Abstract
Amino acid deprivation is sensed by the eIF2α kinase GCN2. Under conditions of essential amino acid limitation, GCN2 phosphorylates eIF2α, inhibiting the formation of new ternary complex and hence mRNA translation initiation. While decreasing global mRNA translation, eIF2α phosphorylation also increases the translation of the integrated stress response (ISR) transcription factor ATF4, which increases the expression of many stress response genes that contain a C/EBP-ATF response element (CARE), including ATF4, 4EBP1, ASNS and CHOP. Using wild-type as well as GCN2 knockout and unphosphorylatable eIF2α mutant MEFs, we characterized a novel GCN2/eIF2α phosphorylation-independent, but ATF4-dependent, pathway that upregulates expression of CARE-containing genes in MEFs lacking GCN2 or phosphorylatable eIF2α when these cells are exposed to methionine-deficient, and to a lesser extent arginine- or histidine-deficient medium. Thus, we demonstrate a GCN2/eIF2α phosphorylation-independent pathway that converges with the GCN2/eIF2α kinase dependent pathway at the level of ATF4, and similarly results in upregulation of CARE-containing genes. We hypothesize that the essential role of methionine-charged initiator tRNA in forming ternary complex is responsible for the robust ability of methionine deficiency to induce ATF4 and the ISR even in the absence of GCN2 or eIF2 kinase activity.
Keywords: Methionine, GCN2, eIF2α, ATF4, CARE
INTRODUCTION
The regulation of gene expression in response to changes in the nutritional environment, including the availability of amino acids, has been well documented in recent decades (reviewed in Baird and Wek 2012). The major signaling pathway for the regulation of mammalian gene expression in response to amino acid availability involves the alpha subunit of eukaryotic initiation factor 2 (eIF2α) kinase pathway. GCN2 (eIF2α kinase 4) is one of four mammalian eIF2α kinases (Donnelly et al. 2013), and GCN2 is specifically activated by “uncharged” tRNA accumulation during amino acid starvation (Wek et al. 1989; Wek et al. 1995; Zhu et al. 1996; Dong et al. 2000; Sood et al. 2000; Qiu et al. 2001; Zhang et al. 2002; Narasimhan et al. 2004; Dever and Hinnebusch 2005; Zaborske et al. 2009). The lack of any essential amino acid can result in GCN2 activation and large changes in gene expression (Jousse et al. 2004; Kilberg et al. 2009). Other types of cellular stresses activate the other three eIF2α kinases, also leading to large changes in gene expression (Dang Do et al. 2009; Walter and Ron 2011; Donnelly et al. 2013). Because different types of stress converge at the level of phosphorylated eIF2α, the eIF2α kinase-mediated response is often called the integrated stress response (Baird and Wek 2012).
Eukaryotic initiation factor 2 (eIF2), which is a trimer of α, β, and γ subunits, is required for the initiation of mRNA translation (Stolboushkina and Garber 2011; Hinnebusch 2014). It mediates the binding of the charged initiator tRNA (Met-tRNAiMet) to the ribosome in a GTP-dependent manner by formation of the ternary complex (eIF2-GTP-Met-tRNAiMet). Upon base pairing of the AUG start codon with Met-tRNAiMet, the GTP is hydrolyzed to GDP and the eIF2-GDP is released (Hinnebusch 2014). For the eIF2 that is released to be used for a new round of translation initiation, the bound GDP must be exchanged for GTP by action of the guanine nucleotide exchange factor, eukaryotic initiation factor 2B (eIF2B) (Webb and Proud 1997; Unbehaun et al. 2004; Van Der Kelen et al. 2009).
The activation of any one of the four mammalian eIF2α kinases results in the phosphorylation of eIF2α (Wek et al. 2006). When eIF2α is phosphorylated, it binds tightly to eIF2B, which inhibits the guanine nucleotide exchange factor and prevents the formation of new eIF2-GTP (Pavitt et al. 1998; Krishnamoorthy and Pavitt 2001). Because eIF2-GTP is required for formation of new a ternary complex, the phosphorylation of eIF2α impacts global mRNA translation initiation (Anthony et al. 2001; Krishnamoorthy and Pavitt 2001; Wek et al. 2006; Baird and Wek 2012). Paradoxically, the lack of sufficient ternary complex, and hence 43S preinitiation complex, also drives an increase in the translation of activating transcription factor 4 (ATF4), a transcription factor that plays a key role in initiating the integrated stress response (Kilberg et al. 2009; Baird and Wek 2012). ATF4 mRNA contains upstream inhibitory open reading frames, and under normal conditions, translation is initiated at a short upstream open reading frame that encodes a small peptide. After translation of this short region of mRNA, the 60S subunit falls off and the 40S continues to slide down the mRNA searching for a new start site. Under nutrient-sufficient conditions with abundant eIF2-GTP, reformation of the ternary complex with Met-tRNAiMet occurs readily, and this allows reinitiation at a subsequent start codon that is still upstream of the ATF4 start codon but has an open reading frame that overlaps (but is out-of-frame with) the open reading frame for ATF4. This results in translation of a functionally inert protein and bypass of the ATF4 start site. However, under stress conditions in which the reformation of eIF2-GTP is inhibited and the abundance of ternary complex is low, the 40S subunit is more likely to bypass the start site for the dummy protein before the 43S preinitiation complex is reformed, facilitating engagement with the downstream ATF4 start site and translation of ATF4 (Vattem and Wek 2004; Wek et al. 2006; Kilberg et al. 2009; Baird and Wek 2012).
When translationally induced, the transcription factor ATF4 enters the nucleus where it forms heterodimers with CCAAT-enhancer-binding proteins (C/EBP) (Kilberg et al. 2009). The heterodimers recognize the consensus sequence of “TGATGXAAX” in gene promoter regions. This sequence is termed the CARE (C/EBP-ATF response element), and C/EBP-ATF4 binding results in the upregulation of genes essential for the protection of cells in a variety of stress conditions (Kilberg et al. 2009; Shan et al. 2009; Dey et al. 2010). These genes include the cationic amino acid transporter (CAT-1), system A neutral amino acid transporter 2 (SNAT2), asparagine synthetase (ASNS), eukaryotic initiator 4E binding protein 1 (4EBP1) and genes that function in the stimulation of autophagy, as well as ATF4 itself (Siu et al. 2002; Yamaguchi et al. 2008; Kilberg et al. 2009; Dey et al. 2010; B’Chir et al. 2013; B’chir et al. 2014).
Despite the common role of eIF2α kinases and ATF4 in the integrated stress response, gene expression in response to various cellular stresses is not identical, suggesting the existence of additional signaling pathways that respond to the various types of stress (Sikalidis et al. 2011). Even with similar stresses, such as the lack of one essential amino acid versus another essential amino acid, there can be a high degree of non-overlap of differentially regulated genes (Shan et al. 2010) or a high degree of variation in the extent of up- or down-regulation (Palii et al. 2009). Deval et al. (2009) investigated the relative roles of GCN2 and mTORC1 in the differential expression of genes in response to leucine starvation and concluded that the GCN2 pathway accounted for the major, but not the total, response of gene transcription to leucine deprivation. Wanders et al. (2016) recently demonstrated that GCN2-null mice were still able to respond to sulfur amino acid deprivation by eIF2α phosphorylation. Inclusion of cysteine in the diet prevented this response, and they attributed it to glutathione deficiency and oxidative stress that activated the PKR-like endoplasmic reticulum kinase (PERK, or eIF2α kinase 3).
To further explore the regulation of gene expression in response to amino acid deprivation, we deprived cells of different single amino acids and then looked at the effect of individual amino acid deprivation on ATF4-induced changes in mRNA levels of CARE-containing genes. We demonstrate the presence of a novel GCN2/eIF2α phosphorylation-independent signaling pathway that upregulates expression of CARE-containing genes and that is particularly sensitive to methionine deprivation.
EXPERIMENTAL PROCEDURES
Cell Culture
Wild-type (GCN2+/+) and GCN2(−/−) MEFs immortalized with SV40 Large T antigen were provided by Dr. David Ron (Cambridge University, Cambridge, UK). Knock-in loss-of-function eIF2α mutant (eIF2α (A/A)) and isogenic wild-type (eIF2α (S/S)) cells were supplied by Dr. Randal Kaufman (Sanford-Burnham Medical Research Institute, La Jolla, CA, USA). Wild-type, GCN2(−/−) and eIF2α(A/A) MEFs were cultured in high glucose Dulbecco’s modified Eagle’s medium (DMEM), pH 7.4, containing 1 mM pyruvate and 4 mM glutamine and supplemented with 1× non-essential amino acid mix (Gibco/Invitrogen), 100 units/mL penicillin and 100 μg/mL streptomycin (pen/strep; Gibco/Invitrogen) and 10% (v/v) fetal bovine serum (FBS, Hyclone). Cells were maintained at 37°C in an atmosphere of 95% air and 5% CO2. Experimental control medium was the same as normal growth medium with the following changes: medium was prepared using custom DMEM that lacked sulfur amino acids, histidine, leucine and arginine (prepared by Gibco/Invitrogen); medium was supplemented by us with 0.4 mM L-cysteine, 0.2 mM L-methionine, 0.2 mM L-histidine, 0.8 mM L-leucine and 0.4 mM L-arginine; and 10% dialyzed FBS (Hyclone) was used in place of standard FBS. Histidine-deficient (–His), methionine-deficient (–Met), leucine-deficient (–Leu) and arginine-deficient (–Arg) media were prepared similarly except the respective amino acids were not added back to the deficient DMEM. Cells were grown to 60-70% confluence before exposure to treatment media in order to ensure logarithmic growth throughout the experiment. Cells were collected for qRT-PCR and western blotting as described below.
RNA Isolation and qRT-PCR
Cells were exposed to amino acid-deficient treatment medium for 12 h. Cells were collected in RNeasy lysis buffer (Qiagen) supplemented with 10% (v/v) β-mercaptoethanol. Cells were lysed by centrifugation through a QIAshredder (Qiagen), and RNA was isolated using an RNeasy isolation column (Qiagen) per the manufacturer’s instructions. For cDNA synthesis, 1 μg RNA was reverse transcribed using the High-Capacity Reverse Transcription kit (Applied Biosystems) per the manufacturer’s instructions. For qRT-PCR, cDNA was diluted in ultrapure H2O to a concentration of 4 ng/μL. Then 7.5 μL of the cDNA mixture was combined with 37.5 μL 2× Power SYBR green master mix (Applied Biosystems) and the desired amount of primers and H2O to make 75 μL of complete PCR mix. Final primer concentrations in the 75 μL complete pCR mix were as follows: 300 nM forward and reverse primers for 4EBP1, ATF4 and β-tubulin; 50 nM forward and 50 nM reverse primers for 4EBP2; 50 nM forward and 300 nM reverse primers for ASNS; and 300 nM forward and 50 nM reverse primers for CHOP. Forward and reverse primer sequences were as follows: 4EBP1 forward 5′-GAAGAGCCTCCCATGCAA-3′ reverse 5′-CCATCTCAAATTGTGACTCTTCA-3′; ATF4 forward 5′-ACAGGAAGCATGCAGTTGG-3′ reverse 5′-AGGTGCACACAGGCTGCT-3′; β-tubulin forward 5′-GCTGGACCGAATCTGTGTGT-3′ reverse 5′-GACCTGAGCGAACGGAGTC-3′; 4EBP2 forward 5′-ACAGGAAGCATGCAGTTGG-3′ reverse 5′-AGGTGCACACAGGCTGCT-3′; ASNS forward 5′-GGCTGTGTGTTCAGAAGCT-3′ reverse 5′-AAGGAAGGGCTCCACTTT-3′; CHOP forward 5′-CATACACCACCACACCTGAAAGCA-3′ reverse 5′-GGTGAAAGGCAGGGACTCAGCT-3′. A 20 μL aliquot of the complete qPCR mix was added to each well of a Roche 480 96-well PCR plate such that each well contained 8 ng of cDNA, with triplicate wells being run for each sample. qPCR was done in a Roche 480 Lightcycler with polymerase activation at 95°C for 10 min followed by denaturation at 95°C for 15s and annealing/elongation at 60°C for 60s. The denaturation and annealing/elongation steps were repeated for a total of 35 cycles. Fluorescence was measured at the end of each annealing/elongation cycle. Following qPCR, melting curves were acquired by a stepwise increase of the temperature from 60°C to 95°C to confirm that a single product had been amplified in the reaction. Fold differences of 4EBP1, 4EBP2, ATF4, ASNS and CHOP mRNA were determined using the ΔΔCp method. The β-tubulin mRNA level was used to normalize values within an experiment, and differences in mRNA levels were calculated as fold the average normalized value for wild-type cells cultured in complete medium.
Protein Isolation and Western Blotting
Cells were exposed to amino acid-deficient or control treatment medium for 24 h, with medium being replaced with fresh treatment medium at 12 h. Cells were collected in lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.5%, v/v, Nonidet NP-40) supplemented with Complete protease inhibitor and PhosSTOP phosphatase inhibitor “cocktails” (Roche). Homogenates were then centrifuged at 14,000 × g for 20 min, and the supernatants were collected. Protein concentrations of the sample supernatants were determined by the bicinchonic acid assay (BCA Protein Assay Kit, Thermo Scientific Pierce), and 30 μg protein per lane were resolved by SDS/PAGE using a 12% (w/v) polyacrylamide gel. Proteins were transferred to a 0.45-μm Immobilon PVDF membrane (Millipore Corp.), and the membrane was incubated for 1 h at room temperature with blocking buffer for near infrared fluorescent westerns (Rockland or Odyssey blocking buffer). The membrane was then incubated with antibodies to 4EBP1 (1:1000, Cell Signaling), ATF4 (1:1000, Cell Signaling) or β-tubulin (1:1000, Santa Cruz Biotech) at 4°C overnight. Membranes were washed for 30 min in TTBS (1× Tris-buffered saline, pH 7.6, + 0.1%,v/v, Tween 20) and then exposed to secondary antibodies conjugated to AlexaFluor 680 or IRDye 800 (1:20,000, Li-COR Biosciences) for 1 h at room temperature. Membranes were again washed for 30 min with TTBS and rinsed in PBS (pH 7.4), and the proteins were visualized and quantified using an Odyssey infrared imagining system (Li-COR Biosciences) and Odyssey software. The relative intensities of the bands of interest were normalized to β-tubulin.
ATF4 siRNA Knockdown
A siRNA oligonucleotide targeting murine ATF4 mRNA (Flexitube siRNA Mm_ATF4_1) and a nonsense negative control (AllStars Negative Control siRNA) were purchased from Qiagen. Cells were transfected using the reverse transcription method. Cells were grown in standard growth medium prepared without pen/strep until they were 80% confluent. The cells were then trypsinized and diluted in fresh growth medium without pen/strep so they would yield a confluence of 50-60% when plated. The siRNA and Lipofectamine-2000 (Invitrogen) were both diluted in optiMEM (Gibco/Invitrogen) and combined according to the manufacturer’s protocol. The cell solution was combined with the siRNA-Lipofectamine-optiMEM solution to produce a final siRNA concentration of 33 nM. The cell/transfection solution was mixed and plated as described by the manufacturer (Invitrogen). After 6 h the transfection medium was removed and replaced with control treatment medium. GCN2(−/−) and eIF2α(A/A) MEFs were allowed to recover in control medium for 12 h and 18 h, respectively, at which time they had returned to a logarithmic growth rate; transfected wild-type (control) MEFs were treated similarly. After recovery, cells were cultured in amino acid-deficient medium and harvested for qRT-PCR and western blot analysis as described above except that cells were exposed to amino acid deficient medium for 12 h instead of 24 h for protein abundance measurements in order to collect cells during logarithmic growth.
Statistical Analysis
All experiments were repeated at least three times with triplicate wells of cells within each experiment. Results were expressed as fold of the mean for the same cell type cultured in control medium. Data were log10 transformed and analyzed using ANOVA and Student’s t-test; differences were accepted at p ≤ 0.05.
RESULTS
The GCN2(−/−) and eIF2a(A/A) MEFs used in these experiments have been characterized previously (Harding et al. 2000; Scheuner et al. 2001). We confirmed the lack of eIF2a phosphorylation in response to amino acid deprivation in these cell lines in a preliminary experiment. Culture of cells in leucine-deficient medium for 3 to 10 h resulted in marked phosphorylation of eIF2 Ser51 in MEFs from GCN2(+/+) mice but not in MEFs from GCN2(−/−) mice. Similarly, culture of cells in leucine-deficient medium for 3 to 12 h resulted in marked phosphorylation of eIF2 Ser51 in eIF2 (S/S) cells but not in isogenic eIF2 (A/A) knockin cells. Furthermore, transfection of GCN2(−/−) cells with a GCN2 expression plasmid restored the ability of the GCN2(−/−) cells to increase phosphorylation of eIF2 in response to leucine deprivation. Thus, the cell lines used in the studies reported here exhibit the expected phenotype for cells lacking GCN2 or a phosphorylatable eIF2 .
As the GCN2/eIF2α kinase pathway signals via upregulation of both ATF4 translation and a subsequent increase in Atf4 gene transcription, we first explored the effect of amino acid deficiency on ATF4 mRNA levels. As expected, ATF4 mRNA levels were significantly increased in GCN2(+/+) MEFs when these cells were cultured in medium deficient in either histidine, leucine, arginine or methionine (Fig. 1a). Also, as expected, arginine and leucine deprivation did not result in an increase in ATF4 mRNA in GCN2(−/−) MEFs (Fig. 1b). However, though reduced compared to GCN2(+/+) cells, a significant increase in ATF4 mRNA was still observed in GCN2(−/−) MEFS exposed to histidine and methionine deficient medium compared to cells grown in sufficient medium. GCN2 knockout had the least effect on the response of MEFs to methionine deficient medium such that ATF4 mRNA levels in GCN2(−/−) cells cultured in methionine-deficient medium were still 3-fold those of cells cultured in sufficient medium.
Fig. 1.
Upregulation of ATF4 mRNA in GCN2(+/+), GCN2(−/−), eIF2α(S/S) and eIF2α(A/A) MEFs in response to essential amino acid deficiency. Cells were grown in sufficient medium (Suff) or in medium deficient in histidine (His-), methionine (Met-), leucine (Leu-) or Arginine (Arg-). ATF4 mRNA levels after 12 h of culture in deficient medium, expressed as fold the mean value for cells cultured in sufficient medium, are shown for a) GCN2(+/+), b) GCN2(−/−), c) eIF2α(S/S) and d) eIF2α(A/A) MEFs. Values are means ± SEM for 3 separate experiments, with each experiment having 3 replicates. Bars not denoted by the same letter are significantly different by Student’s t-test comparison at p≤ 0.05.
In order to determine if the increase in ATF4 expression in GCN2(−/−) cells subjected to essential amino acid deficiency was the result of activation of an eIF2α kinase other than GCN2, nonphosphorylatable eIF2α(A/A) MEFs were used (Fig. 1c, d). As observed in GCN2(+/+) MEFs, ATF4 mRNA levels were also upregulated in control eIF2α(S/S) MEFs under essential amino acid deprivation. Less effect of amino acid deprivation on ATF4 mRNA levels were obtained in the eIF2α(A/A) cells. Though reduced compared to eIF2α(S/S) cells, methionine deprivation still resulted in a relatively large increase in ATF4 mRNA levels, and arginine and histidine deprivation resulted in smaller but significant increases, in the eIF2α(A/A) cells. However, leucine deprivation did not result in a significant increase in ATF4 mRNA levels in eIF2α(A/A) cells. These results suggest that the upregulation of ATF4 expression in MEFs in response to essential amino acid deprivation does not strictly require activation of GCN2 or any other eIF2α kinase. However, as the fold increase of ATF4 mRNA was much lower in GCN2(−/−) and eIF2α(A/A) MEFs than in wild-type MEFs, the GCN2-eIF2α kinase signaling pathway clearly does play a major role in ATF4 mRNA induction in GCN2(+/+) and eIF2α(S/S) MEFs. Methionine deprivation appears to be the strongest activator of the GCN2-/eIF2α phosphorylation-independent pathway leading to increased ATF4 mRNA, whereas ATF4 mRNA induction due to leucine deprivation appears to be totally dependent on GCN2-catalyzed eIF2α phosphorylation.
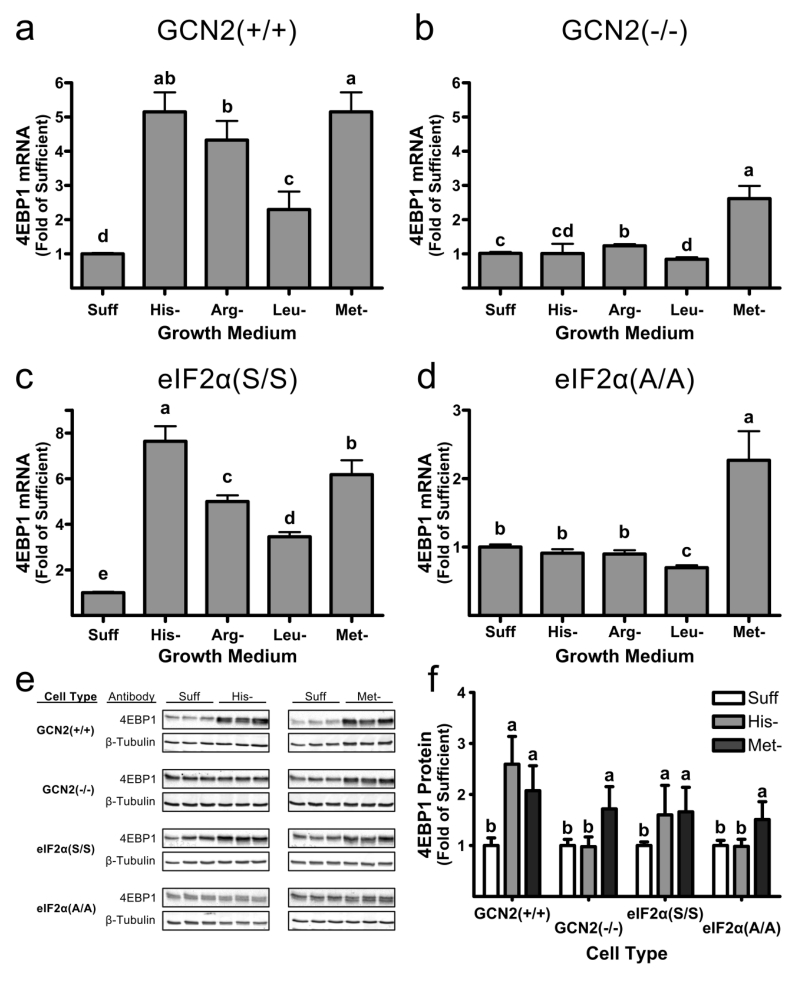
Past studies in our lab have demonstrated an increase in 4EBP1 expression (mRNA and protein) in liver of rats when they were fed diets that were limiting in sulfur amino acids (Sikalidis et al. 2013). Therefore, we were interested in determining if the increases in ATF4 mRNA induction independent of the GCN2-eIF2α kinase signaling pathway resulted in increases in 4EBP1 mRNA and protein. As shown in Figure 2a, histidine, methionine, arginine or leucine deprivation each resulted in a significant increase in the 4EBP1 mRNA level in GCN2(+/+) MEFS. Leucine deprivation resulted in significantly less upregulation than did deprivation of histidine, methionine or arginine, which all had similar effects. In GCN2(−/−) MEFs (Fig. 2b) arginine deprivation resulted in a minimal increase in 4EBP1 mRNA levels, and histidine or leucine derivation resulted in no increase. In contrast, methionine deprivation resulted in a 2.6-fold increase in 4EBP1 mRNA, which was substantial though lower than the 5.2-fold increase observed in GCN2(+/+) MEFs. The results observed for GCN2(+/+) and GCN2(−/−) MEFs were confirmed with eIF2α(S/S) and eIF2α(A/A) MEFs as shown in Figure 2c, d. Histidine, arginine, leucine or methionine deprivation all resulted in increased 4EBP1 mRNA in eIF2α(S/S) MEFs, whereas only methionine deficiency resulted in an increase in 4EBP1 mRNA in eIF2α(A/A) MEFs. These results demonstrate that 4Ebp1 gene expression, like Atf4 gene expression, is regulated in both a GCN2-/eIF2α phosphorylation-dependent and -independent manner. 4EBP1 protein levels were measured in cells cultured in methionine- or histidine-deficient medium (Fig. 2e, f). In GCN2(+/+) and eIF2α(S/S) control MEFs, both histidine- and methionine-deficient medium resulted in a significant increase in 4EBP1 protein abundance. In GCN2(−/−) and eIF2α(A/A) MEFs, 4EBP1 protein levels were not affected by culture in histidine-deficient medium but were still significantly upregulated by culture in methionine-deficient medium, consistent with observations for 4EBP1 mRNA.
Fig. 2.
Upregulation of 4EBP1 mRNA and protein levels in GCN2(+/+), GCN2(−/−), eIF2α(S/S) and eIF2α(A/A) MEFs in response to essential amino acid deficiency. Cells were grown in sufficient medium (Suff) or in medium deficient in histidine (His-) methionine (Met-), leucine (Leu-) or arginine (Arg-). a-d) ATF4 mRNA levels after 12 h of culture in deficient medium, expressed as fold the mean value for cells cultured in complete medium, are shown for experiments in a) GCN2(+/+), b) GCN2(−/−), c) eIF2α(S/S) and d) eIF2α(A/A) MEFs. e,f) 4EBP1 protein abundance in cells after 24 h of culture, shown as e) representative western blots and f) bar graph of the abundance of 4EBP1 protein, expressed as fold of the level in the same cell type cultured in sufficient medium after normalizing values to β-tubulin. All values for mRNA and protein are means ± SEM for 3 separate experiments, with each experiment having 3 replicates. Bars not denoted by the same letter are significantly different by Student’s t-test comparison at p≤ 0.05.
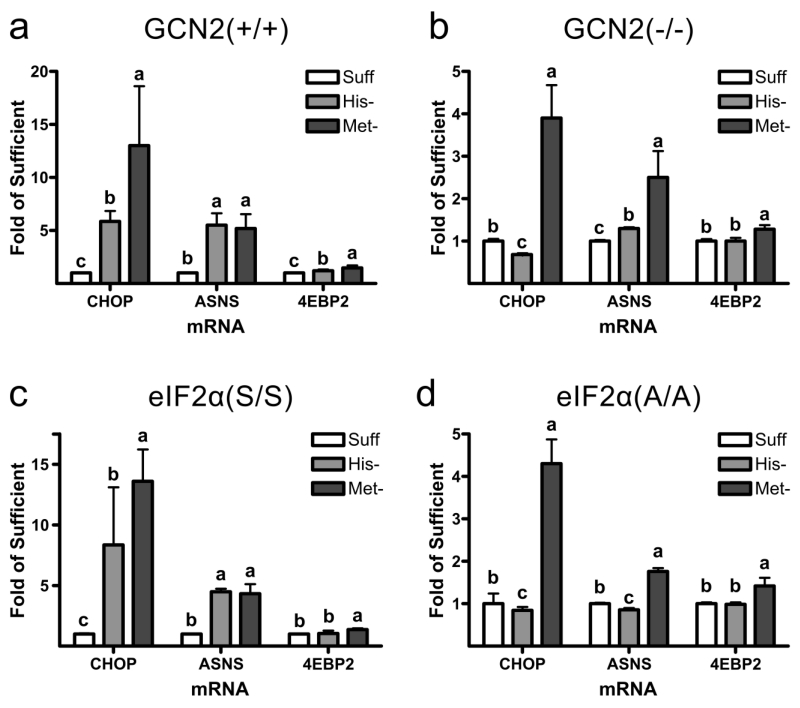
As ATF4 induces the ISR by increasing transcription of genes containing a CARE in their promoter region (Kilberg et al. 2009), we next aimed to determine if other genes that are known to contain CAREs and to be upregulated by GCN2 and other eIF2α kinase pathways would also be upregulated in an amino acid-specific and GCN2/eIF2α-independent manner. Therefore, we measured the changes in the mRNA levels for two other CARE-containing genes, CHOP and ASNS (Fig. 3a, b, c, d). CHOP and ASNS mRNAs were significantly upregulated in GCN2(+/+) and eIF2α(S/S) MEFs exposed to histidine- or methionine-deficient medium. In GCN2(−/−) and in eIF2α(A/A) MEFs, CHOP and ASNS mRNAs still showed a large, through reduced compared to results for GCN2(+/+) and eIF2α(S/S) MEFs, upregulation in response to methionine-deficient medium. GCN2(−/−) and in eIF2α(A/A) cells grown in histidine-deficient medium showed little to no upregulation of ASNS or CHOP mRNA. Thus, the GCN2-/eIF2α-phosphorylation-independent response appears to upregulate CARE-containing genes in addition to 4EBP1 & ATF4, suggesting the response might also target CARE-containing genes.
Fig. 3.
Effect of deprivation of histidine (-His) or methionine (-Met) on expression of genes with or without a C/EBP-ATF4 response element (CARE). GCN2(+/+), GCN2(−/−), eIF2α(S/S) and eIF2α(A/A) MEFs were grown in sufficient medium (Suff) or in medium deficient in histidine (His-) or methionine (Met-). CHOP, ASNS or 4EBP2 mRNA levels after 12 h of culture in deficient medium, expressed as fold the mean value for cells cultured in sufficient medium, are shown for a) GCN2(+/+), b) GCN2(−/−), c) eIF2α(S/S) and d) eIF2α(A/A) MEFs. Values are means ± SEM for 3 separate experiments, with each experiment having 3 replicates. Bars not denoted by the same letter are significantly different by Student’s t-test comparison at p≤ 0.05.
4EBP1 and 4EBP2 are homologous proteins with their genes having similar intron/exon structures with 3 exons, and 4EBP2, like 4EBP1, is expressed in most tissues (Tsukiyama-Kohara et al. 1996). Whereas the 4Ebp1 promoter region, as discussed above, contains two CARE elements located 873 and 899 base pairs upstream of the transcription start site (Kilberg et al. 2009), a search of the 4Ebp2 gene from 1000 base pairs upstream to 1000 base pairs downstream of the transcription start site did not reveal the presence of a CARE. Therefore, we would not expect 4EBP2 abundance to be upregulated by the GCN2/ATF4 pathway. This hypothesis was confirmed in Figure 3a, b, c, d. In GCN2(+/+) MEFs both methionine and histidine deprivation resulted in a minimal increase in 4EBP2 mRNA. In all other cell types only methionine deprivation resulted in a small increase in 4EBP2 mRNA. The increase in 4EBP2 mRNA in GCN2(+/+) or eIF2α(S/S) cells was not on the scale of those observed for CHOP, ASNS or 4EBP1 mRNA levels, leading us to conclude that amino acid deprivation has a minimal effect on 4EBP2 mRNA induction. Therefore, 4EBP2 mRNA levels were used as a representative mRNA for non-CARE-containing genes for the remaining experiments
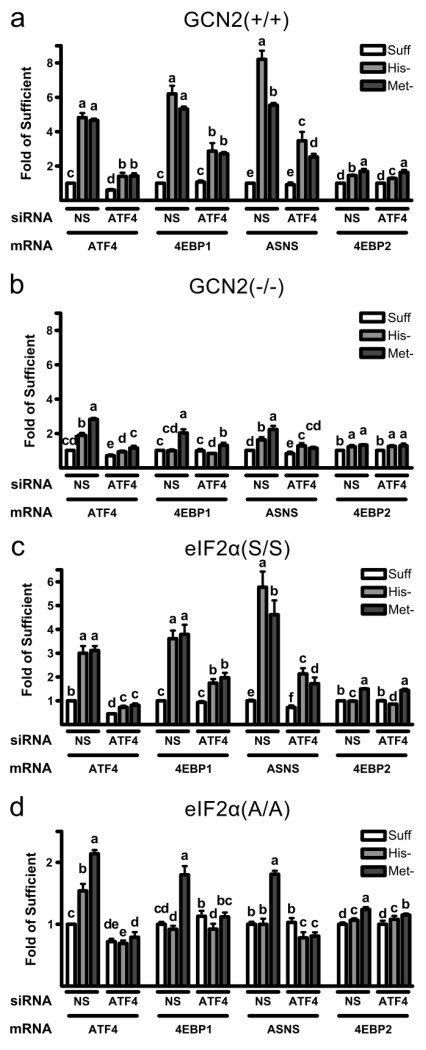
Because the accepted GCN2 signaling pathway, when activated, results in an increase in ATF4 mRNA and because both methionine and histidine deprivation in GCN2(−/−) and eIF2α(A/A) MEFs also resulted in a significant upregulation of ATF4 mRNA, we hypothesized that the GCN2/eIF2α kinase pathway and the GCN2-/eIF2α phosphorylation-independent pathway merge at the level of ATF4. In order to determine if ATF4 induction is essential for the GCN2-/eIF2α phosphorylation-independent upregulation of CARE containing gene expression, ATF4 was knocked down using ATF4-specific siRNA. The ATF4 siRNA knockdown was successful with ATF4 mRNA levels being significantly lower in cells treated with ATF4 siRNA compared to the same cell type treated with nonsense (control) siRNA (Fig. 4 a, b, c, d). Because ATF4 is an essential transcription factor for other cellular processes, we did not attempt to knockdown ATF4 completely but to prevent an increase in ATF4 expression to a level that would inhibit initiation the ISR. As can be seen in Fig. 4, MEFs transfected with nonsense siRNA and hence expressing ATF4 responded normally to either histidine- or methionine-deficiency with an increase in ATF4 mRNA. Consistent with the results shown in Fig. 1, amino acid deficiency was less effective at inducing ATF4 mRNA in the GCN2(−/−) and eIF2α(A/A) MEFs treated with nonsense siRNA than in the GCN2(+/+) and eIF2 (S/S) MEFs. Also consistent with the results shown in Fig. 1, methionine deprivation resulted in a much greater increase in ATF4 mRNA than did histidine deprivation in the GCN2(−/−) and eIF2α(A/A) MEFs treated with nonsense siRNA. Thus, the control nonsense siRNA transfected cells for this experiment behaved similarly to cells that were not transfected with siRNA.
Fig. 4.
Effect of knockdown of ATF4 in GCN2(+/+), GCN2(−/−), eIF2α(S/S) and eIF2α(A/A) MEFs on expression of ATF4, 4EBP1, ASNS and 4EBP2. Nonsense (NS, control) or ATF4 siRNA was transfected into GCN2(+/+), GCN2(−/−), eIF2α(S/S) and eIF2α(A/A) MEFs. After siRNA transfection, MEFs were cultured in sufficient medium (Suff) or in medium deficient in histidine (His-) or methionine (Met-) for 12h. ATF4, 4EBP1, ASNS and 4EBP2 mRNA levels after 12 h of culture in deficient medium after transfection of NS or ATF4 siRNA, expressed as fold the mean value for cells transfected with NS siRNA and cultured in sufficient medium, are shown for a) GCN2(+/+), b) GCN2(−/−), c) eIF2α(S/S) and d) eIF2α(A/A) MEFs. Values are means ± SEM for 3 separate experiments, with each experiment having 3 replicates. Bars not denoted by the same letter are significantly different by Student’s t-test comparison at p≤ 0.05.
In contrast, when ATF4 was knocked down using siRNA, the upregulation in ATF4 mRNA observed during histidine or methionine deprivation was dramatically reduced in all cell types. As can be seen in Fig 4, ATF4 knockdown effectively blocked the increase in ATF4 mRNA levels that occurred in cells treated with nonsense siRNA. This was true for all four cell types: GCN2(+/+), GCN2(−/−), eIF2α(S/S) and eIF2α(A/A) MEFs. In fact, the ATF4 mRNA levels in GCN2(−/−), eIF2α(S/S) and eIF2α(A/A) MEFs transfected with ATF4 siRNA and cultured in amino acid deficient medium were the same or lower than levels in cells transfected with nonsense siRNA and cultured in sufficient medium (Fig 4 b,c,d), and those in GCN2(+/+) cells cultured in amino acid deficient medium were only slightly higher than the control levels (Fig 4a). Therefore, we can conclude that the ATF4 siRNA knockdown was effective at reducing both the GCN2-/eIF2α phosphorylation-dependent and -independent ATF4 mRNA induction due to amino acid deprivation.
4EBP1 and ASNS mRNA levels were measured to determine if expression of CARE-containing genes responds to knockdown of ATF4 (Fig. 4 a, b, c, d). ATF4 knockdown in cells exposed to sufficient medium had no effect on 4EBP1 mRNA levels, regardless of cell type, suggesting that basal ATF4 levels were not high enough to affect CARE-containing gene transcription under amino acid-sufficient conditions. ASNS mRNA however was slightly reduced by ATF4 knockdown in GCN2(−/−) and eIF2α(S/S) MEFs grown in sufficient medium, suggesting some ATF4-mediated gene transcription in these mutant cells even under amino acid sufficient conditions. However, in GCN2(+/+) and eIF2α(S/S) MEFs subjected to histidine or methionine deprivation, the upregulation of 4EBP1 and ASNS mRNA were significantly reduced when ATF4 was knocked down, confirming the necessity of ATF4 for the induction of CARE-containing genes in response to amino acid deprivation. In GCN2(−/−) and eIF2α(A/A) MEFs the knockdown of ATF4 also significantly reduced the upregulation of 4EBP1 and ASNS under methionine deprivation, as well as reducing the smaller increase of ASNS mRNA levels in GCN2(−/−) MEFs under histidine deprivation. These results confirm that the GCN2-/eIF2α phosphorylation-independent pathway, like the GCN2-/eIF2 kinase-dependent pathway, is dependent on ATF4 for the induction of CARE-containing genes
4EBP2 mRNA levels were also measured to determine the effect ATF4 knockdown had on non-CARE containing genes (Fig. 4a, b, c, d). Consistent with the results shown in Figure 3, histidine and methionine deprivation resulted in minimal changes in 4EBP2 mRNA levels in all cell types. Though significantly different than control cells, these changes were not on the scale of the changes observed for 4EBP1 and ASNS mRNAs. As expected due to the absence of a CARE, ATF4 knockdown had minimal effect on 4EBP2 induction. Therefore, the small increases in 4EBP2 mRNA levels might be due to activation of other stress signaling pathways under amino acid deprivation conditions.
In total these results demonstrate that upregulation of ATF4 is an essential component of both the GCN2/eIF2α kinase-dependent pathway and the GCN2-/eIF2α phosphorylation-independent pathway for upregulation of expression of CARE-containing genes.
DISCUSSION
Our results confirm that transcriptional responses to amino acid deprivation occur predominantly, but not totally, by the canonical GCN2/eIF2α/ATF4 pathway
GCN2 is well-known to be a sensor of amino acid deficient conditions in cells via the binding of uncharged tRNAs to a regulatory domain of GCN2, causing activation of GCN2’s kinase domain. In turn, GCN2 phosphorylates the alpha subunit of eIF2 at Ser51, and this leads to depletion of eIF2-GTP and a lack of ternary complex formation (Baird and Wek 2012). Although global protein synthesis is negatively impacted by the lack of ternary complex, the translation of ATF4 is promoted. ATF4, in turn, upregulates the transcription of various stress response genes that contain elements that bind C/EBP-ATF4 heterodimers, resulting in elevated expression of particular proteins that facilitate the cell’s response to lack of amino acids or other stresses.
The mRNA levels for 4EBP1, ASNS, CHOP and ATF4 itself, all of which contain CAREs in their promoter regions, were upregulated in wild-type control cells by deprivation of any one of the four amino acids tested (histidine, arginine, leucine or methionine). Furthermore, the upregulation of mRNAs for the four genes was markedly reduced and in many cases eliminated by knockout of GCN2, mutation of the Ser51 residue of eIF2α to Ala, or knockdown of ATF4. The effects of GCN2 knockout and of eIF2α mutation were generally very similar, indicating that amino acid starvation induces eIF2α phosphorylation almost entirely via activation of GCN2 and not via activation of another eIF2α kinase. These results support the predominant role of the GCN2/eIF2α kinase/ATF4 pathway in mediating the cell’s transcriptional responses to amino acid insufficiency, even of one amino acid. However, our results also clearly show that knockout of GCN2 or mutation of Ser51 of eIF2α to a residue that cannot undergo phosphorylation does not totally eliminate the response of cells to amino acid deficiency, indicating the presence of a GCN2- and eIF2α phosphorylation-independent pathway.
Histidine, arginine, leucine and methionine starvation resulted in somewhat different degrees of transcriptional upregulation of the genes we studied. In general, methionine starvation gave the largest fold changes in mRNA levels, leucine starvation gave the lowest fold changes in mRNA levels, and histidine and arginine gave intermediate fold changes. Patterns were not identical for the different CARE-containing mRNAs analyzed, consistent with the presence of additional minor regulatory mechanisms that differ depending on which amino acid is deficient.
Methionine, and to a lesser extent histidine and arginine, activates a GCN2- and eIF2 phosphorylation-independent but ATF4-dependent pathway that leads to transcriptional upregulation of CARE-containing genes
The transcriptional upregulation of CARE-containing genes is especially sensitive to methionine deficiency in both control cells and in GCN2(−/−) or eIF2α (A/A) cells. The residual upregulation of CARE-containing genes in GCN2(−/−) or eIF2α (A/A) cells was most robust for the case of methionine starvation. A smaller degree of residual upregulation was observed for histidine and arginine-starved cells, but none was observed for leucine-starved cells. The more robust responses to methionine starvation in control cells and in GCN2(−/−) or eIF2α (A/A) cells than to starvation of leucine, histidine or arginine suggests that methionine starvation is being sensed by an additional mechanism that is not dependent upon GCN2 or eIF2α phosphorylation. The requirement for ATF4, however, appears to be common to both the GCN2/eIF2α kinase-dependent and the GCN2-/eIF2α phosphorylation-independent pathways of transcriptional upregulation of CARE-containing genes based on the suppression of both responses by knockdown of ATF4. It should be noted that the PERK-mediated response to sulfur amino acid deficiency reported by Wanders et al. (2016) cannot explain the eIF2 phosphorylation-independent, ATF4-dependent response to methionine that we observed because we included cysteine in our cell culture medium so that cells are not deficient in glutathione and also because the effect was still observed in eIF2 (A/A) cells that cannot be phosphorylated by PERK.
4EBP1 but not 4EBP2 is a CARE-containing gene, and only 4EBP1 is upregulated by GCN2 activation, eIF2α phosphorylation or ATF4 upregulation
Although ATF4, CHOP and ASNS are well-established targets for transcriptional upregulation following activation of eIF2α kinases, 4EBP1 is a less-well established target (Dey et al. 2010; Sikalidis et al. 2011; B’Chir et al. 2013; Sikalidis et al. 2013). We were especially interested in 4EBP1 because of its involvement in the regulation of cap-dependent mRNA translation and its regulation by mTORC1 which is also responsive to amino acid availability. Our results confirm that 4EBP1 expression is upregulated in response to essential amino acid deficiency and that this occurs by the canonical GCN2/eIF2α kinase/ATF4 pathway as well as by the methionine-sensitive ATF4-dependent pathway that is independent of GCN2 and eIF2α phosphorylation. In the case of 4EBP1 mRNA upregulation, we also show that this results in an increase in 4EBP1 protein abundance.
Because 4EBP2 is a close homolog of 4EBP1 that differs from 4EBP1 in that its promoter region does not contain a CARE, we also looked at the transcriptional regulation of 4EBP2 in response to amino acid starvation. Although 4EBP2, like 4EBP1, is expressed in most tissues, the physiological role of 4EBP2 and the regulation of 4EBP2 expression and activity have been much less studied than those of 4EBP1. Grolleau et al. (1999) reported a differential regulation of 4EBP1 and 4EBP2 expression in a promyelocytic leukemia cell line (HL-60) during granulocytic differentiation, and Tsukiyama-Kohara et al. (2001) observed a stronger phosphorylation of 4EBP1 than of 4EBP2 in response to insulin treatment. These results suggested the possibility that the two homologs may have different functions and be regulated by different signals. This was confirmed in the current study where expression of 4EBP2, in contrast to 4EBP1 and other CARE-containing genes, was only minimally upregulated in response to histidine or methionine deprivation and this upregulation was not reduced by knockout of GCN2, mutation of Ser51 in eIF2α or knockdown of ATF4. Thus, 4EBP1, but not 4EBP2, appears to be a canonical target of GCN2/eIF2α kinase/ATF4 stress response pathways.
A possible mechanism for the methionine-sensitive, GCN2- and eIF2α phosphorylation-independent, ATF4-dependent pathway that leads to transcriptional upregulation of CARE-containing genes is a lack of ternary complex due to a decrease in the concentration of Met-charged tRNAiMet
The ISR has been shown to be highly dependent on the increased translation of ATF4 by a mechanism that depends on a low abundance of ternary complex (eIF2-GTP-Met-tRNAiMet) due to eIF2 phosphorylation blocking the regeneration of eIF2-GTP for new ternary complex formation. Additionally, after ATF4 is translated, it in turn induces its own gene transcription, further increasing ATF4 abundance and initiating the ISR. Our finding that methionine deprivation uniquely induced ATF4 and the ISR in MEFs that did not express GCN2 or a phosphorylatable eIF2α suggests that methionine deprivation maybe able to reduce the available amount of ternary complex by a mechanism not dependent upon GCN2 signaling or Ser51 phosphorylation of eIF2α. A likely alternative mechanism by which a deficiency of methionine might uniquely decrease the amount of ternary complex is that methionine deficiency could lead to a lack of methionine-charged initiator tRNA that is necessary for ternary complex formation. In eukaryotes, both the initiator and elongater tRNAMet are aminoacylated with methionine by methionyl-tRNA synthetase with no unique specificity (Kolitz and Lorsch 2010). Structural differences between the initiator and elongator tRNAMet prevent the initiator tRNAMet from participating in translation elongation, and promote its binding to eIF2 (Francis and Rajbhandary 1990; Drabkin et al. 1998). Therefore, under methionine deprivation we would expect the amounts of both charged initiator and elongator tRNAMet to decrease. In addition Met-tRNAiMet binding to eIF2-GTP is facilitated by the presence of methionine on the tRNAiMet as non-acylated tRNAiMet has a Kd value of roughly 130 nM for eIF2-GTP while Met-tRNAiMet binding to eIF2-GTP has a Kd value of 9 nM (Kapp and Lorsch 2004). Thus, because they are both essential components of the ternary complex, a lack of methionine-charged initiator tRNA would be expected to act somewhat similarly to a lack of eIF2α-GTP in reducing formation of ternary complex.
This mechanism would be consistent with our observation that methionine deficiency was especially robust in inducing ATF4 expression and the ISR in wild-type cells, and that methionine deficiency could still induce ATF4 expression and the ISR in GCN2 knockout cells and in cells with nonphosphorylatable eIF2α. A delay in ternary complex formation, regardless of the cause, would be expected to result in a delay in reformation of the 43S preinitiation complex and an increase in ATF4 synthesis. Although efforts to verify this hypothesis by overexpression of the initiator tRNA versus elongator tRNA for methionine were unsuccessful, due to the high efficiency at which cells charge these tRNAs with methionine, our results clearly demonstrate the existence of GCN2- and eIF2α-phosphorylation-independent mechanisms for upregulation of ATF4 that are highly responsive to methionine deprivation. Furthermore, the hypothesis that initiator tRNA plays a critical role in regulating the abundance of ternary complex is supported by previous studies in yeast. Dever et al. (1995) demonstrated that a reduction in the copy number of the initiator tRNA genes resulted in an increase in GCN4, the yeast homolog of ATF4, in GCN2-knockout yeast. This induction of GCN4 in yeast with reduced expression of the initiator tRNAMet was further shown to be dependent on the uORF in the GCN4 mRNA. It is reasonable to hypothesize that both the initiator tRNAMet and methionine-charging to the initiator tRNAMet are necessary for formation of functional ternary complex. Our results are the first to demonstrate the existence of a regulatory mechanism controlling mRNA translation that is uniquely sensitive to methionine status and suggest that there may be a tighter control of protein synthesis and expression of stress-responsive genes in response to methionine deprivation than in response to other amino acid deficiencies.
In summary, we demonstrate the existence of a GCN2- and eIF2α-phosphorylation-independent, but ATF4-dependent, pathway that is highly sensitive to methionine deprivation. We hypothesize this new pathway is due to reduced levels of Met-tRNAiMet for ternary complex formation and, thus, supplements the canonical pathway whereby eIF2 Ser51 phosphorylation regulates ternary complex formation and availability. CARE-containing genes are upregulated in response to amino acid deprivation by the canonical GCN2/eIF2α phosphorylation/ATF4 pathway and, in addition, by the newly demonstrated GCN2- and eIF2 -phosphorylation-independent pathway. In addition, we demonstrate that 4EBP1, a major cap-binding protein thought to promote mRNA translation, but not its 4EBP2 homolog, is regulated by the ATF4/ISR pathway.
Acknowledgments
Funding: This work was supported by Grants DK-083473 and Grant DK-056649 from the National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
Conflict of interest: The authors declare that they have no conflict of interest.
COMPLIANCE WITH ETHICAL STANDARDS
Ethical approval: This article does not contain any studies with human participants or animals performed by any of the authors.
REFERENCES
- Anthony TG, Reiter a K, Anthony JC, et al. Deficiency of dietary EAA preferentially inhibits mRNA translation of ribosomal proteins in liver of meal-fed rats. Am J Physiol Endocrinol Metab. 2001;281:E430–E439. doi: 10.1152/ajpendo.2001.281.3.E430. [DOI] [PubMed] [Google Scholar]
- B’chir W, Chaveroux C, Carraro V, et al. Dual role for CHOP in the crosstalk between autophagy and apoptosis to determine cell fate in response to amino acid deprivation. Cell Signal. 2014;26:1385–1391. doi: 10.1016/j.cellsig.2014.03.009. [DOI] [PubMed] [Google Scholar]
- B’Chir W, Maurin AC, Carraro V, et al. The eIF2α/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 2013;41:7683–7699. doi: 10.1093/nar/gkt563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird T, Wek R. Eukaryotic initiation factor 2 phosphorylation and translational control in metabolism. Adv Nutr. 2012;3:307–321. doi: 10.3945/an.112.002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Do AN, Kimball SR, Cavener DR, Jefferson LS. eIF2alpha kinases GCN2 and PERK modulate transcription and translation of distinct sets of mRNAs in mouse liver. Physiol Genomics. 2009;38:328–341. doi: 10.1152/physiolgenomics.90396.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deval C, Chaveroux C, Maurin AC, et al. Amino acid limitation regulates the expression of genes involved in several specific biological processes through GCN2-dependent and GCN2-independent pathways. FEBS J. 2009;276:707–718. doi: 10.1111/j.1742-4658.2008.06818.x. [DOI] [PubMed] [Google Scholar]
- Dever TE, Hinnebusch AG. GCN2 whets the appetite for amino acids. Mol Cell. 2005;18:141–142. doi: 10.1016/j.molcel.2005.03.023. [DOI] [PubMed] [Google Scholar]
- Dever TE, Yang W, Aström S, et al. Modulation of tRNA(iMet), eIF-2, and eIF-2B expression shows that GCN4 translation is inversely coupled to the level of eIF-2.GTP.Met-tRNA(iMet) ternary complexes. Mol Cell Biol. 1995;15:6351–6363. doi: 10.1128/mcb.15.11.6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey S, Baird TD, Zhou D, et al. Both transcriptional regulation and translational control of ATF4 are central to the integrated stress response. J Biol Chem. 2010;285:33165–33174. doi: 10.1074/jbc.M110.167213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Qiu H, Garcia-Barrio M, et al. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol Cell. 2000;6:269–279. doi: 10.1016/s1097-2765(00)00028-9. [DOI] [PubMed] [Google Scholar]
- Donnelly N, Gorman AM, Gupta S, Samali A. The eIF2alpha kinases: Their structures and functions. Cell Mol Life Sci. 2013;70:3493–3511. doi: 10.1007/s00018-012-1252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drabkin HJ, Estrella M, Rajbhandary UL. Initiator-elongator discrimination in vertebrate tRNAs for protein synthesis. Mol Cell Biol. 1998;18:1459–1466. doi: 10.1128/mcb.18.3.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis M a, Rajbhandary UL. Expression and function of a human initiator tRNA gene in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:4486–4494. doi: 10.1128/mcb.10.9.4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grolleau A, Sonenberg N, Wietzerbin J, Beretta L. Differential regulation of 4E-BP1 and 4E-BP2, two repressors of translation initiation, during human myeloid cell differentiation. J Immunol. 1999;162:3491–3497. [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, et al. Regulated Translation Initiation Controls Stress-Induced Gene Expression in Mammalian Cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem. 2014;83:779–812. doi: 10.1146/annurev-biochem-060713-035802. [DOI] [PubMed] [Google Scholar]
- Jousse C, Averous J, Bruhat A, et al. Amino acids as regulators of gene expression: Molecular mechanisms. Biochem Biophys Res Commun. 2004;313:447–452. doi: 10.1016/j.bbrc.2003.07.020. [DOI] [PubMed] [Google Scholar]
- Kapp LD, Lorsch JR. GTP-dependent recognition of the methionine moiety on initiator tRNA by translation factor eIF2. J Mol Biol. 2004;335:923–936. doi: 10.1016/j.jmb.2003.11.025. [DOI] [PubMed] [Google Scholar]
- Kilberg MS, Shan J, Su N. ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol Metab. 2009;20:436–443. doi: 10.1016/j.tem.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolitz SE, Lorsch JR. Eukaryotic initiator tRNA: Finely tuned and ready for action. FEBS Lett. 2010;584:396–404. doi: 10.1016/j.febslet.2009.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamoorthy T, Pavitt G. Tight binding of the phosphorylated α subunit of initiation factor 2 (eIF2α) to the regulatory subunits of guanine nucleotide exchange factor eIF2B is required for inhibition of translation. Mol Cell Biol. 2001;21:5018–5030. doi: 10.1128/MCB.21.15.5018-5030.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan J, Staschke K a., Wek RC. Dimerization is required for activation of eIF2 kinase Gcn2 in response to diverse environmental stress conditions. J Biol Chem. 2004;279:22820–22832. doi: 10.1074/jbc.M402228200. [DOI] [PubMed] [Google Scholar]
- Palii SS, Kays CE, Deval C, et al. Specificity of amino acid regulated gene expression: Analysis of genes subjected to either complete or single amino acid deprivation. Amino Acids. 2009;37:79–88. doi: 10.1007/s00726-008-0199-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavitt GD, Ramaiah KVA, Kimball SR, Hinnebusch AG. eIF2 independently binds two distinct eIF2b subcomplexes that catalyze and regulate guanine-nucleotide exchange. Genes Dev. 1998;12:514–526. doi: 10.1101/gad.12.4.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Dong J, Hu C, et al. The tRNA-binding moiety in GCN2 contains a dimerization domain that interacts with the kinase domain and is required for tRNA binding and kinase activation. EMBO J. 2001;20:1425–1438. doi: 10.1093/emboj/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuner D, Song B, McEwen E, et al. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell. 2001;7:1165–1176. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- Shan J, Lopez M-C, Baker HV, Kilberg MS. Expression profiling after activation of amino acid deprivation response in HepG2 human hepatoma cells. Physiol Genomics. 2010;41:315–327. doi: 10.1152/physiolgenomics.00217.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan J, Ord D, Ord T, Kilberg MS. Elevated ATF4 Expression, in the Absence of Other Signals, Is Sufficient for Transcriptional Induction via CCAAT Enhancer-binding Protein-activating Transcription Factor Response Elements. J Biol Chem. 2009;284:21241–21248. doi: 10.1074/jbc.M109.011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikalidis AK, Lee JI, Stipanuk MH. Gene expression and integrated stress response in HepG2/C3A cells cultured in amino acid deficient medium. Amino Acids. 2011;41:159–171. doi: 10.1007/s00726-010-0571-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikalidis AK, Mazor KM, Kang M, et al. Total 4EBP1 Is Elevated in Liver of Rats in Response to Low Sulfur Amino Acid Intake. J Amino Acids. 2013;2013:864757. doi: 10.1155/2013/864757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu F, Bain PJ, Leblanc-Chaffin R, et al. ATF4 is a mediator of the nutrient-sensing response pathway that activates the human asparagine synthetase gene. J Biol Chem. 2002;277:24120–24127. doi: 10.1074/jbc.M201959200. [DOI] [PubMed] [Google Scholar]
- Sood R, Porter AC, Olsen D, et al. A mammalian homologue of GCN2 protein kinase important for translational control by phosphorylation of eukaryotic initiation factor-2α. Genetics. 2000;154:787–801. doi: 10.1093/genetics/154.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolboushkina E a, Garber MB. Eukaryotic type translation initiation factor 2: structure-functional aspects. Biochemistry (Mosc) 2011;76:283–294. doi: 10.1134/s0006297911030011. [DOI] [PubMed] [Google Scholar]
- Tsukiyama-Kohara K, Poulin F, Kohara M, et al. Adipose tissue reduction in mice lacking the translational inhibitor 4E-BP1. Nat Med. 2001;7:1128–1132. doi: 10.1038/nm1001-1128. [DOI] [PubMed] [Google Scholar]
- Tsukiyama-Kohara K, Vidal SM, Gingras C, et al. Tissue distribution, genomic structure, and chromosome mapping of mouse and human eukaryotic initiation factor 4E-binding proteins 1 and 2. Genomics. 1996;38:353–363. doi: 10.1006/geno.1996.0638. [DOI] [PubMed] [Google Scholar]
- Unbehaun A, Borukhov SI, Hellen CUT, Pestova TV. The 40S subunit in 48S complexes formed at the initiation codon of mRNA is bound to eukaryotic initiation factor (eIF) 3, eIF1, eIF1A, and an eIF2/GTP/Met-tRNA. Genes Dev. 2004:3078–3093. doi: 10.1101/gad.1255704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Kelen K, Beyaert R, Inzé D, De Veylder L. Translational control of eukaryotic gene expression. Crit Rev Biochem Mol Biol. 2009;44:143–168. doi: 10.1080/10409230902882090. [DOI] [PubMed] [Google Scholar]
- Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci U S A. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Wanders D, Stone KP, Forney LA, Cortez CC, Dille KN, Simon J, Xu M, Hotard EC, Nikonorova IA, Pettit AP, Anthony TG, Gettys TW. Role of GCN2-independent signaling through a noncanonical PERK/NRF2 pathway in the physiological responses to dietary methionine restriction. Diabetes. 2016;65:1499–1510. doi: 10.2337/db15-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb BLJ, Proud CG. Eukaryotic initiation factor 2B (eIF2B) Int J Biochem Cell Biol. 1997;29:1127–1131. doi: 10.1016/s1357-2725(97)00039-3. [DOI] [PubMed] [Google Scholar]
- Wek RC, Jackson BM, Hinnebusch AG. Juxtaposition of domains homologous to protein kinases and histidyl-tRNA synthetases in GCN2 protein suggests a mechanism for coupling GCN4 expression to amino acid availability. Proc Natl Acad Sci U S A. 1989;86:4579–4583. doi: 10.1073/pnas.86.12.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek RC, Jiang H-Y, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans. 2006;34:7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- Wek SA, Zhu S, Wek RC. The histidyl-tRNA synthetase-related sequence in the eIF-2 alpha protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol Cell Biol. 1995;15:4497–4506. doi: 10.1128/mcb.15.8.4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Ishihara H, Yamada T, et al. ATF4-mediated induction of 4E-BP1 contributes to pancreatic beta cell survival under endoplasmic reticulum stress. Cell Metab. 2008;7:269–276. doi: 10.1016/j.cmet.2008.01.008. [DOI] [PubMed] [Google Scholar]
- Zaborske JM, Narasimhan J, Jiang L, et al. Genome-wide analysis of tRNA charging and activation of the eIF2 kinase Gcn2p. J Biol Chem. 2009;284:25254–25267. doi: 10.1074/jbc.M109.000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, McGrath BC, Reinert J, et al. The GCN2 eIF2α kinase is required for adaptation to amino acid deprivation in mice. Mol Cell Biol. 2002;22:6681–6688. doi: 10.1128/MCB.22.19.6681-6688.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Sobolev AY, Wek RC. Histidyl-tRNA synthetase-related sequences in GCN2 protein kinase regulate in vitro phosphorylation of eIF-2. J Biol Chem. 1996;271:24989–24994. doi: 10.1074/jbc.271.40.24989. [DOI] [PubMed] [Google Scholar]