Abstract
PEGylation is an important strategy for enhancing the pharmacokinetic properties of protein therapeutics. The development of chemoselective side-chain modification reactions has enabled researchers to PEGylate proteins with high selectivity at defined locations. However, aside from avoiding active sites and binding interfaces, there are few guidelines for the selection of optimal PEGylation sites. Because conformational stability is intimately related to the ability of a protein to avoid proteolysis, aggregation, and immune responses, it is possible that PEGylating a protein at sites where PEG enhances conformational stability will result in PEG-protein conjugates with enhanced pharmacokinetic properties. However, the impact of PEGylation on protein conformational stability is incompletely understood. This review describes recent advances toward understanding the impact of PEGylation on protein conformational stability, along with the development of structure-based guidelines for selecting stabilizing PEGylation sites.
Introduction
Peptides and proteins are attractive targets for treatment of many human diseases [1,2]. However, their benefits are limited because of fast degradation by proteases, filtration through the kidneys, aggregation, and recognition/neutralization by antibodies [3,4]. In the late 1970s, Davis and Abuchowsky [5,6] demonstrated that non-specific covalent modification of a protein with a polyethylene glycol (PEG) electrophile resulted in a less immunogenic PEG-protein conjugate with extended serum half-life. Subsequently, PEGylation was found to reduce protein aggregation and proteolysis, and to increase protein shelf-life [4,7–9]. Accordingly, PEGylation is now a widely used strategy for enhancing the pharmacokinetic properties of therapeutically relevant proteins, and several PEGylated proteins are now clinically available, including bovine adenosine deaminase, L-asparaginase, interferon-α, granulocyte colony stimulating factor, growth hormone receptor antagonist, urate oxidase, and the Fab fragment of a monoclonal antibody against tumor necrosis factor-α [10].
The extended serum half-life of PEGylated proteins is thought to derive from the large hydrodynamic radius of PEG, which decreases renal clearance by preventing large PEG-protein conjugates from being filtered through the pores of the glomerular wall. The large PEG is also thought to shield the protein from proteases and antibodies via simple steric occlusion. In many cases, a therapeutic PEG-protein conjugate is less active than its non-PEGylated counterpart in in vitro biochemical assays, presumably because the attached PEG also limits the access of substrates or binding partners to the protein surface/active site. However, in many cases, the extended serum half-life of the PEG-protein conjugate compensates for this modest loss of activity, resulting in a therapeutic protein that can be administered less frequently and with fewer side effects than its non-PEGylated counterpart.
Site-specific PEGylation
Initial efforts to generate PEG-protein conjugates depended on reactions with limited chemoselectivity: protein surface nucleophiles (i.e. cysteines, lysines, etc.) were non-specifically modified with various PEG electrophiles, including chlorotriazines [6], succinimides [11], maleimides [12], and aldehydes [13], generally resulting in a heterogeneous mixture of PEG-protein conjugates that differed in the number, location, and occupancy of PEGylation sites. However, the discovery of new bioorthogonal reactions in parallel with the development of strategies for incorporating correspondingly functionalized unnatural amino acids into proteins now allows relatively good control of the site and degree of PEGylation.
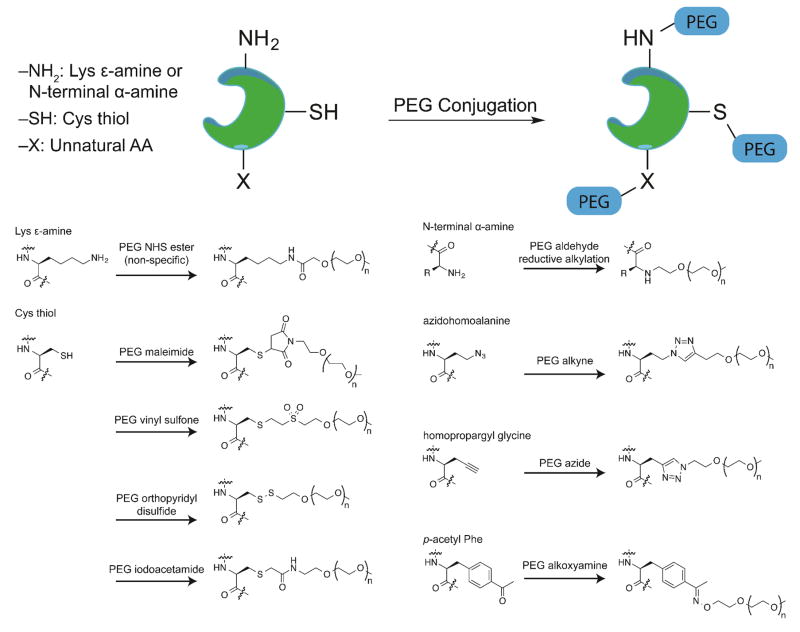
For example (see figure 1), under mildly acidic conditions, a PEG aldehyde can be appended selectively to the N-terminal α-amino group of a protein via reductive alkylation, owing to pKa differences between the N-terminal 7agr;-ammonium and Lys ε-ammonium groups [14]. Cys residues can be targeted selectively via a number of approaches owing to the high nucleophilicity of the Cys thiol: these include conjugate addition to PEG maleimides and PEG vinyl sulfones; disulfide formation with PEG ortho-pyridyl disulfides; and nucleophilic substitution with PEG iodoacetamides [15]. Alternatively, bisalkylation of the Cys side-chain thiol, followed by spontaneous elimination gives dehydroalanine, which can be subsequently modified via conjugate addition with a PEG thiol, following the elegant approach of Davis and coworkers [16]. One potential complication of the conjugate addition approaches is that the products are generally diastereomeric mixtures that differ in the stereochemistry at one of the atoms of the original Michael acceptor; however the significance of this stereochemical hetereogeneity has not been explored experimentally.
Figure 1.
Examples of natural and unnatural amino acids used in protein-PEG conjugation.
In addition, unnatural amino acids bearing orthogonally reactive functional groups (i.e. azides, alkynes, ketones, and alkenes) can be incorporated into expressed proteins as methionine surrogates in auxotrophic bacterial strains (e.g. azidohomoalanine, homopropargylglycine, etc.) [17–19] or via amber suppression (e.g. p-azidophenylalanine, p-propargyloxyphenylalanine, p-acetylphenylalanine, etc.) [20–26]. These unnatural side chains can then be selectively modified with an appropriate PEG reagent to give a PEG-protein conjugate with PEGs installed at defined positions. For example (see figure 1), an azide or alkyne-functionalized side-chain can be modified with an alkyne- or azide-PEG, respectively, via the copper-catalyzed azide/alkyne cycloaddition to give a triazole-linked PEG-protein conjugate [27]. Alternatively, a ketone-functionalized side-chain can be modified with a PEG alkoxyamine to give an oxime-linked PEG-protein conjugate [24].
Alternatively, efforts to incorporate PEGylated unnatural amino acids directly into proteins have experienced some recent success. Shozen and Zang [28,29] recently incorporated PEGylated p-aminophenylalanine (PEGnAF) and lysine (PEGnLys) derivatives having 4, 8, and 12 PEG units into streptavidin using a frameshift suppression method based on a four-base codon-anticodon pair. A longer 24-unit PEG could also be incorporated, but at significantly reduced yields. These early results are encouraging, but if this method is to be used more broadly, it will need to provide access to the larger PEGs (~20–40 kDa) that are more typically used in therapeutic contexts.
Identifying Optimal PEGylation Sites
With access to many methods for site-specific PEGylation, researchers now face the prospect of deciding which of the many possible PEGylation sites is likely to provide optimal pharmacokinetic benefits. Aside from avoiding enzyme active sites or protein-protein binding interfaces, the current empirical approach involves scanning through the many possible PEGylation sites and picking the one that best balances enhanced pharmacokinetic properties with protein function. However, many of the pharmacokinetic challenges encountered by proteins are related to protein conformational stability: a protein aggregates [30], elicits immune responses [31], and is proteolyzed more readily [32] if its folding energy landscape allows a significant population of unfolded, misfolded, or partially folded conformations. Therefore, it is possible that PEGylation at locations where the PEG enhances protein conformational stability will provide the resulting PEG-protein conjugate with enhanced protection from aggregation, proteolysis, and immunogenicity. If such sites could be selected rationally using structure-based criteria, it might be possible to eliminate a substantial amount of time-consuming trial and error from the development of PEGylated protein therapeutics.
How PEG Enhances Protein Conformational Stability
Previous observations indicate that PEGylation can increase [33–40], decrease [41,42], or have no effect on conformational stability [33,43,44]; and in some cases, conflicting reports indicate different outcomes for the same protein [45–48]. The molecular basis for these differences is unclear. However, recent experiments and computational simulations have begun to shed light on how and in what structural context PEGylation can lead to substantial increases in protein conformational stability.
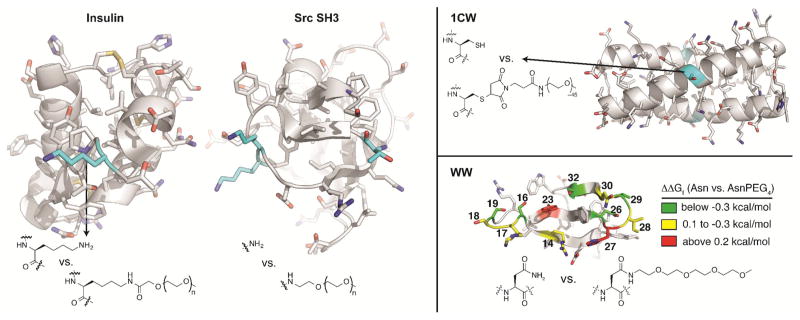
Yang et al [49] used molecular dynamics simulations to investigate the impact of PEGylation on the conformational stability of insulin. In these simulations, PEGs of various sizes (i.e., 10, 50, 100, and 200 ethylene oxide units) were appended to insulin via an amide bond with the ε-amino group of LysB29 (see figure 2). PEG did not substantially change the secondary structure of the simulated PEG-insulin conjugate relative to unmodified insulin, but did substantially reduce the solvent accessible surface area (SASA) of insulin. Analysis of the radial distribution function of water around PEGylated vs. non-PEGylated insulin indicated that PEG excludes water from the surface of insulin, due to PEG-protein interactions, including hydrogen bonds and hydrophobic interactions. These effects were dependent on PEG length to a point, but increasing PEG molecular weight beyond 4000 Da did not result in any additional changes.
Figure 2.
Examples of PEGylated proteins. PEGylated insulin at postion LysB29. The tri-PEGylated SH3 at positions Lys19, Lys20, and N-terminus. PEGylated 1CW at position Cys14. Ribbion diagram of the WW with side-chains shown as sticks. Positions where we incorporated Asn vs AsnPEG4 are highlighted with color, according to the impact of PEGylation on conformational stability.
Xu and coworkers [50] functionalized the trimeric α-helical coiled coil 1CW with a 2 kDa PEG maleimide at a solvent exposed Cys located midway between the N- and C-termini of 1CW(see figure 2). CD experiments demonstrated that PEGylation enhances the helicity of 1CW and does not disrupt its ability to form a coiled coil. Small-angle X-ray scattering studies revealed that the PEG within PEGylated 1CW is more compact than would be expected for a corresponding free PEG [39,51]. Molecular dynamics simulations provided evidence for stabilizing interactions of the PEG oxygen atoms with solvent exposed Lys residues on 1CW, along with interactions between PEG and hydrophobic residues on the coiled-coil surface, with accompanying decreases in SASA [37,52]. However, careful analysis of variable temperature CD data for 1CW and its PEGylated counterpart by Pandey et al [53] found that PEGylation actually destabilizes the 1CW coiled coil by a small but measurable amount. Alanine mutagenesis experiments confirmed the presence of favorable interactions between the Cys-linked PEG and Lys residues at positions 15 and 21 of 1CW, though such interactions are not favorable enough to overcome the intrinsically destabilizing impact of PEGylation on 1CW [53]. However, installing additional Lys residues closer to the PEGylation site (i.e., at the i+3 and i+4 positions relative to the PEGylated Cys) results in a 1CW variant that is stabilized by PEGylation (ΔΔGf = −0.28 kcal/mol) owing to a favorable three-way interaction between the two Lys residues and the PEGylated Cys. Interestingly, this interaction can occur even when Cys is modified with a maleimide derivative that lacks PEG, indicating that the three-way interaction involves the maleimide group and not PEG [53].
Meng et al [54] used stopped-flow and equilibrium denaturation experiments to study the stability of two PEGylated SH3 variants relative to their non-PEGylated counterpart. A mono-PEGylated SH3 variant was generated selectively via reductive alkylation of the N-terminal α-amine with a PEG-aldehyde. A tri-PEGylated SH3 variant was prepared under non-selective reductive alkylation conditions with the same PEG-aldehyde, which modified the N-terminal α-amine along with two Lys ε-amines (see figure 2). PEGylation at the N-terminus did not substantially change SH3 stability; in contrast, tri-PEGylated SH3 was −0.93 kcal/mol more stable than its non-PEGylated counterpart. This increase in stability is accompanied by a decrease in unfolding rate, along with a decrease in the SASA of the SH3 folded conformation, in agreement with previous studies that PEG excludes water from the protein surface.
Rodriguez-Martinez et al [34,35] found that PEGylation leads to slower global hydrogen/deuterium exchange within α-chymotrypsin, presumably due to PEG-based reductions in structural dynamics. This effect could be consistent with PEG-based desolvation of the protein surface (i.e., a decrease in SASA) resulting in a more compact and less dynamic folded conformation.
It is interesting to consider whether any specific sequence or structural patterns result in these stabilizing PEG-based decreases in SASA. Previous simulations suggested that PEG can engage in both hydrophobic and hydrogen-bonding interactions with groups on the protein surface, though the limited number of atomic-resolution structures of PEGylated proteins make it difficult to know whether these interactions have significant structural or thermodynamic consequences. For example, a small angle x-ray scattering study reported by Svergun et al.[55] suggests that PEG partially covers the surface of PEGylated hemolglobin, and is presumably involved in interactions with surface residues. In contrast, the NMR and crystallography studies of Cattni et al[56] on PEGylated plastocyanin suggest that the PEG and the protein behave as independent domains without structural evidence for strong interactions.
We recently showed that attaching a short PEG (comprising four ethylene oxide units) to a single Asn side chain at position 19 in the WW domain of the human protein Pin 1 increases WW conformational stability by −0.70 ± 0.04 kcal mol−1 due to accelerated folding and slowed unfolding [57] (see figure 2). Shorter PEG chains impart less stability to WW than the four-unit PEG, whereas longer PEG chains provide similar stability. PEG-based stabilization depends highly on the orientation of the side chain at position 19: D-Asn is well tolerated in place of L-Asn at this position, but PEGylation of the D-residue has no effect on WW conformational stability. In principle, such an orientation dependent result could indicate the presence of key PEG-protein interactions that are accessible to PEGylated L-Asn, but not to PEGylated D-Asn. Mutagenesis experiments point to OH groups within nearby Ser, Thr, and Tyr residues as important mediators of PEG-based stabilization, though the experimental results are difficult to rationalize in terms of direct PEG-OH interactions, which are also largely absent from atomistic simulations of PEGylated WW variants.
Asn-PEGylation stabilizes WW at some locations and is destabilizing at others. No specific secondary structural motif appears to be generally amenable to PEG-based stabilization: stabilizing and destabilizing positions occur within the reverse turns of WW along with its β-strands. However, the observed stabilizing positions do have a few things in common. First variable temperature CD experiments reveal that PEG-based stabilization of WW is entropic in origin. Assessments of the radial density function of water near stabilizing PEGylation sites of simulated PEG-WW conjugates suggest that PEGylation lowers the density and increases the disorder of water around side-chains that are near the PEGylation site, resulting in partial side-chain desolvation accompanied by the entropically favorable release of water to bulk solvent. Solvent isotope experiments are also consistent with localized disruption of solvent-shell structure by PEG.
A second common feature of stabilizing PEGylation sites within WW is that the wild-type side chains of such stabilizing positions in the crystal structure of the WW tend to be more closely oriented toward nearby OH groups than are side chains at non-stabilizing positions. To quantify this observation, we defined an angle θ, which describes how directly a side chain at a prospective PEGylation site points toward the nearest neighboring OH group. We found that Asn-PEG-based stabilization is more substantial at positions that have smaller values of θ. It is possible that PEG-based surface desolvation is more extensive in the presence of nearby water-binding OH groups, resulting in more entropically favorable release of water to bulk solvent, leading to the observed correlation between θ and PEG-based stabilization.
We next used θ to identify a prospective stabilizing site within the chicken Src SH3 domain. The side chain at position 20 is oriented toward nearby OH-containing Thr22 with θ = 77°. Positions in WW with θ values in this range frequently experienced PEG-based stabilization; therefore, we predicted that Asn-PEGylation at position 20 would enhance SH3 conformational stability. In agreement with this prediction, variable temperature CD experiments demonstrated that the SH3 variant with a four-unit Asn-PEG at position 20 is −1.2 ± 0.1 kcal mol−1 more stable than its non-PEGylated counterpart.
We also found that PEG-based protection from proteolysis is greater for WW variants where PEG strongly increases conformational stability. This relationship holds even for variants modified with the 45-unit PEG, indicating that the conformational stability can play an important role, even for proteins modified with larger PEG oligomers. Because the angle θ is correlated to PEG-based stabilization, which is in turn associated with enhanced resistance to proteolysis, we anticipate that θ will be useful for identifying Asn-PEGylation sites that provide optimal proteolytic protection to β-sheet proteins and protein drugs. However, Asn-PEG is not genetically encodable: Asn-PEGylated peptides and proteins can be prepared via solid-phase peptide synthesis [58] and/or native chemical ligation [59], but not via biological expression. It will be interesting to see whether the correlation between PEG-based stabilization and side-chain orientation relative to OH groups holds for PEGs incorporated site-specifically [60] via chemoselective reactions [61] with genetically encodable amino acids whose structures differ substantially from that of Asn.
Conclusions
Diverse studies agree that PEG-based changes to protein surface solvation play a key role in whether or not PEGylation results in substantial increases to protein conformational stability. Because such increases are associated with enhanced resistance to proteolysis, structure-based mastery of these effects remains an important goal. Early work along these lines is promising and provides hope that such a goal is reasonably attainable. However, it is still unclear whether the predictable trends observed for Asn-PEGylation will hold for PEGylation of genetically encodable side chains that can be functionalized chemoselectively. It will also be interesting to see whether PEG-based increases in conformational stability can provide measurable extensions of serum half-life in vivo, along with decreased immunogenicity.
Highlights.
Chemoselective reactions enable site-specific PEGylation
PEG-based conformational stabilization provides enhanced proteolytic resistance
PEG-based protein stabilization associated with local protein surface desolvation
Structure-based criteria identify stabilizing PEGylation sites in certain contexts
Acknowledgments
This work was supported by the NIH NIGMS (1R15GM116055-01).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Leader B, Baca QJ, Golan DE. Protein therapeutics: a summary and pharmacological classification. Nat Rev Drug Discov. 2008;7:21–39. doi: 10.1038/nrd2399. [DOI] [PubMed] [Google Scholar]
- 2.Caravella J, Lugovskoy A. Design of next-generation protein therapeutics. Curr Opin Chem Biol. 2010;14:520–528. doi: 10.1016/j.cbpa.2010.06.175. [DOI] [PubMed] [Google Scholar]
- 3.Veronese FM, Mero A. The Impact of PEGylation on Biological Therapies. Biodrugs. 2008;22:315–329. doi: 10.2165/00063030-200822050-00004. [DOI] [PubMed] [Google Scholar]
- 4.Jevsevar S, Kunstelj M, Porekar VG. PEGylation of therapeutic proteins. Biotechnol J. 2010;5:113–128. doi: 10.1002/biot.200900218. [DOI] [PubMed] [Google Scholar]
- 5.Abuchowski A, van Es T, Palczuk NC, Davis FF. Alteration of immunological properties of bovine serum albumin by covalent attachment of polyethylene glycol. J Biol Chem. 1977;252:3578–3581. [PubMed] [Google Scholar]
- 6.Abuchowski A, Mccoy JR, Palczuk NC, Vanes T, Davis FF. Effect of Covalent Attachment of Polyethylene-Glycol on Immunogenicity and Circulating Life of Bovine Liver Catalase. J Biol Chem. 1977;252:3582–3586. [PubMed] [Google Scholar]
- 7.Harris JM, Chess RB. Effect of pegylation on pharmaceuticals. Nat Rev Drug Discov. 2003;2:214–221. doi: 10.1038/nrd1033. [DOI] [PubMed] [Google Scholar]
- 8.Fishburn CS. The pharmacology of PEGylation: Balancing PD with PK to generate novel therapeutics. J Pharm Sci. 2008;97:4167–4183. doi: 10.1002/jps.21278. [DOI] [PubMed] [Google Scholar]
- 9.Veronese FM, editor. PEGylated Protein Drugs: Basic Sience and Clinical Applications. Basel: Birkhauser Verlag; 2009. [Google Scholar]
- 10.Li W, Zhan P, Clercq E, Lou H, Liu X. Current drug research on PEGylation with small molecular agents. Prog Polym Sci. 2013;38:421–444. [Google Scholar]
- 11.Tsutsumi Y, Kihira T, Tsunoda S, Kanamori T, Nakagawa S, Mayumi T. Molecular design of hybrid tumour necrosis factor alpha with polyethylene glycol increases its anti-tumour potency. Br J Cancer. 1995;71:963–968. doi: 10.1038/bjc.1995.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marshall D, Pedley RB, Boden JA, Boden R, Melton RG, Begent RHJ. Polyethylene glycol modification of a galactosylated streptavidin clearing agent: effects on immunogenicity and clearance of a biotinylated anti-tumour antibody. Br J Cancer. 1996;73:565–572. doi: 10.1038/bjc.1996.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kinstler O, Molineux G, Treuheit M, Ladd D, Gegg C. Mono-N-terminal poly(ethylene glycol)–protein conjugates. Adv Drug Deliv Rev. 2002;54:477–485. doi: 10.1016/s0169-409x(02)00023-6. [DOI] [PubMed] [Google Scholar]
- 14.Soderquist RG, Milligan ED, Sloane EM, Harrison JA, Douvas KK, Potter JM, Hughes TS, Chavez RA, Johnson K, Watkins LR. PEGylation of brain derived neurotrophic factor for preserved biological activity and enhanced spinal cord distribution. J Biomed Mat Res Part A. 2009;91:719–729. doi: 10.1002/jbm.a.32254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunstelj M, Fidler K, Škrajnar Š, Kenig M, Smilović V, Kusterle M, Caserman S, Zore I, Porekar VG, Jevševar S. Cysteine-Specific PEGylation of rhG-CSF via Selenylsulfide Bond. Bioconjugate Chem. 2013;24:889–896. doi: 10.1021/bc3005232. [DOI] [PubMed] [Google Scholar]
- 16.Chalker JM, Lercher L, Rose NR, Schofield CJ, Davis BG. Conversion of Cysteine into Dehydroalanine Enables Access to Synthetic Histones Bearing Diverse Post-Translational Modifications. Angew Chem Int Ed. 2012;51:1835–1839. doi: 10.1002/anie.201106432. [DOI] [PubMed] [Google Scholar]
- 17.van Hest JC, Tirrell DA. Efficient introduction of alkene functionality into proteins in vivo. FEBS Lett. 1998;428:68–70. doi: 10.1016/s0014-5793(98)00489-x. [DOI] [PubMed] [Google Scholar]
- 18.van Hest JCM, Kiick KL, Tirrell DA. Efficient Incorporation of Unsaturated Methionine Analogues into Proteins in Vivo. J Am Chem Soc. 2000;122:1282–1288. [Google Scholar]
- 19.Kiick KL, Saxon E, Tirrell DA, Bertozzi CR. Incorporation of azides into recombinant proteins for chemoselective modification by the Staudinger ligation. Proc Natl Acad Sci USA. 2002;99:19–24. doi: 10.1073/pnas.012583299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L, Zhang ZW, Brock A, Schultz PG. Addition of the keto functional group to the genetic code of Escherichia coli. Proc Natl Acad Sci USA. 2003;100:56–61. doi: 10.1073/pnas.0234824100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deiters A, Cropp TA, Summerer D, Mukherji M, Schultz PG. Site-specific PEGylation of proteins containing unnatural amino acids. Bioorg Med Chem Lett. 2004;14:5743–5745. doi: 10.1016/j.bmcl.2004.09.059. [DOI] [PubMed] [Google Scholar]
- 22.Goerke AR, Swartz JR. High-level cell-free synthesis yields of proteins containing site-specific non-natural amino acids. Biotechnol Bioeng. 2009;102:400–416. doi: 10.1002/bit.22070. [DOI] [PubMed] [Google Scholar]
- 23.Bundy BC, Swartz JR. Site-specific incorporation of p-propargyloxyphenylalanine in a cell-free environment for direct protein-protein click conjugation. Bioconjugate Chem. 2010;21:255–263. doi: 10.1021/bc9002844. [DOI] [PubMed] [Google Scholar]
- 24.Cho H, Daniel T, Buechler Y, Litzinger DC, Maio Z, Putnam A-M, Kraynov VS, Sim B-C, Bussell S, Javahishvili T, et al. Optimized clinical performance of growth hormone with an expanded genetic code. Proc Natl Acad Sci USA. 2011;108:9060–9065. doi: 10.1073/pnas.1100387108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen H, Lu Y, Fang Z, Liu J, Tian H, Gao X. High-level production of uricase containing keto functional groups for site-specific PEGylation. Biochem Eng J. 2011;58–59:25–32. [Google Scholar]
- 26.Tada S, Andou T, Suzuki T, Dohmae N, Kobatake E. Genetic PEGylation. PLoS One. 2012;7:e49235. doi: 10.1371/journal.pone.0049235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Q, Chan TR, Hilgraf R, Fokin VV, Sharpless KB, Finn MG. Bioconjugation by Copper(I)-Catalyzed Azide-Alkyne [3 + 2] Cycloaddition. J Am Chem Soc. 2003;125:3192–3193. doi: 10.1021/ja021381e. [DOI] [PubMed] [Google Scholar]
- 28.Shozen N, Iijima I, Hohsaka T. Site-specific incorporation of PEGylated amino acids into proteins using nonnatural amino acid mutagenesis. Bioorg Med Chem Lett. 2009;19:4909–4911. doi: 10.1016/j.bmcl.2009.07.105. [DOI] [PubMed] [Google Scholar]
- 29.Zang Q, Tada S, Uzawa T, Kiga D. Two site genetic incorporation of varying length polyethylene glycol into the backbone of one peptide. Chem Commun. 2015;51:14385–14388. doi: 10.1039/c5cc04486c. [DOI] [PubMed] [Google Scholar]
- 30.Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 31.Ohkuri T, Nagatomo S, Oda K, So T, Imoto T, Ueda T. A Protein’s Conformational Stability Is an Immunologically Dominant Factor: Evidence That Free-Energy Barriers for Protein Unfolding Limit the Immunogenicity of Foreign Proteins. J Immunol. 2010;185:4199–4205. doi: 10.4049/jimmunol.0902249. [DOI] [PubMed] [Google Scholar]
- 32.Parsell DA, Sauer RT. The structural stability of a protein is an important determinant of its proteolytic susceptibility in Escherichia coli. J Biol Chem. 1989;264:7590–7595. [PubMed] [Google Scholar]
- 33.Monfardini C, Schiavon O, Caliceti P, Morpurgo M, Harris JM, Veronese FM. A Branched Monomethoxypoly(Ethylene Glycol) for Protein Modification. Bioconjugate Chem. 1995;6:62–69. doi: 10.1021/bc00031a006. [DOI] [PubMed] [Google Scholar]
- 34.Rodriguez-Martinez JA, Solá RJ, Castillo B, Cintron-Colon HR, Rivera-Rivera I, Barletta G, Griebenow K. Stabilization of alpha-Chymotrypsin Upon PEGylation Correlates With Reduced Structural Dynamics. Biotechnol Bioeng. 2008;101:1142–1149. doi: 10.1002/bit.22014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodríguez-Martínez J, Rivera-Rivera I, Solá R, Griebenow K. Enzymatic activity and thermal stability of PEG-α-chymotrypsin conjugates. Biotechnol Lett. 2009;31:883–887. doi: 10.1007/s10529-009-9947-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiu K, Agoubi LL, Lee I, Limpar MT, Lowe JW, Goh SL. Effects of Polymer Molecular Weight on the Size, Activity, and Stability of PEG-Functionalized Trypsin. Biomacromolecules. 2010;11:3688–3692. doi: 10.1021/bm1006954. [DOI] [PubMed] [Google Scholar]
- 37.Jain A, Ashbaugh HS. Helix Stabilization of Poly(ethylene glycol)–Peptide Conjugates. Biomacromolecules. 2011;12:2729–2734. doi: 10.1021/bm2005017. [DOI] [PubMed] [Google Scholar]
- 38.Yang C, Lu DN, Liu Z. How PEGylation Enhances the Stability and Potency of Insulin: A Molecular Dynamics Simulation. Biochemistry. 2011;50:2585–2593. doi: 10.1021/bi101926u. [DOI] [PubMed] [Google Scholar]
- 39.Shu JY, Lund R, Xu T. Solution Structural Characterization of Coiled-Coil Peptide–Polymer Side-Conjugates. Biomacromolecules. 2012;13:1945–1955. doi: 10.1021/bm300561y. [DOI] [PubMed] [Google Scholar]
- 40.Meng W, Guo X, Qin M, Pan H, Cao Y, Wang W. Mechanistic Insights into the Stabilization of srcSH3 by PEGylation. Langmuir. 2012;28:16133–16140. doi: 10.1021/la303466w. [DOI] [PubMed] [Google Scholar]
- 41.Garcia-Arellano H, Valderrama B, Saab-Rincon G, Vazquez-Duhalt R. High temperature biocatalysis by chemically modified cytochrome c. Bioconjugate Chem. 2002;13:1336–1344. doi: 10.1021/bc025561p. [DOI] [PubMed] [Google Scholar]
- 42.Plesner B, Fee CJ, Westh P, Nielsen AD. Effects of PEG size on structure, function and stability of PEGylated BSA. Eur J Pharm Biopharm. 2011;79:399–405. doi: 10.1016/j.ejpb.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 43.Plesner B, Westh P, Nielsen AD. Biophysical characterisation of GlycoPEGylated recombinant human factor VIIa. Int J Pharm. 2011;406:62–68. doi: 10.1016/j.ijpharm.2010.12.034. [DOI] [PubMed] [Google Scholar]
- 44.Rodríguez-Martínez JA, Rivera-Rivera I, Griebenow K. Prevention of benzyl alcohol-induced aggregation of chymotrypsinogen by PEGylation. J Pharm Pharmacol. 2011;63:800–805. doi: 10.1111/j.2042-7158.2011.01288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nodake Y, Yamasaki N. Some properties of a macromolecular conjugate of lysozyme prepared by modification with a monomethoxypolyethylene glycol derivative. Biosci Biotechnol Biochem. 2000;64:767–774. doi: 10.1271/bbb.64.767. [DOI] [PubMed] [Google Scholar]
- 46.Gokarn YR. PhD Thesis: Hydrodynamic Behavior and Thermal Stability of a PEGylated Protein: Studies with Hen Egg Lysozyme. University of New Hampshire; 2003. [Google Scholar]
- 47.Popp MW, Dougan SK, Chuang TY, Spooner E, Ploegh HL. Sortase-catalyzed transformations that improve the properties of cytokines. Proc Natl Acad Sci USA. 2011;108:3169–3174. doi: 10.1073/pnas.1016863108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Natalello A, Ami D, Collini M, D’Alfonso L, Chirico G, Tonon G, Scaramuzza S, Schrepfer R, Doglia SM. Biophysical Characterization of Met-G-CSF: Effects of Different Site-Specific Mono-Pegylations on Protein Stability and Aggregation. PLoS One. 2012;7:e42511. doi: 10.1371/journal.pone.0042511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang C, Lu D, Liu Z. How PEGylation enhances the stability and potency of insulin: a molecular dynamics simulation. Biochemistry. 2011;50:2585–2593. doi: 10.1021/bi101926u. Molecular dynamics simulations suggest that PEGylation of LysB29 does not substantially disrupt the seconary structure of insulin, but does decrease its solvent-accessible surface area. [DOI] [PubMed] [Google Scholar]
- 50.Shu JY, Tan C, DeGrado WF, Xu T. New Design of Helix Bundle Peptide–Polymer Conjugates. Biomacromolecules. 2008;9:2111–2117. doi: 10.1021/bm800113g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lund R, Shu J, Xu T. A Small-Angle X-ray Scattering Study of α-helical Bundle-Forming Peptide-Polymer Conjugates in Solution: Chain Conformations. Macromolecules. 2013;46:1625–1632. Small-angle x-ray scattering experiments reveal that the PEG appended to an α-helical coiled coil is more compact than would be expected for a free PEG. [Google Scholar]
- 52.Hamed E, Xu T, Keten S. Poly(ethylene glycol) Conjugation Stabilizes the Secondary Structure of α-Helices by Reducing Peptide Solvent Accessible Surface Area. Biomacromolecules. 2013;14:4053–4060. doi: 10.1021/bm401164t. [DOI] [PubMed] [Google Scholar]
- 53.Pandey BK, Smith MS, Price JL. Cys(i)-Lys(i+3)-Lys(i+4) Triad: A General Approach for PEG-Based Stabilization of alpha-Helical Proteins. Biomacromolecules. 2014;15:4643–4647. doi: 10.1021/bm501546k. Variable temperature CD experiments confirm the presence of favorable PEG-Lys interactions in a previously characteried PEGylated α-helical coiled coil, though these interactions are not strong enough to overcome the intrinstically destabilizing impact of PEG on the coiled coil. [DOI] [PubMed] [Google Scholar]
- 54.Meng W, Guo X, Qin M, Pan H, Cao Y, Wang W. Mechanistic insights into the stabilization of srcSH3 by PEGylation. Langmuir: the ACS journal of surfaces and colloids. 2012 doi: 10.1021/la303466w. [DOI] [PubMed] [Google Scholar]
- 55.Svergun DI, Ekström F, Vandegriff KD, Malavalli A. Solution structure of poly (ethylene) glycol-conjugated hemoglobin revealed by small-angle X-ray scattering: implications for a new oxygen therapeutic. Biophysical journal. 2008 doi: 10.1529/biophysj.107.114314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cattani G, Vogeley L, Crowley PB. Structure of a PEGylated protein reveals a highly porous double-helical assembly. Nature chemistry. 2015 doi: 10.1038/nchem.2342. NMR studies on PEGylated plastocyanin suggests that the PEG and the protein behave as independent domains. This PEG is sufficiently disordered that it is not resolved in the x-ray crystal structure of this PEG-protein conjugate. [DOI] [PubMed] [Google Scholar]
- 57.Pandey BK, Smith MS, Torgerson C, Lawrence PB, Matthews SS, Watkins E, Groves ML, Prigozhin MB, Price JL. Impact of Site-Specific PEGylation on the Conformational Stability and Folding Rate of the Pin WW Domain Depends Strongly on PEG Oligomer Length. Bioconjugate Chem. 2013;24:796–802. doi: 10.1021/bc3006122. [DOI] [PubMed] [Google Scholar]
- 58.Lawrence PB, Gavrilov Y, Matthews SS, Langlois MI, Shental-Bechor D, Greenblatt HM, Pandey BK, Smith MS, Paxman R, Torgerson CD, et al. Criteria for Selecting PEGylation Sites on Proteins for Higher Thermodynamic and Proteolytic Stability. J Am Chem Soc. 2014;136:17547–17560. doi: 10.1021/ja5095183. PEG-based stabilization of the WW domain depends on local protein surface desolvation by PEG. Proximity of a PEGylation site and orientation relative to a nearby OH-containing side chain is a reasonable predictor of PEG-based stabilization within WW and can be used to predict the location of a stabilizing site within the Src SH3 domain. PEG-based stabilization is associated with enhanced resistance to proteolysis. [DOI] [PubMed] [Google Scholar]
- 59.Dawson PE, Muir TW, Clarklewis I, Kent SBH. Synthesis of Proteins by Native Chemical Ligation. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 60.Nischan N, Hackenberger CPR. Site-specific PEGylation of Proteins: Recent Developments. J Org Chem. 2014;79:10727–10733. doi: 10.1021/jo502136n. [DOI] [PubMed] [Google Scholar]
- 61.Sletten EM, Bertozzi CR. Bioorthogonal Chemistry: Fishing for Selectivity in a Sea of Functionality. Angew Chem Int Ed. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]