Abstract
Sugar beet (Beta vulgaris ssp. vulgaris) is a biennial, sucrose-storing plant, which is mainly cultivated as a spring crop and harvested in the vegetative stage before winter. For increasing beet yield, over-winter cultivation would be advantageous. However, bolting is induced after winter and drastically reduces yield. Thus, post-winter bolting control is essential for winter beet cultivation. To identify genetic factors controlling bolting after winter, a F2 population was previously developed by crossing the sugar beet accessions BETA 1773 with reduced bolting tendency and 93161P with complete bolting after winter. For a mapping-by-sequencing analysis, pools of 26 bolting-resistant and 297 bolting F2 plants were used. Thereby, a single continuous homozygous region of 103 kb was co-localized to the previously published BR1 QTL for post-winter bolting resistance (Pfeiffer et al., 2014). The BR1 locus was narrowed down to 11 candidate genes from which a homolog of the Arabidopsis CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR 73-I (CPSF73-I) was identified as the most promising candidate. A 2 bp deletion within the BETA 1773 allele of BvCPSF73-Ia results in a truncated protein. However, the null allele of BvCPSF73-Ia might partially be compensated by a second BvCPSF73-Ib gene. This gene is located 954 bp upstream of BvCPSF73-Ia and could be responsible for the incomplete penetrance of the post-winter bolting resistance allele of BETA 1773. This result is an important milestone for breeding winter beets with complete bolting resistance after winter.
Keywords: sugar beet, winter beet, flowering time, mapping-by-sequencing, vernalization
Introduction
Sugar beet (Beta vulgaris ssp. vulgaris var. altissima) is the only sucrose-storing crop cultivated in temperate regions. It accounts for nearly 30% of the world's annual sugar production (http://faostat3.fao.org, 2015), whereby pulp and molasses are used for animal feeding and methane production. Sugar beets are conventionally sown in spring and harvested in the vegetative stage before winter. Cultivation of sugar beet as a winter crop instead, by sowing in autumn and harvesting in the next year, might increase the beet yield up to 26% due to the pre-winter development and the accelerated growth in spring (Jaggard and Werker, 1999; Hoffmann and Kluge-Severin, 2011). The advanced development of winter beets will also allow an earlier harvest and start of beet campaigns. Therefore, winter beet production is one of the major aims in sugar beet breeding.
One challenge of winter cultivation is the control of bolting after winter. Sugar beets are biennials that grow vegetatively during the first season. A prolonged exposure to cold during winter results in vernalization and the plants acquire floral competence. Under long-day conditions of the next season, the plants start bolting which is indicated by stem elongation and is followed by flower development. Bolting drastically reduces the beet and sugar yield and hampers harvesting processes (Wood and Scott, 1975; Jaggard et al., 1983; Hoffmann and Kluge-Severin, 2011). Thus, bolting is completely undesired for farming although necessary for seed production. To transform sugar beet from a summer into a winter crop, bolting control is an obligate requirement.
Sugar beets were bred for obligate vernalization requirement, whereas wild beets include annual, biennial, and even perennial plants (Hautekeete et al., 2001; Van Dijk, 2009). In Arabidopsis thaliana, the gene FLOWERING LOCUS C (FLC) is a major regulator of vernalization requirement. Although the FLC-homolog of beet, BvFL1, could complement Arabidopsis flc mutants, it does not play a major role in controlling the vernalization response of biennial beets (Reeves et al., 2007; Vogt et al., 2014). Instead, the gene BOLTING TIME CONTROL 1 (BTC1) that resides within the B locus of B. vulgaris is a major regulator of vernalization requirement (Pin et al., 2012). Beets with a dominant BTC1 allele behave as annuals and require only long-day conditions for floral transition. Beets homozygous for the recessive btc1 alleles behave as biennials and require cold exposure followed by long-days to bolt. A second gene, BvBBX19, was recently identified from the B2 locus, which controls the vernalization requirement of beets epistatically to BTC1 (Dally et al., 2014). BTC1 and BvBBX19 promote annual growth through repression of the B. vulgaris bolting repressor gene BvFT1 and activation of the floral activator gene BvFT2, both homologs of the floral integrator gene FLOWERING LOCUS T (FT) of Arabidopsis. Plants with homozygous recessive alleles of either BTC1 or BvBBX19 express BvFT1 and suppress BvFT2 resulting in bolting resistance before winter. Subsequent vernalization leads to BvFT1 down-regulation and BvFT2 activation and bolting is promoted (Pin et al., 2012; Dally et al., 2014). Vernalization and subsequent bolting is also epigenetically controlled in beets. Recent studies showed that cold exposure alters the DNA and RNA methylation in shoot apical meristems of sugar beets resulting in gene expression patterns specific for bolting sensitive and resistant genotypes. Thereby, the DNA methylation correlates negatively with the bolting rate and DNA hypermethylation treatments delay or even inhibit bolting in vernalized sugar beets (Trap-Gentil et al., 2011; Hébrard et al., 2013, 2016).
An obligate bolting tolerance or resistance after winter is required for winter beet cultivation. Biennial beets, that over-express BvFT1 or repress btc1 expression, show bolting resistance after vernalization (Pin et al., 2010, 2012). Within the Beta gene pool, natural variation for post-winter bolting resistance is also available. Kirchhoff et al. (2012) identified beet accessions with low bolting tendencies (e.g., BETA 1773) after growing wild and cultivated beet accessions over winter. Recently, a major quantitative trait locus (QTL) for post-winter bolting resistance (BR1) was determined using a F3 mapping population derived from a cross between BETA 1773 and a sugar beet which is regularly bolting after winter (Pfeiffer et al., 2014). Both parents were homozygous for the biennial btc1 allele and the annual BBX19 allele. BTC1 and BvBBX19 control early flowering resulting in an annual or biennial life cycle. In contrast, BR1 controls bolting after winter. Plants homozygous for the recessive allele (br1) cannot bolt even after winter which in the following will be termed “never-bolting” or “post-winter bolting resistance.” Within the F2 and F3 generations, post-winter bolting resistance showed a non-Mendelian segregation. Thus, post-winter bolting resistance was determined as a quantitative trait which was measured as bolting rates of F3 families. The QTL was mapped to chromosome 9 with a confidence interval of 4 cM and explained 65% of the phenotypic variation. Thereby, the BETA 1773 allele caused a reduced bolting rate in a partially recessive manner (Pfeiffer et al., 2014). F3 families that were homozygous for the BETA 1773 allele at the BR1 locus showed an average bolting rate of 0.33, indicating an incomplete penetrance of the bolting resistance allele. Although the BR1 locus was genetically mapped and quantitatively characterized, the genetic factor which underlies BR1 and its interaction with other bolting genes are unknown.
As shown recently for different organisms, genetic regions controlling discrete phenotypes can be mapped by applying Next Generation Sequencing (NGS) on pooled DNA of individuals that were bulked by their discrete phenotypes. By aligning NGS reads of the distinct sequence bulks to a reference genome and comparing read allele frequencies between both sequence bulks, trait-related loci can be identified and screened for candidate genes (Schneeberger et al., 2009; Laitinen et al., 2010; Qi et al., 2013; Takagi et al., 2013; Lu et al., 2014; Mascher et al., 2014). The publication of the sugar beet genome now allows the application of mapping-by-sequencing also for B. vulgaris (Dohm et al., 2014).
In this study, we aimed to physically map the BR1 locus and to identify BR1 candidate genes by applying mapping-by-sequencing. To achieve this, we used the same F2 population that was developed to genetically map the BR1 QTL (Pfeiffer et al., 2014). This population segregated for bolting behavior after cold-treatment. DNA of bolting-resistant and bolting F2 plants were bulked to produce a bolting-resistant and a bolting sequence pool. We hypothesized that bolting-resistant plants were homozygous for br1, whereas bolting plants were either heterozygous or homozygous for the bolting or even bolting resistance allele due to the incomplete penetrance of br1 (BR1BR1, br1br1). Accordingly, the bolting sequence pool was expected to be polymorphic at each position that has a cross-specific sequence variation, even at the BR1 locus. In contrast, the bolting-resistant sequence pool was expected to be monomorphic at the br1 locus whereas any other genomic region, which does not control the bolting-resistant phenotype, was expected to be polymorphic. Applying a sliding window analysis, we physically mapped the BR1 locus to a 103 kb region on chromosome 9. This region encompasses 11 genes that we analyzed for sequence polymorphisms and characterized for putative functions. A putative function of the most promising BR1 candidate gene, a CPSF73-I homolog of Arabidopsis, is discussed.
Materials and methods
Plant material and growth conditions
According to Pfeiffer et al. (2014), a F2 population consisting of 410 plants was derived from a single F1 plant that was produced by a hand-cross of the biennial sugar beets BETA 1773 and 93161P (Figure S1). Seeds of BETA 1773 and 93161P were provided by the Leibniz Institute of Plant Genetics and Crop Plant Research, IPK (Gatersleben, Germany) and the breeding company Saatzucht Dieckmann (Nienstädt, Germany), respectively. BETA 1773 segregates for bolting and never-bolting plants, whereas 93161P plants are regularly bolting after winter. Subsequently we named BETA 1773 “Pbr” and the bolting parent 93161P “Pb.” Both accessions are homozygous for the biennial btc1 allele and the dominant BvBBX19 allele (btc1a/btc1a, BvBBX19/BvBBX19). As described in detail by Pfeiffer et al. (2014), the F2 plants were pre-cultivated in the greenhouse from November 1 to December 20, 2010 under 16 h light at 20°C. Then they were cold-treated for 16 weeks in a cold chamber at 5°C and under 22 h light. After 1 week acclimatization under 22 h light and 8°C, the F2 plants were planted on April 19, 2011 to a field nursery in Kiel, Germany. Bolting F2 plants were bag-isolated to produce F3 seeds. On December 9, 2011, seeds of 254 F3 families were sown in soil in quickPot-plates96T (Hermann Meyer KG, Germany) and cultivated at 20°C and 16 h light in the greenhouse for 4 weeks. Subsequent cold treatment was done for 16 weeks at 5°C and 16 h light in a climate chamber. At the beginning of May 2012, 248 F3 families were planted to field nurseries in Kiel, Germany (Pfeiffer et al., 2014). The bolting phenotype was recorded on October 11, 2011 for F2 plants and on October 15, 2012 for F3 plants. Bolting was scored, when stem elongation was visible. Furthermore, the existence of flowers and floral buds was recorded as an indicator for complete floral transition.
For climate chamber experiments, 93161P and two F3 families were used. The F3 families were homozygous for the recessive br1 allele as determined previously for the corresponding parental F2 plant using marker CAU3903. The plants were cultivated in soil in 9 × 9 × 9.5 cm3 pots in the greenhouse at 16 h light and 22°C. After 4.5 weeks, 45–47 plants per accession were transferred to cold chambers and kept under 16 h light at 4 and 6°C, respectively. After 14 weeks of cold-treatment, 22–24 plants from the 4 and 6°C cold-treatment were kept in climate chambers under 22 or 16 h light at 20°C for 3 months.
DNA extraction and sequencing
Genomic DNA was extracted from freeze-dried leaf samples of the parental, F1 and F2 plants following a slightly modified CTAB protocol (Saghai-Maroof et al., 1984). DNA amount and quality were determined on 1% agarose gels against λ DNA standards (http://www.thermofisher.com) and using the NanoDrop 2000 spectrophotometer (http://www.thermofisher.com). DNA concentration was adjusted to 10 ng/μl.
NGS and analysis of NGS data
DNA was pooled based on the plant's phenotype (bolting or bolting-resistant after cold-treatment). Four NGS libraries (B0679–B0682) with DNA from 6 to 7 bolting-resistant plants and two NGS libraries (B0683, B0684) with DNA from 148 to 149 bolting plants, respectively, were prepared for whole genome sequencing to generate a bolting-resistant (“br pool”) and bolting sequence pool (“b pool”). Each library was sequenced with the Illumina HiSeq2000 system (http://www.illumina.com) at one lane as 101 bp paired-end reads (Table S1). The raw sequence data have been deposited at the NCBI in the Short Read Archive (SRA) database under the accession number SRP078892. Quality control was performed with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The raw data were trimmed using Sickle (https://github.com/najoshi/sickle) using the paired end mode with 25 as threshold for trimming based on average quality in a window and 60 nucleotides as minimal length for accepted reads after removing low quality bases. High quality paired-end reads were mapped to the sugar beet reference genome RefBeet-1.1 (Dohm et al., 2014) using Bowtie 2 version 2.1.0 (Langmead and Salzberg, 2012) with the parameters—sensitive-local—X 800–I 100. To reduce ambiguity in the alignment, reads with multiple mappings were filtered out. An InDel realignment and duplicate removal was performed on uniquely mapped reads using GATK (McKenna et al., 2010). Afterwards, a multi-sample SNP/InDel and genotype calling was done with GATK's UnifiedGenotyper version 2.8 (DePristo et al., 2011; Van der Auwera et al., 2013). Only SNPs and InDels (≤24 nucleotides) at positions with at least 50-fold coverage over all DNA pools were kept for further analysis. A SNP or InDel position was considered as candidate if it was heterozygous within the b pool and homozygous within the br pool. The first condition was necessary to exclude sequence polymorphisms due to differences to the reference genome derived from the sugar beet genotype KWS2320. For the sliding window analysis, we used a window size of 200 kb and a step size of 100 kb. Per window, the number of monomorphic positions of the br pool out of polymorphic positions of the b pool was computed using in-house R scripts.
A second computational processing of the same NGS raw data was performed using the software CLC Genomics Workbench 6.5.1 (http://www.clcbio.com) to validate specific sequence polymorphisms of BR1 candidate genes based on two independently generated read mapping sets. The 101 bp paired-end reads were imported with a paired-end distance of 50–1000 nucleotides. After quality control, 2 nucleotides were removed from the 5′ site of all reads. Reads were only accepted with maximum 2 ambiguous nucleotides and a minimum length of 80 nucleotides. Trimmed paired and broken reads were mapped against the reference genome RefBeet-1.1 with mismatch cost 2, insertion and deletion cost 1, length fraction 0.5 and similarity fraction 0.9. Only uniquely mapped reads were considered.
Translation of nucleotide sequences was done using the CLC software Main Workbench 7.5 or Genomics Workbench 6.5.1 (http://www.clcbio.com). For candidate genes, a gene ontology analysis was performed using Blast2GO (https://www.blast2go.com). The miRNA target prediction was done using the web server TAPIR (Bonnet et al., 2010). BLAST analyses of DNA and protein sequences were performed using the bl2seq function of NCBI BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Multiple sequence alignments were performed using Clustal Omega 1.2.1 (Sievers et al., 2011).
Molecular marker analysis
Molecular markers were developed based on sequence polymorphisms detected in NGS read mapping data. Primers flanking these polymorphic regions were designed using the software tool OligoCalc (Kibbe, 2007). All primers were obtained from Eurofins Genomics (http://www.eurofinsgenomics.eu). Polymerase chain reaction (PCR) assays were run in a total volume of 15 μl consisting of 1 × PCR buffer without Mg2+, 1.5 mM MgCl2, 0.2 mM dNTP mix, 0.2 mM of forward and reverse primer, 0.3 U Taq DNA Polymerase (Invitrogen™, http://www.thermofisher.com) and up to 10 ng template DNA. PCR products were separated by agarose gel electrophoresis or Sanger sequenced (Institute for Clinical Molecular Biology/IKMB, University of Kiel, Germany). All markers including the primer sequences are listed in Table S2.
Results
A physical map of the post-winter bolting resistance locus
For mapping post-winter bolting resistance, a sugar beet F2 population was used that segregated for bolting and bolting resistance after cold-treatment. This population consisted of 410 F2 individuals and was derived from a cross between BETA 1773 (Pbr) and 93161P (Pbr, Figure S1). After cold-treatment, the F2 population segregated into 26 bolting-resistant, 365 regularly bolting, and 19 plants with incomplete bolting. Plants, that showed incomplete bolting, produced stem-like structures from 5 to 50 cm without developing flowers (Figure 1). The 26 bolting-resistant plants were classified as post-winter bolting resistant, whereas the 19 plants with incomplete bolting were classified as bolting together with the 365 regular bolting beets.
Figure 1.
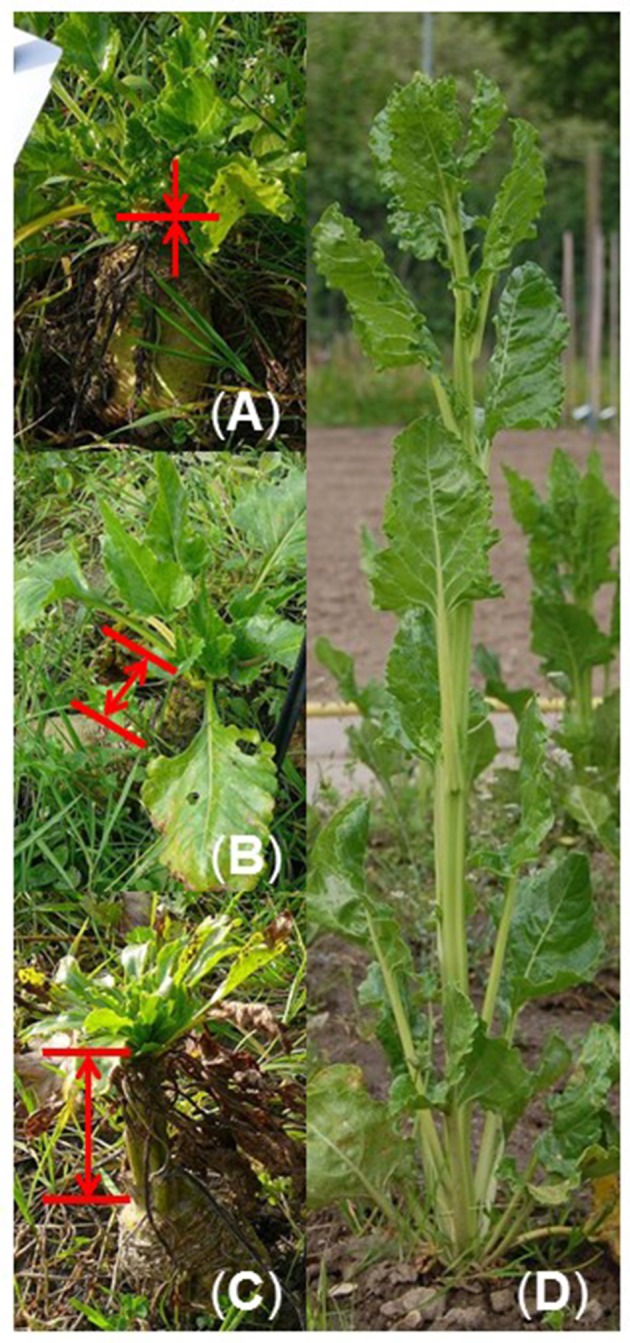
Bolting and bolting-resistant F2 beets in the field after cold-treatment. (A) Bolting-resistant beet growing vegetatively without stem elongation. (B,C) Beets with incomplete bolting producing stem-like structures of 5–50 cm (marked by arrows) but lacking inflorescences and flowers. (D) Normally bolting beet from the bolting accession 93161P with elongated stem, regular inflorescence, and flower development.
To physically map post-winter bolting resistance, we selected the 26 bolting-resistant F2 plants and produced 4 NGS libraries (B0679–B0682, br pool) each containing DNA of 6–7 bolting-resistant plants (Table S3). In addition, we produced two more NGS libraries (B0683, B0684, b pool) which contained DNA of 148 and 149 randomly selected bolting F2 plants. Each library was subjected to high-throughput sequencing on an Illumina HiSeq system, which resulted in 6 sequence sub-pools that contained in total 1140 million 2 × 101 bp paired-end reads corresponding to 230 Gbp of sequence data. Trimmed reads of each sub-pool were mapped separately to the sugar beet reference genome RefBeet-1.1 (Dohm et al., 2014). For each library, between 31 and 33% of the trimmed reads mapped uniquely (including paired and single reads), resulting in 14–25-fold coverage of the published genome sequence (Table S1). In sum, a 72-fold and 34-fold coverage of the RefBeet-1.1 sequence were obtained for the br and b pool, respectively. Through multi-sample SNP/InDel and genotype calling of the uniquely mapped reads of each sub-pool against RefBeet-1.1, we detected in total 3,576,287 positions with sequence polymorphisms in at least one of the 6 sequence sub-pools.
Next, the data of the two b sub-pools were combined and used to identify cross-specific sequence polymorphisms. On Refbeet-1.1 positions with cross-specific sequence polymorphisms, the read allele frequencies were about 0.5, because the b pool contained a genome-wide mixture of Pbr and Pb sequences at each genome position due to bulking F2 plants. Positions with cross-specific polymorphisms were computed as heterozygous by genotype calling. In contrast, monomorphic positions of the b pool indicated a sequence variation to RefBeet-1.1 with identical sequences of the crossing parents. In total, 2,105,427 positions with heterozygous, cross-specific polymorphisms were identified.
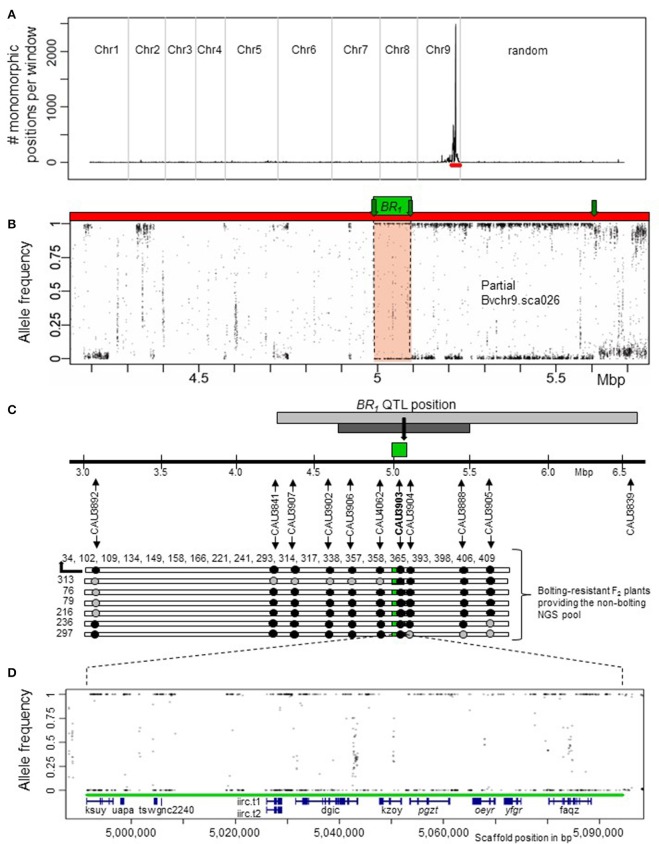
Subsequently, we searched within the br pool for continuous homozygous regions. For this, we applied a sliding window approach over all scaffolds and contigs of RefBeet-1.1. In this way, we analyzed all positions that were called heterozygous in the b pool and called homozygous in the br pool. As a result, only a single peak on chromosome 9 at scaffold Bvchr9.sca026 was detected (Figure 2A), indicating that only one major locus controls post-winter bolting resistance. Bvchr9.sca026 is located at the bottom of chromosome 9, where the BR1 QTL had been previously mapped (Pfeiffer et al., 2014). To locate the candidate region on Bvchr9.sca026, we plotted the average read allele frequency of the br pool at each position that was called heterozygous in the b pool (Figure 2B). Next, the graphic was screened for continuous regions showing allele frequencies of 1 and 0, which indicate homozygous-different or homozygous-identical positions compared to RefBeet-1.1. A continuous homozygous region with a length of 103 kb was detected between scaffold positions 4,991,549 and 5,094,401 (Figure 2B). The BR1 QTL flanking markers CAU3841 and CAU3839 are located 0.75 Mbp upstream and 1.47 Mbp downstream of the identified 103 kb-region (Figure 2C). Thus, the QTL interval covers the complete region. Combining physical and genetic mapping data, we reason that we have physically mapped the BR1 locus.
Figure 2.
Localization of the BR1 locus by a mapping-by-sequencing strategy. (A) A major peak was detected on chromosome 9 at scaffold Bvchr9.sca026 (red bar) by plotting the number of monomorphic positions of the br pool out of polymorphic positions of the b pool within a 200 kb sliding window and a step size of 100 kb. (B) Allele frequencies in the br pools based on polymorphic positions in the b pool at scaffold Bvchr9.sca026 (red). The genome-wide screening had revealed only a single, 103 kb region (green box) that was completely monomorphic in the br pool (allele frequency values 1 and 0 indicate monomorphic positions different and identical to RefBeet-1.1, respectively) at positions that were polymorphic in the b pool. This region is located at Bvchr9.sca026 between position 4,991,549 and 5,094,401. For the remaining genome positions, the allele frequencies differed from 1 and 0, whereby positions at unlinked regions showed on average allele frequencies of about 0.5 (not shown). Green arrows indicate recombination sites of bolting-resistant F2 plants. (C) Co-localization of the BR1 QTL and the physically mapped BR1 locus (green). Localization of the BR1 QTL (light gray), its confidence interval (dark gray) and QTL position (black arrow) is based on the sequence of the QTL flanking markers CAU3841 and CAU3839 (Pfeiffer et al., 2014) in relation to RefBeet-1.1. Genotypic data derived from 10 codominant CAU markers at the BR1 locus indicate crossover events around the BR1 locus in 6 out of 26 bolting-resistant F2 plants. The plant IDs of 20 plants homozygous for BR1: 34, 102, 109, 134, 149, 158, 166, 221, 241, 293, 314, 317, 338, 357, 358, 365, 393, 398, 406, 409. Black arrows show marker positions, black dots indicate marker positions that are homozygous for the allele derived from the bolting-resistant parent BETA 1773, gray dots indicate heterozygous positions. (D) Location of RefBeet-1.1 gene models (blue) within the physically mapped BR1 locus (green): ksuy.t1, uapa.t1, tswg.t1, nc2240, iirc.t1/t2, dgic.t1, kzoy.t1, pgzt.t1, oeyr.t1, yfgr.t1, faqz.t1. Details are given in Table 1. Genes in reverse orientation are written in italics.
Following, we confirmed the physically mapped BR1 region and the flanking recombination sites by marker analysis. We selected the BR1-specific marker CAU3903 and 9 flanking markers (Figure 2C), and genotyped the 26 bolting-resistant F2 plants. All 26 plants were homozygous for the bolting resistance allele br1 derived from Pbr (Figure 2D, Table S3). By using the flanking markers, we detected several crossover events within this population. The bolting-resistant F2 plant 313 carried the first crossover at the left side of the BR1 locus, followed by crossovers of plant 76, 79, and 216. At the right side, the first and second crossovers were detected for plant 297 and 236, respectively (Figure 2D). These results confirmed the map position of the BR1 locus and proved that post-winter bolting resistance was derived from the bolting-resistant accession BETA 1773.
Low penetrance of the BR1 allele for post-winter bolting resistance
The physical mapping of the BR1 locus was based on 26 bolting-resistant F2 plants. To determine the distribution of the bolting resistance allele br1 within the whole F2 population, we genotyped all 410 F2 plants using the co-dominant marker CAU3903 which is specific for the BR1 locus. We detected 95 plants that were homozygous for br1, 201 heterozygous plants and 114 plants homozygous for the dominant bolting allele BR1 (Table S4). The marker genotypes segregated 1:2:1 (X2 = 1.917, non-significant at α = 0.05). This segregation ratio is typical for F2 populations and shows that no artificial selection had occurred favoring one of the alleles. Out of 95 F2 plants that were genotyped as homozygous for br1, only 26 plants were bolting-resistant after cold treatment, whereas 13 and 56 plants showed incomplete or regular bolting, respectively. In contrast, all plants with a dominant BR1 allele were bolting. Thus, the genotype at the BR1 locus predicted 83% of the phenotype. However, 73% of the F2 plants that were homozygous for br1 were bolting which could be explained by further loci which might be involved in bolting control after winter. Thus, we performed additional genome wide screenings under various parameter settings using the sequence information of the 26 bolting-resistant F2 plants. However, we did not detect other loci that could explain the bolting resistance phenotype assuming that the BR1 locus might interact with various minor loci that are below the detection level.
The F2 phenotypic data are based on single plants. Next, we evaluated the post-winter bolting resistance of 248 F3 families that were derived from selfed F2 plants as described by Pfeiffer et al. (2014). After cold treatment, the F3 plants grew under field conditions from May until October 2012, together with control plants of the bolting-resistant and bolting accessions. Post-winter bolting resistance was recorded and the bolting rate was calculated for each F3 family as the number of bolting plants out of the total plant number of this family. The bolting-resistant control showed a bolting rate of 0.12, whereas the bolting control bolted completely. Under the same growing conditions, 209 F3 families with 6–31 plants per family showed bolting rates between 0 and 1. Depending on the F2 genotype at the BR1 locus, the average bolting rates of these F3 families varied significantly (Figure 3). F3 families derived from F2 plants that were homozygous for the br1 allele showed an average bolting rate of 0.27 ± 0.216. F3 families derived from heterozygous or homozygous-dominant F2 plants showed average bolting rates of 0.74 ± 0.146 and 0.96 ± 0.075, respectively. These results demonstrate a clear dominant-recessive inheritance of post-winter bolting resistance which is controlled primarily by the BR1 locus. Only regularly bolting F2 plants could be used to produce F3 families. Noteworthy, 3 out of 21 regularly bolting F2 plants that were homozygous for br1, generated completely bolting-resistant F3 families (in sum 71 plants). This suggests a reduced penetrance of the br1 allele rather than additional BR loci.
Figure 3.
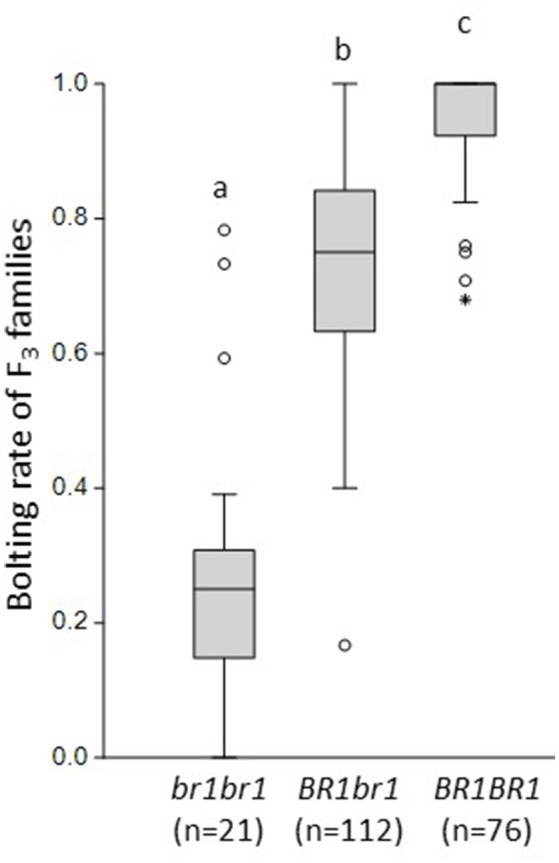
Bolting rates of F3 families grouped by the BR1 genotypes of their F2 parents. Marker CAU3903 was used for genotyping. The mean bolting rates of F3 families differed significantly between all BR1 genotypes. Statistical analysis is based on Tukey's pairwise test with α = 0.05. The letters a, b, c indicate significant differences.
Moreover, we used F3 phenotypic data to explore if the BR1 locus also controls incomplete bolting of sugar beets resulting in stem-like structures without producing flowers. 183 out of 209 F3 families contained 1–18 plants that had bolted incompletely until the end of the season. When grouping the F3 families according to the BR1 genotype, then 71.4 (homozygous br1), 97.3 (heterozygous) and 77.6% (homozygous BR1) of the F3 families included incomplete bolting plants. Thus, the incomplete bolting phenotype cannot be explained through the presence of a certain BR1 allele and genetic factors unlinked to BR1 seem to control this phenotype.
Subsequently we aimed to determine the effect of vernalization temperature and photoperiod on the bolting rate of plants which were homozygous for br1. We selected two F3 families (112206, 112210) that were derived from br1 homozygous F2 plants. These plants were cold-treated together with bolting control plants at 4° or 6°C, to test whether different vernalization temperatures affect the bolting rate. Subsequently, the plants were kept at 20°C under 22 h or 16 h light, to test whether different photoperiods affect the bolting rate. After 3 months, we determined the bolting rate per accession and treatment. The bolting rates of the F3 families ranged from 0.52 to 0 (Figure 4). They were significantly reduced compared to the bolting control in each treatment. Under 22 h light, the bolting rates ranged from 0.52 to 0.35 after vernalization at 4°C and from 0.38 to 0.04 after 6°C vernalization, respectively. Complete bolting resistance was obtained under 16 h light in both families for both cold treatments. The bolting control showed also a reduced bolting rate under sub-optimal conditions of decreased day-length and increased vernalization temperature. Thus, environmental factors such as day length and vernalization intensity affect bolting after vernalization, whereby the impact of the photoperiod was significantly stronger than the impact of vernalization temperature. Although both F3 families responded with varying bolting rates to different growth conditions, they never reached a bolting rate of 1 due to the br1-mediated bolting resistance.
Figure 4.
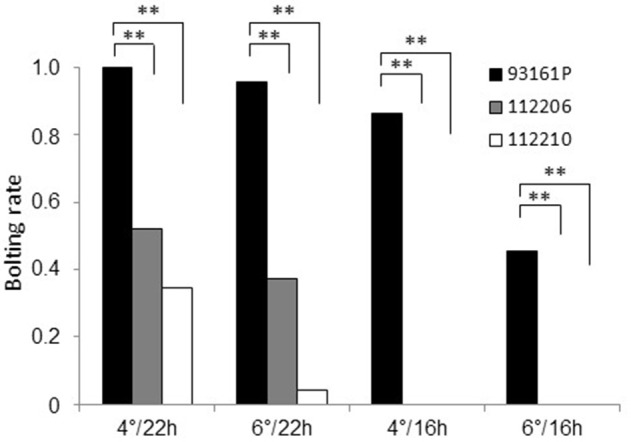
Temperature and day-length dependence of bolting rates of F3 families 112206 and 112210 which are homozygous for br1. The parental accession 93161P was used as bolting control. Plants were vernalized at 4°C or 6°C under 16 h light and subsequently grown under 22 or 16 h light at 20°C. Asterisks indicate significant differences at p.adjust < 0.05 for each growth condition based on Pearson's chi-squared tests.
The BR1 locus houses 11 genes
Based on the RefBeet-1.1 annotation, we found 11 gene models with RNA evidence within the BR1 locus including 10 protein-coding and one miRNA gene (Table 1). However, the gene ontology (GO) analysis identified no genes or miRNA target genes related to floral transition genes or flowering time genes of Arabidopsis. We expected for the BR1 gene a Pbr specific mutation that results in a reduced or complete loss-of-function. Furthermore, the BR1 gene should be expressed in leaves and/or shoot apices where plants perceive floral inducing signals (Corbesier and Coupland, 2006). In a first step, we screened the coding sequences of the 11 genes for nucleotide variations between the parents and RefBeet-1.1. For this, we used the NGS read mapping sets of the bolting-resistant and bolting sequence pools and a more stringent read mapping of the sequence sub-pools B0679 and B0680 (bolting-resistant) and B0683 (bolting). No sequence variation was found for the miRNA gene. For the protein-coding genes, we detected 7 genes with Pbr specific amino acid changes, which resulted in 3 genes (uapa.t1, tswg.t1, yfgr.t1) with premature STOP codons (Table 1, Figure S2). Thereby, yfgr.t1 showed the most severe mutation through a 2 bp deletion that causes a frameshift. This frameshift results in a truncated protein with a length of 203 instead of 633 amino acids. The Pbr specific sequence polymorphisms in uapa.t1 and tswg.t1 resulted in less truncated proteins with lengths of 226 instead of 237 and 190 instead of 204 amino acids.
Table 1.
Characterization of B. vulgaris genes located within the BR1 locus interval (102,853 bp) on chromosome 9 on scaffold Bvchr9.sca026.
| Gene models based on RefBeet-1.1 (abbreviation) | Pbr (BETA 1773) specific amino acid changes compared to RefBeet-1.1 and Pb (93161P) | GO protein description/putative function | Homologous Arabidopsis gene, annotation according to TAIR database |
|---|---|---|---|
| Bv9_227290_ksuy.t1 (ksuy)a,b,c | No | Curvature thylakoid protein, chloroplastic | AT1G52220, CURVATURE THYLAKOID 1C (CURT1C), protein located in the chloroplast thylakoid membrane |
| Bv9_227300_uapa.t1 (uapa)a | aa51 H D, aa119 T D, aa119 T S, aa208 T S, aa208 T A, premature STOP codon for Pbr and Pb resulting in a protein with 223 instead of 235aa (RefBeet-1.1) A, premature STOP codon for Pbr and Pb resulting in a protein with 223 instead of 235aa (RefBeet-1.1) |
DNA-directed RNA polymerase II subunit RPB1-like | No significant hit |
| Bv9_227310_tswg.t1 (tswg)a | aa84 T K, aa128 H K, aa128 H -, aa151 P -, aa151 P PS, aa182 P PS, aa182 P L, aa187 F L, aa187 F L, aa188 D L, aa188 D F, aa189 H F, aa189 H STOP codon resulting in a protein with 188 instead of 202aa STOP codon resulting in a protein with 188 instead of 202aa |
Hypothetical protein BVRB_9g221070 | No significant hit |
| Bv9_227320_iirc.t1/t2 (iirc)a,b | aa204 E Q, aa255 A Q, aa255 A E, aa300 E E, aa300 E G, aa319 Q G, aa319 Q R, aa332 A R, aa332 A V, aa339 R V, aa339 R S, aa407/373 K S, aa407/373 K N, aa454/420 V N, aa454/420 V I, aa467/433 F I, aa467/433 F S specific for t1 and t2, aa384 I S specific for t1 and t2, aa384 I V specific for t1 V specific for t1 |
Serpin-ZX-like protein (F: serine-type endopeptidase inhibitor activity, transferase activity) | AT1G47710, SERPIN, protein involved in the negative regulation of endopeptidase activity in the apoplast |
| Bv9_227330_dgic.t1 (dgic)a,b,c | aa95 E G, aa113 V G, aa113 V L, aa152 E L, aa152 E D, aa223 T D, aa223 T S, aa248 G S, aa248 G E, aa252 K E, aa252 K N, aa266 V N, aa266 V I, aa302 I I, aa302 I T, aa309 Q T, aa309 Q L, aa324 A L, aa324 A V, aa772 E V, aa772 E D, aa787 A D, aa787 A S S |
Golgin candidate 5 | AT1G79830, GOLGIN CANDIDATE 5 (GC5), structural component of the Golgi apparatus |
| Bv9_227340_kzoy.t1 (kzoy)a,b,c | No | Basic-leucine zipper transcription factor family protein isoform 1 (F:sequence-specific DNA binding transcription factor activity; P:regulation of transcription, DNA-dependent) | AT1G58110, basic-leucine zipper (bZIP) transcription factor, protein involved in DNA-templated transcription in the nucleus |
| Bv9_227350_pgzt.t1 (pgzt)a,b,c | No | Magnesium transporter mrs2-4 (C: membrane; F:magnesium ion transmembrane transporter activity; P:transmembrane transport, magnesium ion transport) | AT3G58970, MAGNESIUM TRANSPORTER 6 (MGT6), integral component of the plasma membrane |
| Bv9_227360_oeyr.t1 (oeyr)a,b,c | aa618 E K, aa629 A K, aa629 A V V |
Cleavage and polyadenylation specificity factor subunit 3-i-like isoform (F: hydrolase activity) | AT1G61010, CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR 73-I (CPSF73-I), protein involved in the mRNA polyadenylation in the nucleus |
| Bv9_227370_yfgr.t1 (yfgr)a,b,c | aa144 T A, from aa191 changed sequence by a frame shift mutation, at aa204 A, from aa191 changed sequence by a frame shift mutation, at aa204 STOP codon resulting in a protein with 203 instead of 633aa STOP codon resulting in a protein with 203 instead of 633aa |
Cleavage and polyadenylation specificity factor subunit 3-i-like isoform (F: hydrolase activity) | AT1G61010, CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR 73-I (CPSF73-I), protein involved in the mRNA polyadenylation in the nucleus |
| Bv9_227380_faqz.t1 (faqz)a,b,c | aa31 L I I |
Probable plastidic glucose transporter 3 (C: trans-Golgi network, endosome, integral to membrane; P: negative regulation of biological process, carbohydrate transmembrane transport, determination of bilateral symmetry, xylem and phloem pattern formation, flower morphogenesis; F: sugar:hydrogen symporter activity) | AT1G79820, SUPPRESSOR OF G PROTEIN BETA1 (SGB1), protein involved in the carbohydrate transport through membranes |
| Non-coding genes based on RefBeet-1.1 | Pbr specific nucleotide changes compared to RefBeet-1.1 and Pb | Homology to published miRNAs based on mirBase | Number of putative target genes based on RefBeet-1.1 gene models |
| ncRNA_2240/MIR1122 (nc2240)a | No | Brachypodium distachyon miR5181d, involved in cold stress response; Brachypodium distachyon miR5174e stem-loop, involved in reprogramming leaf growth during drought stress | 12 (non-related to flowering) |
RNA evidence according to RNAseq and EST data of RefBeet-1.1 (Dohm et al., 2014).
RNA evidence according to RNAseq data derived from shoot apices of annual sugar beet accession 001684 during floral transition (Tränkner et al. unpublished).
RNA evidence according to SuperSAGE data derived from leaves of non-vernalized and vernalized KWS2320 sugar beets (Vogt et al. unpublished).
Furthermore, we investigated which of these genes are transcribed in leaves and shoot apices of beet. We screened two unpublished transcriptome data sets derived from shoot apices of an annual beet and from leaves of vernalized and non-vernalized plants of a biennial sugar beet. Yfgr.t1 is transcribed in shoot apices of the annual beet during floral transition and upregulated during vernalization in the biennial genotype (Figure S3), whereas transcripts of uapa.t1 and tswg.t1 were not detected. Thus, we excluded both of the non-expressed genes from being a BR1 candidate and assumed yfgr.t1 as the best candidate gene for BR1. For completeness, transcripts of iirc.t1/t2 were detectable at a very low level only in shoot apices of annual beets, whereas transcripts of ksuy.t1, dgic.t1, pgzt.t1, oeyr.t1, faqz.t1, and kzoy.t1 were clearly detectable in shoot apices of annual beets as well as leaves of biennial beets (Figure S3).
The BR1 candidate gene yfgr.t1 shares 81% identity at the protein level with the A. thaliana CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR 73-I (AtCPSF73-I) gene. In contrast to Arabidopsis, B. vulgaris contains a second CPSF73-I homologous gene, oeyr.t1, which shows 83% identity with AtCPSF73-I at protein sequence level. Following we named yfgr.t1 BvCPSF73-Ia and oeyr.t1 BvCPSF73-Ib. BvCPSF73-Ib is also located on the BR1 locus, 954 bp upstream of BvCPSF73-Ia (Figure 2D, oeyr/yfgr). Both genes show a sequence identity of 97% at coding sequence and protein sequence level. Furthermore, both genes show a similar transcription pattern in annual and biennial sugar beets (Figure S3). Thus, we suggest that both genes originated from a common ancestor and possess a similar or same function.
Discussion
Post-winter bolting resistance is controlled by a major gene on chromosome 9
Post-winter bolting resistance is essential for prospective winter beet cultivars. While the bolting loci BTC1 and BvBBX19 control vernalization requirement, which distinguishes annual and biennial beets, the major QTL BR1 controls bolting after vernalization and distinguishes between biennial and never-bolting beets (Pfeiffer et al., 2014). Never-bolting is an absolute requirement to transform sugar beet from spring into a winter crop. In this study, we physically mapped the BR1 locus to a single region on chromosome 9. This region is located at RefBeet-1.1 scaffold Bvchr9.sca026 and includes 11 genes.
However, our F2 population showed a non-Mendelian segregation of the bolting-resistant phenotype with a clear over-representation of bolting plants. A natural selection of F2 plants against post-winter bolting resistance was excluded, because codominant markers of the BR1 locus segregated in a 1:2:1 ratio which is typical for F2 populations. Thus, additional loci might be expected. However, there was no evidence for another locus. Mapping-by-sequencing allows the detection of candidate sequence polymorphisms on a genome-wide level. Thereby, homozygosity is extended around the causal locus of the recessive trait due to genetic linkage (Schneeberger, 2014). In this study, we had detected chromosome 9 scaffolds 26, 25, and 24 which displayed the highest numbers of monomorphic positions per mega base pair. A considerably lower number or even no monomorphic positions were detected for all other scaffolds covering the whole genome of sugar beet. Scaffolds 24 and 25 are located upstream of scaffold 26 which is located in the telomeric region of chromosome 9 (Dohm et al., 2014). Both scaffolds had a clearly reduced number of candidate positions compared to scaffold 26. They point to scaffold 26 as the precise location of the BR1 gene due to the gradual enrichment of br1 alleles around the BR1 locus resulting in homozygosity at BR1. Previously, Pfeiffer et al. (2014) had detected only one QTL for post-winter bolting resistance using bolting rate data of 186 F3 families of the same population. The interval of the BR1 QTL completely covers the identified 103 kb BR1 region, showing a perfect co-localization. In conclusion, mapping-by-sequencing in combination with QTL mapping pointed exclusively to the BR1 locus as major locus controlling post-winter bolting resistance in beet.
Incomplete penetrance of the bolting resistance phenotype
Surprisingly, 72.6% of the F2 plants that were homozygous for the br1 allele exhibited a bolting phenotype. F3 families derived from these plants after selfing showed bolting rates from 0 to 0.78 with an average of 0.27. Since the bolting-resistant phenotype of this mapping population was clearly linked to the recessive br1 allele, these findings suggest an incomplete penetrance of the br1 allele derived from the bolting-resistant accession BETA 1773. Incomplete penetrance of br1 was also implied through varying bolting rates of BETA 1773 in previous experiments. For example, Pfeiffer et al. (2014) reported bolting rates of 0.12 and 0.5 under field conditions in Germany. Under the synonyms “Kaweaa” or “Kleinwanzleben AA/Klein AA” (IPK, 2006; JKI, 2012), BETA 1773 showed bolting rates from 0.14 to 0.85 after over-winter cultivation in Spain (Lasa and Medina, 1978) and an average bolting rate of 0.57 after over-winter cultivation in England in 1971 that varied from 0.78 to 0.92 in 1972 depending on the sowing date in autumn. Progenies of bolting-resistant plants that had flowered after a second vernalization showed bolting rates between 0.63 and 0.77 after over-winter cultivation (Wood and Scott, 1975). Thus, a high variation for post-winter bolting resistance is observable within the original accession, its subsequent generations and within the crossing population used here.
Lasa and Medina (1978) determined for BETA 1773 a positive correlation between bolting and the number of days with minimum air temperatures between 3 and 10°C. In biennial beets, bolting is controlled by photothermal induction, requiring periods of cold followed by long-day conditions (Owen et al., 1940). Thereby, additional light units can replace temperature units (Steinberg and Garner, 1936; Stout, 1946). Photothermal induction is genotype-specific concerning vernalization temperature, duration of cold exposure and day length after cold-treatment. Temperatures from 2 to 10°C are mostly sufficient for thermal induction, whereby the highest bolting rates are obtained at about 4°C (Stout, 1946). Our climate chamber experiments confirmed that the bolting-resistant phenotype is strongly affected by photothermal induction. Two F3 families that were homozygous for the recessive br1 allele reached higher bolting rates after 4°C cold treatment and subsequent 22 h light exposure than under suboptimal bolting induction conditions at higher vernalization temperatures or with less light units. Since the bolting rates of both F3 families varied depending on the photoperiod as well as the vernalization temperature, BR1 seems to integrate signals from the photoperiodic and vernalization pathway assuming a functional position downstream of both flowering pathways. Additionally, the bolting-resistant phenotype might be also regulated by endogenous factors because F3 family 112206 showed in the climate chamber experiments higher bolting rates than 112210, whereas under 16 h artificial light both families were completely bolting-resistant.
The results of our study indicate incomplete penetrance of the bolting resistance phenotype. Incomplete penetrance is a widespread feature which appears in a wide range of organisms affecting diverse traits. For example, a major locus controlling in situ gynogenesis of maize shows incomplete penetrance (Barret et al., 2008). In tomato, mutants of the TERMINATING FLOWER (TMF) gene exhibit a single-flower phenotype instead of multi-flowered inflorescences with 50% penetrance in the original mutant background and varying levels up to 100% in the 3rd backcross generation (MacAlister et al., 2012). Thereby, tmf mutants developed solitary flowers through precocious activation of a floral specification complex. The authors proposed the presence of unknown modifiers to explain the varying penetrance levels, but they did not give further information about the modifier's type or mode of action. Reasons for incomplete penetrance are diverse and different regulatory mechanisms are involved. As reviewed by Cooper et al. (2013), the penetrance of a phenotype can be affected by the specific mutation itself, copy number variations, or differential allelic expression. Furthermore, unlinked genes, environmental or developmental (age-dependent) factors or epigenetic regulation of expression can affect the penetrance of a phenotype. The role of DNA methylation in bolting has been described on a whole genome scale (Trap-Gentil et al., 2011; Hébrard et al., 2016). Thus, conditional epigenetic modification of the BR1 locus could result in altered gene activities giving rise to a non-Mendelian segregation of the bolting phenotype. In addition, mutations in genes which act redundantly can result in phenotypes with incomplete penetrance as described for the auxin influx carrier genes aux1, lax1, and lax2 of Arabidopsis. The aux1, lax1, and lax2 single mutants and aux1 lax2 and lax1 lax2 double mutants showed no obvious defects during embryo development, whereas aux1 lax1 double mutants and triple mutants showed incomplete penetrance of 4–22.1%. Thereby the penetrance level and severity of defects was increased in triple mutants, clearly demonstrating a functional redundancy of these auxin influx carrier genes (Robert et al., 2015). The regulatory mechanism that controls penetrance of the bolting resistance phenotype of sugar beet is unknown until now.
Indications that a cleavage and poly-adenylation specificity factor 73-I homolog underlies the BR1 phenotype
Out of the 11 genes located within the physically mapped BR1 interval, we propose yfgr.t1 (BvCPSF73-Ia) as BR1 candidate. BvCPSF73-Ia fulfilled all criteria that we expected from a hypothetical BR1 gene: (i) It is located at the BR1 locus, (ii) it is transcribed during floral transition in leaves and shoot apices of regularly bolting beet genotypes, (iii) the BETA 1773 allele encodes a severely truncated protein because of a BETA 1773-specific 2 bp deletion within the coding sequence, and (iv) a loss-of-function can be assumed. BvCPSF73-Ia is a homolog of the Arabidopsis gene CPSF73-I. CPSF proteins are essential for the polyadenylation of mRNA and splicing of terminal introns in eukaryotes. In Arabidopsis, the CPSF proteins CPSF160, CPSF100, CPSF73, and CPSF30 form a complex which recognizes and directly binds to polyadenylation sites of pre-mRNAs. After recruitment of further factors, the cleavage endonuclease CPSF73 removes the RNA 3' end and the synthesis of the poly(A) tail follows (Shi and Manley, 2015). The Arabidopsis genome contains 5 CPSF genes including two CPSF73 homologs, AtCPSF73-I and AtCPSF73-II. Both AtCPSF73 genes are essential for plant development because knockout and knockdown mutants are lethal (Xu et al., 2004, 2006). Mutations within 3′-end processing factors can affect flowering through alternative polyadenylation of mRNAs or deficient mRNA processing which induces gene silencing (Herr et al., 2006; Zhang et al., 2015). Previous studies showed also in vivo associations between AtCPSF proteins and the FY protein. FY controls floral transition through down-regulation of the floral repressor gene FLC, which is a major regulator of vernalization requirement (Simpson et al., 2003; Manzano et al., 2009). If BvCPSF73-Ia has a similar function like its Arabidopsis homolog, a loss-of-function of BvCPSF73-Ia can result in bolting resistance after vernalization.
Interestingly, sugar beet contains three CPSF73 homologs (gene models yfgr.t1, oeyr.t1, and gfgf.t1). While BvCPSF73-Ia (yfgr.t1) and BvCPSF73-Ib (oeyr.t1) show highest identity to AtCPSF73-I, gfgf.t1 is the homolog of AtCPSF73-II. BvCPSF73-Ia and BvCPSF73-Ib are located next to each other within the BR1 locus. The high sequence identity between both BvCPSF73-I homologs suggests an ancient gene duplication. Due to the same origin and the similar transcription pattern during floral transition, we assume that BvCPSF73-Ia and BvCPSF73-Ib have a similar function and might act redundantly. Accordingly, we hypothesize that beets with null alleles of BvCPSF73-Ia will express intact BvCPSF73-I protein due to BvCPSF73-Ib, but that the total BvCPSF73-I protein amount will be lower than in wild-type plants. However, the expression level of BvCPSF73-I might be crucial for plant development because the overexpression, knockout, or knockdown of AtCPSF73-I in Arabidopsis is lethal, whereas a slightly increased expression results in normal looking but male sterile plants (Xu et al., 2006). Based on our hypothesis, bolting resistance might be caused when the concentration of BvCPSF73-I protein falls below a certain threshold. In contrast, the incomplete phenotype of BETA 1773 plants can result from an environment-dependent or genotype-specific higher expression of oeyr.t1. Otherwise, incomplete penetrance might be the result of a threshold-specific expression of a putative BvCPSF73-I targeted flowering gene like the floral promoter BvFT2.
Although BvCPSF73-Ia is the most promising candidate gene, other genes of the BR1 locus cannot be fully excluded. For example, the putative bZIP transcription factor kzoy.t1 contains a Pbr specific 7 bp deletion in the promoter region that might affect post-winter bolting behavior by altered gene expression levels. Thus, further research is necessary to ultimately identify the BR1 gene and to unravel the molecular mechanism that leads to post-winter bolting resistance. Understanding the mechanism of incomplete penetrance of BR1-mediated post-winter bolting resistance will allow identifying BR1 modifier genes that can subsequently be used for winter beet breeding. Thus, functional analysis of the BR1 gene like spatial and temporal expression analysis, complementation of BR1 homozygous plants, knockout or knockdown of functional alleles by mutagenesis or targeted genome editing must follow. Moreover, the genes targeted by BR1 must be identified. If yfgr.t1 underlies the BR1 locus, then targeted genes might have altered polyadenylation sites or they show a reduced expression through posttranscriptional regulation. If kzoy.t1 is the BR1 gene, then homozygous dominant and homozygous recessive plants will show different transcript levels of target genes that can be identified by RNAseq approaches.
Relevance for breeding winter beet
Due to the monogenic inheritance of the BR1-mediated post-winter bolting resistance, post-winter bolting resistance of BETA 1773 can easily be introgressed into winter beet genotypes. Thereby the BR1-specific InDel marker CAU3903 or a marker specific for the 2 bp deletion in yfgr.t1 will allow the selection of progenies with the recessive br1 allele. Modern sugar beet cultivars are mostly hybrids. Thus, the introgression of the recessive br1 allele into both parental lines is necessary to obtain homozygous recessive hybrids. Since BR1-mediated post-winter bolting resistance shows incomplete penetrance, further loci for bolting resistance or bolting delay must be combined in a genotype to acquire complete bolting resistance after winter. For this, the natural variation of post-winter bolting resistance in the B. vulgaris gene pool must be explored further. Furthermore, the Beta gene pool should be scanned for BR1 haplotypes to identify br1 alleles with increased penetrance. However, haplotypes causing complete bolting resistance will be rare because bolting-resistant beets cannot be multiplied and were lost during previous breeding and propagation processes. Simultaneously to the identification and pyramiding of bolting resistance alleles, systems have to be developed that allow controlled bolting induction of bolting-resistant beets for seed production.
Author contributions
CT supervised the project, designed and performed experiments and wrote the manuscript. IL and IG designed and performed computational analyses and wrote the manuscript. MS performed NGS. NE, NP, ST and FK provided phenotypic and genotypic data. SV and AM provided SuperSAGE-data. CJ supervised the experiments and revised the manuscript. All authors read and approved the final manuscript.
Funding
This study was funded by the DFG Priority Program SPP1530 grant numbers TR1088/1-1 and GR 3526/2-1, and the German Federal Ministry of Education and Research (BMBF) program grant numbers FKZ 0315465B and 0315058A.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Claudia Havel for excellent technical assistance, Maria Albani, Martina Blümel, Gina Capistrano-Großmann, Siegbert Melzer, and Claus Weinholdt for help and intensive discussions, and thank Christopher Somes for reading the manuscript.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01662/full#supplementary-material
References
- Barret P., Brinkmann M., Beckert M. (2008). A major locus expressed in the male gametophyte with incomplete penetrance is responsible for in situ gynogenesis in maize. Theor. Appl. Genet. 117, 581–594. 10.1007/s00122-008-0803-6 [DOI] [PubMed] [Google Scholar]
- Bonnet E., He Y., Billiau K., Van de Peer Y. (2010). TAPIR, a web server for the prediction of plant microRNA targets, including target mimics. Bioinformatics 26, 1566–1568. 10.1093/bioinformatics/btq233 [DOI] [PubMed] [Google Scholar]
- Cooper D. N., Krawczak M., Polychronakos C., Tyler-Smith C., Kehrer-Sawatzki H. (2013). Where genotype is not predictive of phenotype: towards an understanding of the molecular basis of reduced penetrance in human inherited disease. Hum. Genet. 132, 1077–1130. 10.1007/s00439-013-1331-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L., Coupland G. (2006). The quest for florigen: a review of recent progress. J. Exp. Bot. 57, 3395–3403. 10.1093/jxb/erl095 [DOI] [PubMed] [Google Scholar]
- Dally N., Xiao K., Holtgräwe D., Jung C. (2014). The B2 flowering time locus of beet encodes a zinc finger transcription factor. Proc. Natl. Acad. Sci. U.S.A. 111, 10365–10370. 10.1073/pnas.1404829111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo M. A., Banks E., Poplin R., Garimella K. V., Maguire J. R., Hartl C., et al. (2011). A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498. 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohm J. C., Minoche A. E., Holtgräwe D., Capella-Gutiérrez S., Zakrzewski F., Tafer H., et al. (2014). The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature 505, 546–549. 10.1038/nature12817 [DOI] [PubMed] [Google Scholar]
- Hautekeete N. C., Piquot Y., Van Dijk H. (2001). Investment in survival and reproduction along a semelparity-iteroparity gradient in the Beta species complex. J. Evol. Biol. 14, 795–804. 10.1046/j.1420-9101.2001.00322.x [DOI] [Google Scholar]
- Hébrard C., Peterson D. G., Willems G., Delaunay A., Jesson B., Lefèbvre M., et al. (2016). Epigenomics and bolting tolerance in sugar beet genotypes. J. Exp. Bot. 67, 207–225. 10.1093/jxb/erv449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hébrard C., Trap-Gentil M.-V., Lafon-Placette C., Delaunay A., Joseph C., Lefèbvre M., et al. (2013). Identification of differentially methylated regions during vernalization revealed a role for RNA methyltransferases in bolting. J. Exp. Bot. 64, 651–663. 10.1093/jxb/ers363 [DOI] [PubMed] [Google Scholar]
- Herr A. J., Molnar A., Jones A., Baulcombe D. C. (2006). Defective RNA processing enhances RNA silencing and influences flowering of Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 14994–15001. 10.1073/pnas.0606536103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann C. M., Kluge-Severin S. (2011). Growth analysis of autumn and spring sown sugar beet. Eur. J. Agron. 34, 1–9. 10.1016/j.eja.2010.09.001 [DOI] [Google Scholar]
- IPK (2006). GBIS/I-Genebank Information System of the Leibniz Institute of Plant Genetics and Crop Plant Research (IPK). Gatersleben. [Google Scholar]
- Jaggard K. W., Werker A. R. (1999). An evaluation of the potential benefits and costs of autumn-sown sugarbeet in NW Europe. J. Agric. Sci. 132, 91–102. 10.1017/S002185969800608X [DOI] [Google Scholar]
- Jaggard K. W., Wickens R., Webb D. J., Scott R. K. (1983). Effects of sowing date on plant establishment and bolting and the influence of these factors on yields of sugar-beet. J. Agric. Sci. 101, 147–161. 10.1017/S0021859600036479 [DOI] [Google Scholar]
- JKI (2012). International Database for Beta (IDBB). Available online at: http://idbb.jki.bund.de (Accessed April 20, 2016).
- Kibbe W. A. (2007). OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res. 35, W43–W46. 10.1093/nar/gkm234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhoff M., Svirshchevskaya A., Hoffmann C., Schechert A., Jung C., Kopisch-Obuch F. J. (2012). High degree of genetic variation of winter hardiness in a panel of Beta vulgaris L. Crop Sci. 52, 179–188. 10.2135/cropsci2011.04.0185 [DOI] [Google Scholar]
- Laitinen R. A., Schneeberger K., Jelly N. S., Ossowski S., Weigel D. (2010). Identification of a spontaneous frame shift mutation in a nonreference Arabidopsis accession using whole genome sequencing. Plant Physiol. 153, 652–654. 10.1104/pp.110.156448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasa J. M., Medina B. (1978). Multiplication of bolting resistant sugar-beet (Beta vulgaris L.). Anales Estacion Experimental Aula Dei 14, 163–172. [Google Scholar]
- Lu H., Lin T., Klein J., Wang S., Qi J., Zhou Q., et al. (2014). QTL-seq identifies an early flowering QTL located near Flowering Locus T in cucumber. Theor. Appl. Genet. 127, 1491–1499. 10.1007/s00122-014-2313-z [DOI] [PubMed] [Google Scholar]
- MacAlister C. A., Park S. J., Jiang K., Marcel F., Bendahmane A., Izkovich Y., et al. (2012). Synchronization of the flowering transition by the tomato TERMINATING FLOWER gene. Nat. Genet. 44, 1393–1398. 10.1038/ng.2465 [DOI] [PubMed] [Google Scholar]
- Manzano D., Marquardt S., Jones A. M., Bäurle I., Liu F., Dean C. (2009). Altered interactions within FY/AtCPSF complexes required for Arabidopsis FCA-mediated chromatin silencing. Proc. Natl. Acad. Sci. U.S.A. 106, 8772–8777. 10.1073/pnas.0903444106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher M., Jost M., Kuon J.-E., Himmelbach A., Aßfalg A., Beier S., et al. (2014). Mapping-by-sequencing accelerates forward genetics in barley. Genome Biol. 15:R78. 10.1186/gb-2014-15-6-r78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., et al. (2010). The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen F., Carsner E., Stout M. (1940). Photothermal induction of flowering in sugar beets. J. Agric. Res. 61, 101–124. [Google Scholar]
- Pfeiffer N., Tränkner C., Lemnian I., Grosse I., Müller A. E., Jung C., et al. (2014). Genetic analysis of bolting after winter in sugar beet (Beta vulgaris L.). Theor. Appl. Genet. 127, 2479–2489. 10.1007/s00122-014-2392-x [DOI] [PubMed] [Google Scholar]
- Pin P. A., Benlloch R., Bonnet D., Wremerth-Weich E., Kraft T., Gielen J. J. L., et al. (2010). An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science 330, 1397–1400. 10.1126/science.1197004 [DOI] [PubMed] [Google Scholar]
- Pin P. A., Zhang W., Vogt S. H., Dally N., Büttner B., Schulze-Buxloh G., et al. (2012). The role of a pseudo-response regulator gene in life cycle adaptation and domestication of beet. Curr. Biol. 22, 1095–1101. 10.1016/j.cub.2012.04.007 [DOI] [PubMed] [Google Scholar]
- Qi J., Liu X., Shen D., Miao H., Xie B., Li X., et al. (2013). A genomic variation map provides insights into the genetic basis of cucumber domestication and diversity. Nat. Genet. 45, 1510–1515. 10.1038/ng.2801 [DOI] [PubMed] [Google Scholar]
- Reeves P. A., He Y., Schmitz R. J., Amasino R. M., Panella L. W., Richards C. M. (2007). Evolutionary conservation of the FLOWERING LOCUS C-mediated vernalization response: evidence from the sugar beet (Beta vulgaris). Genetics 176, 295–307. 10.1534/genetics.106.069336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert H. S., Grunewald W., Sauer M., Cannoot B., Soriano M., Swarup R., et al. (2015). Plant embryogenesis requires AUX/LAX-mediated auxin influx. Development 142, 702–711. 10.1242/dev.115832 [DOI] [PubMed] [Google Scholar]
- Saghai-Maroof M. A., Soliman K. M., Jorgensen R. A., Allard R. W. (1984). Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. U.S.A. 81, 8014–8018. 10.1073/pnas.81.24.8014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger K. (2014). Using next-generation sequencing to isolate mutant genes from forward genetic screens. Nat. Rev. Genet. 15, 662–676. 10.1038/nrg3745 [DOI] [PubMed] [Google Scholar]
- Schneeberger K., Ossowski S., Lanz C., Juul T., Petersen A. H., Nielsen K. L., et al. (2009). SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat. Methods 6, 550–551. 10.1038/nmeth0809-550 [DOI] [PubMed] [Google Scholar]
- Shi Y., Manley J. L. (2015). The end of the message: multiple protein–RNA interactions define the mRNA polyadenylation site. Genes Dev. 29, 889–897. 10.1101/gad.261974.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson G. G., Dijkwel P. P., Quesada V., Henderson I., Dean C. (2003). FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell 113, 777–787. 10.1016/S0092-8674(03)00425-2 [DOI] [PubMed] [Google Scholar]
- Steinberg R. A., Garner W. (1936). Response of certain plants to length of day and temperature under controlled conditions. J. Agric. Res. 52, 943–960. [Google Scholar]
- Stout M. (1946). Relation of temperature to reproduction in sugar beets. J. Agric. Res. 72, 49–68. [Google Scholar]
- Takagi H., Abe A., Yoshida K., Kosugi S., Natsume S., Mitsuoka C., et al. (2013). QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 74, 174–183. 10.1111/tpj.12105 [DOI] [PubMed] [Google Scholar]
- Trap-Gentil M. V., Hébrard C., Lafon-Placette C., Delaunay A., Hagege D., Joseph C., et al. (2011). Time course and amplitude of DNA methylation in the shoot apical meristem are critical points for bolting induction in sugar beet and bolting tolerance between genotypes. J. Exp. Bot. 62, 2585–2597. 10.1093/jxb/erq433 [DOI] [PubMed] [Google Scholar]
- Van der Auwera G. A., Carneiro M. O., Hartl C., Poplin R., del Angel G., Levy-Moonshine A., et al. (2013). From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinformatics 11, 11.10.1–11.10.33. 10.1002/0471250953.bi1110s43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk H. (2009). Evolutionary change in flowering phenology in the iteroparous herb Beta vulgaris ssp. maritima: a search for the underlying mechanisms. J. Exp. Bot. 60, 3143–3155. 10.1093/jxb/erp142 [DOI] [PubMed] [Google Scholar]
- Vogt S. H., Weyens G., Lefèbvre M., Bork B., Schechert A., Müller A. E. (2014). The FLC-like gene BvFL1 is not a major regulator of vernalization response in biennial beets. Front. Plant Sci. 5:146. 10.3389/fpls.2014.00146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood D., Scott R. (1975). Sowing sugar beet in autumn in England. J. Agri. Sci. 84, 97–108. 10.1017/S0021859600071926 [DOI] [Google Scholar]
- Xu R., Ye X., Li Q. Q. (2004). AtCPSF73-II gene encoding an Arabidopsis homolog of CPSF 73 kDa subunit is critical for early embryo development. Gene 324, 35–45. 10.1016/j.gene.2003.09.025 [DOI] [PubMed] [Google Scholar]
- Xu R., Zhao H., Dinkins R. D., Cheng X., Carberry G., Li Q. Q. (2006). The 73 kD subunit of the cleavage and polyadenylation specificity factor (CPSF) complex affects reproductive development in Arabidopsis. Plant Mol. Biol. 61, 799–815. 10.1007/s11103-006-0051-6 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Gu L., Hou Y., Wang L., Deng X., Hang R., et al. (2015). Integrative genome-wide analysis reveals HLP1, a novel RNA-binding protein, regulates plant flowering by targeting alternative polyadenylation. Cell Res. 25, 864–876. 10.1038/cr.2015.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.