Abstract
Progressive multifocal leukoencephalopathy (PML) is a debilitating disease resulting from infection of oligodendrocytes by the JC polyomavirus (JCPyV). Currently, there is no anti-viral therapeutic available against JCPyV infection. The clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein 9 (Cas9) system (CRISPR/Cas9) is a genome editing tool capable of introducing sequence specific breaks in double stranded DNA. Here we show that the CRISPR/Cas9 system can restrict the JCPyV life cycle in cultured cells. We utilized CRISPR/Cas9 to target the noncoding control region and the late gene open reading frame of the JCPyV genome. We found significant inhibition of virus replication and viral protein expression in cells recipient of Cas9 together with JCPyV-specific single-guide RNA delivered prior to or after JCPyV infection.
The JC polyomavirus (JCPyV), a member of the Polyomaviridae family, is the causative agent of a rare and debilitating demyelinating disease termed progressive multifocal leukoencephalopathy (PML) caused by infection and destruction of oligodendrocytes in the central nervous system (CNS)1,2. PML can develop in the context of immunodeficiency and treatment with immunomodulatory drugs3. Indeed, it is one of the most common CNS-related diseases in AIDS, affecting 5% of the HIV-1 positive patients. The incidence of PML has also risen to 0.2–0.4% in patients receiving immunomodulatory therapies4,5.
JCPyV infection is common in the general population infecting approximately 50% to 80% of the population6. JCPyV persists asymptomatically in the kidney and is shed in urine in about one third of the infected population7,8. The route of viral dissemination from the initial site of infection to the CNS in the context of immunosuppression remains unknown9,10,11,12, but it has been proposed to be mediated by hematopoietic cells crossing the blood brain barrier13.
JCPyV has a nonenveloped, icosahedral capsid containing a circular double-stranded DNA genome divided into three regions; the early region encoding the regulatory T antigens (small t antigen, large T antigen and T’ proteins); the late region encoding the capsid proteins; and the non-coding control region (NCCR) which contains the viral origin of DNA replication and the transcriptional promoters of the early and late genes14,15. The regulatory T antigens play critical roles in initiation of viral DNA replication and the transcription of the late genes (Agno and capsid proteins VP1, VP2 and VP3)16.
The type II bacterial clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated 9 (Cas9) system (CRISPR/Cas9) is a powerful tool for editing cellular genomes17,18,19. The Cas9 endonuclease together with a small guide RNA (gRNA) can introduce double strand breaks in targeted DNA in a sequence-specific manner20. Similar to the original function of CRISPR, a microbial nuclease system serving as a defense mechanism against invading phages and plasmids, CRISPR/Cas9 has now been employed to specifically target mammalian viral genomes, including those of human papillomaviruses (HPV), Hepatitis B virus (HBV), Epstein-Barr virus (EBV) and HIV-1 (Reviewed in refs 21 and 22). In these cases, the CRISPR/Cas9 system introduced double strand breaks in the viral DNA genome associated with mutational inactivation of viral genes resulting in the inhibition of viral protein production and viral DNA replication. The CRISPR/Cas9 system was used to target viral genomes in cells latently infected with viruses, such as EBV and HIV23,24,25 or in cells with integrated viral genomes, such as HBV, HPV or HIV provirus26,27,28,29,30,31,32,33. In these cases, the copy number of targeted viral genomes in the cells were relatively low compared to actively replicating viral DNAs23,26,27,28,29,30,31,32,33. When inhibition of de novo virus infection by the CRISRP/Cas9 system was demonstrated for HBV, virus infection was done in cells stably expressing CRISPR/Cas9 system27. Similarly, cells expressing Cas9 and gRNAs targeting the T antigen region of JCPyV genome were less permissive to JCPyV infection34. In both cases, however, CRISPR/Cas9 together with specific gRNAs were already present in the cells prior to the introduction of viral genomes; it is therefore possible that the viral genome was targeted before initiation of DNA replication thus being able to restrict viral growth with high efficiency.
Here we extended these studies and explored whether the CRISPR/Cas9 system introduced into cells after acute viral infection could also be effective to restrict virus replication. We used cells acutely infected with JCPyV and then exposed them to CRISPR/Cas9 specifically targeted to the genome of JCPyV. We identified regions in the JCPyV genome susceptible to DNA cleavage by CRISPR/Cas9 and found that when these cleavages failed to be properly repaired by the host cell, JCPyV replication was significantly reduced. This observation suggests that CRISRP/Cas9 together with gRNAs specific for the JCPyV genome can be used to inhibit JCPyV infection when introduced into cells already infected with JCPyV.
Results
Cells stably expressing JCPyV specific gRNAs are resistant to Mad1 JCPyV infection
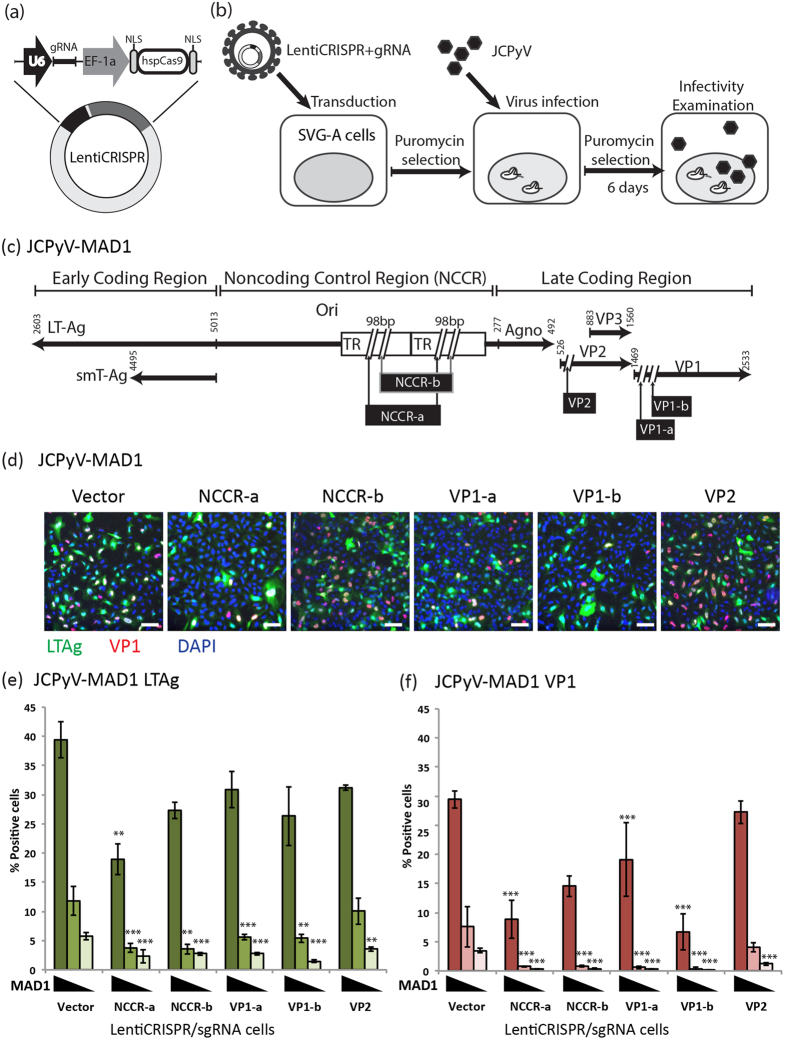
To identify regions of the JCPyV genome susceptible to targeted genome editing by CRISPR/Cas9, we designed 5 different gRNAs specific to the JCPyV genome (Table 1) within the NCCR (NCCR-a and NCCR-b) and the open reading frame encoding the capsid proteins VP1 (VP1-a and VP1-b) and VP2 (Fig. 1c), respectively. The NCCR of the rearranged JCPyV strain MAD1 contains a 98 bp tandem repeat, therefore the gRNA target sequence is present twice for both NCCR targets (NCCR-a and NCCR-b) (Fig. 1c). These regions were chosen to target the origin and regulatory region for viral gene expression contained in the NCCR region and the open reading frames encoding two structural proteins essential for successful generation of virus particles. The target sequences were selected based on highly evaluated scores for predicted specificity and low off-target potential using the online CRISPR design tool (http://www.genome-engineering.org/crispr/).
Table 1. The sequences and location of gRNA targeting JCPyV genome.
| Name | JCPyV genomic target | Sense or anti-sense | Target location |
|---|---|---|---|
| NCCR-a | CAAGCATGAGCTCATACCTA | Sense | NCCR TandemRepeat (50, 148) |
| NCCR-b | TTTACTGGCTGTTAGCTGGT | Anti-sense | NCCR TandemRepeat (78, 177) |
| VP1-a | AAACCCCTAAGATGCTCATC | Anti-sense | VP1 ORF (1622) |
| VP1-b | GAAGTTTTGGAACTTGCACG | Anti-sense | VP1 ORF (1507) |
| VP2 | GGTGCCGCACTTGCACTTTT | Sense | VP2 ORF (529) |
Numbering is according to the Mad-1 strain of JCPyV (GeneBank accession no. NC_001699).
Figure 1. Expression of Cas9 and JCPyV specific gRNA inhibited JCPyV infection.
(a) Depiction of the LentiCRISPR/gRNA plasmid design. EF-1a: Human elongation factor 1 alpha promoter, NLS: nuclear localization signal, hspCas9: human codon-optimized streptococcus pyogenes Cas9. (b) Experimental outline. SVG-A cells transduced with LentiCRISPR/gRNA virus with indicated gRNA were selected with puromycin and then infected with JCPyV-MAD1. (c) JCPyV specific gRNA target sites in JCPyV-MAD1 genome. (d) Immunofluorescence images showing the expression of LTAg (green) and VP1 (red) in SVG-A cells expressing Cas9 and the indicated gRNA at day 6 post-infection (highest virus inoculation dose in (e,f)). Scale bar = 100 μm (e) Quantification of the percentages of LTAg positive cells. (f) Quantification of the percentages of VP1 positive cells. Triangle represents the inoculation dose of JCPyV-MAD1 virus (5 × 107 genome copies/ml, 2.5 × 107 genome copies/ml, 1.25 × 107 genome copies/ml). **One-way ANOVA, p value < 0.01; ***one-way ANOVA, p value < 0.001. N = 3.
We first determined whether the JCPyV genome could be successfully targeted by CRISPR/Cas9 in SVG-A cells, first transduced with LentiCRISPR/gRNA viruses (schematically represented in Fig. 1b) and then infected with JCPyV-MAD1. Virus infectivity, indirectly estimated by using immunofluorescence staining to determined the number of cells expressing the viral proteins large T antigen (LTAg) and VP1, was used to assess if Cas9 and the JCPyV-specific gRNAs already present in the cells could target incoming JCPyV genomes and reduce virus infection (schematically shown in Fig. 1c). As expected, the number of cells positive for LTAg was dependent on the dose of input JCPyV-MAD1 in control cells stably expressing CRISPR/Cas9 without gRNA (Fig. 1e, vector: 39 ±3%, 12 ± 3% and 5% ±1% of cells). LTAg expression in cells exposed to the lowest two doses of JCPyV-MAD1 was susceptible to the inhibitory effect of all LentiCRISPR/gRNAs tested. When we increased the amount of virus inoculated, we observed a significant decrease in the number of cells expressing LTAg only in cells transduced with LentiCRISPR/gRNA-NCCR-a (representative images in Fig. 1d and quantitative data in Fig. 1e), suggesting that gRNA-NCCR-a was most potent amongst the other gRNAs tested (Fig. 1e). A similar analysis carried out to follow the effect of LentiCRISPR/gRNAs on VP1 instead of LTAg expression also showed that targeting the NCCR, VP1 and VP2 regions hindered late viral protein production, with gRNA-NCCR-a and gRNA-VP1-b being the most potent (Fig. 1f). From both set of results, we concluded that the extent of the inhibitory effects of the LentiCRISPR/gRNAs inversely correlated with the amount of JCPyV used for infection.
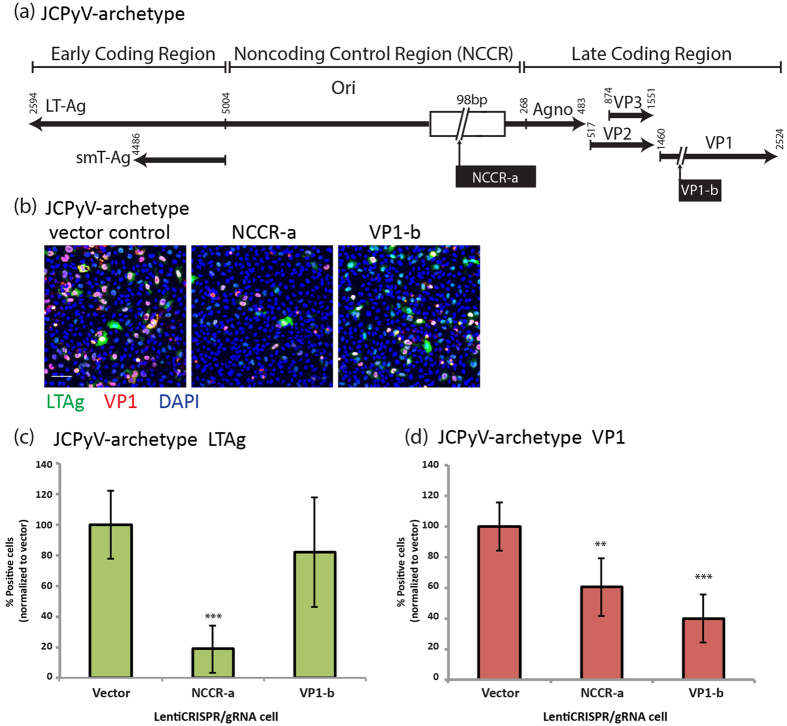
A similar decrease in the number of cells infected with JCPyV-archetype containing a non-rearranged NCCR (Fig. 2a) was achieved by expression of LentiCRISPR/gRNA-NCCR-a or LentiCRISPR/gRNA-VP1-b (Fig. 2b–d). As with JCPyV-MAD1 infection, gRNA-NCCR-a was more potent than gRNA-VP1-b in inhibiting the expression of LTAg for JCPyV-archetype virus (Fig. 2b,c).
Figure 2. Expression of Cas9 and JCPyV specific gRNA inhibit JCPyV-archetype virus infection.
(a) Positions of JCPyV specific gRNA target sites in the JCPyV -archetype genome. (b) SVG-A cells puromycin selected for Cas9 and gRNA expressions were infected with JCPyV -archetype virus (7.5 × 108 genome copies/ml) for 6 days. Immunofluorescence images showing LTAg (green), VP1 (red) positive cells are presented. Cell nuclei were stained with DAPI (blue). Scale bar = 100 μm (c) Quantification of the percentages of LTAg and (d) VP1 positive cells are shown. **One-way ANOVA, p value < 0.01. ***One-way ANOVA, p value < 0.001. N = 2.
Effect of JCPyV specific gRNAs on the production of JCPyV transcripts and viral proteins
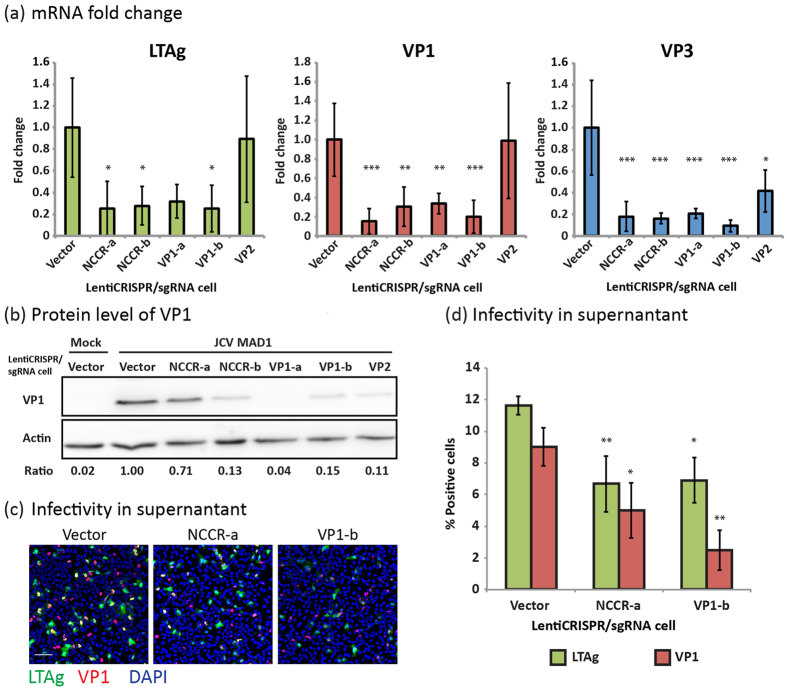
Except for the LentiCRISPR/gRNA targeting VP2, SVG-A cell expression of all the other LentiCRISPR/gRNAs significantly reduced the amount of mRNA of LTAg, VP1 and VP3 as determined by quantitative PCR (qPCR) (Fig. 3a). LentiCRISPR/gRNA targeting VP2 partially reduced the amount of VP3 transcript; this was the expected result given that alternative splicing from a common mRNA generates VP2 and VP3 mRNAs. VP1 protein production determined by western blot analysis was also reduced in LentiCRISPR/gRNA cells (Fig. 3b). These results agreed with the outcome of the infectivity experiments described above (Fig. 1).
Figure 3. Expression of Cas9 and JCPyV specific gRNA inhibited productive JCPyV infection.
(a) Real-time PCR analysis showing the mRNA level of LTAg, VP1 and VP3 genes in different LentiCRISPR/gRNA stably expressing cells at day 3 post-infection. Fold changes of the mRNA is normalized to the vector control. *One-way ANOVA, p value < 0.05; **one-way ANOVA, p value < 0.01; ***one-way ANOVA, p value < 0.001. N=3 (b) Western blot analysis showing the level of VP1 in different LentiCRISPR/gRNA stably expressing cells at day 5 post-infection. Intensity Ratio between Actin and VP1 were measured using ImageJ and presented below the blots. (c,d) SVG-A cells were infected with supernatant collected from JCPyV-MAD1 infected LentiCRISPR/gRNA cells for 6 days. (c) Immunofluorescence images showing the expression of LTAg (green) and VP1 (red) in SVG-A cells and (d) the Quantification of the percentages of LTAg, VP1 positive SVG-A cells are shown. **One-way ANOVA, p value < 0.01.
Effect of JCPyV specific gRNAs on the production of infectious viral particles
SVG-A cells were incubated with supernatant containing newly released virus particles collected from cells first transduced with either LentiCRISPR/gRNA-NCCR-a or LentiCRISPR/gRNA-VP1-b and then infected with JCPyV-MAD1. Infectivity was assayed by counting the number of LTAg and VP1 positive cells 6 days post infection. LentiCRISPR/ gRNA-NCCR-a or LentiCRISPR/gRNA-VP1-b cells generated 50% less infectious particles (Fig. 3c,d). Viruses produced from LentiCRISPR/gRNA-VP1-b cells yielded fewer VP1 positive cells as compared to those from LentiCRISPR/gRNA-NCCR-a or control cells, suggesting the existence of defective viral genomes that could express LTAg normally but not VP1. It is also possible that a fraction of JCPyV genomes in the LentiCRISPR/gRNA-VP1-b cells contained VP1 frame-shifts or deletions generated by aberrant DNA repair of Cas9/gRNA-VP1-b cleavage that nevertheless could still be amplified by replication and then packaged into the progeny virus particles.
Cleavages of the JCPyV genome mediated by LentiCRISPR/gRNAs system resulted in incorrect genome repair
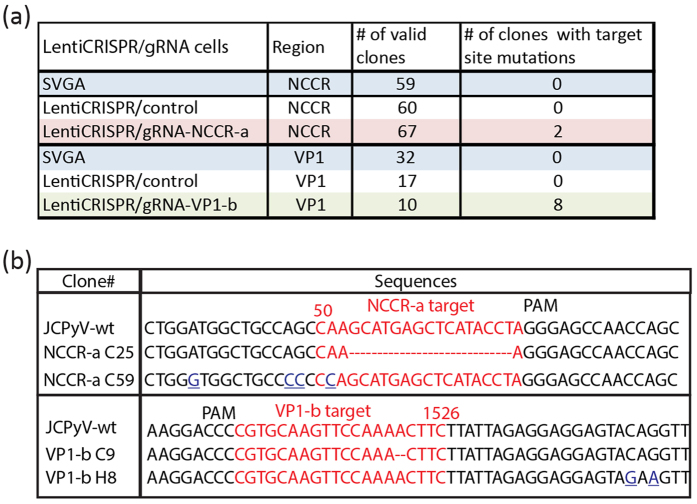
We used DNA sequence analysis to confirm that inhibition of JCPyV infectivity was a direct consequence of the combined effect of the specific targeting of the Cas9 endonuclease to the JCPyV genome and its incorrect repair by the host cell. JCPyV genome was introduced into LentiCRISPR/gRNA cells by transfection of re-circularized JCPyV genome amplified in E.coli. At 5-day post transfection, DNA was extracted from the cells and digested with DpnI enzyme to eliminate transfected genome plasmids of bacterial origin and preserve the amplified JCPyV genome. DNA sequencing of the replicated JCPyV genome was carried out with DNA extracted from SVG-A, SVG-A-LentiCRISPR-control, SVG-A-LentiCRISPR/gRNA-NCCR-a and SVG-A-LentiCRISPR/gRNA-VP1-b cells. Since the NCCR region is vital for the replication of viral genome, the cleavage and aberrant repair of the NCCR will abrogate the synthesis and amplification of the mutant genome in the cells, resulting in a population of viral genome bias towards the intact ones. Indeed, JCPyV genome sequence reads from the SVG-A LentiCRISPR/gRNA-NCCR-a showed that 2 out of a total of 67 contained a deletion or a point mutation in the NCCR-a target region (data summarized Fig. 4a and DNA sequence shown in Fig. 4b). JCPyV genomes with mutations in the VP1 ORF are likely to be able to replicate and accumulate in the cells, making the mutations prominent to detect. As expected, 8 of 10 sequence reads contained deletions or point mutations in the VP1-b region from JCPyV genomes extracted from SVG-A-LentiCRISPR/gRNA-VP1-b cells (data summarized in Fig. 4a and examples of DNA sequence shown in Fig. 4b). These mutations were specific and required presence of the appropriate gRNAs as shown by absence of mutations in control SVG-A cells either alone or just transduced with LentiCRISPR (data summarized in Fig. 4a). Thus, Cas9 together with gRNA-NCCR-a or gRNA-VP1-b mediated targeted cleavage of the JCPyV genome that when incorrectly repaired by the host cells caused reduced viral gene expression, infectivity and virus production.
Figure 4. Expression of Cas9 and JCPyV specific gRNA mediated targeted cleavage in JCPyV genome.
(a) JCPyV genome from LentiCRISPR/gRNA stably expressing cells was extracted and the regions around the indicated target were amplified by PCR reactions. PCR products were clones and 50–70 colonies from each condition were sequenced. The chart showing the number of mutant colonies identified from the sequencing analysis. (b) Examples of deletions or mutations identified from the sequencing analysis for JCPyV genome from LentiCRISPR/gRNA-NCCR-a and LentiCRISPR/gRNA-VP1-b cells.
Inhibition of established JCPyV infection by CRISPR/Cas9
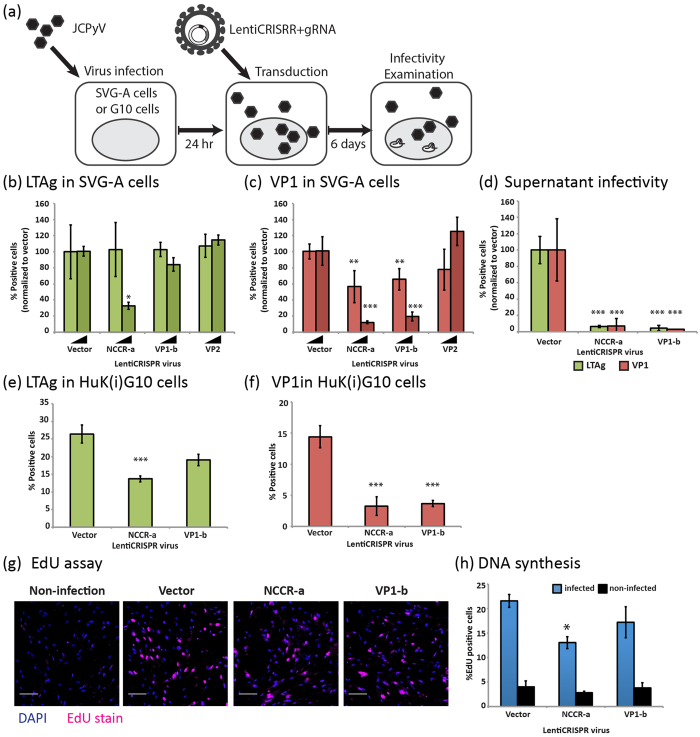
We then tested whether CRISPR/Cas9 targeted to the viral genome could prevent further viral growth in cells already infected with JCPyV. As described below, we found that transduction of LentiCRISPR/gRNAs specific for the JCPyV genome in glial derived SVG-A cells or human fetal kidney derived hTERT transformed HuK(i)G10 cells already infected with JCPyV prevent viral replication and subsequent expression of LTAg and VP1. These cells were first infected for 24 hours with JCPyV-MAD1, allowing for virus replication and then transduced with LentiCRISPR/gRNAs specific for NCCR-a, VP1-b or VP2. The effect on JCPyV infectivity was assessed six days post-infection using immunofluorescence by determining the number of cells expressing LTAg or VP1 (schematic protocol depicted in Fig. 5a).
Figure 5. JCPyV specific CRISPR-Cas9 system inhibited established JCPyV infection.
(a) Experimental scheme. SVG-A cells or HuK(i)G10 cells were infected with JCPyV-MAD1 for 24 hours and then were transduced with LentiCRISPR/gRNA viruses expressing different gRNAs. The level of virus infection was examined at day 6 post-infection. (b,c) Infected SVG-A cells were transduced with two different doses of LentiCRISPR/gRNA viruses, the percentages of (b) LTAg cells and (c) VP1 positive cells are shown. The inoculated genome copies concentration ratio of JCPyV-MAD1 and LentiCRISPR/gRNA virus were 10:1 and 40:1. The inoculation dose of JCPyV-MAD1 was 5 × 107 genome copies/ml. (d) SVG-A cells were infected with supernatant collected from JCPyV-MAD1 infected cells transduced with LentiCRISPR/gRNA viruses (genome copy concentration ratio of JCPyV-MAD1: LentiCRISPR/gRNA = 10:1) for 6 days. The Quantification of the percentages of LTAg, VP1 positive SVGA cells are shown. ***One-way ANOVA, p value < 0.001. N = 2 (e,f) HuK(i)G10 cells were infected with JCPyV-MAD1 as illustrated in (a), the percentages of (e) LTAg and (f) VP1 positive cells are shown. ***One-way ANOVA, p value < 0.001. (g,h) DNA replication efficiency of the JCPyV virus genome is measured using EdU assay. (g) Fluorescent images of virus infected cells after Click-iT reactions are shown. Scale bar = 50 μm (h) Quantification of the percentages of EdU positive cells are shown for infected and non-infected cells. Non-infected HuK(i)G10 was included in the assay as a control indicating the background signal of EdU assay due to cellular DNA replication. *t-test, p value < 0.05.
As shown above with SVG-A cells first transduced with LentiCRISPR/gRNA-NCCR-a (Fig. 1) and then infected with JCPyV-MAD1, we also found reduction in the number of SVG-A or HuK(i)G10 cells expressing LTAg when exposed to LentiCRISPR/gRNA-NCCR-a after JCPyV-MAD1 infection (Fig. 5b,e) while expression of LTAg was not influenced by LentiCRISPR/gRNA-VP1-b (Fig. 5d,f). Similarly, cells targeted with either LentiCRISPR/gRNA-NCCR-a or LentiCRISPR/gRNA-VP1-b displayed a significant reduction in the number of cells expressing VP1 (Fig. 5c,f). These inhibitory effects on LTAg, VP1 expression also strongly reduced the release of infectious virus particles (Fig. 5d). In all cases, the extent of inhibition reflected the amount of LentiCRISPR/gRNAs used for transduction.
We confirmed that LentiCRISPR/gRNA transduction of HuK(i)G10 cells already subjected to JCPyV-MAD1 infection also inhibited viral DNA synthesis, by determining the extent of EdU incorporation into newly synthesized viral DNA. As shown in Fig. 5g,h, lentiCRISPR/gRNA-NCCR-a virus transduction significantly reduced the amount of newly synthesized viral DNA when compared to cells transduced with the LentiCRISPR control virus, analyzed 7-day post JCPyV infection. In contrast, lentiCRISPR/gRNA-VP1-b transduction reduced virus DNA synthesis to a lesser extent consistent with the modest reduction in LTAg expression (Fig. 5g,h). We observed diffuse EdU incorporation in non-infected HuK(i)G10 cells indicating that the punctate EdU signal detected in JCPyV infected cells originated from viral DNA synthesis (Fig. 5g,h).
Discussion
It has been suggested that DNA double strand breaks introduced by the CRISPR/Cas9 system leading to subsequent mutations due to unfaithful DNA repair by the host cells can be applied in the treatment of diseases, including interference with virus infections21,35. Specifically, it has recently been shown that the use of virus specific gRNAs can result in the mutational inactivation of viral genomes of viruses such as HPV, HBV, HSV, EBV and HIV-1, mostly by targeting the integrated viral genomes or viral DNA in latently infected cells21,22,23,24,25,26,27,28,29,30,31,32,34,36,37,38,39. Here we show that it is also possible to use the CRISPR/Cas9 system to inhibit acute virus infection. Our results with specific targeting of the JCPyV genome demonstrated that Cas9 expression alongside virus genome specific gRNAs significantly reduced viral infectivity not only prior JCPyV infection but importantly, also following JCPyV infection.
We showed that amongst the five different gRNAs selected, two gRNAs targeting the NCCR (NCCR-a) and VP1 (VP1-b) displayed strong anti-JCPyV effects. The effective gRNA-NCCR-a target sites are located within the tandem repeat region of the prototype JCPyV-MAD1 genome. Since gRNA-NCCR-a can also restrict the infection of JCPyV-archetype virus, containing one instead of two gRNA-NCCR-a target sites, it is likely that recognition and cleavage of one of the two target sites in JCPyV-MAD1 by the CRISPR/Cas9 is sufficient to mediate JCPyV inhibition. Since the NCCR region contains all the promoters and enhancer elements required for viral gene transcription, disruption of this region should affect both the early and late gene expression. Indeed, we found that gRNA-NCCR-a decreased expression of both the early gene LTAg and the late gene VP1 (Fig. 1e,f).
Presently, there is no therapeutic available to specifically treat JCPyV infection and most of the reported JCPyV inhibitors generically target the host cell such as retrograde trafficking inhibitors that can inhibit early entry steps of the virus40. Since JCPyV persists asymptomatically in an estimated 50–80% of the adult population41 and causes PML by active infection of oligodendrocytes in the CNS, it is of importance to develop strategies to diminish viral genomes already present in the host cells. Of importance, we uncovered that transduction of LentiCRISPR/gRNA viruses 24-hours post JCPyV infection significantly reduced JCPyV infectivity. Thus, the CRISPR/Cas9 system is able to inhibit an established JCPyV infection. Previously, Wollebo et al., showed that stable expression of Cas9 and gRNAs targeting the N-terminal region of small-T antigen is able to inhibit viral DNA replication and they also demonstrated the ability of Cas9/gRNA to edit viral DNA integrated in the genome34. We now show that the CRISPR/Cas9 system can also be used to inhibit on-going viral DNA replication in acutely JCPyV infected cells.
We confirmed the specificity of the inhibition by showing the ability of the CRISPR/Cas9 system to cleave incoming JCPyV genome, which is not integrated into the cellular genome (Fig. 4). The ability of the CRISPR/Cas9 system to restrict the JCPyV life cycle when delivered after virus infection has occurred, demonstrating the potential of this type of approach as a possible anti-viral strategy to fight against latent and active JCPyV infection. An efficient way of CRISPR/Cas9/gRNA delivery remains to be developed before further clinical consideration. For example, it has been proposed that adeno-associated virus (AAV) with optimized Cas9 expression vectors may be another feasible way to deliver CRISPR/Cas9/gRNA in vitro and in vivo21,42,43,44. Nevertheless, the use of the CRISPR/Cas9 system may be limited by the potential to generate JCPyV mutants of enhanced infectivity due to aberrant DNA double strand repair. Given that JCPyV strains isolated from PML patients contain genomic rearranged NCCR regions, demonstrated to increase viral gene expression and replication45,46, the application of CRISPR/Cas9 to JCPyV infection must be approached with caution.
In summary, we developed a CRISPR/Cas9 system capable of inhibiting already established JCPyV infection. We identified several gRNAs with particular two gRNAs (NCCR-a and VP1-b) able to mediate efficient anti-viral effects. We showed that pre-existence of CRISPR/Cas9 and appropriate gRNAs in cells stably transduced with LentiCRISPR/gRNA viruses rendered the cells resistant to initial JCPyV infection. Importantly, actively replicating JCPyV was also restricted upon the delivery of CRISPR/Cas9 and gRNAs with LentiCRISPR/gRNA viruses at 24-hours post JCPyV infection. Together, these data provide evidence that a JCPyV specific CRISPR/Cas9 platform might serve as a valuable research tool to study JCPyV genomics as well as a tool to restrict the JCPyV life cycle.
Material and Methods
Cell lines and antibodies
SVG-A cells kindly provided by Walter J. Atwood were maintained in minimum essential medium (MEM; 10–010-CV, Mediatech) supplemented with 10% fetal bovine serum (FBS), incubated at 37 °C and 5% CO2 in humidified incubators. 293FT cells used for lentivirus production were cultured in DMEM (10565–042, Life Technologies) supplemented with 10% FBS and 1 mg/ml G418, incubated at 37 °C and 5% CO2 in humidified incubators. HuK(i)G10 cell line was derived by transduction of human fetal kidney cells with a lentiviral vector expressing human telomerase reverse transcriptase (hTERT) (Applied Biological Materials #G200). HuK(i)G10 was selected from single-cell colonies isolated by limiting dilution for permissivity to JCPyV-archetype virus infection. HuK(i)G10 cells were maintained in RenaLife Complete media at 37 °C and 5% CO2 in humidified incubators. The antibodies used for immunofluorescence staining are: PAB2000 or PAB2003 specific for JCPyV large T antigen (gift from Richard Frisque), BIIB048 and chPAB597 (gift from Ed Harlow) specific for JCPyV VP1. Secondary antibodies: anti-mouse IgG Alexa Fluor 647 (A21236, Life technologies); Alexa Fluor 594 goat anti-human IgG (A11014, Invitrogen, Molecular Probes).
Viruses
JCPyV-MAD1 virus (type 1Av (accession #J02226.1)) was produced by transfection of 293FT cells with pMAD1 JCPyV genome (Richard Frisque, Pennsylvania State University) circularized by ligation after EcoR1 digestion. JCPyV-archetype virus was produced using the modified pMAD1 plasmid in which the prototype NCCR sequence was replaced by the NCO1 fragment of an archetype NCCR (accession # AY121915).
LentiCRISPR/gRNA viruses targeting JCPyV genome
Cas9 and gRNA was delivered into JCPyV permissive cells with high efficiency using a lentiviral vector (LentiCRISPR)47 encoding Cas9, a gRNA and a puromycin selection marker (Fig. 1a) was kindly provided by Dr. Feng Zhang.
The gRNAs target sequences in the JCPyV genome were designed using an online CRISPR design tool (http://www.genome-engineering.org/crispr/). Insertion of the gRNA targeting nucleotide sequence was performed as described47. LentiCRISPR viruses were produced according to protocols accessible online at http://www.bu.edu/dbin/stemcells/protocols.php with minor modifications. Briefly, 12 μg of LentiCRISPR plasmid was co-transfected with 1.2 μg of VSV-g, 0.6 μg of gag/pol, 0.6 μg of rev and 0.6 μg of tat expression plasmids into 293FT cells in 10 cm dish using 45 μl of lipofectamine 2000 (11668027, Invitrogen). Supernatant containing the LentiCRISPR virus was collected 72 hours post transfection and centrifuged at 3000 rpm, at 4 °C for 10 min to remove cell debris. The supernatant was then filtered through 0.45 µm filter (28144–007, Acrodisc Syringe Filter, VWR) and aliquots were stored at −80 °C before use.
Virus infections and immunofluorescence analysis of JCPyV infection
SVG-A cells stably transduced with LentiCRISPR/gRNA viruses were plated in 384 well plates with 1500 cells per well 24 hours before JCPyV infection. The cells were then infected with JCPyV-MAD1 (5 × 107 genome copies/ml) or JCPyV-archetype viruses (7.5 × 108 genome copies/ml) in MEM supplemented with 2% FBS. The cells were fixed at the indicated times with 3.7% formaldehyde in 1× PBS for 10 minutes. Cells were then washed and permeablized with 0.1% TritonX-100 in 1× PBS for 5 minutes followed by a brief wash with 1× PBS, then blocked with 1× PBS supplemented with 5% bovine serum albumin (BSA) for 30 minutes and stained with BIIB048 or chPAB597 and PAB2003 for 1 hour at room temperature. Cells were washed 3 times with 5% BSA in 1× PBS and then incubated with goat anti-human Alexa-Fluor 594 (A11014, Invitrogen, Molecular Probes) and rabbit anti-mouse Alex-Fluor 647, Samples were washed 3 times with 5% BSA in 1× PBS and stained with DAPI for 2 minutes before imaging. The number of infected cells were counted in samples imaged using a spinning disc confocal microscope48. Images were acquired at 10x magnification and the analysis was performed using ImageJ. Binary masks were created for the DAPI, LTAg and the VP1 channels, respectively. The masks from the LTAg and DAPI channels were then logically multiplied, resulting in new binary image representing LTAg positive nuclei. A similar procedure was performed to obtain binary images to identify VP1 positive nuclei. The Analyze Particle plugin module of ImageJ was then used to determine the total numbers of nuclei, LTAg positive nuclei, and VP1 positive nuclei in a total of 25 imaged fields per well for each sample.
Detection of JCPyV gene expression by Quantitative PCR
SVG-A cells stably expressing LentiCRISPR/gRNAs were infected with JCPyV-MAD1 (5 × 107 genome copies/ml) for 3 days. The viral mRNA was extracted using RNeasy Plus Mini Kit (Qiagen). Real-time PCR analyses were performed using the TaqMan system with Express One-step Superscript qRT-PCR-kit (Invitrogen), following the manufacturer’s protocol. In brief, each 20 μl reaction contained 40 ng of RNA template; 10 μl of qPCR SuperMix Universal; 0.25 μl of 10 μM forward and reverse primers; 0.25 μl of 5 μM TaqMan probe and 0.4 μl ROX reference dye. Thermal cycling began with a step of 30 min at 48 °C followed by 95 °C for 10 min and 40 cycles of 95 °C 15 sec and 60 °C for 1 min, using Applied Biosystems StepOnePlus real-time PCR system. Three biological replicates were done for each condition and the level of GAPDH was analyzed for normalization. The sequences of the primer used were: (1) VP1 forward: 5′ GGGACATGCTTCCTTGTTACAGT 3′; (2) VP1 reverse: 5′ ATTTCCACAGGTTAGATCCTCATTTAG 3′; (3) VP1 FAM probe: 5′ TGGCCAGAATTCCACT 3′; (4) LTAG forward: 5′ AGGCAGCAAGCAATGAATCC 3′; (5) LTAG reverse: 5′ATGGCAATGCTGTTTTAGAGCAA 3′; (6) LTAG FAM probe: 5′ CCACCCCAGCCATAT 3′; (7) VP3 forward: 5′ CCAGGAGGTGCAAATCAAAGA 3′; (8) VP3 reverse: 5′ CCCGTACAACCCTAAAAGTAAAGG 3′; (9) VP3 NED probe: 5′ CTGCTCCTCAATGGA 3′; (10) GAPDH forward: 5′ GAAGGTGAAGGTCGGAGTC 3′; (11) GAPDH reverse: 5′ GAAGATGGTGATGGGATTTC 3′; (12) GAPDH FAM probe: 5′ TGGAATCATATTGGAACATG 3′.
Western blot analysis
SVG-A cells stably expressing LentiCRISPR/gRNAs were infected with JCPyV-MAD1 (5 × 107 genome copies/ml) for 5 days. Equal amount of cell lysates for each condition was resolved using 12.5% SDS-PAGE and wet transferred onto a PVDF membrane (Pall). The membrane was blocked with 5% nonfat milk in 1× PBS with 0.05% Tween-20 (PBST) for 30 min and incubated with anti-VP1 PAB597 mouse monoclonal antibody and rabbit anti-cytoskeletal actin affinity purified antibody (Bethyl) in 5% nonfat milk in PBST overnight at 4 °C, followed by washes and incubation with horseradish peroxidase-labeled secondary antibody against mouse or rabbit IgG (GE Healthcare), for detection of VP1 or actin, respectively. The signal was developed using LumiGLO Chemiluminescent Substrate Kit (KPL) and imaged using the Alphaimager 2000.
EdU assay to measure JCPyV DNA replication
HuK(i)G10 cells infected with JCPyV-MAD1 (2.6 × 108 genome copies/ml) virus for 7 days were incubated with medium containing 10 μM EdU for 20 min and the newly synthesized DNA was determined by EdU incorporation using the Click-it PLUS EdU imaging kit (Molecular Probes). At least 15 fields per sample were imaged using the high content imaging platform Operetta (Perkin Elmer) at 20x magnification and images analyzed using the Columbus software (Perkin Elmer).
JCPyV sequence analysis
The JCPyV-archetype genome was transfected into SVG-A-LentiCRISPR control cells, SVG-A- LentiCRISPR/gRNA-NCCR-a and SVG-A-LentiCRISPR/gRNA-VP1-b cells. At day 5-post transfection, viral DNA was extracted using the QlAmp MiniElute Virus Spin Kit (Qiagen), DpnI digested, and subjected to PCR and cloning using procedures described for Guide-it Indel Indentification Kit (Clontech Laboratories). The NCCR-a target regions were amplified using NCCR forward (5′CGGTACCCGGGGATCCATGACAGGAATGTTCCCCCATG3′) and reverse primers (5′CGACTCTAGAGGATCTTTTCCCGTCTACACTGTCTTCAC3′); PCR reactions for the VP1-b target region were performed using VP1-t forward (5′CGGTACCCGGGGATC CCAAATGTGCAATCTGGTGAATTT3′) and reverse primers (5′CGACTCTAGAGGATCAGTTGCTTGCCCATTAGAGTGC3′). For each sample, 50–70 colonies were picked for colony PCR using Colony PCR forward (5′ACGTTGTAAAACGACGGCCAGTGA3′) and reverse primers (5′CAATTTCACACAGGAAACAGCTATGACC3′) and the PCR products were purified using the Zymogen DNA Clean& Concentrator-5 (Zymo Research) system and sequenced using colony PCR forward primer. Valid viral sequences contained at least 100 base pair (bp) matches with the wild type JCPyV sequences at both the 5′ and 3′ ends.
Additional Information
How to cite this article: Chou, Y.-y. et al. Inhibition of JCPyV infection mediated by targeted viral genome editing using CRISPR/Cas9. Sci. Rep. 6, 36921; doi: 10.1038/srep36921 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Acknowledgments
We thank Walter J. Atwood for providing the SVG-A cells and we thank Wayne Marasco for collaborating to generate the HuK(i)G10 cell line. This work was supported in part by grants to T.K. from Biogen and NIH CETR U19-Al109740.
Footnotes
This work is partially funded by Biogen. A.K., C.K., E.CM., receive compensation as members of Biogen and own stocks in the company. Y.C., M.M., R.G., T.K., declare no potential conflict of interest.
Author Contributions Y.C., A.K., E.CM. and T.K. designed the experiment and drafted the main manuscript, Y.C. performed experiments for Figure 1, 2, 3, 4 and 5 and A.K. performed experiments for Figure 4d–f. C.K. generated the HuK(i)G10 cell line and prepared virus samples. Y.C. prepared all the figures and table. M.M. and R.G. designed and constructed the LentiCRISPR plasmids. All authors reviewed the manuscript.
References
- Jiang M., Abend J. R., Johnson S. F. & Imperiale M. J. The role of polyomaviruses in human disease. Virology 384, 266–273 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kean J. M., Rao S., Wang M. & Garcea R. L. Seroepidemiology of Human Polyomaviruses. PLoS Pathog. 5, e1000363 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomgren G. et al. Risk of natalizumab-associated progressive multifocal leukoencephalopathy. N. Engl. J. Med. 366, 1870–1880 (2012). [DOI] [PubMed] [Google Scholar]
- Major E. O. Progressive Multifocal Leukoencephalopathy in Patients on Immunomodulatory Therapies*. Annu. Rev. Med. 61, 35–47 (2010). [DOI] [PubMed] [Google Scholar]
- Ferenczy M. W. et al. Molecular biology, epidemiology, and pathogenesis of progressive multifocal leukoencephalopathy, the JC virus-induced demyelinating disease of the human brain. Clin. Microbiol. Rev. 25, 471–506 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brew B. J., Davies N. W. S., Cinque P., Clifford D. B. & Nath A. Progressive multifocal leukoencephalopathy and other forms of JC virus disease. Nat Rev Neurol 6, 667–679 (2010). [DOI] [PubMed] [Google Scholar]
- Reid C. E. et al. Sequencing and Analysis of JC Virus DNA From Natalizumab-Treated PML Patients. Journal of Infectious Diseases 204, 237–244 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogo Y. et al. Isolation of a Possible Archetypal JC Virus DNA Sequence from Nonimmunocompromised Individuals. J. Virol. 64, 3139–3143 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörries K., Vogel E., Günther S. & Czub S. Infection of Human Polyomaviruses JC and BK in Peripheral Blood Leukocytes from Immunocompetent Individuals. Virology 198, 59–70 (1994). [DOI] [PubMed] [Google Scholar]
- Dubois V. et al. Latency and reactivation of JC virus in peripheral blood of human immunodeficiency virus type 1-infected patients. J. Clin. Microbiol. 35, 2288–2292 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois V. et al. Detection of JC virus DNA in the peripheral blood leukocytes of HIV-infected patients. AIDS 10, 353–358 (1996). [DOI] [PubMed] [Google Scholar]
- Houff S. A. et al. Involvement of JC Virus–Infected Mononuclear Cells from the Bone Marrow and Spleen in the Pathogenesis of Progressive Multifocal Leukoencephalopathy. N. Engl. J. Med. 318, 301–305 (1988). [DOI] [PubMed] [Google Scholar]
- Van Loy T. et al. JC Virus Quasispecies Analysis Reveals a Complex Viral Population Underlying Progressive Multifocal Leukoencephalopathy and Supports Viral Dissemination via the Hematogenous Route. J. Virol. 89, 1340–1347 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaprio J. A., Imperiale M. J. & Major E. O. Polyomaviruses. (Lippincott Williams & Wilkins, 2015). [Google Scholar]
- Frisque R. J., Bream G. L. & Cannella M. T. Human polyomavirus JC virus genome. J. Virol. 51, 458–469 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj G. V. E. A. Transcriptional Regulation: Lessons from the Human Neurotropic Polyomavirus. JCV. 1–9 (1995). [DOI] [PubMed] [Google Scholar]
- Mali P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Wei J. J., Sabatini D. M. & Lander E. S. Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell M. R. et al. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature 1–15, doi: 10.1038/nature13769 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy E. M. & Cullen B. R. Bacterial CRISPR/Cas DNA endonucleases: A revolutionary technology that could dramatically impact viral research and treatment. Virology 479–480C, 213–220 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saayman S., Ali S. A., Morris K. V. & Weinberg M. S. The therapeutic application of CRISPR/Cas9 technologies for HIV. Expert Opin Biol Ther 15, 819–830 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebina H., Misawa N., Kanemura Y. & Koyanagi Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci. Rep. 3, 2510 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen K.-S. et al. CRISPR/Cas9-mediated genome editing of Epstein-Barr virus in human cells. J. Gen. Virol. 96, 626–636 (2015). [DOI] [PubMed] [Google Scholar]
- Kennedy E. M. et al. Suppression of hepatitis B virus DNA accumulation in chronically infected cells using a bacterial CRISPR/Cas RNA-guided DNA endonuclease. Virology 476, 196–205 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.-R. et al. The CRISPR/Cas9 System Facilitates Clearance of the Intrahepatic HBV Templates In Vivo. Mol Ther Nucleic Acids 3, e186 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan V. et al. CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis B virus. Sci. Rep. 5, 10833 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen S. et al. Harnessing the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated Cas9 system to disrupt the hepatitis B virus. Gene Therapy 22, 404–412 (2015). [DOI] [PubMed] [Google Scholar]
- Liu X., Hao R., Chen S., Guo D. & Chen Y. Inhibition of Hepatitis B Virus by CRISPR/Cas9 System via Targeting the Conserved Regions of Viral Genome. J. Gen. Virol. vir.0.000159, doi: 10.1099/vir.0.000159 (2015). [DOI] [PubMed] [Google Scholar]
- Hu Z. et al. Disruption of HPV16-E7 by CRISPR/Cas System Induces Apoptosis and Growth Inhibition in HPV16 Positive Human Cervical Cancer Cells. Biomed Res Int 2014, 612823–612829 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy E. M. et al. Inactivation of the human papillomavirus E6 or E7 gene in cervical carcinoma cells using a bacterial CRISPR/Cas RNA-guided endonuclease. J. Virol. JVI. 01879–14, doi: 10.1128/JVI.01879-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dampier W., Nonnemacher M. R., Sullivan N. T., Jacobson J. M. & Wigdahl B. HIV Excision Utilizing CRISPR/Cas9 Technology: Attacking the Proviral Quasispecies in Reservoirs to Achieve a Cure. MOJ Immunol 1, doi: 10.15406/moji.2014.01.00022 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebina H. et al. A high excision potential of TALENs for integrated DNA of HIV-based lentiviral vector. PLoS ONE 10, e0120047 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollebo H. S. et al. CRISPR/Cas9 System as an Agent for Eliminating Polyomavirus JC Infection. PLoS ONE 10, e0136046–20 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lijuan Y., Siqi H. & Fei G. The application of CRISPR-Cas9 gene editing technology in viral infection diseases. Yi Chuan 37, 412–418 (2015). [DOI] [PubMed] [Google Scholar]
- Dong C. et al. Targeting hepatitis B virus cccDNA by CRISPR/Cas9 nuclease efficiently inhibits viral replication. Antiviral Res. 118, 110–117 (2015). [DOI] [PubMed] [Google Scholar]
- Liao H.-K. et al. Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat Comms 6, 6413–10 (2015). [DOI] [PubMed] [Google Scholar]
- Seeger C. & Sohn J. A. Targeting Hepatitis B Virus With CRISPR/Cas9. Mol Ther Nucleic Acids 3, e216 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. & Quake S. R. RNA-guided endonuclease provides a therapeutic strategy to cure latent herpesviridae infection. Proc. Natl. Acad. Sci. USA 111, 13157–13162 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C. D. S. et al. A retrograde trafficking inhibitor of ricin and Shiga-like toxins inhibits infection of cells by human and monkey polyomaviruses. mBio 4, e00729–13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles W. A. et al. Population-based study of antibody to the human polyomaviruses BKV and JCV and the simian polyomavirus SV40. J. Med. Virol. 71, 115–123 (2003). [DOI] [PubMed] [Google Scholar]
- Howes R. & Schofield C. Genome engineering using Adeno-Associated Virus (AAV). Methods Mol. Biol. 1239, 75–103 (2015). [DOI] [PubMed] [Google Scholar]
- Senís E. et al. CRISPR/Cas9-mediated genome engineering: an adeno-associated viral (AAV) vector toolbox. Biotechnol J 9, 1402–1412 (2014). [DOI] [PubMed] [Google Scholar]
- Deverman B. E. et al. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol. 34, 204–209 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch H. H., Kardas P., Kranz D. & Leboeuf C. The human JC polyomavirus (JCPyV): virological background and clinical implications. APMIS 121, 685–727 (2013). [DOI] [PubMed] [Google Scholar]
- Gosert R., Kardas P., Major E. O. & Hirsch H. H. Rearranged JC Virus Noncoding Control Regions Found in Progressive Multifocal Leukoencephalopathy Patient Samples Increase Virus Early Gene Expression and Replication Rate. J. Virol. 84, 10448–10456 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O. et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cureton D. K., Massol R. H. & Kirchhausen T. The Length of Vesicular Stomatitis Virus Particles Dictates a Need for Actin Assembly during Clathrin-Dependent Endocytosis. PLoS Pathog. 6, e1001127 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]