Abstract
Tubby-like proteins (TLPs), which have a highly conserved β barrel tubby domain, have been found to be associated with some animal-specific characteristics. In the plant kingdom, more than 10 TLP family members were identified in Arabidopsis, rice and maize, and they were found to be involved in responses to stress. The publication of the apple genome makes it feasible to systematically study the TLP family in apple. In this investigation, nine TLP encoding genes (TLPs for short) were identified. When combined with the TLPs from other plant species, the TLPs were divided into three groups (group A, B, and C). Most plant TLP members in group A contained an additional F-box domain at the N-terminus. However, no common domain was identified other than tubby domain either in group B or in group C. An analysis of the tubby domains of MdTLPs identified three types of conserved motifs. Motif 1 and 2, the signature motifs in the confirmed TLPs, were always present in MdTLPs, while motif 3 was absent from group B. Homology modeling indicated that the tubby domain of most MdTLPs had a closed β barrel, as in animal tubby domains. Expression profiling revealed that the MdTLP genes were expressed in multiple organs and were abundant in roots, stems, and leaves but low in flowers. An analysis of cis-acting elements showed that elements related to the stress response were prevalent in the promoter sequences of MdTLPs. Expression profiling by qRT-PCR indicated that almost all MdTLPs were up-regulated at some extent under abiotic stress, exogenous ABA and H2O2 treatments in leaves and roots, though different MdTLP members exhibited differently in leaves and roots. The results and information above may provide a basis for further investigation of TLP function in plants.
Keywords: tubby-like protein, bioinformatics, signature motifs, abiotic stress, apple
Introduction
Tubby-like proteins (TLPs) are present in all eukaryotes, from single-celled to multicellular organisms (Liu, 2008), including Caenorthabditis elegans, Drosophila, Arabidopsis, rice, maize, chicken, and mouse (North et al., 1997; Heikenwalder et al., 2001; Ronshaugen et al., 2002; Figlewicz et al., 2004). TLPs have a typical tubby domain that forms a closed β barrel with 12 anti-parallel strands and a central hydrophobic α helix (Boggon et al., 1999). The number of TLPs ranges from 4 in humans to 15 in maize (Stone and Callis, 2007; Yulong et al., 2016). The distribution of this family across many species suggests that these proteins have a basic function. In mammals, the TLP genes play important roles in the maintenance and functioning of neuronal cells, and mutating these genes can result in obesity, loss of vision and hearing, infertility, and insulin resistance (Coleman and Eicher, 1990; Heckenlively et al., 1995; Kleyn et al., 1996; Noben-Trauth et al., 1996). Tubby proteins may function as bipartite transcriptional regulators by binding to double-stranded DNA and activating transcription in the nucleus (Boggon et al., 1999; Santagata et al., 2001). Despite tubby domains being highly conserved, different animal TLP members are unable to compensate for one another in function (Ikeda et al., 1999).
Plant TLPs contain a conserved C-terminal tubby domain; however, unlike animal TLPs, most plant TLPs also contain highly conserved F-box domains in their N-terminus (Gagne et al., 2002; Lai et al., 2004). F-box containing proteins are involved in protein ubiquitination by acting as bridges between specific substrates and generic components of the SCF-type (Skp1-Cullin-F-box) or ECS-type (ElonginC-cullin-SOCS-box) E3 ubiquitin ligase complexes (Kile et al., 2002). Compared with the wide array of cellular functions identified for animal TLPs, the functions and mechanisms of plant TLPs are relatively unknown. In Arabidopsis, AtTLP9 and AtTLP3 are involved in ABA signaling during germination (Lai et al., 2004). AtTLP9 also plays a role in responses to salt and drought stress (Bao et al., 2014). Overexpression of a TLP from chickpea was recently demonstrated to confer increased tolerance to salt, drought and oxidative stress (Wardhan et al., 2012). Furthermore, two studies in rice indicated that TLPs play important roles in host-pathogen interactions (Cai et al., 2008; Kou et al., 2009). Nevertheless, the highly conserved evolution of tubby (or tubby-like) proteins and their redundancy suggests that they play an indispensable role in plants.
As one of the most widely cultivated fruit trees, apple (Malus domestica) is also one of the most economically important woody plants (Hummer and Janick, 2009). The completion of the apple (Malus domestica) genome map offers the possibility of investigating the TLP gene family in this species (Velasco et al., 2010). A previous study showed that the expression of an apple tubby protein (TLP7) can enhance stress tolerance (Du et al., 2014). In this study, we performed a genome-wide search of TLP genes in the apple genome and analyzed their chromosomal distributions, functional domains, and expression patterns in different organs and processes. The three-dimensional structure of the tubby domain and conserved motifs were further modeled and characterized. Transcriptional profiling demonstrated organ-specific expression patterns for individual TLP genes. The expression of some MdTLPs were found to be sensitive to abiotic stress, which suggested that TLP family genes might be vital for the response and adaptation to abiotic stresses in apple. This study provides a foundation for the further functional analysis of plant TLPs.
Materials and Methods
Identification of MdTLPs in the Apple Genome
Two different approaches were used to identify MdTLPs in the apple genome. For the first approach, the published Arabidopsis TLP protein sequences were retrieved from the TIGR database1 and used as queries in BLASTP searches against the Malus domestica genome database (Malus domestica Genome v1.02). Previous studies failed to detect AtTLP4 in Arabidopsis using several experimental approaches, suggesting that it might be a pseudogene (Lai et al., 2004; Bao et al., 2014). Thus, we used 10 Arabidopsis TLP protein sequences as queries. To avoid excluding any possible candidates, the E-value cut-off was set to 0.001, as was done previously in a similar investigation (Li et al., 2011; Jia et al., 2013; Cui et al., 2015). Redundant sequences with the same accession numbers were removed from the data set. The retrieved sequences were then queried against the InterPro3 database to ensure the presence of tubby domains (Mitchell et al., 2015). For the second approach, the tubby-domain Hidden Markov Model Profile, which was downloaded from the Pfam4 database, was used as a query to search for all of the annotated proteins in the complete apple genome database2 using HMMER 3.0 (Eddy, 1998). The candidate proteins’ sequences were extracted by a Perl script and then examined for the tubby domain using the InterPro database. All of the sequences identified through these two methods were submitted to SMART5 to ensure the integrity of the tubby domain sequence. A BLASTN search (E-value ≤E-100) against the apple EST dataset was conducted to find the corresponding expression record for each putative family member6.
Multiple Alignments and Phylogenetic Analysis
Multiple sequence alignments of the TLP amino acid sequences in apple (MdTLP1-9), Arabidopsis (AtTLP1-3, AtTLP5-11), rice (OsTLP1-14), and maize (ZmTLP1-15) were performed with ClustalX (Version 2.17) (Saitou and Nei, 1987; Larkin et al., 2007). To make sure that all sequences were properly aligned, the sequences of the tubby domain were adjusted manually first, then let the other regions realigned, using software CLC Sequence Viewer 78. An unrooted phylogenetic tree was constructed from the alignments of the full-length protein sequences according to the neighbor-joining method with 1,000 replications, and the phylogenetic tree was drawn with the MEGA5 program (Tamura et al., 2011).
Chromosomal Location and Determination of the Exon/Intron Structure
Chromosomal location data were retrieved from apple genome annotations downloaded from the Genome Database for Rosaceae9. The chromosome map showing the physical location of all of the MdTLP genes was generated using a revised version of MapDraw (Liu and Meng, 2003). To explore the diverse exon–intron organizations of MdTLPs and AtTLPs (AtTLP1-3, AtTLP5-11), we compared the predicted coding sequences of MdTLPs and AtTLPs with their corresponding genomic sequences using GSDS software10 (Hu et al., 2015). The molecular weight and isoelectric point (pI) of the proteins were calculated with the ExPASY Compute pI/Mw Program11 (Wilkins et al., 1999).
Identification of Additional Domains and Protein Subcellular Locations
To identify potential protein motifs and detect any additional domains outside the apple tubby domains, sequences of full-length MdTLPs were queried against the InterPro database12 (Mitchell et al., 2015). Protein subcellular locations were predicted using WoLF PSORT13, an extension of the PSORT II program (Horton et al., 2007).
Motif Analysis and Homology Modeling of the Tubby Domain
All putative MdTLPs were analyzed by MEME (version 4.11.114), a motif search tool for identifying conserved motifs of tubby domains (Bailey et al., 2009). To obtain the most significant conserved motifs in the nine MdTLPs, different numbers of motifs were tried with default parameters in normal mode. The identified motifs were annotated using SMART protein analysis software15.
The three-dimensional structure of the tubby domain was obtained by homology modeling using the website CPHmodels 3.2 Server16. Images were generated in the modeling package PyMOL v1.517 (Nielsen et al., 2010).
Searching for cis-Acting Elements in the Promoters of MdTLPs
To investigate cis-acting elements in the promoter sequences of MdTLPs, 1,500 bp of genomic DNA sequence upstream of the transcriptional start sites was obtained from the apple genome. The upstream sequences were subsequently scanned in the PlantCARE database18 for the presence of various cis-acting elements (Lescot et al., 2002).
Sample Preparation and Total RNA Extraction
To investigate the expression of MdTLPs under abiotic stress, 3-year-old apple (Malus sieversii) seedlings were treated with either chilling at 4°C, 20% PEG6000, 100 mM H2O2, or 100 μM ABA for 0, 1, 3, 6, and 9 h. The leaves and roots from five individual plants were collected, placed into liquid nitrogen and stored at -80°C until further use. With respect to the samples used for organ-specific expression, different organs were collected and also stored at -80°C. Total RNA was isolated from the leaves using the CTAB procedure (Gasic et al., 2004). The RNA concentrations and A260/A280 ratios were determined using a NanoDrop Spectrophotometer (ND-1000 Spectrophotometer, Peqlab). The integrity of the RNA samples was examined with an Agilent 2100 Bioanalyzer (RNA Nano Chip, Agilent, Santa Clara, CA, USA). Suitable RNA was used for cDNA synthesis and qRT-PCR.
Quantitative Real-Time PCR (qRT-PCR) Analysis
cDNA fragments were synthesized from total RNA using TransScriptTM One-step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China). To ensure the cDNA samples obtained were qualified, two stress-specific genes (DREB genes: MDP0000147009 and MDP0000218344) in apple genome were selected as marker genes, which expressions have been demonstrated to be up-regulated under stress treatments (Zhao et al., 2012). Gene-specific primers for amplification from cDNA were designed based on target gene sequences using the Beacon Designers 8.10 software. The primer sequences used in this investigation are listed in Supplementary Table S1. qRT-PCR was performed with a CFX96 real-time system (Bio-Rad, USA) in a final volume of 20 μl containing 0.8 μl of cDNA, 10 μl of 2 × SYBR Premix Ex Taq (SYBR Green; Tiangen, China) and 0.8 μl of (10 μM) primers. The thermal cycling conditions were as follows: 44 cycles of 95°C denaturation for 15 s, 55°C annealing for 30 s and 72°C extension for 15 s. The apple actin gene was used as an internal control. The real-time PCR experiment was carried out at least three times under identical conditions. The relative expression levels were calculated as 2-(ΔCtoftreatment-ΔCtofcontrol). The relative expression levels of MdTLPs in stressed samples (1, 3, 6, and 9 h) were compared to the controls (0 h) with parametric one-way ANOVA at significance levels of P ≤ 0.05 and P ≤ 0.01.
Results
Genome-Wide Identification and Phylogenetic Analysis of MdTLPs in Apple
To identify the TLP protein-coding genes in apple, two approaches were used. For the first strategy, the peptide sequences of the TLPs of Arabidopsis were used as BLAST queries against the apple genome (Malus domestica Genome v1.0). To ensure that potential TLPs were not excluded and to obtain credible results, the E-value was set to 0.001, as done in a similar investigation (Li et al., 2011; Jia et al., 2013; Cui et al., 2015). Using this approach, 25 potential TLPs were identified in the apple genome. To determine whether these proteins contained tubby domains, the sequences were compared against the InterPro Database. Using this approach, eight potential TLPs were identified from the apple genome. For the second strategy, the tubby domain Hidden Markov Model Profile (Pfam01167) from the Pfam database19 was used to search the apple genome. A total of 10 putative TLPs were obtained. These sequences were also analyzed using the InterPro Database, and eight potential TLPs contained a tubby domain. All of the potential TLPs obtained by the above two strategies were submitted to the SMART database20 to verify the integrity of the tubby domain sequence and confirm the presence of apple TLP genes. Finally, eight genes were identified.
In our previous study, we cloned a TLP gene (named MdTLP7) from apple, whose expression increased significantly under cold stress (Du et al., 2014). However, the MdTLP7 gene sequence could not be identified in the published apple genome. To determine whether MdTLP7 was present in the apple genome and to check the expression of this and other identified apple TLP genes, the gene sequences were searched against the apple EST database at NCBI21. In BLASTN analysis, each of the nine gene sequences hit several apple ESTs sequences (Supplementary Table S2), which indicated that these nine genes were genuinely expressed in apple. Therefore, we believe that the apple genome contains nine MdTLP genes (Table 1). To remain consistent with our previous studies, the MdTLP7 gene name was retained, and the other eight MdTLP genes were named MdTLP1-6 and MdTLP8-9 based on their distribution in the phylogenetic tree (Figure 1). The MdTLP peptides in length ranged from 269 to 693 amino acids, with predicted molecular weights between 29.9 and 78 kDa (Table 1).
Table 1.
TLP family information for apple.
| Group | Name | Gene identifier | ORF (bp) | Protein (aa) | theoretical Mw(kDa)/pI | Subcellular localization |
|---|---|---|---|---|---|---|
| A1 | MdTLP1 | MDP0000912429 | 810 | 269 | 29.9/9.43 | Nuclear |
| MdTLP2 | MDP0000184528 | 1635 | 544 | 60.5/9.58 | Chloroplast | |
| MdTLP3 | MDP0000264408 | 1209 | 402 | 45.2/9.40 | Chloroplast | |
| A2 | MdTLP4 | MDP0000122158 | 1302 | 433 | 48.8/9.47 | Nuclear |
| MdTLP5 | MDP0000303852 | 2082 | 693 | 78.0/9.44 | Nuclear | |
| A4 | MdTLP6 | MDP0000237033 | 1038 | 345 | 38.7/9.29 | Nuclear |
| MdTLP7 | HM122708.1 | 1245 | 414 | 46.6/9.36 | Nuclear | |
| B | MdTLP8 | MDP0000175577 | 1338 | 445 | 49.6/9.49 | Nuclear |
| MdTLP9 | MDP0000320802 | 1431 | 475 | 52.8/9.53 | Nuclear |
Gene identifiers with prefix MDP are from the Genome Database for Rosaceae (GDR, http://www.rosaceae.org/) and Gene identifiers with prefix HM from NCBI (National Center for Biotechnology Information). Protein subcellular locations were predicted using WoLF PSORT (http://psort.nibb.ac.jp/).
FIGURE 1.
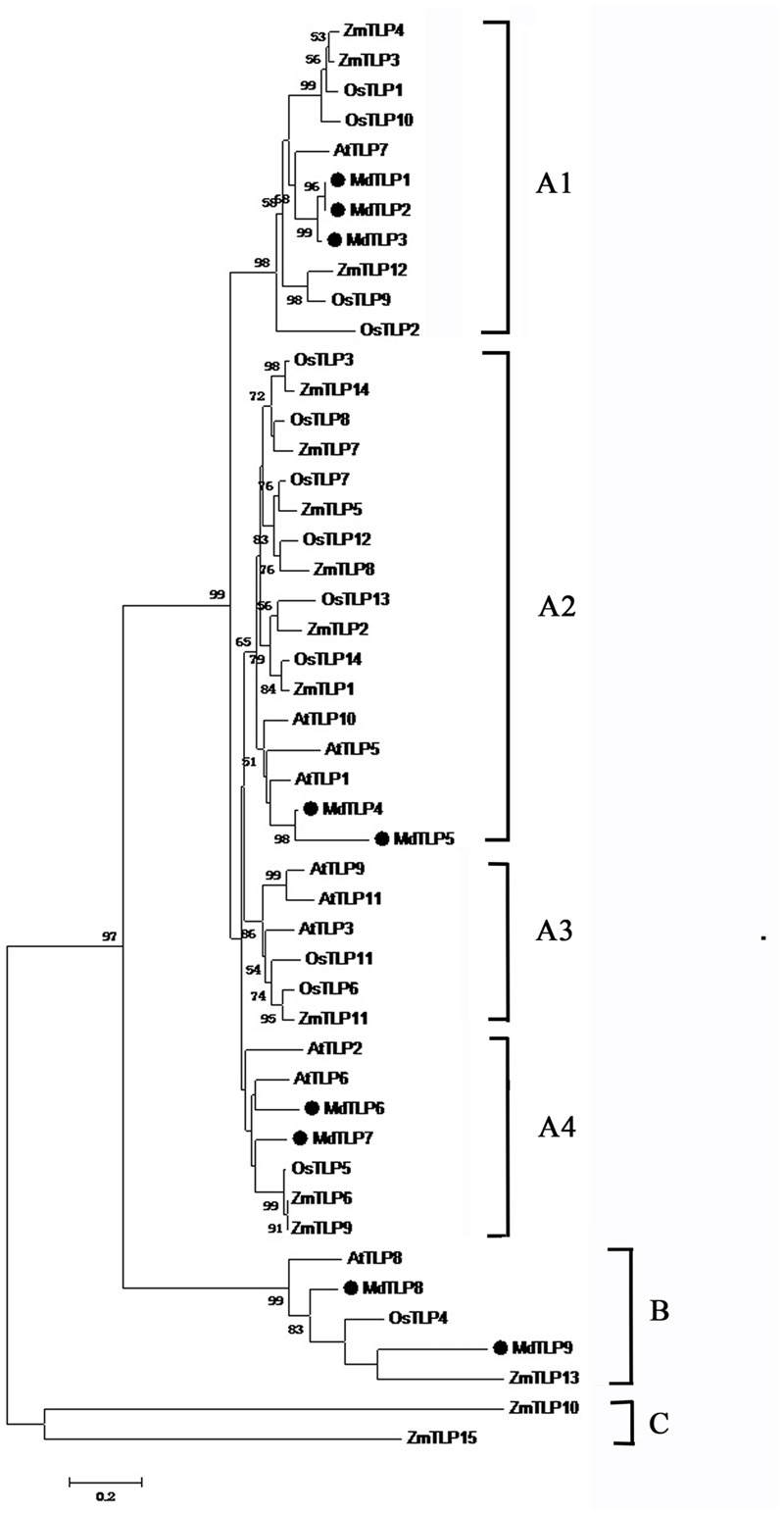
Phylogenetic tree of putative MdTLPs and the TLPs from Arabidopsis, rice and maize constructed by ClustalX (Version 2.1). Branch lengths indicate distance. The bootstrap support value of lower than 50% was hidden.
Multiple sequence alignment showed that all plant TLPs had a highly conservative tubby domain at C-terminal almost with a conservative proline residue at the beginning (Supplementary Figure S1). To gain insight into the evolutionary relationships among all plant TLP proteins, a phylogenetic tree was constructed based on the full-length amino acid sequences from apple, Arabidopsis, rice, and maize (Lai et al., 2004; Yang et al., 2008; Yulong et al., 2016). All of the TLP encoding genes from the above four plant species were divided into three distinct groups: A, B, and C (Figure 1). Of 48 members, 41 plant TLPs belonged to group A and were further divided into the four subgroups A1–A4. Group B contained five members, and group C only contained two maize TLPs. Group A and group B contained TLP members from both dicot and monocot plants. The results showed that in some cases, the evolutionary relationship of plant TLPs between dicot and monocot was closer than that among dicot or monocot plants (Figure 1). For example, MdTLP9 was closer with ZmTLP13 than with MdTLP8 in group B (Figure 1). To check whether this result was valid, a detailed sequence alignment was checked (Supplementary Figure S1). Indeed, MdTLP9 had higher similarity with ZmTLP13 than MdTLP8 in sequences, especially in the segment of core α helix in tubby domain (indicated in the box of Supplementary Figure S1). These results suggested that the main characteristics of plant TLPs in group A and group B had been established before the dicot–monocot plants split. The TLPs from apple (MdTLPs) were distributed into two groups, group A, including A1, A2, and A4, and group B (Figure 1). The members in subgroup A3, AtTLP3 and AtTLP9 from Arabidopsis, have been studied in detail, and it has been indicated that they are involved in the stress response (Lai et al., 2004; Reitz et al., 2012, 2013; Bao et al., 2014). Whether MdTLPs in the different subgroups or different groups have similar functions requires further investigation.
MdTLPs Chromosomal Location, Gene Structure, Additional Functional Domains, and Subcellular Localization Analysis
Based on the chromosomal distribution map of MdTLP genes generated in this study, all of the MdTLP genes were distributed across six of the seventeen apple chromosomes, including 8, 9, 10, 11, 15, and 17 (Supplementary Figure S2). Only chromosomes 8 and 11 contained two MdTLPs, while other MdTLPs were mapped to different chromosomes. MdTLP1 and MdTLP2 were both found on chromosomes 8, located near each other. They were also clustered together in the phylogenetic tree. To understand the possible structural evolution of MdTLPs, the intron-exon structure of MdTLPs and AtTLPs was analyzed in this study (Supplementary Figure S3). Intron/exon organizations for all MdTLPs were determined based on their exon position and gene length. The number of introns in the MdTLPs varied, ranging from 2 to 9 (Supplementary Figure S3). Unlike other MdTLPs, MdTLP5 had much longer introns, although the number of introns (8) was similar to that of other MdTLPs. The MdTLPs in group B shared similar intron-exon structure distribution characteristics.
Most MdTLPs in group A contained an additional functional domain, an F-box domain at the N-terminus (Figure 2). F-box-containing proteins constitute a large family in eukaryotes, which are characterized by a conserved F-box domain consisting of approximately 50 amino acids at their N-terminus. The appearance of both domains in one protein suggested that an interplay between tubby and F-box domains may play roles in physiological processes. Of nine MdTLPs, three members had no F-box domain (MdTLP1, MdTLP8, and MdTLP9) (Figure 2). Although these members were quite similar in patterns of protein structure, multiple alignments, and phylogenetic analysis showed that MdTLP1 was far from MdTLP8 and MdTLP9 (Figure 1; Supplementary Figure S1).
FIGURE 2.
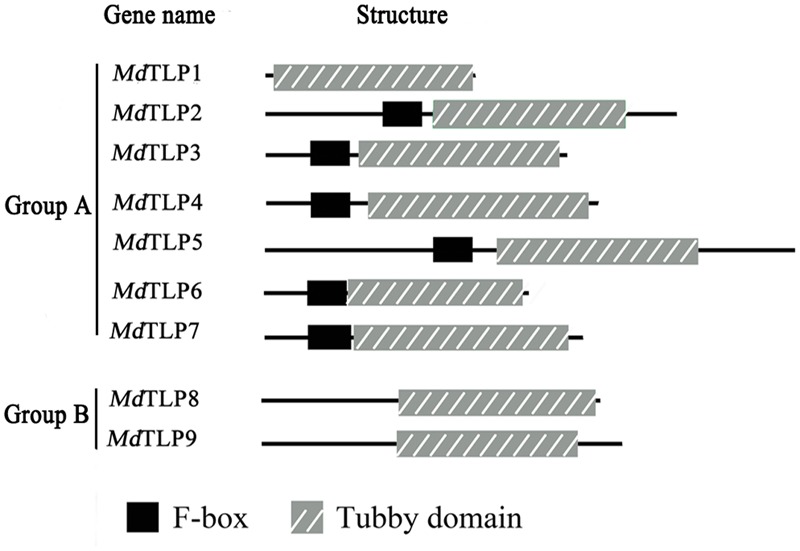
Structure and organization of MdTLPs in apple. The position of the signature tubby domain and F-box domain are shown. The gene name is indicated on the left.
The subcellular location of MdTLPs was predicted, and the results are shown in Table 1. Most MdTLPs were predicted to be located in the nucleus. Only MdTLP2 and MdTLP3 in subgroup A1 were predicted to be located in chloroplasts. In mice, TLPs were mainly localized within the nucleus as transcription factors (Boggon et al., 1999). Here, we speculate that the nuclear-localized MdTLPs may also function as transcription factors. Future investigations are needed to experimentally confirm their location and transcription factor activity.
Motif and Three-Dimensional Structure Analysis of the Tubby Domain
All of the MdTLP peptide sequences were submitted to MEME22, and three types of motifs were identified in the tubby domain of MdTLPs (Figure 3A). The consensus sequences of motifs are shown in Figure 3B, with the height of each stacked letter representing the probability of that amino acid appearing at each position. Motif 1 and 2 were more highly conserved and usually located near the C-terminus of the polypeptide, harboring significant tubby domain characteristics similar to other plant and animal TLPs. Motif 3 often existed in the middle of the protein with a highly conserved amino acid sequence (RGPRRM), suggesting that this sequence may have an important biological function.
FIGURE 3.
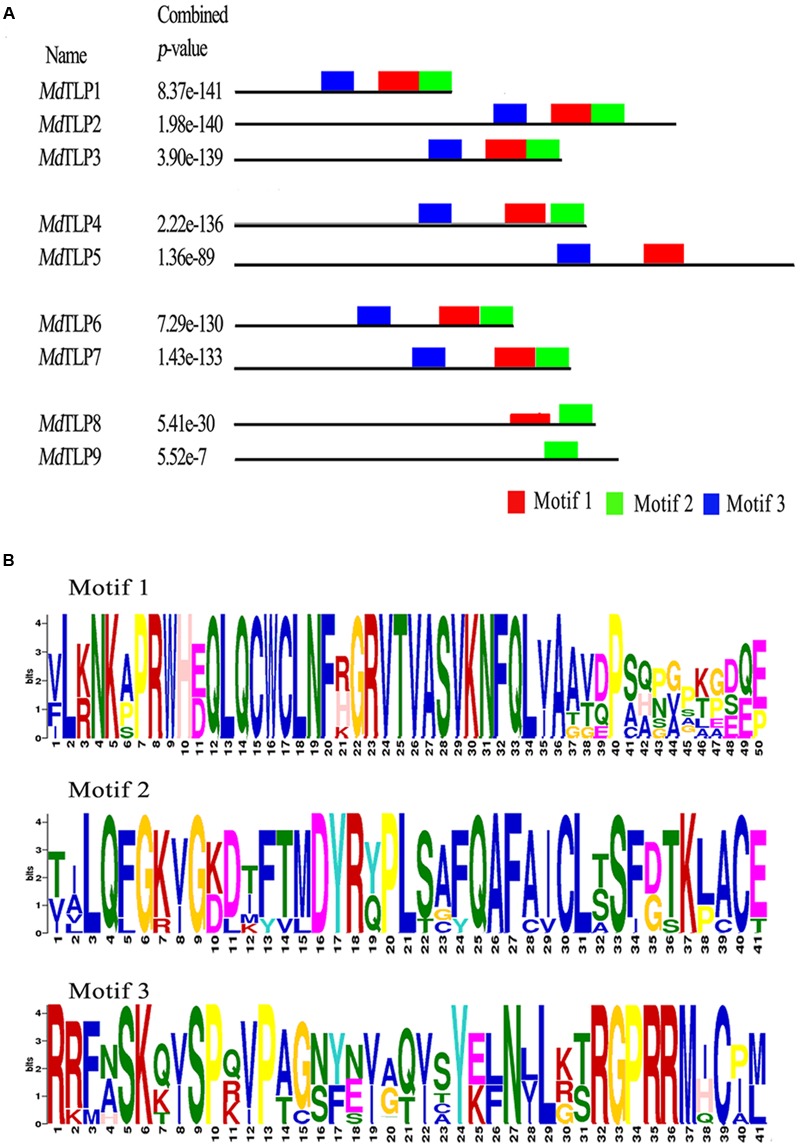
Conserved motifs of nine predicted tubby domains of MdTLPs identified with the MEME program. (A) Distribution of the identified motifs in the tubby domain of nine MdTLPs. (B) Consensus sequence for putative motifs.
Three-dimensional structures of MdTLP tubby domains were established by homology modeling of a central α helix surrounded by a β barrel (Figure 4). Some MdTLP members had a typical tubby architecture with a closed β barrel formed by 12 anti-parallel strands and a central α helix, for example, MdTLP1, MdTLP2, MdTLP7, and MdTLP8. Other members contained an incomplete β barrel (less than 12 anti-parallel strands) and a central α helix, for example, MdTLP3, MdTLP4, and MdTLP6, which had 10, 11, and 6 anti-parallel sheets, respectively. MdTLP9 consisted of a complete β barrel without a central α helix, while MdTLP5 had an incomplete β barrel without a central α helix. Differences in the three-dimensional structures may lead to the functional diversification of different MdTLPs.
FIGURE 4.
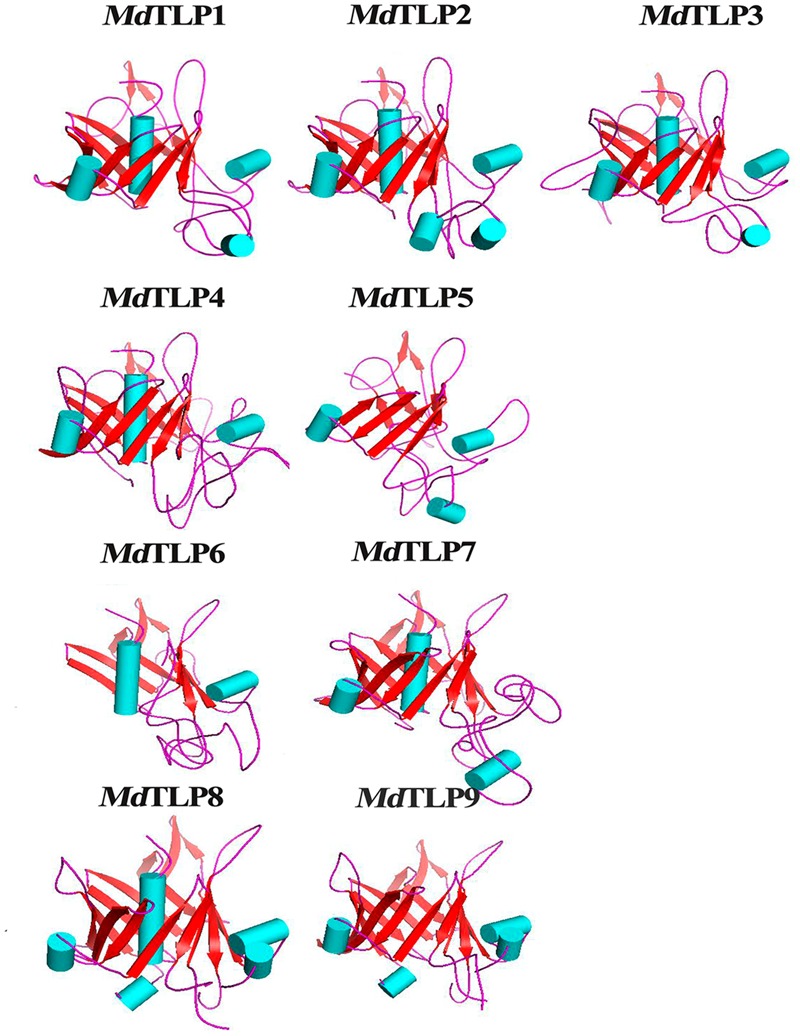
Homology modeling of the tubby domain of MdTLPs. The alpha helix is shown in light blue, and the beta fold is shown in red.
Analysis of Promoter Sequences of MdTLPs
Transcriptional control of stress-responsive genes is a crucial part of the plant response to a range of abiotic and biotic stresses. Transcription factors have the potential to activate or repress genes through cis-acting sequences in promoter regions that respond to specific stresses (Singh et al., 2002). In plants, some TLPs play a role in responses to abiotic stress (Wardhan et al., 2012; Bao et al., 2014; Du et al., 2014). A search for putative cis-acting elements within the 1,500 bp of the genomic sequence upstream of the MdTLP 5′-UTRs was performed. Many stress-response related cis-acting elements were found in the promoter regions of MdTLPs, including ABREs, DRE, LTRE and MYB and MYC transcription factor elements (Table 2). MYB elements exist in all of the MdTLP promoters. MYB has been demonstrated to be involved in stress-induced drought, low temperature, salt, and ABA responses (Xiong et al., 2002; Dai et al., 2007). ABRE responds to drought and ABA via ABRE binding proteins in Arabidopsis (Hobo et al., 1999; Li et al., 2012). DRE binding proteins participate in drought, salt, low temperature, and ABA responses in rice (Zhang et al., 2009). LTRE primarily contributes to the regulation of low temperature responses in poplar (Jiang et al., 1996; Maestrini et al., 2009). The presence of abiotic stress-responsive elements suggests that MdTLPs may be regulated by various stresses. In addition to the cis-acting elements mentioned above, other types of cis-acting elements were also detected and are listed in Supplementary Table S3. Two promoters of identified MdTLPs were not analyzed because their promoter sequences could not be found in the apple genome database.
Table 2.
Distribution of ABRE, DRE, LTRE, MYB, and MYC cis-acting elements in MdTLPs promoters.
| Group | Gene | ABRE | DRE | LTRE | MYB | MYC | Total |
|---|---|---|---|---|---|---|---|
| A | MdTLP1 | 0 | 0 | 0 | 2 | 0 | 2 |
| MdTLP2 | 3 | 0 | 0 | 21 | 19 | 43 | |
| MdTLP3 | 5 | 0 | 1 | 17 | 12 | 35 | |
| MdTLP4 | 3 | 0 | 2 | 13 | 13 | 31 | |
| MdTLP5 | 2 | 1 | 3 | 17 | 10 | 33 | |
| B | MdTLP8 | 1 | 3 | 3 | 24 | 22 | 53 |
| MdTLP9 | 3 | 2 | 2 | 22 | 20 | 49 |
Expression of MdTLPs under Abiotic Stress, ABA, and H2O2 Treatments
Based on the promoter analysis results, MdTLPs may be associated with the abiotic stress response. Thus, to further investigate the potential functions of MdTLPs under abiotic stress conditions, cDNA samples were obtained using apple seedlings exposed to either PEG, H2O2, exogenous ABA, or cold stress for 0, 1, 3, 6, and 9 h. To detect the quality of cDNA samples, two stress-sensitive genes (DREB genes: MDP0000147009 and MDP0000218344) were selected as marker genes. Similar with the previous report (Zhao et al., 2012), their expressions were significantly up-regulated under different stresses both in leaves and roots (Supplementary Figure S4). As to the expression of MdTLPs, almost all of the MdTLPs were also up-regulated in both leaves and roots under different stresses and exogenous ABA. During PEG treatment, six MdTLPs (MdTLP1-5, MdTLP9) were up-regulated significantly in leaves (Figure 5A). For example, the relative transcript levels of MdTLP1 and MdTLP9 were both up-regulated by approximately 35-fold compared with the control condition. Only three MdTLPs (MdTLP6, MdTLP7, and MdTLP8) were down-regulated under PEG stress. In the roots, five MdTLPs (MdTLP3, MdTLP4, and MdTLP7-9) were up-regulated significantly in response to PEG treatment (Figure 5B). Among them, the expression of MdTLP4 changed maximum, reaching nearly 30-fold. Under cold stress, almost all MdTLPs in leaves and in roots showed significantly up-regulated transcript levels, suggesting that all MdTLPs are cold-responsive genes. Among these genes, MdTLP4 expression increased nearly 70-fold in leaves in 6 h and about 150-fold in roots in 3 h. In response to H2O2 or ABA treatment, five members (MdTLP1-5) were significantly up-regulated in leaves, while in roots, the expression of MdTLP4, MdTLP6, MdTLP8, and MdTLP9 showed a significant increase. Notably, MdTLP4 was up-regulated in all the treatments both in leaves and roots.
FIGURE 5.
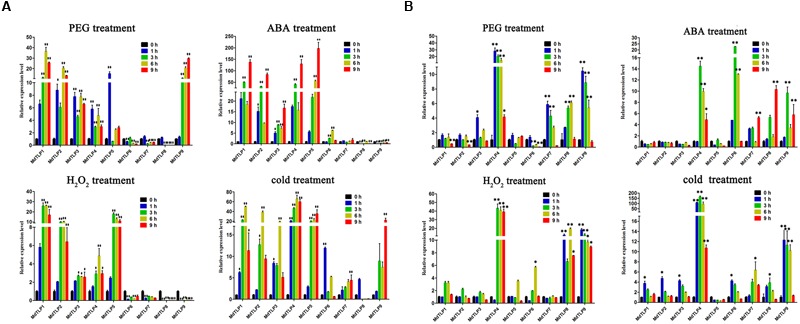
Quantitative real-time PCR (qRT-PCR) for apple under PEG, H2O2, ABA or cold stress and exposure to 20 % PEG6000, 100 mM H2O2, and 100 μM ABA at 4°C for 0, 1, 3, 6, and 9 h. Data were normalized to the expression level of actin gene. The mean expression value was calculated from three independent replicates. Vertical bars indicate the standard error of the mean. ∗∗P ≤ 0.01 and ∗P ≤ 0.05 compared with 0 h. (A) The expression levels of nine MdTLPs in apple leaves under different treatments. (B) The expression levels of nine MdTLPs in apple roots under different treatments.
Expression of MdTLPs in Different Organs
To provide further clues to the putative roles of the MdTLP genes in apple development, the expression patterns of all 9 MdTLP genes were investigated by qRT-PCR in six different organs (leaves, roots, stems, flowers, seeds, and buds), and the results are indicated in Figure 6. Relatively high levels of MdTLPs were found in roots, stems, and leaves; in flowers, the transcript levels of MdTLPs were rather low. Compared with other MdTLPs, MdTLP8, and MdTLP9 had low expression levels in most organs. MdTLP1, MdTLP2, MdTLP3, and MdTLP4 were highly expressed in roots, stems, and leaves, with lower levels of expression in flowers, seeds and buds. Conversely, the MdTLP6, MdTLP7, MdTLP8, and MdTLP9 transcript levels were relatively high in buds, which suggest that these genes play an important role in the sprouting process.
FIGURE 6.
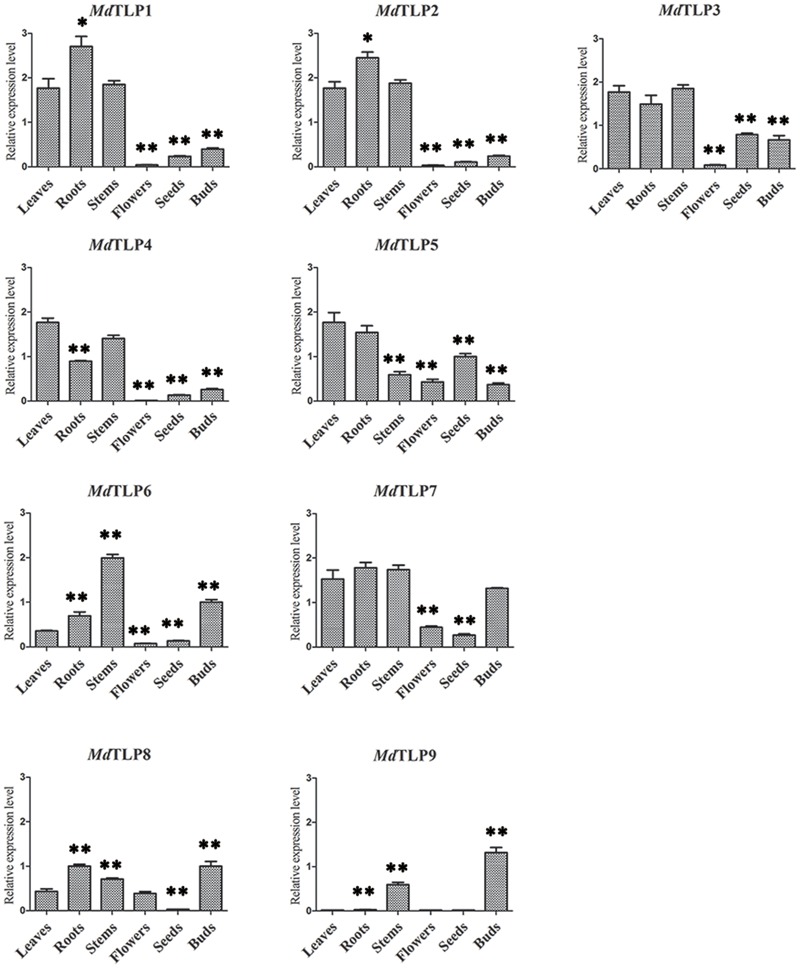
Organ-specific expression pattern of MdTLPs. The relative expression levels of MdTLPs in different organs were compared to the leaves with T-test at significance levels of ∗∗P ≤ 0.01 and ∗P ≤ 0.05.
Discussion
Tubby-like protein genes are members of a conserved gene family that has been identified in many species. Compared to animals, few TLPs have been functionally studied in plants. In the plant kingdom, TLPs have only been reported for a few species, such as Arabidopsis (Lai et al., 2004), rice (Liu, 2008), and maize (Yulong et al., 2016). The publication of the apple genome provided the opportunity to study the characteristics of this family in apple (Velasco et al., 2010). In this study, nine genes encoding TLP proteins were identified in apple, which is similar to the numbers reported in other plants, 11 for Arabidopsis, 14 for rice, and 15 for maize.
Phylogenetic analysis of TLPs from four plant species showed that all of the TLP genes were divided into three groups (Figure 1). The results in this study are similar to those of a previous report comparing the evolutionary relationship of Arabidopsis and rice TLPs (Yang et al., 2008). Here, MdTLPs were distributed into groups A and B. Group A included four subgroups, and group B contained five proteins: MdTLP8 and MdTLP9 in apple, AtTLP8 in Arabidopsis, OsTLP4 in rice, and ZmTLP13 in maize. One apparent feature of all of the members of group B is that there is no F-box domain fused with the tubby domain at the N terminus. The members of this group may have originated from one gene in an ancestral species. Three motifs were identified in MdTLPs, and all three of these motifs could be found in all of the members of group A, except MdTLP5, which only possessed motifs 1 and 3. Motif 3 was not found in group B, MdTLP9 had only motif 2 and MdTLP8 had motifs 1 and 2. Motifs 1 and 2 were also found in the tubby domain of other species, such as Arabidopsis and rice (Lai et al., 2004; Kou et al., 2009). These two motifs were highly conserved among the TLPs from various organisms, suggesting that the two motif sequences were signatures of the tubby domain. However, motif 3 may be unique to apple.
Modeling of the three-dimensional structures suggested that the typical tubby domain of MdTLPs had a central α helix surrounded by a closed β barrel consisting of 12 anti-parallel sheets, as found in MdTLP1, MdTLP2, MdTLP7, and MdTLP8. In mammals, the crystal structure of the tubby domain was found to consist of a 12-stranded β barrel with a central hydrophobic α helix. The tubby domain binds readily to double-stranded DNA due to the β strands (5, 6, 7, 8, 9, and 10) of the β barrel (Boggon et al., 1999). A membrane-bound animal tubby protein translocates from the plasma membrane to the nucleus acting as a transcription factor to regulate the expression of genes when the cell receives a signal (Santagata et al., 2001; Carroll et al., 2004). However, some MdTLP members had an incomplete structure, such as an incomplete β barrel, either with or without the central α helix, which may contribute to different functions in apple.
Unlike animal TLPs, most plant TLPs have an additional F-box domain in the N-terminal protein sequence (Lai et al., 2004; Yang et al., 2008; Yulong et al., 2016) and are therefore also considered to be F-box proteins. F-box proteins have been reported to play roles in responding to abiotic stress (Stone and Callis, 2007; Yan et al., 2011; Maldonado-Calderon et al., 2012; Chen et al., 2014; Cui et al., 2015). The stress responses of plants are regulated by multiple signaling pathways, and there is a significant overlap between the patterns of gene expression that are induced by different stresses (Durrant et al., 2000; Schenk et al., 2000; Glazebrook, 2001; Knight and Knight, 2001; Seki et al., 2001; Chen et al., 2002). Gene induction by stress primarily occurs at the level of transcription and regulates the temporal and spatial expression patterns (Rushton and Somssich, 1998). Many transcription factors are involved in the expression of stress-related genes in plants. Often, several closely related transcription factors have the potential to activate or repress genes through cis-acting sequences that respond to specific stresses (Singh et al., 2002). In this study, the analysis of MdTLP promoter regions revealed a frequent occurrence of cis-acting elements, such as MYB/MYC, ABRE, DRE, and/or LTRE. These elements basically participate in drought, low temperature, and exogenous ABA responses (Xiong et al., 2002; He et al., 2012; Li et al., 2012).
To further study the response of MdTLPs to abiotic stress, qRT-PCR was used to determine the expression patterns of nine putative MdTLPs in apple. MdTLP genes were induced to varying degrees under different treatments, including drought, oxidative, exogenous ABA, and cold stresses (Figure 5). In particular, MdTLP4, MdTLP6, MdTLP8, and MdTLP9 were up-regulated by more than 10-fold under exogenous ABA treatment at 3 h in roots. Four genes (MdTLP1, MdTLP2, MdTLP4, and MdTLP5) were up-regulated by more than 100-fold under exogenous ABA treatment at 9 h in leaves. Most MdTLP genes were also significantly up-regulated under drought, oxidative, and cold stresses in leaves and roots. Of MdTLPs, MdTLP1, MdTLP8, and MdTLP9 do not contain an F-box domain in the N-terminus. Therefore, we speculate that proteins containing an F-box domain or tubby domain may function in response to stress. When these two domains occur together, a synergistic effect may occur. In Arabidopsis, AtTLP3 and AtTLP9 were found to be up-regulated under abiotic stresses or ABA treatment (Lai et al., 2004), which two members belongs to subgroup A3 in phylogenetic tree. In this study, we found that the MdTLPs in subgroup A1, A2, A4, and group B were also sensitive to abiotic stresses. Especially, MdTLP4 was significantly up-regulated under different stresses both in leaves and roots. Our result suggested that plant TLP members may participate in response to abiotic stress. Compared with the previous studies about plant TLP family, especially Arabidopsis TLPs, all the members of MdTLPs were comprehensive analyzed and confirmed to be sensitive to stresses in this investigation.
Author Contributions
Designed the experiments: X-YW, S-SX, and J-NX. Performed the experiments: S-SX, J-NX, and Z-RZ. Analyzed the data: S-SX, J-NX, Z-RZ, and X-SC. Wrote the paper: X-YW, J-NX, and S-SX.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by Special Research Fund of Public Welfare of China Agricultural Ministry (201303093).
Abbreviations
- ABA
abscisic acid
- ABRE
ABA-response element
- DRE
dehydration-response element
- DREB
dehydration responsive element binding protein
- LTRE
low-temperature responsive element
- qRT-PCR
Quantitative real-time PCR
- TLP
tubby-like protein
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01693/full#supplementary-material
Multiple alignments of the full-length proteins sequence of all plant TLPs. The black asterisk over one amino acid indicates the start of the tubby domain. The locations of F-box domain is indicated with single solid lines in black above the sequences. The segment of core α helix in tubby domain was enclosed with a red rectangle.
Locations of MdTLPs on apple chromosomes.
Intron patterns of TLPs from apple and Arabidopsis.
Expression of the DREB genes (MDP0000147009 and MDP0000218344) in leaves (A) and roots (B) under different treatments. Data were normalized to the expression level of actin gene. Vertical bars indicate the standard error of the mean. **P ≤ 0.01 and *P ≤ 0.05 compared with 0 h.
Sequences of the primers used in the quantitative real-time PCR analysis.
The BLAST search result of each MdTLP against apple EST assemblies.
Cis-acting element analysis of MdTLPs using the PLACE database.
References
- Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., Clementi L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37 W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y., Song W. M., Jin Y. L., Jiang C. M., Yang Y., Li B., et al. (2014). Characterization of Arabidopsis tubby-like proteins and redundant function of AtTLP3 and AtTLP9 in plant response to ABA and osmotic stress. Plant Mol. Biol. 86 471–483. 10.1007/s11103-014-0241-6 [DOI] [PubMed] [Google Scholar]
- Boggon T. J., Shan W. S., Santagata S., Myers S. C., Shapiro L. (1999). Implication of tubby proteins as transcription factors by structure-based functional analysis. Science 286 2119–2125. 10.1126/science.286.5447.2119 [DOI] [PubMed] [Google Scholar]
- Cai M., Qiu D., Yuan T., Ding X., Li H., Duan L., et al. (2008). Identification of novel pathogen-responsive cis-elements and their binding proteins in the promoter of OsWRKY13, a gene regulating rice disease resistance. Plant Cell Environ. 31 86–96. [DOI] [PubMed] [Google Scholar]
- Carroll K., Gomez C., Shapiro L. (2004). Tubby proteins: the plot thickens. Nat. Rev. Mol. Cell Biol. 5 55–63. 10.1038/nrm1278 [DOI] [PubMed] [Google Scholar]
- Chen R., Guo W., Yin Y., Gong Z. H. (2014). A novel F-box protein CaF-box is involved in responses to plant hormones and abiotic stress in pepper (Capsicum annuum L.). Int. J. Mol. Sci. 15 2413–2430. 10.3390/ijms15022413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Provart N. J., Glazebrook J., Katagiri F., Chang H. S., Eulgem T., et al. (2002). Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14 559–574. 10.1105/tpc.010410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman D., Eicher E. (1990). Fat (fat) and tubby (tub): two autosomal recessive mutations causing obesity syndromes in the mouse. J. Heredity 81 424–427. [DOI] [PubMed] [Google Scholar]
- Cui H. R., Zhang Z. R., Lv W., Xu J. N., Wang X. Y. (2015). Genome-wide characterization and analysis of F-box protein-encoding genes in the Malus domestica genome. Mol. Genet. Genomics 290 1435–1446. 10.1007/s00438-015-1004-z [DOI] [PubMed] [Google Scholar]
- Dai X., Xu Y., Ma Q., Xu W., Wang T., Xue Y., et al. (2007). Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol. 143 1739–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du F., Xu J. N., Zhan C. Y., Yu Z. B., Wang X. Y. (2014). An obesity-like gene MdTLP7 from apple (Malus x domestica) enhances abiotic stress tolerance. Biochem. Biophys. Res. Commun. 445 394–397. 10.1016/j.bbrc.2014.02.005 [DOI] [PubMed] [Google Scholar]
- Durrant W. E., Rowland O., Piedras P., Hammond-Kosack K. E., Jones J. D. (2000). cDNA-AFLP reveals a striking overlap in race-specific resistance and wound response gene expression profiles. Plant Cell 12 963–977. 10.2307/3871222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S. R. (1998). Profile hidden Markov models. Bioinformatics 14 755–763. 10.1093/bioinformatics/14.9.755 [DOI] [PubMed] [Google Scholar]
- Figlewicz D. P., Zavosh A., Sexton T., Neumaier J. F. (2004). Catabolic action of insulin in rat arcuate nucleus is not enhanced by exogenous “tub” expression. Am. J. Physiol. Endocrinol. Metab. 286 E1004–E1010. 10.1152/ajpendo.00427.2003 [DOI] [PubMed] [Google Scholar]
- Gagne J. M., Downes B. P., Shiu S. H., Durski A. M., Vierstra R. D. (2002). The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 99 11519–11524. 10.1073/pnas.162339999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasic K., Hernandez A., Korban S. S. (2004). RNA extraction from different apple tissues rich in polyphenols and polysaccharides for cDNA library construction. Plant Mol. Biol. Report. 22 437–438. 10.1007/BF02772687 [DOI] [Google Scholar]
- Glazebrook J. (2001). Genes controlling expression of defense responses in Arabidopsis–2001 status. Curr. Opin. Plant Biol. 4 301–308. 10.1016/S1369-5266(00)00177-1 [DOI] [PubMed] [Google Scholar]
- He Y., Li W., Lv J., Jia Y., Wang M., Xia G. (2012). Ectopic expression of a wheat MYB transcription factor gene, TaMYB73, improves salinity stress tolerance in Arabidopsis thaliana. J. Exp. Bot. 63 1511–1522. [DOI] [PubMed] [Google Scholar]
- Heckenlively J. R., Chang B., Erway L. C., Peng C., Hawes N. L., Hageman G. S., et al. (1995). Mouse model for Usher syndrome: linkage mapping suggests homology to Usher type I reported at human chromosome 11p15. Proc. Natl. Acad. Sci. U.S.A. 92 11100–11104. 10.1073/pnas.92.24.11100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikenwalder M. F., Koritschoner N. P., Pajer P., Chaboissier M. C., Kurz S. M., Briegel K. J., et al. (2001). Molecular cloning, expression and regulation of the avian tubby-like protein 1 (tulp1) gene. Gene 273 131–139. 10.1016/S0378-1119(01)00578-9 [DOI] [PubMed] [Google Scholar]
- Hobo T., Asada M., Kowyama Y., Hattori T. (1999). ACGT-containing abscisic acid response element (ABRE) and coupling element 3 (CE3) are functionally equivalent. Plant J. 19 679–689. 10.1046/j.1365-313x.1999.00565.x [DOI] [PubMed] [Google Scholar]
- Horton P., Park K. J., Obayashi T., Fujita N., Harada H., Adams-Collier C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35 W585–W587. 10.1093/nar/gkm259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Jin J., Guo A. Y., Zhang H., Luo J., Gao G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31 1296–1297. 10.1093/bioinformatics/btu817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummer K. E., Janick J. (2009). Rosaceae: taxonomy, economic importance, genomics. Genet. Genom. Rosaceae 6 1–17. 10.1007/978-0-387-77491-6_1 [DOI] [Google Scholar]
- Ikeda S., He W., Ikeda A., Naggert J. K., North M. A., Nishina P. M. (1999). Cell-specific expression of tubby gene family members (tub, Tulp1,2, and 3) in the retina. Invest. Ophthalmol. Vis. Sci. 40 2706–2712. [PubMed] [Google Scholar]
- Jia F., Wu B., Li H., Huang J., Zheng C. (2013). Genome-wide identification and characterisation of F-box family in maize. Mol. Genet. Genomics 288 559–577. 10.1007/s00438-013-0769-1 [DOI] [PubMed] [Google Scholar]
- Jiang C., Iu B., Singh J. (1996). Requirement of a CCGAC cis-acting element for cold induction of the BN115 gene from winter Brassica napus. Plant Mol. Biol. 30 679–684. 10.1007/BF00049344 [DOI] [PubMed] [Google Scholar]
- Kile B. T., Schulman B. A., Alexander W. S., Nicola N. A., Martin H. M., Hilton D. J. (2002). The SOCS box: a tale of destruction and degradation. Trends Biochem. Sci. 27 235–241. 10.1016/S0968-0004(02)02085-6 [DOI] [PubMed] [Google Scholar]
- Kleyn P. W., Fan W., Kovats S. G., Lee J. J., Pulido J. C., Wu Y., et al. (1996). Identification and characterization of the mouse obesity gene tubby: a member of a novel gene family. Cell 85 281–290. 10.1016/S0092-8674(00)81104-6 [DOI] [PubMed] [Google Scholar]
- Knight H., Knight M. R. (2001). Abiotic stress signalling pathways: specificity and cross-talk. Trends Plant Sci. 6 262–267. 10.1016/S1360-1385(01)01946-X [DOI] [PubMed] [Google Scholar]
- Kou Y., Qiu D., Wang L., Li X., Wang S. (2009). Molecular analyses of the rice tubby-like protein gene family and their response to bacterial infection. Plant Cell Rep. 28 113–121. 10.1007/s00299-008-0620-z [DOI] [PubMed] [Google Scholar]
- Lai C. P., Lee C. L., Chen P. H., Wu S. H., Yang C. C., Shaw J. F. (2004). Molecular analyses of the Arabidopsis TUBBY-like protein gene family. Plant Physiol. 134 1586–1597. 10.1104/pp.103.037820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Lescot M., Déhais P., Thijs G., Marchal K., Moreau Y., Van de Peer Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. 10.1093/nar/30.1.325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Cui X., Meng Z., Huang X., Xie Q., Wu H., et al. (2012). Transcriptional regulation of Arabidopsis MIR168a and argonaute1 homeostasis in abscisic acid and abiotic stress responses. Plant Physiol. 158 1279–1292. 10.1104/pp.111.188789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wu B., Yu Y., Yang G., Wu C., Zheng C. (2011). Genome-wide analysis of the RING finger gene family in apple. Mol. Genet. Genomics 286 81–94. 10.1007/s00438-011-0625-0 [DOI] [PubMed] [Google Scholar]
- Liu Q. (2008). Identification of rice TUBBY-like genes and their evolution. FEBS J. 275 163–171. 10.1111/j.1742-4658.2007.06186.x [DOI] [PubMed] [Google Scholar]
- Liu R. H., Meng J. L. (2003). MapDraw: a microsoft excel macro for drawing genetic linkage maps based on given genetic linkage data. Yi Chuan 25 317–321. [PubMed] [Google Scholar]
- Maestrini P., Cavallini A., Rizzo M., Giordani T., Bernardi R., Durante M., et al. (2009). Isolation and expression analysis of low temperature-induced genes in white poplar (Populus alba). J. Plant Physiol. 166 1544–1556. 10.1016/j.jplph.2009.03.014 [DOI] [PubMed] [Google Scholar]
- Maldonado-Calderon M. T., Sepulveda-Garcia E., Rocha-Sosa M. (2012). Characterization of novel F-box proteins in plants induced by biotic and abiotic stress. Plant Sci. 18 208–217. 10.1016/j.plantsci.2011.10.013 [DOI] [PubMed] [Google Scholar]
- Mitchell A., Chang H. Y., Daugherty L., Fraser M., Hunter S., Lopez R., et al. (2015). The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 43 D213–D221. 10.1093/nar/gku1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M., Lundegaard C., Lund O., Petersen T. N. (2010). CPHmodels-3.0–remote homology modeling using structure-guided sequence profiles. Nucleic Acids Res. 38 W576–W581. 10.1093/nar/gkq535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noben-Trauth K., Naggert J. K., North M. A., Nishina P. M. (1996). A candidate gene for the mouse mutation tubby. Nature 380 534–538. 10.1038/380534a0 [DOI] [PubMed] [Google Scholar]
- North M. A., Naggert J. K., Yan Y., Noben-Trauth K., Nishina P. M. (1997). Molecular characterization of TUB, TULP1, and TULP2, members of the novel tubby gene family and their possible relation to ocular diseases. Proc. Natl. Acad. Sci. U.S.A. 94 3128–3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitz M. U., Bissue J. K., Zocher K., Attard A., Huckelhoven R., Becker K., et al. (2012). The subcellular localization of tubby-like proteins and participation in stress signaling and root colonization by the mutualist Piriformospora indica. Plant Physiol. 160 349–364. 10.1104/pp.112.201319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitz M. U., Pai S., Imani J., Schafer P. (2013). New insights into the subcellular localization of Tubby-like proteins and their participation in the Arabidopsis-Piriformospora indica interaction. Plant Signal. Behav. 8:E25198 10.4161/psb.25198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronshaugen M., McGinnis N., Inglis D., Chou D., Zhao J., McGinnis W. (2002). Structure and expression patterns of Drosophila TULP and TUSP, members of the tubby-like gene family. Mech. Dev. 117 209–215. 10.1016/S0925-4773(02)00211-3 [DOI] [PubMed] [Google Scholar]
- Rushton P. J., Somssich I. E. (1998). Transcriptional control of plant genes responsive to pathogens. Curr. Opin. Plant Biol. 1 311–315. 10.1016/1369-5266(88)80052-9 [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. [DOI] [PubMed] [Google Scholar]
- Santagata S., Boggon T. J., Baird C. L., Gomez C. A., Zhao J., Shan W. S., et al. (2001). G-protein signaling through tubby proteins. Science 292 2041–2050. 10.1126/science.1061233 [DOI] [PubMed] [Google Scholar]
- Schenk P. M., Kazan K., Wilson I., Anderson J. P., Richmond T., Somerville S. C., et al. (2000). Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. U.S.A. 97 11655–11660. 10.1073/pnas.97.21.11655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M., Narusaka M., Abe H., Kasuga M., Yamaguchi-Shinozaki K., Carninci P., et al. (2001). Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13 61–72. 10.1105/tpc.13.1.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K., Foley R. C., Onate-Sanchez L. (2002). Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 5 430–436. 10.1016/S1369-5266(02)00289-3 [DOI] [PubMed] [Google Scholar]
- Stone S. L., Callis J. (2007). Ubiquitin ligases mediate growth and development by promoting protein death. Curr. Opin. Plant Biol. 10 624–632. 10.1016/j.pbi.2007.07.010 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco R., Zharkikh A., Affourtit J., Dhingra A., Cestaro A., Kalyanaraman A., et al. (2010). The genome of the domesticated apple (Malus x domestica Borkh.). Nat. Genet. 42 833–839. 10.1038/ng.654 [DOI] [PubMed] [Google Scholar]
- Wardhan V., Jahan K., Gupta S., Chennareddy S., Datta A., Chakraborty S., et al. (2012). Overexpression of CaTLP1, a putative transcription factor in chickpea (Cicer arietinum L.), promotes stress tolerance. Plant Mol. Biol. 79 479–493. 10.1007/s11103-012-9925-y [DOI] [PubMed] [Google Scholar]
- Wilkins M. R., Gasteiger E., Bairoch A., Sanchez J. C., Williams K. L., Appel R. D., et al. (1999). Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112 531–552. [DOI] [PubMed] [Google Scholar]
- Xiong L., Schumaker K. S., Zhu J. K. (2002). Cell signaling during cold, drought, and salt stress. Plant Cell 14 S165–S183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y. S., Chen X. Y., Yang K., Sun Z. X., Fu Y. P., Zhang Y. M., et al. (2011). Overexpression of an F-box protein gene reduces abiotic stress tolerance and promotes root growth in rice. Mol. Plant 4 190–197. 10.1093/mp/ssq066 [DOI] [PubMed] [Google Scholar]
- Yang Z., Zhou Y., Wang X., Gu S., Yu J., Liang G., et al. (2008). Genomewide comparative phylogenetic and molecular evolutionary analysis of tubby-like protein family in Arabidopsis, rice, and poplar. Genomics 92 246–253. 10.1016/j.ygeno.2008.06.001 [DOI] [PubMed] [Google Scholar]
- Yulong C., Wei D., Baoming S., Yang Z., Qing M. (2016). Genome-wide identification and comparative analysis of the TUBBY-like protein gene family in maize. Genes Genom. 38 25–36. 10.1007/s13258-015-0338-6 [DOI] [Google Scholar]
- Zhang Y., Chen C., Jin X. F., Xiong A. S., Peng R. H., Hong Y. H., et al. (2009). Expression of a rice DREB1 gene, OsDREB1D, enhances cold and high-salt tolerance in transgenic Arabidopsis. BMB Rep. 42 486–492. [DOI] [PubMed] [Google Scholar]
- Zhao T., Liang D., Wang P., Liu J., Ma F. (2012). Genome-wide analysis and expression profiling of the DREB transcription factor gene family in Malus under abiotic stress. Mol. Genet. Genomics 287 423–436. 10.1007/s00438-012-0687-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multiple alignments of the full-length proteins sequence of all plant TLPs. The black asterisk over one amino acid indicates the start of the tubby domain. The locations of F-box domain is indicated with single solid lines in black above the sequences. The segment of core α helix in tubby domain was enclosed with a red rectangle.
Locations of MdTLPs on apple chromosomes.
Intron patterns of TLPs from apple and Arabidopsis.
Expression of the DREB genes (MDP0000147009 and MDP0000218344) in leaves (A) and roots (B) under different treatments. Data were normalized to the expression level of actin gene. Vertical bars indicate the standard error of the mean. **P ≤ 0.01 and *P ≤ 0.05 compared with 0 h.
Sequences of the primers used in the quantitative real-time PCR analysis.
The BLAST search result of each MdTLP against apple EST assemblies.
Cis-acting element analysis of MdTLPs using the PLACE database.


