Abstract
The present data are related to the article entitled “Insights into ligand stimulation effects on gastro-intestinal stromal tumors signaling” (C. Bahlawane, M. Schmitz, E. Letellier, K. Arumugam, N. Nicot, P.V. Nazarov, S. Haan, 2016) [1]. Constitutive and ligand-derived signaling pathways mediated by KIT and PDGFRA mutated proteins found in gastrointestinal stromal tumors (GIST) were compared. Expression of mutant proteins was induced by doxycycline in an isogenic background (Hek293 cells). Kit was identified by FACS at the cell surface and found to be quickly degraded or internalized upon SCF stimulation for both Kit Wild type and Kit mutant counterparts. Investigation of the main activated pathways in GIST unraveled a new feature specific for oncogenic KIT mutants, namely their ability to be further activated by Kit ligand, the stem cell factor (scf). We were also able to identify the MAPK pathway as the most prominent target for a common inhibition of PDGFRA and KIT oncogenic signaling. Western blotting and micro-array analysis were applied to analyze the capacities of the mutant to induce an effective STATs response. Among all Kit mutants, only Kit Ex11 deletion mutant was able to elicit an effective STATs response whereas all PDGFRA were able to do so.
Keywords: c-KIT, PDGFRα, MAPK, PI3K, Gastro-intestinal stromal tumours, PD0325901, Stem cell factor
Specification Table
| Subject area | Cancer Research |
| More specific subject area | Signal transduction AND Receptor Tyrosine Kinases |
| Type of data | Western blot; qPCR, FACS, micro-array, Computational modeling |
| How data was acquired | Fusion-FX7 chemiluminescence detection device (Vilber) for Western blotting, |
| CLARIOstar microplate reader (BMG LABTECH) for fluorescence measurements of cell viability | |
| GeneChip Human Gene ST 2.0 arrays (Affymetrix) for micro-array | |
| FACS CantoII Instrument (Becton Dickinson, Heidelberg, Germany) for flow cytometry | |
| Model building and refinement with CHARMM | |
| Experimental features | KIT and PDGFRA mutants were expressed in hek293 cell lines upon doxycycline addition |
| Experiments were performed with/without ligand induction | |
| Data source location | LIH, Luxembourg and university of Luxembourg, Luxembourg |
| Data accessibility | Data are available in the article, and at ArrayExpress E-MTAB-4548 |
Value of the data
-
•
Expression of different mutant proteins (Kit and PDGFRA) in an isogenic background to allow a direct comparison of their signalling capacities, without the complex patient specific- background. The constructs and/or the Hek293 cell lines, could be used for further molecular characterization, protein-protein interaction experiments or protein localization studies. Identified mutations in GIST could easily be investigated by insertion of the newly discovered mutation in the WT constructs by site-directed mutagenesis. Plasmids are available upon request.
-
•
Identification of a new feature specific to Kit mutants: their ability to be further stimulated by their natural ligand, in addition to their constitutive activation derived from the mutations observed in GIST.
-
•
A new MEK inhibitor (PD0325901) was identified to be efficient in inhibiting GIST cell proliferation in the nanomolar range.
1. Data
The data presented here derived mainly from western blot analysis for the quantification of signaling pathways activated by KIT and PDGFRA mutants, from micro-array analysis for quantification of changes in gene expression levels induced by the different mutations and from flow cytometry analysis for the detection of KIT at the cell surface.
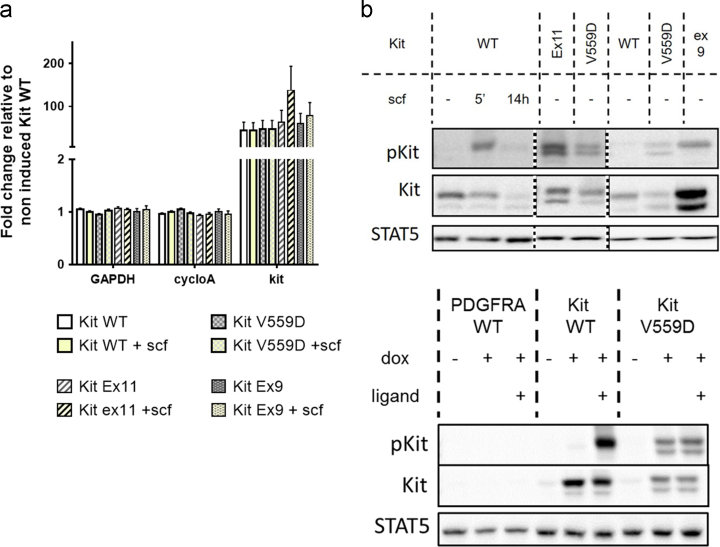
Doxycycline induces Kit expression by a factor 100 in all cell lines and comparable mRNA expression levels were observed for all mutants (Fig. 1a). However, some differences in the protein expression levels were observed between the different constructs (Fig. 1b).
Fig. 1.
KIT RNA and protein expression levels in the stable transfected cells. (a) KIT mRNA expression level as assessed by qPCR in Hek293 cells expressing KIT WT, KIT Ex11, KIT Ex9 and KIT V559D 14 h after induction with doxycycline (5 ng/ml). SCF was added at 100 ng/ml for the time of induction. Data represent the means of 3–6 biological replicates and are normalized using Genorm following the MIQE guidelines [2]. (b) Western blot analysis indicating KIT expression level as well as phosphorylation status in Hek293 cells expressing KIT WT, KIT Ex11, KIT Ex9 and KIT V559D 14 h after induction with doxycycline (5 ng/ml). SCF was added at 100 ng/ml for the time of induction or 5 min before cell harvesting as indicated on the figure. Representative data of 3 biological replicates. STAT5 is used as loading control.
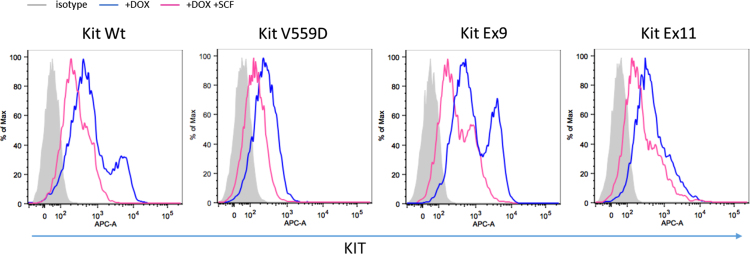
The ratio of surface (Fig. 1a in [1]) to total KIT expression (Fig. 1b in [1]) indicates that KIT WT is expressed almost exclusively at the surface, while this is the case for 70% of KIT Ex9. This value drops to 50% for both Ex11 mutants. The results of the FACS analysis after SCF stimulation (Fig. 2) indicate the decrease of KIT expression at the cell surface for all KIT mutants and wild type following stimulation with KIT ligand, SCF.
Fig. 2.
Effect of ligand stimulation on KIT expression at the cell surface.
The Mean Fluorescence Intensity (MFI) of Kit staining in the different Kit mutants are indicated in Table 1.
Table 1.
Mean Fluorescence Intensity (MFI) of Kit staining in the different Kit mutants.
| MFI | Kit WT | Kit V559D | Kit Ex9 | Kit ex11 | ||||
|---|---|---|---|---|---|---|---|---|
| −SCF | +SCF | −SCF | +SCF | −SCF | +SCF | −SCF | +SCF | |
| Surface | 2099 | 111 | 172 | 11 | 1435 | 284 | 562 | 222 |
| Overall expression | 2122 | 642 | 374 | 319 | 2106 | 1355 | 1008 | 852 |
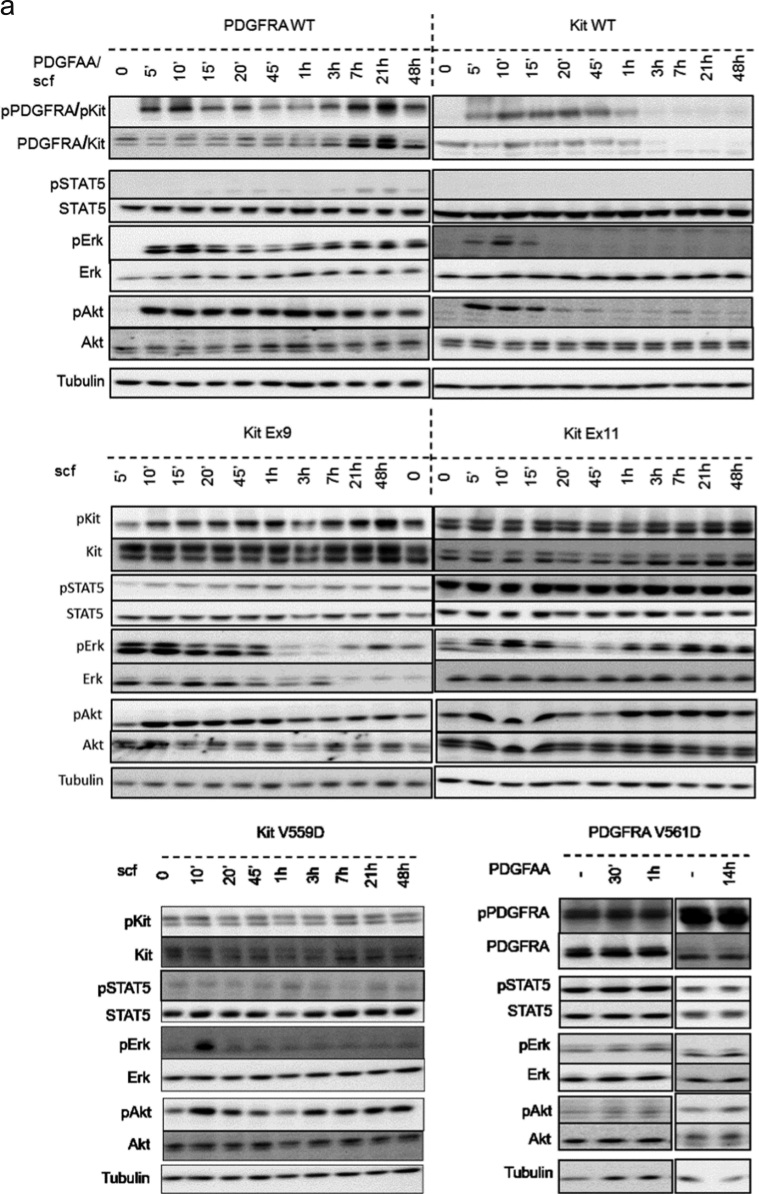
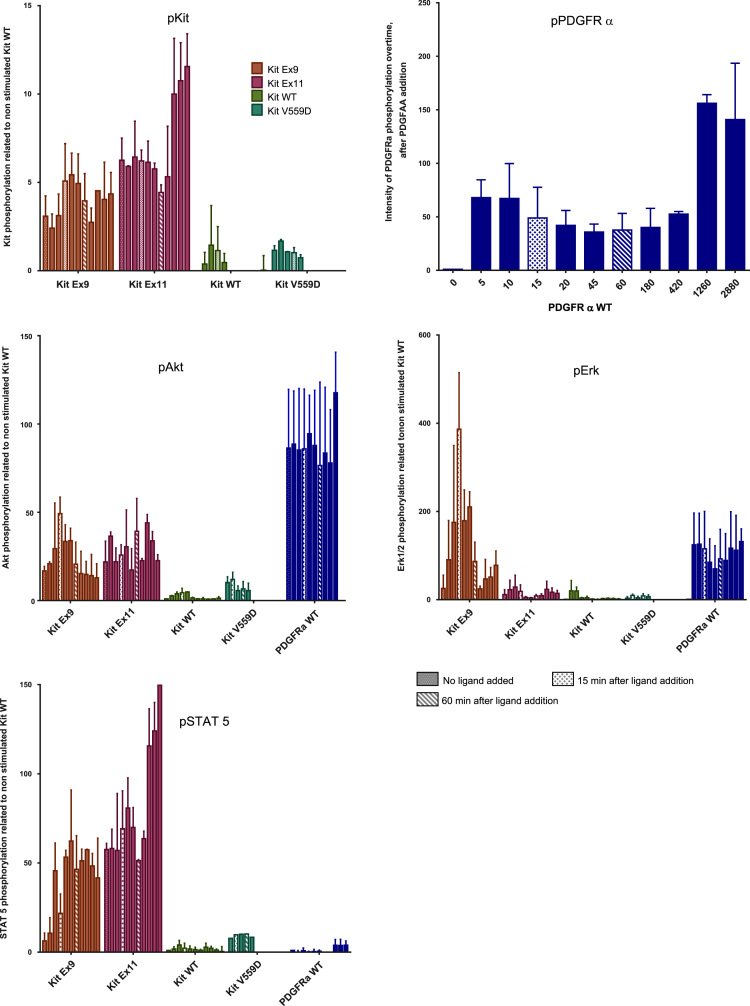
While PDGFRα, Akt, and Erk phosphorylation were induced by PDGFAA in PDGFRaWT, phosphorylation of PDGFRα, Akt, Erk and STAT5 remained identical to the non-stimulated control for the PDGFRA mutant V591D, as previously shown [3]. In contrast, Kit mutants exhibit all constitutive phosphorylation of kit at tyrosine 703 but the signal intensities for Erk, Akt and STAT5 phosphorylation was further increased upon SCF stimulation (Fig. 3a and Fig. 3b).
Fig. 3.
Effect of ligand stimulation on downstream signaling in GIST mutants. (a) Western blot analysis indicating the phosphorylation status of KIT/PDGFRα, STAT5, Akt and Erk in Hek293 cells expressing PDGFRα WT, PDGFRα V561D, KIT WT, KIT Ex11, KIT Ex9 and KIT V559D after PDGFAA or SCF addition. Representative blots of 3 biological replicates. Tubulin is used as loading control. (b) Quantification of the signal intensities from the western blots shown in a. Data were calibrated using the sample “KIT WT non-stimulated” (background level), except for PDGFRA phosphorylation where PDGFRa WT non-stimulated was used. Each dot represents the mean of biological triplicates and the error bars the standard error of the mean. From left to right, bars represent the signal intensities after ligand addition (1st bars correspond to no ligand, 15 and 60 min after ligand addition are marked for better visibility).
STATs translocation to the nucleus was identified for Kit Ex11 deletion mutant and for Kit Ex9 duplication mutant to a lower extend. Induction of gene expression known to be part of the STAT pathway was found for Kit Ex11 deletion mutant after SCF stimulation as well as for Kit Ex9 duplication mutant upon SCF stimulation to a lower extend (Fig. 4a and Fig. 4b).
Fig. 4.
Activation of STAT species by GIST mutants. (a) Nuclear translocation of STAT species. Nuclear extracts were prepared as previously done [4], diluted in 4 times Laemmli buffer and subjected to Western blot analysis. Phosphorylation status of STAT5, STAT3 and STAT1 is shown for nuclear extract prepared from Hek293 cells expressing KIT WT, KIT Ex11, KIT Ex9 and KIT V559D. (b) Induction of known STAT target genes by GIST mutants. Gene expression level of known STAT target genes, previously identified to be induced in PDGFRA GIST mutants [3], was retrieved from micro-array data and presented as heat map. Grey boxes indicate that the genes are not part of the DEG list for the corresponding mutant (FDR>0.05 or AbsFC<0.5). The intensity of the red color corresponds to the value of the ratio to the background (Hek293 KIT WT non-stimulated for KIT WT and mutants and Hek293 PDGFRα non-stimulated for PDGFRα mutants). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article) .
We investigated KIT Ex11 specific gene signature, comparing KIT Exon 11 deletion mutant regulated genes to other GIST mutants (PDGFRA mutants and KIT Exon 9). 277 genes (Table 3) were differentially expressed in KIT Ex11 deletion mutant only. As noted in [1], these genes were associated with “cell cycle” and “insulin signalling pathway”.
Table 3.
KIT Ex11 deletion mutant specific DEG, with FDR<0.05 and absolute logFC>0.5.
| ACER2 | CCNYL1 | EYA1 | IRX4 | LOC401321 | NKX3-1 | RN5S217 | SNORA10 | UTRN |
| ADAMTS20 | CD109 | FAHD1 | ITGA2 | LOC642838 | NLRP1 | RN5S335 | SNORA16B | VRK3 |
| ADCK3 | CD3EAP | FAM46A | JAM2 | LOC645166 | NOA1 | RND2 | SNORA2A | WDR63 |
| ADCY1 | CD68 | FAM47A | JMY | LPAL2 | NR1D2 | RNF167 | SNORD111 | WDR77 |
| AKAP8 | CDC25B | FAM59A | KCNA3 | LPCAT2 | NRN1 | RNFT1 | SNORD126 | WWP2 |
| ALDH1L2 | CDT1 | FAM64A | KDELR1 | LPHN3 | OLIG2 | RNU2-7P | SNORD3C | ZC3H6 |
| ALG12 | CES3 | FAM83B | KDELR3 | LPPR4 | PABPC1L | RNU7-6P | SNORD60 | ZFP14 |
| AMIGO2 | CHEK2 | FBXO32 | KIAA0355 | LRFN1 | PAMR1 | RPL13P5 | SNORD71 | ZNF140 |
| ANKRD18DP | CITED1 | FLJ44342 | KIAA1430 | LURAP1L | PAQR4 | RYR2 | SNORD91A | ZNF17 |
| ANKRD20A12P | CKMT1A | FLJ45248 | KIAA1609 | LYSMD2 | PARK2 | SAMD15 | SRCRB4D | ZNF239 |
| ANKRD20A5P | CNN2 | FRAT2 | KLF11 | MALAT1 | PARM1 | SCARA5 | TAS2R31 | ZNF280B |
| ANKRD27 | CNTNAP3B | GALNT6 | KLHL24 | MCL1 | PARP14 | SCARNA21 | TBX15 | ZNF296 |
| ARHGAP20 | CPEB4 | GGA2 | KLHL36 | MCM5 | PARP4 | SEMA6A | TCF25 | ZNF347 |
| ARHGAP35 | CPPED1 | GLTSCR2 | KLRAP1 | MDM2 | PBX1 | SERPINB1 | TFAP4 | ZNF485 |
| ARHGEF1 | CRKL | GRAMD4 | LCMT1 | MED12L | PCDH7 | SGK1 | THOC6 | ZNF502 |
| ARNT2 | CRYBB2P1 | GSPT2 | LIG4 | MGARP | PDP1 | SGSM3 | TIGD4 | ZNF574 |
| ASRGL1 | CSDA | GYPC | LIN37 | MGST1 | PDPR | SHKBP1 | TMEM143 | ZNF581 |
| BACE1-AS | CYP2S1 | GYS1 | LINC00282 | MIR22HG | PHKB | SHMT2 | TMEM159 | ZNF70 |
| BBIP1 | CYP4×1 | H2BFXP | LNP1 | MIR3143 | PKIA | SLC35B2 | TMEM185B | ZNF738 |
| BCL6 | DDTL | HIST1H2BG | LOC100130776 | MIR338 | PLA2G7 | SLC44A2 | TMEM238 | ZNF836 |
| BEX5 | DEDD2 | HIST1H3A | LOC100132439 | MIR3671 | PLXNA2 | SLC45A3 | TNFRSF10D | ZSCAN12P1 |
| BRD2-IT1 | DLC1 | HIST1H3H | LOC100133985 | MIR4263 | PMFBP1 | SLC6A6 | TNFRSF13C | |
| BZW1 | DMRTA1 | HLA-DRB5 | LOC100272228 | MIR4324 | PPP1R13L | SLC7A6OS | TNRC6B | |
| C10orf10 | DYNLL2 | HSDL1 | LOC100287628 | MIR4530 | PPP1R14C | SLC8A1 | TOM1 | |
| C10orf25 | EBF3 | IER5 | LOC100288018 | MIR4773-2 | PROX1 | SLC9A1 | TP53I13 | |
| C19orf54 | ECH1 | IFITM3 | LOC100288520 | MIR548A2 | PRPH | SLC9A2 | TRBV23OR9-2 | |
| C22orf13 | EID3 | IGHD2-21 | LOC100507299 | MRI1 | PRR12 | SLC9A9 | TRBV6-9 | |
| C2orf77 | EIF1 | IL11 | LOC147670 | MTRF1L | PSEN2 | SMOC1 | TRPC1 | |
| CABIN1 | ELAC1 | ILDR2 | LOC147727 | MTSS1 | RAB39B | SMPD1 | TRPS1 | |
| CABLES1 | EMR2 | IMPA2 | LOC284648 | NFIX | RBL2 | SNAR-D | TSC2 | |
| CAMLG | EPHX4 | INPP5D | LOC399815 | NIPSNAP1 | RHBDF1 | SNAR-H | TSSK3 | |
| CAP2 | EPS15L1 | IQCH | LOC400927 | NIPSNAP3A | RN5S180 | SNN | TXNDC17 |
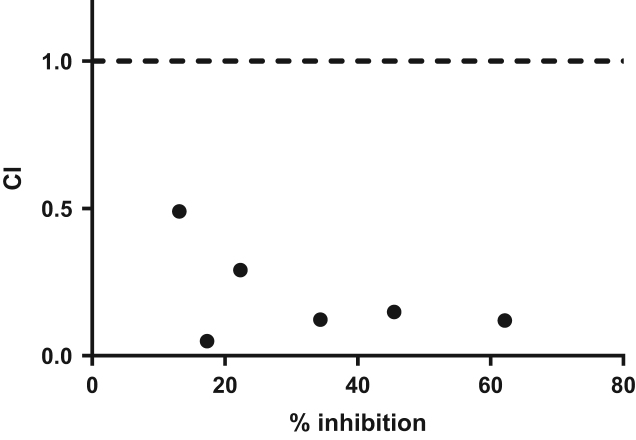
Both inhibitors were added to the medium 24 h after seeding for 30 h. Both drugs were added individually and in combination with a constant ratio of 1:1 (Fig. 5). Viability was assessed using PrestoBlue following the manufacturer׳s recommendation. Results were analysed using the Compusyn software [5] and the combination Index (CI) values are represented as a function of the percentage of inhibition. CI values below 1 indicates a synergy between the two compounds, while CI values above 1 indicates antagonism.
Fig. 5.
Synergistic assessment for PD0325901 and XL184 inhibitors in GIST882.
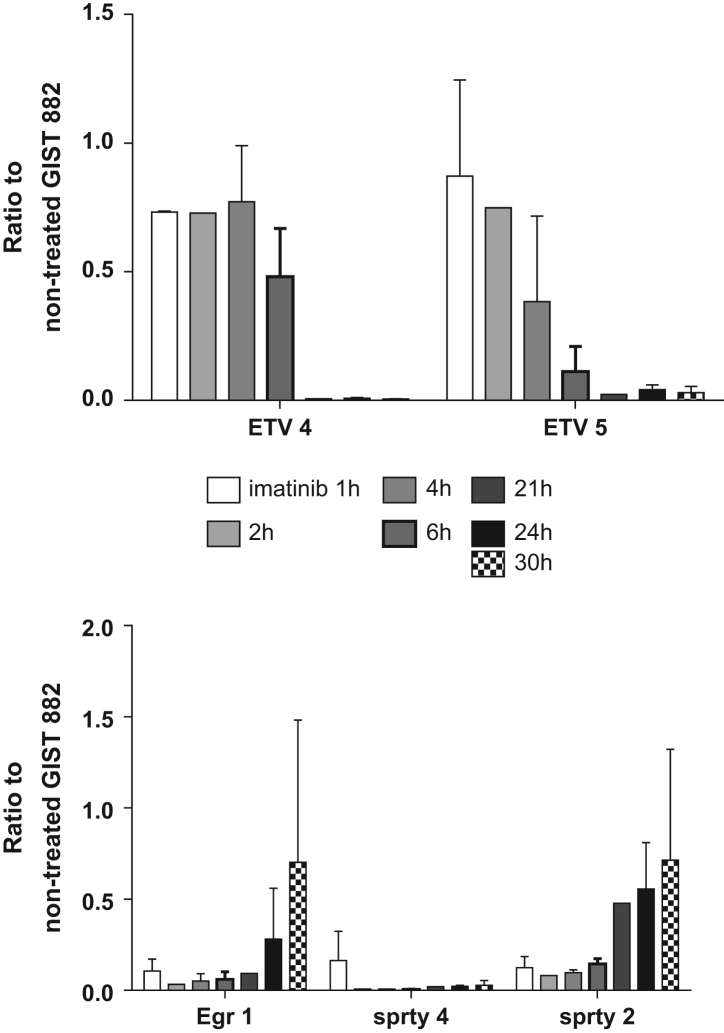
GIST882 were treated with 100 nM imatinib for different times and mRNA expression level of ETV4, ETV5, egr1, FosL2 and FosB were assessed by qPCR. n= 3, Mean±SEM (Fig. 1, Fig. 2, Fig. 3, Fig. 4, Fig. 5, Fig. 6).
Fig. 6.
Imatinib effect on MAPK gene expression level over time.
qPCR primers are listed in Table 2.
Table 2.
primers used for qPCR analysis.
| Targeted gene | Primers sequences |
| GAPDH | gtccttccacgataccaaagt |
| atgagaagtatgacaacagcct | |
| HPRT | tggacaggactgaacgtctt |
| gagcacacagagggctacaa | |
| PPIA/cycloA | cagacaaggtcccaaagaca |
| ccattatggcgtgtgaagtc | |
| Tubulin | agatcggtgccaagttctg |
| ccacctgtggcttcattgta | |
| ETV4 | gcccctcgactctgaagat |
| tggaaatcaggaacaaactgc | |
| ETV5 | atccccgattatactttgacg |
| agaagggtgaccaggaactg | |
| Kit | acaaagagcaaatccatccc |
| tgtaggtcagaatcatcacaataat | |
| egr1 | agtggtttggctggggtaa |
| ctacgagcacctgaccgc | |
| sprty2 | ttgcacatcgcagaaagaag |
| ggtcactccagcaggcttag | |
| sprty4 | gggagccactgagaacagag |
| tggctcctaaatccatcctg |
2. Experimental design, materials and methods
2.1. Flow cytometry analysis
Cell surface expression of KIT wild type and mutants was analyzed by flow cytometry using a FACS CantoII Instrument (Becton Dickinson, Heidelberg, Germany) either without ligand (blue lines) or 15 minutesmin after stimulation with 100ng/ml100 ng/ml SCF (pink lines). Cells were then either incubated with 10 μL KIT primary antibody (anti-CD117-APC conjugated; C7244; Dako, Belgium) for cell surface expression. Specificity was controlled using an isotype-matched/ APC conjugated antibody (grey).
2.2. Western blot analysis
Cell lysis was performed on ice, using 1x Laemmli buffer. Proteins were subjected to SDS-PAGE, transferred to polyvinylidene difluoride membranes (Roth) and probed with primary antibodies. Primary antibodies against PLCγ and phosphospecific antibodies against STAT1 (pTyr701), STAT3 (pTyr705), ERK1/2 (pThr202/pTyr204), PDGFRA (pTyr849)/β(pTyr857), AKT (pSer473) and Mek1/2 were purchased from Cell Signalling Technology. Anti-STAT1 and anti-STAT3 antibodies and phosphospecific antibodies for STAT5 (pTyr694) and PLCγ1 (pTyr783) were obtained from BD Biosciences. Antibodies against STAT5 (C-17), PDGFRα (C-20), ERK1 (C-16), ERK2 (c-14), AKT1/2 (N-19) and tubulin (DM1A) were bought from Santa Cruz. Anti-CD117 (KIT) antibody was obtained from Dako and horseradish peroxidase-conjugated secondary antibodies from Cell Signalling Technology. Signals were detected on a Fusion-FX7 chemiluminescence detection device (Vilber) using a home-made ECL (Enhanced ChemiLuminescence) solution containing 2.5 mM luminol, 2.6 mM hydrogenperoxide, 100 mM Tris/HCl pH 8.8 and 0.2 mM para-coumaric acid [6]. Signal intensities were quantified using the Bio1D analysis package (Vilber).
2.3. Microarray analysis
293FR cells expressing KIT-WT, KIT Ex11 deletion mutant and KIT Ex9 mutant were incubated with 5 ng/ml doxycycline and 100 ng/ml SCF for 21 h in DMEM with 1%FCS. Cells were then starved for 3 h (without FCS) and further stimulated with SCF. RNAs of three biological replicates were isolated using the miRNeasy Mini KIT (Qiagen) according to manufacturer׳s instructions with additional on-column DNase I digestion. RNA quality and purity were assessed using a Nanodrop Spectrophotometer (Thermo Scientific) and Agilent 2100 Bioanalyzer (Nano KIT). Gene Expression analysis was performed using GeneChip Human Gene ST 2.0 arrays (Affymetrix). Quality control and data normalization were performed as previously reported [3], [7]. We focused on differentially expressed genes (DEG) across the mutants comparing with non-stimulated KIT-WT. To exclude non-relevant lowly expressed transcript clusters, only those showing log2 expression above 4.5 were kept for further analysis. Transcript clusters were further summarized in order to obtain a single expression value for each gene in each experiment. The differentially expressed genes were statistically evaluated by two-factor linear model with empirical Bayes statistics approach using limma package of R/Bioconductor [8]. In order to correct for the false discovery rate (FDR), the Benjamini and Hochberg step-up method correction was applied. Probe-sets with FDR <0.05 and absolute fold change >0.5 were considered to be significantly differentially expressed (DEGs). Microarray data are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-4548. KIT Ex11 specific gene signature was determined by comparing KIT Exon 11 deletion mutant regulated genes to other GIST mutants (PDGFRA mutants and KIT Exon 9). 277 genes.
2.4. Nuclear extract preparation
Nuclear extracts were prepared as previously done [4]. In brief, cells were washed with ice-cold PBS, harvested gently with cell scraper and centrifuged at 4 °C for 5 min, 4000 rpm. The pellet was then resuspend in Buffer A (10 mM Hepes/KOH pH7.9, 1.5 mM MgCl2 and 10 mM KCl). Following 10 min incubation on ice, samples were centrifuged at maximum speed, 4 °C for 5 min. The operation was performed a second time and the pellets were finally resuspended in Buffer C for nuclear extraction (20 mM Hepes/KOH pH7.9, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA and 25% glycerol). Protein concentrations were determined using Bradford reagent before analysis by western blotting.
2.5. Synergy analysis
Synergy between Receptor Tyrosine kinase (RTK) and MEK inhibitors were performed at constant ratio, as recommended by Chou et al. [5], [9]. GIST primary cells were seeded in 96 well plates 24 h prior treatement. Inhibitors were added at concentration between 5 nM and 13 μM, either alone or in combination to a final volume of 90 μL. Endpoint viability was assessed using PrestoBlue (Thermofischer), by adding 10 μL of reagent to each well. Following 30 min incubation, fluorescence intensities were recorded on a CLARIOstar microplate reader (BMG LABTECH). Inhibition was then calculated as a ratio to the non-treated samples.
Acknowledgements
This work was supported by the grants F1R-LSC-PUL-09PDGF and F1R-LSC-PUL-11PDGF of the University of Luxembourg. The authors thank the Fondation Cancer for supporting this study via the grant F1R-LSC-PAU-13PLAT. We thank Sebastien Plançon for advice regarding the flow cytometry experiments and Demetra Philippidou for the preparation of Mel501 cDNAs.
Footnotes
Transparency data associated with this article can be found in the online version at 10.1016/j.dib.2016.10.026.
Transparency document. Supplementary material
Supplementary material
References
- 1.Bahlawane C., Schmitz M., Letellier E., Arumugam K., Nicot N., Nazarov P.V., Haan S. Insights into ligand stimulation effects on gastro-intestinal stromal tumors signalling. Cell. Signal. 2017;29 doi: 10.1016/j.cellsig.2016.10.009. 138–149. [DOI] [PubMed] [Google Scholar]
- 2.Bustin S.A., Beaulieu J.-F., Huggett J., Jaggi R., Kibenge F.S.B., Olsvik P.A., Penning L.C., Toegel S. MIQE precis. Practical implementation of minimum standard guidelines for fluorescence-based quantitative real-time PCR experiments. BMC Mol. Biol. 2010:11. doi: 10.1186/1471-2199-11-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bahlawane C., Eulenfeld R., Wiesinger M.Y., Wang J., Muller A., Girod A., Nazarov P.V., Felsch K., Vallar L., Sauter T. Constitutive activation of oncogenic PDGFR alpha-mutant proteins occurring in GIST patients induces receptor mislocalisation and alters PDGFR alpha signalling characteristics. Cell Commun. Signal. 2015;13:21. doi: 10.1186/s12964-015-0096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haan S., Haan C. Detection of activated STAT species using electrophoretic mobility shift assay (EMSA) and potential pitfalls arising from the use of detergents. Methods Mol. Biol. (Clifton, NJ) 2013;967:147–159. doi: 10.1007/978-1-62703-242-1_10. [DOI] [PubMed] [Google Scholar]
- 5.Chou T.-C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70(2):440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 6.Haan C., Behrmann I. A cost effective non-commercial ECL-solution for Western blot detections yielding strong signals and low background. J. Immunol. Methods. 2007;318(1-2):11–19. doi: 10.1016/j.jim.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 7.Haan S., Bahlawane C., Wang J., Nazarov P.V., Muller A., Eulenfeld R., Haan C., Rolvering C., Vallar L., Satagopam V.P. The oncogenic FIP1L1-PDGFRalpha fusion protein displays skewed signaling properties compared to its wild-type PDGFRalpha counterpart. Jak.-Stat. 2015;4(1) doi: 10.1080/21623996.2015.1062596. (e1062596-e1062596) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7) doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ashton J.C. Drug combination studies and their synergy quantification using the Chou-Talalay method-letter. Cancer Res. 2015;75(11) doi: 10.1158/0008-5472.CAN-14-3763. (2400-2400) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material