Abstract
Purpose
Rett Syndrome (RTT) is a neurodevelopmental disorder caused primarily by de novo mutations (DNMs) in MECP2 and sometimes in CDKL5 and FOXG1. However, some RTT cases lack mutations in these genes.
Methods
Twenty-two RTT cases without apparent MECP2, CDKL5, and FOXG1 mutations were subjected to both whole exome sequencing and single nucleotide polymorphism array-based copy number variant (CNV) analyses.
Results
Three cases had MECP2 mutations initially missed by clinical testing. Of the remaining 19 cases, 17 (89.5%) had 29 other likely pathogenic intragenic mutations and/or CNVs (10 cases had two or more). Interestingly, 13 cases had mutations in a gene/region previously reported in other NDDs, thereby providing a potential diagnostic yield of 68.4%. These mutations were significantly enriched in chromatin regulators (corrected p = 0.0068) and moderately in postsynaptic cell membrane molecules (corrected p = 0.076) implicating glutamate receptor signaling.
Conclusion
The genetic etiology of RTT without MECP2, CDKL5, and FOXG1 mutations is heterogeneous, overlaps with other NDDs, and complex due to high mutation burden. Dysregulation of chromatin structure and abnormal excitatory synaptic signaling may form two common pathological bases of RTT.
Keywords: Rett syndrome, chromatin regulation, glutamate signaling, exome sequencing, CNV
Introduction
Rett Syndrome (RTT, MIM #312750) is a neurodevelopmental disorder that mostly affects girls and is primarily caused by de novo mutations in the Methyl-CpG-binding Protein 2 (MECP2) gene on the X chromosome.1 The disorder affects about 1 out of 10,000 live female births2 and is characterized by apparently normal early development in the first 6-18 months of life followed by psychomotor regression involving loss of speech and hand use, and development of gait problems and characteristic repetitive hand stereotypies.3 RTT cases that satisfy all the revised diagnostic criteria of the disease are classified as typical RTT and almost 97% of these carry de novo mutations in MECP2.3,4 Cases that satisfy some but not all of the diagnostic criteria are classified as atypical RTT, which are further divided based on overall severity or profile of symptoms. Up to 86% of atypical cases with mild symptoms, including the “preserved speech” variant of RTT, can be accounted for by mutations in MECP2.4,5 Some atypical cases with an early onset of seizures before regression (“early seizure” variants) are due to de novo mutations in CDKL5, whereas those that regress earlier and have a gross early abnormal development (“congenital” variants of RTT) are caused by mutations in FOXG1.6,7 However, mutations in the latter two genes account for a substantially smaller proportion of atypical RTT cases when compared to mutations in MECP2.
The primary RTT gene MECP2 codes for a methyl-CpG binding protein that binds to chromatin and both activates and represses gene transcription, as demonstrated by studies of gene expression changes in brains of knockout mice and of those over-expressing MECP2 where reciprocal changes in expression were observed for many genes.8 Attempts have been made to show that MECP2, CDKL5, and FOXG1 share some common pathways.9 For instance, MeCP2 can regulate the expression of CDKL5 whose protein product can in turn phosphorylate MeCP2. Some similarity has also been suggested between MECP2 and FOXG1 based on their overlapping domains of expression in the brain.6 Despite these observations, it remains unclear as to which specific biological functions or pathways may be affected in RTT. More recently, mutations in a few additional genes have been found in a handful of cases of RTT-like disorders. These genes include MEF2C,10 WDR45,11 STXBP1.12,13 Other genes found to be mutated in a few RTT cases, but which have primarily been associated with non-RTT neurodevelopmental disabilities, are IQSEC2,13 SCN8A,13 and SMC1A,14 suggesting that they might impact some shared biological pathways important to brain development and/or maintenance of proper brain function.
In this study we hypothesized that genes other than MECP2, CDKL5, and FOXG1 could contribute to RTT. We used genomic approaches to identify some of the genetic causes of both typical and atypical RTT cases that lack mutations in MECP2, CDKL5, and FOXG1, anticipating that at least some of the causes will be due to mutations in genes already implicated in other neurodevelopmental disorders involving epilepsy, intellectual disability, and autism spectrum disorder (ASD) due to their phenotypic overlap with RTT. We carried out a combination of exome sequencing and high density single nucleotide polymorphism (SNP) array-based copy number variant (CNV) analyses on a total of 22 RTT cases lacking mutations in the above three genes.
Materials and Methods
Patient Cohort and Clinical Diagnosis
Written, informed consent was obtained from all parents for participation in this study, which was approved by the Institutional Review Board of the Baylor College of Medicine. All of the participants were enrolled in the Rett Syndrome Natural History Study (U54HD061222, clinicaltrials.gov NCT00299312), and enrollment in this study requires either a clinical diagnosis of RTT or a pathogenic mutation in MECP2. Diagnosis of either typical or atypical RTT was made by expert clinicians (JLN, DGG, WEK, SAS, AKP) following the recently revised diagnostic criteria3. The requirements for a diagnosis of typical RTT is evidence of a period of regression followed by stabilization, loss of acquired hand skills, loss of acquired spoken language, gait abnormalities, and stereotyped hand movements. These are considered the “main criteria” for diagnosis and the presence of these features in the participants in this study are presented in detail in Table S1. The diagnosis of atypical RTT requires the period of regression followed by stabilization and two of the four remaining main criteria, plus 5/11 supportive criteria. This is also outlined in Table S1. Genomic DNA was extracted from peripheral blood at Baylor Molecular Genetics Diagnostic Laboratory according to standard, CLIA approved methods. Clinical efforts to arrive at a molecular diagnosis included Sanger-sequencing of coding regions of known genes (MECP2, CDKL5, FOXG1) and assessing structural variations through a combination of methods including Multiplex Ligation-Dependent Probe Amplification (MLPA), Southern blotting, and BAC or oligonucleotide array comparative genomic hybdridization.
SNP Genotyping and Copy Number Variation (CNV) Analysis
Genome-wide copy number variation (CNV) analysis was performed by genotyping probands on the Illumina Omni 2.5m single nucleotide polymorphism (SNP) array using standard procedures in the Laboratory for Translational Genomics at Baylor College of Medicine. PennCNV was used to identify CNVs from arrays that had >99% call rate, standard deviation of log R ratio <0.3, and GC wave factor between -0.04 and +0.04. All samples satisfied these criteria. Two samples (cases 102000 and 101329) each resulted in over 800 CNV calls and were removed. CNVs from remaining samples were filtered to retain those that were at least 30-kilobases in length with 10 or more SNPs and a confidence score of at least 10 and which impacted at least one exon of at least one protein coding gene.
Exome Sequencing and Variant Identification
Genomic DNA of probands and parents was processed for paired-end whole exome sequencing on Illumina HiSeq 2000 at the Baylor-Hopkins Center for Mendelian Genomics. Exome capture was achieved either by the Baylor College of Medicine-developed Human Genome Sequencing Center (HGSC) Core reagent or Nimblegen's VCRome 2.1 reagent.15 Over 6Gb of uniquely aligned sequence was produced per individual with at least 85% of bases covered by >=20× and overall average coverage of 87×. Alignments were made using Burrows-Wheeler Aligner (BWA v0.5.9) to the hg19 reference human genomes and duplicates flagged by Picard v1.98. Variants were identified by following the best practice work-flow of the Genome Analysis Toolkit (GATK v2.5-2) and annotated using ANNOVAR (v2014Sept09).
Variants were filtered to select only those whose inheritance appeared to be consistent with dominant or recessive models of disease (de novo, homozygous, compound heterozygous). Since RTT results in a clinically obvious and severe phenotype, it is extremely unlikely to be caused by variants present in control populations or in populations with other non-neurodevelopmental diseases even at low frequencies. Thus, for de novo variants, we prioritized only those that were not found in dbSNP138, 1000 Genomes, ESP6500, and ExAC databases. For compound heterozygous variants the frequency of each individual variant had to be less than 0.005 (with no homozygotes reported for both variants) so as to be consistent with a reasonable combined incidence of typical and atypical RTT cases not caused by mutations in MECP2, CDKL5, and FOXG1 of about 0.000025 which is 25% of the total incidence of RTT of 1 out of 10,000. The total read depth cutoff was set at 10, and for heterozygous variants at least 2 reads had to carry the variant. Additionally, the proportion of reads with the heterozygous variant had to be between 15-85%. Missense variants were prioritized based on their predicted deleteriousness as determined by 12 tools (SIFT, Polyphen2_HDIV, Polyphen2_HVAR, LRT, MutationTaster, MutationAssessor, FATHMM, RadialSVM, LR, VEST3, and conservation scores from GERP++_RS and CADD). These additional criteria were used to select likely pathogenic variants from RTT cases for whom DNA samples of one or both parents were unavailable: occurrence in genes previously reported to have de novo mutations in epileptic encephalopathies,16,17 Autism Spectrum Disorders (ASD),18-22 intellectual disability (ID),23 and unexplained developmental delays;24,25 and an observation of a nervous system phenotype in mouse (phenotype code MP:0003631 from Mouse Genome Informatics (http://www.informatics.jax.org/phenotypes.shtml).
Sanger Validation of Candidate Variants from Exome Data
Standard polymerase chain reaction (PCR) was used to amplify products between 300-800 base pairs for Sanger sequencing. Briefly, between 20-30ng of genomic DNA template and KAPA HiFi Hotstart DNA polymerase (KAPA Biosystems, Woburn MA) were used for amplification in a 30ul reaction as per the manufacturer's instructions. All forward and reverse primers were respectively designed to have M13F-41 (GGTTTTCCCAGTCACGAC) and M13R-27 (GGAAACAGCTATGACCATG) universal sequences at their 5-prime ends. PCR products were cleaned with a clean-up kit (Qiagen, Valencia CA or Bioneer Inc, Alameda CA) and sequenced at at SeqWright, LoneStar Sequencing (both Houston TX) or Eton Bioscience (San Diego CA).
Results
Overview of Genetic Findings
Of the 22 cases examined, 11 had a clinical diagnosis of typical and 11 of atypical RTT (Table S1), as defined by the consensus criteria which is outlined in Table S13. Notably, all cases showed regression followed by stabilization, specifically lost either hand skills or spoken language, had gait abnormalities, and developed characteristic repetitive hand stereotypies. Exomes of both unaffected parents of 6 typical and 7 atypical RTT cases were also sequenced. All variants considered to be likely pathogenic are in Table 1. This table also lists all de novo mutations identified from exome analysis, regardless of whether they were considered likely pathogenic or not. All CNVs and exome variants that were selected for Sanger-validation per case are listed in Table S2. Sanger sequence of one mosaic de novo mutation is presented in Fig. S1. The intensity and B-allele frequency plots of CNVs are provided as Figs. S2-S10. Three cases were found to have causative MECP2 mutations that were initially missed during clinical testing. One was a 5 base-pair (bp) frameshift deletion (p.E50fs) in the third exon of MECP2 not present in the unaffected mother and which was eventually detected in the clinic upon resequencing. The second was a de novo 17bp frameshift duplication c.41_57dup17 (p.R20fs) initially undetected by clinical sequencing as this exon was not routinely sequenced. However, a revised sequencing report was able to detect this mutation. Our exome sequence data could not detect this mutation due to the high GC content of the first exon of MECP2, a molecular feature that can decrease capture efficiency in the hybridization-based capture step of exome sequencing. In light of this, we sequenced by Sanger the first exon of MECP2 in all the remainder of our cases and found one de novo mutation (M1V) in the initiation codon in case. This exact mutation has been reported in a typical RTT patient and is expected to abolish the normal translation of the MeCP2_e1 transcript which is the more abundant isoform in the nervous system.26
Table 1.
List of all de novo and other likely pathogenic variants contributing towards the RTT phenotype.
| Case ID | Gene or CNV cytoband |
Mutation type | cDNA (and protein) changes or CNVs |
SIFT | PP2 HDIV |
Inheritance | Domain/region of mutation |
Known disorders and other information |
|---|---|---|---|---|---|---|---|---|
| 100935-t | MECP2 | Frameshift del | c.148_152del (p.E50fs) | na | na | Not maternal | - | Rett syndrome (MIM #312750). |
| 106000-t | MECP2 | Frameshift dup | c.41_57dup17 (p.R20fs) | na | na | de novo | - | Rett syndrome (MIM #312750). |
| PWP2* | Missense | c.G2169T (p.E723D) | T(0.07) | P(0.9) | de novo | 14th WD repeat | - | |
| SCG2* | Missense | c.A269G (p.Q90R) | T(0.8) | B(0.193) | de novo | FANCD2 interaction domain | - | |
| IZUMO4* | Missense | c.G392A (p.R131H) | T(1) | B(0) | de novo | - | ||
| XAB2* | In-frame del | c.2487_2492del (p.829_831del) | na | na | de novo | 26 residues from C-terminus | - | |
| ZSCAN12* | Nonsense | c.C1603T (p.R535X) | T(0.21) | na | de novo | 10th C2H2-type zinc finger domain (disrupts only the last 2 of 10 such domains) | - | |
| 101073-t | MECP2 | Missense | c.A1G (p.M1V) | na | na | de novo | Initiation methionine | Rett syndrome (MIM #312750). |
| 108286-t | IQSEC2 | In-frame del | c.83_85del (p.28_29del) | na | na | de novo | Coiled-coil | XLID in males (MIM #309530); RTT-like disorder reported in one female (ref. 13). Mutation may have a dominant negative effect since IQSEC2 escapes X-inactivation. |
| 100182-t | FAM151A | Missense | c.G512T (p.W171L) | D(0.02) | D(1) | de novo | - | Expressed in many of the same human brain structures as MECP2 (BrainSpana data); female knockout mice have a neurological phenotype of impaired sensorimotor gating/attentionb. |
| SYNE2 (mosaic)* | Missense | c.A5479G (p.T1827A) | T(0.31) | B(0.021) | de novo | 14th spectrin repeat | Emery-Dreifuss muscular dystrophy-5 (MIM #612999); DNMs found in one EE (ref. 17) and one DD patient (ref. 25). This is an apparent mosaic with 14% variant-to-reference read ratio in exome data and Sanger peak height of variant allele no more than 20% of reference allele (Figure S1). | |
| 100976-t | SMC1A | Nonsense | c.C2161T (p.Q721X | T(0.95) | na | de novo | 3rd coiled-coil domain | Missense/in-frame mutations cause Cornelia de Lange Syndrome type 2 (MIM #300590). Loss-of-function mutations reported in a case with RTT-like disorder (ref. 14). |
| ARHGEF10L* | Missense | c.G3040A (p.V1014M) | T(0.24) | D(0.973) | de novo | - | One de novo missense mutation found in an ASD case (ref. 18). | |
| HDAC1 | Missense | c.G208A (p.D70N) | D(0.03) | D(0.998) | de novo | Histone deacetylase domain | Required for normal nervous system development (ref. 33). | |
| TAF1B* | Missense | c.G619A (p.V207M) | na | P(0.811) | de novo | N-terminal cyclin fold | - | |
| 107526-t | KCNJ10 | Missense | c.A1061G (p.K354R) | T(0.3) | D(1) | Paternal | Cytoplasmic | Recessive SESAME syndrome (MIM #612780) with epilepsy, ataxia, and deafness. dbSNP ID rs142596580; highest control population frequency of 0.0006. |
| KCNJ10 | Missense | c.C811T (p.R271C) | D(0.02) | D(0.999) | Maternal | Cytoplasmic | Recessive SESAME syndrome (MIM #612780) with epilepsy, ataxia, and deafness. dbSNP ID rs1130183; highest control population frequency of 0.047; listed as “probable-non-pathogenic” in the CLINVAR database; because this channel forms homomers, a partial loss-of-function of channels formed by the two mutations in aggregate within the same cell is possible. | |
| 3p25.3 (0.68Mb) | Deletion CNV | chr3:10923941-11606490 | na | na | de novo | - | Associated with myoclonic epilepsy (ref. 34). Affects 6 genes including the GABA transporter SLC6A1; determined to be de novo by comparing exome coverage in proband and parents (Figure S2). | |
| 100217-t | CHD4 | Missense | c.C5083T (p.R1695C) | D(0) | D(1) | Not maternal | Pericentrin binding domain | De novo missense mutations reported in two cases of unexplained DD (ref. 25) and one of EE (ref. 16). |
| LRRC40 | Missense | c.A689G (p.N230S) | D(0) | D(1) | Not maternal | 7th leucine-rich repeat region | A de novo missense mutation reported in the 19th leucine-rich repeat in an ASD case (ref. 18). | |
| 102000-t | LAMB2 | Missense | c.T2635A (p.C879S) | D(0) | D(0.999) | Not present in mother and sister | 8th laminin EGF-like domain | Recessive Pierson syndrome (MIM #609049); a de novo missense mutation in this gene (and other lamin genes) was reported in an ASD case (ref. 18). Pierson syndrome mutations are clustered in exons 2-7 (ref. 35), whereas the ASD mutation is in exon 14 and the mutation in the RTT case listed here is in exon 20. |
| 102475-t | 22q13.2-q13.33 (8.9Mb) | Deletion CNV | chr22:42287454-51184884 | na | na | Very likely de novo | - | Phelan-McDermid syndrome (MIM #606232). Affects 136 genes including SHANK3. This case was not subjected to exome sequencing. |
| 100696-at-c | GRIN2B | In-frame del | c.1687_1695del (p.563_565del) | na | na | de novo | 1st transmembrane domain | Early infantile epileptic encephalopathy-27 (MIM #616139). Mutation may destabilize the receptor. |
| 101223-at-c | BRAF | Missense | c.T1914G (p.D638E) | D(0) | D(1) | de novo | Kinase domain | Dominant cardiofaciocutaneous (MIM #115150), Noonan type 7 (MIM #13706), and LEOPARD type 3 (MIM #613707) syndromes. This exact mutation occurred de novo in two individuals with cardiofaciocutaneous syndrome (ref. 36). |
| IMPDH2 | Missense | c.G1439A (p.R480Q) | T(0.17) | D(0.992) | de novo | Last residue of the catalytic domain | A de novo missense mutation reported in an ASD case (ref. 22). IMPDH2 is involved in maintaining the normal cellular concentrations of guanine deoxy- and ribonucleotides which are required for many small GTP-binding proteins, mutations in some of which cause NDDs including ID when mutated. | |
| 112626-at-c | SAFB2* | Missense | c.G2305A (p.D769N) | T(0.07) | D(0.999) | de novo | Arginine-rich region | - |
| ACTL6B | Homozygous stoploss | c.1279delT (p.X427D) | na | na | Both heterozygous parents | - | A de novo missense mutation reported in an ASD case (ref. 25). Mutation adds at least 17 extra residues beyond the normal C-terminus; this gene codes for a neuron-specific subunit of the Brg1-associated factor (BAF) chromatin remodeling complex and mouse knockouts have abnormal hippocampal synaptic plasticity and proper dendritic arborization (Mouse Genome Informatics phenotype database). | |
| 3p26.3 (0.98Mb) | Duplication CNV | chr3:269740-1255822 | na | na | Paternal | - | ID, ASD features, epilepsy and global DD (ref. 37). | |
| 100146-at-c | STXBP1 | Missense | c.C1216T (p.R406C) | D(0) | D(1) | Not maternal | - | Early infantile epileptic encephalopathy-4 (MIM #612164); typical and atypical RTT (refs. 12, 13). An atypical RTT case had a different missense mutation (R406H) in this same residue listed here. |
| 22q11.23 (1.29Mb) | Duplication CNV | chr22:23725886-25013346 | na | na | ND | - | Seizures and other congenital abnormalities (ref. 38). | |
| Xp22.31 (1.64Mb) | Duplication CNV | chrX:6494073-8138088 | na | na | ND | - | Cognitive deficits, seizures, motor delays, and ASD (ref. 39). | |
| 102477-at-c | TRRAP | Missense | c.G6809A (p.G2270E) | na | D(0.993) | Not maternal | p53-binding domain | Multiple de novo missense mutations reported in ASD (ref. 18), unexplained DD (ref. 25), and EE (ref. 17). |
| WDR45 | Missense | c.G439C (p.G147R) | na | D(1) | Not maternal | 4th WD repeat | Neurodegeneration with brain iron accumulation (MIM #300894) and RTT-like features (ref. 11). | |
| 100047-at-c | SLC39A13 | Missense | c.C860T (p.A287V) | D(0) | D(1) | ND | 6th transmembrane domain | Recessive Ehlers-Danlos syndrome-like spondylocheirodysplasia (MIM #612350). Expressed in almost all regions of the developing human brain (BrainSpana data). |
| FAT3 | Missense | c.G12557C (p.R4186P) | D(0.04) | D(1) | ND | 4th EGF-like calcium-binding domain | Three ASD cases reported with missense de novo mutations (refs. 18, 22). Localized to dendrites of retinal amacrine cells; mutant mice have more neurites (Mouse Genome Informatics phenotype database). | |
| IQGAP3 | Missense | c.A4778G (p.Y1593C) | D(0.01) | D(1) | ND | - | Two ASD cases reported with de novo missense mutations (ref. 18). | |
| NCOR2 | Missense | c.C1940T (p.S647L) | D(0) | D(1) | ND | SANT2 domain involved in chromatin interactions | Codes for a nuclear receptor co-repressor which represses transcription of some nuclear receptors by modulating chromatin structure; MeCP2 is has a domain that binds to NCOR2 (UniProt). | |
| 133932-at-esz | GABRB2 | Missense | c.C911T (p.A304V) | D(0) | D(1) | de novo | 3rd transmembrane domain (first residue entering the cell membrane from extracellular side | A de novo mutation reported in a case with epilepsy and intellectual disability (ref. 40). This case was not genotyped on SNP arrays. |
| 144945-at-esz | TCF4 | Splice | c.923-1G>- | na | na | de novo | Dominant Pitt-Hopkins Syndrome (MIM #610954). This mutation affects a constitutive exon and is predicted to result in a frameshift due to out of phase exon skipping. | |
| 122215-at-ps | GRIN2A | Missense | c.G904A (p.A302T) | na | D(1) | ND | Extracellular | Focal epilepsy with speech disorder and with or without mental retardation (MIM #245570). |
| 7q11.22 (1.81Mb) | Duplication CNV | chr7:70153859-71969713 | na | na | ND | - | Autosomal dominant mental retardation-26 (MIM #615834). This particular CNV affects six genes including partially duplicating the last 14 exons of AUTS2. |
t, typical RTT; at-c, atypical congenital RTT; at-esz, atypical early seizure RTT; at-ps, atypical preserved speech RTT; XLID, X-linked intellectual disability; ASD, Autism Spectrum Disorder; EE, epileptic encephalopathy; DD, developmental delay; NDD, neurodevelopmental disorder. Mode of inheritance not determined (ND) for some due to lack of parental DNA samples. Asterisks indicate mutations not likely to be pathogenic and which were not included in enrichment analysis of biological functions. While predictions of deleteriousness only by SIFT and Polyphen2 (PP2_HDIV) are shown here, predictions by additional algorithms are in Table S2. Plots of LogR-ratios and B-allele frequency for CNVs listed here are in Figures S2-S10. All coordinates are from the human genome haploid reference sequence version hg19.
BrainSpan: Atlas of the Developing Human Brain available at www.brainspan.org
From the exome data of the remaining 19 cases we selected 78 variants for Sanger-based confirmation of which 13 (16.7%) were loss-of-function (nonsense, splice, and frameshift insertions or deletions), 4 were in-frame insertions or deletions, one was a stoploss mutation, and 60 were missense mutations. From these, a total of 15 de novo mutations were confirmed in 11 trios, giving a rate of 1.36 such mutations per trio. One de novo mutation was apparently mosaic. Three (25%) de novo mutations were loss-of-function. One de novo deletion CNV was also identified. At least one likely pathogenic mutation was found in 17 of the 19 cases (89.5%) with 13 having mutations previously associated with other NDDs, thereby providing a potential molecular diagnostic yield of 68.4%. This suggests that severe neurodevelopmental disorders are more likely than not caused by genetic defects due to new mutations.
An Increased Mutation Burden Potentially Contributes to RTT Phenotype
Ten of 19 cases (52.6%) had more than one likely pathogenic mutation identified either from exome sequence data, CNV analysis, or both. This is a high proportion of cases with multiple likely causal variants, and suggests that a high burden of mutation may contribute to the final disease phenotype in these cases. Even though not all individual de novo mutations were considered to contribute to disease, we note that 4 out of the 11 cases that were part of complete trios carried two or more such mutations from exome sequence data. We therefore determined the overall rate of such protein altering de novo mutations in RTT cases and compared it with the same rate reported in controls.27 As there were 15 confirmed de novo mutations identified out of a total of 695,695,712 high quality bases sequenced at a depth of at least 10×, the rate was 1.36 DNMs per trio, or 2.16×10-8 per base per generation. While this rate is higher, it is statistically not significantly different from the reported27 control rate of 1.47×10-8 (binomial p = 0.15), which likely reflects the small sample size of 11 trios. However, when the two cases with confirmed de novo mutations in MECP2 (one of whom also had five additional de novo mutations all of which are listed in Table 1) were included, then the observed rate of such mutations was 1.70 per trio (22 de novo mutations in 13 trios with 829,661,092 high quality bases sequenced at a depth of at least 10×), or 2.57×10-8 per base per generation, which is significantly higher than the reported rate in controls (binomial p = 0.009). Hence, a high burden of de novo mutations may be a feature of RTT in general which, when combined with CNVs, results in an increased overall mutation burden that contributes to RTT.
Enrichment of Chromatin Regulators and Glutamate Receptor Signaling Molecules in Genes with Likely Pathogenic Mutations
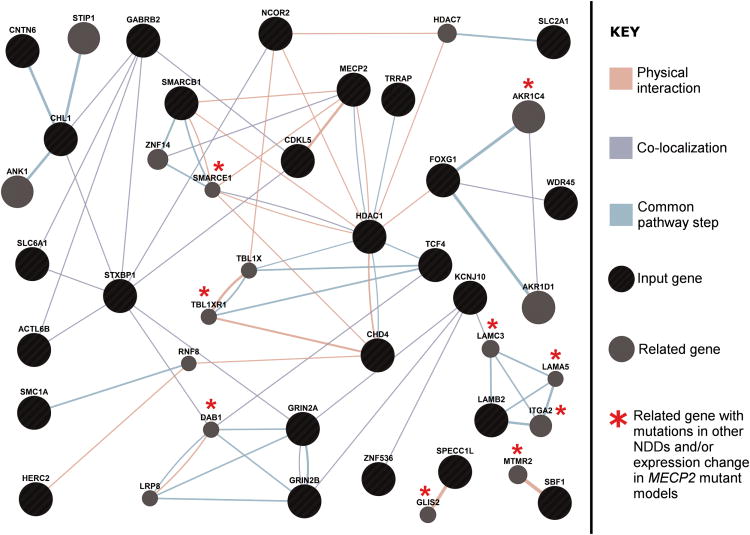
We asked whether the genes with likely pathogenic mutations in our patients were significantly enriched for those that code for proteins with common biological functions. We compiled a list of 46 genes (Table 2) comprising those with likely pathogenic intragenic mutations identified from exome sequencing as well as select genes impacted by CNVs in our patients in which intragenic de novo mutations had been reported in at least one patient in large-scale exome sequencing studies of ASD, intellectual disability, epilepsy, and other developmental disorders. Using the DAVID functional annotation tool (https://david.ncifcrf.gov/) we found that there was a highly significant enrichment of the term “chromatin regulator” (uncorrected p = 0.00011; Benjamini-Hochberg corrected p = 0.0068) of the Protein Information Resource (PIR) database. The six genes within this term were ACTL6B, BRD1, CHD4, HDAC1, SMARCB1, TRRAP. There was also a moderate enrichment of the term “postsynaptic cell membrane” (uncorrected p = 0.002; Benjamini-Hochberg corrected p = 0.076). The four genes within this term were GABRB2, GRIN2A, GRIN2B, and SHANK3, the latter three being members of the glutamate receptor signaling pathway. We next asked whether the protein products of genes listed in Table 2, when analyzed together with the known RTT genes MECP2, CDKL5, and FOXG1, physically interact with each other, co-localize, or participate in the same step of a given pathway. Using GeneMania (http://www.GeneMANIA.org/) there were 23 out of the 46 genes (50%) that interacted with each other (or with other genes reported to have mutations in NDDs and/or with genes showing an expression change in MECP2 mutant model system) either directly or indirectly through at least one of these three ways (Figure 1). To determine whether the enrichment discovered from these cases is not spurious, a similar analysis using 65 genes with de novo loss of function and missense mutations predicted to be deleterious observed in control individuals from several studies did not yield significant results (Table S3 and Figure S11).18,20,27,41 Functional annotation using DAVID showed an enrichment of a broad term “phosphoprotein” comprising of 41 genes and a corrected p=0.0093 which was not considered a highly specific enrichment. These analyses support the contention that many genes mutated in our RTT patients share some common features with the other known RTT genes, further implicating them as having a role in this disease.
Table 2.
List of 46 genes with de novo and likely pathogenic mutations contributing to RTT identified from either exome sequencing or CNV analysis used for enrichment testing of biological functions.
| Gene | Source |
|---|---|
| ACTL6B | Exome |
| AUTS2 | CNV |
| BRAF | Exome |
| BRD1 | CNV |
| CELSR1 | CNV |
| CHD4 | Exome |
| CHL1 | CNV |
| CNTN6 | CNV |
| FAM151A | Exome |
| FAT3 | Exome |
| GABRB2 | Exome |
| GRIN2A | Exome |
| GRIN2B | Exome |
| HDAC1 | Exome |
| HERC2 | CNV |
| IMPDH2 | Exome |
| IQGAP3 | Exome |
| IQSEC2 | Exome |
| KCNJ10 | Exome |
| LAMB2 | Exome |
| LRRC40 | Exome |
| NAGA | CNV |
| NCOR2 | Exome |
| NDNL2 | CNV |
| OTUD7A | CNV |
| PLXNB2 | CNV |
| PPP6R2 | CNV |
| SBF1 | CNV |
| SCO2 | CNV |
| SCUBE1 | CNV |
| SHANK3 | CNV |
| SLC2A1 | CNV |
| SLC39A13 | Exome |
| SLC6A1 | CNV |
| SMARCB1 | CNV |
| SMC1A | Exome |
| SPECC1L | CNV |
| STXBP1 | Exome |
| TCF20 | CNV |
| TCF4 | Exome |
| TRPM1 | CNV |
| TRRAP | Exome |
| TUBGCP6 | CNV |
| WDR45 | Exome |
| WNT7B | CNV |
| ZNF536 | Exome |
Genes from CNVs were selected if they had intragenic de novo mutations reported previously by large scale exome sequencing studies of ASD, intellectual disability, epilepsy, or developmental delays (see text for appropriate references). All 46 genes served as input to determine enriched terms using the DAVID functional annotation tool as well as to generate the interaction network in Figure 1 except that in the latter case four RTT genes were also included (MECP2, CDKL5, and FOXG1).
Figure 1.
An interaction network of genes with likely pathogenic mutations contributing to RTT in our cases. Black circles are input genes and gray circles are genes highly related to the input genes chosen by the network-building algorithm to maximize connectivity. The network was generated by using an input list of 46 genes with likely pathogenic mutations listed in Table 2 as well as the 3 known RTT genes MECP2, CDKL5, and FOXG1. Of the 46 genes, 23 were found to interact amongst each other either directly or indirectly through at least one of three ways: physical interactions (orange lines), co-localization of protein products (light blue lines), and participating in the same step of a given pathway (light green lines). Asterisks indicate genes related to input genes that have been reported to either carry de novo mutations in at least one patient with other NDDs (TBL1XR1, MTMR2, AKR1C4) or whose expression has been reported to be significantly altered in a MECP2 mutant model system (DAB1, ITGA2, LAMA5), or both (GLIS2, LAMC3, SMARCE1). Network weighting was assigned based on query genes so as to maximize connectivity among input genes, and at most 20 related genes and 10 related attributes were allowed to be incorporated in the network.
Discussion
It is well known that MeCP2, the product of the primary RTT gene, has the capacity to alter chromatin structure. Notable lines of evidence include abnormal organization of heterochromatin during neural differentiation of a Mecp2-deficient mouse embryonic stem cell line, the inability of MeCP2 containing many RTT-causing missense mutations to cause heterochromatin to cluster, and the requirement of MeCP2 binding to chromatin to form loops that bring distal regions into close proximity for the proper transcription of specific genes.8 Because RTT is commonly considered to be part of ASD, it is not surprising that other studies have also uncovered mutations in chromatin regulators in ASD.28 Given that we observed a significant enrichment of mutations in genes coding for chromatin regulators despite our small sample size compared to those of ASD studies, it is possible that dysregulation of normal chromatin architecture plays a more important role in the etiology of RTT which is more severe than ASD. Nevertheless, our results demonstrate the presence of a shared biological function that is disrupted more often in both RTT and ASD and raises the possibility of further research into discovering overlapping treatment options for these two related, but yet distinct, disorders.
Dysregulation of neuronal excitation is one factor that leads to RTT as many patients have seizures. Interestingly, previous studies utilizing Mecp2 mutant mice have implicated both the glutamate and GABA signaling pathways. For instance, MeCP2-deficient hippocampal glutamatergic neurons exhibit a significant reduction in synaptic response whereas those that over-express MECP2 display a higher response.29 Additionally, ablating Mecp2 function in cortical excitatory neurons but not inhibitory forebrain neurons leads to spontaneous seizures in mice.30 The glutamate signaling pathway is also dysregulated in other neurodevelopmental disorders related to RTT such as ASD31 and intellectual disability.23 In light of this, our results reinforce the role of abnormal glutamatergic signaling in RTT and, given its importance in other disorders, warrant further research to explore the possibility of treatment options that modulate this neuronal pathway as is being done for ASD.31 Our exome and CNV data did not reveal likely pathogenic variants in many genes from the GABA signaling pathway potentially due to the small size of our patient cohort.
Because of the presence of some shared clinical features between RTT and other neurodevelopmental disorders such as ASD, Pitt-Hopkins Syndrome, and CdLS, it is not surprising that most of our cases had likely pathogenic variants in genes and CNVs that had previously been reported in patients with other disorders. However, what was surprising was that 10 out of the 19 cases (52.6%) carried two or more likely pathogenic mutations including a combination of intragenic variants and CNVs, suggesting the importance of increased mutation burden in causing disease. Even though 4 out of the 11 RTT probands that were part of complete trios who lacked mutations in the three known RTT genes carried at least two de novo mutations, the overall burden of such mutations was not significantly different from the reported rate in control trios. Interestingly, including just two additional trios who had de novo MECP2 mutations revealed that this rate was significantly higher. Thus, it is possible that there is an increased burden of de novo intragenic mutations in RTT in general and it will be interesting to assess these, as well as CNVs, in a larger cohort of patients particularly those that also harbor causal MECP2 mutations and carry out detailed genotype-phenotype correlations. Similar increases in mutation burden have been observed in other neurologic disorders such as Charcot-Marie-Tooth disease, a peripheral neuropathy in which a high burden of rare variants was shown to contribute towards variable expressivity by possibly destabilizing different pathways and protein networks which could in turn modulate the phenotype.32 This underscores the importance of using both exome sequencing and CNV analyses to identify a specific combination of likely causal variants that could help explain the variability of phenotypes in individual patients who otherwise meet the overall diagnostic criteria of particular disorder.
Our study shows that the genetic etiology of RTT cases without mutations in MECP2, CDKL5, and FOXG1 is heterogeneous as we did not find any recurrent pathogenic variants. While our cohort of RTT patients is the largest of its kind reported to date subjected to both exome sequencing and CNV analysis, recurrence will undoubtedly be observed as larger cohorts are analyzed. A particular focus on genes involved in chromatin remodeling and glutamate signaling in additional patients could help identify recurrence and/or novel RTT genes as these pathways were over-represented by the genes found mutated in our cohort. We also note the usefulness of many large scale exome sequencing studies of ASD, intellectual disability, epileptic encephalopathy, and unexplained development delay as these have revealed de novo mutations in many genes in single patients. Smaller scale studies with more phenotypic information can potentially bolster the evidence supporting the involvement of many of these genes in neurological disease as these smaller cohorts may also find single patients with deleterious de novo mutations in those same genes. The challenge will be to compare in detail the clinical phenotypes of the respective patients from disparate studies and cohorts, keeping in mind that any phenotypic differences may not necessarily exclude the gene in question as being causal because there could be additional variants elsewhere modifying the phenotype.
Supplementary Material
Acknowledgments
We thank all subjects and their families for participating in this study, and the local coordinators who helped facilitate sample collection and processing. This work was funded by grants from the U.S. National Institutes of Health Grants U54HG006542 and U54HD061222, a grant from the International Rett Syndrome Foundation, and the Cynthia and Anthony Petrello Scholar fund at the Jan and Dan Duncan Neurological Research Institute, Texas Children's Hospital. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Eunice Kennedy Shriver Child Health and Human Development Institute (NICHD).
Footnotes
Supplementary information is available at the Genetics in Medicine website.
References
- 1.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23(2):185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 2.Laurvick CL, de Klerk N, Bower C, et al. Rett syndrome in Australia: a review of the epidemiology. J Pediatr. 2006;148(3):347–352. doi: 10.1016/j.jpeds.2005.10.037. [DOI] [PubMed] [Google Scholar]
- 3.Neul JL, Kaufmann WE, Glaze DG, et al. Rett syndrome: revised diagnostic criteria and nomenclature. Ann Neurol. 2010 Dec;68(6):944–950. doi: 10.1002/ana.22124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neul JL, Lane JB, Lee HS, et al. Developmental delay in Rett syndrome: data from the natural history study. J Neurodev Disord. 2014;6(1):20. doi: 10.1186/1866-1955-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Percy AK, Lane JB, Childers J, et al. Rett syndrome: North American database. J Child Neurol. 2007 Dec;22(12):1338–1341. doi: 10.1177/0883073807308715. [DOI] [PubMed] [Google Scholar]
- 6.Ariani F, Hayek G, Rondinella D, et al. FOXG1 is responsible for the congenital variant of Rett syndrome. Am J Hum Genet. 2008 Jul;83(1):89–93. doi: 10.1016/j.ajhg.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bahi-Buisson N, Nectoux J, Rosas-Vargas H, et al. Key clinical features to identify girls with CDKL5 mutations. Brain. 2008 Oct;131(Pt 10):2647–2661. doi: 10.1093/brain/awn197. [DOI] [PubMed] [Google Scholar]
- 8.Lombardi LM, Baker SA, Zoghbi HY. MECP2 disorders: from the clinic to mice and back. J Clin Invest. 2015 Aug 3;125(8):2914–2923. doi: 10.1172/JCI78167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guerrini R, Parrini E. Epilepsy in Rett syndrome, and CDKL5- and FOXG1-gene-related encephalopathies. Epilepsia. 2012 Dec;53(12):2067–2078. doi: 10.1111/j.1528-1167.2012.03656.x. [DOI] [PubMed] [Google Scholar]
- 10.Lambert L, Bienvenu T, Allou L, et al. MEF2C mutations are a rare cause of Rett or severe Rett-like encephalopathies. Clin Genet. 2012 Nov;82(5):499–501. doi: 10.1111/j.1399-0004.2012.01861.x. [DOI] [PubMed] [Google Scholar]
- 11.Ohba C, Nabatame S, Iijima Y, et al. De novo WDR45 mutation in a patient showing clinically Rett syndrome with childhood iron deposition in brain. J Hum Genet. 2014 May;59(5):292–295. doi: 10.1038/jhg.2014.18. [DOI] [PubMed] [Google Scholar]
- 12.Romaniello R, Saettini F, Panzeri E, Arrigoni F, Bassi MT, Borgatti R. A de-novo STXBP1 gene mutation in a patient showing the Rett syndrome phenotype. Neuroreport. 2015 Mar 25;26(5):254–257. doi: 10.1097/WNR.0000000000000337. [DOI] [PubMed] [Google Scholar]
- 13.Olson HE, Tambunan D, LaCoursiere C, et al. Mutations in epilepsy and intellectual disability genes in patients with features of Rett syndrome. Am J Med Genet A. 2015 Apr 25; doi: 10.1002/ajmg.a.37132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilissen C, Hehir-Kwa JY, Thung DT, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014 Jul 17;511(7509):344–347. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- 15.Lupski JR, Gonzaga-Jauregui C, Yang Y, et al. Exome sequencing resolves apparent incidental findings and reveals further complexity of SH3TC2 variant alleles causing Charcot-Marie-Tooth neuropathy. Genome Med. 2013;5(6):57. doi: 10.1186/gm461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Epi KC, Epilepsy Phenome/Genome P. Allen AS, et al. De novo mutations in epileptic encephalopathies. Nature. 2013 Sep 12;501(7466):217–221. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Euro E-RESCEae-RESuab, Epilepsy Phenome/Genome P, Epi KC, Euro E-RESC, Epilepsy Phenome/Genome P, Epi KC. De Novo Mutations in Synaptic Transmission Genes Including DNM1 Cause Epileptic Encephalopathies. Am J Hum Genet. 2014 Oct 2;95(4):360–370. doi: 10.1016/j.ajhg.2014.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iossifov I, O'Roak BJ, Sanders SJ, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014 Nov 13;515(7526):216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Roak BJ, Deriziotis P, Lee C, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011 Jun;43(6):585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Roak BJ, Vives L, Girirajan S, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012 May 10;485(7397):246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Roak BJ, Stessman HA, Boyle EA, et al. Recurrent de novo mutations implicate novel genes underlying simplex autism risk. Nat Commun. 2014;5:5595. doi: 10.1038/ncomms6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang YH, Yuen RK, Jin X, et al. Detection of clinically relevant genetic variants in autism spectrum disorder by whole-genome sequencing. Am J Hum Genet. 2013 Aug 8;93(2):249–263. doi: 10.1016/j.ajhg.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamdan FF, Srour M, Capo-Chichi JM, et al. De novo mutations in moderate or severe intellectual disability. PLoS Genet. 2014 Oct;10(10):e1004772. doi: 10.1371/journal.pgen.1004772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright CF, Fitzgerald TW, Jones WD, et al. Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet. 2014 Dec 16; doi: 10.1016/S0140-6736(14)61705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deciphering Developmental Disorders. Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015 Mar 12;519(7542):223–228. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saunders CJ, Minassian BE, Chow EW, Zhao W, Vincent JB. Novel exon 1 mutations in MECP2 implicate isoform MeCP2_e1 in classical Rett syndrome. Am J Med Genet A. 2009 May;149A(5):1019–1023. doi: 10.1002/ajmg.a.32776. [DOI] [PubMed] [Google Scholar]
- 27.Rauch A, Wieczorek D, Graf E, et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012 Nov 10;380(9854):1674–1682. doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- 28.Lasalle JM. Autism genes keep turning up chromatin. OA Autism. 2013 Jun 19;1(2):14. doi: 10.13172/2052-7810-1-2-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chao HT, Zoghbi HY, Rosenmund C. MeCP2 controls excitatory synaptic strength by regulating glutamatergic synapse number. Neuron. 2007 Oct 4;56(1):58–65. doi: 10.1016/j.neuron.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang W, Peterson M, Beyer B, Frankel WN, Zhang ZW. Loss of MeCP2 from forebrain excitatory neurons leads to cortical hyperexcitation and seizures. J Neurosci. 2014 Feb 12;34(7):2754–2763. doi: 10.1523/JNEUROSCI.4900-12.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rojas DC. The role of glutamate and its receptors in autism and the use of glutamate receptor antagonists in treatment. J Neural Transm. 2014 Aug;121(8):891–905. doi: 10.1007/s00702-014-1216-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gonzaga-Jauregui C, Harel T, Gambin T, et al. Exome Sequence Analysis Suggests that Genetic Burden Contributes to Phenotypic Variability and Complex Neuropathy. Cell Rep. 2015 Aug 18;12(7):1169–1183. doi: 10.1016/j.celrep.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Foti SB, Chou A, Moll AD, Roskams AJ. HDAC inhibitors dysregulate neural stem cell activity in the postnatal mouse brain. Int J Dev Neurosci. 2013;31(6):434–447. doi: 10.1016/j.ijdevneu.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 34.Carvill GL, McMahon JM, Schneider A, et al. Mutations in the GABA Transporter SLC6A1 Cause Epilepsy with Myoclonic-Atonic Seizures. Am J Hum Genet. 2015;96(5):808–815. doi: 10.1016/j.ajhg.2015.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matejas V, Hinkes B, Alkandari F, et al. Mutations in the human laminin beta2 (LAMB2) gene and the associated phenotypic spectrum. Hum Mutat. 2010;31(9):992–1002. doi: 10.1002/humu.21304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarkozy A, Carta C, Moretti S, et al. Germline BRAF mutations in Noonan, LEOPARD, and cardiofaciocutaneous syndromes: molecular diversity and associated phenotypic spectrum. Hum Mutat. 2009;30(4):695–702. doi: 10.1002/humu.20955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kashevarova AA, Nazarenko LP, Schultz-Pedersen S, et al. Single gene microdeletions and microduplication of 3p26.3 in three unrelated families: CNTN6 as a new candidate gene for intellectual disability. Mol Cytogenet. 2014;7(1):97. doi: 10.1186/s13039-014-0097-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coppinger J, McDonald-McGinn D, Zackai E, et al. Identification of familial and de novo microduplications of 22q11.21-q11.23 distal to the 22q11.21 microdeletion syndrome region. Hum Mol Genet. 2009;18(8):1377–1383. doi: 10.1093/hmg/ddp042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Esplin ED, Li B, Slavotinek A, et al. Nine patients with Xp22.31 microduplication, cognitive deficits, seizures, and talipes anomalies. Am J Med Genet A. 2014;164A(8):2097–2103. doi: 10.1002/ajmg.a.36598. [DOI] [PubMed] [Google Scholar]
- 40.Srivastava S, Cohen J, Pevsner J, et al. A novel variant in GABRB2 associated with intellectual disability and epilepsy. Am J Med Genet A. 2014;164A(11):2914–2921. doi: 10.1002/ajmg.a.36714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanders SJ, Murtha MT, Gupta AR, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485(7397):237–41. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.