Summary
Many RNA binding proteins (RBPs) bind specific RNA sequence motifs, but only a small fraction (~15-40%) of RBP motif occurrences are occupied in vivo. To determine what contextual features discriminate between bound and unbound motifs, we performed an in vitro binding assay using 12,000 mouse RNA sequences with the RBPs MBNL1 and RBFOX2. Surprisingly, the strength of binding to motif occurrences in vitro was significantly correlated with in vivo binding, developmental regulation and evolutionary age of alternative splicing. Multiple lines of evidence indicate that the primary context effect that impacts binding in vitro and in vivo is RNA secondary structure. Large-scale combinatorial mutagenesis of unfavorable sequence contexts revealed a consistent pattern whereby mutations that increased motif accessibility improved protein binding and regulatory activity. Our results indicate widespread inhibition of motif binding by local RNA secondary structure and suggest that mutations that alter sequence context commonly impact RBP binding and regulation.
Graphical abstract
Introduction
RBPs regulate many steps in gene expression. Their influence is often directed to specific sites within the transcriptome through interaction with specific RNA sequence motifs. A particularly widespread form of RNA-based regulation is alternative splicing (AS). AS expands proteomic diversity through the expression of multiple transcript isoforms for a single gene. Splicing is carried out in a step-wise fashion by a large ribonucloeprotein complex, termed the spliceosome. This complex recognizes the 5′ and 3′ splice sites, a polypyrimidine tract and a branchpoint sequence. The decision to use or skip a splice site within the pre-mRNA is commonly influenced by short cis-acting sequence elements usually ~4-6 nt in length that bind trans-acting RBPs to stabilize or inhibit nearby spliceosome formation (Gerstberger et al., 2014; Glisovic et al., 2008).
Most RBPs require the presence of a particular sequence motif to efficiently bind RNA (Ray et al., 2013). However, the presence of a cognate motif is generally not sufficient for effective binding in vivo (Van Nostrand et al., 2016). Even for an RBP that binds RNA with high affinity and specificity, the presence of an optimal motif does not guarantee binding, either in vivo or in vitro (Hiller et al., 2006). Comprehensive analyses of binding have found that a majority of motifs present in expressed transcripts are not bound by their cognate RBP in vivo (Li et al., 2010) (and Fig. 1A below). This presents a central mystery of RBP function – why are most occurrences of high affinity RBP motifs not bound? What contextual features beyond primary motif sequence influence RBP binding?
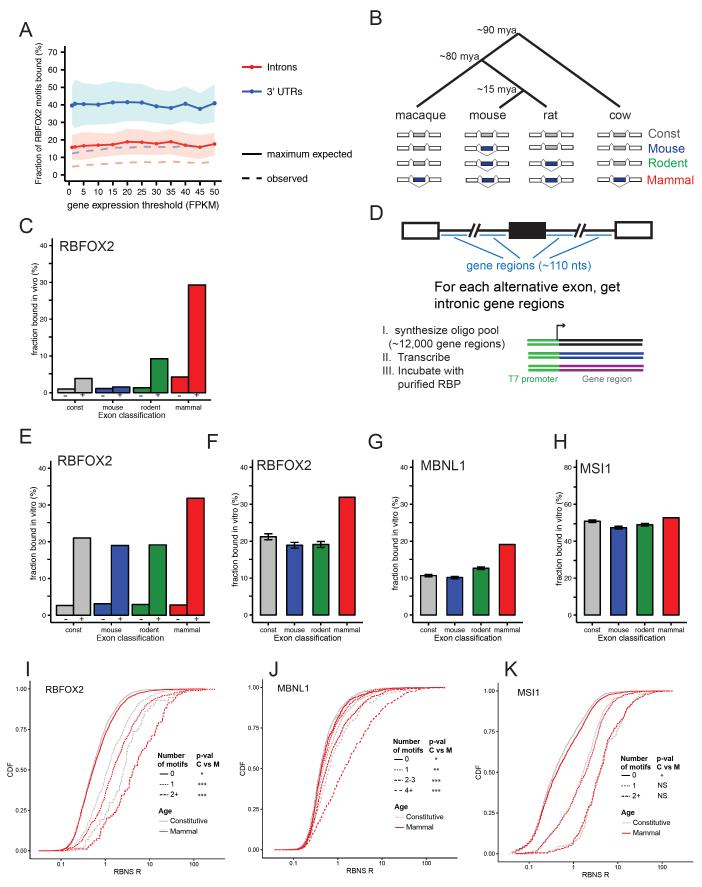
Figure 1. RBP/RNA interaction measured in vitro is influenced by both RNA motif content and contextual features.
A) The fraction of RBFOX2 RNA motifs (UGCAUG) identified as occupied in vivo by RBFOX2 using eCLIP, using genes binned by expression level (x-axis). The dotted line represents the observed fraction of RBFOX2 motifs that were bound in expressed introns and 3' UTRs. This fraction was then corrected to take into account the estimated sensitivity of the eCLIP assay to create a maximum expected fraction as described in supplementary Methods. Lines and shaded areas represent mean and standard deviation, respectively. B) Phylogenetic tree shows the relative evolutionary age of mouse, rat, cow, and macaque. Cassette exons (blue) were classified according to their evolutionary age of alternative splicing (Merkin et al., 2012) – see Supplemental Methods. C) Considering intronic regions downstream of cassette exons in each evolutionary age group, the percent of introns that show significant interaction with RBFOX2 in vivo in mESCs are shown. Tandem bars show introns without (−, left) or with (+, right) an RBFOX2 motif. D) Experimental design of natural sequence RBNS experiment. E) As in C, except that the y-axis signifies the fraction of oligos that were bound by RBFOX2 in vitro. F) Intronic regions corresponding to exons of younger evolutionary ages were subsampled to match the RBFOX2 motif counts in the mammalian-wide set. The average fraction of introns with RBFOX2 CLIP peaks in 50 independent subsamples is shown. Error bars show standard deviation. G) As in F, but shows MBNL1 binding of oligos subsampled to match the MBNL motif count in the mammalian-wide set. H) As in F, but showing MSI1 binding of oligos subsampled to match the MSI1 motif number in the mammalian-wide set. I-K) The cumulative distribution function of RBNS R scores is shown for intronic regions flanking constitutive (gray) and mammalian (red) exons for I) RBFOX2, J) MBNL1 and K) MSI1 RBNS experiments. Distinct line types correspond to different motif numbers for the indicated RBP. See also Figure S1.
An important goal of the splicing field is to develop a splicing “code” that predicts the splicing patterns of transcripts based on presence of splice site and RBP binding motifs and other features (Barash et al., 2010; Wang et al., 2004; Xiong et al., 2015). However, such approaches must typically assume that each occurrence of a motif is equivalent in its ability to bind its cognate RBP. Since the majority of RBP motif occurrences, as assayed by CLIP-seq and related methods, are unoccupied in vivo, the need to make this assumption introduces many false positives and may limit the accuracy of such approaches. Defining contextual features that allow discriminating predictions between bound and unbound motifs is therefore essential to the development of more accurate splicing codes.
A number of different features may impact whether or not an RBP occupies any specific occurrence of its cognate motif. Considering intronic binding, several features may be relevant, and examples of each are known (listed below). These features may include: 1) whether the local concentration or activity of the RBP (van der Houven van Oordt et al., 2000), or of its binding partners (Damianov et al., 2016), near the transcribed locus is sufficient; 2) whether or not access to the motif is blocked by local RNA structure (Kazan et al., 2010; Li et al., 2010); 3) whether the motif occurs in a sequence context that has other (non-structural) features favorable for binding (Agarwal et al., 2015); or 4) whether or not access to the motif is sterically blocked by other RBPs (HafezQorani et al., 2016; Liu et al., 2015; Zarnack et al., 2013). Other factors such as RNA modifications may influence binding in some cases, but pre-mRNA modifications are thought to be fairly rare (Carlile et al., 2014; Geula et al., 2015).
One clue to this puzzle of motif discrimination emerged from an analysis of MBNL1 binding to exons alternatively spliced for different lengths of evolutionary time. Independent of transcript expression level, identical motifs near exons with evolutionarily ancient alternative splicing have a several fold higher chance of being bound in vivo (Merkin et al., 2012). This trend indicates that evolution can sculpt a locus to impact RBP binding and suggests that we might be able to learn these features by studying properties of exons of different evolutionary ages. If certain intronic motifs are more bound because of where they are expressed in the nucleus or differences in the presence of competing RNA-bound factors, for example, then these differences would not be reproducible from interaction of an individual RBP with RNA in vitro. However, if evolutionarily ancient AS exons are more often bound because they occur in a favorable sequence or structural context, then we might hope to recapitulate this trend with recombinant RBP and RNA in vitro. RNA structure has been implicated in modulating protein-RNA interactions (Kazan et al., 2010; Li et al., 2010), but some recent studies (e.g., (Rouskin et al., 2014)) have suggested that there is much less structure in vivo than in vitro raising questions about whether structure is a major determinant of protein-RNA interaction in vivo.
To help understand the causes of differences binding between identical motifs in different transcripts, we performed an RNA Bind-n-Seq (RBNS) assay (Lambert et al., 2014) using naturally occurring intronic RNA sequences. Surprisingly, we observed a highly significant correlation with the extent of binding observed in vivo. These observations, supplemented by further experiments and analyses, provide strong support for the model that the extent to which an RNA motif is occluded by RNA secondary structure is a major determinant of RBP binding both in vitro and in vivo (Gosai et al., 2015). This model has implications for the impact of genetic variation on RNA-based regulation, the manipulation of RBP-interactions, and predictive models of RNA processing and regulation.
Results
Sequences flanking conserved alternative exons are more often bound by RBPs in vitro and in vivo
For many RBPs, a preferentially bound RNA motif is known. However, studies of in vivo RNA/RBP interactions have generally observed that RBPs bind only to a small subset of the occurrences of even their highest affinity RNA sequence motifs. As an example, we examined in vivo binding data for RBFOX2 (Jangi et al., 2014), which is well known to bind with high affinity (Kd ~10 nM) to the RNA motif UGCAUG (Auweter et al., 2006; Lambert et al., 2014). Analyzing crosslinking data generated using the eCLIP protocol, which yields much more comprehensive in vivo protein-RNA interaction data than other CLIP protocols (Van Nostrand et al., 2016), we estimated that no more than ~15% to 40% of UGCAUG motifs present in RNAs expressed in HepG2 cells (where RBFOX2 is highly expressed) are bound. This analysis corrects for the estimated sensitivity of the eCLIP assay; observed binding fractions were substantially lower in both introns and 3' UTRs (Fig. 1A, Supp. Methods). This small proportion indicates that presence of even a high affinity motif in an expressed RNA is not sufficient for in vivo binding, consistent with previous studies using earlier generations of RIP and CLIP protocols (Licatalosi et al., 2008; Sugimoto et al., 2012).
These observations raise the puzzle of why most motif occurrences are not bound in vivo, while others are. We sought to resolve this puzzle using a variety of biochemical, computational and evolutionary approaches. Exons that have undergone alternative splicing for tens of millions of years often have conserved patterns of tissue-specific regulation, and extensive sequence conservation in flanking introns (Merkin et al., 2012), suggesting the presence of selection related to splicing regulation. We classified exons based on their patterns of constitutive or alternative splicing across nine tissues in four mammalian species (Fig. 1B), as previously (Merkin et al., 2012). We then examined interactions of flanking intronic sequences with the splicing regulator RBFOX2 using a high-quality iCLIP dataset from mouse embryonic stem cells (mESCs) (Jangi et al., 2014). Strikingly, RBFOX motifs flanking exons alternatively spliced in all four mammals (“mammalian-wide AS exons”) were several times more likely to be bound in vivo by RBFOX2 than identical motifs located adjacent to constitutive exons (Fig. 1C, Fig. S1A). Compared to constitutive exons, rodent-specific AS exons were also more likely to be bound by RBFOX2 in vivo, though to a lesser extent than mammalian-wide AS exons, suggesting that sequence context that promotes binding by RBFOX proteins evolves gradually over many millions of years. We have also observed greatly increased binding by MBNL1 of Mbnl motifs flanking mammalian-wide AS exons (Merkin et al., 2012). Together, these observations support that evolution of favorable binding context for RBPs may commonly occur as exons acquire and maintain regulated splicing.
The specific binding of RNA by an RBP in vivo may be influenced by factors related to the cellular environment such as where in the nucleus the RNA is expressed or presence/absence of competing RNA-bound factors which will not occur when RBP and RNA are isolated in vitro, or by features intrinsic to the RNA such as local RNA structure (Li et al., 2014). To test the hypothesis that intrinsic features play a prominent role in determining RBP binding, we employed a high-throughput biochemical approach. We used the RNA Bind-n-Seq (RBNS) in vitro binding method (Lambert et al., 2014) to assess the sequence and structural specificity of RBP interactions with 110 nt natural sequences flanking ~3000 constitutive and alternative exons of varying evolutionary ages (Fig. 1D). This design reflected that introns are thought to be the major binding locations of the MBNL and RBFOX family splicing factors studied, and the 110 nt size was the largest that could be practically synthesized at this scale (Supp. Methods). Using oligonucleotide synthesis, we placed these sequences downstream of T7 promoter sequences to enable in vitro transcription. We then incubated this pool of approximately 12,000 sequences with recombinant MBNL1, RBFOX2, or Musashi-1 (MSI1) protein. Intronic sites are not expected to have evolved efficient binding to MSI1 because this protein is primarily cytoplasmic in mammals (Katz et al., 2014), so this factor serves as a type of negative control.
In standard RBNS with pools of random oligos, the ratio of the abundance of an RNA motif in the bound pool to its abundance in the input pool, termed the “raw enrichment” or R value, is used to assess binding affinity, but the sequence pool is too diverse to calculate R values for individual oligos. However, in this “natural sequence” (ns) RBNS experiment the reduced diversity enabled measurement of an R value for each individual oligonucleotide (top and bottom 100 sequences listed in Table S1). RNA oligos were then classified based on whether they were bound in vitro by MBNL1, RBFOX2 or MSI1 in RBNS experiments. Oligos were classified as “bound” or “unbound” based on their RBNS R value (Supp. Methods). R values of individual oligos derived using different RBP concentrations were highly concordant (Fig. S1D-G).
Oligonucleotides containing canonical motifs for each of these RBPs were several fold more likely to be bound in vitro than those lacking such motifs (Fig. 1E, S1B,C), mirroring CLIP-seq data for RBFOX2 (Fig. 1C) and MBNL1 (Merkin et al., 2012). However, only a moderate fraction (~20% to at most 50%) of oligos containing canonical motifs for these factors were bound in vitro, indicating the presence of repressive transcript features even in these simplified conditions. Strikingly, oligos from introns flanking conserved mammalian AS exons were ~1.5-fold more likely to bind RBFOX2 in vitro (Fig. 1F) and ~2-fold more likely to bind MBNL1 in vitro (Fig. 1G) than those from constitutive introns, when comparing sets of intronic oligonucleotides with identical motif content. These results suggest the surprising conclusion that some of the evolved features that facilitate RBP interaction in vivo also function in this in vitro system. No such difference in binding was observed for the cytoplasmic RBP MSI1 (Fig. 1H), suggesting that these introns have evolved preferential binding to specific RBPs – presumably those involved in regulation of their splicing (see below) – rather than a generic affinity to all RBPs.
For all three RBPs, the presence of additional motifs was associated with increased in vitro binding of these intronic sequences (Fig. 1I-K, motif counts distinguished by line type). Compared to sequences flanking constitutive exons, sets of regions flanking mammalian-wide AS exons were more frequently bound by RBFOX2 at motif count 1, and for MBNL1 at each specific motif count of one or greater, but such a trend was not observed for MSI1 (Fig. 1I-K, red versus gray). These observations provide more in-depth confirmation that the sequence context of the motifs in ancient AS introns promote their interaction with specific splicing regulatory RBPs.
Intronic motifs bound in vitro are more likely to exhibit developmental regulation
To test the idea that binding in the nsRBNS assay is associated with in vivo regulation of splicing, we assessed splicing changes in differentiation and developmental settings in which RBFOX and MBNL proteins are induced. We focused primarily on a neuronal induction time-course of mESCs into glutamatergic ESC-derived neurons (ESNs) (Hubbard et al., 2013), which had strong up-regulation of both MBNL and RBFOX family genes, and secondarily on a heart development study spanning embryonic day 17 to postnatal day 17 (Giudice et al., 2014), in which MBNLs are upregulated. Both MBNL1 and RBFOX2 are known to exhibit substantial changes in protein abundance during neuronal differentiation and heart development, impacting alternative splicing (Kalsotra et al., 2008; Underwood et al., 2005). We observed that the total levels of Mbnl1/Mbnl2 transcripts increase during both neuronal induction and heart development (Fig. 2A, Fig. S2A, top), while levels of RBFOX family genes increase strongly and then stabilize during neuronal induction (Fig. 2A, Fig. S2A, bottom).
Figure 2. Intronic sequences flanking exons regulated during in vivo differentiation are more often bound in vitro.

A) Expression levels of Mbnl and Rbfox transcripts during the induction of mouse embryonic stem cells into glutamatergic neurons. B) Diagram of Monotonicity z-score (MZ score) definition during neuronal differentiation. Exons with negative MZ scores show consistent, monotonic decrease in inclusion while those with positive MZ scores show consistent, monotonic increase. C) Cumulative distributions of absolute monotonicity scores of mammalian, rodent and mouse skipped exons during in vitro neuronal differentiation show mammalian-wide AS exons are more likely to be developmentally regulated. D) The distribution of monotonicity scores during neuronal induction for exons flanked by in vitro bound and unbound intronic RNAs. Because MBNL sites are most active in the intronic sequence immediately upstream of skipped exons, we considered those regions for this analysis. Introns flanking exons that are regulated in vivo were more likely to be bound by MBNL1 in vitro. See also Figure S2.
To assess the developmental regulation of particular exons in these time courses we calculated monotonicity Z (MZ) scores (Wang et al., 2015) for all expressed exons in these time courses. The MZ score is a permutation-based measure of the direction of change in percent spliced in (PSI) of an exon, expressed in standard deviation units. A positive MZ score (e.g., MZ > 2) indicates monotonic increase in PSI over a time course, while negative MZ indicates monotonic decrease in exon inclusion (Figs. 2B, S2B). The distributions of absolute MZ score (capturing both monotonic increase and decrease in exon inclusion) across both ESN induction and heart development were significantly higher for mammalian-wide alternative exons compared to mouse-specific or rodent-specific alternative exons (Figs. 2C, S2C). MBNL sites are most active immediately upstream of skipped exons (Wang et al., 2012). Comparing the absolute MZ score distributions of these intronic regions bound in vitro by MBNL1 or RBFOX2 versus those not bound, we observed a substantially larger number of exons with large |MZ| values among those bound in vitro (Figs. 2D, S2D). These observations indicate that binding in the nsRBNS assay is predictive of in vivo regulation.
Sequence context effects on motif binding in vitro predict in vivo binding and regulation
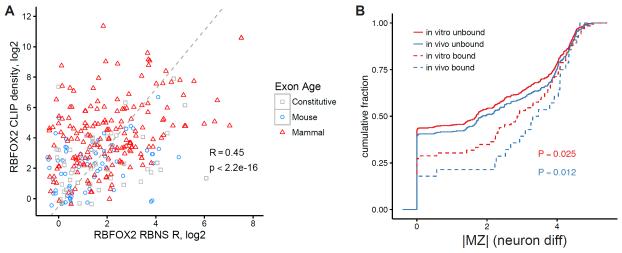
CLIP-seq analysis of RBP binding in vivo to individual intronic regions is reasonably reproducible, with correlations of read densities in intronic regions between technical replicates of MBNL1 and PTB CLIP experiments ranging from 0.31 to 0.61 using the set of intronic regions analyzed here (Licatalosi et al., 2012; Poulos et al., 2013) (Fig. S3). To ask whether the context effects measured by nsRBNS analysis of individual regions relate to in vivo binding, we compared the R values of individual regions to the density of RBFOX2 iCLIP-seq reads in mESCs in these regions, considering only regions containing exactly one RBFOX motif (Fig. 3A). R values from our in vitro binding assay were significantly correlated (RSpearman = 0.45, p < 2.2e-16) with iCLIP read density (controlled for expression level). Because the regions analyzed all contained a single RBFOX motif, differences in binding between regions reflect exclusively contextual effects. The magnitude of correlation observed between in vitro and in vivo binding, almost as high as the correlation between CLIP replicates, suggests that contextual effects on motif binding that occur in vivo are similar to those that occur in vitro.
Figure 3. RBP/RNA interactions measured in vivo are recapitulated in vitro.
A) Scatter plot of the RBNS R scores for RBFOX2 motif-containing introns versus RBFOX2 CLIP-seq density in mESCs. Colors correspond to evolutionary age. B) The regulation of alternative exons during neuronal differentiation was measured and plotted as MZ scores. Introns flanking these exons were classified as bound and unbound both in vitro and in vivo. The cumulative distribution of MZ scores for each class of intronic sequence is shown. P-values are from Wilcoxon rank-sum tests between bound and unbound sequences. See also Figure S3.
We then explored the extent to which measured differences in in vitro and in vivo binding to different motif-containing introns predicted regulation during ESN differentiation. Oligos with binding detected by CLIP-seq were substantially more likely to have large |MZ| values, consistent with more frequent regulation (P = 0.012, Fig. 3B). Furthermore, in vitro binding detected by nsRBNS was also associated with greater |MZ|, to a comparable but slightly lesser extent (P = 0.025, Fig. 3B). These results suggest that a substantial portion of relevant regulatory interactions are captured in vitro.
Introns flanking ancient alternative exons are more conserved and have less RNA secondary structure
To explore the sequence properties that may influence RBP binding, we measured sequence conservation in the assessed oligos using Phastcons scores for genomic coordinates of the oligo (Siepel and Haussler, 2005) in windows of 40 bp surrounding RBP motifs. Introns flanking mammalian-wide AS exons were consistently more conserved than those flanking mouse-specific AS exons (Fig. S4A). Oligos that were bound by MBNL1 and/or RBFOX2 in vitro were also more conserved than unbound sequences (Fig. S4A). This signature was not observed for the cytoplasmic RBP MSI1. These observations provide additional evidence that some intronic regions adjacent to RBP motifs may experience selection to promote effective binding of regulatory RBPs.
We hypothesized that local RNA secondary structure is a primary contextual feature that determines whether an RBP does or does not bind to an occurrence of its cognate RNA motif. Here we used two approaches to assess RNA secondary structure: 1) the thermodynamic-based software RNAstructure (Reuter and Mathews, 2010); and 2) structure analysis using Selective 2′ hydroxyl acylation analyzed by primer extension followed by high-throughput sequencing (SHAPE-seq), which identifies flexible regions of RNA that can be used to constrain possible RNA foldings (Aviran et al., 2011; Mortimer et al., 2012; Reuter and Mathews, 2010). To determine the structural characteristics of the assessed oligos, we performed SHAPE-seq analysis on 588 of our oligos in parallel. From these data, and calculated basepairing probabilities (Ppair) via SHAPE-assisted RNAstructure analysis (Aviran et al., 2011; Mortimer et al., 2012; Reuter and Mathews, 2010). In general, introns flanking mammalian-wide AS exons tended to have more single-stranded character than those flanking mouse-specific alternative exons or constitutive exons (Fig. S4B).
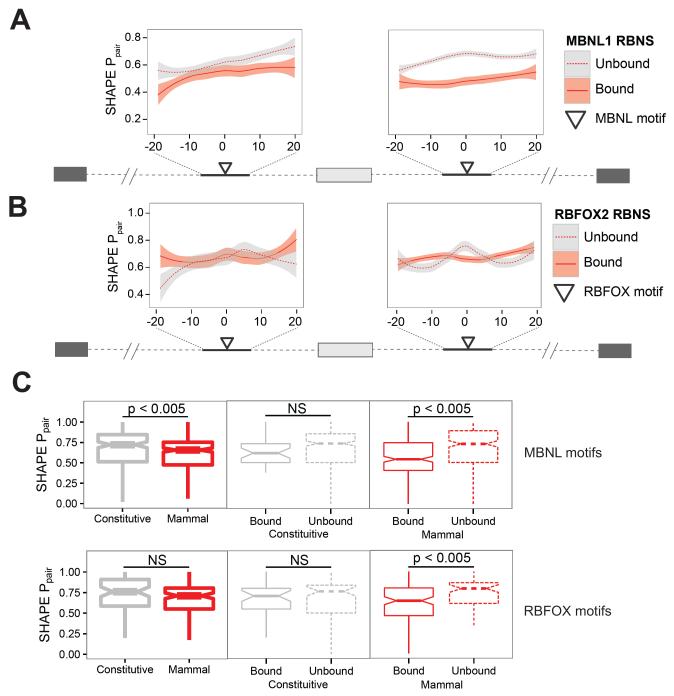
We next considered the RNA structure of sequences immediately flanking RBP motifs. We observed that the regions surrounding MBNL1 motifs had significantly lower SHAPE Ppair values in oligos that were bound by MBNL1 in vitro compared to those that were not bound, particularly in downstream intronic regions (Fig. 4A). We also observed similar, though subtler effects in regions surrounding RBFOX2 motifs (Fig. 4B). In the case of MBNL1, the preferred region of single-strandedness extended at least 20 bases upstream and downstream of the motif, while for RBFOX2 this region was much smaller. Furthermore, we observed that bases in the RBP motifs themselves had significantly lower average SHAPE Ppair values in oligos that were bound in vitro than those that were not bound. This trend held only for the mammalian-wide AS exons, and only with respect to MBNL1 and RBFOX2 (Fig. 4C), and not for MSI1 (not shown), and was robust to changes in the R value cutoff used to define bound oligos (Fig. S4C). MBNL1 and RBFOX2 motifs in mammalian-wide AS oligos that were bound in vitro were also significantly more conserved than those that were not bound (Fig. S4D,E), but no such effect was observed for MSI1 motifs (Fig. S4F). Considering each nucleotide of the MBNL motifs UGCU and UGCC individually, the pyrimidines flanking the central GC dinucleotide had significantly lower SHAPE Ppair values in bound oligos than in unbound oligos (Fig. S4G), while no bias or preference related to pairing of the GC dinucleotides was observed (Lambert et al., 2014). Again, this effect was only seen in introns flanking mammalian-wide AS exons. Perhaps mammalian-wide AS exons contain additional motif-contextual features besides favorable secondary structure, like flanking nucleotide composition, that promote efficient binding.
Figure 4. Reduced basepair probabilities in and around motifs bound by RBPs in vitro.
A, B) Basepair probabilities in intronic RNA oligos surrounding mammalian alternative exons are shown for sequences immediately surrounding MBNL1 (A) and RBFOX2 (B) motifs. RNA oligos have been classified based on whether or not they were bound by the RBP in vitro. Lines represent LOESS fits of the basepair probabilities while shaded areas represent the 95% confidence interval of the fit. C) Basepair probabilities were calculated as in B and then averaged across the nucleotides of each motif occurrence. Motifs were separated based on whether the RNA sequence they are contained within was bound in vitro by the indicated RBP (line type) and the evolutionary age and regulation of the neighboring alternative exon (color). See also Figure S4.
Combinatorial mutagenesis identifies contextual features that promote MBNL1 binding
Our findings indicate that the sequence context in which an RNA motif occurs plays a decisive role in determining whether it will be bound by an RBP, and suggest that repressive RNA secondary structures frequently impede RBP interaction. To test this idea, we selected two RNA sequences from introns within the Myo1b and Dtna genes that harbor a single high affinity MBNL1 motif UGCU, but have weak binding to MBNL1 in vitro (R value ≤ 1.5) or in vivo, despite high expression in cells where MBNL1 CLIP-seq was performed (Fig. S5A). Consistent with the trends observed in Figure 4, RNAfold analysis indicated that bases in the UGCU motifs in these RNAs are predicted to pair with other bases nearby (Fig. 5A, S5H), and that these RNAs display moderate to high overall secondary structure compared to other intronic oligos (Fig. S5B). To ask what specific aspects of the sequence context of these motifs repress MBNL1 binding, we used a combinatorial mutagenesis approach. We synthesized DNA based on the Myo1b and Dtna introns, but incorporating a ~6% substitution rate (2% of each non-native base) at every position except the UGCU motifs, which remained unmutated. The resulting oligonucleotide pool was in vitro transcribed, purified and subjected to RNA sequencing. This approach yielded complex RNA repertoires with the desired substitution rate and with little apparent synthesis bias (Fig. S5C-G).
Figure 5. Distinct patterns of RNA mutations lead to increased RBP/RNA interaction through secondary structure rearrangements.
A) Frequencies of mutations at each position across the randomly mutated Myo1b RNA oligo. The frequencies of mutations in the input, no protein control pulldown, and MBNL1 pulldown RNA pools were calculated for each position in the oligo. The frequencies in each pulldown were compared to the frequencies in the input RNA pool. Adapter and motif regions (shaded) were held constant and thus have mutation frequencies of zero. The MFE structure of the wildtype Myo1b oligo is shown above the sequence. Positions at which the MBNL1 pulldown frequency was greater than the 99th percentile of no protein control frequencies are marked with an asterisk. B) One million randomly selected Myo1b RNA oligos from each of the input, no protein pulldown, and MBNL1 pulldown libraries were computationally folded to assess secondary structure. The difference in mean basepair probabilities at each position between pulldown oligos and input oligos are shown. For each position, the statistical significance (Wilcoxon rank sum) of the basepair probability difference between the pulldown oligos (MBNL1 and no protein) and input oligos was calculated. Positions at which the P value for the MBNL1 pulldown was greater than the 99th percentile of P values for the no protein control across all positions are marked with an asterisk. C) The relative frequencies (MBNL1 pulldown / input) of co-occurring pairs of mutations in the Myo1b oligo are shown in the lower triangle. The most enriched pairs of mutations are shown here. In the upper triangle, the relative frequencies (No protein control pulldown / input) of these co-occurring mutation pairs are shown. D) As in B, one million randomly selected Myo1b RNA oligos from the input and MBNL1 pulldown libraries were computationally folded to assess secondary structure. For each (i, j) pair of positions, the mean probability across those million sequences that bases i and j were paired was calculated. The difference in mean probability between input and MBNL1 pulldown libraries is shown. E) The centroid predicted structure for the wildtype Myo1b oligo (center). Bases that have predicted basepair probabilities of greater than 0.9 or less than 0.1 are outlined in red and blue, respectively. The position of the MBNL motif is indicated with green dots. Introducing the most enriched trio of mutations from the MBNL1 pulldown (blue dots, right) results in a different predicted structure with the MBNL motif less paired. Conversely, introducing the most depleted trio of mutations from the MBNL1 pulldown (orange dots, left), resulted in a predicted structure with the MBNL1 motif more paired. See also Figure S5.
To examine the ability of these RNAs to interact with MBNL1 we carried out RBNS with recombinant MBNL1 or a “no protein” control. In this experiment, termed combinatorial mutagenesis RBNS (“cmRBNS”), a diverse pool of subtle RNA sequence variants of the native sequence compete for protein binding in the same reaction. We obtained 10-20 million reads for each “input”, “no protein” and “MBNL1 pulldown” sample. In these pools, 70-99% of the reads were unique (Fig. S5F-G), indicating a highly diverse sequence pool. For each ancestral sequence, the mutational profiles of the “no protein” and “input” pools were highly similar (Fig. 5A, S5H, red lines), as desired. In contrast, the mutational profiles of the MBNL1 pulldown pools showed much greater variability in substitution frequency, with particular positions enriched or depleted for mutations relative to the input and no protein pools (Fig. 5A, S5H, blue lines). Further analysis revealed that specific nucleotides were enriched in the MBNL1 pulldown pools at these positions (Fig. S5I,J). Furthermore, specific pairs of mutations were enriched to a far greater extent in the MBNL1 pulldown than in the no protein control (Figs. 5C, S5K).
cmRBNS-selected oligos display motif-centric changes in secondary structure
In principle, mutations may improve binding by creating a new MBNL motif, by altering structure around the motif, or by creating some other (non-structural) favorable sequence context. We observed a modest preference for substitutions that created new MBNL1 motifs (discussed below). However, when we excluded reads containing motif-creating substitutions, we observed many biases that reflected alterations in the RNA structural context of MBNL1 motifs present in the native sequence and chose to focus most of our efforts on this class of substitutions. We used in silico RNA folding (Lorenz et al., 2011) to computationally assess the structure of one million randomly selected sequences from the “input”, “no protein”, and “MBNL1 pulldown” pools for each oligo. For each base in each sequence, we calculated the probability that it was paired to another base in the sequence, and the mean basepairing probability (mean Ppair) for that position across all sequences in the sample. We observed a number of positions with significantly lower or higher mean Ppair values in the Myo1b oligo “MBNL1 pulldown” sample relative to the “no protein” control (Fig. 5B). The greatest decreases in mean Ppair occurred at a cluster of six positions (numbers 55-60) centered on the location of the UGCU motif (at 56-59), and at a cluster of five downstream positions from 102-106 that are predicted to form a stem with positions 59-63, overlapping the UGCU motif. On the other hand, basepairing at positions 51-54 just upstream of the UGCU motif and at a few other positions was increased in the pulldown library.
The data were deep enough that they enabled analysis of changes in the mean pairing probabilities of specific pairs of positions. This analysis (Fig. 5D, S5L) confirmed the inferences above, showing significantly reduced probability of pairing between the positions 56-63 (overlapping the UGCU) and partners at 102-108, potentially enhancing accessibility of the UGCU motif. It also revealed significantly increased probability of pairing between positions 49-54 and 59-64, a stem interaction that would place the first three bases of the UGCU motif in a hairpin loop. We also examined particular trios of substitutions that were enriched or depleted in MBNL1-interacting oligos. The most enriched combination of 3 substitutions, [ A42G, G45U, A62C ], results in a predicted secondary structure in which the stem overlapping the UGCU in the native structure is disrupted, and the UGCU motif is instead present in a hairpin loop formed by pairing of positions 49-53 with positions 61-64 (Fig. 5E, right). The most depleted trios of substitutions, [ G47A, G88C, U99C ], also has a substantial effect on predicted RNA structure, pairing all four bases of the UGCU motif as part of an extended stem structure that includes pairing of bases 55-60 with bases 81-86, likely sequestering the UGCU more effectively than in the native structure (Fig. 5E, left).
A similar analysis of the Dtna oligo revealed that the primary MBNL motif occurs in the middle of a strong 16 bp stem, which is essentially impervious to unpairing at a 6% mutation rate (Fig. S5M). Previously, we have observed MBNL1 affinity for a set of GC-containing tetranucleotide motifs, including UGCA, using a random pool RBNS approach (Table S2). Analysis of the pairwise basepair probabilities of the Dtna oligo showed preference for reduced secondary structure overlapping a (“secondary”) UGCA motif but not the “primary” UGCU motif (Fig. S6A-D).
Although reduced basepairing of specific MBNL-associated motifs was observed in both the Myo1b and Dtna oligos, the overall extent of baseparing of the bound pool remained very similar to that of the input and control RNA pools (Fig. S6E-F), suggesting that localized changes to motif structural context rather than wholesale loss of structure may be sufficient to drive binding. Taken together, the cmRBNS experiments support our hypothesis that presence of repressive RNA structures commonly prevents recognition of RBP motifs.
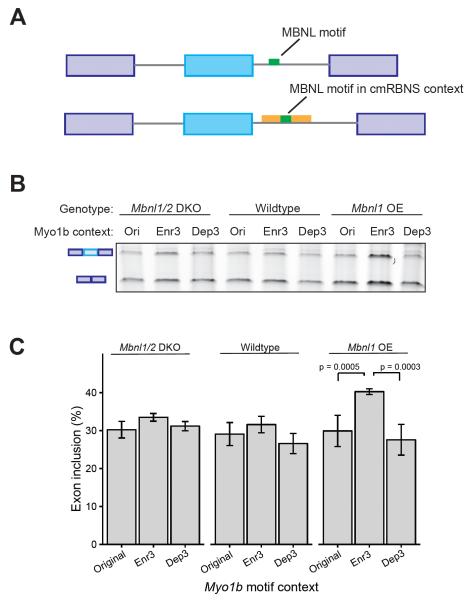
We then sought to test the splicing regulatory activity of MBNL motifs in different contexts highlighted by the cmRBNS data. We used a splicing reporter minigene derived from the mouse Vldlr gene in which MBNL1 enhances inclusion of a skipped exon via binding to a single motif (Du et al., 2010) (Fig. S6G-H). We replaced the existing intronic Mbnl motif with an Mbnl motif in the context of the Myo1b oligo used in the cmRBNS experiment. The Mbnl motif was placed in its original Myo1b context (WT) as well as in contexts reflecting the most depleted (Dep3) and most enriched (Enr3) co-occurring triplets of mutations from the cmRBNS experiment (Fig. 5E, 6A). Compared to the wildtype Myo1b context, the Enr3 context promoted exon inclusion when MBNL was overexpressed, relative to the original (p = 0.0005) or Dep3 (p = 0.0003, t-test) context (Fig. 6B, C). These results are consistent with an increased ability of MBNL proteins to bind the motif in the Enr3 context relative to the original or Dep3 context. This effect occurred only the context of MBNL1 overexpression (Fig. 6B, C), directly linking MBNL1 to increased exon inclusion in the Enr3 context.
Figure 6. cmRBNS-enriched mutations enhance regulatory activity.
A) Schematic design of splicing reporter construct based on mouse Vldlr alternative exon 16. The intronic Mbnl motif was replaced by an Mbnl motif in specific contexts derived from cmRBNS analysis of the Myo1b oligo (Fig. 5). The original Myo1b context was used along with the Enr3 and Dep3 contexts representing the most significantly enriched and depleted triplets of mutations. B) Semi-quantitative RT-PCR analysis of exon inclusion using the original, Enr3 and Dep3 contexts. MBNL levels were modulated by using Mbnl1/Mbnl2 double knockout MEFs, wildtype MEFs, and Mbnl1 overexpression MEFs. C) sqRT-PCR analysis of 3 biological replicate experiments like those in B, with 3 replicates of each PCR assay. Error bars represent standard deviation. P-values by t-test. See also Figure S6.
Some mutations may enhance binding by creating new RBP motifs rather than altering structure of existing motifs (Fig. 7A). To explore the relative impacts of these two phenomena on MBNL1 binding, we compared the enrichment in the bound pool relative to the input pool for the Myo1b oligo of: 1) sequences with Ppair < 0.5 for the primary UGCU motif; 2) sequences containing one or more new MBNL1 motif(s); or 3) both of these features. This analysis revealed that having an unpaired original motif and having a new motif were enriched to a similar extent over input (1.16 versus 1.19, respectively), suggesting that on average these features impact binding to a similar extent (Fig. 7B). Sequences having both of these features were enriched to a greater extent, while sequences with neither feature were depleted in the pulldown fraction. As a control, no enrichment was observed for sequences with Ppair < 0.5 at nonmotif positions.
Figure 7. The appearance of new motifs and increased accessibility of the original motif are approximately equally enriched in the MBNL1 pulldown.
A) Model for acquisition of RBP binding. In principle, RBP binding may arise in two ways in mutated oligos. Mutations could result in the appearance of new MBNL motifs (upper arrow), which may fall in unpaired (top) or paired (below) regions. Mutations may also result in changes to the RNA secondary structure around the original MBNL motif, promoting binding by increasing the accessibility of a pre-existing motif. B) Relative enrichments of the indicated classes of RNA sequences in the MBNL1 pulldown compared to input. Secondary structure change around nonmotif sequences was not enriched (left). However, reduced basepairing (Ppair < 0.5) of the original MBNL motif (yellow) and appearance of new MBNL motifs (blue) were enriched singly and the co-occurrence of both phenomena was strongly enriched (green). C) Appearance of new unpaired motifs (blue) was more strongly enriched than appearance of new motifs that were basepaired (gray). RNA sequences containing both an unpaired original motif and an unpaired new motif (purple) were the most enriched class. Error bars correspond to 95% confidence intervals.
New motifs may not all be created equal. Given the importance of structure observed above, one might expect that new motifs that arise in positions where they are basepaired would be less often bound than those that are unpaired. Consistent with this expectation, we observed substantially higher enrichment for sequences with new motifs with Ppair < 0.5 than for those with higher Ppair (Fig. 7C). This was true both for the subset of sequences where the original motif was paired (1.52 versus 1.10) and for those where the original motif was unpaired (2.11 versus 1.56). Taken together, these data suggest that context effects will strongly influence whether or not a newly arising motif is bound and therefore potentially functional.
Discussion
Most protein-RNA interactions are driven by affinity of an RBP for a specific motif. However, recent crosslinking data indicate that most occurrences of RBP motifs in transcripts expressed with the corresponding RBP are not bound in vivo. Here, using high-throughput binding of recombinant protein to pools of natural RNA sequences of 110 nt, we observed that the context effects on binding to different transcripts containing identical RBP motifs observed in vivo are similar to those that occur in vitro. These effects of motif context on binding observed in vitro also predicted the extent of splicing regulation in cellular differentiation and development, and in a reporter assay. These observations, and the cmRBNS data, argue that local RNA secondary structures (often occurring within ~100 nt) play a decisive role in limiting access of RBPs to a large subset of motifs that would otherwise be occupied, restricting their regulatory activity (Fig. 6).
This finding has important implications and raises further questions. One implication is that efforts to simulate or predict splicing or other aspects of mRNA metabolism regulated by RBPs may be drastically improved if they consider the effects of RNA structure on RBP interaction with cognate motifs. It is also worth reconsidering how genetic variation influences the activity of RBPs, e.g., splicing quantitative trait loci (sQTLs), or mutations that affect splicing, decay or other aspects of mRNA metabolism (Monlong et al., 2014; Pai et al., 2012). Our results indicate that, in addition to variants that create or disrupt RBP motifs, one must be equally cognizant of variants that alter RBP access by changing local RNA structure in the vicinity of motifs in order to capture the full spectrum of variants that change RNA-based regulation (Fig. 7). For example, we observed significant increased regulation by MBNL1 as a result of just 3 contextual mutations near a single Mbnl motif (Fig. 6). Cells may commonly use differences in RNA structure mediated by changes in temperature, concentrations of metal ions or polycationic proteins like spermines, or the activities of RNA chaperones or helicases as a regulatory strategy to alter the composition of mRNPs by changing their secondary structures (Jankowsky, 2001).
Our findings have implications for the evolution of RNA-based gene regulation. A question arises as to whether the “default” state of an arbitrary motif inserted into a natural RNA sequence such as a 3' UTR or intron is accessible or inaccessible to RBPs. Evolutionarily, which usually comes first: presence of the motif or presence of a permissive structural environment? If RNA structure is widespread, as appears to be the case, there is likely a large pool of ‘latent’ regulatory capacity in transcripts in the form of RBP motifs made inaccessible by presence of repressive structures. This perspective emphasizes the potential for use of RNA structure-altering antisense or small molecule approaches for perturbing RBP binding and regulation (Guan and Disney, 2012). Of course, structure is not the whole story; other contextual effects surely also contribute to RBP binding. Understanding the full spectrum of features that impact whether a given motif occurrence is or is not bound by protein remains an important goal for future study.
Data Deposition
All sequencing data has been deposited at SRA under accession number SRP080275.
Experimental Procedures
Natural sequence RNA Bind-N-seq
Recombinant GST-/SBP-tagged MBNL1, RBFOX2, and MSI1 were purified as previously described (Jangi et al., 2014; Lambert et al., 2014). Oligonucleotides containing a T7 promoter and the genomic regions described above were in vitro transcribed using T7 RNA polymerase to produce a pool of approximately 12,000 110 nt RNA sequences. A total of 2957, 2947, and 3517 sequences were synthesized from regions surrounding constitutive, mouse-specific, and mammalian-wide alternative exons, respectively (as well as additional sequences not analyzed here). RNA Bind-nseq was then performed as previously described (Lambert et al., 2014; Merkin et al., 2012). Briefly, 10 nmol of RNA pool was incubated with purified RBP at 3 different protein concentrations (25, 125, 625 nM) for one hour at 22° C. Then RBP was then purified using streptavidin magnetic beads (Invitrogen) and washed. RNA was eluted from the purified protein by incubation at 70° C for 10 minutes in elution buffer (10mM Tris pH 7.0, 1mM EDTA, 1% SDS), reverse transcribed, subjected to 8-10 cycles of PCR, and prepared for Illumina sequencing. For further analyses, MBNL1 motifs were defined as UGCU, CGCU, and GCUU. RBFOX2 motifs were defined as UGCAU and GCAUG. MSI1 motifs were defined as UAGUU, UUAGU, AUUAG, UAGUA, UUUAG, and AUAGU.
Developmental time course analysis
Mouse glutamatergic neuron induction and heart development time-courses (PRJNA185305 and SRP029464 respectively) were mapped to MM10 using STAR v2.4.2a (Dobin et al., 2013). Gene expression TPMs were called using RSEM v1.2.20 (Li and Dewey, 2011) and percent spliced in values of alternative exons were calculated using MISO (Katz et al., 2010). MZ-scores were calculated as described in (Wang et al., 2015).
CLIP / RBNS comparisons
RBFOX2 iCLIP data from mouse ES cells (Jangi et al., 2014) was used to compare CLIP read densities in defined intronic regions to in vitro RBFOX2 binding data to the defined intronic regions used in the RBNS assay. Only regions that were in expressed genes (> 5 rpkm) and contained at least one RBFOX2 motif were used. CLIP read densities were normalized by gene expression values.
For comparisons of technical CLIP replicates, we used high quality CLIP-seq data for MBNL3 and PTBP2 (Licatalosi et al., 2012; Poulos et al., 2013). Reads were mapped to the mouse genome. Read densities in defined intronic regions and defined 3' UTR regions were then calculated and compared between replicates.
Intronic region secondary structure analysis
1m7 SHAPE reagent was provided through kind gifts of Donald Rio, Manny Ares, and Julius Lucks. In order to produce sufficient coverage across the RNA oligos tested, a subset of 588 of the original ~12,000 oligos were subjected to SHAPE-seq analysis. SHAPE-seq was performed as previously reported (Lucks et al., 2011; Mortimer et al., 2012) with the slight modification of reversing the 5' and 3' adapter sequences to facilitate efficient cluster identification during sequencing. Libraries were sequenced on an Illumina HiSeq sequencer and the resulting reads were trimmed using cutadapt (Martin, 2011) to remove 3' adapter sequences and then further trimmed to 25 nt. SHAPE reactivity profiles were calculated using the spats software package (Aviran et al., 2011). These reactivity values, along with thermodynamic predictions of RNA secondary structure based on primary sequence were used to calculate basepair probabilities with the RNAstructure software package (Reuter and Mathews, 2010).
Combinatorial mutagenesis RBNS experiment
DNA oligonucleotides corresponding to exon-proximal intronic regions in the Dtna and Myo1b genes were chemically synthesized (CustomArray). At each position a 6% error rate was introduced during synthesis by spiking in equal amounts of the incorrect nucleotides (e.g. If position 5 is A, then the synthesis of position 5 would proceed with 94% A, 2% G, 2% T and 2% C). The TGCT MBNL motif sequence within each sequence was synthesized with a 0% error rate. These oligos were flanked by priming sites for Illumina sequencing and the T7 promoter sequence. To produce RNA, a T7 oligonucleotide was annealed to the T7 region within the mutagenized oligo (65 degrees for 5 min and then allowed to cool to room temperature) and the resulting partially double-stranded oligos were transcribed in vitro with Ampliscribe T7 RNA transcription kit (Epibio) according to manufacturer’s specifications. RNA products were gel-purified on a 6% TBE-Urea polyacrylamide gel.
MBNL1 protein was purified as described above. The RNA-bind-n-seq assay was performed as described above with the following slight modifications. Briefly, 100 nM of MBNL protein on streptavidin or beads alone were incubated with 1.2 uM of RNA in a 250 uL reaction at 25 C for 30 min. Beads were washed 3 times with binding buffer and MBNL-RNA complexes were eluted two times by incubating with 50 uL of 4mM biotin (in PBS) for 15 minutes at 25 C. Eluted RNA was collected by phenol-chloroform extraction. 50% of RNA recovered was used for RT-PCR and 25% of the resulting cDNA was used for 18 cycles of PCR. 50 picomoles of input RNA were subjected to the same RT-PCR conditions, with 14 cycles of PCR. The resulting PCR products were sequenced on a NextSeq instrument (135 bp pair-end). The analysis of the cmRBNS experiment is described further in Supplemental Methods.
Supplementary Material
Acknowledgements
We thank Donald Rio, Manny Ares, and Julius Lucks for providing the 1m7 RNA modifying molecule used in SHAPE-Seq and Manny Ares for the MBNL-sensitive splicing reporter. MBNL1/2 double knockout MEFs were provided by Maurice Swanson and Eric Wang. We thank Ana Fiszbein, Sean McGeary and M. Swanson for helpful comments on the manuscript.
Footnotes
Author Contributions
The project was conceived by JMT, NJL, JJM and CBB. Experiments were performed by JMT, NJL, DD and CB. Bioinformatic analyses were performed by JMT, NJL, PHS, MSA and JJM. The manuscript was written by JMT, NJL, PHS, DD and CBB.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agarwal V, Bell GW, Nam J-W, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auweter SD, Fasan R, Reymond L, Underwood JG, Black DL, Pitsch S, Allain FH-T. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. Embo J. 2006;25:163–173. doi: 10.1038/sj.emboj.7600918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviran S, Trapnell C, Lucks JB, Mortimer SA, Luo S, Schroth GP, Doudna JA, Arkin AP, Pachter L. Modeling and automation of sequencing-based characterization of RNA structure. Proc Natl Acad Sci USA. 2011;108:11069–11074. doi: 10.1073/pnas.1106541108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ. Deciphering the splicing code. Nature. 2010;465:53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515:143–146. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damianov A, Ying Y, Lin C-H, Lee J-A, Tran D, Vashisht AA, Bahrami-Samani E, Xing Y, Martin KC, Wohlschlegel JA, et al. Rbfox Proteins Regulate Splicing as Part of a Large Multiprotein Complex LASR. Cell. 2016;165:606–619. doi: 10.1016/j.cell.2016.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Cline MS, Osborne RJ, Tuttle DL, Clark TA, Donohue JP, Hall MP, Shiue L, Swanson MS, Thornton CA, et al. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat Struct Mol Biol. 2010;17:187–193. doi: 10.1038/nsmb.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nat Rev Genet. 2014;15:829–845. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- Giudice J, Xia Z, Wang ET, Scavuzzo MA, Ward AJ, Kalsotra A, Wang W, Wehrens XHT, Burge CB, Li W, et al. Alternative splicing regulates vesicular trafficking genes in cardiomyocytes during postnatal heart development. Nat Commun. 2014;5:3603. doi: 10.1038/ncomms4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosai SJ, Foley SW, Wang D, Silverman IM, Selamoglu N, Nelson ADL, Beilstein MA, Daldal F, Deal RB, Gregory BD. Global Analysis of the RNA-Protein Interaction and RNA Secondary Structure Landscapes of the Arabidopsis Nucleus. Mol Cell. 2015;57:376–388. doi: 10.1016/j.molcel.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan L, Disney MD. Recent advances in developing small molecules targeting RNA. ACS Chem. Biol. 2012;7:73–86. doi: 10.1021/cb200447r. [DOI] [PubMed] [Google Scholar]
- HafezQorani S, Lafzi A, de Bruin RG, van Zonneveld AJ, van der Veer EP, Son YA, Kazan H. Modeling the combined effect of RNA-binding proteins and microRNAs in post-transcriptional regulation. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller M, Pudimat R, Busch A, Backofen R. Using RNA secondary structures to guide sequence motif finding towards single-stranded regions. Nucleic Acids Res. 2006;34:e117. doi: 10.1093/nar/gkl544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard KS, Gut IM, Lyman ME, McNutt PM. Longitudinal RNA sequencing of the deep transcriptome during neurogenesis of cortical glutamatergic neurons from murine ESCs. F1000Research. 2013;2:35. doi: 10.12688/f1000research.2-35.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangi M, Boutz PL, Paul P, Sharp PA. Rbfox2 controls autoregulation in RNA-binding protein networks. Genes Dev. 2014;28:637–651. doi: 10.1101/gad.235770.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E. Active Disruption of an RNA-Protein Interaction by a DExH/D RNA Helicase. Science. 2001;291:121–125. doi: 10.1126/science.291.5501.121. [DOI] [PubMed] [Google Scholar]
- Kalsotra A, Xiao X, Ward AJ, Castle JC, Johnson JM, Burge CB, Cooper TA. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc Natl Acad Sci USA. 2008;105:20333–20338. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y, Li F, Lambert NJ, Sokol ES, Tam W-L, Cheng AW, Airoldi EM, Lengner CJ, Gupta PB, Yu Z, et al. Musashi proteins are post-transcriptional regulators of the epithelial-luminal cell state. eLife. 2014;3:23. doi: 10.7554/eLife.03915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan H, Ray D, Chan ET, Hughes TR, Morris Q. RNAcontext: a new method for learning the sequence and structure binding preferences of RNA-binding proteins. PLoS Comp Biol. 2010;6:e1000832. doi: 10.1371/journal.pcbi.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert N, Robertson A, Jangi M, McGeary S, Sharp PA, Burge CB. RNA Bind-n-Seq: Quantitative Assessment of the Sequence and Structural Binding Specificity of RNA Binding Proteins. Mol Cell. 2014;54:887–900. doi: 10.1016/j.molcel.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Kazan H, Lipshitz HD, Morris QD. Finding the target sites of RNA-binding proteins. WIREs RNA. 2014;5:111–130. doi: 10.1002/wrna.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Quon G, Lipshitz HD, Morris Q. Predicting in vivo binding sites of RNA-binding proteins using mRNA secondary structure. Rna. 2010;16:1096–1107. doi: 10.1261/rna.2017210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Yano M, Fak JJ, Mele A, Grabinski SE, Zhang C, Darnell RB. Ptbp2 represses adult-specific splicing to regulate the generation of neuronal precursors in the embryonic brain. Genes Dev. 2012;26:1626–1642. doi: 10.1101/gad.191338.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Ouyang M, Rao JN, Zou T, Xiao L, Chung HK, Wu J, Donahue JM, Gorospe M, Wang J-Y. Competition between RNA-binding proteins CELF1 and HuR modulates MYC translation and intestinal epithelium renewal. Mol Biol Cell. 2015;26:1797–1810. doi: 10.1091/mbc.E14-11-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R, Bernhart SH, Höner Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucks JB, Mortimer SA, Trapnell C, Luo S, Aviran S, Schroth GP, Pachter L, Doudna JA, Arkin AP. Multiplexed RNA structure characterization with selective 2'-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq) Proc Natl Acad Sci USA. 2011;108:11063–11068. doi: 10.1073/pnas.1106501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.Journal. 2011;17:10–12. [Google Scholar]
- Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science. 2012;338:1593–1599. doi: 10.1126/science.1228186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monlong J, Calvo M, Ferreira PG, Guigó R. Identification of genetic variants associated with alternative splicing using sQTLseekeR. Nat Commun. 2014;5:4698. doi: 10.1038/ncomms5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer SA, Trapnell C, Aviran S, Pachter L, Lucks JB. SHAPE-Seq: High-Throughput RNA Structure Analysis. Curr Protoc Chem Biol. 2012;4:275–297. doi: 10.1002/9780470559277.ch120019. [DOI] [PubMed] [Google Scholar]
- Pai AA, Cain CE, Mizrahi-Man O, De Leon S, Lewellen N, Veyrieras J-B, Degner JF, Gaffney DJ, Pickrell JK, Stephens M, et al. The contribution of RNA decay quantitative trait loci to inter-individual variation in steady-state gene expression levels. PLoS Genet. 2012;8:e1003000. doi: 10.1371/journal.pgen.1003000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulos MG, Batra R, Li M, Yuan Y, Zhang C, Darnell RB, Swanson MS. Progressive impairment of muscle regeneration in muscleblind-like 3 isoform knockout mice. Human Molecular Genetics. 2013;22:3547–3558. doi: 10.1093/hmg/ddt209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499:172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter JS, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505:701–705. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Haussler D. Phylogenetic hidden Markov models. Statistical Methods in Molecular Evolution. 2005:326–350. [Google Scholar]
- Sugimoto Y, König J, Hussain S, Zupan B, Curk T, Frye M, Ule J. Analysis of CLIP and iCLIP methods for nucleotide-resolution studies of protein-RNA interactions. Genome Biol. 2012;13:R67. doi: 10.1186/gb-2012-13-8-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Houven van Oordt W, Diaz-Meco MT, Lozano J, Krainer AR, Moscat J, Caceres JF. The MKK(3/6)-p38-signaling cascade alters the subcellular distribution of hnRNP A1 and modulates alternative splicing regulation. Journal of Cell Biology. 2000;149:307–316. doi: 10.1083/jcb.149.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, Blue SM, Nguyen TB, Surka C, Elkins K, et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP) Nat Methods. 2016 doi: 10.1038/nmeth.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Cody NAL, Jog S, Biancolella M, Wang TT, Treacy DJ, Luo S, Schroth GP, Housman DE, Reddy S, et al. Transcriptome-wide Regulation of Pre-mRNA Splicing and mRNA Localization by Muscleblind Proteins. Cell. 2012;150:710–724. doi: 10.1016/j.cell.2012.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Ward AJ, Cherone JM, Giudice J, Wang TT, Treacy DJ, Lambert NJ, Freese P, Saxena T, Cooper TA, et al. Antagonistic regulation of mRNA expression and splicing by CELF and MBNL proteins. Genome Res. 2015;25:858–871. doi: 10.1101/gr.184390.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, Burge CB. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Xiong HY, Alipanahi B, Lee LJ, Bretschneider H, Merico D, Yuen RKC, Hua Y, Gueroussov S, Najafabadi HS, Hughes TR, et al. RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science. 2015;347:1254806. doi: 10.1126/science.1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnack K, König J, Tajnik M, Martincorena I, Eustermann S, Stévant I, Reyes A, Anders S, Luscombe NM, Ule J. Direct Competition between hnRNP C and U2AF65 Protects the Transcriptome from the Exonization of Alu Elements. Cell. 2013;152:453–466. doi: 10.1016/j.cell.2012.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.