Abstract
We report the main characteristics of “Corynebacterium urinapleomorphum” strain Marseille-P2799T (CSURP2799), isolated from a urine sample from a 2-month-old boy with rotavirus gastroenteritis.
Keywords: “Corynebacterium urinapleomorphum”, culturomics, genomics, taxonogenomics, taxonomy
In 2016, as a part of culturomics study [1], [2] of the human microbiome, a bacterial strain that could not be identified by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) screening using a Microflex spectrometer (Bruker Daltonics, Bremen, Germany) [3] was isolated from the urine of a 2-month-old boy with rotavirus gastroenteritis. The patient's parents provided signed informed consent, and the ethics committee of the Institut Fédératif de Recherche IFR48 approved the study under number 09-022.
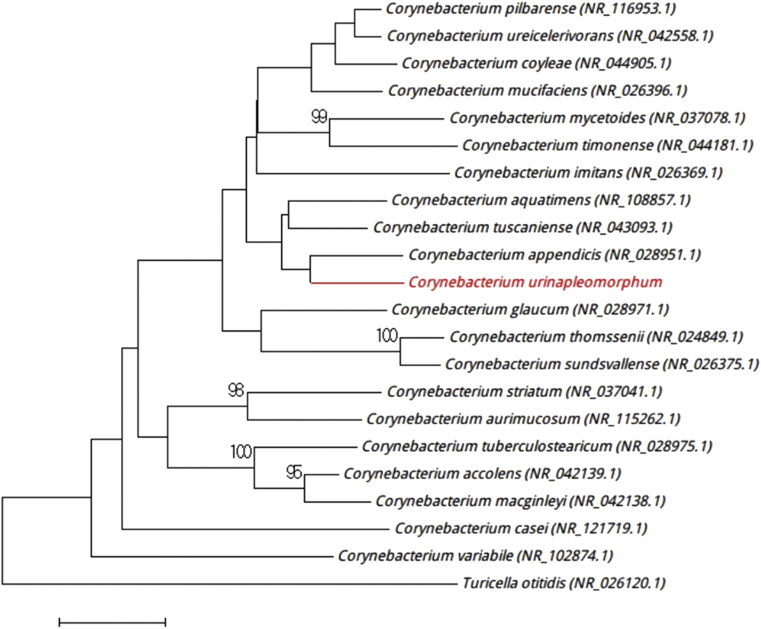
A pure culture of strain Marseille-P2799 was obtained after 72 hours of incubation at 37°C on 5% sheep's blood–antioxidant agar homemade R-medium (Hôpital de la Timone, Marseille, France) in anaerobic atmosphere generated using the GENbag anaer system (bioMérieux, Marcy l'Étoile, France). Agar-grown colonies were pale grey and had a mean diameter of 500 μm. Bacterial cells were nonmotile, Gram-positive, pleomorphic bacilli with a length ranging from 700 to 2000 nm and width ranging from 400 to 600 nm. Strain Marseille-P2799 exhibited catalase activity but no oxydase activity. The 16S rRNA gene was sequenced using the fD1-rP2 primers as previously described [4], using a 3130-XL sequencer (Applied Biosciences, Saint Aubin, France). Strain Marseille-P2799 exhibited 98% sequence similarities to Corynebacterium appendicis strain IMMIB R-3491T (GenBank accession no. AJ314919), its phylogenetically closest species with standing in nomenclature (Fig. 1) [5]. Corynebacterium appendicis strain IMMIB R-3491T was isolated from an abdominal swab of a patient with appendicitis accompanied with abscess formation. Strain IMMIB R-3491T stained Gram positive and consisted of nonmotile, thin, pleomorphic, coryneform cells. On Columbia blood agar, colonies were very small, dry and slightly greyish in color. The strain IMMIB R-3491T was growing in facultative anaerobic atmosphere and was catalase positive. Because the nucleotide sequence of strain identity Marseille-P2799 was lower than the 98.63% cutoff recommended to delineate bacterial species [6], we consider strain Marseille-P2799T to be the type strain of a novel Corynebacterium species, “Corynebacterium urinapleomorphum” sp. nov. (u.ri.na.pleo.morph.um composed of u.ri.na L.N. gen. fem. urina, the Latin word for “urine,” as strain Marseille-P2799 was first found in a paediatric urine sample, and pleo.morph.um. L. neutral. adj. pleomorphum of pleo, “several” or “different,” and morph, “shape,” as cells were bacilli with cytoplasmic inclusion that could make us think that the bacterium was catenary Gram-positive cocci).
Fig. 1.
Phylogenetic tree showing position of “Corynebacterium urinapleomorphum” strain Marseille-P2799 relative to other phylogenetically close neighbours. 16S rRNA sequences were aligned using CLUSTALW and phylogenetic inferences obtained using maximum-likelihood method within MEGA software. Numbers at nodes are percentages of bootstrap values (≥95%) obtained by repeating analysis 500 times to generate majority consensus tree. GenBank accession numbers are indicated in parentheses. Scale bar indicates a 1% nucleotide sequence divergence.
The MALDI-TOF MS spectrum of “Corynebacterium urinapleomorphum” strain Marseille-P2799T is available at http://mediterranee-infection.com/article.php?laref=256&titre=urms-database.
Nucleotide sequence accession number
The 16S rRNA gene sequence was deposited in GenBank under accession number LT576404.
Deposit in a culture collection
Strain Marseille-P2799T was deposited in the Collection de Souches de l'Unité des Rickettsies (CSUR, WDCM 875) under number P2799.
Acknowledgement
This study was funded by the Fondation Méditerranée Infection.
Conflict of Interest
None declared.
References
- 1.Lagier J.C., Hugon P., Khelaifia S., Fournier P.E., La Scola B., Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28:237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hilt E.E., McKinley K., Pearce M.M., Rosenfeld A.B., Zilliox M.J., Mueller E.R. Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J Clin Microbiol. 2014;52:871–876. doi: 10.1128/JCM.02876-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seng P., Abat C., Rolain J.M., Colson P., Lagier J.C., Gouriet F. Identification of rare pathogenic bacteria in a clinical microbiology laboratory: impact of matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol. 2013;51:2182–2194. doi: 10.1128/JCM.00492-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drancourt M., Bollet C., Carlioz A., Martelin R., Gayral J.P., Raoult D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol. 2000;38:3623–3630. doi: 10.1128/jcm.38.10.3623-3630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yassin A.F., Steiner U., Ludwig W. Corynebacterium appendicis sp. nov. Int J Syst Evol Microbiol. 2002;52(Pt 4):1165–1169. doi: 10.1099/00207713-52-4-1165. [DOI] [PubMed] [Google Scholar]
- 6.Kim M., Oh H.S., Park S.C., Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64(Pt 2):346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]