Abstract
Androgen receptor (AR) is expressed in 90% of estrogen receptor alpha positive (ER+) breast tumors, but its role in tumor growth and progression remains controversial. Use of two anti-androgens that inhibit AR nuclear localization, enzalutamide and MJC13, revealed that AR is required for maximum ER genomic binding. Here, a novel global examination of AR chromatin binding found that estradiol induced AR binding at unique sites compared to dihydrotestosterone (DHT). Estradiol-induced AR binding sites were enriched for estrogen response elements and had significant overlap with ER binding sites. Furthermore, AR inhibition reduced baseline and estradiol-mediated proliferation in multiple ER+/AR+ breast cancer cell lines, and synergized with tamoxifen and fulvestrant. In vivo, enzalutamide significantly reduced viability of tamoxifen-resistant MCF7 xenograft tumors and an ER+/AR+ patient-derived model. Enzalutamide also reduced metastatic burden following cardiac injection. Lastly, in a comparison of ER+/AR+ primary tumors versus patient-matched local recurrences or distant metastases, AR expression was often maintained even when ER was reduced or absent. These data provide pre-clinical evidence that anti-androgens that inhibit AR nuclear localization affect both AR and ER, and are effective in combination with current breast cancer therapies. In addition, single agent efficacy may be possible in tumors resistant to traditional endocrine therapy, since clinical specimens of recurrent disease demonstrate AR expression in tumors with absent or refractory ER.
Implications
This study suggests that AR plays a previously-unrecognized role in supporting E2-mediated ER activity in ER+/AR+ breast cancer cells, and that enzalutamide may be an effective therapeutic in ER+/AR+ breast cancers.
Keywords: Androgen Receptor, Estrogen Receptor, Enzalutamide, Breast Cancer
INTRODUCTION
AR is more frequently expressed in breast cancer than estrogen receptor alpha (ER) or progesterone receptor (PR) (1); however the role of AR is complex, dependent on the hormonal milieu, and remains controversial. AR positivity is associated with better prognosis in ER+ breast cancer (2–4), possibly due to the fact that like ER, AR positivity is indicative of a more well-differentiated state. In the presence of estradiol (E2), the androgen dihydrotestosterone (DHT) decreased E2-induced proliferation (2) and ER transcriptional activity (5), leading to the conclusion that AR is protective in breast cancer. However, there is accumulating evidence that androgen signaling and AR are involved in resistance to ER-directed endocrine therapies. De novo or acquired resistance to anti-estrogen therapies is a frequent occurrence, and ultimately all metastatic ER+ breast cancers are resistant (6,7). In ER+ tumors responsive to neoadjuvant aromatase inhibitor (AI) therapy, AR mRNA and nuclear AR protein decreased, whereas in non-responsive tumors it remained elevated (8,9). AR over-expression in breast cancer cell lines resulted in resistance to tamoxifen (tam) and AIs in vitro and in vivo (10,11). One mechanism of resistance to anti-estrogen therapies may therefore be tumor adaptation from estrogen to androgen dependence.
AIs block the conversion of androgens to estrogens, and free testosterone and dehydroepiandrosterone sulfate (DHEA-S) increased in patients on AIs (12). Furthermore, high levels of the adrenal androgen DHEA-S are predictive of failure on AIs, and circulating DHEA-S increased during treatment in patients with tumors that progressed during AI treatment (13). Patients with tumors exhibiting a high ratio of percent cells positive for AR versus ER protein are more likely to have recurrent disease while on tamoxifen and also have a worse overall prognosis compared to those with a more equal ratio of these two receptors, as is found in normal breast epithelium (14). So although AR, like ER, is associated with a better prognosis, anti-androgen therapies may benefit patients with AR+ breast cancers if the tumors are dependent on activated AR.
We previously reported that the new generation AR antagonist enzalutamide (Enza), which inhibits AR nuclear localization, decreased estrogen-induced tumor growth, while the first-generation AR antagonist bicalutamide (bic) did not (14). However, the mechanism by which Enza affected ER activity was not known. Herein, we demonstrate for the first time that in response to E2, nuclear localization of AR supports maximum ER genomic binding, and that AR inhibition with the pure antagonist Enza significantly decreases E2-induced growth of ER+/AR+ cell lines and patient-derived xenografts, as well as tam-resistant tumors in vivo, and also decreases metastatic burden. Importantly, these data suggest that patients with ER+/AR+ breast cancer may benefit from combining anti-androgen therapy with anti-estrogen therapy, and that tumors resistant to traditional ER-directed therapies may be responsive to AR-directed drugs.
Materials and Methods
Cell Lines
All cell lines were authenticated by short tandem repeat analysis using AmpFLSTR Identifiler PCR Amplification Kit (Life Technologies) and tested negative for mycoplasma in January 2015. MCF7 cells were obtained from Dr. Kate Horwitz at the University of Colorado Anschutz Medical Campus. MCF7-TamR cells obtained from Dr. Doug Yee at the University of Minnesota were generated by chronic treatment of MCF7 cells with 100nM tamoxifen. All other cell lines were obtained from the ATCC. Additional cell culture details are included in supplementary material. The BCK4 cell line is an ER+/AR+ breast cancer line recently derived from a pleural effusion (15), and the PT12 breast cancer cell line is ER+/AR+ and created from a patient-derived xenograft (PDX) (16). Originally the PT12 PDX was described as AR-negative (16), but upon staining of the original passage with a more sensitive AR antibody (SP107 from Cell Marque), the PDX was found to be AR+ (Supplementary Figure S6a).
Cellular Assays and Reagents
Cells were treated with 10 nM estradiol (E2, Sigma Aldrich) and 10 nM dihydrotestosterone (DHT, Sigma Aldrich). Androgen concentrations have been previously examined in breast cancer (17) and intratumoral DHT concentrations (249 pg/g) were significantly higher than in blood. The DHT concentration of the present study is consistent with other in vitro studies of DHT in breast cancer (18,19), and approximates levels of circulating testosterone in obese, postmenopausal women (12) as well as DHT levels in FBS used during routine tissue culture. 10 µM Enza (Medivation, San Francisco, CA) approximates the IC50 of the three cell lines studied and is a clinically achievable, well-tolerated treatment concentration (NCT01889238).
Proliferation Assays
Proliferation assays were performed using the IncuCyte ZOOM live cell imaging system (Essen BioSciences) or crystal violet as previously described (20). For synergy experiments, percent inhibition was calculated compared to vehicle control, and the combination index was calculated for each dose combination by Calcusyn (21) (BIOSOFT, Cambridge, UK). Soft agar assays were performed in 6-well plates using 0.5% bottom and 0.25% top layer agar (Difco Agar Noble, BD Biosciences). Wells were photographed and colony number and size was determined by ImageJ software (National Institutes of Health).
Tumor studies
Xenograft experiments were approved by the University of Colorado Institutional Animal Care and Use Committee (IACUC protocol 83614(01)1E) and were conducted in accordance with the NIH Guidelines of Care and Use of Laboratory Animals. 1×106 MCF7-GFP-Luc cells were mixed with growth factor-reduced Matrigel (BD Biosciences) and injected bilaterally into the mammary fat pad of female ovariectomized athymic nu/nu mice (Taconic). E2 pellets (60-day release, 1.5 mg/pellet, Innovative Research of America) were implanted subcutaneously (SQ) at the back of the neck. Once tumors were established, mice were randomized into groups based on total tumor burden as measured by in vivo imaging. Mice received Enza in their chow (~50 mg/kg daily dose). Enza was mixed with ground mouse chow (Research Diets Inc.) at 0.43 mg/g chow. Control mice received the same chow without Enza. All mice were given free access to Enza-formulated chow or control chow during the study. Mice were euthanized by carbon dioxide asphyxiation followed by cervical dislocation, and tumors were harvested. The MCF7-TamR xenograft experiment was performed as described above without estrogen pellets. For the PT-12 xenograft study, 1×106 cells were injected bilaterally into the mammary fat pad of NOD-SCID-IL2Rgc−/− female mice. Mice were implanted with a DHT (8mg) or E2 (2mg) pellet. For the metastasis experiment, 2.5×10^5 GFP-Luciferase labeled PT12 cells were injected intracardially in NOD-SCID-IL2Rgc−/− mice implanted with E2 pellets (2mg). PT12 experiments with DHT were performed in ovariectomized females, while PT12 experiments with E2 were performed in non-ovariectomized females since the E2 pellet overrides the estrus cycle.
Immunoblotting
Whole cell protein extracts (50 µg) were denatured, separated on SDS PAGE gels and transferred to PVDF membranes. After blocking in 3% BSA in TBS-T, membranes were probed overnight at 4°C. Primary antibodies used were: ERalpha (Neomarkers Ab-16, 1:500 dilution), AR (EMD Millipore PG-21, 1:500 dilution), Topo 1 (Santa Cruz C-21, 1:100 dilution) and alpha-tubulin (clone B-5-1-2 from Sigma, 1:30,000 dilution). After incubation with appropriate secondary antibody, results were detected using Western Lightning Chemiluminescence Reagent Plus (Perkin Elmer).
Nuclear-cytoplasmic fractionation
1×106 cells were seeded in 10cm dishes in medium supplemented with 5% charcoal stripped serum (CSS). After three days the cells were pre-treated with vehicle or 10 µM Enza for 3 hr and then co-treated with either DHT for 3 hr plus or minus Enza, or E2 for 1 hr plus or minus Enza. Cells were washed with phosphate buffered saline (PBS) and cellular fractionation was performed using the NE-PER Nuclear and Cytoplasmic Extraction Kit (Life Technologies) as per manufacturer’s instructions.
Proximity ligation assay
PLA was performed using the Duolink kit according to manufacturer’s instructions (Olink Bioscience, Uppsala, Sweden). Briefly, 1.5×104 cells were plated in 8-well chamber slides and hormone starved in phenol red-free media with 5% CSS for 72hrs. Cells were then pre-treated with Veh or Enza for 3hrs, then treated with hormones +/− Enza as described for 1hr. After fixation with 4 % paraformaldehyde, cells were permeabilized then blocked. Samples were then incubated with primary antibodies AR D6F11 (Cell Signaling), and ERα clone 6F11 (Vector Laboratories) overnight at 4 degrees. Samples were then incubated with secondary antibodies linked to PLA probes and ligase was added. Detection reagent red was added and DAPI mounting media was added to visualize nuclei. Images were captured using a 20× objective. DAPI-labeled nuclei and red ERα/AR complexes were quantified using CellProfiler (22).
ChIP-seq
ChIP-seq was performed by Active Motif (Carlsbad, CA). Briefly, 1×106 MCF7 cells were seeded in 15cm dishes in phenol red-free medium supplemented with 5% CSS for 72 hrs. Cells were pre-treated with vehicle, 10 µM Enza, or 30uM MJC13 for 3 hrs. E2 was then added for 1 hr in continued presence of vehicle, Enza, or MJC13. For AR ChIP-seq, an additional sample was treated with DHT for 4 hrs. The cells were washed with PBS then fixed as per manufacturer’s instructions (Active Motif). AR antibody H-280 (Santa Cruz) or ER antibody HC-20 (Santa Cruz) were utilized. Peak calls were made by MACS2 (23) with default parameters using the sequence alignments obtained from Active Motif. Motif discovery was performed on 100 base pairs surrounding the peak summit using BioProspector (24). Patser (25) was used to determine significant matches to AREs and EREs.
RNA-seq
RNA Libraries were constructed using Illumina TruSEQ stranded mRNA Sample Prep Kit (Cat# RS-122-2101). Total RNA was combined with RNA purification beads to bind PolyA RNA to oligodT magnetic beads. mRNA was eluted and converted to double stranded DNA. A Tailing, adapter ligation, and PCR amplification using 15 cycles was used to complete the library construction. Libraries were quantitated via Qubit, analyzed on a Bioanalyzer Tape Station and diluted to appropriate concentration to run on an Illumina HiSEQ 2500 High Throughput Flow Cell. Reads were mapped to the human genome (hg19) by gSNAP, expression (FPKM) derived by Cufflinks, and differential expression analyzed with ANOVA in R (26,27).
Statistical analyses
For most analyses, statistical significance was evaluated using a 2-tailed Student t-test or ANOVA with Bonferroni or Dunnett’s multiple comparisons test or non-parametric equivalents in GraphPad Prism (Ver 6, GraphPad Software, San Diego, CA) or SAS (ver 9.4, SAS Institute, Cary, NC). Test assumptions were check for all analyses. If data distributions were skewed, data transformations were attempted to allow the use of parametric tests. If data transformations failed, than a non-parametric test was used. For the PT12 xenograft experiments, due to unequal time measurements, the repeated measures mixed model approach was used rather than a standard repeated measures ANOVA. The data met the assumption of normality (Shapiro-Wilk test p>0.05 and frequency distribution graphs were symmetrical without evidence of outliers). For the PT12 cardiac injection experiment, we were not able to use a repeated measures approach as the data did not meet the assumptions of a normal distribution despite different data transformations. Therefore a single Wilcoxon rank sum test was used to determine difference between E2 and E2 plus enzalutamide at week 12.P ≤ 0.05 was considered statistically significant, with p-values indicated in figures as :* p≤.05, **p≤ 0.01, *** p≤.001, **** p≤ .0001. Error bars represent SEM unless otherwise noted.
RESULTS
AR inhibition impairs ER+/AR+ breast cancer cell proliferation
The role of AR in ER+/AR+ breast cancer remains controversial, with conflicting data suggesting either proliferative or protective effects on breast cancer cells in vitro (2,5,28–31). Most studies of AR function in breast cancer have focused on the effect of androgen stimulation in the presence of E2 in hormone-depleted media. However, we believe it is more relevant to study the effects of activating or inhibiting AR either 1) in the absence of E2, to model postmenopausal women with breast cancer treated with aromatase inhibitors, or 2) in full serum, which contains androgens as well as sufficient estrogens to induce ER activity and genomic binding (32).
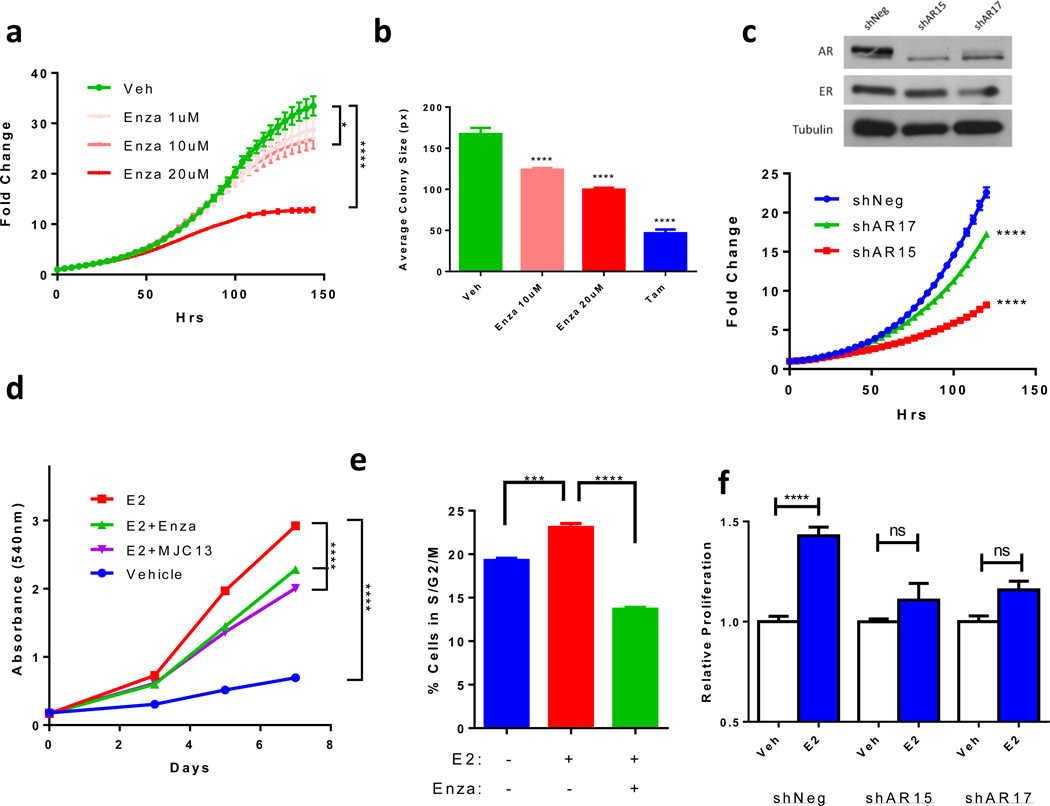
Enza, which inhibits AR nuclear translocation and DNA binding (14,33), significantly decreased growth of MCF7 cells grown in full serum (Figure 1a) as well as two additional ER+/AR+ cell lines, T47D and ZR-75-1, (Supplementary Figure S1a) in a concentration-dependent manner. This shows that AR activity is necessary for ER+/AR+ cell growth under typical culture conditions. Enza also decreased colony size of MCF7 cells (Figure 1b) and T47D cells (Supplementary Figure S1b) grown in soft agar using complete culture media, similar in magnitude to the effect of the anti-estrogen tam. We next decreased AR expression in MCF7 cells using two different shRNA constructs, and AR protein was confirmed to be decreased by western blot (Figure 1c). AR knockdown led to a significant decrease in MCF7 cell growth over the course of 7 days (Figure 1c), further demonstrating that AR is required for baseline proliferation of ER+/AR+ breast cancer cells in hormone-replete conditions.
Figure 1. AR inhibition decreases ER+/AR+ breast cancer growth.
(a) Proliferation of MCF7 cells treated with increasing concentrations of Enza was monitored by IncuCyte. (b) MCF7 cells were grown in soft agar with Enza or tam and colony size was measured by ImageJ. (c) Immunoblotting for AR, ER, and Tubulin in MCF7 cells expressing a non-targeting (shNeg) or AR-targeting (shAR15 and shAR17) shRNA constructs (upper). Proliferation was monitored by IncuCyte (lower). (d) MCF7 cells were grown in media with CSS for 72 hrs then treated with vehicle (Veh), E2, or E2 + Enza or MJC13 and cell number was measured by crystal violet. (e) MCF7 cells were grown in media with CSS for 72hrs then treated with Veh, E2, or E2+Enza for 24hrs followed by cell cycle analysis. (f) MCF7 cells expressing shNeg, shAR15, or shAR17 were cultured in media with CSS for 72 hrs then treated with veh or E2 and growth was measured by crystal violet. Error bars represent standard error of the mean. * p<.05, **** p<.0001 by ANOVA with Dunnett’s Multiple Comparison Test.
New-generation AR inhibitors decrease E2-induced proliferation
We previously showed that Enza, which does not bind to ER by ligand binding assay, inhibits E2-induced growth of ER+/AR+ breast cancer cells in vitro and in vivo (14). To demonstrate that this is AR-dependent and not specific to Enza, we also utilized MJC13, which inhibits AR nuclear localization by targeting ligand-induced dissociation of AR from FKBP52 in the cytosol (34). Both Enza and MJC13 inhibited E2-induced proliferation in MCF7 cells (Figure 1d). Enza decreased E2-induced growth in a concentration-dependent manner in additional luminal cell lines T47D and ZR-75-1, as well as in ER+/AR+ PT12 cells recently created from a patient-derived xenograft (16) (Supplementary Figure S1c). EC50 values for Enza-mediated inhibition of E2-induced growth in MCF7 and T47D cells were determined to be 19.0 µM and 17.1 µM, respectively (Supplementary Figure S1d), which are concentrations readily achieved in patients.
To specifically determine whether Enza affected E2-induced proliferation, cell cycle analysis of E2-treated MCF7 and T47D cells was performed. Enza significantly decreased the percent cells in S and G2/M phases compared to E2 treatment alone (Figure 1e and Supplementary Figure S1e). Silencing AR using shRNA also significantly decreased E2-induced proliferation of MCF7 cells compared to cells transduced with non-targeting shRNA (Figure 1f). Together, these data confirm that pharmacological AR inhibition or AR knockdown similarly diminish E2-driven proliferation of ER+/AR+ breast cancer cells.
AR inhibitors diminish ER genome binding
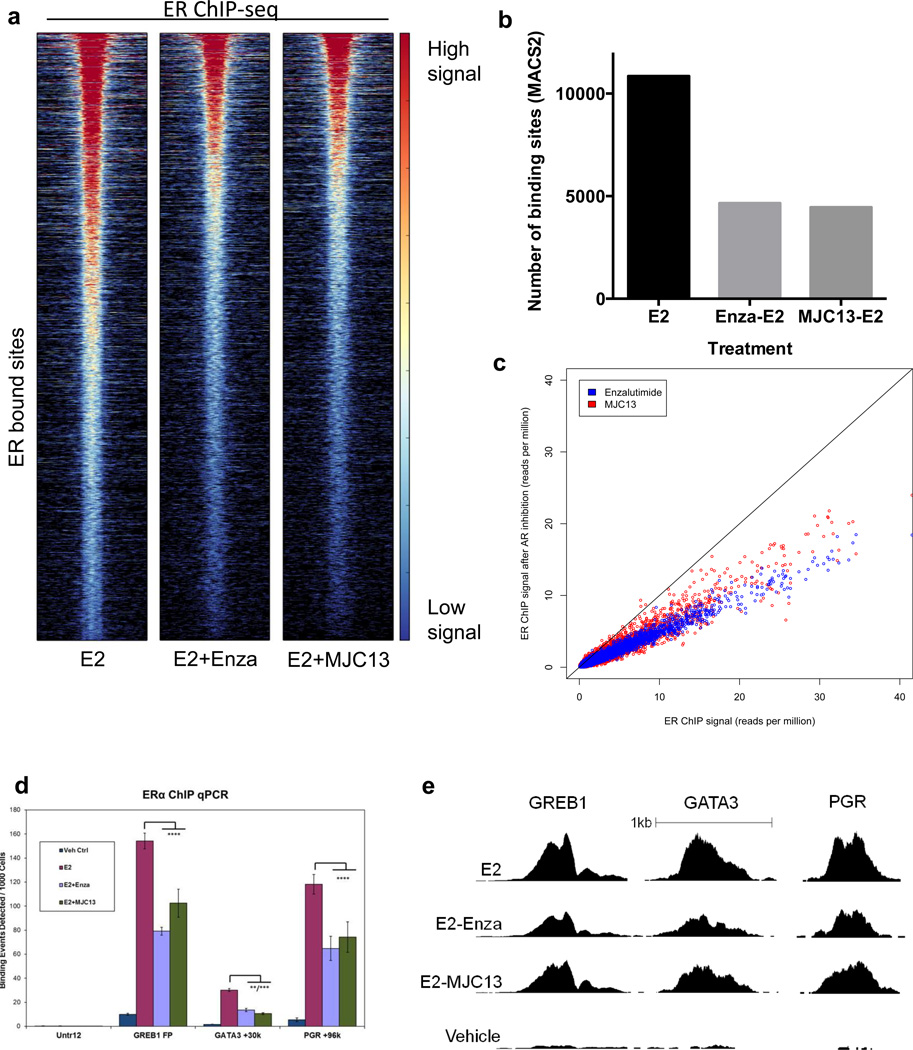
AR is capable of interacting with ER and estrogen response elements (EREs) (2,5,35), thus, we postulated that inhibitors of AR nuclear localization might diminish baseline and E2-induced growth by altering ER genomic binding. To test this hypothesis, MCF7 cells were pretreated for 3 hrs with Veh, Enza, or MJC13 then treated with E2 1 hr and global ER ChIP-seq was performed. Surprisingly, the anti-androgens Enza or MJC13 dramatically decreased E2-induced ER genomic binding (Figure 2a). The majority of sites displayed an approximate 50% decrease in ER binding (Figure 2a–c), with no appreciable shift in the location of ER binding sites upon Enza or MJC13 treatment. The decrease in ER binding intensity by Enza or MJC13 was confirmed by qPCR after ChIP at previously-characterized ER binding sites including GREB1, GATA3, and PGR (Figure 2d–e). Together, this suggests that the interaction of AR and ER is necessary for efficient ER genomic binding in response to E2, and that inhibition of nuclear AR localization decreases E2-induced ER activity by diminishing ER genome binding.
Figure 2. AR inhibitors diminish ER genomic binding.
ChIP-seq for ER in MCF7 cells grown in CSS for 3 days then treated with E2 +/− Enza or MJC-13. (a) Heat map of ER binding. The heat map is shown with a horizontal window of +/− 2kb. (b) The number of binding sites identified by MACS2, using vehicle treatment as the control. (c) The ER ChIP-seq signal at individual sites with E2 alone (x-axis) versus E2 + Enza (blue) or MJC13 (red) (y-axis). (D–E) ChIP-qPCR (d) and ChIP-seq read depth (e) at well-characterized ER binding sites. Error bars represent standard error of the mean. ** p<.01, *** p<.001, **** p<.0001 by ANOVA with Dunnett’s Multiple Comparison Test except.
Enza decreases nuclear localization of both AR and ER
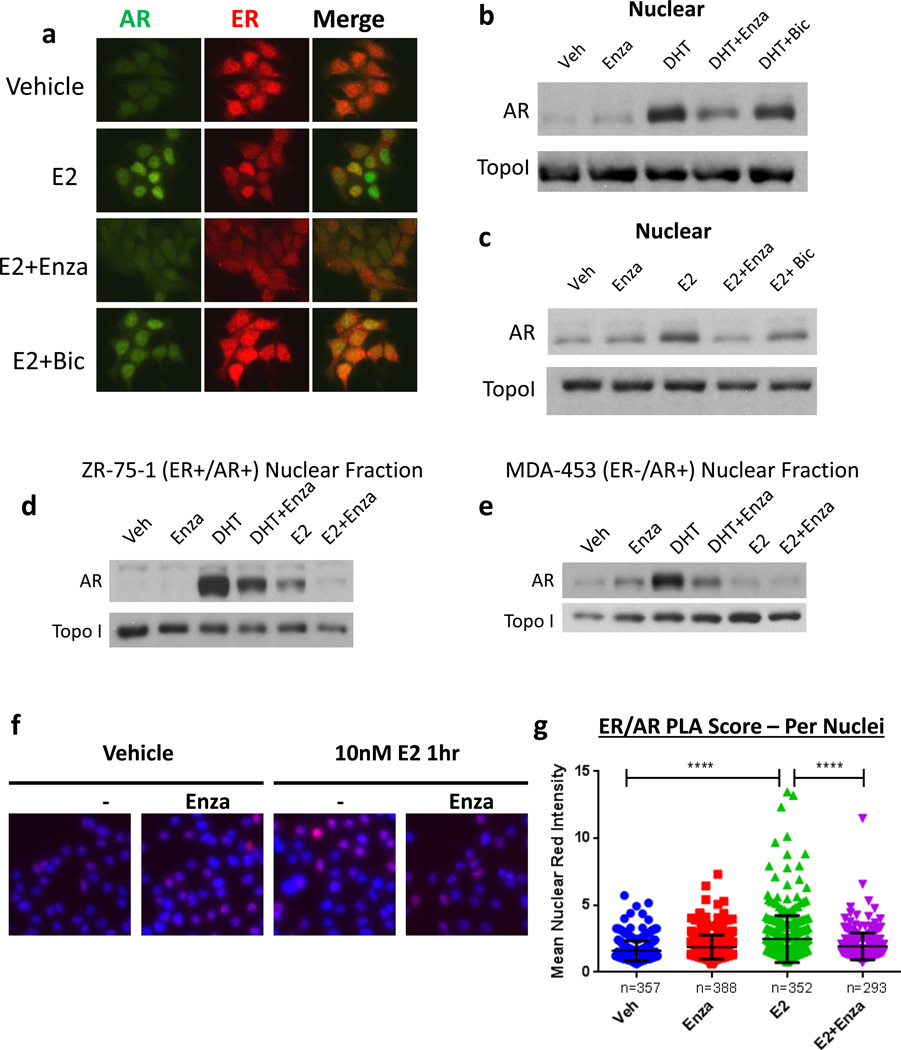
Since E2-induced ER genome binding was globally decreased by anti-androgens, we speculated that ER nuclear localization in response to E2 might be affected. Immunofluorescent staining of MCF7 cells grown in CSS revealed nuclear localization of both ER and AR following E2 treatment (Figure 3a). Notably, treatment with Enza decreased nuclear localization of both receptors (Figure 3a), while bic did not, suggesting that the mechanism by which Enza globally inhibits ER genomic binding may be by decreasing ER nuclear localization. We previously showed that Enza decreased E2-driven growth of MCF7 xenograft tumors equally as well as tamoxifen (14). IHC for ER performed on tumors from mice on Enza-containing chow had significantly decreased nuclear localization of ER compared to tumors from mice on control chow (14).
Figure 3. AR and ER co-localize in the nucleus in response to E2.
(a) MCF7 cells were grown in media with CSS for 72 hr then pre-treated with veh, 10 uM Enza, or 1 uM bicalutamide (bic). Following pre-treatment, cells were treated with veh or 10nM E2 +/− Enza or bic as shown for an additional 3 hr. Cells were then fixed and ICC was performed for AR (green) and ER (red). (b,c) MCF7 cells were grown in media with CSS for 72hrs then treated with the indicated treatment for 3 hrs, and nuclear extracts were immunoblotted for AR and TOPO1. (d) ER+/AR+ ZR-75-1 or (e) ER−/AR+ MDA-453 cells were grown in media with CSS for 72hrs, then pre-treated for 3 hr with Enza or vehicle control. Following pre-treatment, cells were treated with veh, 10nM DHT, or 10nM E2 +/− Enza as shown for 3 additional hrs. Nuclear extracts were then obtained and subjected to western blotting for AR and TopoI. (f) MCF7 cells were grown in media with CSS for 72hrs then treated with E2 +/− Enza for 1 h followed by fixation and PLA staining for AR and ER (red). Nuclei were stained with DAPI (blue). (g) Fluorescent intensity per nuclei was measured by CellProfiler. Error bars represent standard error of the mean. ****p <0.0001 by ANOVA with Dunnett’s Multiple Comparison Test.
To further examine AR nuclear localization in response to E2, MCF7 cells were treated with E2 or DHT plus or minus Enza for 3 hrs, and nuclear and cytoplasmic protein fractions were isolated. DHT induced a strong increase in AR nuclear localization as expected, which was largely blocked by co-treatment with Enza, but not bicalutamide (bic) (Figure 3b). E2 treatment also increased AR nuclear localization, and this effect was blocked by Enza, but not bic (Figure 3c). E2-induced nuclear localization of AR was also observed in ZR-75-1 (Figure 3d). However, E2 did not induce AR nuclear localization in ER−/AR+ MDA-MB-453 cells (Figure 3e) or MDA-MB-231 cells (Supplemental Figure S2a), suggesting that the observed AR nuclear localization is not due to promiscuous binding of E2 to AR, but rather that AR becomes localized to the nucleus in an ER-dependent manner upon E2 stimulation in ER+/AR+ breast cancer cells.
Since both ER and AR were localized to the nucleus following E2 treatment, we next tested whether E2 induced AR and ER to co-localize using the proximity ligation assay (PLA). MCF7 cells treated with 10 nM E2 for 1hr demonstrated a strong increase in PLA signal when probed for ER and AR compared to vehicle control or Enza treatment alone. This E2-induced increase in PLA signal was dramatically inhibited by pre-treatment with Enza (Figure 3f–g). Similar results were observed in T47D cells (Supplementary Figure S2b–d), suggesting that AR co-localizes with ER in the nucleus in response to E2.
E2 induces AR DNA binding distinct from DHT
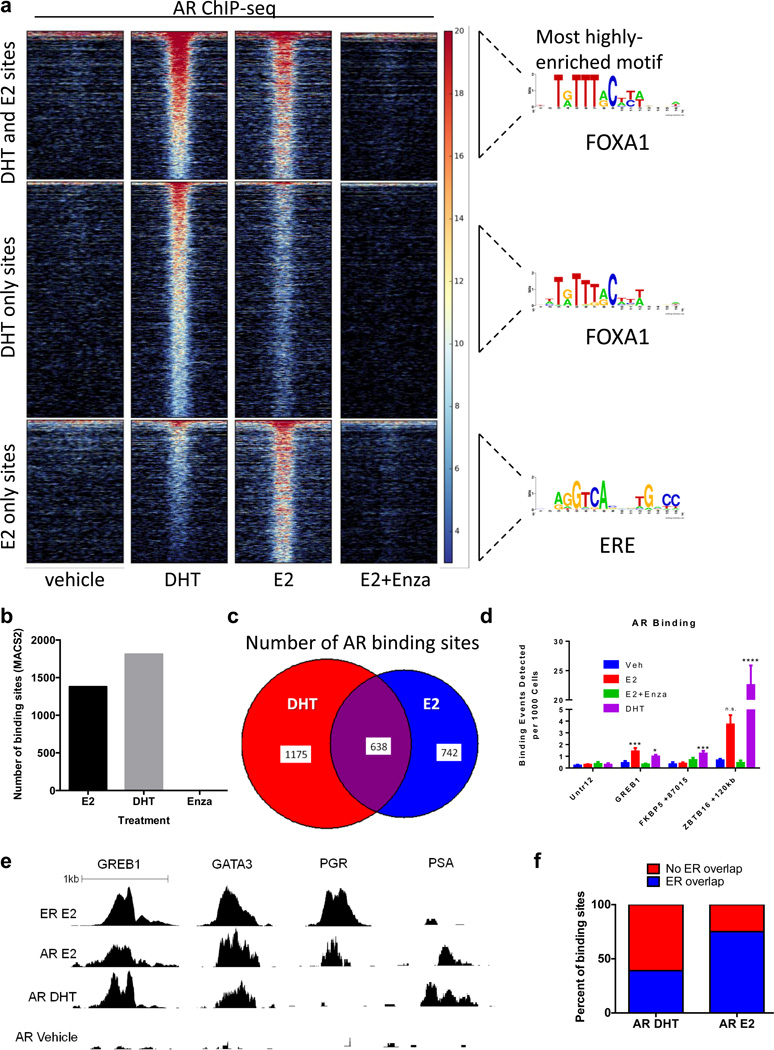
To examine whether the observed nuclear localization of AR in response to E2 was associated with AR genome binding, hormone-deprived MCF7 cells were treated with DHT or E2 followed by global AR ChIP-seq analysis. As expected, DHT treatment induced a significant increase in AR genome binding compared to vehicle treatment (Figure 4a–b). Among the 1,813 DHT-induced AR binding sites identified in MCF7 cells, 49% were previously identified as bound by AR in LNCaP, a prostate cancer cell line, while 73.6% were bound by AR in MDA-MB-453, an ER−/AR+ breast cancer cell line (36) (Supplementary Figure S3a). This indicated that DHT-induced AR binding may be more similar between luminal breast cancer cell lines than between breast and prostate cancer cell lines, and is similar to previously reported findings in ZR-75-1 cells (35).
Figure 4. E2 induces AR genome binding that overlaps with ER binding.
ChIP-seq for AR in MCF7 cells grown in CSS for 3 days then treated with E2 for 1 h or DHT for 4 h. (a) Heat map of binding showing a horizontal window of +/− 2kb and enriched motifs from each category. (b) The number of binding sites identified by MACS2, using vehicle treatment as the control. (c) The number of AR binding sites that are unique to DHT (red), unique to E2 (blue), or shared (overlap) are shown. (d,e) ChIP-qPCR (d) and ChIP-seq read depth (e) results show AR binding at well-characterized ER binding sites following E2 treatment. (f) The percentage of AR binding sites in response to DHT (left) or E2 (right) that were also identified as ER binding sites (blue) is shown.
Surprisingly, E2 also induced AR genome binding, with 1,380 AR binding events identified in E2-treated MCF7 cells (Figure 4a–b). Enza abolished E2-induced AR genomic binding, consistent with inhibition of AR nuclear localization and previously published reports in prostate cancer (33). Only 25% of all AR bound sites overlapped between the two hormone treatments, indicating a large shift in AR genomic binding between DHT and E2 (Figure 4c). For example, qPCR after ChIP demonstrated that DHT, but not E2, induced a robust increase in AR binding at previously-characterized AR targets FKBP5 and ZBTB16 (Figure 4d). Both E2 and DHT treatments resulted in AR binding to previously-characterized ER targets GREB1 and GATA3, but only E2 treatment resulted in AR binding at a different ER target, PGR (Figure 4d–e).
The most highly-enriched motif among AR binding sites in response to DHT was a FOXA1 motif (Figure 4a), consistent with previous studies demonstrating strong overlap between AR and FOXA1 binding sites in breast cancer cells (36). However, the most highly-enriched motif among AR binding sites unique to E2 treatment was a slightly degenerate estrogen response element (ERE) (Figure 4a), suggesting that AR was bound within 200bp of ER binding sites in the presence of E2. Indeed, full palindromic EREs were highly enriched among these sites, compared to sites bound by AR in response to DHT (Supplementary Figure S3b–c). Validated nuclear ERα network was the most highly-enriched pathway among genes near AR binding sites unique to E2 treatment, whereas this network was not enriched among genes near AR binding sites in response to DHT (Supplementary Table S1). Thus, in response to E2, AR binds to many sites correlated with ER regulation.
Finally we compared AR and ER binding following E2 treatment and found that 75% of E2-induced AR binding sites overlapped with ER binding sites (Figure 4f). Notably, ER genome binding was more strongly inhibited by Enza or MJC13 at these overlapping sites compared to non-overlapping sites (Supplementary Figure S3d–e), suggesting that AR might be facilitating ER binding at these loci. Taken together, these data demonstrate that in response to E2, AR and ER bind a significant number of overlapping loci and suggest that new generation anti-androgens which inhibit AR nuclear localization decrease ER activity and E2-mediated tumor growth by diminishing ER genome binding.
Enza synergizes with anti-estrogens
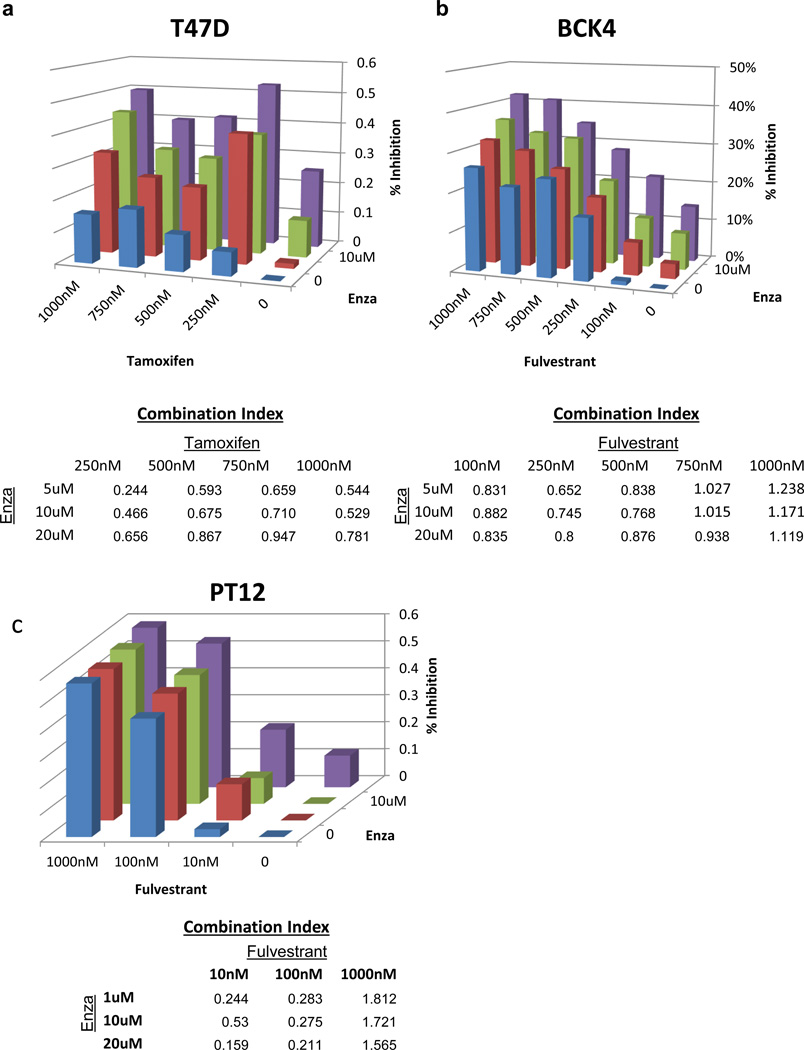
Because Enza inhibited baseline and E2-induced growth by a different mechanism than currently-used anti-estrogens, we hypothesized that it might act synergistically with anti-estrogens such as tam or fulvestrant in ER+/AR+ breast cancer cells. T47D cells were treated with varying concentrations of Enza and/or tam, and all combinations showed synergistic inhibition of E2-induced growth as determined by Calcusyn (Figure 5a). Enza and tam also showed synergy or additive inhibition of E2-induced proliferation in MCF7 cells (Supplementary Figure S4a), and the combination of Enza plus tam reduced MCF7 growth in soft agar more significantly than either drug alone (Supplementary Figure S4b). We also tested for synergy between Enza and fulvestrant. In BCK4 cells, these drugs showed synergy in 10 of 15 dose combinations (Figure 5b), with similar results also observed in PT12 cells (Figure 5c) and ZR-75-1 cells (Supplementary Figure S4c). Together, this shows that Enza effectively synergizes with anti-estrogens to inhibit both baseline and E2-induced growth of ER+/AR+ cells, likely due to the ability of Enza to inhibit AR as well as to indirectly inhibit ER.
Figure 5. Enza synergizes with tam and fulvestrant in vitro.
(a) T47D cells were grown in media with complete serum and Enza and/or tam, and cell number was monitored by IncuCyte. Percent inhibition was compared to vehicle after 5 days, and synergy was calculated using Calcusyn software. A Combination Index (CI) value < 1 is indicative of synergy (yellow). (b–c) BCK4 or PT12 cells were grown in phenol red-free media with CSS for 1 day then treated with E2 with Enza and/or fulvestrant and cell number was monitored by Incucyte.
Enza inhibits Tam-resistant tumor growth
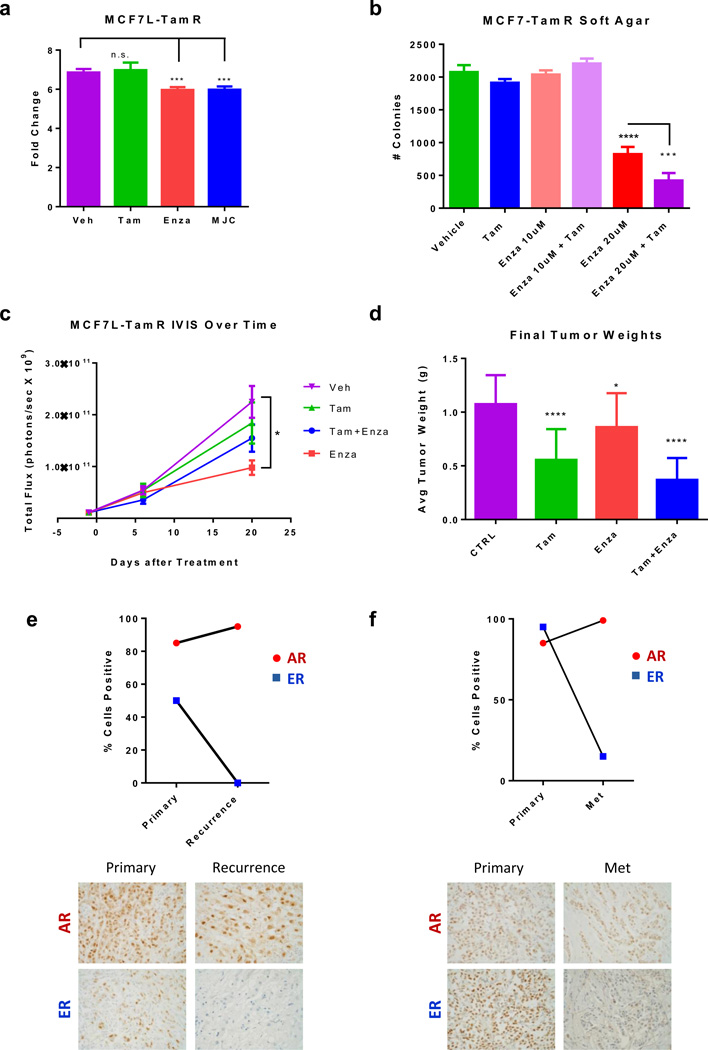
Resistance to currently-used endocrine therapies is a common occurrence facing ER+ breast cancer patients. Therefore, we also tested whether Enza could inhibit growth of tam-resistant MCF7 (MCF7-TamR) cells (37). In vitro, both Enza and MJC13 significantly decreased growth of MCF7-TamR cells (Figure 6a). Enza also decreased growth of MCF7-TamR cells in soft agar, and the combination of Enza+tam was more effective than Enza alone (Figure 6b).
Figure 6. Enza inhibits tamoxifen-resistant tumor growth in vitro and in vivo, and AR is expressed in recurrent breast cancers.
(a) Growth of MCF7-TamR cells treated with vehicle, Tam, Enza, or MJC13 for 7 days. (b) MCF7-TamR cells were plated in soft agar and the number of colonies was counted after 14 days. (c–d) MCF7-TamR cells were implanted into the mammary glands of nude mice with estrogen pellets and were matched into groups to receive either control chow (CTRL), tamoxifen pellets (tam), enzalutamide-containing chow (enza), or both (tam+enza). (c) Tumor growth was measured over time by luminescence. (d) Final tumor weights of mice from each group (lower). (e) IHC for AR and ER in clinical samples of patient-matched primary tumor and recurrence 110 months later. (f) IHC for AR and ER in clinical samples of patient-matched primary tumor and metastasis 167 months later **(400×). * p<.05, *** p<.001, **** p<.0001 by ANOVA with Dunnett’s Multiple Comparison Test.
We next tested whether Enza could inhibit growth of Tam-resistant tumor xenografts in vivo using GFP-luciferase labeled MCF7-TamR cells. Once tumors were established, mice were matched into groups to receive CTRL chow, tam pellets, Enza-containing chow, or Enza+tam. 20 days after beginning treatment, the Enza-treated mice demonstrated a significant decrease in tumor viability by IVIS compared to those in the CTRL group (Figure 6c). Each treatment resulted in a significant decrease in tumor weight compared to CTRL-treated tumors, with Enza+tam resulting in the smallest tumors by weight at the end of the experiment (Figure 6d). TUNEL staining revealed increased apoptosis in each of the treatment groups compared to CTRL (Supplementary Figure S5a). Interestingly, the combination resulted in a significant decrease in ER expression compared to CTRL or either drug alone (Supplementary Figure S5b–c).
AR is expressed in recurrent ER+ breast cancers
To validate the potential clinical utility of anti-androgens as a therapy for advanced ER+ tumors refractory to traditional anti-estrogen directed therapy, we examined AR expression in primary tumors compared to the same patient’s local recurrence or metastatic disease. Sections of formalin-fixed paraffin embedded breast tumors from a cohort of 192 female patients (median age of 68 years) diagnosed with breast cancer at the Massachusetts General Hospital (Partners) between 1977 and 1993, treated with adjuvant tamoxifen and followed through 1998 were stained for AR (14). Of 49 patients with ER+/AR+ primary tumors that developed local recurrence, 96% retained AR positivity (>1% cells positive) in the recurrence. Further, in more than half of these cases the ratio of AR to ER expression (percent cells positive) was higher in the recurrence compared to the primary tumor.
Of 55 patients that developed distant metastasis, 67% retained AR positivity in the metastatic lesion. Notably, one patient with an ER+/AR− primary tumor developed an ER−/AR+ metastasis. Nearly half of these metastases showed an increased ratio of AR to ER expression compared to the primary tumor. Two examples of cases in which the recurrence or metastasis displayed increased percent cells positive for AR, but decreased percent cells positive for ER compared to the primary tumor are shown in Figure 6e. Our findings are consistent with other studies demonstrating that AR status is highly conserved in recurrences and metastases (38), and that AR is more highly expressed in metastases than ER and PR (39). Collectively, this suggests that anti-androgens may be a useful therapeutic strategy for patients with anti-estrogen refractory disease, since AR is frequently expressed in recurrences and metastases, often at even higher levels than in the primary tumor.
Enza Inhibits Primary and Metastatic Tumor Growth in vivo
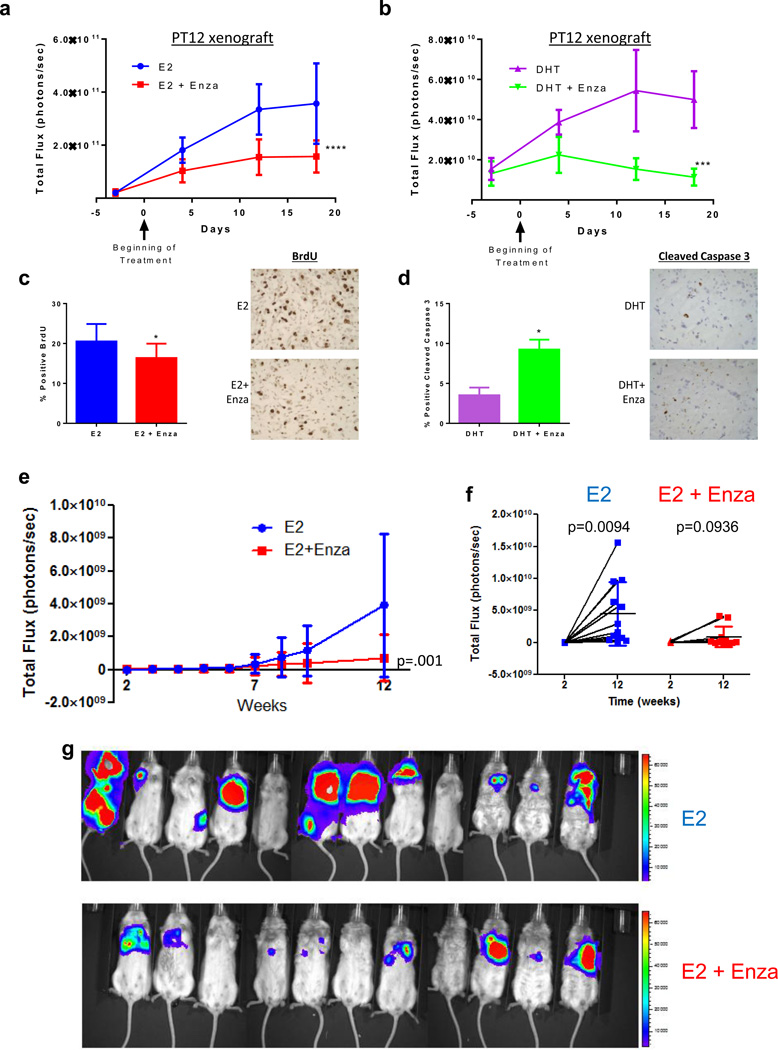
To assess the effect of Enza on E2− and DHT-induced growth in vivo, we utilized AR+/ER+ PT12 cells, recently cultured from a patient-derived xenograft and expressing GFP-luciferase (16). Cells were injected orthotopically in mice implanted with either E2 or DHT pellets, and once tumors were established, mice were matched based on tumor burden to receive either Enza-containing or control chow. Although E2 induced more rapid tumor growth, DHT also stimulated tumor growth (Figure 7a–b). This demonstrates that DHT, in the absence of E2, promotes ER+/AR+ tumor growth in vivo, similar to our previous finding with MCF7 xenografts (14). As shown in Figure 7a, enza significantly reduced the growth rate of E2-driven tumors when compared to E2 alone (difference between treatment groups, F(1,23) = 37.41, p <0.0001; and group*time interaction, F(3,23) = 13.75, p <0.0001). Enza also significantly reduced growth rate of DHT-driven tumors compared to DHT alone (difference between treatment groups, F(1,8) = 27.80, p=0.001; and group*time interaction, F(3,24) = 11.34, p <0.0001). In E2-driven tumors, BrdU staining demonstrated that Enza significantly decreased proliferation (Figure 7c), but had no effect on apoptosis (not shown). Conversely, in DHT-driven tumors Enza significantly increased apoptosis as measured by cleaved caspase 3 staining (Figure 7d), but had no effect on proliferation.
Figure 7. Enza decreases hormone driven growth of PT12 primary tumors and metastases.
(a–d) 1×106 GFP-luciferase expressing PT12 cells were injected orthotopically into the mammary fat pad of NOD-SCID-IL2Rgc−/− mice followed by implantation of either an E2 or DHT pellet. When tumors reached an average of 39 mm3, mice were randomized into the following groups: E2 with control chow (n=10) or enza chow (n=10), or DHT with control chow (n=5) or enza chow (n=5). Tumor viability was measured by IVIS for mice with E2 pellets (a) or DHT pellets (b). (c) BrdU staining of tumors from mice with E2 pellets. (d) Cleaved caspase staining of tumors from mice with DHT pellets. (e) PT12 GFP-luciferase cells were injected intracardially in NOD-SCID-IL2Rgc−/− mice. Metastatic burden of mice treated with E2 or E2+Enza was monitored using IVIS (photons/second) over 12 weeks (total signal supine+prone). (f) IVIS signal from mice in supine position at 2 weeks versus 12 weeks in E2 versus E2+Enza mice. (g) IVIS image of mice in the supine position in the E2 (n=11) or E2+Enza (n=12) groups after 12 weeks, red denotes high IVIS signal. Error bars represent SEM. *p<0.05, ***p<0.001, ****p <0.0001 by repeated measures mixed model approach for (a–b), ANOVA with Bonferroni’s Multiple Comparison Test for (c–d), and Wilcoxon rank sum test for (e).
To identify the molecular mechanisms by which Enza decreased E2-induced tumor growth, we performed RNA-seq on PT12 tumors from E2 treated mice. Enza significantly altered 484 genes (p<.05, fold change > 1.2); 144 upregulated and 340 downregulated compared to E2 alone (Supplementary Table S2). Of these, 107 (22.1%) of the genes affected by Enza were previously identified as regulated by estradiol in the original PT12 xenograft model (Supplementary Figure S6b) (16). Metacore analysis of the 484 genes altered by Enza treatment identified AR and ER as among the transcription factors most highly implicated as upstream regulators (Supplementary Table S3a). Gene set over-representation analysis also identified AR regulation as a highly-enriched pathway, as well as the HIF-1α and HIF-2α networks (Supplementary Table S3b), which have previously been associated with AR in prostate cancer (40). Finally, of the 340 genes downregulated by Enza in PT12 tumors, 56 were also identified in our AR ChIP-seq experiment as being the nearest gene to sites bound by AR in response to E2 treatment in MCF7 cells. Notably, several genes decreased by Enza are reported to be both ER targets and critical for ER activity including GREB1, an E2-responsive ER co-activator (41), and the histone demethylases KDM3A and KDM4B, which mediate ER binding to target gene promoters (42,43). Together, these data confirm our in vitro observations and show that Enza alters expression of ER target genes in vivo.
Finally, since we found that AR is frequently expressed in metastases of ER+ breast cancers, we tested whether Enza could inhibit metastatic growth in vivo. PT12 cells were injected intracardially into mice implanted with E2 pellets, and mice were randomized onto control chow or chow containing Enza. Mice were monitored weekly by IVIS imaging of luciferase over 12 weeks in both the supine and prone positions. Tumors in the enza-treated group were significantly smaller at week 12 (z=−3.82, p=0.0001, two-sided test) (Figure 7e). Next we analyzed IVIS signal of mice at week 2 (first detectable luciferase signal) versus week 12 (Figure 7f). While control mice showed a significant increase in tumor burden over time, there was no significant increase in tumor burden in the Enza treated mice (Figure7f and 7g). This held true whether IVIS signal was measured in the supine (shown) or prone position (not shown) or both added together. These data demonstrate clearly, in a model of ER+/AR+ breast cancer recently derived from a patient, that Enza is effective in reducing the growth of metastatic disease.
DISCUSSION
AR was previously thought to antagonize ER activity because androgens such as DHT diminished the transcriptional and proliferative response of breast cancer cells to E2 (2), likely because AR and ER compete for some of the same binding sites on chromatin. However, our previous and present studies indicate that DHT is proliferative in the context of no E2, as would be the case in a postmenopausal woman treated with aromatase inhibitors (14). The AR antagonist bicalutamide has also been shown to increase E2-induced ER activity (2). But unlike bicalutamide, Enza and MJC13 are newer-generation anti-androgens that inhibit AR nuclear localization, and this is the first study to test the effects of new-generation AR inhibitors on ER chromatin binding. By inhibiting AR nuclear localization or decreasing AR expression by shRNA, we have discovered that AR supports ER genome binding and activity in breast cancer.
In response to E2, AR translocated to the nucleus in ER+ cell lines and bound chromatin at sites that overlap with ER binding sites and are enriched for EREs. Inhibiting nuclear localization of AR with Enza or MJC13 dramatically decreased E2-induced ER chromatin binding, with the greatest effects observed at sites also bound by AR. Both the AR antagonist Enza and AR knockdown decreased baseline and E2-induced breast cancer cell proliferation in vitro, and Enza decreased both DHT- and E2-stimulated growth of ER+/AR+ xenografts as well as metastatic burden in vivo.
These results further underscore the crosstalk between AR and ER in dual-positive breast cancer cells. There is evidence that DHT metabolites can have estrogenic effects and stimulate breast cancer growth through ER activation (44). However, Enza inhibits growth differently in E2-driven tumors, where it decreases proliferation, compared to DHT-driven tumors, where it increases apoptosis. This suggests that DHT-driven growth in ER+/AR+ xenografts is not mediated through ER, but rather directly through AR. Conversely, the expression of the AR cofactor ARA70 can result in E2 having a weak agonist effect on AR (45,46), which could explain how AR is translocated to the nucleus in response to E2 in our studies. Another possibility is that AR is activated by growth factor pathways subsequent to ER activation. However, E2 only drove nuclear localization of AR in ER+ cell lines, suggesting an ER-dependent mechanism. Further studies are ongoing to determine the mechanism of AR nuclear translocation in response to E2, and whether ARA70 is necessary for AR DNA binding in response to E2.
Although our findings may seem contradictory to prior studies, in actuality they are not mutually exclusive. Ligand-bound AR interfered with E2-mediated ER activity (2) and diminished E2-induced upregulation of a subset of ER target genes (35), likely due to competition between AR and ER for some of the same genome binding sites, this is consistent with our observation that AR can bind to ER binding sites in the genome. Additionally, a recent study using AI-resistant MCF7 cells found that ER and AR cooperate on known androgen- and estrogen-responsive gene promoters (10).
We propose that in ER+/AR+ breast cancer cells, AR supports ER nuclear localization and genome binding, possibly by increasing chromatin availability of ER bound loci (47), by stabilizing ER binding to chromatin, and/or interacting directly with ER or as part of an ER-containing complex, as the proximity ligation assay suggests. While this challenges the current view of AR as antagonizing ER activity, a similar effect has been observed with retinoic acid receptor-α (RARα), which interacts with ER binding sites in an ER-dependent manner in ER+ breast cancer cells (48). This interaction is required for E2-induced proliferation and ER transcriptional activity (48). Additionally, glucocorticoid receptor (GR), which is highly similar to AR (49,50) increases chromatin availability and subsequent ER binding at response elements bound by both receptors, a mechanism termed “assisted loading” (47). Similarly, our data show that ER chromatin binding is most inhibited by anti-androgens at sites where E2 induces binding of AR and ER. Thus, even though androgen-bound AR can diminish E2-stimulated ER activity, we show that anti-androgens that prevent AR nuclear translocation have the same effect—suppression of ER activity—via an entirely different molecular mechanism.
Importantly, the assays herein were performed using endogenous AR and ER in cells that naturally express both receptors. In light of recent data from our lab and others suggesting that the ratio of AR:ER protein expression is a predictor of response to traditional ER-directed endocrine therapy (14) and DCIS progression (51), it is likely that the interplay of these receptors may depend on their relative expression level, the levels of their respective ligands in circulation and within tumors, and levels of shared co-factors such as FOXA1. Our data show that across multiple cell lines and preclinical models of ER+/AR+ breast cancer, AR antagonists such as Enza and MJC13 that inhibit AR nuclear translocation also inhibit ER activity indirectly. This combined effect on AR and ER may account for the synergy demonstrated between Enza and the anti-estrogens tam and fulvestrant in vitro. Further analysis of these data is ongoing to determine the mechanisms of this synergy.
Collectively, these data strongly suggest that in the most common form of breast cancer—ER+/AR+ disease—primary tumors and recurrent disease may become reliant on AR and that AR may serve as an effective therapeutic target either in combination with traditional ER-directed therapies (particularly in tumors that have a high AR:ER protein ratio) or upon resistance to ER-directed therapies. Our present and prior (14) studies on the role of AR in ER+ breast cancer contribute to a deeper understanding of the complex molecular interplay between the two most widely expressed hormone receptors in breast cancer (AR and ER), and have already led to clinical trials testing the efficacy of enzalutamide in combination with the aromatase inhibitor exemestane in patients with advanced ER+ disease (NCT02007512) and in combination with fulvestrant (NCT01597193).
Finally, our data demonstrate that Enza effectively inhibits growth of ER+/AR+ metastases in vivo. A recent study of ER− PDX models that metastasize from the orthotopic site found that AR mRNA was increased in circulating tumor cells and micrometastases compared to the primary tumors (52), indicating AR may be an important target for inhibition of metastasis. Likewise, we show in clinical specimens of patient-matched ER+/AR+ primary tumors compared to local or distant recurrences occurring during tamoxifen treatment, AR expression is often maintained, and sometimes increased, in breast cancers refractory to anti-estrogen therapy. We are actively investigating the specific role that AR plays in facilitating the process of metastasis in both ER+ and ER− breast cancer.
Supplementary Material
Acknowledgments
Funded by: BC120183 W81XWH-13-1-0090 DOD BCRP Clinical Translational Award to JKR and ADE; R01 CA187733-01A1 to JKR; American Cancer Society Award 124475-PF-13-314-01-CDD to NCD
The authors thank A. Protter at Medivation Inc. for comments on the manuscript. We thank Dr. Marc Cox for graciously providing MJC13 for these studies. We thank Active Motif Inc. for their assistance with AR and ER ChIPseq experiments. The authors also acknowledge the Genomics and Microarray Core and other Shared Resources of Colorado’s NIH/NCI Cancer Center Support Grant P30CA046934.
Footnotes
Conflicts of Interest: Vernon T. Phan is a fulltime employee of Medivation, Inc.
REFERENCES
- 1.Collins LC, Cole KS, Marotti JD, Hu R, Schnitt SJ, Tamimi RM. Androgen receptor expression in breast cancer in relation to molecular phenotype: results from the Nurses' Health Study. Mod Pathol. 2011;24(7):924–931. doi: 10.1038/modpathol.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peters AA, Buchanan G, Ricciardelli C, Bianco-Miotto T, Centenera MM, Harris JM, et al. Androgen receptor inhibits estrogen receptor-alpha activity and is prognostic in breast cancer. Cancer research. 2009;69(15):6131–6140. doi: 10.1158/0008-5472.CAN-09-0452. [DOI] [PubMed] [Google Scholar]
- 3.Vera-Badillo FE, Templeton AJ, de Gouveia P, Diaz-Padilla I, Bedard PL, Al-Mubarak M, et al. Androgen receptor expression and outcomes in early breast cancer: a systematic review and meta-analysis. Journal of the National Cancer Institute. 2014;106(1):djt319. doi: 10.1093/jnci/djt319. [DOI] [PubMed] [Google Scholar]
- 4.Tsang JY, Ni YB, Chan SK, Shao MM, Law BK, Tan PH, et al. Androgen receptor expression shows distinctive significance in ER positive and negative breast cancers. Annals of surgical oncology. 2014;21(7):2218–2228. doi: 10.1245/s10434-014-3629-2. [DOI] [PubMed] [Google Scholar]
- 5.Panet-Raymond V, Gottlieb B, Beitel LK, Pinsky L, Trifiro MA. Interactions between androgen and estrogen receptors and the effects on their transactivational properties. Molecular and Cellular Endocrinology. 2000;167:139–150. doi: 10.1016/s0303-7207(00)00279-3. [DOI] [PubMed] [Google Scholar]
- 6.Dees EC, Carey LA. Improving endocrine therapy for breast cancer: it's not that simple. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013;31(2):171–173. doi: 10.1200/JCO.2012.46.2655. [DOI] [PubMed] [Google Scholar]
- 7.Giuliano M, Schifp R, Osborne CK, Trivedi MV. Biological mechanisms and clinical implications of endocrine resistance in breast cancer. Breast. 2011;20(Suppl 3):S42–S49. doi: 10.1016/S0960-9776(11)70293-4. [DOI] [PubMed] [Google Scholar]
- 8.Harvell DM, Richer JK, Singh M, Spoelstra N, Finlayson C, Borges VF, et al. Estrogen regulated gene expression in response to neoadjuvant endocrine therapy of breast cancers: tamoxifen agonist effects dominate in the presence of an aromatase inhibitor. Breast Cancer Res Treat. 2008 doi: 10.1007/s10549-008-9923-6. [DOI] [PubMed] [Google Scholar]
- 9.Harvell DM, Spoelstra NS, Singh M, McManaman JL, Finlayson C, Phang T, et al. Molecular signatures of neoadjuvant endocrine therapy for breast cancer: characteristics of response or intrinsic resistance. Breast Cancer Res Treat. 2008 doi: 10.1007/s10549-008-9897-4. [DOI] [PubMed] [Google Scholar]
- 10.Rechoum Y, Rovito D, Iacopetta D, Barone I, Ando S, Weigel NL, et al. AR collaborates with ERalpha in aromatase inhibitor-resistant breast cancer. Breast cancer research and treatment. 2014;147(3):473–485. doi: 10.1007/s10549-014-3082-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Amicis F, Thirugnansampanthan J, Cui Y, Selever J, Beyer A, Parra I, et al. Androgen receptor overexpression induces tamoxifen resistance in human breast cancer cells. Breast cancer research and treatment. 2010;121(1):1–11. doi: 10.1007/s10549-009-0436-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallicchio L, Macdonald R, Wood B, Rushovich E, Helzlsouer KJ. Androgens and musculoskeletal symptoms among breast cancer patients on aromatase inhibitor therapy. Breast cancer research and treatment. 2011;130(2):569–577. doi: 10.1007/s10549-011-1611-2. [DOI] [PubMed] [Google Scholar]
- 13.Morris KT, Toth-Fejel S, Schmidt J, Fletcher WS, Pommier RF. High dehydroepiandrosterone-sulfate predicts breast cancer progression during new aromatase inhibitor therapy and stimulates breast cancer cell growth in tissue culture: a renewed role for adrenalectomy. Surgery. 2001;130(6):947–953. doi: 10.1067/msy.2001.118378. [DOI] [PubMed] [Google Scholar]
- 14.Cochrane DR, Bernales S, Jacobsen BM, Cittelly DM, Howe EN, D'Amato NC, et al. Role of the androgen receptor in breast cancer and preclinical analysis of enzalutamide. Breast cancer research : BCR. 2014;16(1):R7. doi: 10.1186/bcr3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jambal P, Badtke MM, Harrell JC, Borges VF, Post MD, Sollender GE, et al. Estrogen switches pure mucinous breast cancer to invasive lobular carcinoma with mucinous features. Breast Cancer Res Treat. 2013;137(2):431–448. doi: 10.1007/s10549-012-2377-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kabos P, Finlay-Schultz J, Li C, Kline E, Finlayson C, Wisell J, et al. Patient-derived luminal breast cancer xenografts retain hormone receptor heterogeneity and help define unique estrogen-dependent gene signatures. Breast cancer research and treatment. 2012;135(2):415–432. doi: 10.1007/s10549-012-2164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Recchione C, Venturelli E, Manzari A, Cavalleri A, Martinetti A, Secreto G. Testosterone, dihydrotestosterone and oestradiol levels in postmenopausal breast cancer tissues. The Journal of steroid biochemistry and molecular biology. 1995;52(6):541–546. doi: 10.1016/0960-0760(95)00017-t. [DOI] [PubMed] [Google Scholar]
- 18.Peters AA, Ingman WV, Tilley WD, Butler LM. Differential effects of exogenous androgen and an androgen receptor antagonist in the peri- and postpubertal murine mammary gland. Endocrinology. 2011;152(10):3728–3737. doi: 10.1210/en.2011-1133. [DOI] [PubMed] [Google Scholar]
- 19.Ni M, Chen Y, Lim E, Wimberly H, Bailey ST, Imai Y, et al. Targeting androgen receptor in estrogen receptor-negative breast cancer. Cancer cell. 2011;20(1):119–131. doi: 10.1016/j.ccr.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barton VN, D'Amato NC, Gordon MA, Lind HT, Spoelstra NS, Babbs BL, et al. Multiple molecular subtypes of triple-negative breast cancer critically rely on androgen receptor and respond to enzalutamide in vivo. Mol Cancer Ther. 2015;14(3):769–778. doi: 10.1158/1535-7163.MCT-14-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacological reviews. 2006;58(3):621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 22.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome biology. 2006;7(10):R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS) Genome biology. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X, Brutlag DL, Liu JS. BioProspector: discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes; Pacific Symposium on Biocomputing Pacific Symposium on Biocomputing; 2001. pp. 127–138. [PubMed] [Google Scholar]
- 25.Hertz GZ, Stormo GD. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15(7–8):563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- 26.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26(7):873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fioretti FM, Sita-Lumsden A, Bevan CL, Brooke GN. Revising the role of the androgen receptor in breast cancer. Journal of molecular endocrinology. 2014;52:R257–R265. doi: 10.1530/JME-14-0030. [DOI] [PubMed] [Google Scholar]
- 29.Kumar MV, Leo ME, Tindall DJ. Modulation of Androgen Receptor Transcriptional Activity by the Estrogen Receptor. Journal of Andrology. 1994;15:534–542. [PubMed] [Google Scholar]
- 30.Greeve MA, Allan RK, Harvey JM, Bentel JM. Inhibition of MCF-7 breast cancer cell proliferation by 5alpha-dihydrotestosterone; a role for p21(Cip1/Waf1) J Mol Endocrinol. 2004;32(3):793–810. doi: 10.1677/jme.0.0320793. [DOI] [PubMed] [Google Scholar]
- 31.Garay JP, Karakas B, Abukhdeir AM, Cosgrove DP, Gustin JP, Higgins MJ, et al. The growth response to androgen receptor signaling in ERalpha-negative human breast cells is dependent on p21 and mediated by MAPK activation. Breast cancer research : BCR. 2012;14(1):R27. doi: 10.1186/bcr3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nature genetics. 2011;43:27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science. 2009;324(5928):787–790. doi: 10.1126/science.1168175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Leon JT, Iwai A, Feau C, Garcia Y, Balsiger HA, Storer CL, et al. Targeting the regulation of androgen receptor signaling by the heat shock protein 90 cochaperone FKBP52 in prostate cancer cells. Proc Natl Acad Sci U S A. 2011;108(29):11878–11883. doi: 10.1073/pnas.1105160108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Need EF, Selth LA, Harris TJ, Birrell SN, Tilley WD, Buchanan G. Research resource: interplay between the genomic and transcriptional networks of androgen receptor and estrogen receptor alpha in luminal breast cancer cells. Mol Endocrinol. 2012;26(11):1941–1952. doi: 10.1210/me.2011-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson JLL, MacArthur S, Ross-Innes CS, Tilley WD, Neal DE, Mills IG, et al. Androgen receptor driven transcription in molecular apocrine breast cancer is mediated by FoxA1. The EMBO Journal. 2011;30:3019–3027. doi: 10.1038/emboj.2011.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fagan DH, Uselman RR, Sachdev D, Yee D. Acquired resistance to tamoxifen is associated with loss of the type I insulin-like growth factor receptor: implications for breast cancer treatment. Cancer research. 2012;72(13):3372–3380. doi: 10.1158/0008-5472.CAN-12-0684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grogg A, Trippel M, Pfaltz K, Ladrach C, Droeser RA, Cihoric N, et al. Androgen receptor status is highly conserved during tumor progression of breast cancer. BMC cancer. 2015;15:872. doi: 10.1186/s12885-015-1897-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lea OA, Kvinnsland S, Thorsen T. Improved measurement of androgen receptors in human breast cancer. Cancer research. 1989;49(24 Pt 1):7162–7167. [PubMed] [Google Scholar]
- 40.Boddy JL, Fox SB, Han C, Campo L, Turley H, Kanga S, et al. The androgen receptor is significantly associated with vascular endothelial growth factor and hypoxia sensing via hypoxia-inducible factors HIF-1a, HIF-2a, and the prolyl hydroxylases in human prostate cancer. Clin Cancer Res. 2005;11(21):7658–7663. doi: 10.1158/1078-0432.CCR-05-0460. [DOI] [PubMed] [Google Scholar]
- 41.Mohammed H, D'Santos C, Serandour AA, Ali HR, Brown GD, Atkins A, et al. Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep. 2013;3(2):342–349. doi: 10.1016/j.celrep.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gaughan L, Stockley J, Coffey K, O'Neill D, Jones DL, Wade M, et al. KDM4B is a master regulator of the estrogen receptor signalling cascade. Nucleic acids research. 2013;41(14):6892–6904. doi: 10.1093/nar/gkt469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wade MA, Jones D, Wilson L, Stockley J, Coffey K, Robson CN, et al. The histone demethylase enzyme KDM3A is a key estrogen receptor regulator in breast cancer. Nucleic acids research. 2015;43(1):196–207. doi: 10.1093/nar/gku1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sikora MJ, Cordero KE, Larios JM, Johnson MD, Lippman ME, Rae JM. The androgen metabolite 5alpha-androstane-3beta,17beta-diol (3betaAdiol) induces breast cancer growth via estrogen receptor: implications for aromatase inhibitor resistance. Breast cancer research and treatment. 2009;115(2):289–296. doi: 10.1007/s10549-008-0080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thin TH, Kim E, Yeh S, Sampson ER, Chen YT, Collins LL, et al. Mutations in the helix 3 region of the androgen receptor abrogate ARA70 promotion of 17beta-estradiol-induced androgen receptor transactivation. The Journal of biological chemistry. 2002;277(39):36499–36508. doi: 10.1074/jbc.M202824200. [DOI] [PubMed] [Google Scholar]
- 46.Yeh S, Miyamoto H, Shima H, Chang C. From estrogen to androgen receptor: a new pathway for sex hormones in prostate. Proc Natl Acad Sci U S A. 1998;95(10):5527–5532. doi: 10.1073/pnas.95.10.5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Voss TC, Schiltz RL, Sung MH, Yen PM, Stamatoyannopoulos JA, Biddie SC, et al. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell. 2011;146(4):544–554. doi: 10.1016/j.cell.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ross-Innes CS, Stark R, Holmes KA, Schmidt D, Spyrou C, Russell R, et al. Cooperative interaction between retinoic acid receptor-alpha and estrogen receptor in breast cancer. Genes & development. 2010;24:171–182. doi: 10.1101/gad.552910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arora VK, Schenkein E, Murali R, Subudhi SK, Wongvipat J, Balbas MD, et al. Glucocorticoid receptor confers resistance to antiandrogens by bypassing androgen receptor blockade. Cell. 2013;155(6):1309–1322. doi: 10.1016/j.cell.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wright AP, Zilliacus J, McEwan IJ, Dahlman-Wright K, Almlof T, Carlstedt-Duke J, et al. Structure and function of the glucocorticoid receptor. J Steroid Biochem Mol Biol. 1993;47(1–6):11–19. doi: 10.1016/0960-0760(93)90052-x. [DOI] [PubMed] [Google Scholar]
- 51.Tumedei MM, Silvestrini R, Ravaioli S, Massa I, Maltoni R, Rocca A, et al. Role of androgen and estrogen receptors as prognostic and potential predictive markers of ductal carcinoma in situ of the breast. The International journal of biological markers. 2015:0. doi: 10.5301/jbm.5000163. [DOI] [PubMed] [Google Scholar]
- 52.Lawson DA, Bhakta NR, Kessenbrock K, Prummel KD, Yu Y, Takai K, et al. Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature. 2015;526(7571):131–135. doi: 10.1038/nature15260. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.