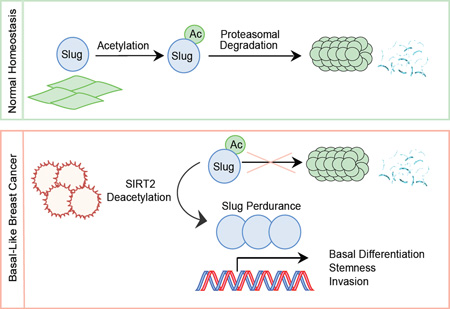
Summary
Overabundance of Slug protein is common in human cancer and represents an important determinant underlying the aggressiveness of basal-like breast cancer (BLBC). Despite its importance, this transcription factor is rarely mutated in BLBC, and the mechanism of its deregulation in cancer remains unknown. Here we report that Slug undergoes acetylation-dependent protein degradation and identify the deacetylase SIRT2 as a key mediator of this post-translational mechanism. SIRT2 inhibition rapidly destabilizes Slug, whereas SIRT2 overexpression extends Slug stability. We show that SIRT2 deacetylates Slug protein at lysine residue K116 to prevent Slug degradation. Interestingly, SIRT2 is frequently amplified and highly expressed in BLBC. Genetic depletion and pharmacological inactivation of SIRT2 in BLBC cells reverse Slug stabilization, cause the loss of clinically relevant pathological features of BLBC, and inhibit tumor growth. Our results suggest that targeting SIRT2 may be a rational strategy for diminishing Slug abundance and its associated malignant traits in BLBC.
Graphical Abstract
Introduction
Over the past decade, large-scale genomic profiling has revealed the molecular landscape of breast cancers (Perou et al., 2000; van ’t Veer et al., 2002), identifying discrete subtypes as well as underlying driver genes. For the majority of breast cancer subtypes, tailored targeted therapies are now available and have significantly improved patient survival (Cuzick et al., 2010; Ignatiadis et al., 2012; Regan et al., 2011; Slamon et al., 2001). The notable exception is one of the deadlier and more aggressive subtypes, called basal-like breast cancer (BLBC) and so-named for its molecular similarities to the basal mammary epithelial cell differentiation program (Harris et al., 2012). Sharing an immunophenotype with triple-negative breast cancer, BLBC is identified clinically by the absence of estrogen receptor, progesterone receptor and HER2, and affects approximately 20% of breast cancer patients (Fan et al., 2006; Rakha et al., 2008). Unfortunately, analyses of somatic mutation profiles of BLBC have not yet revealed promising targets for therapeutic intervention (Foulkes et al., 2010; Gusterson, 2009).
Robust tumorigenic capacity, early dissemination and metastasis, and frequent resistance to conventional chemo- and radiotherapy regimens are central clinical features of BLBC (Foulkes et al., 2010; Harris et al., 2012; Metzger-Filho et al., 2012; Rakha et al., 2008). Recent studies have identified the transcriptional repressor SNAI2/Slug as a critical determinant underlying these malignant phenotypes (DiMeo et al., 2009; Phillips and Kuperwasser, 2014; Proia et al., 2011; Samanta et al., 2016; Storci et al., 2008). In cancer biology, Slug is well known to promote tumor progression and metastasis through the epithelial-mesenchymal transition (EMT), causing loss of cell adhesion and polarity while conferring migratory and invasive properties (Polyak and Weinberg, 2009; Yang and Weinberg, 2008). In addition, studies on mammary gland biology have outlined essential roles for Slug in orchestrating transcriptional programs for stem cell self-renewal and basal mammary epithelial cell differentiation (Cobaleda et al., 2006; Guo et al., 2012; Nassour et al., 2012; Phillips et al., 2014). By suppressing the luminal differentiation program while activating EMT, high Slug expression levels bias tumor development toward stem/basal-like phenotypes and enable the adoption of tumor-initiating and invasive capabilities (DiMeo et al., 2009; Proia et al., 2011; Samanta et al., 2016; Storci et al., 2008). Experimental depletion of Slug diminishes these aggressive traits, and SNAI2 knockout animals are resistant to mammary tumorigenesis (Phillips et al., 2014).
Consistent with Slug playing a central role in the development of BLBC, an overabundance of Slug protein is commonly observed in BLBC tumors (Liu et al., 2013; Proia et al., 2011). However, despite its frequent overabundance, SNAI2 is rarely mutated or amplified in BLBC. While Slug is a shorted-lived and rapidly degraded protein in normal tissue, we have previously observed extended Slug stability in BLBC caused by decreased proteasomal degradation of Slug (Proia et al., 2011). Proteolytic turnover of Slug, like many labile transcription factors, is regulated by post-translational modifications. Phosphorylation mediated by GSK3β primes Slug for ubiquitination (Kao et al., 2014; Wu et al., 2012), and several E3 ligases (FBXL14, β-Trcp1 and CHIP) are involved in the ubiquitin-mediated degradation of Slug (Kao et al., 2014; Vernon and LaBonne, 2006; Wu et al., 2005, 2012). However, GSK3β inactivation does not strongly correlate with Slug overabundance in cancer, and prior studies have demonstrated that Slug undergoes proteasomal degradation independent of GSK3β-mediated phosphorylation (Lander et al., 2011; Montserrat-Sentís et al., 2009). Therefore the mechanism by which Slug escapes proteasomal degradation in BLBC remains unknown.
We reasoned that elucidating the molecular mechanism underlying the phenomenon of extended Slug stability could provide a new target for BLBC therapeutic intervention. Thus, in this study, we endeavored to identify the post-translational mechanism by which Slug protein stability is regulated in breast epithelial cells and evaluate whether components of this mechanism are altered in breast cancer. We found that Slug acetylation represents a major determinant governing its abundance, and deacetylation of the SLUG domain by the mammalian sirtuin SIRT2 regulates Slug stability. Notably, SIRT2 is frequently amplified in BLBC, and experimental manipulation of SIRT2 in BLBC cells antagonized the cancer-associated phenotypes mediated by Slug. Together, these findings unravel an intricate molecular interplay between SIRT2 amplification, Slug stability and the BLBC phenotype.
Results
A combined proteomic and chemical inhibitor approach identifies acetylation in the regulation of Slug protein turnover
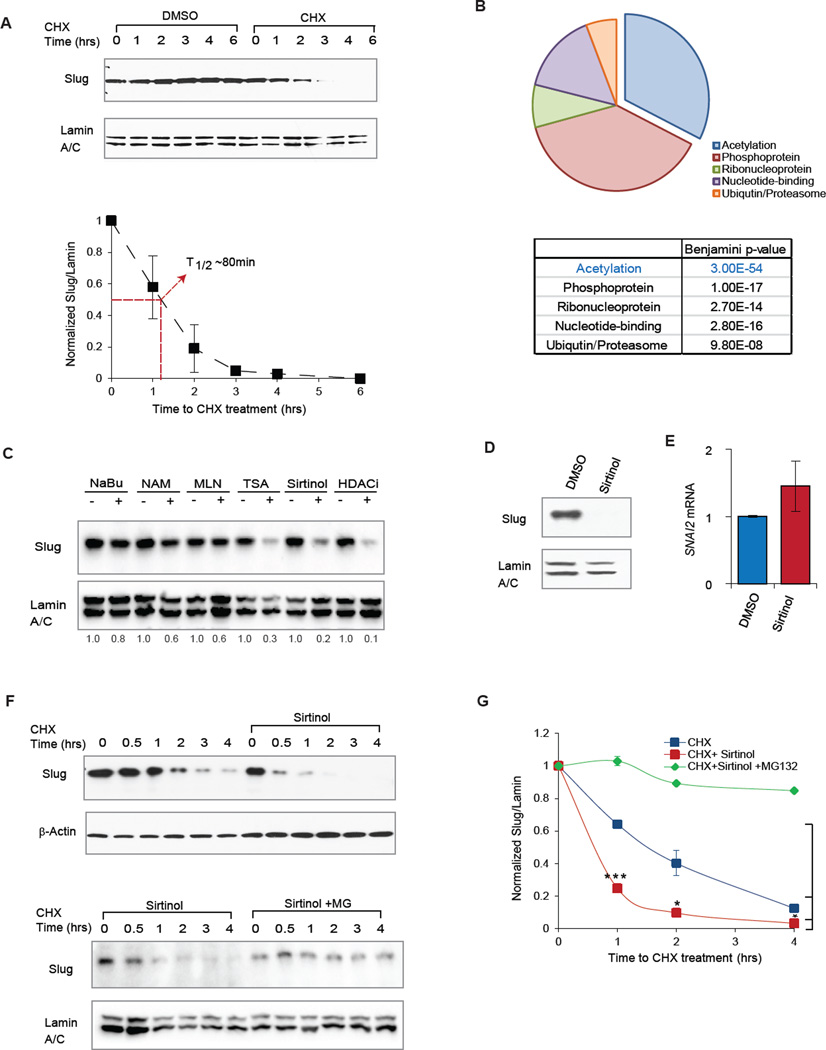
We have previously shown that Slug protein is abundantly expressed yet undergoes rapid turnover in normal mammary epithelial cells (Phillips et al., 2014; Proia et al., 2011). Indeed, in immortalized, non-transformed MCF10A human breast epithelial cells, Slug is rapidly degraded upon cycloheximide (CHX) blockade of de novo protein synthesis, exhibiting a half-life of ~80 min (Fig 1A). In addition, proteasomal inhibition by MG132 treatment completely prevented Slug protein turnover (Fig S1B). To identify proteins that may contribute to the regulation of Slug protein levels, we performed immunoprecipitation of Slug followed by mass spectrometry (co-IP/MS). This proteomic approach identified 287 unique Slug-binding partners (Table S1), several of which have been previously validated (Kao et al., 2014; Phillips et al., 2014; Wu et al., 2012). We interrogated this list of Slug-binding partners for common molecular functions using the DAVID functional annotation tool (Fig 1B & Fig S1A) (http://david.niaid.nih.gov). Consistent with the function of Slug as a DNA-binding transcriptional co-repressor, a significant number of binding partners were nuclear proteins. However, proteins involved in post-translational modification were also highly represented, and surprisingly, acetylation was the most statistically significant functionality associated with Slug binding partners (Fig 1B, FDR = 3.0 × 10−54).
Figure 1. A combinatorial proteomic and chemical inhibitor approach identifies acetylation in the regulation of Slug protein turnover.
(A) (Top) MCF10A cells were treated with cycloheximide (CHX) to prevent de novo protein synthesis at indicated time intervals. Immunoblots for Slug protein and Lamin A/C loading control levels at the indicated time intervals following CHX treatment. (Bottom) Quantification of relative Slug levels from five independent experiments, normalized to Lamin A/C levels. The half-life of Slug protein is indicated (red line). Data shown are mean ± SEM.
(B) Pie chart and table of major functional groups associated with Slug-interacting partners as identified by SLUG coIP/MS in HEK293T cells. False recovery rates are represented as Benjamini-Hochberg p-values in the table.
(C) MCF10A cells were treated with the indicated panel of chemical inhibitors or vehicle control, and Slug protein abundance was assessed by immunoblot. Relative levels of Slug protein were quantified and are shown below the immunoblots.
(D) MCF10A cells were treated with sirtinol (25 µm) for 4 hours, and Slug protein level was assessed by immunoblot.
(E) MCF10A cells were treated with sirtinol (25 µm) for 4 hours and SNAI2 transcript expression was assessed by qRT-PCR. Data are shown as mean ± SEM (n = 3), p=0.08.
(F) MCF10A cells were pre-treated with sirtinol alone (top) or sirtinol plus the proteasome inhibitor MG132 (bottom), and then subsequently treated with cycloheximide (CHX) to prevent de novo protein synthesis at indicated time intervals. Immunoblots of Slug and Lamin A/C protein levels are shown at the indicated time intervals.
(G) Quantification of relative Slug protein levels from three independent experiments performed as in (F), normalized to Lamin A/C levels. The degradation curves of Slug protein are plotted for CHX only (blue), CHX + sirtinol (red) and CHX + sirtinol + MG132 (green). Data shown are mean ± SEM. * p < 0.05.
Based on these findings, we examined whether acetylation might affect Slug protein levels. Indeed, we observed that small molecule inhibitors of deacetylation, targeting class I, II or III HDACs (TSA, sirtinol and HDACi), resulted in dramatic reductions of Slug protein in MCF10A cells, whereas other HDAC inhibitor such as sodium butyrate and a cullin/E3 ligase inhibitor (MLN4924) failed to appreciably affect Slug levels (Fig 1C). Interestingly, the most selective of these HDAC inhibitors – sirtinol, a selective inhibitor of sirtuin 1 and 2 – led to marked Slug depletion in a dose- and time-dependent manner (Fig S1C & D). Furthermore, Slug protein depletion was not a result of transcriptional repression (Fig 1D), as SNAI2 mRNA levels were slightly increased following sirtinol treatment (Fig 1E). Consistent with the post-translational nature of this regulation, sirtinol treatment drastically accelerated Slug protein turnover and led to substantial shortening of Slug protein half-life (Fig 1F & G, * p < 0.05). Moreover, the sirtinol-induced loss of Slug protein also correlated with de-repression of EPCAM and CDH1, two canonical Slug target genes (S1E, ** p <0.01). Taken together, these data identify acetylation as a potential regulator of Slug protein level, and show that combined pharmacological inhibition of the deacetylases SIRT1 and SIRT2 by sirtinol is sufficient to reduce Slug protein half-life and activity.
Protein deacetylase SIRT2 regulates Slug protein abundance, stability and function
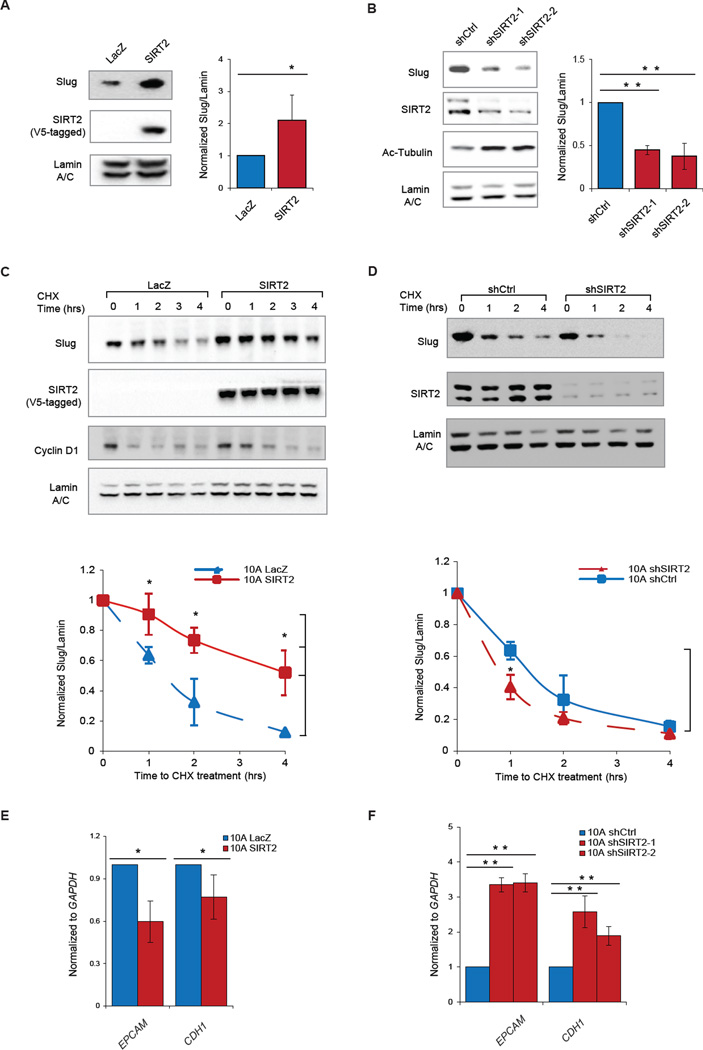
Mammalian sirtuins (SIRT1–7) are a class of NAD+-dependent type III histone and protein deacetylases classically known for their regulatory roles in cellular metabolism and aging (Guarente el al., 2011). Interestingly, a growing body of literature highlights a novel function of the sirtuin family in regulating the stability of short-lived transcriptional factors, including several cancer-relevant substrates such as p53 and FOXO3 (Liu et al., 2013a; Hoffmann et al., 2014). Given our finding that sirtinol substantially reduces Slug protein abundance, we investigated whether regulation of Slug protein stability requires either SIRT1 or SIRT2, or both. To this end, we individually perturbed SIRT1 and SIRT2 expression in MCF10A cells and examined the ensuing effect on Slug. We found that neither overexpression nor depletion of SIRT1 resulted in a significant change in Slug protein levels or stability (Fig S2A & B). In contrast, overexpressing SIRT2 resulted in a striking increase in Slug protein abundance (Fig 2A, * p<0.05), without an observable change in SNAI2 mRNA levels (Fig S2D, * p = 0.03). Conversely, knockdown of SIRT2 by two independent short hairpin RNAi constructs (shRNA) led to significantly diminished Slug protein levels (Fig 2B, ** p < 0.01), without significant decrease in SNAI2 mRNA levels (Fig S2E).
Figure 2. SIRT2 regulates Slug protein abundance and activity.
(A) (Left) Immunoblots showing levels of Slug, V5 tagged SIRT2 (SIRT2-V5) and Lamin A/C in MCF10A cells ectopically expressing SIRT2-V5 or control lacZ. (Right) Quantification of relative Slug levels from five independent experiments, normalized to Lamin A/C. Data are shown as mean ± SEM. * p < 0.05.
(B) (Left) Immunoblots showing levels of Slug, SIRT2, acetylated tubulin (a bona fide SIRT2 substrate) and Lamin A/C in MCF10A cells expressing two different hairpins targeting SIRT2. (Right) Quantification of relative Slug levels from three independent experiments, normalized to Lamin A/C. Data are shown as mean ± SEM. ** p < 0.01.
(C) (Top) Immunoblots showing levels of Slug, SIRT2-V5, Cyclin D1 and Lamin A/C in CHX-treated MCF10A cells overexpressing SIRT2-V5 or control LacZ. (Bottom) Quantification of relative Slug levels from three independent experiments, normalized to Lamin A/C. Data are shown as mean ± SEM. * p < 0.05.
(D) (Top) Immunoblot showing levels of Slug, SIRT2-V5 and Lamin A/C in CHX-treated MCF10A cells expressing either shSIRT2 or a non-silencing shRNA control (shCtrl). (Bottom) Quantification of relative Slug levels from three independent experiments, normalized to Lamin A/C. Data are shown as mean ± SEM. * p < 0.05.
(E) Expression of Slug target genes EPCAM and CDH1 in MCF10A cells overexpressing SIRT2 or LacZ (n = 3) as determined by qRT-PCR. Data are shown as mean ± SEM. * p < 0.05.
(F) Expression of Slug target genes EPCAM and CDH1 in MCF10A cells expressing shSIRT2 or shCtrl (n = 3) as determined by qRT-PCR. Data are shown as mean ± SEM. ** p < 0.01.
We next examined whether SIRT2 regulates Slug protein turnover. MCF10A cells overexpressing SIRT2 exhibited prolonged Slug stability, with high protein levels up to 4 hours following CHX treatment (Fig 2C, * p < 0.05). By contrast, Slug protein abundance was rapidly reduced and became minimally detectable after 4 hours in control LacZ-expressing MCF10A cells (Fig 2C). In addition, SIRT2 knockdown promoted rapid destabilization of Slug, such that the protein promptly disappeared by 1 hour post-CHX treatment (Fig 2D, * p < 0.05). We observed that the half-life of Slug was prolonged from 80 to 240 minutes in SIRT2-overexpressing MCF10A cells, and shortened to 40 minutes in SIRT2-depleted MCF10A cells (Fig 2C & D). Importantly, the differences in Slug stability were specific to genetic manipulation of SIRT2 levels, and not due to a general effect on protein degradation since cyclin D1 was still degraded normally (Fig 2C).
To determine whether Slug repressor activity is affected by the changes in Slug protein levels mediated by SIRT2, we examined the expression of Slug target genes following SIRT2 perturbation. Elevated Slug protein caused by SIRT2 overexpression corresponded to stronger repression of the Slug transcriptional targets, EPCAM and CDH1 (Fig 2E, * p < 0.05). Likewise, in SIRT2-knockdown cells where Slug protein levels were diminished, de-repression of these Slug target genes was observed (Fig 2F, ** p < 0.01). Thus, the ability of SIRT2 to regulate Slug abundance directly influences the bioavailability of Slug protein and the expression of its downstream transcriptional targets. Taken together, these data demonstrate that SIRT2 regulates Slug protein abundance, stability and function.
SIRT2 interacts with and deacetylates Slug
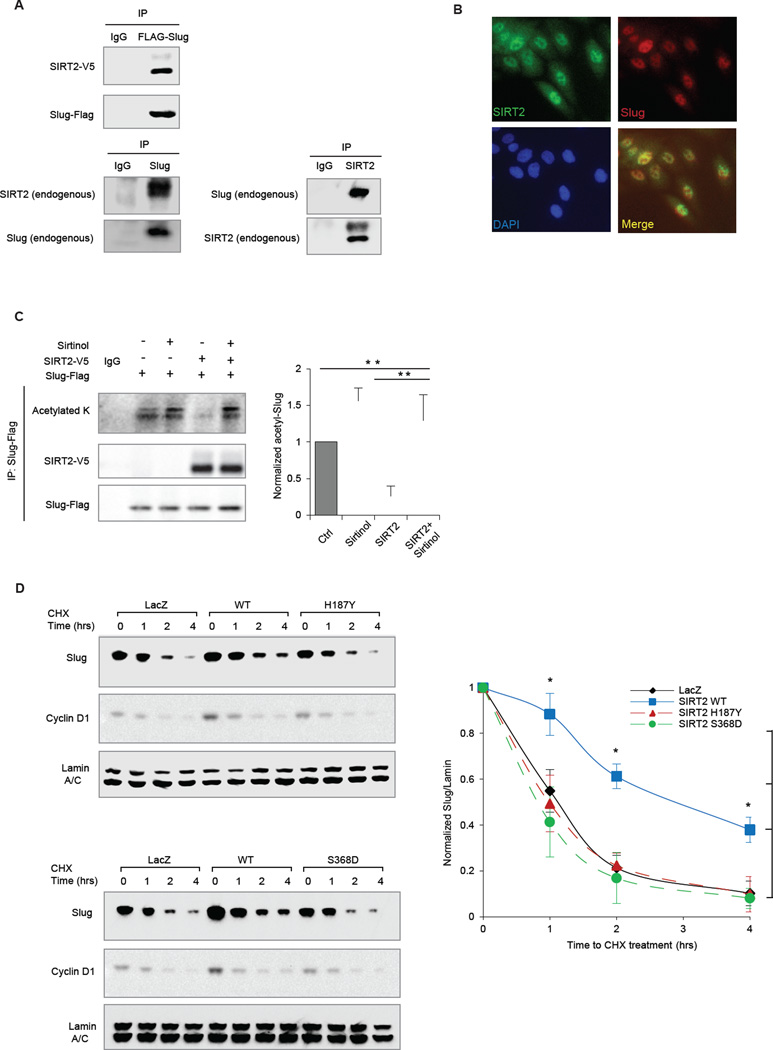
Since experimental manipulation of SIRT2 expression levels specifically alters Slug protein stability, we next sought to determine whether Slug might directly interact with and be a substrate of SIRT2. Accordingly, reciprocal co-immunoprecipitation (co-IP) was performed in HEK293T cells that ectopically expressed Flag-tagged Slug and V5-tagged SIRT2. Indeed, Slug co-immunopurified with SIRT2 (Fig 3A). Likewise, reciprocal co-IPs between endogenous SIRT2 and endogenous Slug in MCF10A were also observed (Fig 3A). We further ruled out the possibility of non-specific interaction between SIRT2 and Slug mediated through chromatin by performing the co-IP in the presence of DNase (Fig S3A & B). Notably, Slug failed to co-IP with SIRT1, indicating a specific interaction with SIRT2 (Fig S2C). Consistent with these findings, dual-immunofluorescence (IF) staining of endogenous Slug and SIRT2 proteins revealed their nuclear co-localization in MCF10A cells (Fig 3B). Together, these results show that SIRT2 physically interacts with Slug protein, suggesting that Slug may be a substrate for the enzymatic activity of the deacetylase SIRT2.
Figure 3. SIRT2 binds to and deacetylates Slug.
(A) (Top) Lysates from HEK293T cells transfected with Flag-tagged Slug (Flag-Slug) and V5-tagged SIRT2 (SIRT2-V5) were subjected to anti-Flag immunoprecipitation. Immunoblots of Flag-Slug and SIRT2-V5 are shown. (Bottom) Lysates from MCF10A were subjected to anti-Slug immunoprecipitation (left) and anti-SIRT2 immunoprecipitation (right). Immunoblots of endogenous Slug and SIRT2 are shown.
(B) Immunofluorescence of Slug (Red) and SIRT2 (Green) in MCF10A cells. DAPI (Blue) was used to visualize nuclei.
(C) (Left) Anti-Flag immunoprecipitation of lysates from HEK293T cells, overexpressing Flag-Slug and/or SIRT2-V5, and treated with sirtinol or vehicle control were immunoblotted for panacetylated lysine, Flag-Slug and SIRT2-V5. (Right) Relative levels of acetylated Slug protein from three independent experiments, normalized to immunoprecipitated Flag-Slug. Data are shown as mean ± SEM. ** p < 0.01.
(D) (Left) Immunoblots showing levels of Slug, Cyclin D1 and Lamin A/C in CHX-treated MCF10A cells expressing control LacZ, wild-type SIRT2 or catalytically inactive SIRT2 mutants H187Y (top) and S368D (bottom). (Right) The degradation curves of relative Slug protein from three independent experiments are normalized to Lamin A/C and plotted for cells overexpressing LacZ (black), wild-type SIRT2 (blue), or SIRT2 mutants H187Y and S368D (red and green, respectively). Data shown are mean ± SEM. * p < 0.05.
SIRT2, like other members of the mammalian sirtuin family, regulates its substrate proteins by directly modulating their acetylation state. Given our finding that acetylation targets Slug protein for degradation (Fig 1), we asked whether SIRT2 constitutes an integral component of this regulatory mechanism. To determine whether SIRT2 deacetylates Slug, we first examined Slug acetylation status following SIRT2 inhibition by sirtinol. This treatment resulted in the hyper-acetylation of Slug relative to untreated cells (Fig 3C). Similarly, SIRT2 depletion using two independent short hairpin RNAi constructs promoted acetylation of Slug protein (Fig S4B). By contrast, when we overexpressed SIRT2 in untreated cells, a marked decrease in Slug acetylation was evident (Fig 3C, p <0.01). However, treatment of SIRT2-overexpressing cells with sirtinol restored the hyper-acetylated status of Slug (Fig 3C, p <0.01). Since protein acetylation and ubiquitination are often linked and act in concert to influence protein degradation (Jiang et al., 2011; Wang et al., 2012), we next examined whether de-acetylation of Slug by SIRT2 could affect Slug ubiquitination and degradation. Indeed, SIRT2 overexpression markedly reduced Slug ubiquitination (Fig S3C). Conversely, enzymatic inhibition of SIRT2 by sirtinol treatment promoted Slug ubiquitination (Fig S3C).
To specifically determine whether the deacetylase activity of SIRT2 is necessary for regulating Slug protein levels, we created single amino acid substitutions within the catalytic domain of SIRT2 to produce deacetylase-defective mutants (North and Verdin, 2007; North et al., 2003). In cells overexpressing these mutant proteins, Slug acetylation was significantly higher than in cells overexpressing WT SIRT2 (Fig S3D). Importantly, overexpression of the deacetylase-defective SIRT2 H187Y and S368D mutants failed to stabilize Slug (Fig 3F, * p < 0.05). Consistently, overexpression of wild-type SIRT2, but not the deacetylation-defective mutants, resulted in increased Slug abundance at the steady state (Fig S3E). Furthermore, the SIRT2 mutants failed to appreciably alter Slug target gene expression (Fig S3F). Collectively, these data show that SIRT2 deacetylase activity can alter the acetylation state of Slug and coordinate its ubiquitination, thereby controlling Slug stability and activity.
Identification of K116 as an acetylated Slug substrate of SIRT2
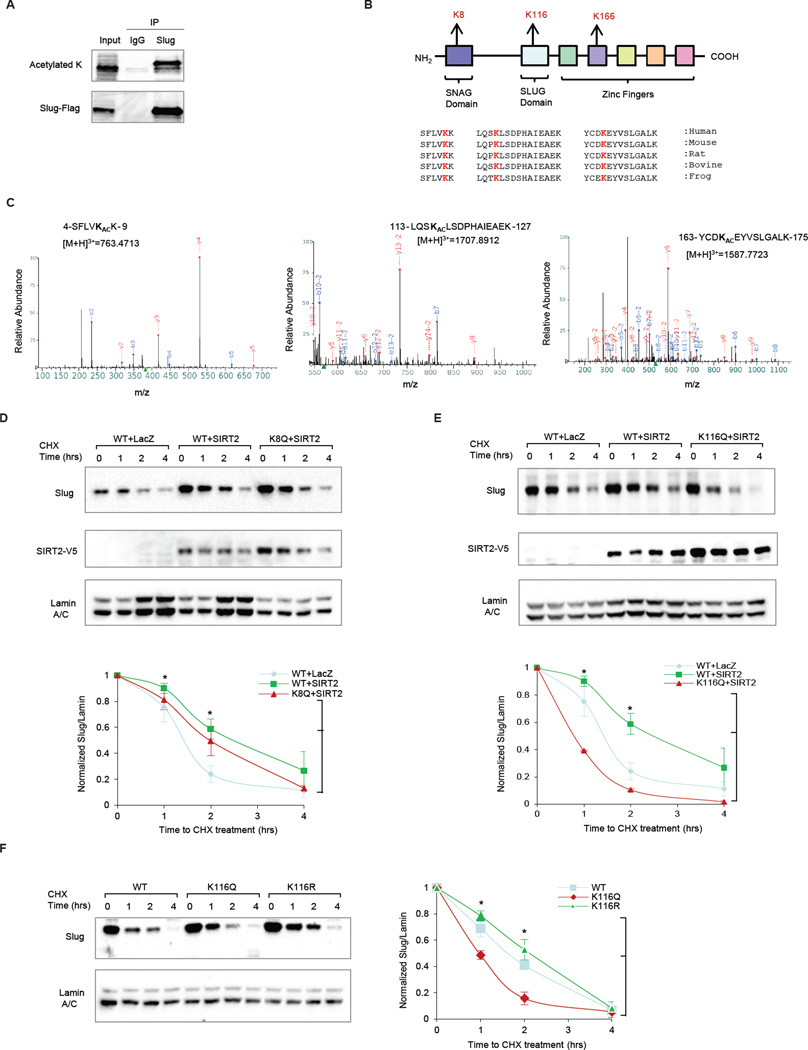
To further validate our findings, we searched for acetylated residues of Slug by immunoprecipitating and subjecting Slug protein to proteolytic digestion and liquid chromatography-tandem mass spectrometry analysis (LC-MS/MS) (Fig 4A & S4A). Using this approach, we identified acetylated residues at K8, K116 and K166 in the SNAG, SLUG and zinc finger domains, respectively (Fig 4A–C). Sequence alignment revealed that all three acetylation sites are highly conserved across species (Fig 4B).
Figure 4. Identification of K116 as an acetylated Slug substrate of SIRT2.
(A) Anti-FLAG immunoprecipitation of HEK293T cells overexpressing FLAG-Slug, followed by pan-acetylated lysine blotting detects acetylated Slug, which was digested and analyzed by mass-spectrometry.
(B) Schematic summarizes the three identified acetylated residues in relationship to functional domains of Slug. Sequence alignment analysis shows that all three acetylated residues are highly conserved across different species.
(C) LC/MS/MS spectrums are displayed for each of the three acetylated Slug residues.
(D) (Top) Immunoblots comparing the effect of SIRT2 overexpression on wild-type (WT) Slug versus an acetylation-mimic K8Q Slug mutant in MCF10A cells. Immunoblot of SIRT2-V5 is included to show SIRT2 overexpression. (Bottom) The degradation curves of relative Slug protein from three independent experiments are normalized to Lamin A/C and plotted for cells overexpressing WT Slug + LacZ control (blue), WT Slug + SIRT2 (green), and K8Q Slug mutant + SIRT2 (red). Data shown are mean ± SEM. * p < 0.05.
(E) (Top) Immunoblots comparing the effect of SIRT2 overexpression in WT versus an acetylation-mimic K116Q Slug mutant in MCF10A cells. Immunoblot of V5-SIRT2 is included to show SIRT2 overexpression. (Bottom) The degradation curves of relative Slug protein from three independent experiments are normalized to Lamin A/C and plotted for cells overexpressing WT Slug + LacZ control (blue), WT Slug + SIRT2 (green), and K116Q Slug mutant + SIRT2 (red). Data shown are mean ± SEM. * p < 0.05.
(F) (Left) Immunoblots showing the stability of the acetylation-mimic K116Q Slug mutant and the acetylation-resistant K116R Slug mutant in MCF10A cells. (Right) The degradation curves of relative Slug protein from three independent experiments are normalized to Lamin A/C and plotted for cells overexpressing WT Slug (blue), K116R Slug mutant (green), and K116Q Slug mutant (red). Data shown are mean ± SEM. * p < 0.05.
By virtue of their locations in the regulatory SNAG and functionally unknown SLUG domains, we decided to focus our investigation on whether K8 or K116 are substrates of SIRT2. We deemed that K166, found in the zinc finger domain that functions in DNA binding, would be unlikely to affect Slug protein degradation. To investigate whether K8 or K116 are substrates of SIRT2, we constructed acetyl-lysine mimic (KQ) Slug mutants at these sites and assessed their impact on Slug stability mediated by SIRT2 in MCF10A cells. SIRT2 overexpression stabilized the K8Q Slug mutant in a similar fashion to wild-type Slug (Fig 4D, * p < 0.05), suggesting that K8 acetylation is not required for the SIRT2-dependent regulation of Slug. In contrast, SIRT2 overexpression failed to stabilize the K116Q Slug mutant (Fig 4E, * p < 0.05). Furthermore, SIRT2 knockdown failed to destabilize a non-acetyl-lysine mimic (K116R) Slug mutant (Fig S4C, * p < 0.05). Together these data suggest that the K116 residue in the SLUG domain represents a critical deacetylation target required for mediating SIRT2-driven Slug stabilization.
To further corroborate this finding, we studied the direct effect of reversible acetylation at K116 on Slug protein stability using acetyl-lysine mimic (K116Q) and non-acetyl-lysine mimic (K116R) Slug mutants, without altering wild-type SIRT2 levels. Compared to wild-type Slug, the acetylation-mimic K116Q mutant was rapidly degraded, whereas the degradation of the acetylation-resistant K116R mutant was attenuated in MCF10A cells (Fig 4F, * p < 0.05). Thus, the mutational analysis of the K116 residue of Slug partially phenocopies the Slug stability results generated using acetylation inhibitors and the genetic manipulation of SIRT2. Taken together, these results suggest that reversible acetylation of K116 is sufficient to regulate Slug proteolytic turnover, and further elucidate the mechanistic framework of how SIRT2 regulates Slug.
SIRT2 is highly amplified and expressed in human BLBC
Given our discovery that SIRT2 is an essential regulator of Slug homeostasis in the normal breast epithelium, we evaluated the relevance of this regulation to human breast cancer. Clinically, breast tumors are commonly classified into distinct subtypes based on their intrinsic molecular or histological features. Slug protein overabundance is frequently observed in the BLBC molecular subtype, which histologically exhibits basal differentiation with infiltrating ductal features and is predominantly associated with triple-negative tumors (estrogen-receptor, progesterone receptor and HER2 negative) (Foulkes et al., 2010; Proia et al., 2011; Rakha et al., 2008; Storci et al., 2008). We therefore assessed whether SIRT2 expression is altered in specific breast cancer subtypes.
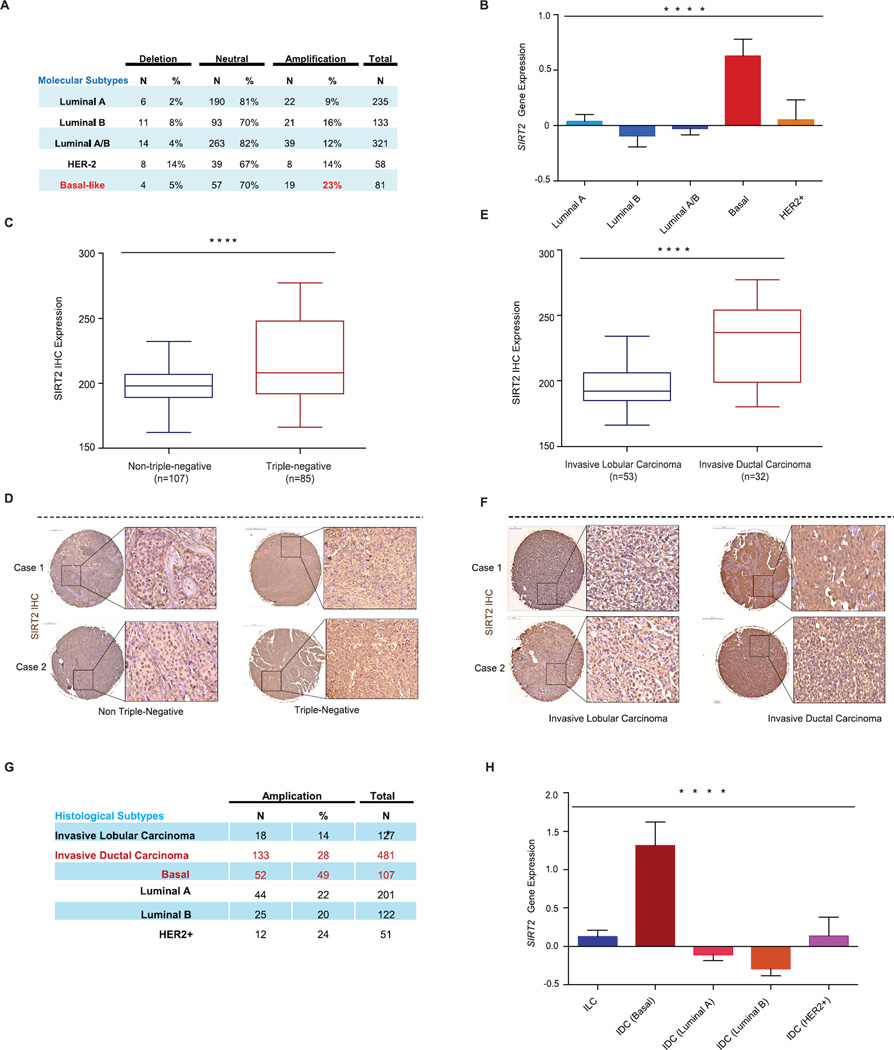
Analyzing The Cancer Genome Atlas (TCGA) dataset, we first examined SIRT2 somatic copy number status and gene expression in tumor samples from nearly 1,000 breast cancer cases (Cerami et al., 2012). Molecular subtype classification revealed that SIRT2 is frequently amplified in BLBCs (23%) compared to luminal A and luminal B tumors (9% and 16%, respectively) (Fig 5A, **** p < 0.001). In addition, SIRT2 mRNA was markedly elevated in basal-like tumors when compared to other subtypes of breast cancer (Fig 5B, **** p < 0.001).
Figure 5. SIRT2 expression is elevated in BLBCs.
(A) SIRT2 somatic copy number analysis by TCGA across the molecular subtypes of breast cancer. Data were analyzed using two-tailed unpaired student’s t-tests, p-values for basal vs luminal A=0.011, basal vs. luminal B=0.031, basal vs. Luminal A/B=0.488, basal vs. HER2+=0.040.
(B) SIRT2 gene expression across the molecular subtypes of breast cancer from the TCGA data set. Data were analyzed using one-way ANOVA, Data shown are mean ± SEM. **** p<0.0001.
(C) Immunohistochemical analysis of SIRT2 protein expression in tissue microarrays containing 192 breast tumor samples. Shown is the comparison of SIRT2 expression between triplenegative and non-triple-negative breast cancer cases. Data were analyzed using two-tailed unpaired student’s t-test. **** p < 0.0001.
(D) Representative SIRT2 immunohistochemical staining from triple-negative and non-triple-negative breast cancer cases are shown.
(E) Triple-negative (TN) cases were further stratified by histological classification, and shown is the comparison of SIRT2 expression between TN-invasive lobular carcinoma and TN-invasive ductal carcinoma. Data were analyzed using two-tailed unpaired student’s t-test. **** p<0.0001.
(F) Representative SIRT2 immunohistochemical staining from TN-invasive lobular carcinoma and TN-invasive ductal carcinoma are shown.
(G) SIRT2 somatic copy number analysis by TCGA in invasive lobular carcinoma (ILCs) and invasive ductal carcinoma (IDCs) subclasses. Data were analyzed using two-tailed unpaired student’s t-tests, p-values for: ILC vs. IDC (basal) =0.001, ILC vs. IDC (luminal A) =0.001, ILC vs. IDC (luminal B) =2.72 × 10−6, ILC vs. IDC (HER2+) =0.001.
(H) SIRT2 gene expression from the TCGA data set for the invasive lobular carcinoma (ILCs) and invasive ductal carcinoma (IDCs) subclasses. Data were analyzed using one-way ANOVA, Data shown are mean ± SEM. **** p<0.0001.
To corroborate to these findings, we examined SIRT2 protein levels in 192 breast cancer cases by immunohistochemistry (IHC). Patient stratification based on receptor status revealed that SIRT2 levels were higher in triple negative breast cancer cases, when compared to the non-triple negative breast cancer cases (estrogen-receptor, progesterone receptor and/or HER2 positive tumors) (Fig 5C–D, **** p < 0.001). Interestingly, among the triple-negative cohort, further stratification based on histological subtypes revealed a striking difference in SIRT2 protein level among invasive lobular carcinoma (ILC) and invasive ductal carcinoma (IDC) (Fig 5E–F, **** p < 0.001). SIRT2 protein was abundantly expressed in triple-negative tumors with IDC features, a poorly differentiated histological subtype most commonly associated with highly expressed Slug (Martin et al., 2005; Prasad et al., 2009). In contrast, triple-negative tumors with ILC features showed lower SIRT2 protein expression, at levels comparable to non-triple-negative tumors (Fig 5E & F). Consistent with the SIRT2 protein level results, further analysis of TCGA data revealed that SIRT2 copy number amplification most frequently occurs in IDC, especially in the basal subtypes (49%) (Fig 5G, p = 0.001). Furthermore, SIRT2 gene expression is overall highly expressed in IDC compared to ILC (Fig 5H, **** p < 0.001).
SIRT2 is necessary for Slug overabundance in BLBC cells
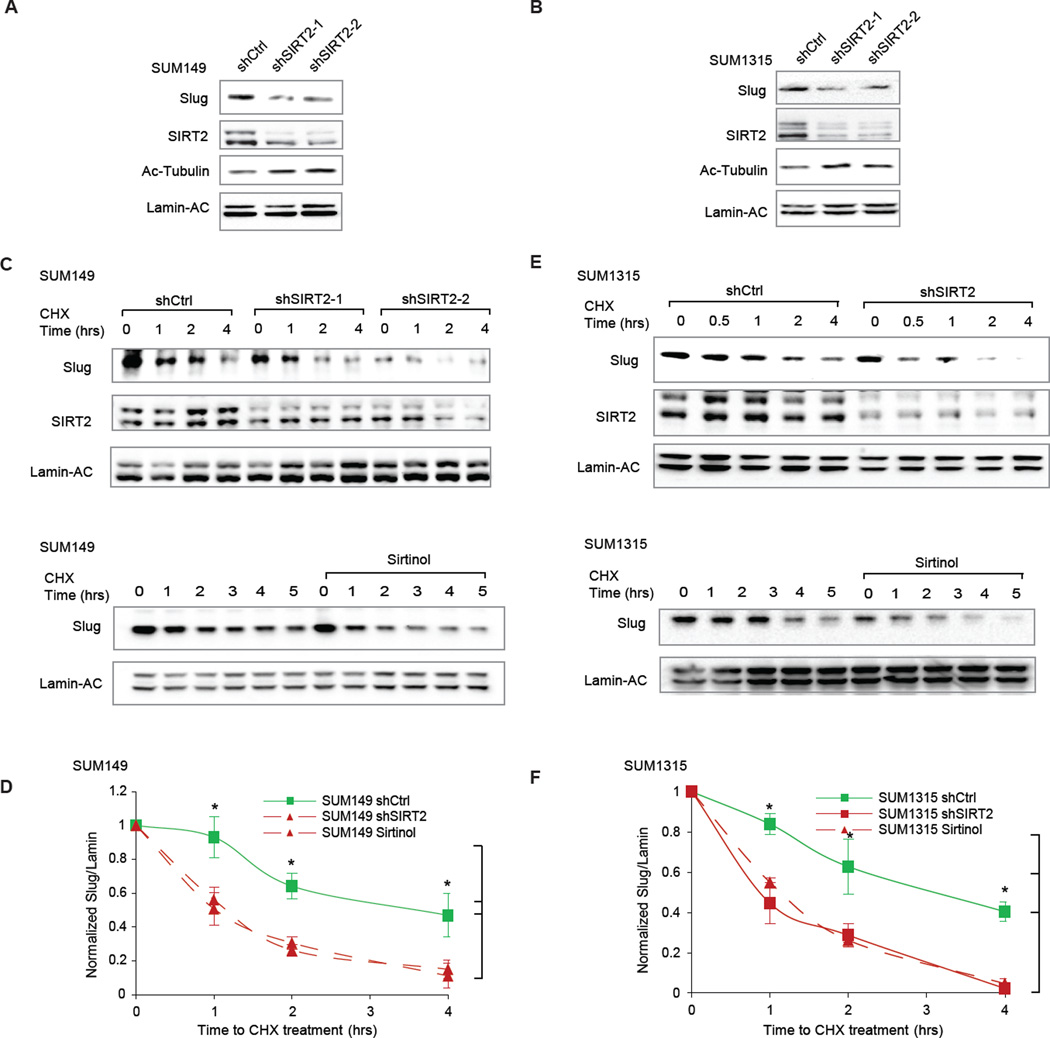
Considering the frequent overexpression of SIRT2 in invasive ductal BLBCs, we asked whether inhibiting SIRT2 would cause Slug protein levels to diminish in BLBC cell lines. For these experiments, we identified two BLBC cancer cells (SUM149 & SUM1315) derived from invasive ductal carcinomas that exhibit robust Slug expression and stability (Proia et al., 2011). Indeed, targeting SIRT2 by two independent short hairpin RNAi constructs (shRNA) greatly diminished Slug protein levels in both cell lines (Fig 6A–B).
Figure 6. SIRT2 is required for Slug stability in BLBCs.
(A) Immunoblots showing the effect of SIRT2 knockdown on Slug protein abundance in SUM149 BLBC cells, compared to a non-targeting hairpin control (shCtrl). Acetylated-tubulin levels are also shown to assess SIRT2 deacetylation activity.
(B) Immunoblots showing the effect of SIRT2 knockdown on Slug protein abundance in SUM1315 BLBC cells, compared to a non-targeting hairpin control (shCtrl). Acetylated-tubulin levels are also shown to assess SIRT2 deacetylation activity.
(C) (Top) Immunoblots showing Slug degradation in SIRT2-depleted SUM149 cells, compared to a non-targeting hairpin control (shCtrl). (Bottom) Immunoblots showing the effect of SIRT2 inhibition by sirtinol on Slug degradation in SUM149 cells, compared to DMSO vehicle treatment.
(D) Quantification from three separate experiments shows the effect of genetic silencing or pharmacological inhibition of SIRT2 in SUM149 cells. Data shown are mean ± SEM. * p < 0.05.
(E) (Top) Immunoblots showing Slug degradation in SIRT2-depleted SUM1315 cells, compared to a non-targeting hairpin control (shCtrl). (Bottom) Immunoblots showing the effect of SIRT2 inhibition by sirtinol on Slug degradation in SUM1315 cells, compared to DMSO vehicle treatment.
(F) Quantification from three separate experiments shows the effect of genetic silencing or pharmacological inhibition of SIRT2 in SUM1315 cells. Data shown are mean ± SEM. * p < 0.05.
Notably, the half-lives of Slug in these BLBC cells are longer than those in normal epithelial cells (Proia et al., 2011) (Fig 6 E–F & Fig 1A); we therefore investigated whether SIRT2 is necessary for the aberrant stabilization of Slug protein using these BLBC cells. SIRT2 knockdown caused rapid degradation of Slug protein, effectively abolishing extended Slug stability (Fig 6C and 6E). Similarly, pharmacological inhibition of SIRT2 also led to higher degradation rates for Slug, resembling the normal Slug turnover kinetics in MCF10A cells (Fig 6D and 6F & Fig 1A). Collectively, these results demonstrate that SIRT2 is necessary for Slug overabundance in BLBC.
Silencing SIRT2 causes the loss of aggressive BLBC features and inhibits tumor growth
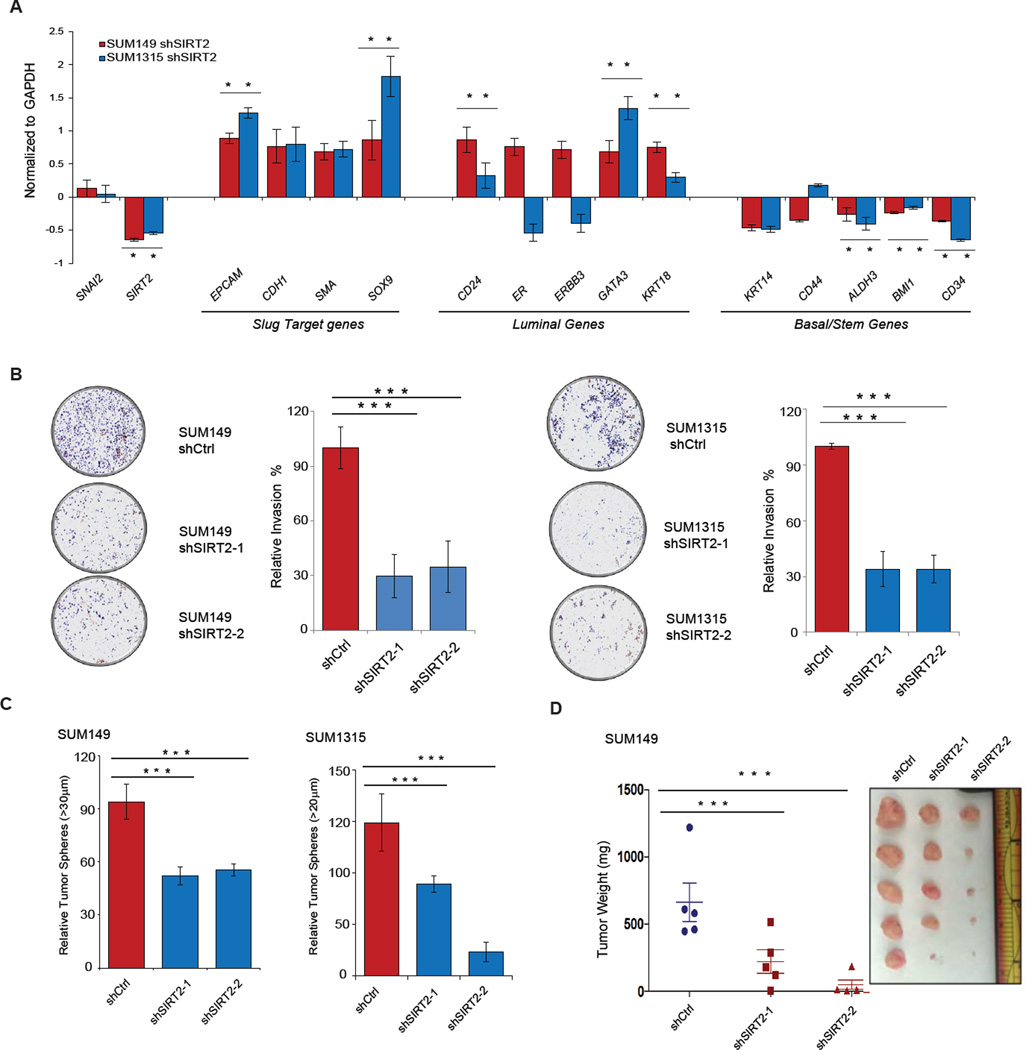
Through regulation of transcriptional programs that orchestrate basal differentiation, stem cell activity, cellular motility and invasiveness, Slug enables aggressive malignant characteristics manifested by BLBCs (Kao et al., 2014; Phillips et al., 2014; Proia et al., 2011; Samanta et al., 2016). We hypothesized that SIRT2 may also regulate the BLBC phenotype through its effect on Slug. Given the importance of high Slug expression as a determinant of basal differentiation status (Phillips et al., 2014; Proia et al., 2011; Storci et al., 2008), we first examined whether perturbation of SIRT2 might alter the differentiation status of BLBC. Following SIRT2 depletion in the BLBC cell lines SUM149 and SUM1315, we interrogated the expression levels of luminal and basal differentiation genes regulated by Slug. Increased expression of several luminal differentiation markers, including GATA3, KRT18 and CD24 were observed upon SIRT2 inhibition, and concomitant repression of basal- and stem cell-associated markers were also apparent (Fig 7A, * p < 0.05).
Figure 7. Silencing SIRT2 causes the loss of aggressive BLBC features in cancer cells.
(A) qRT-PCR analysis of expression levels of Slug target genes as well as luminal and basal differentiation markers following SIRT2 silencing in SUM149 (red) and SUM1315 (blue) BLBCs. Genes differentially expressed are represented as a log2 fold change over the non-targeting shCtrl cell lines. Data shown are mean ± SEM. * p < 0.05.
(B) (Left) Representative images showing the effect of SIRT2 silencing on the invasive capacity of SUM149 and SUM1315 cancer cells, compared to non-targeting hairpin control (shCtrl). (Right) The total number of cells invading through the Matrigel coated transwell were quantified (n = 3 per cell line). Data are shown as mean ± SEM. *** p < 0.001
(C) Tumor sphere-forming ability of SUM149 and SUM1315 cells following SIRT2 depletion, compared to a non-targeting hairpin control (shCtrl). Quantification is from three independent experiments. Data are shown as mean ± SEM. *** p < 0.001.
(D) Quantification of tumor mass (left panel) and photograph (right panel) of SUM149 cells grown as orthotopic xenografts in NOD/SCID mice. Tumors with two different hairpins targeting SIRT2 or a non-targeting hairpin control (shCtrl) are indicated. Data are plotted as individual data points from the five animals of each group. The mean ± SEM are shown, and comparisons between groups were performed using two tailed Student’s t-tests. *** p < 0.001.
The predilection to early metastasis is a key aspect of the poor prognosis in BLBC (Anders and Carey, 2009; Kennecke et al., 2010). Slug is a well-recognized inducer of the epithelial-mesenchymal transition (EMT), enabling the dissemination and invasion of cancer cells (Bolós et al., 2003; DiMeo et al., 2009; Kim et al., 2014; Taube and Herschkowitz, 2010). We therefore examined whether SIRT2 perturbation alters the invasive phenotypes of BLBC. Matrigel invasion assays were performed using SUM149 and SUM1315 cells that were depleted of SIRT2. SIRT2-depleted cells exhibited significantly reduced invasive capabilities (Fig 7B, *** p < 0.001), correlating with the diminished Slug protein levels in these cells (Fig 6). Importantly, the dampening of invasion in SIRT2-depleted cells can be rescued by overexpressing wild-type Slug or its non-acetyl-lysine K116R variant (Fig S5A & B). Surprisingly, we observed that overexpression of the acetyl-lysine mimic K116Q variant also rescues invasion (Fig S5A & B), but one caveat is that driving overexpression of the K116Q variant may outweigh its decrease in stability. At the molecular level, the reduction in invasion in SIRT2-deficient cells was accompanied by an up-regulation of E-Cadherin and down-regulation of Vimentin, two Slug targets that are canonical EMT markers associated with cancer metastasis (Fig S5C & D) (Hajra el al., 2002, Thiery el al., 2012). Indeed, high SIRT2 expression clinically correlates with metastatic recurrence (Vijver et al., 2002) (Fig S5E, * p < 0.05).
Another essential feature underlying the aggressiveness of BLBC is robust self-renewal and tumor-initiating capability, contributing to therapeutic resistance and early recurrence (Fillmore and Kuperwasser, 2008b; Foulkes et al., 2010; Metzger-Filho et al., 2012; Rakha et al., 2008). Since Slug is a major regulator of the stem cell state and contributes to the genesis and progression of BLBC, we examined whether the loss of SIRT2 might affect Slug-associated, stem-like features in BLBC (Guo et al., 2012; Proia et al., 2011; Chang et al., 2016). Indeed, SIRT2 silencing in SUM1315 and SUM149 cells markedly inhibited their ability to form tumor spheres in suspension, a phenotypic assay commonly employed to assess the self-renewal and tumor-initiating characteristics of cancer cells (Ponti et al., 2005) (Fig 7C, *** p < 0.001).
We have previously shown that SNAI2/Slug knockout mice are highly resistant to mammary tumorigenesis (Cobaleda et al., 2006; Guo et al., 2012; Nassour et al., 2012; Phillips et al., 2014b). Given our finding that SIRT2 depletion and the consequent abrogation of Slug stabilization causes the loss of tumor sphere-forming capability of BLBC cells, we addressed the in vivo relevance of SIRT2 inhibition in BLBC by examining tumorigenesis. SIRT2-depleted BLBC cancer cells (SUM149) were orthotopically implanted into the mammary fat pad of female NOD/SCID mice, and tumor growth was monitored over a sixteen-week timespan. Over this period, SIRT2 inhibition significantly impaired the growth of tumor xenografts (Fig 7D, *** p < 0.001). In aggregate, these data demonstrate that SIRT2 inhibition dampens the cancer-associated activities mediated by Slug in BLBC
Discussion
Slug/SNAI2 is a labile protein that is strictly regulated in normal tissues; however, disruption of this regulation appears to be a widespread phenomenon in human epithelial malignancies, and manifests as Slug overabundance in BLBC. In this study, we identified acetylation as a regulatory mechanism that governs Slug stability in human mammary epithelial cells. We elucidated the role of SIRT2 as a major regulator of Slug stability via its ability to deacetylate the K116 residue of the SLUG domain. Together with frequent SIRT2 overexpression in human BLBC, our findings describe a molecular pathway that contributes to the abnormal Slug stabilization frequently observed in this aggressive subtype of breast cancer and therefore may provide a rational therapeutic avenue for targeting BLBC tumors.
Our study has shown that Slug is acetylated and that post-translational acetylation of Slug acts to promote degradation and thereby limit Slug abundance. While the paralogous protein Snail has been shown to be acetylated in the zinc finger region to limit its transcriptional activity (Hsu el al., 2014), acetylation has not been reported for Slug. Interestingly, our mutational analysis pinpointed the K116 residue of the SLUG domain, a region not shared with Snail, as the principle regulatory site for Slug stability and SIRT2-mediated deacetylation. Thus, our results elucidate an important functionality of the SLUG domain. Accordingly, we speculate that other factors such as acetyltransferases might also be involved in, and cooperate with, SIRT2 to regulate Slug protein stability. Collectively, these data expand upon the known regulatory repertoire by which phosphorylation and ubiquitination coordinately regulate Slug protein stability (Wu 2012, Kao 2014), raising intriguing questions regarding the complex interplay among these post-translational modifications (PTMs) and how they collectively regulate Slug protein abundance. For example, do acetylation and multiple PTMs operate in a serial or parallel fashion? Is the interaction between acetylation and other PTMs cooperative or antagonistic in nature? Previous work has shown that acetylation can promote E3 ligase-mediated ubiquitination in multiple proteins subject to rapid turnover (Jiang et al., 2011; Liu et al., 2013b; Wang et al., 2012). In some contexts, acetylation also regulates subcellular localization to facilitate proteasomal degradation of the target protein (Fujita et al., 2015; Song et al., 2015). Future studies are needed to fully understand how acetylation is integrated into the regulatory network regulating Slug abundance and its downstream biological effects.
The transcriptional repressor Slug/SNAI2 is frequently deregulated in tumor cells, resulting in extended Slug stability. While this ultimately drives transcriptional programs that confer tumor-initiating and invasive capabilities, resistance to apoptosis and therapeutic failure (Chang et al., 2011; Guo et al., 2012; Kurrey et al., 2009; Wang et al., 2009; Wu et al., 2005), the molecular basis of Slug protein stabilization in cancer has remained unclear. Here we have elucidated a mechanism by which the protein deacetylase SIRT2 acts as a critical mediator of Slug protein stability. Although the oncogenic role of SIRT2 remains controversial and earlier evidence showed that SIRT2 loss mildly predisposed aged animals to mammary tumors (Kim el al., 2011), SIRT2 is highly expressed in multiple types of human cancer, and inhibition of SIRT2 generally exhibits anti-cancer effects (Zhang et al., 2009; Rotili et al., 2012; McCarthy et al., 2013; Hoffmann et al., 2014; Heltweg et al., 2006 ; Chen et al., 2013; Dan et al., 2012; Grbesa et al., 2015; Singh et al., 2015). Our data support this latter evidence, as we have found that SIRT2 is frequently overexpressed in the aggressive basal subtype of breast cancer, which is most strongly associated with the phenomenon of Slug overabundance. Notably, when we destabilized Slug by genetic depletion or pharmacological inhibition of SIRT2 in Slug-stabilizing BLBC cells, the cancer cells lost key aspects of BLBC character, including basal differentiation genes expression, tumor sphere-forming ability, and invasion. Collectively, these data connect the basic mechanistic regulation of Slug by SIRT2 to the context of Slug overabundance in BLBC tumors. Furthermore, in line with the emerging role of sirtuins in the proteolytic turnover of labile proteins (Jiang et al., 2011; Liu et al., 2013b; Wang et al., 2012), co-opting this molecular mechanism to aberrantly stabilize cancer drivers could be a general feature of cancer cells for adopting malignant behaviors.
From a therapeutic standpoint, our work presents important insights into molecular strategies to dampen Slug stabilization in BLBC. We provide evidence that SIRT2 inhibition accelerates proteasomal degradation of Slug, even in BLBC cells that exhibit extended Slug stability (Proia et al., 2011). Importantly, SIRT2 inhibition in these BLBC cells caused the loss of the basal differentiation phenotype, colony-forming ability, and invasiveness characteristic of aggressive BLBC, ultimately resulting in diminished tumorigenic capability. Interestingly, a recent study demonstrated that selective SIRT2 inhibition has broad anti-cancer effects, in part by promoting the proteasomal degradation of the c-Myc oncoprotein (Jing et al., 2016). Notably in this study, several BLBC cancer cells also exhibited sensitivity to SIRT2 inhibition despite no observable decrease of c-Myc, suggesting that destabilization of other cancer-relevant substrates of SIRT2, such as Slug, may contribute to this anti-cancer effect in BLBCs. Taken together with our findings, the pharmacological inhibition of SIRT2 may owe its remarkable anticancer property in BLBCs to its potent effect on essential tumorigenic substrates such as Slug and c-Myc. Overall, these findings motivate future preclinical studies to more rigorously address the in vivo efficacy of SIRT2 inhibition against BLBC formation and progression. Because SIRT2 regulates a number of protein substrates involved in diverse biological processes (Black et al., 2008; Liu et al., 2013b; North et al., 2003; Zhang et al., 2016; Zhao et al., 2014), an alternative targeting strategy such as peptide inhibitors that selectively block SIRT2 and Slug interaction may be more useful for these purposes. This rational approach, if successful, would significantly reshape our abilities to combat BLBC and improve the associated poor disease prospects.
Experimental Procedures
Cell Lines, Tissue Cultures and Chemical Treatments
Cell lines used in this study were purchased from ATCC (MCF10A and HEK293T), or obtained from Dr. Stephen Ethier (SUM149 and SUM1315). Additional tissue culture details are provided in Supplemental Experimental Procedures. For analysis of protein stability, cells were treated with the protein synthesis inhibitor cycloheximide (4 µg/ml, Sigma-Aldrich) followed by pulse-chase at indicated time points. Deacetylation inhibitors sodium butyrate (10 mM, Sigma-Aldrich), nicotinamide (5 µM, Sigma-Aldrich), trichostatin A (10 µM, Sigma-Aldrich), sirtinol (12.5 µM, Sigma-Alrich), deacetylation inhibitor cocktail (Santa Cruz) and Nedd8-activating enzyme inhibitor MLN4924 (10 mM, Calbiochem) were added to cells 6 hrs before harvest.
Cell Transfections, Immunoblotting and Immunoprecipitation
Full length SIRT1, SIRT2 and SNAI2 were cloned into plenti6.2-V5/DEST destination vector or pPGS-Flag vector. Point mutations for SIRT2 and Slug were generated by site-directed mutagenesis. Plasmid transfections were carried out using Fugene HD (Promega) according to the manufacturer’s protocol. To isolate protein from whole-cell lysis for immunoblotting, cells were lysed in RIPA buffer (10 mM Tris, 150 mM NaCl, 1 mM EDTA, 0.1% SDS) supplemented with protease inhibitor cocktail (Roche). For immunoprecipitation experiments, whole-cell protein lysates were pre-cleared by incubating with either Dynabeads (Life Technologies) or FLAG-M2 agarose beads (Sigma-Aldrich) for one hour, then incubated with antibodies overnight. Complexes were then bound to beads for two hours, washed three times with lysis buffer, and eluted by boiling in SDS loading buffer. Protein samples were separated by SDS-PAGE according to standard procedures, transferred onto a nitrocellulose membrane and blocked with 5% milk. Immunoblotting was performed according to standard procedures, and membranes were developed with either West Pico or West Dura ECL substrate (Pierce).
Acetylation Assay and Mass Spectrometry Analysis
Cultured cells were treated with 10 µm MG132 proteasome inhibitor (Sigma-Aldrich) and 1:100 dilution of deacetylation inhibitor cocktail (Santa Cruz) for 4 hours before harvest, then cells were lysed and washed in RIPA buffer supplemented with protease inhibitor cocktail (Roche), MG132 proteasome inhibitor and deacetylation inhibitor cocktail. Flag-Slug or endogenous Slug immunoprecipitation was performed and analyzed for acetylation by anti-acetylated lysine antibodies (Cell Signaling). For protein mass spectrometry, Slug IP lysates were separated by SDS-PAGE, fixed in the gel, and stained with Coomassie Blue. To identify acetylated residues in Slug protein, the gel slice containing the full-length Slug protein was excised, trypsin digested and analyzed by liquid chromatography-tandem mass spectrometry (Taplin Mass Spectrometry Facility, Harvard Medical School). For the identification of Slug-binding partner proteins, the gel was partitioned into five sections for trypsin digestion and downstream analysis.
Patient Tumor Arrays Analysis
The breast cancer tissue microarrays (TMAs) containing 192 cases of primary breast tumors with annotated receptor status were purchased from US Biomax (BR1921 and BR487). Each specimen consisted of a 5 µM thick core section which was immunostained with anti-SIRT2 antibodies (Sigma). Immunohistochemistry was performed using the avidin-biotin complex method (Vector Laboratories). Additional immunohistochemistry details are provided in the Supplemental Experimental Procedures. IHC staining was analyzed in a pathologist blinded fashion using automated quantitative imaging method (Panoramic scanner, 3D HISTECH). The two-tailed unpaired student’s t-test was used for statistical analysis of SIRT2 expression between patient cohorts.
Tumor Sphere, invasion and orthotopic tumor xenograft assays
For tumor sphere assays, 1 × 103 cells each of SUM149 and SUM1315 were seeded in 6-well Ultra-Low Attachment Surface tissue culture plates (Corning Life Sciences) and cultured for one week. Floating tumor spheres were collected and diluted in Isoton II (Beckman Coulter) and glycerol, and analyzed using Multisizer 3 cytometer (Beckman Coulter). Invasion assays were carried out in 24-well plate with 8 µm pre-coated Matrigel chamber inserts (Corning Life Sciences). 2 × 105 SUM cells were plated in serum free medium on top of the insert with SUM media containing 10% FBS at the bottom of the insert, and cultured for 24 hours. Chamber inserts were then washed, scraped, methanol fixed and stained with Crystal Violet. Invaded cells were visualized under the microscope and counted using ImageJ software (NIH). The invaded cells were normalized with 24-hr cell proliferation as measured by the CellTiter-Glo assay (Promega). For orthotopic tumor xenograft assays, all procedures were performed in accordance with the animal protocol approved by the Tufts University Institutional Animal Care and Use Committee. Prior to surgery, 10-week old, female NOD/SCID mice (The Jackson Laboratory) were anesthetized by isoflurane. An incision was made along the right flank to expose the inguinal mammary gland, and 2.0 × 106 cells in a total volume of 50µl 1:1 Matrigel: phosphate-buffered saline were injected into the gland. Post-operative analgesic and monitoring were provided. Animals were sacrificed 16 weeks after the surgery, and tumors were dissected and weighted.
Statistical Analysis
Data were analyzed and compared between groups using two-trailed Student’s t-tests and oneway ANOVA, and p < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank R.Tomaino and the Taplin Mass Spectrometry Facility at Harvard Medical School for mass spectrometry analysis; L. Arendt for technical assistance; and S.E. Herwald and G. Gill for critical reading of the manuscript. This work was supported by funding from the Raymond & Beverly Sackler Convergence Laboratory and grants from ArtBeCAUSE (to C.K.), American Cancer Society #PF-14-046-01-DMC (to T.K.N.), the Breast Cancer Research Foundation (to C.K.), and the NIH/NICDH HD073035 and NIH/NCI CA170851 (to C.K.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
W.Z., T.K.N. and C.K. conceptualized and designed the research; W.Z., A.W., B.G., A.S. performed the research; W.Z., T.K.N. and C.K. wrote the paper.
References
- Anders CK, Carey LA. Biology, metastatic patterns, and treatment of patients with triple-negative breast cancer. Clinical Breast Cancer. 2009 doi: 10.3816/CBC.2009.s.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black JC, Mosley A, Kitada T, Washburn M, Carey M. The SIRT2 deacetylase regulates autoacetylation of p300. Mol. Cell. 2008;32:449–455. doi: 10.1016/j.molcel.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolós V, Peinado H, Pérez-Moreno MA. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J. Cell. Sci. 2003 doi: 10.1242/jcs.00224. (2003) [DOI] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discovery. 2012 doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T-HH, Tsai M-FF, Su K-YY, Wu S-GG, Huang C-PP, Yu S-LL, Yu Y-LL, Lan C-CC, Yang C-HH, Lin S-BB, et al. Slug confers resistance to the epidermal growth factor receptor tyrosine kinase inhibitor. Am. J. Respir. Crit. Care Med. 2011;183:1071–1079. doi: 10.1164/rccm.201009-1440OC. [DOI] [PubMed] [Google Scholar]
- Chen J, Chan AW, To K-FF, Chen W, Zhang Z, Ren J, Song C, Cheung Y-SS, Lai PB, Cheng S-HH, et al. SIRT2 overexpression in hepatocellular carcinoma mediates epithelial to mesenchymal transition by protein kinase B/glycogen synthase kinase-3β/β-catenin signaling. Hepatology. 2013;57:2287–2298. doi: 10.1002/hep.26278. [DOI] [PubMed] [Google Scholar]
- Cobaleda C, Pérez-Caro M, Vicente-Dueñas C, Sánchez-García I. Function of the zinc-finger transcription factor SNAI2 in cancer and development. Annual Review of Genetics. 2006;41:41–61. doi: 10.1146/annurev.genet.41.110306.130146. [DOI] [PubMed] [Google Scholar]
- Cuzick J, Sestak I, Baum M, Buzdar A, Howell A. Effect of anastrozole and tamoxifen as adjuvant treatment for early-stage breast cancer: 10-year analysis of the ATAC trial. The Lancet Oncology. 2010 doi: 10.1016/S1470-2045(10)70257-6. [DOI] [PubMed] [Google Scholar]
- Dan L, Klimenkova O, Klimiankou M, Klusman J-HH, van den Heuvel-Eibrink MM, Reinhardt D, Welte K, Skokowa J. The role of sirtuin 2 activation by nicotinamide phosphoribosyltransferase in the aberrant proliferation and survival of myeloid leukemia cells. Haematologica. 2012;97:551–559. doi: 10.3324/haematol.2011.055236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMeo TA, Anderson K, Phadke P, Fan C, Feng C, Perou CM, Naber S, Kuperwasser C. A novel lung metastasis signature links Wnt signaling with cancer cell self-renewal and epithelial-mesenchymal transition in basal-like breast cancer. Cancer Res. 2009;69:5364–5373. doi: 10.1158/0008-5472.CAN-08-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Oh DS, Wessels L, Weigelt B. Concordance among gene-expression–based predictors for breast cancer. N. Engl. J. Med. 2006;355(6):560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008a doi: 10.1186/bcr1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008b;10:R25. doi: 10.1186/bcr1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N. Engl. J. Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- Fujita Y, Fujiwara K, Zenitani S, Yamashita T. Acetylation of NDPK-D Regulates Its Subcellular Localization and Cell Survival. PLoS ONE. 2015;10:e0139616. doi: 10.1371/journal.pone.0139616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbesa I, Pajares MJJ, Martínez-Terroba E, Agorreta J, Mikecin A-MM, Larráyoz M, Idoate MA, Gall-Troselj K, Pio R, Montuenga LM. Expression of sirtuin 1 and 2 is associated with poor prognosis in non-small cell lung cancer patients. PLoS ONE. 2015;10:e0124670. doi: 10.1371/journal.pone.0124670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, Itzkovitz S, Noske A, Zürrer-Härdi U, Bell G, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusterson B. Do “basal-like” breast cancers really exist? Nature Reviews. Cancer. 2009;9:128–134. doi: 10.1038/nrc2571. [DOI] [PubMed] [Google Scholar]
- Harris JR, Lippman ME, Osborne CK, Morrow M. Diseases of the Breast. 2012 [Google Scholar]
- Heltweg B, Gatbonton T, Schuler AD, Posakony J, Li H, Goehle S, Kollipara R, Depinho RA, Gu Y, Simon JA, et al. Antitumor activity of a small-molecule inhibitor of human silent information regulator 2 enzymes. Cancer Res. 2006;66:4368–4377. doi: 10.1158/0008-5472.CAN-05-3617. [DOI] [PubMed] [Google Scholar]
- Hoffmann G, Breitenbücher F, Schuler M, Ehrenhofer-Murray AE. A novel sirtuin 2 (SIRT2) inhibitor with p53-dependent pro-apoptotic activity in non-small cell lung cancer. J. Biol. Chem. 2014;289:5208–5216. doi: 10.1074/jbc.M113.487736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatiadis M, Singhal SK, Desmedt C. Gene modules and response to neoadjuvant chemotherapy in breast cancer subtypes: a pooled analysis. Journal of clinical Oncology. 2012 doi: 10.1200/JCO.2011.39.5624. (2012) [DOI] [PubMed] [Google Scholar]
- Jiang W, Wang S, Xiao M, Lin Y, Zhou L, Lei Q, Xiong Y, Guan K-LL, Zhao S. Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol. Cell. 2011;43:33–44. doi: 10.1016/j.molcel.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing H, Hu J, He B, Negrón Abril YL, Stupinski J, Weiser K, Carbonaro M, Chiang Y-LL, Southard T, Giannakakou P, et al. A SIRT2-Selective Inhibitor Promotes c-Myc Oncoprotein Degradation and Exhibits Broad Anticancer Activity. Cancer Cell. 2016;29:297–310. doi: 10.1016/j.ccell.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao S-HH, Wang W-LL, Chen C-YY, Chang Y-LL, Wu Y-YY, Wang Y-TT, Wang S-PP, Nesvizhskii AI, Chen Y-JJ, Hong T-MM. GSK3β controls epithelial-mesenchymal transition and tumor metastasis by CHIP-mediated degradation of Slug. Oncogene. 2014;33:3172–3182. doi: 10.1038/onc.2013.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennecke H, Yerushalmi R, Woods R. Metastatic behavior of breast cancer subtypes. Journal of clinical Oncology. 2010 doi: 10.1200/JCO.2009.25.9820. (2010) [DOI] [PubMed] [Google Scholar]
- Kim S, Yao J, Suyama K, Qian X, Qian BZ. Slug promotes survival during metastasis through suppression of Puma-mediated apoptosis. Cancer Research. 2014 doi: 10.1158/0008-5472.CAN-13-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurrey NK, Jalgaonkar SP, Joglekar AV, Ghanate AD, Chaskar PD, Doiphode RY, Bapat SA. Snail and slug mediate radioresistance and chemoresistance by antagonizing p53-mediated apoptosis and acquiring a stem-like phenotype in ovarian cancer cells. Stem Cells. 2009;27:2059–2068. doi: 10.1002/stem.154. [DOI] [PubMed] [Google Scholar]
- Lander R, Nordin K, LaBonne C. The F-box protein Ppa is a common regulator of core EMT factors Twist, Snail, Slug, and Sip1. The Journal of Cell Biology. 2011;194:17–25. doi: 10.1083/jcb.201012085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Zhang X, Shang M, Zhang Y, Xia B. Dysregulated expression of Slug, vimentin, and E-cadherin correlates with poor clinical outcome in patients with basal-like breast cancer. Journal of Surgical Oncology. 2013a doi: 10.1002/jso.23240. (2013) [DOI] [PubMed] [Google Scholar]
- Liu PY, Xu N, Malyukova A, Scarlett CJ, Sun YT, Zhang XD, Ling D, Su S-PP, Nelson C, Chang DK, et al. The histone deacetylase SIRT2 stabilizes Myc oncoproteins. Cell Death Differ. 2013b;20:503–514. doi: 10.1038/cdd.2012.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin TA, Goyal A, Watkins G, Jiang WG. Expression of the transcription factors snail, slug, and twist and their clinical significance in human breast cancer. Ann. Surg. Oncol. 2005;12:488–496. doi: 10.1245/ASO.2005.04.010. [DOI] [PubMed] [Google Scholar]
- McCarthy AR, Sachweh MC, Higgins M, Campbell J, Drummond CJ, Leeuwen IM, Pirrie L, Ladds M, Westwood NJ, Laín S. Tenovin-D3, a novel small-molecule inhibitor of sirtuin SirT2, increases p21 (CDKN1A) expression in a p53-independent manner. Molecular Cancer Therapeutics. 2013;12:352–360. doi: 10.1158/1535-7163.MCT-12-0900. [DOI] [PubMed] [Google Scholar]
- Menssen A, Hydbring P, Kapelle K, Vervoorts J, Diebold J, Lüscher B, Larsson L-GG, Hermeking H. The c-MYC oncoprotein, the NAMPT enzyme, the SIRT1-inhibitor DBC1, and the SIRT1 deacetylase form a positive feedback loop. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E187–E196. doi: 10.1073/pnas.1105304109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger-Filho O, Tutt A, de Azambuja E, Saini KS, Viale G, Loi S, Bradbury I, Bliss JM, Azim HA, Ellis P, et al. Dissecting the heterogeneity of triple-negative breast cancer. J. Clin. Oncol. 2012;30:1879–1887. doi: 10.1200/JCO.2011.38.2010. [DOI] [PubMed] [Google Scholar]
- Montserrat-Sentís B, Baulida J, lix Bonilla F. The Hypoxia-controlled FBXL14 Ubiquitin Ligase Targets SNAIL1 for Proteasome Degradation. J. Biol. Chem. 2009 Feb 5;285(6):3794–3805. doi: 10.1074/jbc.M109.065995. (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassour M, Idoux-Gillet Y, Selmi A, Côme C, Faraldo M-LML, Deugnier M-AA, Savagner P. Slug controls stem/progenitor cell growth dynamics during mammary gland morphogenesis. PLoS ONE. 2012;7:e53498. doi: 10.1371/journal.pone.0053498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North BJ, Verdin E. Mitotic regulation of SIRT2 by cyclin-dependent kinase 1-dependent phosphorylation. J. Biol. Chem. 2007;282:19546–19555. doi: 10.1074/jbc.M702990200. [DOI] [PubMed] [Google Scholar]
- North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- Perou CM, Sørlie T, Eisen MB, van de Rijn M. Molecular portraits of human breast tumours. Nature. 2000 doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Phillips S, Kuperwasser C. SLUG: Critical regulator of epithelial cell identity in breast development and cancer. Cell Adhesion & Migration. 2014 doi: 10.4161/19336918.2014.972740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips S, Prat A, Sedic M, Proia T, Wronski A. Cell-state transitions regulated by SLUG are critical for tissue regeneration and tumor initiation. Stem Cell Reports. 2014a doi: 10.1016/j.stemcr.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips S, Prat A, Sedic M, Proia T, Wronski A, Mazumdar S, Skibinski A, Shirley SH, Perou CM, Gill G, et al. Cell-state transitions regulated by SLUG are critical for tissue regeneration and tumor initiation. Stem Cell Reports. 2014b;2:633–647. doi: 10.1016/j.stemcr.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nature Reviews Cancer. 2009 doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- Ponti D, Costa A, Zaffaroni N, Pratesi G, Petrangolini G. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Research. 2005 doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- Prasad CP, Rath G, Mathur S, Bhatnagar D, Parshad R, Ralhan R. Expression analysis of E-cadherin, Slug and GSK3beta in invasive ductal carcinoma of breast. BMC Cancer. 2009;9:325. doi: 10.1186/1471-2407-9-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proia TA, Keller PJ, Gupta PB, Klebba I, Jones AD, Sedic M, Gilmore H, Tung N, Naber SP, Schnitt S, et al. Genetic predisposition directs breast cancer phenotype by dictating progenitor cell fate. Cell Stem Cell. 2011;8:149–163. doi: 10.1016/j.stem.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakha EA, Reis-Filho JS, Ellis IO. Basal-like breast cancer: a critical review. J. Clin. Oncol. 2008;26:2568–2581. doi: 10.1200/JCO.2007.13.1748. [DOI] [PubMed] [Google Scholar]
- Regan MM, Neven P, Giobbie-Hurder A, Goldhirsch A, Ejlertsen B, Mauriac L, Forbes JF, Smith I, Láng I, Wardley A. Assessment of letrozole and tamoxifen alone and in sequence for postmenopausal women with steroid hormone receptor-positive breast cancer: the BIG 1–98 randomised clinical trial at 8· 1 years median follow-up. The Lancet Oncology. 2011;12:1101–1108. doi: 10.1016/S1470-2045(11)70270-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotili D, Tarantino D, Nebbioso A, Paolini C, Huidobro C, Lara E, Mellini P, Lenoci A, Pezzi R, Botta G, et al. Discovery of salermide-related sirtuin inhibitors: binding mode studies and antiproliferative effects in cancer cells including cancer stem cells. J. Med. Chem. 2012;55:10937–10947. doi: 10.1021/jm3011614. [DOI] [PubMed] [Google Scholar]
- Samanta S, Sun H, Goel HL, Pursell B, Chang C, Khan A, Greiner DL, Cao S, Lim E, Shultz LD, et al. IMP3 promotes stem-like properties in triple-negative breast cancer by regulating SLUG. Oncogene. 2016;35:1111–1121. doi: 10.1038/onc.2015.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Kumar PU, Thakur S, Kiran S, Sen B, Sharma S, Rao VV, Poongothai AR, Ramakrishna G. Expression/localization patterns of sirtuins (SIRT1, SIRT2, and SIRT7) during progression of cervical cancer and effects of sirtuin inhibitors on growth of cervical cancer cells. Tumour Biol. 2015;36:6159–6171. doi: 10.1007/s13277-015-3300-y. [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Leyland-Jones B, Shak S. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. New England Journal of Medicine. 2001 doi: 10.1056/NEJM200103153441101. (2001) [DOI] [PubMed] [Google Scholar]
- Song EH, Oh W, Ulu A, Carr HS, Zuo Y, Frost JA. Acetylation of the RhoA GEF Net1A controls its subcellular localization and activity. J. Cell. Sci. 2015;128:913–922. doi: 10.1242/jcs.158121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storci G, Sansone P, Trere D, Tavolari S, Taffurelli M, Ceccarelli C, Guarnieri T, Paterini P, Pariali M, Montanaro L, et al. The basal-like breast carcinoma phenotype is regulated by SLUG gene expression. J. Pathol. 2008;214:25–37. doi: 10.1002/path.2254. [DOI] [PubMed] [Google Scholar]
- Taube JH, Herschkowitz JI. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proceedings of the National Academy of Sciences. 2010 doi: 10.1073/pnas.1004900107. (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van ’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- Vernon AE, LaBonne C. Slug stability is dynamically regulated during neural crest development by the F-box protein Ppa. Development. 2006;133:3359–3370. doi: 10.1242/dev.02504. [DOI] [PubMed] [Google Scholar]
- Vijver VM, He YD, Veer, van’t L. A gene-expression signature as a predictor of survival in breast cancer. New England Journal of Medicine. 2002 doi: 10.1056/NEJMoa021967. (2002) [DOI] [PubMed] [Google Scholar]
- Wang F, Chan C-HH, Chen K, Guan X, Lin H-KK, Tong Q. Deacetylation of FOXO3 by SIRT1 or SIRT2 leads to Skp2-mediated FOXO3 ubiquitination and degradation. Oncogene. 2012;31:1546–1557. doi: 10.1038/onc.2011.347. [DOI] [PubMed] [Google Scholar]
- Wang S-PP, Wang W-LL, Chang Y-LL, Wu C-TT, Chao Y-CC, Kao S-HH, Yuan A, Lin C-WW, Yang S-CC, Chan W-KK, et al. p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of Slug. Nat. Cell Biol. 2009;11:694–704. doi: 10.1038/ncb1875. [DOI] [PubMed] [Google Scholar]
- Wu W-SS, Heinrichs S, Xu D, Garrison SP, Zambetti GP, Adams JM, Look AT. Slug antagonizes p53-mediated apoptosis of hematopoietic progenitors by repressing puma. Cell. 2005;123:641–653. doi: 10.1016/j.cell.2005.09.029. [DOI] [PubMed] [Google Scholar]
- Wu Z-QQ, Li X-YY, Hu CY, Ford M, Kleer CG, Weiss SJ. Canonical Wnt signaling regulates Slug activity and links epithelial-mesenchymal transition with epigenetic Breast Cancer 1, Early Onset (BRCA1) repression. Proc. Natl. Acad. Sci. U.S.A. 2012;109:16654–16659. doi: 10.1073/pnas.1205822109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Developmental Cell. 2008 doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Zhang H, Head PE, Daddacha W, Park S-HH, Li X, Pan Y, Madden MZ, Duong DM, Xie M, Yu B, et al. ATRIP Deacetylation by SIRT2 Drives ATR Checkpoint Activation by Promoting Binding to RPA-ssDNA. Cell Rep. 2016;14:1435–1447. doi: 10.1016/j.celrep.2016.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Au Q, Zhang M, Barber JR, Ng SC, Zhang B. Identification of a small molecule SIRT2 inhibitor with selective tumor cytotoxicity. Biochem. Biophys. Res. Commun. 2009;386:729–733. doi: 10.1016/j.bbrc.2009.06.113. [DOI] [PubMed] [Google Scholar]
- Zhao D, Mo Y, Li M-TT, Zou S-WW, Cheng Z-LL, Sun Y-PP, Xiong Y, Guan K-LL, Lei Q-YY. NOTCH-induced aldehyde dehydrogenase 1A1 deacetylation promotes breast cancer stem cells. J. Clin. Invest. 2014;124:5453–5465. doi: 10.1172/JCI76611. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.