Abstract
Background
The kinesin family member C1 (KIFC1, also known as HSET) is a kinesin superfamily protein (KIFs). Although KIFC1 acts as a crucial role in the development of several human cancers, the KIFC1 expression profile and functional remain unclear in non-small cell lung cancer (NSCLC).
Methods
We collected the fresh NSCLC samples and paired normal lung tissue in patients with lung cancer operation, and detected KIFC1 expression using quantitative reverse-transcription polymerase chain reaction (qRT-PCR) and Western blotting. To expand on previous smaller-scale studies, NSCLC tissue microarrays (TMA) were analyzed by IHC. Finally, cell lines were employed to further probe the potential mechanisms,
Results
In this study, we described that KIFC1 was significantly upregulated in NSCLC tissues compared with the corresponding normal tissues. Moreover, KIFC1 overexpression was associated with the poor overall survival (OS) of NSCLC patients, and siRNA-mediated knockdown of KIFC1 significantly suppressed tumor cell proliferation in vitro. Further verification showed that inhibition of KIFC1 gene expression caused the upregulation of the cyclin-dependent kinases inhibitor p21 and downregulation of the cell cycle driver protein cdc2, which arrested cells in the G2-M phase.
Conclusions
we report that increased KIFC1 expression may promote cell proliferation and identified it as a biomarker of unfavorable prognosis in NSCLC patients.
Keywords: Kinesin family member C1 (KIFC1), non-small cell lung cancer (NSCLC), cyclin-dependent kinase inhibitor 1A (p21), cell division cycle protein 2 homolog (cdc2), prognostic
Introduction
Lung cancer is currently the leading cause of cancer death throughout the world. The latest statistics show that lung cancer is the most common cancer in China (1). Lung cancer includes two main types, namely, small cell lung cancer (SCLC) and non-SCLC (NSCLC). NSCLC accounts for approximately 85% of all cases of lung cancer (2,3). Survival rates for lung cancer remain abysmal, with nearly 70% new cases diagnosed at advanced stages. The 5-year survival rate in the United States for lung cancer is 15.6% (4), the data of age-standardized 5-year relative survival is 15.4% in China (5) and has been unchanged for decades despite surgical advances and new antitumor drugs. Therefore, the importance of further understanding the molecular mechanisms underlying tumor cell proliferation cannot be overemphasized.
Cell proliferation divide by mitosis, and mitosis is a part of the cell cycle in which chromosomes in a cell nucleus are separated into two identical sets of chromosomes, finally, each set ends up in its own nucleus. During mitosis, the chromosomes, which have already duplicated, condense and attach to spindle fibers that pull one copy of each chromosome to opposite sides of the cell (6). Dysregulation of mitosis can create aneuploid cells that have too few or too many of one or more chromosomes, a condition associated with cancer (7). In addition, other errors during mitosis can induce apoptosis (programmed cell death) or cause mutations. Certain types of cancer can arise from such mutations (8).
Kinesin superfamily proteins (KIFs) is a protein belonging to a class of motor proteins which can attach to microtubules and move along them to transport organelles, protein complexes, and messenger RNAs (mRNAs). In recent years, it has been found that a number of KIFs have a role in mitosis (cell division) through taking part in chromosomal and spindle movements (9,10); Thus, alterations in KIFs expression might cause cancer formation. The KIFs superfamily consists of 45 members, including 39 N-kinesins, three M-kinesins, and three C-kinesins (11). Kinesin family member 14 (KIF14) is distinguished from other kinesins because it has minus-end-directed motility (12). Several studies have shown that the KIF14 protein is overexpressed in retinoblastomas and primary lung tumors compared with their respective normal tissues (13), and KIF14 expression is an independent prognostic factor for disease-free survival in lung cancer.
KIFC1 is the only member of the KIF14 family that has minus-end-directed motility in mammalian cells (14). It is located on the short arm of chromosome 6 with the 2nd District 1 band 3rd sub-band second subzone sites (http://www.genenames.org) and has four transcripts containing a total of 20 exons on the forward strand (http://www.ebi.ac.uk/uniprot). Research shows that KIFC1 participates many physiological and pathological processes. Such as, KIFC1 is associated with the nuclear membrane and acrosome in round and elongating spermatids (15) and is required for chromosome congression and alignment during mitosis (16). Farina et al. reported that depletion of KIFC1 significantly decreases double-stranded DNA transport, which might provide new insight for gene therapy and DNA-based therapy (17). The exchange of intracellular substances is the driving force for the survival of cells, even more so for cancer cells. Previous studies have shown that KIFC1 is considered to be a potential cancer biomarker and molecular target for cancer therapy. For instance, KIFC1 is overexpressed in ovarian cancer and metastases compared with the respective normal tissue (18), overexpression of KIFC1 may mediate Docetaxel resistance in breast cancer (19), and the expression levels of KIFC1 are highly predictive of brain metastasis in early and advanced lung cancer (20). However, the expression profile and biological role of KIFC1 in NSCLC remain largely incomprehension.
The present study was designed to detect the expression of KIFC1 in fresh NSCLC samples and paired normal lung tissue through quantitative reverse-transcription polymerase chain reaction (qRT-PCR) and Western blotting, and NSCLC tissue microarrays (TMA) were analyzed by IHC. Our results show that KIFC1 is overexpression in lung cancer tissues. Meanwhile, KIFC1 knockdown induce growth inhibition by arresting the cell cycle in human lung cancer cell lines in vitro, furthermore, the p21 and cdc2 proteins seem an essential part of this process.
Methods
Tissue collection
Forty paired lung adenocarcinoma and corresponding normal lung tissue samples were collected from patients who underwent surgery at the Department of Thoracic Surgery, Jinling Hospital, Nanjing, China between 2014 and 2015. All of the final diagnoses were confirmed based on histopathological examination. No enrolled patients had received any prior chemotherapy or radiotherapy before surgery. All of the obtained tissue samples were immediately snap-frozen in liquid nitrogen and stored at −80 °C until RNA extraction. RNA extraction was performed using a non-enzymatic procedure. Informed consent to use the data-use agreement was obtained from all patients before surgery.
Cell lines and culture conditions
Human NSCLC cell lines A549, H1299 and SPC-A1 were purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). A549 and H1299 were maintained in RPMI 1640 basic medium (GIBCO-BRL), and SPC-A1 was cultured in Dulbecco’s modified Eagle’s medium (DMEM) (GIBCO-BRL) supplemented with 10% fetal bovine serum (FBS) (Gibco), 100 U/L penicillin and 0.1 mg/mL streptomycin (all of which were from Invitrogen, Carlsbad, California). All cells were maintained at 37 °C in a humidified incubator with 5% CO2.
RNA isolation and qRT-PCR analyses
Total RNA was extracted from the cultured cells or tissues using TRIZOL reagent (Invitrogen) according to the manufacturer’s instructions. For qRT-PCR, the isolated RNA was reverse transcribed to cDNA using a Reverse Transcription Kit (Takara, Dalian, China). qRT-PCR analyses were conducted with SYBR Premix Ex TaqTM (Tli RNaseH Plus) (Takara, Dalian, China) according to the manufacturer’s instructions. The results were normalized to the expression of β-actin. The primers were as follows: β-actin sense 5'-TGACGTGGACATCCGCAAAG-3', reverse 5'- CTGGAAGGTGGACAGCGAGG-3'; KIFC1 sense 5'-TGAGCAACAAGGAGTCC CAC-3', reverse 5'-TCACTTCCTGTTGGCCTGAG-3'. The PCR reaction was conducted at 95 °C for 30 s followed by 40 cycles of 95 °C for 5 s and 60 °C for 34 s in an ABI 7500 real-time PCR system (Applied Biosystems, Foster City, CA, USA). The results are presented as the mean ± standard deviation (SD) for duplicate runs. The relative quantification of KIFC1 expression was calculated using the 2−△△CT method relative to β-actin.
Transfection of cell lines
The siRNA (small interfering RNA) sequences were as follows: siRNA- control sense 5'-UUC UCC GAA CGU GUC ACG UTT-3', antisense 5'-ACG UGA CAC GUU CGG AGA ATT-3'; si-KIFC1-1 sense 5'-CCU GGA GCC UGA GAA GAA ATT -3', antisense 5'-UUU CUU CUC AGG CUC CAG GTT -3'; si-KIFC1-2 sense 5'-GGU CAG UUA UGU GAC CUA ATT -3', antisense 5'-UUA GGU CAC AUA ACU GAC CTT -3'. A549, H1299 and SPC-A1 cells were seeded and cultured in growth media until the cell density reached 60–70% prior to transfection with small interfering RNAs (siRNAs). Lipofectamine® 2000 reagent (Invitrogen, 1070962) was used for siRNA transfection following the manufacturer’s instructions, and the media were replaced 6 h post-transfection. The cells were harvested 48 h after transfection, and the total RNA or protein was isolated according to the manufacturer’s instructions. Specific silencing of KIFC1 expression was assessed using qRT-PCR and western blot.
MTT cell growth assay
KIFC1 siRNA-transfected and control siRNA-transfected A549, H1299 and SPC-A1 cells (3,000/well) were allowed to grow in 96-well plates 24 h after siRNA transfection. Total cell numbers were assessed every 24 h after incubation with 20 µL of MTT (5 mg/mL) for 4 h. Following incubation, the cell supernatants were carefully aspirated from each well and discarded; then, 200 µL of DMSO was added to each well. The plates were then oscillated in the dark for 10 min until the crystals were fully dissolved. The optical density (OD) of each well was then measured with a microplate reader at 490 nm, and the OD values were reported as the means ± SD.
Colony formation assays
Cells were transfected in six-well plates for 24 h with siRNA, placed in six-well plates (500 cells per well) and maintained in media containing 10% FBS. The medium was replaced every 4 days. After 14 days, colonies were fixed with methanol and stained with 0.1% crystal violet (Sigma, St. Louis, MO). Visible colonies were then manually counted. For each treatment group, wells were counted in triplicate.
Western blotting analysis
Cells were lysed using a lysis buffer containing the mammalian protein extraction reagent RIPA (Beyotime China), a protease inhibitor cocktail (Roche) and PMSF (Roche). Protein concentrations were determined using the Bradford method with a Bio-Rad Protein Assay kit (KeyGEN Biotech). The protein samples (30 µg) were assessed by 12% or 10% SDS-PAGE and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, USA). The membranes were blocked with PBST buffer (PBS plus 0.05% Tween-20) containing 5% w/v nonfat milk, incubated overnight with several antibodies at 4 °C, and then incubated with a specific secondary antibody for 2 h at room temperature. Protein bands were visualized using an ECL detection system (Tanon, China), and β-actin (Sigma, USA) was used as a control. Additionally, an antibody (1:1,000 dilution) against KIFC1 was purchased from Santa Cruz Biotechnology, Inc. (Shang Hai, China).
Flow-cytometric analysis of the cell cycle
The three cell lines transiently transfected with siRNAs of KIFC1 or negative control were harvested by trypsinization and washed twice with cold phosphate buffered saline (PBS) 48 h after transfection. For cell cycle analysis, the cells were stained with PI using the Cycle TESTTM PLUS DNA reagent kit (BD Biosciences) according to the manufacturer’s protocol. The ratio of cells in the G0/G1, S, and G2/M phases were counted and compared. All of the samples were assayed in triplicate.
TMA and IHC
A TMA from Shanghai Outdo Biotech Co., LTD was used to analyze the expression of KIFC1 by IHC. For the evaluation of KIFC1 staining, a semiquantitative scoring criterion that notes the signal strength was used: 0 (no signal), 1 (weak), 2 (moderate), and 3 (strong). Percentage scores were assigned as 0: 0%; 1: 1% to 25%; 2: 26% to 50%; 3: 51% to 75% and 4: 76% to 100%. The staining index of each sample was derived by multiplying the signal score by the percentage score to give a final score from 0 to 12. The average of all of the scores served as the optimal cutoff value that was used to classify high or low expression and analyze the related clinical characteristics.
Statistical analysis
Statistical analysis was performed using SPSS 19.0 and GraphPad Prism5 software. The χ2 test was used to evaluate the possible correlations between KIFC1 expression and the baseline clinical characteristics of patients with NSCLC. The differences in the expression of KIFC1 between NSCLC and relevant normal lung tissue were examined by the paired-sample t-test. The Kaplan-Meier method was used to plot the patient survival probability and was compared with the log-rank test. A Cox regression model was used for the analysis of independent prognostic risk factors. All data were presented as the mean ± standard error of the mean. Two-sided P values <0.05 were considered significant.
Results
KIFC1 is overexpressed in lung cancer tissues and correlates with poor prognosis
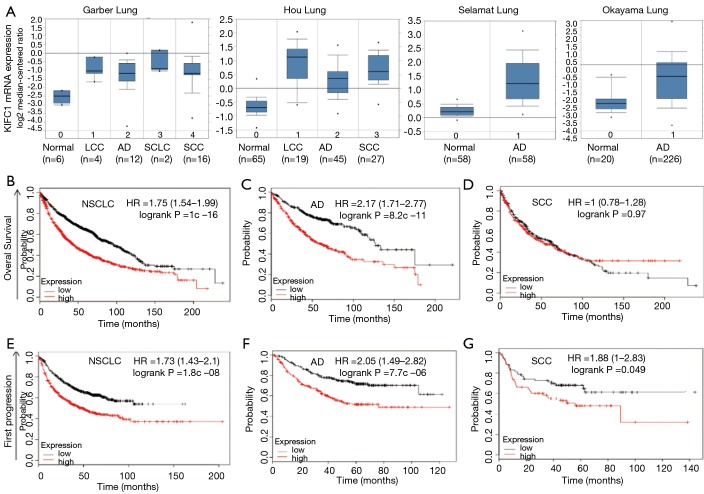
To validate the relationship between the KIFC1 expression status and lung cancer development, we mined the data from the publicly available Oncomine (www.oncomine.org) database. The validation data confirmed that KIFC1 mRNA expression levels were overexpressed in lung cancer tissues compared with the corresponding normal tissues (Figure 1A). Moreover, the browser’s Kaplan-Meier plot (http://kmplot.com/analysis/index.php?p=background) suggested that the patients in the high KIFC1 mRNA expression subgroup (red line) had worse overall survival (OS) (Figure 1B,C,D) and progression-free survival (PFS) (Figure 1E,F,G) compared to the low expression subgroup (black line), especially in lung adenocarcinomas (Figure 1C,F).
Figure 1.
The expression of KIFC1 and the survival curves in lung cancer patients from the publicly available database. (A) KIFC1 mRNA expression of lung cancer tissues is higher than normal tissues due to four databases. (B-D) KIFC1 overexpression is associated with worse overall survival (OS) and progression-free survival (PFS) (E-G). Red line: the high KIFC1 expression subgroup; black line: the low expression subgroup. The letters (B,E), (C,F) and (D,G) represent NSCLC, lung adenocarcinoma and lung squamous carcinoma, respectively. KIFC1, kinesin family member C1; NSCLC, non-small cell lung cancer.
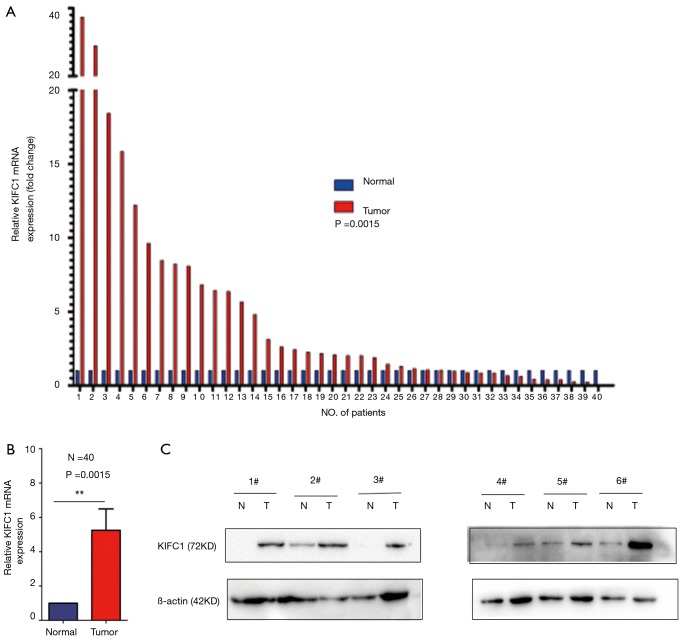
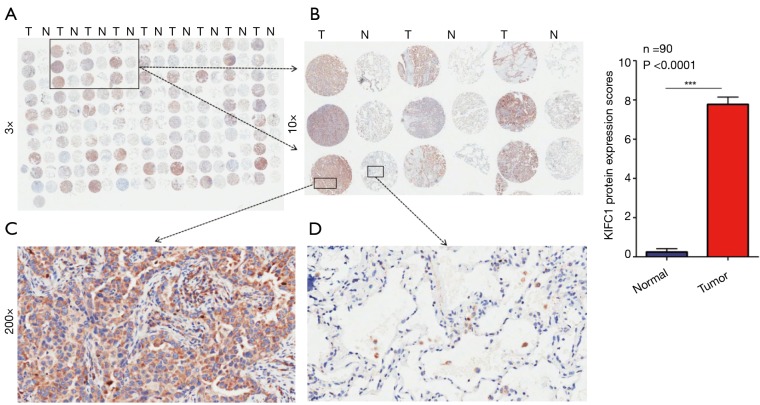
To confirm whether KIFC1 was differentially expressed in lung cancer tissues, we analyzed KIFC1 mRNA expression in 40 clinical lung cancer tissues and paired adjacent normal lung tissues using qRT-PCR. KIFC1 mRNA expression levels were significantly higher in lung cancer tissues than in adjacent normal tissues (P<0.001; Figure 2A,B). Western blotting analysis also showed that KIFC1 protein expression was upregulated in lung cancer tissues compared with the adjacent normal tissues (Figure 2C). To further expand the sample validation, IHC was performed using a NSCLC TMA that was constructed according to a method described previously (21). Using the above criteria, KIFC1 expression in normal lung tissue was significantly lower than that in lung cancer tissues (P<0.001; Figure 3). As shown in Table 1, KIFC1 overexpression was observed in 59/90 (65.6%) of NSCLC tissues (Table 1). These data demonstrate that KIFC1 may be a good diagnostic biomarker for NSCLC.
Figure 2.
Relative KIFC1 expression in NSCLC tissues. (A) The log2 relative KIFC1 mRNA expression in NSCLC tissues (n=40) and in paired adjacent normal tissues (n=40) was determined by qRT-PCR. KIFC1 mRNA expression was normalized to β-actin expression; (B) the statistical results of A; (C) Western blot of KIFC1 protein expression in NSCLC tissues (n=6) and in paired adjacent normal tissues (n=6). KIFC1, kinesin family member C1; NSCLC, non-small cell lung cancer; qRT-PCR, quantitative reverse-transcription polymerase chain reaction. **, P<0.01.
Figure 3.
KIFC1 protein expression in lung cancer and corresponding normal tissues with tissue microarray (TMA). (A) Immunohistochemical (IHC) staining of KIFC1 in lung cancer and corresponding normal tissue of 90 lung cancer patients; (B) typical IHC staining of KIFC1 in nine pairs of tissues; (C) high IHC staining of KIFC1 in lung cancer sample; (D) low IHC staining of KIFC1 in non-cancerous tissue sample. Orange arrow shows negative staining of non-cancerous cells. Original magnification ×3 in (A); ×10 in (B); ×200 in (C,D). (E) KIFC1 protein expression scores are presented as the mean ± SD in lung cancer and corresponding normal tissue. The percentage and intensity scores were multiplied to obtain a total score (range, 0–12).
Table 1. Relationship of KIFC1 protein expression with clinicopathological characteristics in lung cancer patients.
| Characteristics | Number of patients | KIFC1 protein expression | ||
|---|---|---|---|---|
| Low (﹤8) | High (≥8) | P | ||
| All patients | 90 | 31 | 59 | |
| Gender | 0.403 | |||
| Male | 49 | 15 | 34 | |
| Female | 41 | 16 | 25 | |
| Age (years) | 0.608 | |||
| ﹤63 | 46 | 17 | 29 | |
| ≥63 | 44 | 14 | 30 | |
| Size of tumor (cm) | 0.121 | |||
| ≤3 | 33 | 8 | 25 | |
| >3 | 57 | 23 | 34 | |
| Grade | 0.354 | |||
| Low | 21 | 9 | 12 | |
| High | 69 | 22 | 47 | |
| p-N status | 0.8 | |||
| N0 | 39 | 14 | 25 | |
| N1–3 | 51 | 17 | 34 | |
| p-TNM stages | 0.771 | |||
| I + II | 57 | 19 | 38 | |
| III + IV | 33 | 12 | 21 | |
p-N, Lymph node metastasis; *, P﹤0.05.
Association between KIFC1 expression and clinical characteristics
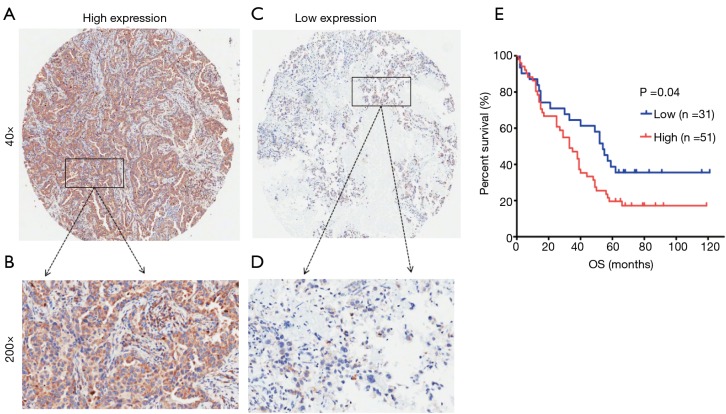
The TMA consisted of 90 pairs of non-squamous-NSCLC patients who underwent surgical treatment in July 2004 to June 2009 and were followed up until August 2014. The follow-up strategies and treatment to the patients in the TMA were conducted according to the time of clinical guidelines. The primary clinical information of these patients is listed in Table 1. The average age was 62.5 years (range 30 to 84 years) for the group of 49 men and 41 women. Eighty-two common lung adenocarcinomas, 7 bronchioloalveolar carcinomas and 1 muco-epidermoid carcinoma were included in this study. One patient had distant metastasis, while 51 patients had lymph node metastasis. Regarding the clinical TNM stage, 33 patients were in advanced stages III–IV, while the other 57 patients were in stages I–II. The Kaplan-Meier survival analysis showed that the OS of patients with low KIFC1 staining was significantly longer than that of patients with high KIFC1 staining (log-rank test P=0.04; Figure 4), while no significant associations between KIFC1 overexpression and age, gender, size of tumor, p-N status and p-TNM stage were found (Table 1). Furthermore, a Cox multivariate regression analysis also showed that lymph node metastasis (HR =2.133; 95% CI: 1.153 to 3.944; Table 2) and advanced stage indicated poor prognosis (HR =1.794; 95% CI: 1.001 to 3.219; Table 2). Taken together, these results demonstrate that KIFC1 is overexpressed in lung cancer and associated with poor survival.
Figure 4.
KIFC1 overexpression is associated with poor prognosis in lung adenocarcinomas. (A,B) High expression of KIFC1 in lung adenocarcinomas; (C,D) low expression of KIFC1 in lung adenocarcinomas. Original magnification ×40 in (A) and (C); ×200 in (B) and (D). (E) Survival curves of common lung adenocarcinomas patients with low expression versus high expression of KIFC1 (P<0.05, log-rank test).
Table 2. Multivariate analyses of KIFC1 protein expression and other clinical prognostic factors for OS in lung cancer patients.
| Factors | OS (over survival) | |
|---|---|---|
| HR (95% CI) | P | |
| Age (≥63/<63 years) | 0.979 (0.565–1.698) | 0.941 |
| Gender (female/male) | 0.912 (0.539–1.542) | 0.730 |
| Size of tumor (>3/≤3 cm) | 1.417 (0.781–2.573) | 0.252 |
| Grade (low/high) | 0.724 (0.387–1.352) | 0.311 |
| N stage (N1–3/N0) | 2.133 (1.153–3.944) | 0.016* |
| p-TNM stage (III + IV/I + II) | 1.794 (1.001–3.219) | 0.049* |
| KIFC1 expression (high/low) | 1.020 (0.565–1.843) | 0.311 |
HR, hazard ration; 95% CI, 95% confidence interval. *, P﹤0.05.
KIFC1 siRNA efficiently silences KIFC1 expression in NSCLC cell lines
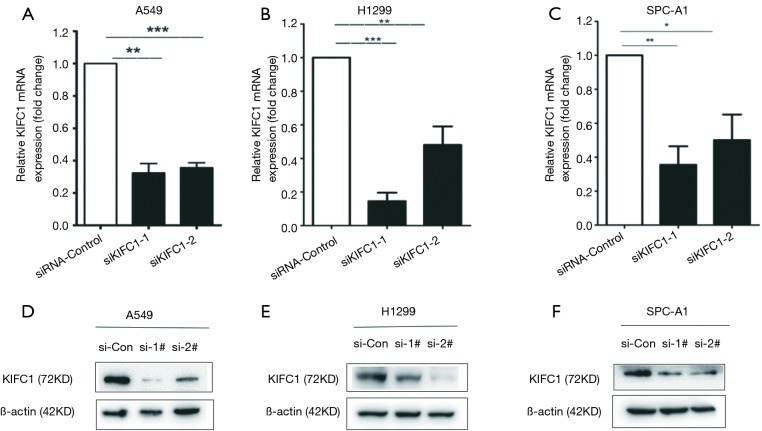
To explore the role of KIFC1 in lung cancer, three NSCLC cell lines, including A549, H1299, and SPC-A1, were utilized for in vitro investigations. We designed a siRNA that targeted human KIFC1 and transfected it into the cells. Then, we tested the gene silencing efficiency in these cell lines. Compared to the empty vector (siRNA-control) transfected cells, si-KIFC1-1 and si-KIFC1-2 efficiently silenced KIFC1 expression at the mRNA level (Figure 5A,B,C) and reduced the protein production (Figure 5D,E,F).
Figure 5.
KIFC1 expression could be inhibited by KIFC1 siRNA. (A-C) The expression of KIFC1 mRNA level was measured following the treatment of A549, H1299 and SPC-A1 cells with siRNA by qRT-PCR; (D-F) Western blot analyze KIFC1 expression after the treatment of A549, H1299 and SPC-A1 cells with siRNA in protein level. The letters (A,D), (B,E) and (C,F) represent A549, H1299 and SPC-A1 cell lines, respectively. *, P<0.05; **, P<0.01; ***, P<0.001. KIFC1, kinesin family member C1; qRT-PCR, quantitative reverse-transcription polymerase chain reaction.
Downregulated KIFC1 represses proliferation in NSCLC cell lines
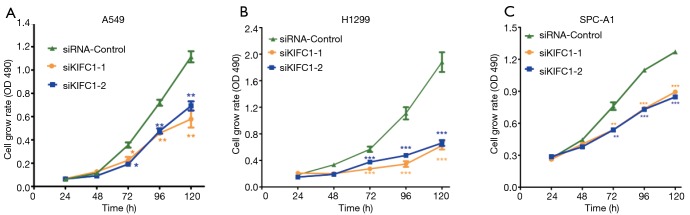
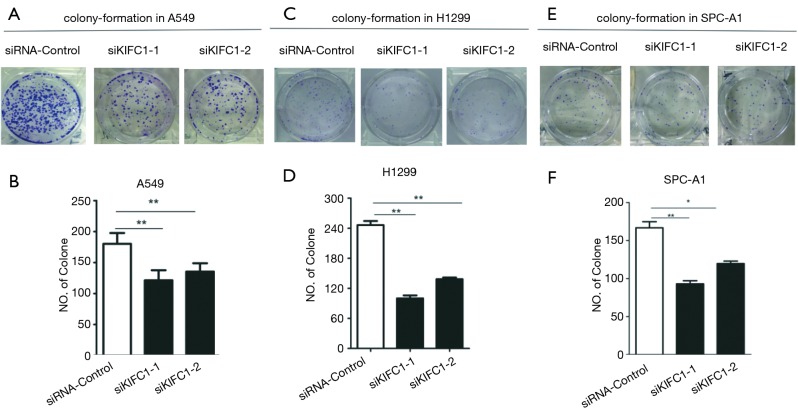
We examined the impact of KIFC1 knockdown on the growth of human NSCLC cells. Compared with the siRNA-control transfected cells, transfection with si-KIFC1-1 and si-KIFC1-2 resulted in a significant decrease in A549, H1299 and SPC-A1 cell viability, as monitored by the MTT assay (Figure 6). Moreover, the colony formation assay results also revealed that KIFC1 knockdown greatly attenuated clonogenic survival (Figure 7). Thus, these data suggest that lower KIFC1 expression inhibits the proliferation of NSCLC cells.
Figure 6.
Downregulation of KIFC1 decreases A549, H1299 and SPC-A1 cell proliferation by MTT assay. The letters (A), (B) and (C) represent A549, H1299 and SPC-A1 cell lines, respectively. *, P<0.05; **, P<0.01; ***, P<0.001. KIFC1, kinesin family member C1; MTT, 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay.
Figure 7.
The cell clone formation ability be suppressed when KIFC1 or control siRNA-transfected A549, H1299 and SPC-A1 cells. (A,C,E) The colonies were captured; (B,D,F) the count was showed and the data represent the mean ± SD from three independent experiments. The letters (A,B), (C,D) and (E,F) represent A549, H1299 and SPC-A1 cell lines, respectively. *, P<0.05; **, P<0.01. KIFC1, kinesin family member C1.
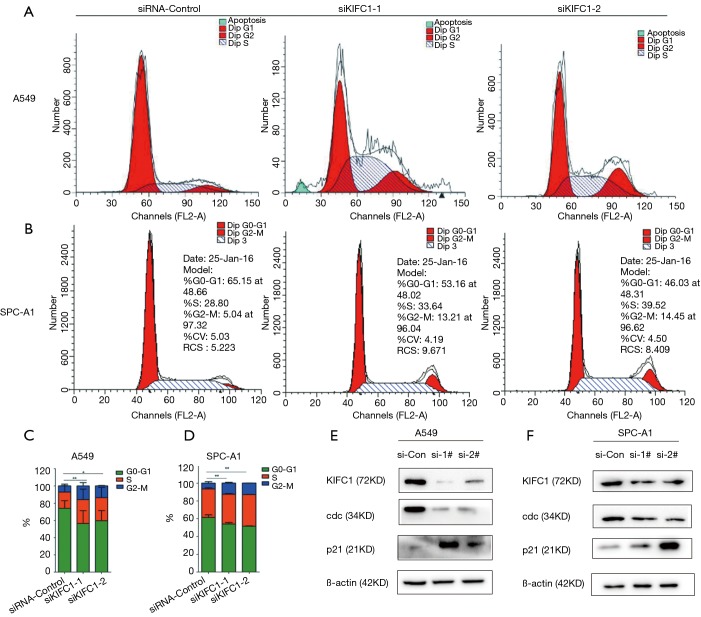
Depletion of KIFC1 expression leads to cell cycle arrest in the G2/M phase
As KIFC1 expression can affect the growth of cells, we investigated the role of KIFC1 in the regulation of cell cycle progression in cancer cells using flow cytometry. The cell cycle analyses indicated that growth inhibition of the A549 and SPC-A1 cell lines was accompanied by a corresponding increase in the proportion of cells in the G2/M and a decrease in the proportion of cells in the G0/G1 phases (Figure 8A,B,C,D). These findings demonstrated that low expression of KIFC1 could arrest cells in the G2/M phase, suggesting that KIFC1 plays a role in cancer development through the regulation of cell cycle progression in the G2/M phase.
Figure 8.
Knockdown of KIFC1 expression induced G2/M phase arrest, upregulates expression of p21 and suppresses activation of cdc2 in A549 and SPC-A1. The letters (A,B) (C,D) and (E,F) represent the result of FCM, statistical views and Western blotting analysis, the letters (A,C,E) and (B,D,F) represent A549 and SPC-A1 cell lines, respectively. *, P<0.05; **, P<0.01; ***, P<0.001. KIFC1, kinesin family member C1; p21, cyclin-dependent kinase inhibitor 1A; cdc2, cell division cycle protein 2 homolog.
KIFC1 knockdown upregulates the expression of p21 and suppresses activation of cdc2
Considerable evidence indicates that cdc2 plays an important role in arresting cells in the G2/M phase (22,23). We detected the expression levels of cdc2 in the A549, and SPC-A1 cell lines. The results were similar to the cell cycle data in that knockdown of KIFC1 in A549 and SPC-A1 cells can reduce the expression of cdc2 (Figure 8E,F). Several proteins upstream of cdc2 were also tested. The results showed that the inhibition of KIFC1 significantly affected the expression of p21 in A549 and SPC-A1 cells (Figure 8E,F). Comprehensive analysis of the results found that the downregulation of KIFC1 can inhibit cdc2 expression and arrest cells in G2/M via induction of p21 in NSCLC cells.
Discussion
Although there is a great deal of information from the last few decades about lung cancer at the genetic and molecular level, the importance of exploring the mechanisms of lung cancer cannot be overemphasized such that new molecular parameters can be identified for the prediction of prognosis and treatment of patients with lung cancer. In this study, we proved that the expression of KIFC1 was frequently high in many of the NSCLC specimens based on the multicenter of the data for the first time; in contrast, KIFC1 expression in all of the normal lung tissue specimens was absent or at low levels. Therefore, our observations from this study indicated that KIFC1 might act as an oncogene in human tumor progression. Moreover, overexpression of KIFC1 in lung cancer is associated with a adverse prognosis, especially in lung adenocarcinomas.
PRC1 also referred to as protein regulator of cytokinesis 1, contains two Cdk phosphorylation motifs, and phosphorylation is possibly important to mitotic suppression of bundling (24), which overexpression extensively bundles interphase microtubules (24). PRC1 also involved in the formation of cancer, research shows that the expression of PRC1 is upregulated in the development of breast cancer (25). Similar to KIFC1, endogenous PRC1 levels was significant increase in G2/M phase using breast cancer cell lines (25), and KIF14 targets to the central spindle via its interaction with PRC1 and has an essential function in cytokinesis (26). So, KIFC1 and PRC1 may interact together to accelerate cell cycle period, leading to tumor cell proliferation.
The roles of KIFC1 in NSCLC prognostic have not been previously reported. Although the expression of KIFC1 was a considerable factor in survival univariate analysis (P=0.04) and the higher gene expression of KIFC1 showed trends of poorer outcome in the large sample (Kaplan-Meier plot), multivariate analyses showed expression of KIFC1 has no correlation with prognosis of NSCLC (P=0.311) in TMA data; One of the reasons for this is the small sample size. By the previous studies, KIFC1 can promote lung cancer brain metastases (20), and Pawar et al. found the worse prognosis of ovarian cancer is presented in the high expression of KIFC1 (18). PTP1B (protein tyrosine phosphatase 1B) promotes cell proliferation and metastasis, so it is viewed as an index of prognosis of tumor (27), PP2A (protein phosphatase 2A) had some effects on tumor cell proliferation that could eventually be harmful to a patient, so research suggests that it plays an important role in NSCLC progression (28). The same principles apply to our data, which revealed that knockdown of KIFC1 could suppress the growth of lung cancer cells, and patients with higher KIFC1 expression have shorter survival time than those with lower KIFC1 expression from the point of large sample data. These suggested that KIFC1 could serve as a potential prognosis marker for NSCLC patients.
Cancer cells differ from normal cells in ways that allow for their unregulated growth and proliferation; thus, restraining the growth of cancer cells can significantly reduce the volume of a tumor (29). The role of KIFC1 in cancer cell proliferation has not been previously reported. Zou et al. found that estrogen stimulation strongly induces expression of KIFC1 in breast cancer cells (30). Our study shows that a clear decrease in cell growth and colony formation ability could be induced by the silencing of KIFC1 with siRNAs in NSCLC cell lines. A novel integrated high-throughput synthesis and screening method has shown that AZ82 is a potent KIFC1 inhibitor and that it is safe in mice after intraperitoneal injection (31). Thus, it is possible that KIFC1 inhibition can be exploited therapeutically for human cancers combining with our data.
KIFC1 involves the growth of tumor and prognosis (18-21,32), but these were based on clinical specimens. Our experiment is validated at a cellular level for the first time and the further explore the mechanisms was also first expounded. The dysregulation of cell cycle components may lead to tumor formation (33). The cell cycle can be arrested at the G1, S or G2/M phase by many cytotoxic agents and/or DNA-damaging agents (34). Cdc2 plays a main role in the G2/M phase transition, and this effect depends on the formation of the cdc2-cyclin B1 complex (35). Cdc2-cyclin B1 kinase activity can be regulated by cdc25C (36); in addition, p21 has an inhibitory effect on the expression of cdc2 (37). We detected cdc2, cdc25C (data not shown) and p21 expression in three NSCLC cell lines in which the KIFC1 gene had been silenced by RNA interference, as shown in Figure 8C. P21 was overexpressed and cdc2 expression was reduced in cell lines with cell cycle alterations. These results verified that downregulation of KIFC1 could arrest NSCLC cells in the G2/M phase and that this effect was dependent on the suppression of cdc2 expression due to the increase in p21 expression rather than cdc25C expression.
Our study did have some limitations that affected the overall results. First, it is an in vitro study, so these phenomena are only validated in cell biological terms; thus, animal tests are needed. Second, the tissue specimens that compose the TMA are considerably old, which could affect the detection of protein expression.
Conclusions
In summary, we show for the first time that KIFC1 can be identified as a prognostic factor in NSCLC, especially in lung adenocarcinomas. We also show that the low expression of KIFC1 can suppress NSCLC proliferation by arresting the cell cycle in the G2/M phase by upregulating p21 and subsequently inhibiting cdc2. These data provide a theoretical basis for future applications of KIFC1 inhibitors in the treatment of NSCLC.
Acknowledgements
Funding: This work was supported by the Foundation of Jiangsu Lung cancer diagnostic and treatment research in Medical Science of China (BL2013026). The National Natural Science Foundation of China (No. 81572273) and the National Natural Science Foundation of China (No. 81572937).
Ethical Statement: The study was approved by Jingling Hospital’s Institutional Review Committee on Human Research (No. 2015 GJJ-036a). Informed consent to use the data-use agreement was obtained from all patients before surgery.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 2.Navada S, Lai P, Schwartz AG, et al. Temporal trends in small cell lung cancer: Analysis of the national Surveillance, Epidemiology, and End-Results (SEER) database. J Clin Oncol 2016;24:abstract 7082. [Google Scholar]
- 3.Sher T, Dy GK, Adjei AA. Small cell lung cancer. Mayo Clin Proc 2008;83:355-67. 10.4065/83.3.355 [DOI] [PubMed] [Google Scholar]
- 4.Dela Cruz CS, Tanoue LT, Matthay RA. Lung cancer: epidemiology, etiology, and prevention. Clin Chest Med 2011;32:605-44. 10.1016/j.ccm.2011.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng R, Zeng H, Zhang S, et al. National estimates of cancer prevalence in China, 2011. Cancer Lett 2016;370:33-8. 10.1016/j.canlet.2015.10.003 [DOI] [PubMed] [Google Scholar]
- 6.O'Connor C. Cell Division: Stages of Mitosis. Nature Education 2008;1:188. [Google Scholar]
- 7.Draviam VM, Xie S, Sorger PK. Chromosome segregation and genomic stability. Curr Opin Genet Dev 2004;14:120-5. 10.1016/j.gde.2004.02.007 [DOI] [PubMed] [Google Scholar]
- 8.Kops GJ, Weaver BA, Cleveland DW. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat Rev Cancer 2005;5:773-85. 10.1038/nrc1714 [DOI] [PubMed] [Google Scholar]
- 9.Hirokawa N, Noda Y, Tanaka Y, et al. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol 2009;10:682-96. 10.1038/nrm2774 [DOI] [PubMed] [Google Scholar]
- 10.Sharp DJ, Rogers GC, Scholey JM. Microtubule motors in mitosis. Nature 2000;407:41-7. 10.1038/35024000 [DOI] [PubMed] [Google Scholar]
- 11.Miki H, Setou M, Kaneshiro K, et al. All kinesin superfamily protein, KIF, genes in mouse and human. Proc Natl Acad Sci U S A 2001;98:7004-11. 10.1073/pnas.111145398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fink G, Hajdo L, Skowronek KJ, et al. The mitotic kinesin-14 Ncd drives directional microtubule-microtubule sliding. Nat Cell Biol 2009;11:717-23. 10.1038/ncb1877 [DOI] [PubMed] [Google Scholar]
- 13.Corson TW, Huang A, Tsao MS, et al. KIF14 is a candidate oncogene in the 1q minimal region of genomic gain in multiple cancers. Oncogene 2005;24:4741-53. 10.1038/sj.onc.1208641 [DOI] [PubMed] [Google Scholar]
- 14.Dagenbach EM, Endow SA. A new kinesin tree. J Cell Sci 2004;117:3-7. 10.1242/jcs.00875 [DOI] [PubMed] [Google Scholar]
- 15.Yang WX, Jefferson H, Sperry AO. The molecular motor KIFC1 associates with a complex containing nucleoporin NUP62 that is regulated during development and by the small GTPase RAN. Biol Reprod 2006;74:684-90. 10.1095/biolreprod.105.049312 [DOI] [PubMed] [Google Scholar]
- 16.Gordon MB, Howard L, Compton DA. Chromosome movement in mitosis requires microtubule anchorage at spindle poles. J Cell Biol 2001;152:425-34. 10.1083/jcb.152.3.425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farina F, Pierobon P, Delevoye C, et al. Kinesin KIFC1 actively transports bare double-stranded DNA. Nucleic Acids Res 2013;41:4926-37. 10.1093/nar/gkt204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pawar S, Donthamsetty S, Pannu V, et al. KIFCI, a novel putative prognostic biomarker for ovarian adenocarcinomas: delineating protein interaction networks and signaling circuitries. J Ovarian Res 2014;7:53. 10.1186/1757-2215-7-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De S, Cipriano R, Jackson MW, et al. Overexpression of kinesins mediates docetaxel resistance in breast cancer cells. Cancer Res 2009;69:8035-42. 10.1158/0008-5472.CAN-09-1224 [DOI] [PubMed] [Google Scholar]
- 20.Grinberg-Rashi H, Ofek E, Perelman M, et al. The expression of three genes in primary non-small cell lung cancer is associated with metastatic spread to the brain. Clin Cancer Res 2009;15:1755-61. 10.1158/1078-0432.CCR-08-2124 [DOI] [PubMed] [Google Scholar]
- 21.Kononen J, Bubendorf L, Kallioniemi A, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med 1998;4:844-7. 10.1038/nm0798-844 [DOI] [PubMed] [Google Scholar]
- 22.Frey RS, Li J, Singletary KW. Effects of genistein on cell proliferation and cell cycle arrest in nonneoplastic human mammary epithelial cells: involvement of Cdc2, p21(waf/cip1), p27(kip1), and Cdc25C expression. Biochem Pharmacol 2001;61:979-89. 10.1016/S0006-2952(01)00572-X [DOI] [PubMed] [Google Scholar]
- 23.Taylor WR, Stark GR. Regulation of the G2/M transition by p53. Oncogene 2001;20:1803-15. 10.1038/sj.onc.1204252 [DOI] [PubMed] [Google Scholar]
- 24.Mollinari C, Kleman JP, Jiang W, et al. PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J Cell Biol 2002;157:1175-86. 10.1083/jcb.200111052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimo A, Nishidate T, Ohta T, et al. Elevated expression of protein regulator of cytokinesis 1, involved in the growth of breast cancer cells. Cancer Sci 2007;98:174-81. 10.1111/j.1349-7006.2006.00381.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gruneberg U, Neef R, Li X, et al. KIF14 and citron kinase act together to promote efficient cytokinesis. J Cell Biol 2006;172:363-72. 10.1083/jcb.200511061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu H, Wu Y, Zhu S, et al. PTP1B promotes cell proliferation and metastasis through activating src and ERK1/2 in non-small cell lung cancer. Cancer Lett 2015;359:218-25. 10.1016/j.canlet.2015.01.020 [DOI] [PubMed] [Google Scholar]
- 28.Liu H, Gu Y, Wang H, et al. Overexpression of PP2A inhibitor SET oncoprotein is associated with tumor progression and poor prognosis in human non-small cell lung cancer. Oncotarget 2015;6:14913-25. 10.18632/oncotarget.3818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schram FR, Ng PK. What is cancer? Journal of Crustacean Biology 2012;32:665-72. 10.1163/193724012X64065022287760 [DOI] [Google Scholar]
- 30.Zou JX, Duan Z, Wang J, et al. Kinesin family deregulation coordinated by bromodomain protein ANCCA and histone methyltransferase MLL for breast cancer cell growth, survival, and tamoxifen resistance. Mol Cancer Res 2014;12:539-49. 10.1158/1541-7786.MCR-13-0459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang B, Lamb ML, Zhang T, et al. Discovery of potent KIFC1 inhibitors using a method of integrated high-throughput synthesis and screening. J Med Chem 2014;57:9958-70. 10.1021/jm501179r [DOI] [PubMed] [Google Scholar]
- 32.Pannu V, Rida PC, Ogden A, et al. HSET overexpression fuels tumor progression via centrosome clustering-independent mechanisms in breast cancer patients. Oncotarget 2015;6:6076-91. 10.18632/oncotarget.3475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Champeris Tsaniras S, Kanellakis N, et al. Licensing of DNA replication, cancer, pluripotency and differentiation: an interlinked world? Semin Cell Dev Biol 2014;30:174-80. 10.1016/j.semcdb.2014.03.013 [DOI] [PubMed] [Google Scholar]
- 34.Gamet-Payrastre L, Li P, Lumeau S, et al. Sulforaphane, a naturally occurring isothiocyanate, induces cell cycle arrest and apoptosis in HT29 human colon cancer cells. Cancer Res 2000;60:1426-33. [PubMed] [Google Scholar]
- 35.Huang WW, Ko SW, Tsai HY, et al. Cantharidin induces G2/M phase arrest and apoptosis in human colorectal cancer colo 205 cells through inhibition of CDK1 activity and caspase-dependent signaling pathways. Int J Oncol 2011;38:1067-73. [DOI] [PubMed] [Google Scholar]
- 36.Turowski P, Franckhauser C, Morris MC, et al. Functional cdc25C dual-specificity phosphatase is required for S-phase entry in human cells. Mol Biol Cell 2003;14:2984-98. 10.1091/mbc.E02-08-0515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer 2009;9:400-14. 10.1038/nrc2657 [DOI] [PMC free article] [PubMed] [Google Scholar]