Abstract
Protein kinase D (PKD) consists of a family of three structurally related enzymes that are co-expressed in the heart and have important roles in many biological responses. PKD1 is activated by pro-hypertrophic stimuli and has been implicated in adverse cardiac remodeling. Efforts to define the cardiac actions of PKD2 and PKD3 have been less successful at least in part because conventional methods provide a general screen for PKD activation but are poorly suited to resolve activation patterns for PKD2 or PKD3. This study uses Phos-tag SDS-PAGE, a method that exaggerates phosphorylation-dependent mobility shifts, to overcome this technical limitation. Phos-tag SDS-PAGE resolves PKD1 as distinct molecular species (indicative of pools of enzyme with distinct phosphorylation profiles) in unstimulated cardiac fibroblasts and cardiomyocytes; as a result, attempts to track PKD1 mobility shifts that result from agonist activation were only moderately successful. In contrast, PKD2 and PKD3 are recovered from resting cardiac fibroblasts and cardiomyocytes as single molecular species; both enzymes display robust mobility shifts in Phos-tag SDS-PAGE in response to treatment with sphingosine-1-phosphate, thrombin, PDGF, or H2O2. Studies with GF109203X implicate protein kinase C activity in the stimulus-dependent pathways that activate PKD2/PKD3 in both cardiac fibroblasts and cardiomyocytes. Studies with C3 toxin identify a novel role for Rho in the sphingosine-1-phosphate and thrombin receptor-dependent pathways that lead to the phosphorylation of PKD2/3 and the downstream substrate CREB in cardiomyocytes. In conclusion, Phos-tag SDS-PAGE provides a general screen for stimulus-specific changes in PKD2 and PKD3 phosphorylation and exposes a novel role for these enzymes in specific stress-dependent pathways that influence cardiac remodeling.
Keywords: Protein kinase D, Phos-tag SDS-PAGE, cardiomyocytes, cardiac fibroblasts
1. INTRODUCTION
Protein kinase D (PKD) consists of a family of three structurally-related stress-activated enzymes (PKD1 or PKDμ, PKD2, and PKD3 or PKDν) that regulate a large number of fundamental biological processes involved in cell proliferation, differentiation, apoptosis, immune regulation, cardiac contraction, cardiac hypertrophy, angiogenesis, and cancer [1–3]. PKD isoforms share a common modular domain structure consisting of a C-terminal kinase domain and an N-terminal regulatory domain consisting of tandem C1A/C1B motifs that anchor full-length PKD to diacylglycerol-/phorbol ester-containing membranes and a pleckstrin homology (PH) motif that participates in intramolecular autoinhibitory interactions that regulate enzyme activity (Fig 1). PKD activation is generally attributed to growth factor-dependent mechanisms that promote diacylglycerol accumulation and co-localize PKD with allosterically-activated novel protein kinase C (nPKC) isoforms at lipid membranes. nPKCs then trans-phosphorylate PKD at conserved serine residues in the activation loop. However, PKC-independent activation loop autophosphorylation also can contribute to PKD1 activation under specific stimulatory conditions [4, 5]. Activated forms of PKD1 and PKD2 (but not PKD3) then autophosphorylate at a PKD consensus phosphorylation motif (in a PDZ domain-binding motif) at the extreme C-terminus - this modification regulates interactions with scaffolding proteins, trafficking to distinct cellular subdomains, and the amplitude/tempo of PKD signaling responses [6, 7].
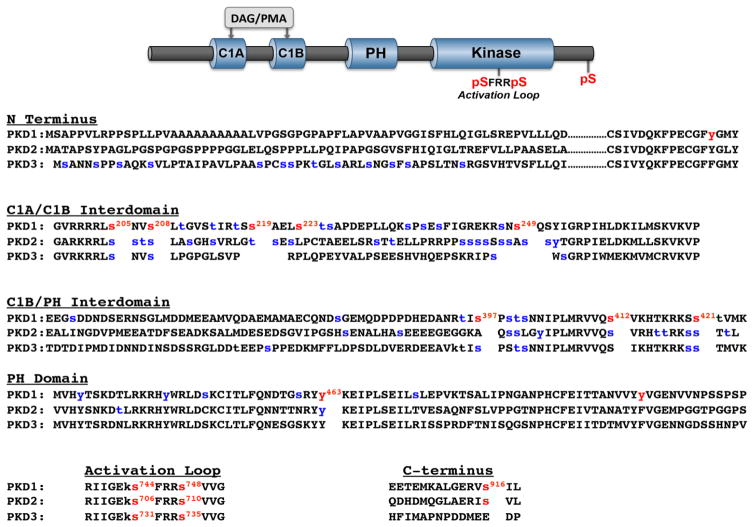
Figure 1. Domain structure and phosphorylation sites on PKD1, PKD2, and PKD3.
Top: The cartoon depicts the common domain structure of PKD isoforms with conserved C1A-C1B and PH domains in the regulatory region, the kinase domain in the catalytic region, the activation loop phosphorylation motif (conserved in all there PKD isoforms), and the C-terminal phosphorylation site (conserved in PKD1 and PKD2, but not PKD3). Bottom: Sequence alignments for PKD isoform the N-termini, C1A-C1B interdomains, C1B-PH interdomains, PH domains, activation loops and C-termini, with phosphorylation sites at the activation loop, the C-terminus, and other sites that have been the focus of studies in the literature in red. Phosphorylation sites identified in high throughput phosphoproteomic studies (curated by Phosphosite [15]) are depicted in blue.
While individual PKD isoforms display similar in vitro substrate specificities and share some overlapping cellular responses, these enzymes are not necessarily functionally redundant under all stimulatory conditions or in all cell types. Some of the major structural differences between these enzymes that could underlie PKD isoform specificity are summarized in Figure 1 and listed below:
PKD1 and PKD2 contain a C-terminal autophosphosphorylation site that is not conserved in PKD3.
PKD isoforms are highly divergent at their extreme N-termini. This region is apolar alanine/proline-rich in PKD1, proline-rich in PKD2, whereas it contains a large number of phosphorylation sites in PKD3.
The N-terminal variable region of PKD1 contains a Tyr residue (Y95) that is flanked by a minimal PKCδ-C2 domain binding motif [pY-(Q/R)-X-(Y/F)] [8]. PKD1 is phosphorylated at this Tyr residue during oxidative stress, generating a docking site for the PKCδ-C2 domain and leading to PKCδ-dependent PKD1 phosphorylation/activation [9]. A tyrosine residue is conserved at this position in PKD2, but not in PKD3; the Y-F substitution at this site in PKD3 prevents redox/PKCδ-dependent activation [9].
All three enzymes contain a large number of Ser/Thr phosphorylation sites in their C1A/C1B interdomains. Autophosphorylation sites at Ser205/Ser208 and Ser219/Ser223 in the C1A/C1B interdomain of PKD1 have been characterized as 14-3-3 protein binding sites that regulate PKD1 activity [10]. The Ser205/Ser208 phosphorylation sites are conserved in PKD3 (Ser219/Ser223 is not), whereas PKD2 contains phosphorylation sites flanked by more variable motifs at this and several other positions in this region of the enzyme.
The PKD1 C1B-PH domain interface contains sites for trans phosphorylation by PKC (Ser412 [11]) and protein kinase A (Ser421 [12]) that are conserved in PKD2 and PKD3. This region also is phosphorylated at Ser397 by p38MAPK [13]; a Ser at this position (with a +1 Pro) is phosphorylated in PKD3, but this sequence is not conserved in PKD2.
A phosphorylation site in the PH domain of PKD1 (Tyr463) disrupts PH domain mediated autoinhibitory regulation and is conserved in PKD2 and PKD3 [14]. Other phosphorylation sites (of unknown significance) also have been identified in the PH domain PKD1, and to a lesser extent in PKD2.
This brief summary serves to emphasize that in addition to the canonical phosphorylation motifs in the activation loop and C-tail, all three PKD isoforms contain a large number of additional phosphorylation sites in other regions of the enzyme. In some cases, phosphorylation at these other sites has been implicated as a mechanism that controls PKD’s cellular actions. However, our current understanding of the controls and consequences of PKD multi-site phosphorylation remains quite limited, at least in part because methods to track phosphorylation at these non-canonical sites remain rather costly and cumbersome. For this reason, we became interested in Phosphate binding tag (Phos-tag) acrylamide gel electrophoresis as a method to separate various phosphorylated and nonphosphorylated forms of individual isoforms of PKD [16–20]. Phos-tag forms complexes with phosphomonoester dianions bound to Ser, Thr, or Tyr residues. Since this slows the electrophoretic migration of phosphorylated forms of a given protein, Phos-tag SDS-PAGE can be used as a general screen for phosphorylation at any residue (Ser, Thr, or Tyr) in individual PKDs.
PKD1, PKD2, and PKD3 are detected at the mRNA level in various cardiac preparations [21–23]. PKD activation has been implicated in both cardioprotection against ischemia/reperfusion (I/R) injury [24] and in adverse cardiomyocyte remodeling, the latter generally attributed to the phosphorylation of proteins that control gene transcription (such as class II histone deacetylases and cAMP response element binding protein [21, 25]) or cardiac contraction (such as cardiac troponin I and cardiac myosin-binding protein C [26]). We previously identified distinct agonist-dependent activation mechanisms for PKD1 and PKD2 in cardiomyocytes [4]. These results could suggest that individual PKD isoforms play distinct roles in different cardiomyocyte effector responses. PKDs could in theory also regulate cardiac remodeling through actions in cardiac fibroblasts, since PKD1 activation has been linked to the induction of procollagen, type I, alpha 2 (Col1a2) gene expression and cardiac fibrosis [27]. However, PKD isoform expression and activation patterns in cardiac fibroblasts have never been examined.
PKD isoform-specific activation profiles typically are resolved using immunoprecipitation/Western blotting methods and/or PKD isoform knockdown strategies. While these methods have been used to considerable advantage in many cell types, they are cumbersome, immunoprecipitation/Western blotting studies are reagent limited (since available phosphorylation site-specific antibodies can only track phosphorylation at the common phosphorylation motifs in the activation loop and C-terminus) and the specificity/efficiency of gene knockdown methods is imperfect; we previously identified an underappreciated effect of PKD1 knockdown to induce a compensatory increase in PKD2 activity in cardiomyocytes [4]. Therefore, this study adopts Phos-tag SDS-PAGE methods to resolve receptor and redox-dependent pathways that activate PKD1, PKD2, and PKD3 in cardiac fibroblasts and cardiomyocytes.
2. MATERIALS AND METHODS
2.1 Reagents
Antibodies were from the following sources: PKD1-Ser(P)744/Ser(P)748 (Cat. #2054), PKD1-Ser(P)916 (Cat. #2051), PKD1 (Cat. #2052), and PKD3 (Cat. #5655) were from Cell Signaling Technologies. PKD2 (Cat. ab57114, a monoclonal antibody raised against residues 1–110 at the N terminus of human PKD2) was from Abcam. C3 exotoxin was from Cytoskeleton, Inc. GF109203X and Gö6976 were from Enzo Life Sciences. S1P was from Avanti Polar Lipids. Thrombin, Endothelin-1, norepinephrine, platelet-derived growth factor (PDGF) and PMA were from Sigma. All other chemicals were regent grade.
2.2 Cell Culture
Cardiomyocytes were isolated from hearts of 2-day-old Wistar rats by a trypsin dispersion procedure that uses a differential attachment procedure followed by irradiation to enrich for cardiomyocytes [28]. Cells were plated on protamine sulfate-coated culture dishes at a density of 5 × 106 cells/100-mm dish and grown in MEM (Invitrogen, BRL) supplemented with 10% fetal calf serum for 4 days and then serum-deprived for 24 h prior to experiment. Nonmyocyte fibroblast-like cells, derived from the cells that adhered to the plastic culture dishes during the preplating step, also were prepared according to methods described previously for some experiments [25]. Primary cardiac fibroblast cultures were maintained in MEM supplemented with 10% fetal calf serum for 6 days and then serum-deprived for 24 h prior to experimental protocols.
2.3 SDS-PAGE and immunoblot analysis
Standard Laemmli SDS-Polyacrylamide gel electrophoresis (PAGE, 8% (w/v) polyacrylamide) was performed according to standard methods as used in our previous studies [25].
The method for Mn2+-Phos-tag SDS-PAGE is virtually identical to the methods used for conventional SDS-PAGE, with the exception that 5 μM of acrylamide-pendant Phos-tag ligand (Phos-tag® Wako Pure Chemical Industries) and 10 mM of MnCl2 are added to 6% (w/v) polyacrylamide separating gels before polymerization [17]. The use of a relatively low concentration of Phos-tag for these studies was based upon preliminary experiments showing that these conditions provide optimal separation of phosphorylated and non-phosphorylated forms of PKD2 [18]. Protein separations were carried out at 4°C for 16 hours (80 V) using an electrophoresis running buffer (pH 8.4) containing 25 mM Tris, 192 mM glycine and 0.1% (w/v) SDS. Mn2+-Phos-tag SDS-PAGE gels were then soaked in a solution containing 25 mM Tris, 192 mM glycine, 10% (v/v) MeOH, and 1.0 mM EDTA for 10 min followed by a second soaking in 25 mM Tris, 192 mM glycine, and 10% (v/v) MeOH for 30 min. The EDTA soaking to chelate manganese ions is a critical additional step since Mn2+-Phos-tag in the gels causes inefficient electroblotting [16–18].
Immunoblotting was performed on cell extracts according to the methods described previously or manufacturer's instructions [25]. In each figure, each panel represents the results from a single gel (exposed for a uniform duration); detection was with enhanced chemiluminescence. All results were replicated in 3 to 6 experiments on separate culture preparations.
3. RESULTS
3.1 PKD isoform regulation in Cardiac Fibroblasts
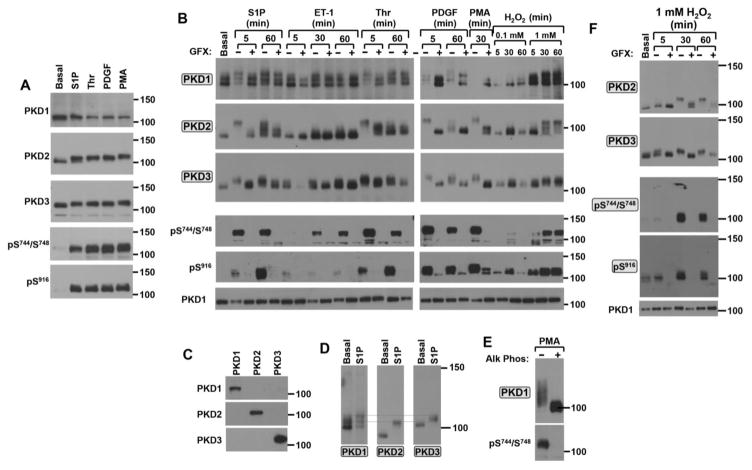
Fig 2 compares the electrophoretic mobilities of phosphorylated and non-phosphorylated forms of PKD1, PKD2, and PKD3 using conventional Laemmli SDS-PAGE (Panel A) and Mn2+-Phos-tag SDS-PAGE (phosphate affinity gel electrophoresis; Panel B). The experiment is performed on lysates prepared from cardiac fibroblasts exposed to a panel of profibrotic agonists that are predicted to activate PKD through distinct molecular mechanisms, including the G protein-coupled receptor agonists sphingosine-1-phosphate (S1P), endothelin-1 (ET-1) and thrombin (Thr) and the tyrosine kinase receptor agonist platelet-derived growth factor (PDGF) [29–32]; phorbol 12-myristate 13-acetate (PMA), which bypasses the receptor machinery and directly activates PKD is included as a control in the experiments. Figure 2A shows that S1P, Thr, PDGF, and PMA induce similar increases in PKD phosphorylation at Ser744/Ser748 and Ser916 and that conventional SDS-PAGE methods resolve a subtle shift in the mobility of PKD2 (and perhaps less convincingly PKD3), but no discernable change in the mobility of PKD1. This is consistent with previous observations that phosphorylation at some (but not all) sites slows the mobility of some proteins in normal SDS-PAGE - and that the magnitude of most phosphorylation-induced mobility shifts in conventional Laemmli SDS-PAGE is modest (even when gel cross-linking and other conditions are adjusted to optimize mobility shifts). It is worth noting that the negative results for PKD1 also may be attributable to the fact that most commercially available anti-PKD1 protein antibodies were raised against a synthetic peptide corresponding to an epitope at the extreme C-terminus (i.e., sequence flanking the Ser916 autophosphorylation site). These antibodies preferentially recognize the inactive form of PKD1 that is not phosphorylated at Ser916 [25, 33]; they are not well suited to detect the phosphorylated/slower migrating forms of PKD1.
Figure 2. Agonist-specific activation patterns for PKD1, PKD2 and PKD3 in cardiac fibroblasts.
Cardiac fibroblasts were challenged with S1P (5 μM), Thr (1 U/ml), or PDGF (50 ng/ml) for 5 min or 200 nM PMA for 20 min (Panel A). Treatment with these agonists as well as ET-1 (100 nM), or H2O2 (at the indicated concentrations) was for the indicated intervals following a pretreatment for 45 min with vehicle or 10 μM GF109203X (GFX) in Panels B and F. Lysates were subjected to Laemmli SDS-PAGE followed by immunoblot analysis for PKD isoform expression and phosphorylation in Panel A. Proteins were separated according to the Mn2+-Phos-tag SDS-PAGE method for the PKD1, PKD2, and PKD3 blots in Panels B and D, for the PKD1 blot in Panel E, and for all blots except PKD1 in Panel F. Labels in shaded boxes in this Figure (as well as in Figs 3–5) denote immunoblots performed following protein separation by Mn2+- Phos-tag SDS-PAGE. In each panel, the results are from a single gel (or a set of gels run in parallel, in Panel B) exposed for a uniform duration. The results are representative of data obtained in separate experiments on three separate culture preparations. Panel C: Lysates from cardiomyocytes infected with adenoviral vectors that drive expression of PKD1, PKD2, or PKD3 were probed with PKD isoform-specific antibodies. Panel F: PKD1 dephosphorylation was accomplished by incubating lysates from PMA-treated cardiac fibroblasts (0.5 mg) in a reaction mixture containing 20 mM Tris HCl, 1 mM MgCl2 and bovine intestinal mucosa alkaline phosphatase (0.5 μg/ml) for 12 hr at 37°C. Lysates were then subjected to immunoblot analysis for PKD1 protein and Ser744/Ser748 phosphorylation.
Fig 2B shows that the Phos-tag SDS-PAGE method exposes additional heterogeneity in the PKD isoform phosphorylation profiles. First, while PKD2 and PKD3 each migrate as a single molecular species in resting cardiac fibroblasts, Phos-tag SDS-PAGE resolves PKD1 as distinct molecular species in resting unstimulated cardiac fibroblasts. The molecular heterogeneity detected by the anti-PKD1 antibody cannot be attributed to cross-reactivity with PKD2 for the following reasons: [1] We previously showed that an Ad-PKD1-RNAi silencing vector (that does not influence the abundance of PKD2) virtually eliminates PKD1 protein immunoreactivity; these results argue that the anti-PKD1 antibody does not cross-react with PKD2 [4]. [2] Western blot experiments performed on lysates from cells that heterologously overexpress PKD1, PKD2, or PKD3 show that the anti-PKD1 antibody specifically recognizes PKD1 and does not cross-react with PKD2 or PKD3, whereas the anti-PKD2 and anti-PKD3 antibodies show similar specificity for their cognate enzymes (Fig 2C). [3] The alignment of the proteins detected by the anti-PKD1, anti-PKD2, anti-PKD3 antibodies (Fig 2D) emphasizes that the protein recognized by the anti-PKD2 antibody in resting cells migrates considerably faster than any protein recognized by antibodies raised against PKD1 or PKD3. These results effectively exclude the trivial explanation that the molecular heterogeneity detected with the anti-PKD1 antibody results from cross-reactivity with PKD2. Rather, the results suggest that the molecular heterogeneity detected with the anti-PKD1 antibody is indicative of pools of enzyme with distinct phosphorylation profiles; the relative abundance of any individual band provides a measure of the stoichiometry of phosphorylation at sites not captured by conventional Western blotting methods with available phosphorylation site-specific antibodies (PSSAs,) since none of these bands are recognized by the PSSAs directed at Ser744/Ser748 or Ser916.
The Phos-tag SDS-PAGE method effectively captures S1P- and Thr-dependent upshifts in PKD1 mobility, but the identification of certain other agonist-dependent changes in PKD1 phosphorylation proved to be more challenging since activation resulted in a marked decrease in PKD1 immunoreactivity. PKD1 immunoreactivity is marked decreased or virtually absent in lysates from cardiac fibroblasts treated with PDGF or PMA, respectively. Similar agonist-dependent decreases in PKD1 band intensity have been noted in studies using normal SDS-PAGE [25]; they are amplified by the Phos-tag SDS-PAGE method that enhances band smearing due to molecular heterogeneity. The loss of PKD1 immunoreactivity also is attributable to the aforementioned limitation of the anti-PKD1 protein antibody, which preferentially recognizes inactive/unphosphorylated enzyme. PKD1 protein immunoreactivity is recovered following sample treatment with alkaline phosphatase (Fig 2E).
The Phos-tag SDS-PAGE method was most effective at capturing agonist-dependent increases in the phosphorylation profiles (i.e., electrophoretic mobility shifts) for PKD2 and PKD3. Fig 2B shows that treatment with S1P, Thr, or PDGF results in a large decrease in the electrophoretic mobility of PKD2. These agonist-dependent mobility shifts are maximal at 5 min and they are sustained (to a variable extent) for up to 60 min of agonist stimulation. Of note, Phos-tag SDS-PAGE resolves a difference in PKD2’s electrophoretic mobility following agonist treatments for 5 versus 60 min that cannot be attributable to gross differences in phosphorylation at Ser744/Ser748 or Ser916 (which is quite high at both time points); these results suggest that PKD2’s phosphorylation profile at other sites is reconfigured with more chronic agonist activation. Fig 2 also shows that endothelin-1 (another pro-fibrotic G protein-coupled receptor agonist that increases collagen production in cardiac fibroblasts [34]) also activates PKD, but in a manner that is quite distinct from S1P and Thr. ET-1 induces a modest decrease in the electrophoretic mobilitiy of PKD3, with no obvious effect on PKD2, at 5 min. ET-1 induces a minor decrease in PKD2 mobility only at late time points (30–60 min).
An alignment of the anti-PKD1, anti-PKD2, anti-PKD3 immunoblots (Fig 2D) also emphasizes the generally underappreciated point that while the inactive/non-phosphorylated form of PKD2 migrates considerably faster than PKD1 or PKD3, all three PKD isoforms migrate quite similarly (even when subjected to Phos-tag SDS-PAGE methods) following agonist activation. This fact may undermine some of the previous conclusions regarding PKD isoform specificity based exclusively on differences in electrophoretic mobility in immunoblots with PSSAs.
Stimulations were performed with GF109203X (a PKC inhibitor that does not inhibit PKD catalytic activity) to resolve PKD phosphorylations that are mediated by (or require) PKC activity. Fig 2B shows that GF109203X pretreatment completely abrogates all agonist-dependent increases in PKD-pSer744/Ser748 immunoreactivity. GF109203X pretreatment also blocks most (but not all) agonist-dependent decreases in PKD isoform electrophoretic mobility. The fact that some S1P- and Thr-dependent PKD2 electrophoretic mobility shifts persist in GF109203X-pretreated cultures indicates that these agonists activate PKD2 phosphorylation through both PKC-dependent and PKC-independent mechanisms (either an autocatalytic reaction that does not require a prior stimulatory phosphorylation at the activation loop or trans-phosphorylation by a GF109203X-insensitive enzyme).
Oxidative stress is a profibrotic stress that also has been implicated as an activator of certain PKD isoforms [9, 35, 36]. Therefore, cardiac fibroblasts also were treated with H2O2. Fig 2B shows that H2O2 induces a dose- and time-dependent activation of all three PKD isoforms. A relatively low H2O2 concentration (0.1 mM) induces a transient increase in PKD phosphorylation at Ser916 (with little-to-no associated increase in PKD-Ser744/Ser748 phosphorylation) in association with a modest decrease in the electrophoretic mobility of PKD2 and PKD3. Since PKD3 lacks the C-terminal autophosphorylation site, the PKD3 band shift detected by Phos-tag SDS-PAGE must be indicative of phosphorylation elsewhere in the enzyme; it is the only readout that tracks activation of PKD3. Responses to 0.1 mM H2O2 are maximal at 30 min and wane with more prolonged stimulations. At a 10-fold higher concentration (1 mM), H2O2 induces time-dependent decreases in the electrophoretic mobilities of PKD1, PKD2, and PKD3. These mobility shifts are associated with marked increases in PKD phosphorylation at the PKC (Ser744/Ser748) and autophosphorylation (Ser916) sites and they are inhibited by pretreatment with GF109203X (Fig 2F).
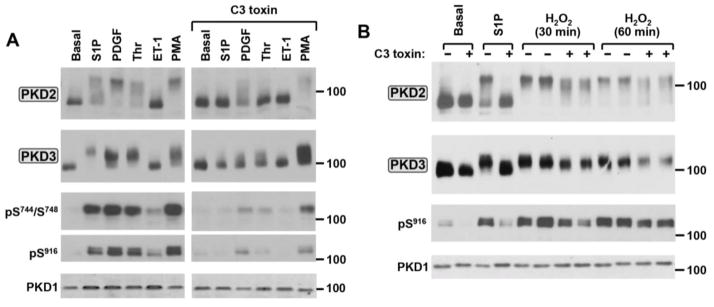
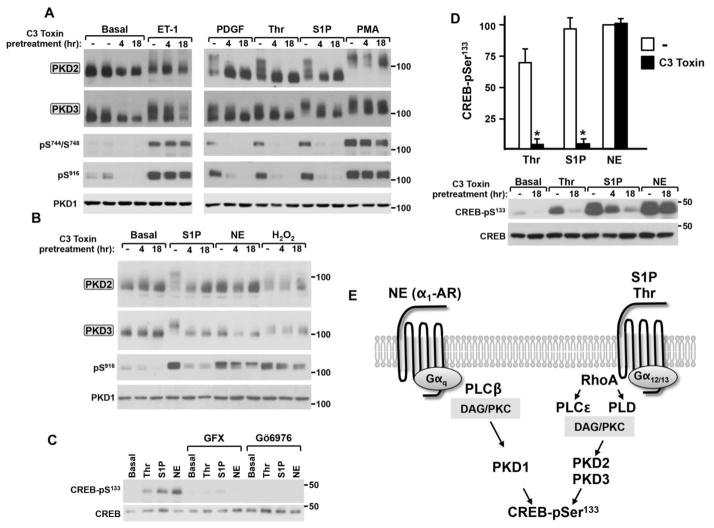
S1P receptors couple to Gα12/13 and activate PKD through a pathway that involves the synergistic actions of PKC and RhoA (a member of the Rho family of small GTPases) in some cell types [37, 38]. While Thr also signals through the Gα12/13-RhoA pathway in many cell types, this mechanism for Thr-dependent PKD activation has never been reported and the notion that the RhoA-dependent pathway might specifically activate only certain isoforms of PKD has never been considered. Therefore, PKD isoform activation was examined in cardiac fibroblasts pretreated with the C. botulinum C3 toxin, which specifically ADP-ribosylates and functionally inactivates Rho proteins [39]. Figure 3 shows that C3 toxin pretreatment completely abrogates S1P- and Thr-dependent activation of PKD2 and PKD3, as evidence by a complete block of the agonist-dependent PKD2 and PKD3 band shifts and increases in Ser744/Ser748 and Ser916 phosphorylation. Of note, PKD isoform activation by PDGF also is attenuated by C3 toxin, whereas PKD isoform activation by H2O2 or PMA is largely preserved in C3 toxin-treated cultures. These results identify a specific role for Rho in the PKD2 and PKD3 activation pathway triggered by Gα12/13-coupled S1P and Thr receptors and show that Rho is not required for PKD2 and PKD3 activation during oxidative stress.
Figure 3. The Rho inhibitor C3 toxin prevents PKD2 and PKD3 activation by S1P and thrombin, but not by PMA or H2O2, in cardiac fibroblasts.
Cardiac fibroblast cultures were pretreated with 2 μg/ml C3 toxin or vehicle for 3 hrs and then challenged for 5 min with S1P (5 μM), Thr (1 U/ml), PDGF (50 ng/ml), or ET-1 (100 nM) or for 30 min with PMA (200 nM; Panel A). Treatment was for 5 min with 5 μM S1P or for the indicated intervals with 1 mM H2O2 in Panel B. Lysates were subjected to Mn2+-Phos-tag SDS-PAGE followed by immunoblot analysis for PKD2 or PKD3, or Laemmli SDS-PAGE followed by immunoblot for PKD1 and PKD isoform phosphorylation at Ser744/Ser748 or Ser916. Similar results were obtained in separate experiments on four separate culture preparations.
3.2 PKD isoform regulation in Cardiomyocytes
We applied a similar approach to resolve agonist-dependent changes in PKD isoform phosphorylation in cardiomyocytes. We previously reported that α1-adrenergic receptor activation with norepinephrine (NE) leads to the selective activation/phosphorylation of PKD1 (and not PKD2), whereas thrombin and PDGFR preferentially active PKD2 and endothelin and PMA are dual activators of PKD1 and PKD2 [4]; the previous studies did not consider possible regulation of PKD3. Lysates from cardiomyocytes stimulated with the aforementioned agonists, as well as S1P and H2O2 (which were not included in the previous study), were subjected to Phos-tag SDS- PAGE and immunoblot analysis to probe for phosphorylation-dependent mobility shifts for PKD1, PKD2, and PKD3.
The Phostag-SDS-PAGE analysis for PKD1 was generally disappointing. While agonist-dependent mobility shifts (particularly for ET-1, PMA, NE and H2O2) were detected in isolated experiments (see Fig 4A), in most experiments PKD1 migrated as a smear and agonist-induced band shifts (including for PMA) were not convincing. We attribute this failure to [1] the properties of the native PKD1 enzyme, which displays a high level of molecular heterogeneity - indicating that PKD1 exists as a mixture of species with a continuum of phosphorylation profiles - even in unstimulated cardiomyocytes), [2] the properties of the anti-PKD1 protein antibody, which preferentially recognizes inactive/unphosphorylated - and not active/phosphorylated – enzyme, and [3] the Phos-tag SDS-PAGE method, which enhances band smearing due to molecular heterogeneity. Since efforts to further optimize the experimental conditions to detect agonist-dependent PKD1 mobility shifts in Phos-tag SDS-PAGE (by changing the source of antibody, the Mn2+-Phos-tag concentration, and/or electrophoresis conditions) proved unsuccessful, Laemmli SDS-PAGE blots of PKD1 protein were obtained and used as loading controls but; further attempts to use Phos-tag SDS-PAGE to track PKD1 mobility shifts (phosphorylation) in cardiomyocytes were abandoned.
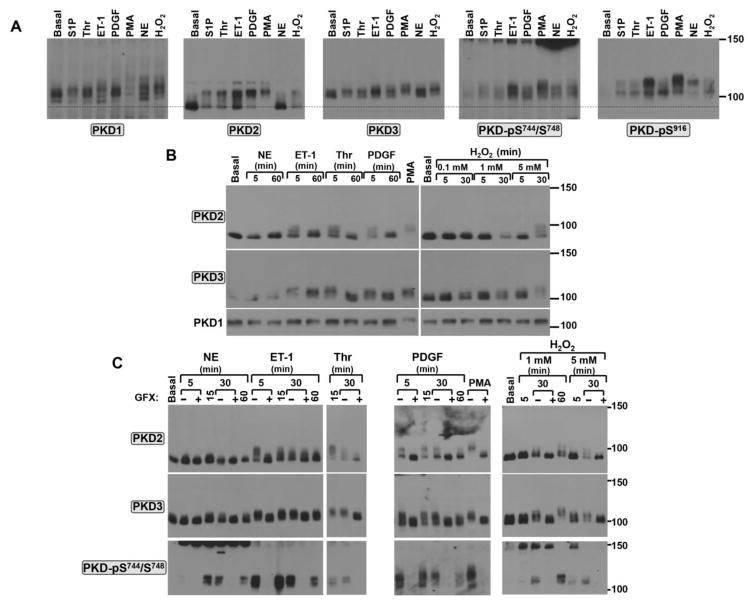
Figure 4. PKD2 and PKD3 are activated in an agonist-specific manner in cardiomyocytes.
Cardiomyocytes were challenged with S1P (5 μM), Thr (1 U/ml), ET-1 (100 nM), NE (10 μM), PDGF (50 ng/ml), PMA (200 nM), or H2O2 (5mM, unless stated otherwise) as indicated in individual panels. Treatments were for 5 min (for S1P, Thr, ET-1, PDGF) or 20 min (for PMA) in Panel A or as indicated in Panels B and C. Agonist treatments followed a pretreatment for 45 min with vehicle or 10 μM GF109203X (GFX) in Panel C. Lysates were subjected to Mn2+-Phos-tag SDS-PAGE followed by immunoblot analysis for PKD isoform expression/mobility and PKD Ser744/Ser748 phosphorylation. In each panel, the results are from a single culture preparation and a single gel (or sets of gels run in parallel, in Panel C) exposed for a uniform duration. Results are representative of data obtained in separate experiments on four separate culture preparations.
Phos-tag SDS-PAGE exposed very robust agonist-dependent changes in the electrophoretic mobility of cardiomyocyte PKD2. PKD2 migrates as a single ~80 kDa species in resting cardiomyocytes, but as a mixture of high (~80-kDa) and low (~105-kDa) mobility species in cardiomyocytes treated with physiologic agonists such as S1P, Thr, ET-1, PDGF, and H2O2 (Figs 4A and 4B). PKD2 is fully (stoichiometrically) converted to the highly phosphorylated (low mobility) species following pharmacologic activation by PMA; it is not influenced by NE, consistent with the previous observation that cardiac α1-ARs activate PKD1 with no associated effect on PKD2 [4]. The striking difference in PKD2’s electrophoretic mobility in response to physiologic (receptor agonists) versus pharmacologic (PMA) stimuli exposed by the Phos-tag SDS-PAGE method (which allows for the simultaneous detection of phosphorylated and dephosphorylated forms of a single protein) emphasizes the frequently overlooked fact that PMA may be an imperfect surrogate for receptor-dependent mechanisms that activate PKD.
Fig 4B shows the time course for the receptor-induced PKD2 mobility shifts. In each case, the receptor-dependent responses are rapid (maximal at 5 min) and wane with more prolonged agonist stimulations, whereas H2O2 responses are concentration- and time-dependent. Of note, a low concentration of H2O2 (0.1 mM), which is associated with a significant upshift in PKD2’s electrophoretic mobility in cardiac fibroblasts, does not alter PKD2 mobility in cardiomyocytes. Rather, PKD2’s phosphorylation profile is altered only with considerably higher concentrations of H2O2 (1–5 mM) in cardiomyocytes, suggesting that the cardiomyocyte PKD2 enzyme requires a higher level of oxidative stress for activation (compared with PKD2 activation in cardiac fibroblasts).
PKD3 has been detected at the mRNA and protein level in mouse heart during embryogenesis [22, 23], but (with the exception of a single recent study that failed to detect PKD3 protein expression in unstimulated neonatal or adult rat cardiomyocytes [40]) PKD3 protein expression and/or actions in postnatal cardiac tissues has not been examined. In this regard, Fig 4 shows that PKD3 protein is readily detected in neonatal rat cardiomyocyte cultures and Phos-tag SDS-PAGE analysis exposes agonist-dependent changes in the electrophoretic mobility of PKD3 that are qualitatively similar (albeit quantitatively more modest) to those identified for PKD2.
Stimulations were performed in the presence of GF109203X to identify the role of PKC activity in agonist-dependent mechanisms that regulate PKD. Fig 4C shows that all of the agonists used in this study (including ET-1, Thr, PDGF, and H2O2 which induce obvious changes in the electrophoretic mobilities of PKD2 and PKD3 and NE which selectively activates PKD1 with little-to-no associated effect on the electrophoretic mobilities of PKD2 or PKD3) induce robust increases in PKD-pSer744/Ser748 immunoreactivity; all agonist-dependent increases in PKD-pSer744/Ser748 immunoreactivity are inhibited by GF109203X. These results indicate that PKC plays a common role in the agonist-dependent pathway that lead to the activation of PKD1, PKD2, and PKD3. GF109203X also blocks all of the rapid agonist-dependent PKD2 and PKD3 electrophoretic mobility shifts. However, the more chronic effect of ET-1 to slow the electrophoretic mobility of PKD2 at the 30 min time point persists in GF109203X-pretreated cultures. This finding confirms our previous conclusion that the more chronic phase of PKD2 regulation by ET-1 is via a PKC-independent mechanism [4].
A previous study showed that overexpression of constitutively active (Q63L) RhoA increases PKD phosphorylation and affords cardioprotection against ischemia/reperfusion injury in neonatal rat cardiomyocyte cultures [24]. Since the PKD isoform(s) downstream of RhoA in this pathway were not identified, agonist treatments also were performed in cardiomyocytes pretreated with C3 toxin. Fig 5 shows that the effects of PDGF, Thr, and S1P to slow the electrophoretic mobility of PKD2 and PKD3 (and to increase PKD-Ser744/Ser748 and Ser916 phosphorylation) are abrogated by the C3 toxin pretreatment, but that C3 toxin has little-to-no effect on responses to ET-1 or PMA. C3 toxin also does not prevent the NE-dependent increase in PKD-Ser916 phosphorylation, which signals activation of PKD1 (Fig 5B). These results identify a specific role for Rho in the agonist-dependent pathways that activate PKD2/PKD3 - and effectively exclude a role for Rho in NE-dependent activation of PKD1 - in cardiomyocytes.
Figure 5. C3 toxin prevents S1P- and thrombin-dependent phosphorylation of PKD2/PKD3 and CREB in cardiomyocytes; C3 toxin does not inhibit the NE-PKD1-CREB-Ser133 phosphorylation pathway.
Panels A, B and D: Cardiomyocytes were pretreated with vehicle or 2 μg/ml C3 toxin for 4 or 18 hrs and then challenged for 5 min with ET-1 (100 nM), PDGF (50 ng/ml), Thr (1 U/ml), S1P (5 μM), NE (10 μM), or H2O2 (5 mM) or for 30 min with PMA (200 nM) as indicated. Lysates were subjected to Mn2+-Phos-tag SDS-PAGE and probed for PKD2/PKD3 protein or Laemmli SDS-PAGE and probed for PKD1 or CREB protein and PKD or CREB-Ser133 phosphorylation as indicated. Results for CREB-Ser133 phosphorylation, corrected for any minor differences in CREB protein levels and normalized to the NE-induced increase in CREB-Ser133 phosphorylation in vehicle-pretreated cultures, are depicted in Panel D (mean +/− SEM, n=3, * p<0.05 compared with corresponding vehicle-pretreated sample). Panel C: Agonist treatments followed a 45 min pretreatment with vehicle, 10 μM GF109203X or 10 μM Gö6976. Panel E: Schematic of agonist- and PKD isoform-specific pathways that increase CREB-Ser133 phosphorylation in cardiomyocytes.
We previously demonstrated that PKD phosphorylates cAMP response element binding protein (CREB) at Ser133 and that PKD mediates the Thr- and α1-adrenergic receptor-dependent increase in CREB-S133 phosphorylation in cardiomyocytes [4, 25]. Fig 5C uses a similar pharmacologic approach to extend these observations and show that Thr, S1P, and NE increase CREB-S133 phosphorylation and that these agonist-dependent increases in CREB-S133 phosphorylation are blocked by GF109203X (an inhibitor of both conventional and novel PKC isoforms) and Gö6976 (an inhibitor of PKD and conventional – but not novel - PKC isoforms). These results suggest that a nPKC-activated PKD enzyme plays a common role to couple NE-, Thr-, and S1P-activated receptors to CREB-Ser133 phosphorylation. However, the observation that Thr and NE induce morphologically distinct forms of cardiac hypertrophy [41] suggests that receptor-specific elements also must be present to distinguish these signaling pathways. Therefore, we examined whether the stimulus-specific differences PKD isoform activation exposed by C3 toxin might translate into differences in the recruitment of the CREB-Ser133 phosphorylation pathway. In fact, Fig 5D shows that Thr and S1P (agonists that effectively activate PKD2/PKD3) promote CREB-S133 phosphorylation through a mechanism that is completely blocked by C3 toxin. In contrast, the NE-dependent CREB-S133 phosphorylation pathway (that involves PKD1, and not PKD2/3) is not blocked by C3 toxin. Collectively, these results identify a specific role for Rho in the S1P/Thr receptor-PKD2/3 pathway, but not the α1-adrenergic receptor-PKD1 pathway, that leads to CREB-S133 phosphorylation in cardiomyocytes (schematized in Fig 5E).
4. DISCUSSION
This study uses Phos-tag SDS-PAGE to identify PKD2 and PKD3 activation mechanisms in cardiac fibroblasts and cardiomyocytes. This method, which involves the incorporation of Phos-Tag into polyacrylamide gel matrices prior to SDS-PAGE and Western blotting, separates phosphoproteins from their nonphosphorylated counterparts. In the case of a protein that is phosphorylated at a single site, the Phos-tag SDS-PAGE method will resolve the monophosphorylated and nonphosphorylated species as two separate bands. However, since PKD family enzymes undergo multi-site phosphorylation, this type of clear separation of inactive (presumably non-phosphorylated) and activated (phosphorylated) species might not necessarily be expected. In this context, we showed that a 5 min with S1P, Thr, or PDGF induced a marked upshift in PKD2 mobility in cardiac fibroblasts - to a single discrete band with an electrophoretic mobility that is markedly slower than the inactive species isolated from unstimulated cells. These results indicate that these receptors trigger a set of synchronized/coordinated phosphorylations at the activation loop, C-terminus and potentially other sites within this enzyme. This contrasts markedly with PKD1, which migrates as multiple bands with distinct electrophoretic mobilities (in cardiac fibroblasts) or a heterogeneous smear (in cardiomyocytes). While this heterogeneity in PKD1’s phosphorylation profile represents a limitation for studies using the Phos-tag SDS method, it provides novel evidence that PKD1 exists as pools of enzyme with distinct phosphorylation profiles even under unstimulated conditions and it provides a rationale for future more detailed MS-based phosphoproteomic analysis.
The Phostag SDS-PAGE method identifies agonist-dependent up-shifts in the electrophoretic mobilities of PKD2 and PKD3 that coincide with phosphorylations at the activation loop and C-terminal phosphorylation sites (phosphorylation sites that can be tracked with commercially available PSSAs). However, the Phos-tag SDS-PAGE method, which provides a more sensitive/unbiased screen for PKD phosphorylation, is uniquely suited to also detect phosphorylations that are not captured by available PSSAs. Our studies identify three examples of the power of this approach:
The Phos-tag SDS-PAGE approach was essential to detect agonist-dependent changes in the phosphorylation of the endogenous PKD3 enzyme. PKD3 has generally been neglected in previous studies, at least in part due to the technical challenges associated with tracking its activation. PKD3 lacks the C-terminal autophosphorylation site. The activation loop phosphorylated (activated) form of PKD3 also is not unambiguously identified in Western blotting studies with the anti-PKD-pSer744/Ser748 PSSA (which recognizes all three PKD isoforms) since the activated form of PKD3 co-migrates with activated forms of PKD1 and PKD2. While the dynamic range for PKD3 electrophoretic mobility shifts are not as impressive as those detected for PKD2, the Phos-tag SDS-PAGE method provides unequivocal evidence that PKD3 is a stimulus-regulated kinase in cardiac fibroblasts and cardiomyocytes.
Phos-tag SDS-PAGE detects slowly migrating/phosphorylated forms of PKD (particularly PKD2) that are not phosphorylated at the activation loop or C-terminal autophosphorylation sites (i.e., site recognized by Western blotting with conventional PSSAs) in agonist-activated cells that have been pretreated with GF109203X. Of note, while these electrophoretic mobility shifts clearly signal changes in PKD phosphorylation profiles, the functional significance of these agonist-dependent changes in PKD phosphorylation (which likely include sites in N-terminal regulatory regions of these enzymes, see Fig 1) is not at all obvious. Any assumptions that these agonist-dependent phosphorylations can be used as general screens (i.e., surrogates) for enzyme activation would be imprudent, since phosphorylations that exert inhibitory controls on catalytic activity have been identified for other enzymes [42] and may also be pertinent to PKDs.
Phos-tag SDS-PAGE provides a sensitive readout for redox-dependent activation of PKD. The fact that H2O2 induces striking up-shifts in the mobilities of all three PKD isoforms indicates that this technique would be particularly useful in cell types where H2O2-dependent increases in PKD activity are associated with relatively modest or nominal increases in phosphorylation at activation loop or C-terminal autophosphorylation sites [43].
Finally, this study shows that PKD1, PKD2 and PKD3 are detected and activated in an isoform-specific manner in both cardiac fibroblasts and cardiomyocytes. Studies in cardiomyocytes confirm (and extend) the previous observation that NE selectively activates PKD1, but not PKD2 or PKD3. This study provides novel evidence that S1P, thrombin, PDGF, and H2O2 effectively activate PKD2 and PKD3 in both cardiomyocytes and cardiac fibroblasts. Of note, all of the heptahelical receptor agonists used in our study recruit the Gαq-dependent pathway that activates phospholipase C-β (PLCβ), but S1P and Thr are unique in that they also couple to Gα12/13-RhoA and the activation of phosphatidylcholine-selective phospholipase D (PLD) or phospholipase C-ε (PLCε) [37]. This study identifies a role for Rho in the S1P and thrombin receptor-dependent pathways that selectively activate PKD2 and PKD3 in both cardiac fibroblasts and cardiomyocytes. We also show that this Rho-PKD2/3 pathway leads to an increase in CREB-S133 phosphorylation in cardiomyocytes. These results suggest that PKD isoforms are not entirely functionally redundant in the heart, and that PKD1 cannot substitute for PKD2 and PKD3 in certain agonist-dependent pathways that influence cardiac remodeling. These results provide the rationale to target the complex cellular machinery that contributes to this PKD isoform signaling specificity for therapeutic advantage.
Highlights.
Phostag SDS-PAGE can be used to resolve protein kinase D isoform activation.
S1P, thrombin, PDGF, and H2O2 activate PKD2 and PKD3 in cardiac cells.
Rho links S1P and thrombin to PKD2/3-dependent CREB activation in cardiomyocytes.
Acknowledgments
Funded by NIH RO1 HL112388.
Abbreviations
- CREB
cAMP response element binding protein
- ET-1
endothelin-1
- NE
norepinephrine
- PMA
phorbol 12-myristate 13-acetate
- Phos-tag
Phosphate binding tag
- PDGF
platelet-derived growth factor
- PH domain
pleckstrin homology domain
- PAGE
polyacrylamide gel electrophoresis
- PKC
protein kinase C
- PKD
protein kinase D
- S1P
sphingosine-1-phosphate
- Thr
thrombin
Footnotes
Disclosures: none
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rozengurt E. Protein kinase D signaling: multiple biological functions in health and disease. Physiology. 2011;26:23–33. doi: 10.1152/physiol.00037.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avkiran M, Rowland AJ, Cuello F, Haworth RS. Protein kinase D in the cardiovascular system: emerging roles in health and disease. Circ Res. 2008;102:157–63. doi: 10.1161/CIRCRESAHA.107.168211. [DOI] [PubMed] [Google Scholar]
- 3.Steinberg SF. Regulation of protein kinase D1 activity. Mol Pharmacol. 2012;81:284–91. doi: 10.1124/mol.111.075986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo J, Gertsberg Z, Ozgen N, Sabri A, Steinberg SF. Protein kinase D isoforms are activated in an agonist-specific manner in cardiomyocytes. J Biol Chem. 2011;286:6500–9. doi: 10.1074/jbc.M110.208058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacamo R, Sinnett-Smith J, Rey O, Waldron RT, Rozengurt E. Sequential protein kinase C (PKC)-dependent and PKC-independent protein kinase D catalytic activation via Gq-coupled receptors: differential regulation of activation loop Ser744 and Ser748 phosphorylation. J Biol Chem. 2008;283:12877–87. doi: 10.1074/jbc.M800442200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kunkel MT, Garcia EL, Kajimoto T, Hall RA, Newton AC. The protein scaffold NHERF-1 controls the amplitude and duration of localized protein kinase D activity. J Biol Chem. 2009;284:24653–61. doi: 10.1074/jbc.M109.024547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanchez-Ruiloba L, Cabrera-Poch N, Rodriguez-Martinez M, Lopez-Menendez C, Martin Jean-Mairet R, Higuero AM, et al. Protein kinase D intracellular localization and activity control kinase D-interacting substrate of 220-kDa traffic through a PDZ-binding motif. J Biol Chem. 2006;281:18888–900. doi: 10.1074/jbc.M603044200. [DOI] [PubMed] [Google Scholar]
- 8.Benes CH, Wu N, Elia AE, Dharia T, Cantley LC, Soltoff SP. The C2 domain of PKCδ is a phosphotyrosine binding domain. Cell. 2005;121:271–80. doi: 10.1016/j.cell.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 9.Doppler H, Storz P. A novel tyrosine phosphorylation site in protein kinase D contributes to oxidative stress-mediated activation. J Biol Chem. 2007;282:31873–81. doi: 10.1074/jbc.M703584200. [DOI] [PubMed] [Google Scholar]
- 10.Hausser A, Storz P, Link G, Stoll H, Liu YC, Altman A, et al. Protein kinase C μ is negatively regulated by 14-3-3 signal transduction proteins. J Biol Chem. 1999;274:9258–64. doi: 10.1074/jbc.274.14.9258. [DOI] [PubMed] [Google Scholar]
- 11.Phan D, Stratton MS, Huynh QK, McKinsey TA. A novel protein kinase C target site in protein kinase D is phosphorylated in response to signals for cardiac hypertrophy. Biochem Biophys Res Commun. 2011;411:335–41. doi: 10.1016/j.bbrc.2011.06.143. [DOI] [PubMed] [Google Scholar]
- 12.Smith FD, Samelson BK, Scott JD. Discovery of cellular substrates for protein kinase A using a peptide array screening protocol. Biochem J. 2011;438:103–10. doi: 10.1042/BJ20110720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sumara G, Formentini I, Collins S, Sumara I, Windak R, Bodenmiller B, et al. Regulation of PKD by the MAPK p38δ in insulin secretion and glucose homeostasis. Cell. 2009;136:235–48. doi: 10.1016/j.cell.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Storz P, Toker A. Protein kinase D mediates a stress-induced NF-κB activation and survival pathway. EMBO J. 2003;22:109–20. doi: 10.1093/emboj/cdg009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Research. 2015;43:D512–20. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Molecular & cellular proteomics. 2006;5:749–57. doi: 10.1074/mcp.T500024-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Kinoshita E, Kinoshita-Kikuta E, Koike T. Advances in Phos-tag-based methodologies for separation and detection of the phosphoproteome. Biochim Biophys Acta. 2015;1854:601–8. doi: 10.1016/j.bbapap.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 18.Kinoshita E, Kinoshita-Kikuta E, Koike T. Separation and detection of large phosphoproteins using Phos-tag SDS-PAGE. Nature Protocols. 2009;4:1513–21. doi: 10.1038/nprot.2009.154. [DOI] [PubMed] [Google Scholar]
- 19.Candasamy AJ, Haworth RS, Cuello F, Ibrahim M, Aravamudhan S, Kruger M, et al. Phosphoregulation of the titin-cap protein telethonin in cardiac myocytes. J Biol Chem. 2014;289:1282–93. doi: 10.1074/jbc.M113.479030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Messer AE, Gallon CE, McKenna WJ, Dos Remedios CG, Marston SB. The use of phosphate-affinity SDS-PAGE to measure the cardiac troponin I phosphorylation site distribution in human heart muscle. Proteomics Clinical applications. 2009;3:1371–82. doi: 10.1002/prca.200900071. [DOI] [PubMed] [Google Scholar]
- 21.Harrison BC, Kim MS, van Rooij E, Plato CF, Papst PJ, Vega RB, et al. Regulation of cardiac stress signaling by protein kinase D1. MolCell Biol. 2006;26:3875–88. doi: 10.1128/MCB.26.10.3875-3888.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oster H, Abraham D, Leitges M. Expression of the protein kinase D (PKD) family during mouse embryogenesis. Gene Expr Patterns. 2006;6:400–8. doi: 10.1016/j.modgep.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 23.Ellwanger K, Pfizenmaier K, Lutz S, Hausser A. Expression patterns of protein kinase D 3 during mouse development. BMC developmental biology. 2008;8:47. doi: 10.1186/1471-213X-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiang SY, Vanhoutte D, Del Re DP, Purcell NH, Ling H, Banerjee I, et al. RhoA protects the mouse heart against ischemia/reperfusion injury. J Clin Invest. 2011;121:3269–76. doi: 10.1172/JCI44371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ozgen N, Obreztchikova M, Guo J, Elouardighi H, Dorn GW, Wilson BA, et al. Protein kinase D links Gq-coupled receptors to cAMP response element-binding protein (CREB)-Ser133 phosphorylation in the heart. J Biol Chem. 2008;283:17009–19. doi: 10.1074/jbc.M709851200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bardswell SC, Cuello F, Rowland AJ, Sadayappan S, Robbins J, Gautel M, et al. Distinct sarcomeric substrates are responsible for protein kinase D-mediated regulation of cardiac myofilament Ca2+ sensitivity and cross-bridge cycling. J Biol Chem. 2010;285:5674–82. doi: 10.1074/jbc.M109.066456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fielitz J, Kim MS, Shelton JM, Qi X, Hill JA, Richardson JA, et al. Requirement of protein kinase D1 for pathological cardiac remodeling. Proc Natl Acad Sci. 2008;105:3059–63. doi: 10.1073/pnas.0712265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuznetsov V, Pak E, Robinson RB, Steinberg SF. Beta2-adrenergic receptor actions in neonatal and adult rat ventricular myocytes. Circ Res. 1995;76:40–52. doi: 10.1161/01.res.76.1.40. [DOI] [PubMed] [Google Scholar]
- 29.Snead AN, Insel PA. Defining the cellular repertoire of GPCRs identifies a profibrotic role for the most highly expressed receptor, protease-activated receptor 1, in cardiac fibroblasts. FASEB J. 2012;26:4540–7. doi: 10.1096/fj.12-213496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Means CK, Brown JH. Sphingosine-1-phosphate receptor signalling in the heart. Cardiovasc Res. 2009;82:193–200. doi: 10.1093/cvr/cvp086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kong P, Christia P, Frangogiannis NG. The pathogenesis of cardiac fibrosis. Cellular and Molecular Life Sciences. 2014;71:549–74. doi: 10.1007/s00018-013-1349-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sabri A, Short J, Guo J, Steinberg SF. Protease-activated receptor-1-mediated DNA synthesis in cardiac fibroblast is via epidermal growth factor receptor transactivation: distinct PAR-1 signaling pathways in cardiac fibroblasts and cardiomyocytes. Circ Res. 2002;91:532–9. doi: 10.1161/01.res.0000035242.96310.45. [DOI] [PubMed] [Google Scholar]
- 33.Matthews SA, Rozengurt E, Cantrell D. Characterization of Ser916 as an in vivo autophosphorylation site for protein kinase D/Protein kinase Cμ. J Biol Chem. 1999;274:26543–9. doi: 10.1074/jbc.274.37.26543. [DOI] [PubMed] [Google Scholar]
- 34.Leask A. Getting to the heart of the matter: new insights into cardiac fibrosis. Circ Res. 2015;116:1269–76. doi: 10.1161/CIRCRESAHA.116.305381. [DOI] [PubMed] [Google Scholar]
- 35.Storz P. Mitochondrial ROS-radical detoxification, mediated by protein kinase D. Trends Cell Biol. 2007;17:13–8. doi: 10.1016/j.tcb.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 36.Lijnen P, Papparella I, Petrov V, Semplicini A, Fagard R. Angiotensin II-stimulated collagen production in cardiac fibroblasts is mediated by reactive oxygen species. Journal of Hypertension. 2006;24:757–66. doi: 10.1097/01.hjh.0000217860.04994.54. [DOI] [PubMed] [Google Scholar]
- 37.Xiang SY, Dusaban SS, Brown JH. Lysophospholipid receptor activation of RhoA and lipid signaling pathways. Biochim Biophys Acta. 2013;1831:213–22. doi: 10.1016/j.bbalip.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan J, Slice LW, Rozengurt E. Activation of protein kinase D by signaling through Rho and the α subunit if heterotrimeric G protein G13. J Biol Chem. 2001;276:38619–27. doi: 10.1074/jbc.M105530200. [DOI] [PubMed] [Google Scholar]
- 39.Aktories K, Just I. In vitro ADP-ribosylation of Rho by bacterial ADP-ribosyltransferases. Methods Enzymol. 1995;256:184–95. doi: 10.1016/0076-6879(95)56023-8. [DOI] [PubMed] [Google Scholar]
- 40.Li C, Li J, Cai X, Sun H, Jiao J, Bai T, et al. Protein kinase D3 is a pivotal activator of pathological cardiac hypertrophy by selectively increasing the expression of hypertrophic transcription factors. J Biol Chem. 2011;286:40782–91. doi: 10.1074/jbc.M111.263046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sabri A, Muske G, Zhang H, Pak E, Darrow A, Andrade-Gordon P, et al. Signaling properties and functions of two distinct cardiomyocyte protease-activated receptors. Circ Res. 2000;86:1054–61. doi: 10.1161/01.res.86.10.1054. [DOI] [PubMed] [Google Scholar]
- 42.Gong J, Yao Y, Zhang P, Udayasuryan B, Komissarova EV, Chen J, et al. The C2 domain and altered ATP-binding loop phosphorylation at Ser359 mediate the redox-dependent increase in protein kinase C-δ activity. Mol Cell Biol. 2015;35:1727–40. doi: 10.1128/MCB.01436-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Storz P, Doppler H, Toker A. Activation loop phosphorylation controls protein kinase D-dependent activation of nuclear factor κB. Mol Pharm. 2004;66:870–9. doi: 10.1124/mol.104.000687. [DOI] [PubMed] [Google Scholar]