Abstract
Myocardial infarction (MI) leads to loss and degradation of contractile cardiac tissue followed by sterile inflammation of the myocardium through activation and recruitment of innate and adaptive cells of the immune system. Recently, it was shown that cardiac myosin binding protein-C (cMyBP-C), a protein of the cardiac sarcomere, is degraded following MI, releasing a predominant N-terminal 40-kDa fragment (C0C1f) into myocardial tissue and the systemic circulation. We hypothesized that early release of C0C1f contributes to the initiation of inflammation and plays a key role in recruitment and activation of immune cells. Therefore, we investigated the role of C0C1f on macrophage / monocyte activation using both mouse bone marrow-derived macrophages and human monocytes. Here we demonstrate that C0C1f leads to macrophage / monocyte activation in vitro. Furthermore, C0C1f induces strong upregulation of pro-inflammatory cytokines (interleukin-6 (IL-6), tumor necrosis factor α (TNFα), and interleukin-1β (IL-1β)) in cultured murine macrophages and human monocytes, resulting in a pro-inflammatory phenotype. We identified the toll-like receptor 4 (TLR4), toll-like receptor 2 (TLR2), and Advanced Glycosylation End Product-Specific Receptor (RAGE) as potential receptors for C0C1f whose activation leads to mobilization of the NFκB signaling pathway, a central mediator of the pro-inflammatory signaling cascade. Thus, C0C1f appears to be a key player in the initiation of inflammatory processes and might also play an important role upon MI.
Keywords: Cardiac Myosin Binding Protein-C, C0C1f, Myocardial infarction, Inflammation, Cell signaling/signal transduction, Ischemic biology - basic studies
1 Introduction
Acute myocardial infarction (MI) is a major cause of morbidity and mortality in Western countries. MI caused by a diminished blood supply and hypoxia of the affected area leads to loss and degradation of contractile cardiac tissue, which is followed by a complex program of inflammatory processes, replacement fibrosis, and cardiac remodeling [1]. Early upon cardiac injury monocytes migrate into the infarct zone and proceed with the clearance of necrotic cells and cellular debris [2–4]. The early initiation of inflammation and the precise mechanisms are still not fully understood. It was shown that damage-associated molecular patterns (DAMPs) trigger the innate immune response and initialize this life-saving remodeling of the damaged cardiac tissue [5]. Often, however, an exaggerated inflammatory response causes a loss of healthy cardiac tissue in the border zone of the infarcted cardiac region [6].
From a clinical point of view, an early diagnosis of MI is essential for the survival of patients, and biomarkers such as cardiac troponin, which are routinely used in clinical practice, help to diagnose MI and initiate personalized therapy. Cardiac myosin binding protein-C (cMyBP-C), a sarcomeric thick filament protein involved in regulating sarcomere structure and function, has recently been identified as novel potential biomarker for myocardial infarction [7]. It was demonstrated that after ischemia/reperfusion injury cMyBP-C is cleaved and its N-terminal fragment is released at high levels into the blood [8]. The N-terminal fragments contain a 40-kDa fragment (C0C1f) consisting of C0 and C1 domains as well as the first 17 residues of the M-domain (amino acids 1–271) [9, 10].
The fact that upon MI, C0C1f is released into the circulation prior to cardiac troponins suggests that it may contribute to the initial sterile inflammation [9, 11, 12]. The mechanism of C0C1f release has been studied extensively, showing a correlation between dephosphorylation of cMyBP-C and its µ-calpain-dependent degradation [9]. This mechanism is target oriented and results in the release of C0C1f even before the cardiomyocyte undergoes apoptosis or necrosis. Furthermore, the role of the peptide is described as being directly connected to cardiomyocyte injury and death: the binding of C0C1f to cardiomyocyte actin competes with the binding of full-length cMyBP-C, resulting in dislocation of the latter and contractile dysfunction [13].
Taken together, these findings prompted us to hypothesize that C0C1f might play a crucial role in the initiation and activation of inflammatory processes upon cardiac injury due to its early release from injured tissue. To this end, we treated murine macrophages and human monocytes with various N-terminal fragments of cMyBP-C and determined inflammatory responses. A precise understanding of the underlying molecular processes could help developing a novel therapeutic approach to inflammation in order to restrict the loss of cardiac tissue and replacement fibrosis to the infarct zone.
2 Methods
2.1 Ethics statements
All animal experiments were performed in accordance with German animal welfare laws and were approved by the Animal Welfare Officer of the Justus Liebig University Giessen (Registration No.: 503_M and 588_M). The animal housing facility was licensed by the local authorities (Az: FD62-§11JLUHumPhys). The methods used to euthanize the mice humanely were consistent with the recommendations of the AVMA Guidelines for the Euthanasia of Animals. All scientific procedures using human material were approved by the ethics committee of the Justus Liebig University Giessen (Az: 05/00) and were therefore performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments.
2.2 Isolation of murine bone marrow-derived stem cells and differentiation into macrophages
The generation of murine macrophages was performed according to previously published methods [14]. Briefly, 8-week-old C57BL/6 mice were purchased from Charles River Laboratories and anesthetized using CO2 or isoflurane before euthanasia by cervical dislocation. Hind limbs were surgically truncated. Tibias and femurs were dissected and placed into ice-cold PBS. Femurs and tibias were separated carefully and fibulas were removed. Proximal and distal ends of the bones were opened. Cells were flushed out from tibiae and femurs with phosphate-buffered saline (PBS) using a 27G cannula and filtered through a cell strainer with a 70-µm cut-off. The resulting cell suspension was mixed with erythrocyte lysis buffer (BD Pharm Lyse™; BD Biosciences, Heidelberg, Germany) and incubated for 15 min at room temperature. The suspension was centrifuged for 5 min at 400 × g and pelleted cells were resuspended in Dulbecco’s modified Eagles medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 100 ng/ml macrophage colony-stimulating factor (M-CSF). For differentiation, 4×106 bone marrow stem cells were seeded on 100-mm dish in DMEM supplemented with 100 ng/ml M-CSF. On Day 3 fresh medium (containing M-CSF) was added and the cells were incubated for 5–7 additional days at 37°C with 5% CO2; thereafter, they were split to the experimental format when 80% confluence was reached. The medium was changed to DMEM containing 2.5% FBS without M-CSF. The quality and purity of macrophages was determined by flow cytometry (Supplementary Fig. 1). For this purpose the cells were stained with CD11b-APC-eFluor780 (eBioscience, CA, USA) and with Aqua Dye (Invitrogen, Darmstadt, Germany) for discrimination between life and dead cells upon Fc-blocking (BioLegend, CA, USA). Analysis was performed using a FACSVerse (BD Biosciences).
2.3 Isolation of human monocytes
Human monocytes were isolated from the blood of healthy donors by double-gradient centrifugation. In brief, buffy coats obtained from a local blood bank were mixed (1:1) with Hank’s balanced salt solution (HBSS; ThermoFisher, Darmstadt, Germany), added to 15 ml Ficoll, and centrifuged at 700 × g without braking for 35 min at room temperature. Peripheral blood mononuclear cells, located in the white ring phase, were isolated. Washing buffer (containing 1% EDTA, 100 mM pyruvate, 20% albumin, and 10% 10× PBS) was added to a volume of 50 ml and centrifuged at 350 × g for 10 min. The pellet was washed again and resuspended in elutriation buffer (washing buffer supplemented with 10% glucose). Afterwards, centrifugal elutriation was performed at 2400 × g at 22°C. The monocyte fraction was centrifuged at 350 × g for 10 min and the cells were pooled in Macrophage Serum-Free Medium (Invitrogen), seeded on culture dishes, and incubated at 37°C with 5% CO2.
2.4 RNA isolation and real-time PCR
Cells or tissues were lysed using the QIAzol lysis reagent (Qiagen, Hilden, Germany) and thereafter processed with the RNeasy mini kit (Qiagen) according to the manufacturer’s instructions. The iScript™ (Bio-Rad Laboratories, Munich, Germany) was used for reverse transcription and cDNA amplification. Real-time PCR was performed using a CFX96 real-time PCR system (Bio-Rad Laboratories). Assays were performed in triplicates in a 20-µl reaction containing 10 nM primer (Table 1), SsoFast™EvaGreen® Supermix (Bio-Rad Laboratories), and 2 µl cDNA (1:5 of 1 µg total RNA). The amount of target mRNA was normalized to glycerinaldehyd-3-phosphate dehydrogenase (GAPDH) mRNA. Fold changes of samples relative to untreated controls were calculated using the ΔΔCT method. The specific primer sequences used are listed in the supplementary material (Table 1).
2.5 Protein analysis by western blotting
To determine the protein levels of macrophages and monocytes, proteins were isolated from cultured monocytes by application of WCE buffer (1 M Hepes, 5 M NaCl, 1 M Na2MoO4, 0.25% glycerin, 0.5 M EDTA). After a 5-min incubation on ice, 10% NP40 was added and the cell suspension was vortexed and sonicated. The suspension was then centrifuged for 10 min and the supernatants were used for further analysis. Proteins (20 to 30 µg) were electrophoresed by SDS-PAGE and subjected to western blotting using specific antibodies against IL-6, TNFα, IL-1β, MRC1, ICAM1, and GAPDH (types and sources listed in Table 2 in supplementary material). Detection was performed using the ChemiDoc MP Imaging System (Bio-Rad Laboratories).
2.6 Peptide production
The recombinant N-terminal fragments of cMyBP-C (C0, C0-L, C0C1, C0C1f and C0C2) were generated using the pET expression system (Novagen, San Diego, CA) as described earlier [8]. Briefly, the cDNA coding for amino acid residues within the C0, C0-L, C0C1, C0C1f and C0C2 regions of cMyBP-C were cloned separately into the pET-28a(+) expression vector (Novagen). The N-terminus of each protein contains the His epitope. Recombinant clones were confirmed by DNA sequencing (ACGT, Inc.). Plasmids were transformed into the BL21 (DE3) strain of Escherichia coli (E. coli) (Cat. No. C6010-03, Invitrogen, Schwerte, Germany), having the gene for the T7 RNA polymerase promoter that is induced by isopropyl-β-D-thiogalactopyranoside (IPTG; Cat. No. 10724815001; Roche, Indianapolis, USA). Expression of the recombinant proteins was confirmed by SDS-PAGE and western blot analysis. Large-scale preparation was conducted for each clone, giving optimal expression of each recombinant protein. Five hundred ml of LB medium was inoculated with 1% E. coli BL21 cells, which were grown for 12 h at 37°C to an optical density (OD) of 0.6.
After induction with 1 mM IPTG, the cultures were incubated for an additional 5 h; this was followed by centrifugation at 4000 × g for 30 min at 4°C to harvest the cells. In accordance with the manufacturer’s suggestion, overexpressed proteins were purified with Ni-NTA agarose chromatography (Cat. No. 1018244, Qiagen). The proteins were dialyzed against 5 liters of 1X PBS (Cat. No. IB70166, MidSci). Buffers were changed twice using 15-kDa molecular weight cut-off membranes (Cat. No. 132124, Spectrum Laboratories, Rancho Dominguez, CA). Protein concentrations were determined following dialysis using the Bradford assay (Bio-Rad Laboratories, Cat No. 500-0205). The quality of the recombinant cMyBP-C fragments was determined by Ponceau S staining (Ponceau S solution; Cat. No. P7170, Sigma-Aldrich, Munich, Germany) and western blot analysis using rabbit polyclonal antibodies against cMyBP-C residues 2–14 (cMyBP-C2–14) [13, 15]. The N-terminal 6× histidine epitope was detected using a mouse anti-His6 monoclonal antibody (Cat. No. 11922416001, Sigma-Aldrich).
2.7 Statistical analysis
Statistical analyses were performed using an unpaired Student’s t test for comparing control and individual peptides values or a one-way analysis of variance (ANOVA) with Tukey’s post-hoc test unless otherwise noted using GraphPad software (GraphPad, La Jolla, CA, USA). Data are presented as mean ± standard error of mean (SEM). Values of p < 0.05 were considered to be statistically significant.
3 Results
3.1 Early biomarker C0C1f is involved in the initiation of inflammation
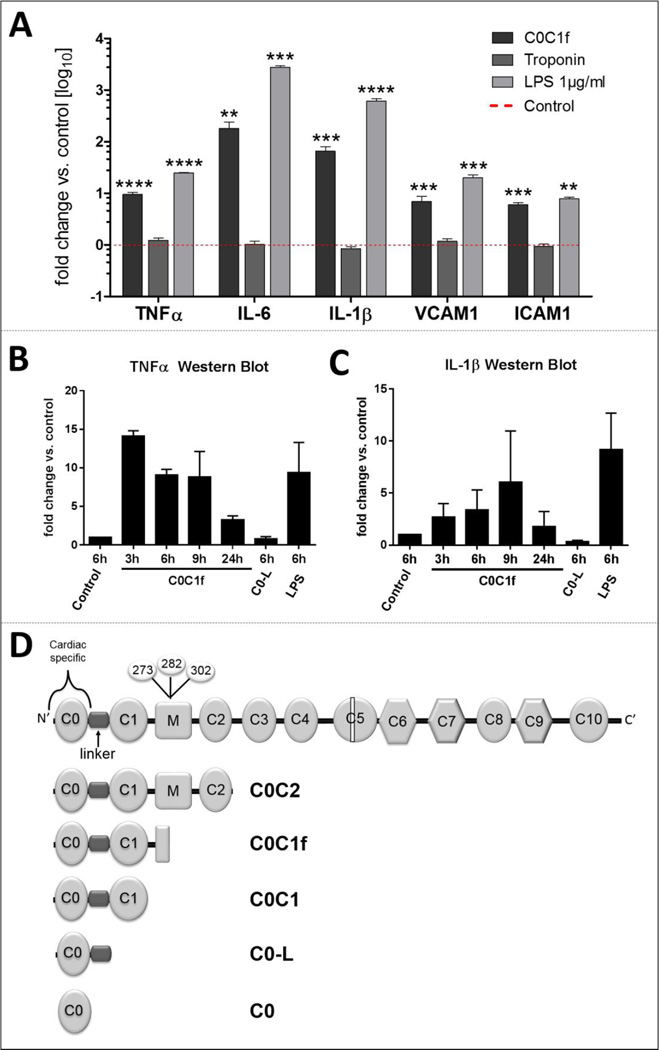
To identify C0C1f as a potential inducer of the inflammatory responses of immune cells upon MI, we used C0C1f to treat murine bone marrow-derived macrophages that had been differentiated by the treatment with M-CSF for 4–7 days. Differentiated cells were treated with cardiac troponin I (cTnI; 500 ng/ml), C0C1f (500 ng/ml), and LPS (1 µg/ml) as positive control for 6 h and this was followed by the isolation of mRNA and analysis by qRT-PCR (Fig. 1). While C0C1f induced significantly pro-inflammatory targets such as interleukin 6 (IL-6), interleukin-1β (IL-1β) and tumor necrosis factor-α (TNFα) (TNFα: 9.5 ± 1.0 fold, IL-6: 1182.42± 57.8 fold, IL-1β: 65.2 ± 15.3 fold) as well as adhesion molecules such as vascular cell adhesion molecule 1 (VCAM1) and intercellular adhesion molecule 1 (ICAM1) (VCAM1: 7.0 ± 1.8 fold, ICAM1: 6.0 ± 0.6 fold), cTnI did not lead to a detectable activation of macrophages compared with untreated controls (0.8- to 1.2-fold difference in TNFα, IL-6, IL-1β, VCAM1, and ICAM1). These results demonstrate that in contrast to cTnI, C0C1f initiates a pro-inflammatory response.
Figure 1. Initiation of inflammatory responses by MI biomarkers.
Murine macrophages were treated with 500 ng/ml C0C1f, 500 ng/ml cTnI, or 1µg/ml LPS for 6 hours. Thereafter, mRNA was isolated and mRNA levels of TNFα, IL-6, IL-1β, VCAM1, and ICAM1 were measured by qRT-PCR. Mean ± SEM; Statistical analysis was performed using Kruskal-Wallis one-way ANOVA with Dunn’s post-hoc test, n.s. non-significant, ** p<0.005, *** p<0.0005, **** p<0.0001 (n=13 for cTnI, n=31 for C0C1f and n=4 for LPS). b–c) Murine macrophages were treated for the indicated lengths of time with 500 ng/ml C0C1f and for 6 h with C0-L or LPS. Western blot analysis was used for determination of protein levels of b) IL-1β and c) TNFα. Depicted is the mean ± SEM of n=3 individual experiments. Statistical analysis was performed using the Mann Whitney U test, comparing each sample individually with control (p>0.07). d) Structure of cardiac MyBP-C. Calpain-dependent cleavage takes place in the M-domain. Various N-terminal fragments were designed.
3.2 Full-length C0C1f is required to induce the expression of pro-inflammatory response genes in vitro
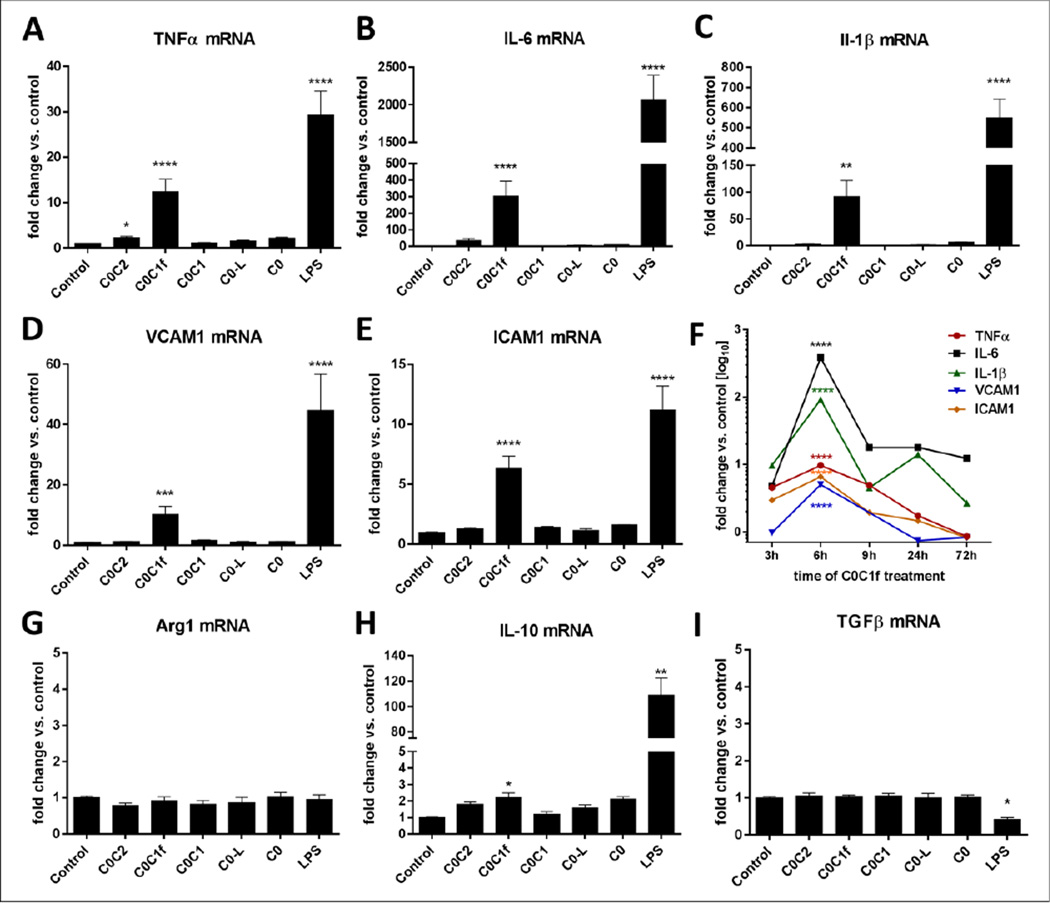
To determine whether the cleaved M-domain of cMyBP-C (C0C1f) specifically leads to the induction of pro-inflammatory responses, various fragments of the N-terminal region of cMyBP-C, in particular recombinant C0C2, C0C1f, C0C1, C0-linker (C0-L), and C0 peptides (Fig. 1d), were used for the treatment (500 ng/ml) of murine bone marrow-derived macrophages. Cells were harvested at different time points (3, 6, 9, 24, or 72 h) and total mRNA was isolated for qRT-PCR analysis to determine the mRNA abundance. Levels of transcripts for inflammatory markers including IL-6, IL-1β, and TNFα as well as the adhesion molecules VCAM1 and ICAM1 were determined. C0C1f significantly induced transcription of pro-inflammatory markers and adhesion markers compared with untreated cells (TNFα: 12.4 ± 2.8 fold, IL-6: 303 ± 90 fold, IL-1β: 91 ± 30 fold, VCAM1: 10.2 ± 2.7 fold, ICAM1: 6.3 ± 1 fold) (Fig 2a–e). C0, C0-L and C0C1 did not significantly increase the abundance of IL-6, IL-1β, or TNFα mRNA. C0C2 significantly induced TNFα mRNA abundance (2.2-fold). After 9 h of treatment the amount of pro-inflammatory and adhesion target transcripts was reduced compared with the 6-h time point. IL-1β transcription was slightly upregulated after 24 h of treatment, and only IL-6 mRNA persisted at increased levels (12–18 fold) for 72 h (Fig. 2f). The induction of a pro-inflammatory response by C0C1f was confirmed at the protein level for IL-1β and TNFα (Fig. 1b, c, Fig. 4h). To determine whether C0C1f affects the expression of anti-inflammatory or pro-fibrotic cytokines in murine macrophages, we also determined the mRNA abundance of IL-10, arginase 1 (Arg1), and transforming growth factor β (TGFβ). Only the expression of IL-10, an anti-inflammatory marker of macrophages that indicates differentiation towards the M2 phenotype, was significantly induced (2-fold increase) by C0C1f. Neither Arg1 nor TGF-β were affected by treatment with any of the N-terminal fragments.
Figure 2. Validation of pro- and anti-inflammatory responses upon treatment with N-terminal cMyBP-C fragments.
a–e) Murine macrophages were treated for 6 h with 500 ng/ml C0C2, C0C1f, C0C1, or C0-L fragments, or 1 µg/ml LPS. mRNA levels of TNFα, IL-6, IL-1β, VCAM1, and ICAM1 were measured by qRT-PCR. f) Murine macrophages were treated with C0C1f peptide for 3, 6, 9, 24, or 72 h. g–i) Murine macrophages were treated for 6 h with C0C2, C0C1f, C0C1, or C0-L fragment and LPS; mRNA levels of Arg1, IL-10, and TGFβ were measured by qRT-PCR. Values shown are mean ± SEM; statistical analysis was performed using Kruskal-Wallis one-way ANOVA with Dunn’s post-hoc test, * p< 0.05, ** p<0.005, *** p<0.0005, **** p<0.0001 (C0: n=4, C0C2 and C0-L: n=11; C0C1: n=18, C0C1f: n=21, LPS: n=6).
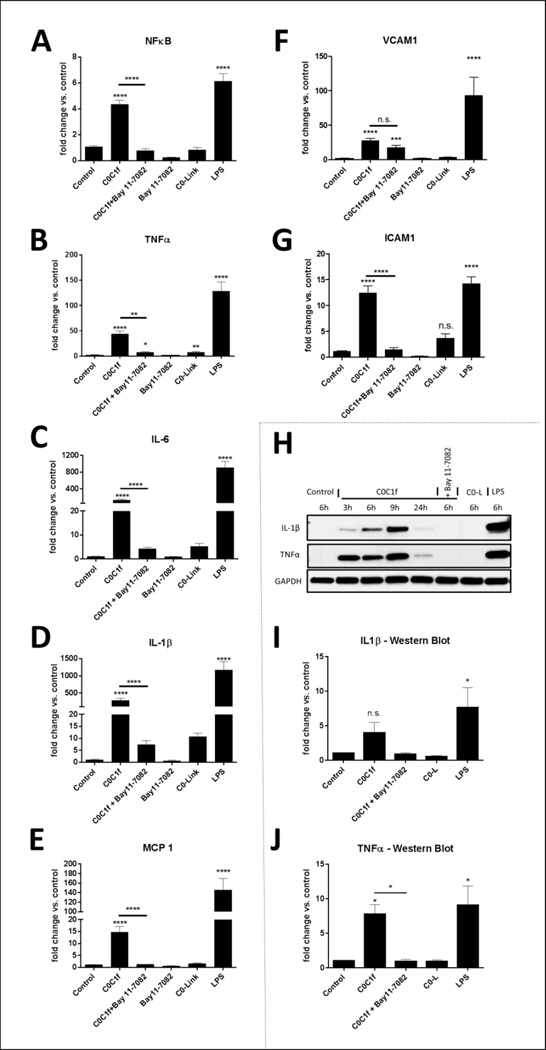
Figure 4. C0C1f induces pro-inflammatory cytokine expression via activation of the NFκB signaling pathway.
Murine macrophages were treated with C0C1f, LPS, or C0-L for 6 h. The NFκB activation was inhibited by 24-h pre-treatment (18 h prior to co-treatment with peptides or LPS) with Bay11–7082. qRT-PCR was performed to determine the gene expression levels of NFκB (a) and the pro-inflammatory cytokines TNFα (b), IL-6 (c), IL-1β (d), and MCP1 (e) as well as adhesion/migration molecules VCAM1 (f) and ICAM1 (g). Values shown are mean ± SEM; statistical analysis was performed using one-way ANOVA with Tukey’s post-hoc test, (Bay11–7082: n=9, LPS, C0C1f+ Bay11–7082 and C0-L: n=28, C0C1f: n=38, n.s. not significant, *** p<0.005, **** p <0.0001). Protein expression of IL-1β (h–i) and TNFα (h, j) with or without 24-h Bay 11–7082 treatment was analyzed by western blotting. Depicted are the quantified data for 6-h treatment with C0C1f, C0C1f + Bay11–7082, LPS, and C0-L (n=4 independent experiments) (i, j). Values shown are mean ± SEM; statistical analysis was performed using the Mann Whitney U test, comparing each sample individually with control, * p<0.05.
3.3 C0C1f induces pro-inflammatory but not anti-inflammatory responses in human monocytes
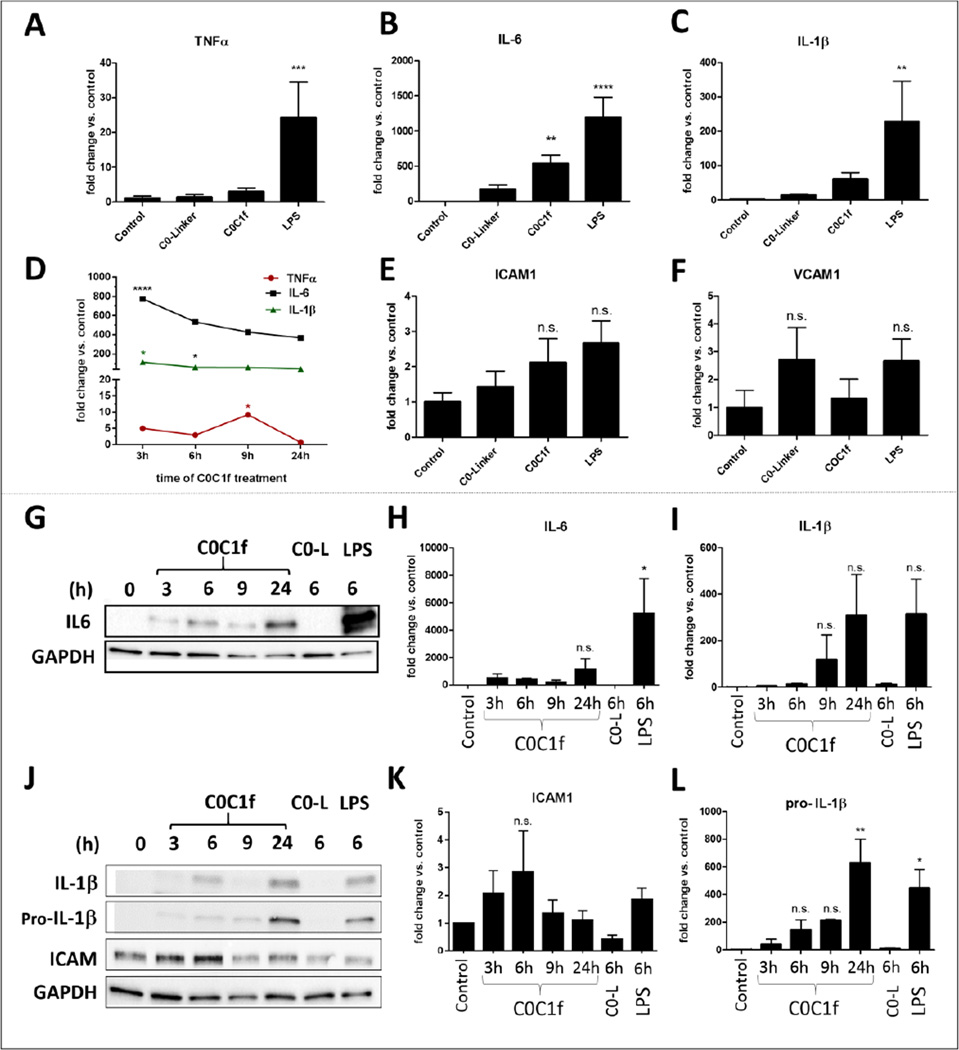
To determine whether there is an effect of C0C1f on human monocytes that is comparable to the observations made in murine macrophages, similar experiments were performed using monocytes isolated from buffy coats of healthy human individuals. One day after isolation, the cells were treated with 500 ng/ml C0C1f or C0-L for 3, 6, 9, and 24 h. Treatment with lipopolysaccharide (LPS) from Escherichia coli served as positive control. Total mRNA was isolated and analyzed by qRT-PCR. Six hours after treatment with C0C1f or LPS transcription of IL-6 was significantly upregulated (for C0C1f: 533 ±124 fold) as were transcripts of IL-1β and TNFα (for C0C1f: IL-1β: 61 ± 20 fold, TNFα: 3 ± 1 fold) (Fig. 3a–c). Lower concentrations of C0C1f, e.g. 50 ng/ml, also induced the transcriptions of IL-6 (Supplementary Figure 6). Treatment with C0-L did not significantly induce the upregulation of any pro-inflammatory target (Fig. 3a–c). C0C1f induced pro-inflammatory upregulation within 3 h of treatment and the abundance of IL-6 and IL-1β transcripts persisted over the 24-h experimental period. A weak induction of TNFα was observed after 3 h with a maximum of 9-fold increase after 9 h of treatment (Fig. 3d). The abundance of transcripts of the adhesion molecules ICAM1 and VCAM1 was not significantly affected by LPS, C0C1f, or C0-L treatment.
Figure 3. C0C1f induces pro-inflammatory responses in human monocytes.
a–e) Human monocytes were isolated from buffy coats of healthy donors and were treated with C0-L, C0C1f, or LPS for 6 h. Thereafter, mRNA was isolated and levels of TNFα, IL-6, IL-1β, VCAM1, and ICAM1 transcripts were measured by qRT-PCR. Values shown are mean ± SEM; statistical analysis was performed using the one-way ANOVA with Tukey’s post-hoc test, n= 10, * p<0.05, ** p<0.005, *** p<0.0005 and **** p<0.0001. f) Human monocytes were treated with C0C1f peptide for the indicated time periods and levels of TNFα, IL-6, IL-1β, VCAM1, and ICAM1 were measured by qRT-PCR. (n=3 donors) g–i) Protein levels of pro-inflammatory cytokines were detected by western blot analysis (g, j) and quantified (h, i, k, l). Values shown are mean ± SEM; statistical analysis was performed using one-way ANOVA with Tukey’s post-hoc test, * p<0.05, ** p<0.005
For further validation, the markers were determined at the protein level by western blot analysis. IL-6 protein levels increased within 3 h after initial treatment of C0C1f and remained elevated until 24-h time point (Fig. 3g, h). Treatment with C0-L did not induce IL-6 protein expression (Fig. 3g, h). The levels of pro-IL-1β and IL-1β protein were increased over time, with the highest levels being observed after 24 h of C0C1f treatment (Fig. 3i, l). The protein levels of IL6 and IL-1β were also upregulated; however, significant upregulation of proteins was observed only upon LPS stimulation for IL-6 (Fig. 3g–l). The adhesion protein ICAM1 was slightly (~3-fold) upregulated upon 6 h of treatment with C0C1f (Fig. 3k). The expression of IL-10 was significantly upregulated (15-fold) by treatment with LPS. Treatment with C0C1f induced IL-10 expression slightly (5-fold increase). C0-L also caused a weak increase in IL-10 expression (~3-fold increase) (Supplementary Fig. 2a). TGF-β protein levels were not affected by the treatment with C0C1f, C0-L, or LPS (Supplementary Fig. 2b).
3.4 The C0C1f-triggered pro-inflammatory response is mediated via NFκB signaling
To investigate the extra- and intracellular signaling of macrophages upon C0C1f stimulation and to determine whether the inflammatory phenotype induced upon C0C1f treatment is dependent on NFκB signaling, we inhibited the NFκB signaling pathway. NFκB plays a crucial role for the expression of pro-inflammatory genes [16]. Under normal conditions NFκB is bound to inhibitory molecules such as IκB that are phosphorylated upon activation of pro-inflammatory signals releasing NFκB which translocates into the nucleus. Murine macrophages differentiated from freshly isolated bone marrow cells were treated with Bay 11–7082 (InvivoGen, San Diego, USA), an irreversible inhibitor of IKKα and the phosphorylation of IκBα resulting in a stable inhibition of NFκB activation and nuclear translocation [17]. The treatment was performed for 16–18 h prior to the co-treatment with C0C1f or C0-L; co-treatment with LPS served as a positive control. The cells were harvested at different time points (0.5, 1, 1.5, 2, 3, 4, 5, and 6 h) after addition of the peptides and mRNA and protein levels were analyzed. We investigated the expression levels of NFκB itself and the expression of pro-inflammatory cytokines TNFα, IL-6, and IL-1β.
At the mRNA level we observed that NFκB expression was induced after 1 h of C0C1f treatment and peaked at 3 h, at which time there was a 6-fold response (Supplementary Fig. 3). At 6 h NFκB was still significantly upregulated by 4-fold (Fig. 4a). In addition, the abundance of IL-6, IL-1β, and TNFα mRNA was significantly increased upon C0C1f treatment (TNFα: 44 ± 6.1 fold, IL-6: 121 ± 30 fold, Il-1β: 282.5 ± 60 fold) (Fig. 4b–d and cf. Fig. 2). The upregulation of these cytokines was abrogated upon treatment with the IκBα phosphorylation inhibitor Bay11–7082 (Fig. 4b–d). The induction of ICAM1 by C0C1f was significantly diminished by the inhibitor (Fig. 4g). Transcription of MCP1 and VCAM1 were not significantly induced by C0C1f treatment, so that an inhibitory effect by Bay11–7082 was not observed (Fig. 4e, f). These observations were confirmed at the protein level: TNFα and IL-1β were upregulated upon 3 h of treatment with C0C1f and the upregulation was observed for all time points investigated (Fig. 1b, c). Upregulation of both IL-1β and TNFα proteins was abrogated upon co-treatment with Bay11–7082 (Fig. 4h–j).
3.5 C0C1f elicits its response via the TLR2, TLR4, and RAGE signaling pathway
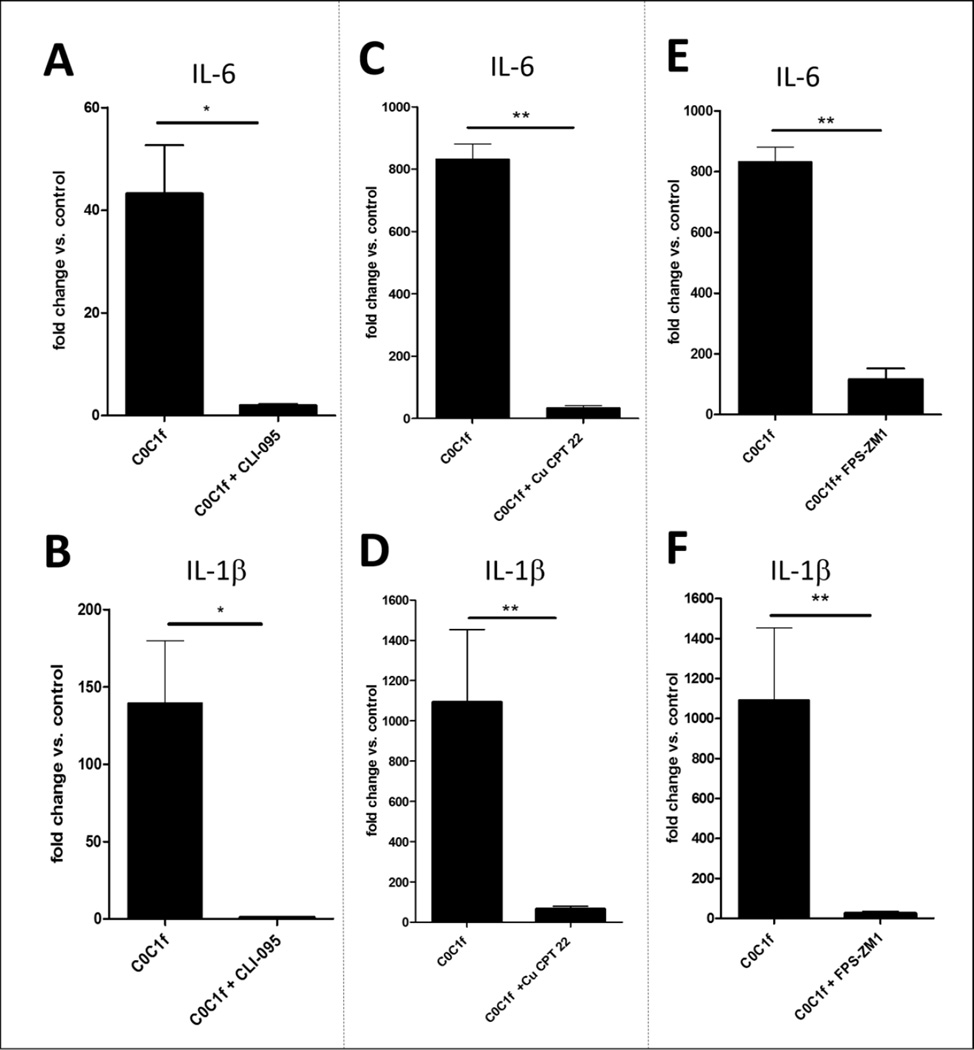
Various pathogen-associated molecular patterns (PAMPs) or DAMPs induce pro-inflammatory responses via TLRs. To investigate NFκB upstream signaling we inhibited different TLRs and RAGE. For validation, cells were treated with LPS and the various inhibitors (Supplementary Fig. 7). TLR4 was inhibited by treatment of the cells using CLI-095 (InvivoGen), a TLR4-specific inhibitor [18], 16–18 h prior to the stimulation with C0C1f. CLI-095 inhibited the stimulatory effect of C0C1f on transcript levels of IL-6 and IL-1β, an effect that was observed for both murine macrophages and human monocytes (Fig. 5). Induction of IL-6 (43.2 ± 19 fold) and IL-1β (140 ± 80 fold) by C0C1f was almost completely abrogated by the treatment of murine macrophages with CLI-095 (IL-6: 1.9 ± 0.6 fold; IL-1β: 0.95 ± 0.4 fold) (Fig. 5a, b). The data show a reduction of more than 95% for IL-6 and more than 99% for IL-1β induction, which is consistent with a TLR4-dependent activation of NFκB and pro-inflammatory signaling upon C0C1f treatment. Although human monocytes showed a less homogeneous cytokine transcription profile, reduction of C0C1f-dependent induction of IL-6 (50%) and IL-1β (62%) was observed (Supplementary Fig. 4). Further, TLR2 was inhibited by treatment of the cells with 5 µM of a TLR2/TLR1-specific inhibitor, Cu CPT22 (Tocris Bioscience, Bristol, UK), and RAGE was inhibited by treatment of the cells with 10 µg/ml RAGE antagonist, FPS-ZM1 (Calbiochem, Darmstadt, Germany) 16–18 h prior to the stimulation with C0C1f. Induction of both IL-6 and IL-1β by C0C1f were diminished significantly by the treatment with FPS-ZM1 or CU CPT 22 (Fig. 5c–f).
Figure 5. C0C1f elicits its response via TLR4, TLR2, and RAGE.
Murine macrophages were treated with various inhibitors. IL-6 and IL-1β mRNA expression was analyzed by qRT-PCR. Cells were treated with a–b) CLI-095, a TLR4-specific inhibitor (n=4), c–d) FPS-ZM1, a RAGE inhibitor (n=6), or e–f) CU CPT22, a TLR2-specific inhibitor (n=6) for 16 h prior to the 6-h treatment with C0C1f. Values shown are mean ± SEM; statistical analysis was performed using the Mann-Whitney U test, * p<0.05, ** p<0.005.
4 Discussion
Here we demonstrate that C0C1f is an inducer of pro-inflammatory signaling in macrophages from murine and monocytes from human origin. The induced transcription of pro-inflammatory genes upon C0C1f treatment was dependent on NFκB and TLR 2, TLR4, and RAGE. Therefore we conclude that C0C1f, which is released early upon cardiac injury, plays a role in the activation of inflammation and might be responsible for the early innate immune response after myocardial infarction.
The innate immune response is an essential process for wound healing following MI. This process, which is triggered shortly after hypoxia and damage of the affected myocardial tissue, is required in order to prevent ventricular rupture and replace the degraded contractile tissue by a resilient scar; however, inflammatory cells often invade the border zone and locally affect the health of myocardial tissue [19, 20].
The exact molecular mechanism that initiates inflammation following MI is not fully understood. DAMPs are described to play a pivotal role during the activation of inflammatory responses. They represent intracellular molecules that are released from apoptotic or necrotic cells as well as extracellular matrix molecules that are upregulated or degraded during tissue damage [21]. Molecules like high mobility box 1 (HMGB1), heat shock proteins (HSP)s, and ATP are released from apoptotic or necrotic cells and have been shown to be involved in wound healing and scar formation in the damaged myocardium (reviewed in [5]). Necrosis of cardiomyocytes starts as early as 20 min after total occlusion, and DAMPs like HMGB1 can be detected 30 min after ischemia at the mRNA level, with a peak occurring after 6 h [22]. Furthermore, hypoxia-triggered apoptosis of cardiomyocytes peaks at 4.5 h after diminished blood supply, and necrosis peaks at 24 h [23]. An increase in the amounts of infiltrating monocytes, however, can be observed as early as 2 h after coronary artery occlusion [24]. This leads to the notion that mechanisms other than a passive release of DAMPs to the circulation account for the very early innate immune response. For the most part DAMPs (e.g. HMBG1) are released passively into the circulation during the early phase of cardiac injury. In contrast, C0C1f is actively released early upon cardiac injury by a calpain-dependent cleavage and can be detected in blood within 30 min of MI [12].
cMyBP-C is a biomarker of MI and is degraded during MI in association with increased calpain activity, causing proteolytic release of C0C1f fragments into the bloodstream. This event may result in contractile dysfunction [8, 10], leaving the released C0C1f fragments in the cardiomyocytes in the border zone of the ischemic area and in the circulation. The C0C1f region consisting of C0, a Pro-Ala region, the C1 domain, and the first 17 residues of the M domain interacts with actin at the C1 domain and the first 17 residues of the M domain, but it does not interact with the myosin S2 region [8, 10]. Although transgenic expression of recombinant C0C1f in vivo results in heart failure (HF) [8, 13, 25], the impact of C0C1f release still needs to be elucidated at the whole-heart level, as several hypertrophic cardiomyopathy mutations localize in the N’-region [26]. It follows that understanding the mechanisms of cMyBP-C regulation and the release of its fragments into the blood is crucial to the development of agents that promote contractile function in HF. It is important to point out that C0C1f was initially identified as a biomarker of MI [11, 12, 27].
Furthermore, the notion that circulating autoimmune antibodies to some cardiac proteins, including released cMyBP-C, can induce acute myocarditis followed by chronic dilated cardiomyopathy has been well studied in animal models [28, 29]. The cMyBP-C antibodies detected by Kasahara et al. were highly reactive to residues 205–916 and weakly reactive with residues 945–1270 of murine cMyBP-C [30]. The immunogenic region of cMyBP-C that could induce myocarditis in immunized mice was found to span residues 205–916 but not the more C-terminal residues [30]. However, the immunogenicity of the N-terminal residues from domains C0 through an early portion of C1 in murine cMyBP-C was not addressed. Whereas the C0–C1f region (residues 1–271) of cMyBP-C could be immunogenic and a target for autoantibodies following ischemic injury, our study focused on the innate response of the immune system to C0C1f and not the adaptive immune response. The detection of C0C1f-targeted autoantibodies and the role such antibodies may play following ischemic injury as part of the adaptive immune response will be the focus of our future studies.
In contrast to many other biomarkers (e.g cTnI), the release of C0C1f is a controlled process involving the dephosphorylation of cMyBP-C and its calpain-dependent degradation upon cardiac injury [9]. Furthermore, it was shown that C0C1f release affects cardiomyocytes by competing with full-length cMyBP-C for binding to actin, disrupting the interaction of native cMyBP-C with the thin filament and leading to decreased Ca2+ transients and impaired contractility [13]. Only the dephosphorylated form of cMyBP-C is cleaved by calpain at S273. Cleavage results in the loss of S273 and all PKA phosphorylation sites on C0C1f; thus phosphorylation of C0C1f would not be expected to play a role in the modulation of immune responses.
To elucidate the functional role of C0C1f we focused on primary cells in vitro. Even though this bears the limitation of transferability to the in vivo situation and the isolation procedure might be responsible for experimental artefacts, primary cells are still considered the best standardized method for validating complex signaling pathways, as they still display many native features due to their proximity to the native environment [31]. To identify the immune-modulating role of C0C1f we focused on murine macrophages derived from bone marrow stem cells and on human blood monocytes; these primary cultures, however, would not be expected to recapitulate the heterogeneity of these cell populations observed in vivo. Thus, the diversity of macrophage and monocyte subsets [32, 33] in heart and their functional heterogeneity must be addressed in further studies.
Upon treatment of macrophages with C0C1f, we observed a strong upregulation of pro-inflammatory genes which was not detected by applying other N-terminal fragment of cMyBP-C. One could speculate that the cleavage at S273 leads to specific structural changes that are responsible for these targeted inflammatory responses. We demonstrated that C0C1f activates NFκB in murine macrophages and human monocytes, leading to the induction of pro-inflammatory cytokine expression. Interestingly, the significant induction of VCAM1 and ICAM1 observed in murine macrophages was not observed in monocytes of human origin. One can speculate that in cells of human origin additional factors are required for the activation of these important adhesion and migration proteins. In addition we observed significant induction of pro-inflammatory markers on mRNA levels and clear tendency of induction on protein levels (Fig. 1b,c and Fig. 2a,c).
The upregulation of pro-inflammatory cytokines was abrogated by the addition of the IKBα-inhibitor Bay11–7082 (Fig. 4), which has been shown to inhibit the TNFα-dependent phosphorylation of IκBα, resulting in reduced translocation of p65 and therefore decreased nuclear NFκB levels [17]. NFκB signaling is dependent on the activation of TLR2, TLR4 and RAGE. Using inhibitors of these proteins we demonstrated that TLR signaling is responsible for the C0C1f-dependent activation of pro-inflammatory signaling (Fig. 5). This observation supports the idea of an interaction between C0C1f with various other cell types expressing TLR4. In primary rat cardiomyocytes, however, we did not find any upregulation of pro-inflammatory cytokines upon C0C1f treatment (Supplementary Fig. 5a). We also investigated the effect of C0C1f on fibroblasts and endothelial cells and did not detect an upregulation of pro-inflammatory cytokines (Supplementary Fig. 5b, c). From these results it appears that C0C1f does not trigger an immune response in these other cell types. The first cells to infiltrate the tissue are neutrophils that, in addition to macrophages, may also be responsive to C0C1f and trigger early pro-inflammatory reactions. Due to the lack of adequate primary cultures of neutrophils this needs to be addressed in future in vivo studies.
An anti-inflammatory role of C0C1f was not detected in any cell type; however, we observed a minor upregulation of IL-10 (Fig. 2, Supplementary Fig. 2), which has been described as participating in a negative feedback loop [34]. The fact that the inhibition of TLR2 or RAGE abrogated the C0C1f induced pro-inflammatory response might indicate that C0C1f interacts directly or in-directly with various receptors to induce pro-inflammatory signaling via the NFκB pathway. Similar findings were made for HMGB1, which is able to interact with various TLRs and RAGE and can also from complexes with TLR ligands such as Pam3CSK4 or CpG-A [35]. Unfortunately, these interactions are still not well understood and further investigations are needed.
5 Conclusions
Taken together, our results demonstrate that C0C1f triggers the TLR2-, TLR4-, and RAGE-dependent activation of NFκB in macrophages, which leads to the upregulation of pro-inflammatory cytokines (IL-1β, IL-6, TNFα). The active calcium/calpain-dependent release of C0C1f into the circulation very early after hypoxia during MI suggests that C0C1f plays a role during myocardial inflammatory and remodeling processes. If C0C1f proves to be essential for the innate immune response after MI, this could serve as the basis for immune-modulating therapies after MI.
Supplementary Material
Highlights.
Pro-inflammatory response to released N-terminal fragment of cMyBP-C (C0C1f).
Monocyte activation by C0C1f is highly dependent on NFκB activation.
C0C1f induces pro-inflammatory responses via TLR2, TLR4, and RAGE.
C0C1f as novel key player in inflammatory processes upon myocardial infarction?
Acknowledgments
The authors were supported by German Center for Cardiovascular Disease (DZHK) (BMBF 81Z1200201), National Institutes of Health grants R01HL130356, R01HL105826 and K02HL114749 (S.S.) and American Heart Association Midwest Affiliate Research Programs; Cardiovascular Genome-Phenome Study (15CVGPSD27020012), Grant-in-Aid (14GRNT20490025) and Predoctoral Fellowships 15PRE22430028 (T.L.L) and the Kerckhoff Institute for Heart Research (KHFI). The authors would like to thank Behnoush Parviz for her expert assistance in the laboratory and Elizabeth Martinson for the editing of the manuscript.
Glossary
- cMyBP-C
cardiac Myosin Binding Protein-C
- cTnI
cardiac Troponin I
- DAMPs
Damage-Associated Molecular Patterns
- HF
Heart Failure
- MI
Myocardial Infarction
- PAMPs
Pathogen-Associated Molecular Patterns
- RAGE
Advanced Glycosylation End Product-Specific Receptor
- TLR
Toll-Like Receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: On behalf of all other authors, the corresponding author states that there is no conflict of interest
References
- 1.Frangogiannis NG. The immune system and the remodeling infarcted heart: cell biological insights and therapeutic opportunities. J Cardiovasc Pharmacol. 2014;63:185–195. doi: 10.1097/FJC.0000000000000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dutta P, Nahrendorf M. Monocytes in myocardial infarction. Arterioscler Thromb Vasc Biol. 2015;35:1066–1070. doi: 10.1161/ATVBAHA.114.304652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hulsmans M, Sam F, Nahrendorf M. Monocyte and macrophage contributions to cardiac remodeling. J Mol Cell Cardiol. 2015 doi: 10.1016/j.yjmcc.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Troidl C, Jung G, Troidl K, Hoffmann J, Mollmann H, Nef H, et al. The temporal and spatial distribution of macrophage subpopulations during arteriogenesis. Curr Vasc Pharmacol. 2013;11:5–12. [PubMed] [Google Scholar]
- 5.de Haan JJ, Smeets MB, Pasterkamp G, Arslan F. Danger signals in the initiation of the inflammatory response after myocardial infarction. Mediators Inflamm. 2013;2013:206039. doi: 10.1155/2013/206039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Land WG. The Role of Damage-Associated Molecular Patterns (DAMPs) in Human Diseases: Part II: DAMPs as diagnostics, prognostics and therapeutics in clinical medicine. Sultan Qaboos Univ Med J. 2015;15:e157–e170. [PMC free article] [PubMed] [Google Scholar]
- 7.Jacquet S, Yin X, Sicard P, Clark J, Kanaganayagam GS, Mayr M, et al. Identification of cardiac myosin-binding protein C as a candidate biomarker of myocardial infarction by proteomics analysis. Mol Cell Proteomics. 2009;8:2687–2699. doi: 10.1074/mcp.M900176-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Govindan S, Sarkey J, Ji X, Sundaresan NR, Gupta MP, de Tombe PP, et al. Pathogenic properties of the N-terminal region of cardiac myosin binding protein-C in vitro. J Muscle Res Cell Motil. 2012;33:17–30. doi: 10.1007/s10974-012-9292-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Govindan S, McElligott A, Muthusamy S, Nair N, Barefield D, Martin JL, et al. Cardiac myosin binding protein-C is a potential diagnostic biomarker for myocardial infarction. J Mol Cell Cardiol. 2012;52:154–164. doi: 10.1016/j.yjmcc.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weith A, Sadayappan S, Gulick J, Previs MJ, Vanburen P, Robbins J, et al. Unique single molecule binding of cardiac myosin binding protein-C to actin and phosphorylation-dependent inhibition of actomyosin motility requires 17 amino acids of the motif domain. J Mol Cell Cardiol. 2012;52:219–227. doi: 10.1016/j.yjmcc.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker JO, Tyther R, Liebetrau C, Clark J, Howarth R, Patterson T, et al. Cardiac myosin-binding protein C: a potential early biomarker of myocardial injury. Basic Res Cardiol. 2015;110:23. doi: 10.1007/s00395-015-0478-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lynch TL, Sadayappan S. Surviving the infarct: A profile of cardiac myosin binding protein-C pathogenicity, diagnostic utility, and proteomics in the ischemic myocardium. Proteomics Clin Appl. 2014;8:569–577. doi: 10.1002/prca.201400011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Witayavanitkul N, Ait Mou Y, Kuster DW, Khairallah RJ, Sarkey J, Govindan S, et al. Myocardial infarction-induced N-terminal fragment of cardiac myosin-binding protein C (cMyBP-C) impairs myofilament function in human myocardium. J Biol Chem. 2014;289:8818–8827. doi: 10.1074/jbc.M113.541128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zanoni I, Ostuni R, Capuano G, Collini M, Caccia M, Ronchi AE, et al. CD14 regulates the dendritic cell life cycle after LPS exposure through NFAT activation. Nature. 2009;460:264–268. doi: 10.1038/nature08118. [DOI] [PubMed] [Google Scholar]
- 15.Copeland O, Sadayappan S, Messer AE, Steinen GJ, van der Velden J, Marston SB. Analysis of cardiac myosin binding protein-C phosphorylation in human heart muscle. J Mol Cell Cardiol. 2010;49:1003–1011. doi: 10.1016/j.yjmcc.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 16.Lawrence T. The Nuclear Factor NF-κB Pathway in Inflammation. Cold Spring Harb Perspect Biol. 2009;1:a001651. doi: 10.1101/cshperspect.a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pierce JW, Schoenleber R, Jesmok G, Best J, Moore SA, Collins T, et al. Novel inhibitors of cytokine-induced IkappaBalpha phosphorylation and endothelial cell adhesion molecule expression show anti-inflammatory effects in vivo. J Biol Chem. 1997;272:21096–21103. doi: 10.1074/jbc.272.34.21096. [DOI] [PubMed] [Google Scholar]
- 18.Ii M, Matsunaga N, Hazeki K, Nakamura K, Takashima K, Seya T, et al. A novel cyclohexene derivative, ethyl (6R)-6-[N-(2-Chloro-4-fluorophenyl)sulfamoyl]cyclohex-1-ene-1-carboxylate (TAK-242), selectively inhibits toll-like receptor 4-mediated cytokine production through suppression of intracellular signaling. Mol Pharmacol. 2006;69:1288–1295. doi: 10.1124/mol.105.019695. [DOI] [PubMed] [Google Scholar]
- 19.Frangogiannis NG. Regulation of the inflammatory response in cardiac repair. Circ Res. 2012;110:159–173. doi: 10.1161/CIRCRESAHA.111.243162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jivraj N, Phinikaridou A, Shah AM, Botnar RM. Molecular imaging of myocardial infarction. Basic Res Cardiol. 2013;109:1–16. doi: 10.1007/s00395-013-0397-2. [DOI] [PubMed] [Google Scholar]
- 21.Piccinini AM, Midwood KS. DAMPening inflammation by modulating TLR signalling. Mediators Inflamm. 2010 doi: 10.1155/2010/672395. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andrassy M, Volz HC, Igwe JC, Funke B, Eichberger SN, Kaya Z, et al. High-Mobility Group Box-1 in Ischemia-Reperfusion Injury of the Heart. Circulation. 2008;117:3216–3226. doi: 10.1161/CIRCULATIONAHA.108.769331. [DOI] [PubMed] [Google Scholar]
- 23.Chiong M, Wang ZV, Pedrozo Z, Cao DJ, Troncoso R, Ibacache M, et al. Cardiomyocyte death: mechanisms and translational implications. Cell Death Dis. 2011;2:e244. doi: 10.1038/cddis.2011.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liebetrau C, Hoffmann J, Dorr O, Gaede L, Blumenstein J, Biermann H, et al. Release kinetics of inflammatory biomarkers in a clinical model of acute myocardial infarction. Circ Res. 2015;116:867–875. doi: 10.1161/CIRCRESAHA.116.304653. [DOI] [PubMed] [Google Scholar]
- 25.Razzaque MA, Gupta M, Osinska H, Gulick J, Blaxall BC, Robbins J. An endogenously produced fragment of cardiac myosin-binding protein C is pathogenic and can lead to heart failure. Circ Res. 2013;113:553–561. doi: 10.1161/CIRCRESAHA.113.301225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris SP, Lyons RG, Bezold KL. In the thick of it: HCM-causing mutations in myosin binding proteins of the thick filament. Circ Res. 2011;108:751–764. doi: 10.1161/CIRCRESAHA.110.231670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marjot J, Liebetrau C, Goodson RJ, Kaier T, Weber E, Heseltine P, et al. The development and application of a high-sensitivity immunoassay for cardiac myosin-binding protein C. Transl Res. 2015 doi: 10.1016/j.trsl.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richardson P, McKenna W, Bristow M, Maisch B, Mautner B, O'Connell J, et al. Report of the 1995 World Health Organization/International Society and Federation of Cardiology Task Force on the Definition and Classification of cardiomyopathies. Circulation. 1996;93:841–842. doi: 10.1161/01.cir.93.5.841. [DOI] [PubMed] [Google Scholar]
- 29.Caforio AL. Role of autoimmunity in dilated cardiomyopathy. Br Heart J. 1994;72:S30–S34. doi: 10.1136/hrt.72.6_suppl.s30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kasahara H, Itoh M, Sugiyama T, Kido N, Hayashi H, Saito H, et al. Autoimmune myocarditis induced in mice by cardiac C-protein. Cloning of complementary DNA encoding murine cardiac C-protein and partial characterization of the antigenic peptides. J Clin Invest. 1994;94:1026–1036. doi: 10.1172/JCI117416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lipps C, May T, Hauser H, Wirth D. Eternity and functionality - rational access to physiologically relevant cell lines. Biol Chem. 2013;394:1637–1648. doi: 10.1515/hsz-2013-0158. [DOI] [PubMed] [Google Scholar]
- 32.Lavine KJ, Epelman S, Uchida K, Weber KJ, Nichols CG, Schilling JD, et al. Distinct macrophage lineages contribute to disparate patterns of cardiac recovery and remodeling in the neonatal and adult heart. Proc Natl Acad Sci U S A. 2014;111:16029–16034. doi: 10.1073/pnas.1406508111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cohen Heather B, Mosser David M. Cardiac Macrophages: How to Mend a Broken Heart. Immunity. 2014;40:3–5. doi: 10.1016/j.immuni.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 34.Platzer C, Meisel C, Vogt K, Platzer M, Volk H-D. Up-regulation of monocytic IL-10 by tumor necrosis factor-α and cAMP elevating drugs. Int Immunol. 1995;7:517–523. doi: 10.1093/intimm/7.4.517. [DOI] [PubMed] [Google Scholar]
- 35.Ibrahim ZA, Armour CL, Phipps S, Sukkar MB. RAGE and TLRs: Relatives, friends or neighbours? Mol Immunol. 2013;56:739–744. doi: 10.1016/j.molimm.2013.07.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.