free Radic Biol Med. 2009;47:1401–1407. doi: 10.1016/j.freeradbiomed.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zielonka J, Sikora A, Hardy M, Joseph J, Dranka BP, Kalyanaraman B. Boronate probes as diagnostic tools for real time monitoring of peroxynitrite and hydroperoxides. Chem Res Toxicol. 2012;25:1793–1799. doi: 10.1021/tx300164j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zielonka J, Zielonka M, Sikora A, Adamus J, Joseph J, Hardy M, Ouari O, Dranka BP, Kalyanaraman B. Global profiling of reactive oxygen and nitrogen species in biological systems: high-throughput real-time analyses. J Biol Chem. 2012;287:2984–2995. doi: 10.1074/jbc.M111.309062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zielonka J, Sikora A, Joseph J, Kalyanaraman B. Peroxynitrite is the major species formed from different flux ratios of co-generated nitric oxide and superoxide: direct reaction with boronate-based fluorescent probe. J Biol Chem. 2010;285:14210–14216. doi: 10.1074/jbc.M110.110080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michalski R, Zielonka J, Gapys E, Marcinek A, Joseph J, Kalyanaraman B. Real-time measurements of amino acid and protein hydroperoxides using coumarin boronic acid. J Biol Chem. 2014;289:22536–22553. doi: 10.1074/jbc.M114.553727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sikora A, Zielonka J, Lopez M, Dybala-Defratyka A, Joseph J, Marcinek A, Kalyanaraman B. Reaction between peroxynitrite and boronates: EPR spin-trapping, hplc analyses, and quantum mechanical study of the free radical pathway. Chem Res Toxicol. 2011;24:687–697. doi: 10.1021/tx100439a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smulik R, Dębski D, Zielonka J, Michałowski B, Adamus J, Marcinek A, Kalyanaraman B, Sikora A. Nitroxyl (HNO) reacts with molecular oxygen and forms peroxynitrite at physiological pH. Biological implications. J Biol Chem. 2014;289:35570–35581. doi: 10.1074/jbc.M114.597740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zielonka J, Zielonka M, VerPlank L, Cheng G, Hardy M, Ouari O, Ayhan MM, Podsiadly R, Sikora A, Lambeth JD, Kalyanaraman B. Mitigation of NADPH oxidase 2 activity as a strategy to inhibit peroxynitrite formation. J Biol Chem. 2016;291:7029–7044. doi: 10.1074/jbc.M115.702787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chalier F, Tordo P. 5-diisopropoxyphosphoryl-5-methyl-1-pyrroline N-oxide, DIPPMPO, a crystalline analog of the nitrone DEPMPO: Synthesis and spin trapping properties. J Chem Soc Perkin Trans. 2002;2:2110–2117. [Google Scholar]
- 21.Alouane A, Labruère R, Le Saux T, Schmidt F, Jullien L. Self-immolative spacers: Kinetic aspects, structure-property relationships, and applications. Angew Chem Int Ed Engl. 2015;54:7492–7509. doi: 10.1002/anie.201500088. [DOI] [PubMed] [Google Scholar]
- 22.Toteva MM, Richard JP. The generation and reactions of quinone methides. Adv Phys Org Chem. 2011;45:39–91. doi: 10.1016/B978-0-12-386047-7.00002-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao H, Joseph J, Fales HM, Sokoloski EA, Levine RL, Vasquez-Vivar J, Kalyanaraman B. Detection and characterization of the product of hydroethidine and intracellular superoxide by HPLC and limitations of fluorescence. Proc Natl Acad Sci U S A. 2005;102:5727–5732. doi: 10.1073/pnas.0501719102. Erratum in: Proc Natl Acad Sci U S A. 2005; 102:9086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michalski R, Zielonka J, Hardy M, Joseph J, Kalyanaraman B. Hydropropidine: A novel, cell-impermeant fluorogenic probe for detecting extracellular superoxide. Free Radic Biol Med. 2013;54:135–147. doi: 10.1016/j.freeradbiomed.2012.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zielonka J, Kalyanaraman B. Hydroethidine- and MitoSOX-derived red fluorescence is not a reliable indicator of intracellular superoxide formation: Another inconvenient truth. Free Radic Biol Med. 2010;48:983–1001. doi: 10.1016/j.freeradbiomed.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zielonka J, Hardy M, Kalyanaraman B. HPLC study of oxidation products of hydroethidine in chemical and biological systems: ramifications in superoxide measurements. Free Radic Biol Med. 2009 Feb 1;46(3):329–338. doi: 10.1016/j.freeradbiomed.2008.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sikora A, Zielonka J, Adamus J, Dębski D, Dybala-Defratyka A, Michalowski B, Joseph J, Hartley RC, Murphy MP, Kalyanaraman B. Reaction between peroxynitrite and triphenylphosphonium-substituted arylboronic acid isomers – Identification of diagnostic marker products and biological implications. Chem Res Toxicol. 2013;26:856–867. doi: 10.1021/tx300499c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zielonka J, Sikora A, Adamus J, Kalyanaraman B. Detection and differentiation between peroxynitrite and hydroperoxides using mitochondria-targeted arylboronic acid. Methods Mol Biol. 2015;1264:171–181. doi: 10.1007/978-1-4939-2257-4_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carroll V, Michel BW, Blecha J, VanBrocklin H, Keshari K, Wilson D, Chang CJ. A boronate-caged [18F]FLT probe for hydrogen peroxide detection using positron emission tomography. J Am Chem Soc. 2014 Oct 22;136(42):14742–14745. doi: 10.1021/ja509198w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roy I, McAllister DM, Gorse E, Dixon K, Piper CT, Zimmerman NP, Getschman AE, Tsai S, Engle DD, Evans DB, Volkman BF, Kalyanaraman B, Dwinell MB. Pancreatic cancer cell migration and metastasis is regulated by chemokine-biased agonism and bioenergetic signaling. Cancer Res. 2015 Sep 1;75(17):3529–3542. doi: 10.1158/0008-5472.CAN-14-2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.