Abstract
The hexanucleotide repeat in the first intron of the C9orf72 gene is the most significant cause of amyotropic lateral sclerosis as well as some forms of fronto‐temporal dementia. The C9orf72 protein has been previously reported to be expressed in post‐mortem human brain as well as in late embryonic and some postnatal stages in mice. Herein, we present a detailed study of the distribution of C9orf72 protein in the embryonic, postnatal and adult mouse brain, spinal cord as well as during the differentiation of P19 embryonal carcinoma cells to neurons including motor neurons. We show that the expression levels of the C9orf72 transcripts in the developing and adult mouse brain as well as in differentiating neurons, are dynamic. Besides the strong expression in the cerebellum and motor cortex reported previously, we show for the first time that C9orf72 is expressed strongly in the olfactory bulb and also in the hippocampus. Our immunostaining data also reveal a hitherto unreported switch in the cellular distribution of C9orf72 from a predominantly cytoplasmic to a nucleo‐cytoplasmic distribution during corticogenesis. This switch in distribution was also observed during differentiation of the pluripotent embryonal carcinoma P19 cell line to mature neurons. Our findings have implications for interpreting the pathophysiology caused by the repeat expansions in C9orf72 in mouse models.
Keywords: brain development, C9orf72, expression, neuronal differentiation, spinal cord
Introduction
Amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) are two neurodegenerative diseases that have overlapping genetic causes and pathological features (Ling et al. 2013). In ALS, upper and lower motor neurons (MNs) die rapidly and progressively, with fatal consequences. In FTD, which is the second most common form of dementia, the frontal and temporal lobes of the brain are affected (Van Langenhove et al. 2012). At least 15% of patients diagnosed with either disease have been shown to have symptoms characteristic of the other (Ringholz et al. 2005; Wheaton et al. 2007). Common cellular pathologies include aberrant RNA processing and protein homeostasis (Ling et al. 2013). Genetic causes underlying these diseases have been identified within these pathways, such as mutations in ubiquilin‐2, FUS, TDP‐43 and C9orf72 (Kabashi et al. 2008; Kwiatkowski et al. 2009; DeJesus‐Hernandez et al. 2011; Deng et al. 2011; Renton et al. 2011).
Mutations in C9orf72 were first identified by DeJesus‐Hernandez et al. (2011) and Renton et al. (2011) in ALS and ALS‐FTD patients. The mutation in C9orf72 takes the form of G4C2 hexanucleotide repeat expansions (HREs) in the first intron of the C9orf72 gene. Over 30 or more studies have reported the frequency of the HREs in C9orf72 in European and North American populations (reviewed in van Blitterswijk et al. 2012). From an analysis of this data, van Blitterswijk et al. (2012) concluded that the C9orf72 HREs accounted for approximately 34.2% of inherited familial ALS cases and 5.9% of sporadic ALS cases. In the case of FTD, C9orf72 HREs accounted for 25.9% of the familial and 5.1% of the sporadic FTD cases. In comparison, only 0.17% of the control subjects carried the C9orf72 HREs. These data suggest that C9orf72 HREs are currently the major genetic cause of ALS and/or FTD (van Blitterswijk et al. 2012).
Typically, up to 20 HRE are found in the normal allele of C9orf72, whereas pathological alleles carry many additional repeats, ranging from ~30 to thousands. The HRE has multiple consequences within the cell. The three main pathological manifestations of the expansion are the appearance of RNA foci, formation of dipeptide proteins and reduced levels of the C9orf72 protein. The stable G‐quadruplex formed by the HRE DNA and RNA, results in RNA foci which have been shown to sequester factors involved transcription and splicing, leading to dysfunction (Fratta et al. 2012; Lee et al. 2013). Repeat associated non‐ATG translation results in the synthesis of dipeptide repeats (DPR), which have been shown to form intracellular inclusions. These aggregates are suggested to be cytotoxic and are found in degeneration‐associated neuronal subtypes (Schludi et al. 2015), as are RNA foci. Other reports find no evidence for the cytotoxicity of the DPRs (Mackenzie et al. 2015). However, both DPRs and RNA foci are not necessarily co‐incidental (Zu et al. 2013), indicating that both mechanisms may contribute separately to the aetiology of the disease. Further evidence for this can be seen in the spinal cord of ALS subjects, which contain RNA foci but rarely DPRs (Cooper‐Knock et al. 2015; Gomez‐Deza et al. 2015). Reduced C9orf72 expression has been found in ALS‐FTD patients by multiple groups; however, conditional ablation of C9orf72 in mouse does not lead to a pathological phenotype, arguing against haplo‐insufficiency as a major contributor to pathophysiology (Koppers et al. 2015).
FTLD is best characterised by degeneration of the frontal and temporal lobes. However, MRI studies have also identified atrophy in the hippocampus, entorhinal cortex and cerebellum in FTLD (Laakso et al. 2000; Hartikainen et al. 2012; Tan et al. 2014). In addition to characteristic abnormalities in the corticospinal tracts and cerebellar white matter (Bede et al. 2015), C9orf72 ALS‐FTD patients also show increased atrophy in the anterior temporal, parietal and occipital lobes, as well as the cerebellum, when compared with phenotypes caused by mutations in other FTD‐causative genes (Whitwell et al. 2012; Bede et al. 2013).
The C9orf72 protein has been detected in the cortex and cerebellum in human postmortem brain lysates. Besides these two regions, a few studies have looked at other regions of the human brain (Gijselinck et al. 2012; Hsiung et al. 2012; Stewart et al. 2012; Waite et al. 2014; Xiao et al. 2015). Both Gijselinck et al. (2012) and Waite et al. (2014) have shown decreased levels of C9orf72 in ALS‐FTD brains. Using isoform‐specific antibodies, Xiao et al. (2015) have shown the strongest expression of the C9orf72 human long isoform is in the motor cortex and cerebellum, whereas the short isoform is detectable in the motor, frontal and temporal cortices, with strongest expression in the cerebellum. Interestingly, they also show the long isoform is present at reduced levels in ALS‐FTD brains with C9orf72 HRE but not in ALS‐FTD brains carrying mutations in other ALS‐causative genes, and the converse for the short human C9orf72 isoform.
If the mouse is to serve as a model for ALS‐FTD, it is important to ascertain whether the expression domains of the mouse orthologue of C9orf72 are equivalent to those found in the human brain. In mice with a lacZ knock‐in at the endogenous C9orf72 locus, the mouse orthologue of C9orf72 (3110043O21Rik, referred to as mC9orf72) was reported to be expressed throughout the adult mouse brain (Suzuki et al. 2013). LacZ expression was undetectable in pre‐natal stages, most likely due the low detection sensitivity of the reporter. Atkinson et al. (2015) studied mC9orf72 expression in the mouse cortex at E18, P1, P14, P28 and P56 by immunohistochemistry and observed changes in distribution during neurite outgrowth and synaptogenesis throughout the cortex and hippocampus. Although expression of mC9orf72 has been demonstrated in the mouse brain, it remains unclear whether the protein is present in domains similar to human C9orf72 and regions affected in ALS‐FTD.
We sought to study in detail the distribution C9orf72 in the mouse brain together with neuronal and brain region‐specific markers, and whether its distribution was regulated both temporally and spatially during embryonic and postnatal brain development as well as during neuronal differentiation of pluripotent stem cells.
Herein, we show that mC9orf72 distribution in the developing mouse brain in embryonic (E12.5 to E18.5) and post‐natal (up to P35) stages is dynamic in location and intracellular distribution. We also describe changes in the distribution of mC9orf72 during the development of the brain, with notable changes seen in the olfactory bulb, cerebral cortex, hippocampus and cerebellum. Using mixed cultures of cortical neurons and astrocytes from P0 cortices we stained for the expression of C9orf72 and cortical layer as well as astrocyte markers and show that C9orf72 is predominantly in the neuronal cells and virtually undetectable in the astrocytes. Cortical neurons from all the cortical layers that we identified by staining for layer‐specific markers expressed C9orf72. We detected a distinct switch in the distribution of C9orf72 from a predominantly cytoplasmic to a nucleo‐cytoplasmic distribution during corticogenesis. This switch in distribution was also observed during differentiation of pluripotent stem cells to mature neurons.
Methods
Cell culture and differentiation
Embryonal Carcinoma P19 cells (Passage 8–9) were cultured in P19 growth medium containing α‐minimum essential medium (α‐MEM; Gibco Life Technologies) supplemented with 10% fetal calf serum (FCS; Biosera) and 1% non‐essential amino acid (NEAA; Gibco Life Technologies) at 37 °C with 5% CO2. When cells reached confluency of 70%, approximately every 2 days, they were passaged using 0.05% trypsin/EDTA (Gibco Life Technologies). Embryonic bodies (EBs) were made from P19 cells with the following method. Cells were trypsinised using 0.05% trypsin/EDTA and seeded in 90‐mm bacteriological dishes at a density of 2.5 × 105 mL−1, where they spontaneously formed aggregates. The medium used for the EBs was α‐MEM supplemented with 10% KnockOut serum replacement (KOSR; Gibco Life Technologies) and 1% NEAA. To differentiate the EBs into motor neurons we added 0.5 μM retinoic acid (Sigma) after 24 h and after 48 h they were plated onto matrigel (BD)‐coated coverslips. For coating the coverslips we added 200 μL of matrigel per coverslip and after 2 h at room temperature matrigel was aspirated and the EBs were added. The EBs were seeded using differentiation medium containing α‐MEM, 1% NEAA, 1% KOSR and 0.5 μM retinoic acid to promote differentiation of the EC cells. The differentiation medium was changed every other day for 6 days and on the 6th day the medium was changed to neurobasal media (Gibco Life Technologies), 1× B27 (Life Technologies), 1% KOSR and 1% glutamax (Life Technologies). We continued changing the medium every 2 days to sustain the neuronal culture.
Western blot
CD1 mouse brains (P10) were dissected into cerebral cortex and cerebellum, minced and then snap‐frozen in liquid nitrogen. Tissue from two mice were thawed with the addition of 2% SDS in 60 mM Tris‐Cl pH 6.8, pooled and homogenised by Dounce homogeniser. Lysates were sheared through 21G and 30G needles (BD), then centrifuged at 16,000Xg in a benchtop microfuge. Supernatants were stored at −70 °C. Protein concentration was determined by BCA microplate assay following the manufacturer's instructions (Pierce). Each sample (10 μg of protein) was mixed with loading buffer [to final concentrations of 2% sodium dodecyl sulphate (SDS), 10% glycerol, 50 mM dithiothreitol (DTT) in 60 mM Tris‐Cl pH 6.8], boiled for 5 min and proteins resolved on a 10% polyacrylamide gel. Proteins were transferred to 0.45 μm polyvinylidene fluoride (PVDF) at 50 V for 90 min in transfer buffer (25 mm Tris, 192 mm glycine, pH 8.3, 20% methanol).
PVDF membranes with the transferred proteins were blocked in 5% Marvel (dried skimmed milk powder) in phosphate‐buffered saline (PBS) for an hour, then incubated overnight in primary antibody (1 : 1000) diluted in 5% Marvel in PBS at 4 °C. After four 5‐min washes in PBS with 0.1% Tween20 (PBSTw) the blots were incubated with appropriate Horseradish peroxidase conjugated secondary antibody (1 : 10000, Abcam) diluted in 5% Marvel in PBS. After further 4× 5‐min washes in PBSTw, blots were incubated with ECL reagent and exposed to film (Amersham hyperfilm). Blots were imaged on a Fusion SL (Vilber Lourmat) using the associated fusion‐capt software.
qRT‐PCR
P19 cells were lysed in Trizol (Invitrogen) for RNA at indicated time points throughout two independent differentiation time courses on matrigel in the presence of retinoic acid (RA). Tissue was dissected from CD1 embryos to isolate heads at E12.5, whole brains at E13.5, E14.5 and E16.5, fore and mid/hind brain at E18.5. Brains was also dissected from postnatal mice at P5, P10 and P15 and split into cerebrum and cerebellum. Tissue was snap‐frozen in Trizol and pooled from four individuals from the same litter. Tissue in Trizol was thawed and disrupted by a Dounce homogeniser and RNA was prepared following the standard Trizol method. RNA was DNased and reverse‐transcribed using OligodT primers. qPCR was performed using primers specific to transcripts 1, 2 and 7 and normalised to Actb. Reactions were carried out in a BioRad iQ5 cycler. Dynamic well factors were collected for 2 min 30 s, then 40 cycles at 60 °C and 95 °C for 20 s, each followed by a melt curve. Expression levels were determined relative to Actb from baseline‐subtracted curves and corrected using primer efficiencies determined previously from serial dilutions of PCR product. See Table 1 for primer sequences.
Table 1.
qRT‐PCR primers
| Transcript (Accession) | Primer Name | Sequence 5′‐3′ | Product (bp) |
|---|---|---|---|
| Transcript 1 (3110043O21Rik‐001) | mC9T1 F | AGCGGCGAGTGGCTATTG | 162 |
| mC9T1 R | CAAAGGTAGCCGCCAACAAG | ||
| Transcript 2 (3110043O21Rik‐002) | mC9T2 F | GGCGAGTGGGAAAGACAAGA | 104 |
| mC9T2 R | ACTTCCCCAGTAAGCATGGG | ||
| Transcript 7 (3110043O21Rik‐007) | mC9T7 F | AGTGAAAGCCTTCCTGGATCA | 190 |
| mC9T7 R | GGGAACGAACAGCAACTGTC | ||
| ActB (NM_001101.3) | mActB‐F | ACCATGGATGATGATATCGC | 281 |
| mActB‐R | TCATTGTAGAAGGTGTGGTG |
Immunohistochemistry
Pregnant dams were killed by cervical dislocation prior to dissection for isolating embryonic stages. E12.5 and E14.5 embryos were fixed whole. E16.5 embryos were decapitated prior to fixation. Brains were dissected from E18.5 through post‐natal stages and bisected prior to fixation. Dissected embryos and brains were fixed overnight in cold 4% paraformaldehyde (PFA), washed twice for 1 h with PBS, then cryo‐protected in 20% sucrose before embedding in optimal cutting temperature compound (OCT). Frozen blocks were sectioned by Leica cryostat and air‐dried for 1 h before storage at –20°C.
Frozen sections were incubated in PBS twice to remove OCT prior to antigen retrieval in boiling 10 mM sodium citrate buffer, pH 6.0 with 0.05% Tween 20 for 20 min. Sections were allowed to cool for 20 min, washed twice with PBS and blocked with 0.1% gelatin in PBS supplemented with 0.5% Triton X‐100 and 1% fetal bovine serum (FBS) for 1 h at room temperature.
Cells on coverslips were washed with PBS and fixed with 4% PFA for 15 min. After two PBS washes, cells were dehydrated with serial ethanol washes. Prior to immunostaining, cells were rehydrated, washed with PBS and blocked as for frozen sections. Both cells on coverslips and sections were incubated with appropriate primary antibody at 4 °C overnight. The sections and fixed cells were then washed four times with 0.1% Triton X‐100 in PBS and then incubated for 1 h with the appropriate secondary antibody together with 4′,6‐diamidino‐2‐phenylindole (DAPI; Invitrogen) to stain cell nuclei. See Table 2 for a list of the antibodies used. Stained sections and cells on coverslips were mounted using Mowiol. Imaging was done with a Leica DMRB5500 epifluorescent microscope. Images were captured and deconvoluted where required using LAS AF (Leica) and figures composed in photoshop CS3 (Adobe). Brain anatomy was identified using the Allan Brain Atlas (http://mouse.brain-map.org/).
Table 2.
Antibodies used in this study
| Target | Dilution | Supplier | Catalogue no. |
|---|---|---|---|
| C9orf72 | 1 : 500 | ProteinTech | 22637‐1‐AP |
| C9orf72 | 1 : 500 | SantaCruz | SC138763 |
| Nestin | 1 : 50 | DSHB | Rat 401 |
| Neurofilament | 1 : 5 | DSHB | 2H3 |
| Pax6 | 1 : 500 | Covance | PRB‐278P |
| Tbr2 | 1 : 500 | Abcam | Ab23345 |
| Nurr1 | 1 : 100 | R&D | AF2156 |
| Tbr1 | 1 : 500 | Abcam | Ab31940 |
| Satb2 | 1 : 100 | Abcam | Ab51502 |
| Ctip2 | 1 : 500 | Abcam | Ab18465 |
| Cux1 | 1 : 50 | SantaCruz | SC13024 |
| GFAP | 1 : 500 | Sigma | A2052 |
| Calbindin | 1 : 4000 | Swant | 300 |
| SV2 | 1 : 5 | DSHB | SV2 |
| PCNA | 1 : 2000 | Cell Signalling | 2586 |
| PSD95 | 1 : 200 | Neuro‐mAb | K28/43 |
| Islet1/2 | 1 : 5 | DSHB | 40.2D6 |
| Alexa Fluor 488, Goat anti‐mouse | 1 : 1000 | Invitrogen | A‐11001 |
| Alexa Fluor 488, Goat anti‐rabbit | 1 : 1000 | Invitrogen | A‐11035 |
| Alexa Fluor 568, Goat anti‐mouse | 1 : 1000 | Invitrogen | A‐11004 |
| Alexa Fluor 594, Goat anti‐rabbit | 1 : 1000 | Invitrogen | A‐11012 |
| Alexa Fluor 488, Goat anti‐rat | 1 : 1000 | Invitrogen | A‐11006 |
Results
Antibody characterisation
Several commercially antibodies are available for researchers seeking to detect C9orf72 protein. As several reports comment about their general unsuitability (Waite et al. 2014; Xiao et al. 2015), we proceeded to characterise the specificity of two popular C9orf72 antibodies both by Western blot using purified recombinant human C9orf72 protein and also comparing them by immunostaining adjacent sections of adult mouse brain.
The mouse C9orf72 orthologue (3110043O21Rik) encodes three transcripts (1, 2 and 7) producing different protein isoforms, unlike the human C9orf72 which only produces two (1 and 3) transcripts. Transcript 1 of both mouse and human code for a 481‐amino acid protein that has 98% homology at the amino acid level. Mouse transcript 7 encodes only the first 420 amino acids of transcript 1 (Supporting Information Fig. S1A). The second protein coding transcript is substantially different from the first, and also differs dramatically between the two species. Human C9orf72 transcript 2 contains only the first 222 amino acids of transcript 1, whereas mouse C9orf72 transcript 2 encodes only the last 317amino acids of the homologous mouse transcript 1.
We found that the ProteinTech and SantaCruz antibodies detect both the 481‐amino acid and 222‐amino acid human C9orf72 isoforms. The ProteinTech epitope, however, is present only in the 481‐ and 420‐amino acid isoforms (transcript 1 and 7) of mouse C9orf72, whereas the SantaCruz antibody was detected all three mouse isoforms (Fig. S1A).
In Western blots, both antibodies recognised the pure recombinant human C9orf72 long isoform (kind gift from S. Iyer and R. Acharya, University of Bath). The SantaCruz C9orf72 antibody produces a more intense signal as compared with the ProteinTech antibody when used at the same concentration at similar exposure times in Western blots (Fig. S1B, ii and iv compared with i and iii). Additional weak bands seen with the SantaCruz antibody are absent in the blots probed with the ProteinTech antibody. These additional bands could represent degraded C9orf72 protein at lower masses and multimers at higher masses (Fig. S1B, II and IV), as we have used C9orf72 protein which has been stored frozen after purification for our Western blot analysis. Such additional bands are frequently seen upon storage of the purified C9orf72 protein (S. Iyer and K. Achraya, pers. comm. – see lines 363–368 for confirmation).
However, comparison of adjacent sections of adult mouse brain immunostained using both antibodies showed very little observable difference (representative areas are shown in Supporting Information Fig. S2; A: cerebellum, B: hind brain and C: cortex). As in Western blots the Santa Cruz antibody gave a stronger signal, all our immunostaining as well as further Western blot studies were carried out using the Santa Cruz C9orf72 antibody.
C9orf72 isoform expression in the cerebral cortex and cerebellum
Protein extracts from postnatal cerebellum and cortex were analysed for expression using the SantaCruz anti‐C9orf72 antibody. The dominant isoform in the brain appears to be that derived from transcript 1 (481 amino acids, 54 kDa), whereas the isoforms encoded by transcript 7 (420 amino acids, 47 kDa) and transcript 2 (222 amino acids, 36 kDa) in both the cortex and cerebellum (Fig. S1C) were significantly lower. The cerebellum showed higher levels of C9orf72 isoform 1 as compared with the cortex.
Crude lysates of bacteria expressing recombinant C9orf72 isoform 1, loaded on the PAGE gel directly without purification, identified a single band corresponding to C9orf72 upon immunoblotting (Fig. S1C). We did not observe any additional non‐specific bands or cross‐reactivity with any bacterial proteins in these samples. This suggests that the additional bands seen with the frozen pure recombinant C9orf72 are degradation or aggregation products rather than non‐specific bands.
C9orf72 expression in the developing mouse cerebral cortex
Cortical neurogenesis occurs in the last 8 days of gestation in the developing mouse. The neural progenitors found in the ventricular zone at E12.5 progressively mature, divide and migrate radially outwards from the cerebral ventricle. By E16.5 this has resulted in the formation of the distinct sub‐ventricular zone, sub‐plate and cortical plate proper. As cortical neurons are born they migrate outwards through the pre‐existing cortical plate neurons, resulting in an inside‐out pattern of early‐born deep layers and late‐born superficial layers. Concomitantly, the sizes of the ventricular and sub‐ventricular progenitor pools progressively diminish by E18.5 (Dehay & Kennedy, 2007). We investigated the pattern of expression of C9orf72 in the developing mouse cortex from E12.5 though to E18.5. At early embryonic stages (E12.5–16.5) we studied the expression of C9orf72 in combination with nestin, the neural progenitor marker and with the 160‐kd neurofilament marker. At later embryonic stages (E16.5–18.5) we studied the co‐localisation of C9orf72 with well characterised markers for each cortical layer.
In the E12.5 developing mouse cerebral cortex (Fig. 1A–D) expression of C90rf72 is seen in regions adjacent to the ventricles both in the ventral and dorsal pallium (Fig. 1A). C9orf72 expression is seen in radially organised cells that co‐express the neuronal progenitor marker nestin. This is seen specifically at the most medial zones in coronal sections both dorsal (Fig. 1B) and ventral (Fig. 1C), but is strongest at the dorsal side. C9orf72 is also detectable in tangentially orientated cells at the superficial surface of the dorsal pallium (Fig. 1A,B, dotted lines). C9orf72 staining appears to be specific to neural tissue, as observed at the interface between the strongly stained pre‐optic area and adjacent non‐neural tissue (Fig. 1D). Staining appears most intense in developing forebrain structures but does not appear in the sub‐pallium or pre‐thalamic regions.
Figure 1.
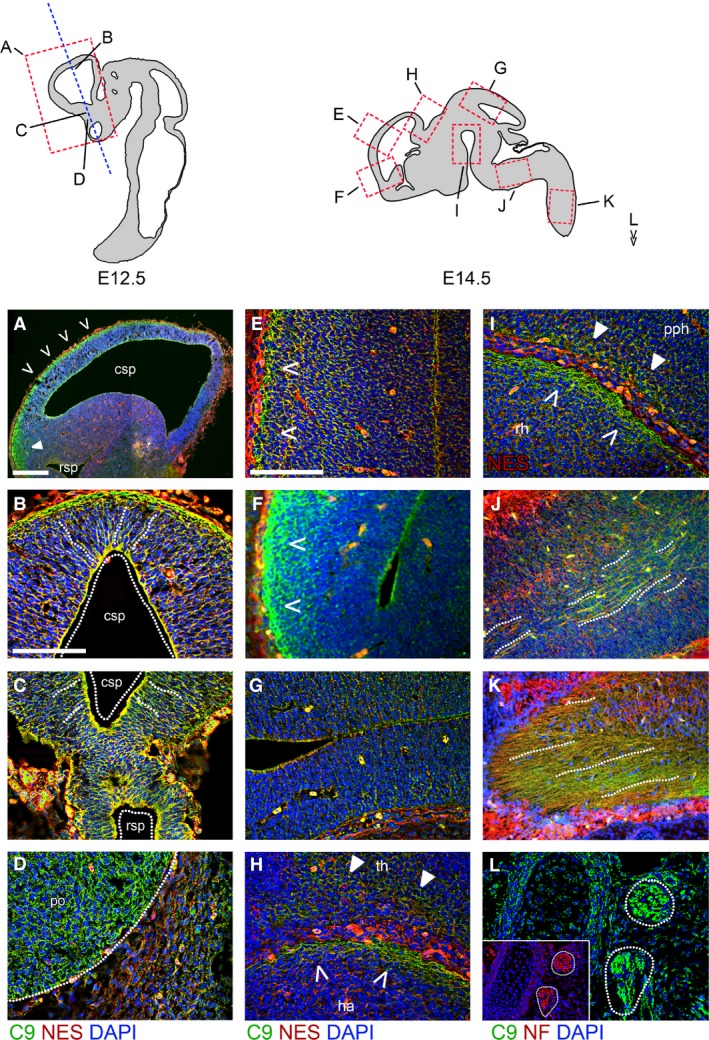
C9orf72 is present in the early embryonic brain and spinal cord. (A–D) Immunostaining for C9orf72 and nestin in E12.5 embryos in sagittal (A) and coronal (B–D) orientations. Strong C9orf72 staining can be seen in the dorsal pallium of the telecephalon (A), in the superficial layer of the rostral pallium (B) as well as in regions adjacent to the caudal secondary proencephalic ventricle (csp) in a radial arrangement spanning outwards (dotted lines). A similar pattern is seen in the caudal subpallium (C) adjacent to the rostral secondary proencephalic ventricle (rsp). C9orf72 is strongest in neuronal tissues, as shown by expression in the pre‐optic area (D, po) adjacent to non‐neuronal tissue. Immunostaining for C9orf72 and nestin (E–I) or neurofilament (J–L) in E14.5 brain. Expression is strongest in the superficial layers of the caudal rostral pallium (E) and pre‐olfactory pallium (F). This pattern of expression is not seen in the caudal midbrain tectum (G). Strong staining again is found in the superficial layer of the hippocampal allocortex (H, ha, open arrows) and spread in deeper layers of the adjacent thalamic tissue (th, closed arrows). Strong staining is also seen in the superficial layer of the rostral hypothalamus (rh) and prepontine hindbrain (pph) (I). C9orf72 can be found aligned with neurofilament‐positive tracts in the medulla (J) and spinal cord (K). C9orf72 and neurofilament are found together in the spinal ganglia (L). Scale bars: 100 μm.
The expression of C9orf72 no longer appears concentrated at the ventricular side of the dorsal pallium at E14.5, but staining is stronger at the superficial surface of both dorsal pallium (Fig. 1E) and olfactory bulb (Fig. 1F). This pattern of staining is not observed in the more anterior medial pallium or midbrain (Fig. 1G). Stronger expression is seen within the superficial stratum of the habenula and the pre‐thalamic structures in areas of reduced cell density directly adjacent to the convolution separating it from the pallium (Fig. 1H, closed arrows). C9orf72 expression at E14.5 is observed in the superficial zones where the hypothalamus and hindbrain abut (Fig. 1I, open and closed arrows). C9orf72 is also expressed in the axonal tracts, which are neurofilament‐positive in the medulla (Fig. 1J, dotted lines) and entering the spinal cord (Fig. 1K, dotted lines). Neurofilament‐positive spinal ganglia can also be seen co‐expressing C9orf72 (Fig. 1L).
As development proceeds to E16.5 (Fig. 2A), increased expression of C9orf72 is no longer seen around the ventricles, but remains around the tangentially orientated cells at the pial surface similar to that observed at E14.5. A second zone of increased expression is present below the cortical plate mid‐pallium in the intermediate zone between the ventricular zone and the cortical plate. This second zone does not overlap with markers such as Tbr2 in the subventricular zone (SVZ) or Nurr1 in the subplate (SP). This pattern of staining is not seen in the subiculum, sub‐pallium, thalamus or developing hippocampus.
Figure 2.
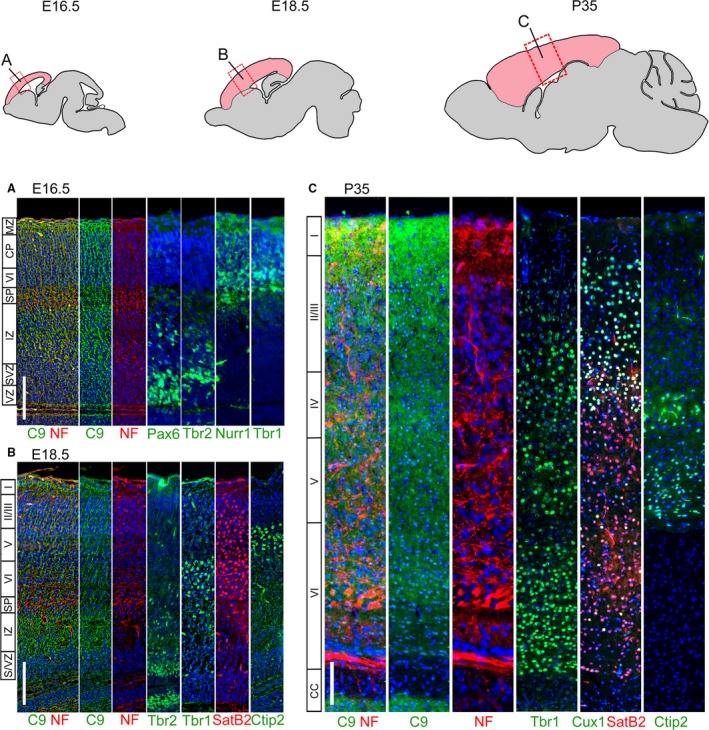
Layer‐specific C9orf72 distribution changes in the developing and adult cortex. Immunostaining for C9orf72, neurofilament and layer specific markers in adjacent sections of the brain. At E16.5 (A) the cortex is divided into the Pax6+ ventricular zone (vz), Tbr2+ subventricular zone (svz), intermediate zone (iz), Nurr1+ subplate (SP), Tbr1+ layer VI/cortical plate (CP) and finally the marginal zone (mz) at the pial surface. C9orf72 expression appears throughout but is strongest in the marginal and intermediate zones. At E18.5 (B), further layers have formed in the cortical plate (layers II/II–VI, marked by SatB2 and Ctip2). Strong expression continues in the marginal zone and in a narrower region of the intermediate zone. By P35 all the layers of the cortex have formed (C) and the intermediate zone is absent. C9orf72 staining remains strongest in layer I at the pial surface and is also seen strongly in upper portions of layer II, throughout layer IV and the middle of layer VI. C9orf72 has also switched from a mainly cytoplasmic distribution at embryonic stages to a more uniform distribution, with staining seen throughout the cytoplasm and nucleus. Scale bars: 100 μm.
C9orf72 expression extends through the entire rostro‐caudal axis of the pallium at E18.5 (Fig. 2B). Expression of late cortical markers such as SatB2 and Ctip2 indicates specification of the upper layers of the cortex in addition to the lower layers present at E16.5, as shown by the staining for Tbr1. Increased expression of C9orf72 persists in both the intermediate zone and superficial marginal zone of C9orf72. The expression domains of cortical layer markers confirm that the internal band of C9orf72 is not part of the cortical plate, as it is below the deep layer marker Tbr1 and is not in the Tbr2‐positive SVZ. At this stage, increased C9orf72 expression can also be seen in the superficial region of the midbrain tectum, similar to the expression in the cortical marginal zone.
The intermediate zone of the cortex dramatically decreases in size at P5 and P10 accompanied by the loss of C9orf72 expression. A gradient of C9orf72 expression can be seen extending from the marginal zone inwards particularly at early postnatal stages (P5 and P10 data not shown) than at embryonic stages. At P35 (Fig. 2C), the cortex layering is complete. C9orf72 expression is detected throughout the adult cortex but is strongest in layer I with a gradual decrease in intensity through to the base of layer II/II. Further bands of increased C9orf72 expression can be found throughout layers IV and VI. C9orf72 also appears to have transitioned from being predominantly cytoplasmic to having a more even nucleo‐cytoplasmic distribution.
In summary, we find C9orf72 expression throughout the developing cortex, strongest closer to the ventricle and superficially at E12.5 and E14.5, the intermediate and superficial marginal zones at E16.5 and E18.5, and then superficially in layers I, IV and VI in P35 mice. While predominantly cytoplasmic during embryonic stages, C9orf72 takes on a nucleocytoplasmic distribution postnatally (Table 3).
Table 3.
Expression of C9orf72 in the developing mouse brain and spinal cord
| Region | Stage | ||||||
|---|---|---|---|---|---|---|---|
| E12.5 | E14.5 | E16.5 | E18.5 | P5 | P10 | P35 | |
| Olfactory bulb | |||||||
| Superficial stratum | +++ | +++ | +++ | +++ | ++ | ++ | ++ |
| Internal stratum | + | ++ | ++ | ++ | ++ | ++ | ++ |
| Internal plexiform layer | +++ | +++ | +++ | ||||
| Mitral cell layer | +++ | +++ | +++ | ||||
| External plexiform layer | ++ | ++ | ++ | ||||
| Glomeruli | +++ | +++ | +++ | ||||
| Cerebral cortex | |||||||
| Cortical plate | |||||||
| I | +++ | +++ | +++ | +++ | +++ | ||
| II/III | ++ | ++ | ++ | ++ | ++ | ||
| IV | + | ++ | +++ | +++ | +++ | ||
| V | + | ++ | ++ | ++ | ++ | ||
| VI | + | ++ | +++ | +++ | +++ | ||
| Preplate | + | +++ | +++ | ||||
| Subplate | + | ++ | ++ | ++ | ++ | ||
| Intermediate zone | +++ | + | +++ | ++ | ++ | ++ | |
| Ventricular zone | +++ | ++ | + | ++ | ++ | ++ | ++ |
| Hippocampus | |||||||
| Dentate gyrus | |||||||
| Granule layer | + | + | + | + | |||
| Hilus | + | +++ | +++ | +++ | |||
| CA3 | |||||||
| S. oriens | + | + | + | + | |||
| S. pyramidale | + | +++ | +++ | +++ | |||
| S. radiatum | + | ++ | ++ | ++ | |||
| CA2 | |||||||
| S. oriens | + | ++ | ++ | ++ | |||
| S. pyramidale | + | ++ | ++ | ++ | |||
| S. radiatum | + | ++ | ++ | ++ | |||
| CA1 | |||||||
| S. oriens | + | + | + | + | |||
| S. pyramidale | + | + | + | + | |||
| S. radiatum | + | + | + | + | |||
| Subiculum | + | + | + | + | |||
| Cerebellum | |||||||
| Anlage | + | + | + | + | |||
| External granule layer | ++ | +++ | ++ | ++ | |||
| Molecular layer | ++ | +++ | +++ | ||||
| Purkinje cell layer | ++ | ++ | ++ | ||||
| Internal granule layer | + | + | + | ||||
| Spine | |||||||
| Dorsal | |||||||
| Horn | + | + | + | ||||
| Funiculus | ++ | ++ | ++ | ||||
| Root ganglion | ++ | +++ | +++ | ||||
| Lateral | |||||||
| Horn | + | ++ | ++ | ||||
| Funiculus | +++ | +++ | ++ | ||||
| Ventral | |||||||
| Horn | ++ | +++ | +++ | ||||
| Funiculus | +++ | +++ | ++ | ||||
S: Stratum. +/++/+++ indicates expression intensity. Nucleocytoplasmic expression in bold, otherwise cytoplasmic expression.
C9orf72 expression in cortical neuronal cultures
We also studied the expression of C9orf72 in the cortical neurons and astrocytes isolated from P0 cortex and co‐expression with transcription factor markers for cortical layers as well as the astrocyte marker GFAP. The primary cortical neurons in culture stained positive for neurofilament and the synaptic vesicle marker SV2. All the cortical neurons showed robust C9orf72 expression in neurofilament‐positive axons, whereas GFAP+ astrocytes express barely detectable levels in comparison (Fig. 3A, arrows). We observed no differences in expression or localisation of C9orf72 in cortical neurons from the different cortical layers as seen by co‐staining for calbindin, SatB2 or Ctip2 (Fig. 3B).
Figure 3.
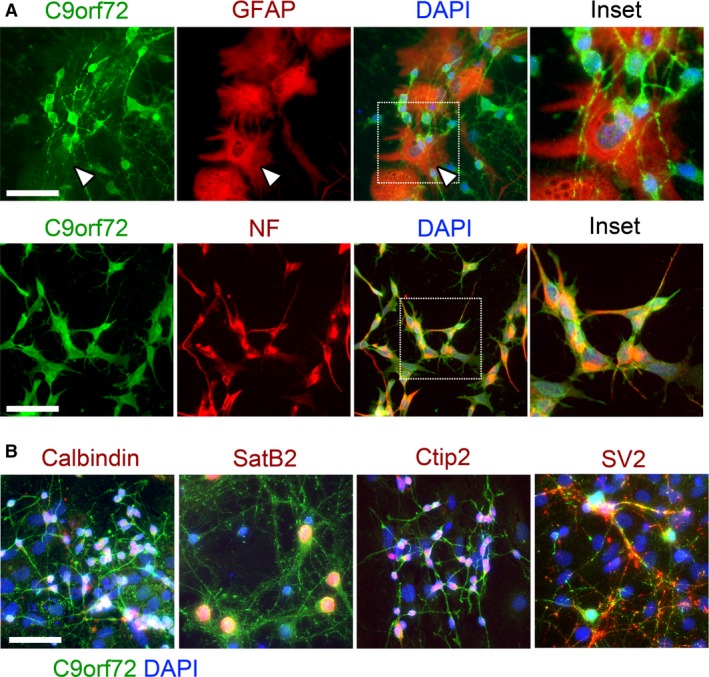
C9orf72 is highly expressed in primary cortical neurons in comparison with glia. Strong expression of C9orf72 is seen in the cell body and neurites of neurofilament‐positive cultured primary cortical neurons isolated at P0 (A). C9orf72 expression in GFAP‐positive glial cells is largely undetectable (A, arrows). C9orf72 expression is found consistently in all cortical neuron subtypes identified (B). Scale bars: 50 μm.
C9orf72 expression in the developing cerebellum
The anlage of the cerebellum emerges at at E11.5; however, much of the development – i.e. expansion and organisation – occurs postnatally until P15. By E18.5, granule cells precursor cells have migrated tangentially from the rhombic lip to form the external granule layer (EGL). The Purkinje cells (PC) are born and migrate radially from the ventricular zone to form the Purkinje cell layer (PCL). As the development of the cerebellum progresses, granule cells from the ECL descend radially to below the PCL to form the internal granule cell layer (IGL). By P15 the thickness of the EGL has substantially reduced, along with the loss of proliferative precursor cells, whereas the IGL has increased in size. The thickness of the molecular layer between the PCL and EGL also increases and contains the extensive PC dendritic arborisation and granule cell parallel fibres or climbing fibres. The expansion of the cerebellum also results in its foliation and characteristic lobes (Martinez et al. 2013).
Expression of C9orf72 can be seen between the developing EGL of the cerebellum at E18.5 and the internal neurofilament‐positive areas (Fig. 4A). This band of C9orf72 expression is expanded at P5 and overlaps with the calbindin‐positive Purkinje cells in the molecular layer (ML) between the EGL and IGL (more defined at this stage), but staining remains stronger on the interior edge of the EGL. Both the IGL and ML thicken by P10 when Purkinje cells have taken on their characteristic positioning on the superficial side of the IGL (Fig. 4A, pc). This pattern is consistent throughout the lobes and is maintained in the fully developed cerebellum at P35. The C9orf72 staining is strongest around the cells directly adjacent to proliferating PCNA‐positive EGL granule cells (Fig. 4B). This persists up to P10, and although by P35 the proliferation and migration in the EGL has ceased, the expression of C9orf72 remains strongest at the most superficial part of the molecular layer.
Figure 4.
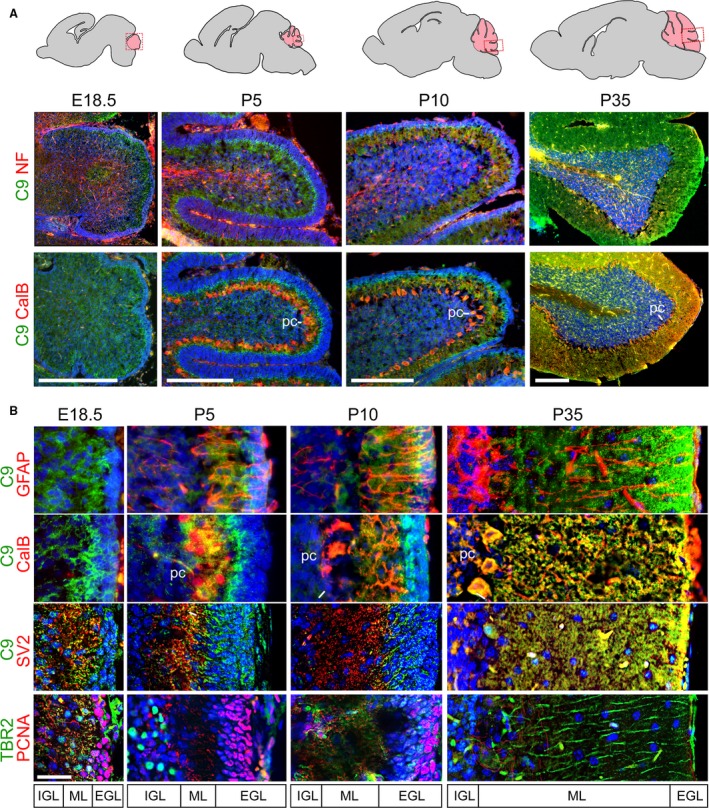
C9orf72 is strongly expressed in the molecular layer of the developing and adult cerebellum. Expression of C9orf72 in the developing cerebellum from E18.5 to P35 (A). C9orf72 staining is not seen in the neurofilament‐positive internal structures at any stage, but is found strongly at the inner edge of the developing external granule layer (EGL) at E18.5, which then goes onto to expand as the molecular layer (ML) expands, between the EGL and calbindin (CalB)‐positive Purkinje cell bodies (pc) adjacent to the forming inner granule layer (IGL). Scale bars: 500 μm. Throughout cerebellar development (B) C9orf72 expression in the molecular layer is not associated with Bergmann glia (GFAP) or proliferating granule cells (PCNA) but does associate with Purkinje dendrites (CalB) and synapses (SV2). C9orf72 appear strongest at the most superficial part of the molecular layer. Scale bars: 50 μm.
C9orf72 staining is not associated with the tangential GFAP‐positive glial fibres extending through the ML at any stage, or with the calbindin‐positive Purkinje cell dendrites at E18.5 to P10. At P35 it does appear to be strongest towards the ends of Purkinje dendrites and to overlap partially with the most superficial synapses stained by SV2 from E18.5 to P35. Additionally, it does not appear associated with Tbr2‐positive granule cell parallel fibres or proliferating EGL, which is strongly positive for PCNA (Fig. 4B).
Closer inspection of the cerebellar molecular layer at P35 showed no overlap or association between C9orf72‐ and GFAP‐positive glial fibres (Fig. 5A). C9orf72 can, however, be seen in the cell body of the Purkinje cells (Fig. 5A, arrows) and also clustering along the length of calbindin‐positive Purkinje cell dendrites, both proximal to the cell body and at distal tips. As one would expect from this, no co‐localisation is seen with the post‐synaptic marker PSD95, but overlap can be seen between C9orf72 and the synaptic vesicle marker SV2 (Fig. 5B).
Figure 5.
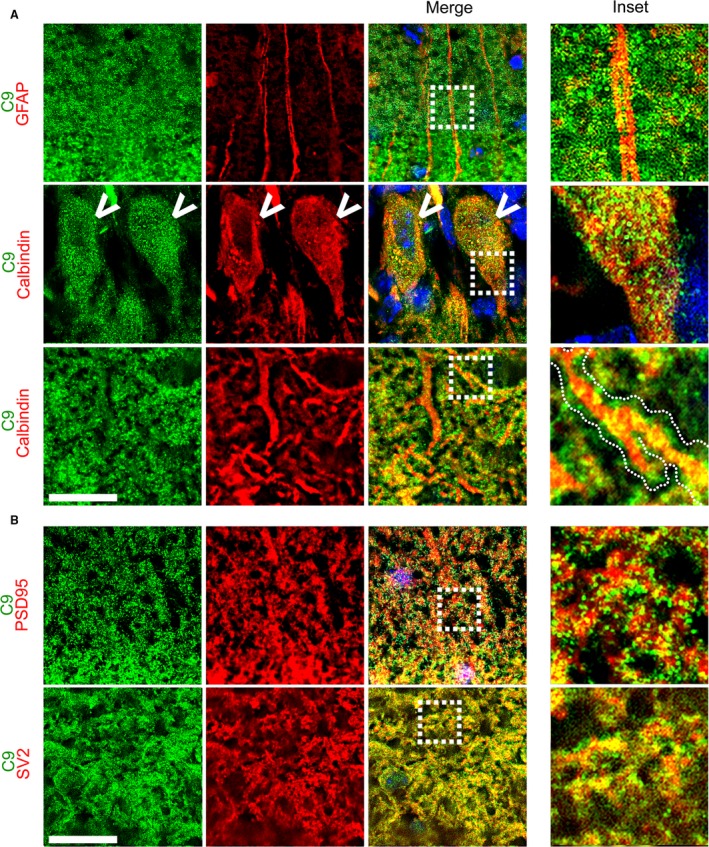
C9orf72 in the adult cerebellar molecular layer is pre‐synaptic. The molecular layer in the adult cerebellum (p35) shows no association of C9orf72 with Bergman glia fibres (GFAP), but clustered staining can be found directly adjacent to calbindin‐positive Purkinje dendrites (A, inset, dotted outline). C9orf72 expression can also be seen within the Purkinje cell body (arrow). C9orf72 shows no overlap with the post‐synaptic marker PSD95 but co‐localises with the synaptic vesicle marker SV2 (B). Scale bars: 20 μm.
To summarise, cerebellar C9orf72 expression is strongest in the regions underlying the EGL at E18.5. Expression at P5 and P10 overlaps with the PCL and ML which is adjacent to the EGL. This distribution pattern is maintained through to P35 when very strong C9orf72 expression is seen in the soma and nuclei of PCs as well as the pre‐synaptic terminals in the ML (Table 3).
C9orf72 expression in the developing olfactory bulb
The olfactory bulb (OB) can first be identified at E12.5 as a protrusion of the rostral telencephalon. After neurogenesis between E11–13, mitral and tufted cells (M/T cells) migrate radially into the OB then orientate tangentially prior to dendritic arborisation to form the external plexiform layer (EPL) between E14 and E16. From E17 onwards these dendrites begin to associate with proto‐glomerular structures. Olfactory glomeruli found in the superficial adult olfactory bulb, where there are sensory neuron axons from the olfactory epithelium, form synapses. Considerable remodelling and pruning of the M/T cell dendrites occurs post‐natally alongside further glomerulogenesis (Treloar et al. 2010).
The olfactory bulb showed distinct and high levels of C9orf72 expression. At E16.5 and E18.5, strong expression of C9orf72 is seen in the superficial stratum of the olfactory bulb compared with the deeper layers (Fig. 6A). By P5 the olfactory bulb has delineated into distinct regions and strong expression is seen in the olfactory glomeruli, and individual C9orf72‐expressing cells can be identified in the external and internal plexiform areas (Fig. 6Bii and iii). Staining in the olfactory glomeruli at the superficial edge of the olfactory bulb increases between P5 and P35 (Fig. 6Bi, glm). C9orf72 staining in the olfactory glomeruli at P35 does not overlap with the pre‐synaptic marker PSD95 (Fig. 6C, inset). C9orf72 expression in the external plexiform area is spread throughout the cells at P5, predominantly at the internal edge of the mitral cell layer (mcl) and in the molecular layer (ml) (Fig. 6Bii). By P10, the diffuse cytoplasmic staining has transitioned to a much more defined nucleo‐cytoplasmic staining, as is seen in the developing cortex. Strong nucleo‐cytoplasmic staining is found in the internal plexiform area from P5 through to P35 and continues to remain at the same intensity as the olfactory bulb expands (Fig. 6Biii). See Table 3 for summary of expression pattern.
Figure 6.
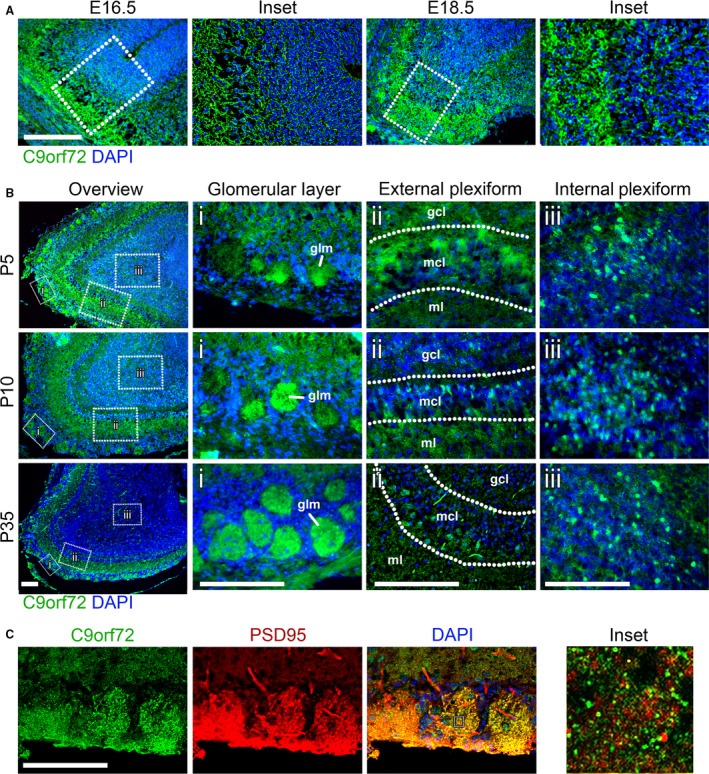
C9orf72 is strongly expressed in the olfactory bulb. Between E16.5 and E18.5, expression is seen in the superficial layer of the olfactory bulb (A). Scale bars: 200 μm. In post‐natal mice from P5 onwards, C9orf72 expression is found in three distinct olfactory layers (B); the glomerular (i), internal (ii) and external plexiform areas (iii). Between P5 and P10, the distribution of C9orf72 in the mitral cell layer of the external plexiform area (mcl) changes from a predominantly cytoplasmic to one with increased nuclear staining. C9orf72 staining in the olfactory glomeruli (glm) becomes more distinct as the size and frequency of glomeruli increases with age. C9orf72 in the glomeruli (C) is strongly co‐incident with PSD95 but without overlap similar to the cerebellum. Scale bars: 250 μm.
C9orf72 expression in the hippocampus
The hippocampus forms from the dorso‐medial telencephalon. The CA fields form sequentially from the outside‐in. The CA3 field is first defined at the dentate pole at E14.5, and then CA1 at the subicular pole from E15.5. Finally, between them, CA2 takes on its identity post‐natally. The dentate gyrus granule cell layer becomes visible by E18.5, with stratification of the different cell types finalising by P1‐2. We have studied C9orf72 expression in the P35 adult hippocampus (Khalaf‐Nazzal & Francis, 2013).
C9orf72 is not detectable in the nuclei of cells in the granular layer of the dentate gyrus or the CA fields (Fig. 7A, dg and CA1‐3). Strong cytoplasmic expression is seen extending from the hilus through to the CA3 stratum radiatum (Fig. 7Bi‐iv). In the CA3 stratum radiatum, C9orf72 can be seen distributed radially from the granular layer, indicating association with the CA3 neuron dendrites (Fig. 7Biv). This distribution extends further round to CA2 (Fig. 7Bv) but is not present in CA1 (Fig. 7Bvi) or in the subiculum. C9orf72 staining also appears to be stronger in the efferent side of CA2 in the stratum oriens adjacent to the granular layer (Fig. 7Bv). This can also be seen to a lesser degree in CA1 (Fig. 7Bvi). See Table 3 for summary of expression pattern.
Figure 7.
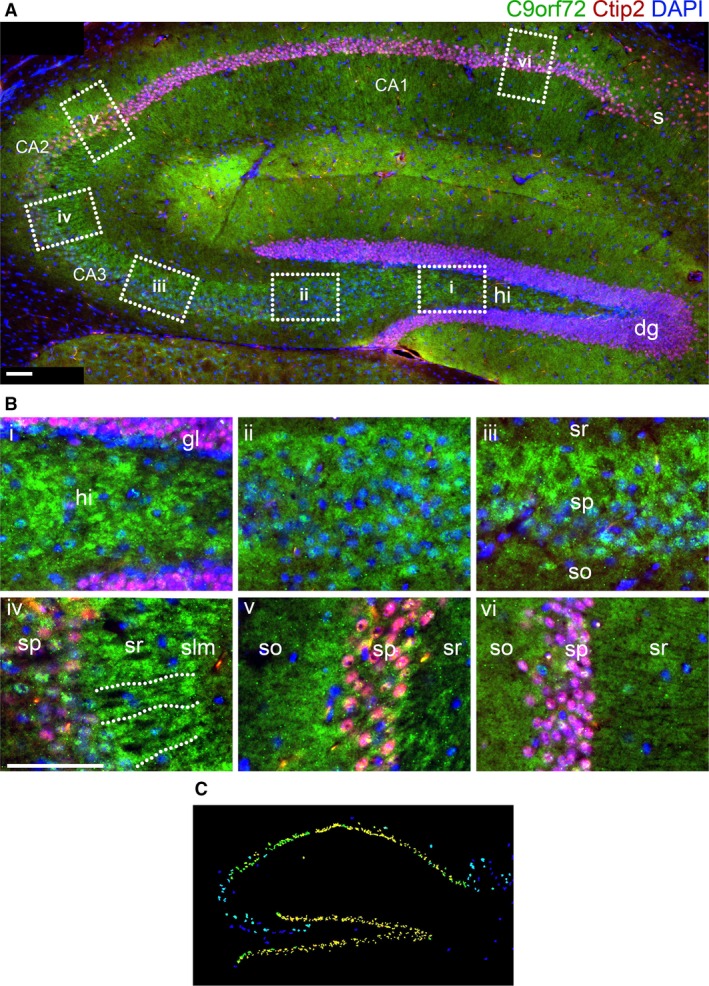
C9orf72 in the hippocampus is found in areas associated with the mossy fibre tracts. An intense band of C9orf72 can be seen extending from the hilus (hi) of the dentate gyrus (dg) to the stratum radiatum (sr) of the CA3 field and part of CA2 (A). It does not appear as tracts between the granule layer (gl) and CA3sr (B, i–iii), but can be seen organised radially from the CA3 stratum pyramidale (sp) (B, iv). Further C9orf72 staining can be seen at the efferent stratum oriens (so) adjacent to the CA2 sp (B, v). C9orf72 staining is not as distinct in the CA1 or the subiculum (s) (B, vi). Allen Brain Atlas expression data (C) show strong expression in the dg but not CA3 granule layers. Scale bars: 100 μm.
Expression of C9orf72 in the developing spinal cord
The anterior‐posterior patterning of the neural tube leads to the specification of the pro‐, mes‐ and rhomb‐encephalon (the early structures which form the fore‐, mid‐ and hind‐brain) and the spinal cord. In addition, dorso‐ventral patterning occurs between E9.5 and E11 in response to BMP/Shh signalling, resulting in progenitor domains that will generate ventral motorneurons, dorsal sensory neurons and interneurons (Lu et al. 2015). Migration and differentiation of the neural crest‐derived sensory neurons to the dorsal root ganglia occurs from E9.5 with migration reduced by E11, after which subtype specification continues to occur (Marmigère & Ernfors, 2007).
We analysed the C9orf72 expression in the spinal cord and dorsal root ganglia (DRG) in mid‐thoracic sections of mouse embryos. We stained for neurofilament to identify axons and Islet1/2 to identify post‐mitotic motor neurons and Islet1+ dorsal root ganglion neurons (Sun et al. 2008).
Expression of C9orf72 is seen throughout the spinal cord but is most intense in the transverse fibres of the corticospinal and spinothalamic tracts in the lateral and ventral funiculus at E12.5 (Fig. 8B). C9orf72 expression is similar to the surrounding tissue where Islet1/2+ motor neurons are found in the ventral horn (Fig. 9A) or in the more lateral pools (Fig. 9B) at E12.5. Increased expression of C9orf72 is observed in Islet1+ motor neurons at E14.5 and E16.5 (Fig. 9A). Strong C9orf72 expression can also be seen in Islet1+ neuronal cell bodies in the dorsal root ganglia at E12.5 with marked increases in staining intensity at E14.5 continuing until E16.5 (Fig. 9B).
Figure 8.
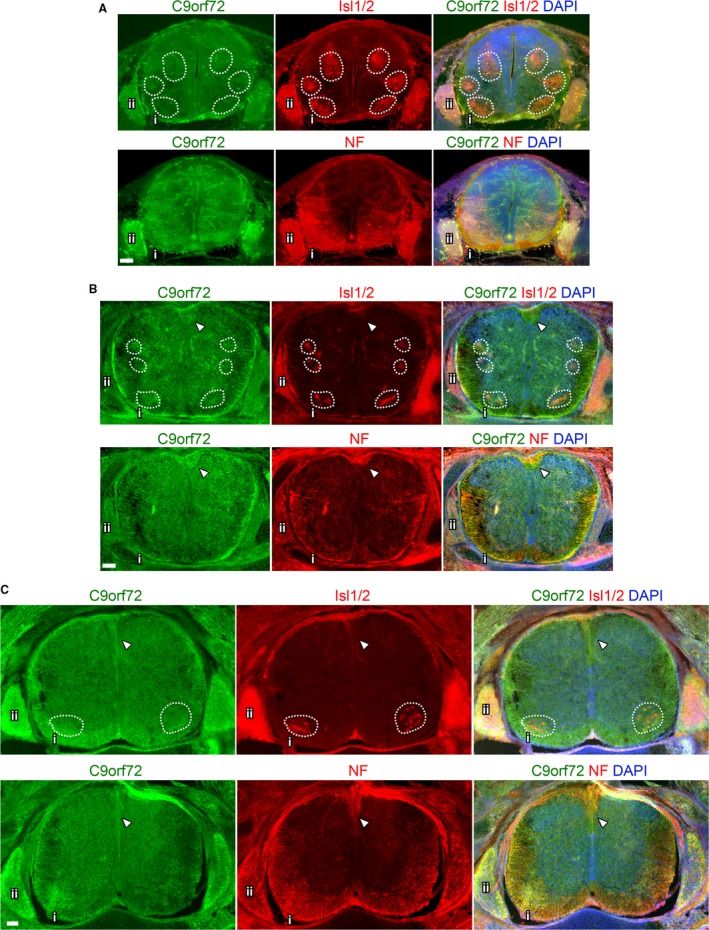
Overview of C9orf72 expression in the developing spinal cord. Transverse sections of mid‐thoracic spinal cord from the indicated embryonic stages. Comparison of adjacent sections immunostained with either Isl1/2 or neurofilament (NF) and C9orf72, counterstained with DAPI at E12.5 (A), E14.5 (B) and E16.5 (C). Motor pools highlighted by dashed boxes. Ventral spinal cord (i) shown in detail in Fig. 8A and Supporting Information Fig. S4A. Dorsal root ganglion shown in detail in Figures 8B and S4B. Arrows indicate dorsal corticospinal tracts. Scale bars: 100 μm.
Figure 9.
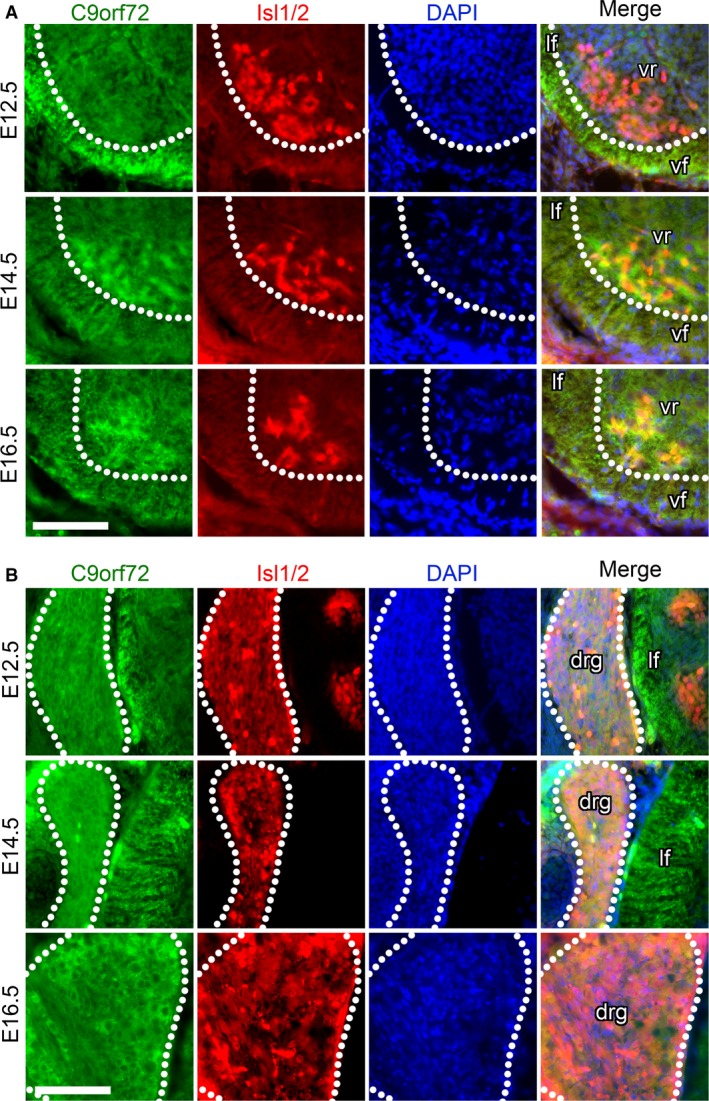
C9orf72 expression increases in ventral root motor‐neurons and dorsal root ganglion neurons during development. Transverse sections of mid‐thoracic region of indicated embryonic stages, stained with anti‐C9orf72 and and anti‐Islet1/2 (Isl1/2) antibodies, counterstained with DAPI. C9orf72 expression in the ventral spinal cord is strongest in the transverse tracts of the lateral (lf) and ventral funiculus (vf) at E12.5. Increased expression can be seen in the motor neuron pool in the ventral root (vr) from E14.5 to E16.5 (A). C9orf72 expression is also found in the dorsal root ganglia with expression increasing from E12.5 to E16.5. Scale bars: 100 μm.
The intense C9orf72 staining found in the funiculus, co‐stains with neurofilament in the descending tracts, particularly at the outermost edge of the spinal cord. At E12.5 staining appears strongest at the ventral funiculus (Supporting Information Fig. S3A), whereas it appears evenly distributed between lateral and ventral tracts at E14.5 and E16.5 (Figs S3B and 8B,C). Additionally, at E14.5 and E16.5 stronger C9orf72 staining can be found in the dorsal corticospinal tracts (Fig. 8B,C, arrows). See Table 3 for a summary of the expression pattern.
C9orf72 expression during in vitro neuronal differentiation from stem cells
To better observe the changes in distribution and expression of C9orf72 during cell differentiation, we monitored the expression of C9orf72 during in vitro differentiation of P19 embryonal carcinoma cell line to mature motor neurons.
Undifferentiated P19 grow in nests and C9orf72 is seen uniformly in all cells of the colony (Fig. 10A). C9orf72 expression is predominantly and strongly cytoplasmic in a granular punctate manner in the undifferentiated P19 cells (Fig. 10Ai). After 8 days of differentiation to neurons, C9orf72 is present in both the nucleus and cytoplasm in the soma in a highly speckled pattern, and can very clearly be seen concentrated at the membranes in neurite extensions (Fig. 10Aii and iii). To identify when this transition in cellular distribution occurs, we looked at intervening time points and co‐immunostained with markers characteristic of the different stages of differentiation. Early speckled cytoplasmic staining is found on day 2 and day 4 (Fig. 10B–D, white arrows). A small proportion of cells on day 4 have begun to show the later speckled C9orf72 distributed more evenly between nucleus and cytoplasm (Fig. 10B–D, black arrows). By day 8 the majority of cells exhibit this nucleo‐cytoplasmic staining in the cell body.
Figure 10.
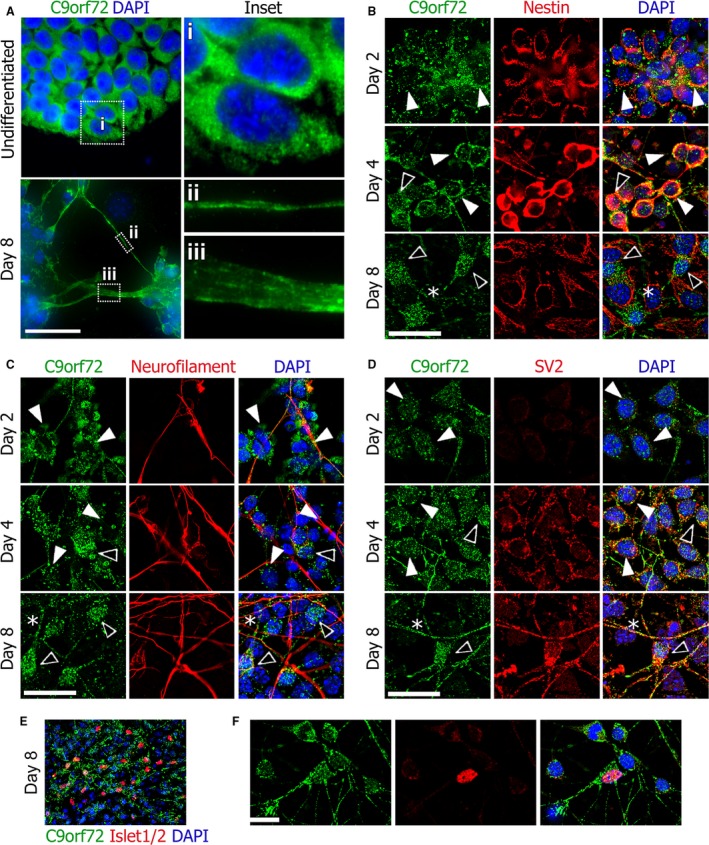
Neurons differentiated from pluripotent P19 cell line including motor neurons show a nucleo‐cytoplasmic change in C9orf72 distribution during differentiation. Undifferentiated P19 cells show a predominantly cytoplasmic distribution of C9orf72 but after differentiation to neurons, distribution is more nuclear and C9orf72 can also be seen on the cell membranes in neurites (A). The early cytoplasmic distribution of C9orf72 is found in nestin‐positive neuronal precursors seen on days 2 and 4, whereas nestin‐negative mature neurons display increased nuclear C9orf72 (B). C9orf72 is found weakly within nestin‐positive processes (B, asterisk). Neurofilament expression follows nestin and, as expected, increased nuclear C9orf72 is seen in mature neurofilament‐positive neurons (C). C9orf72 expression also appears to be found at higher levels in neurofilament‐positive neurites (C, asterisk). The presence of synaptic vesicle marker SV2 indicates mature axons (D); the strongest C9orf72 staining is found in SV2‐positive neurites (D, asterisk). The motor neuron (MN) marker Islet1/2 expression can be seen on day 8 (E) and C9orf72 expression in MNs is similar to other neurons present in the culture (F). Scale bars: 25 μm.
Expression of nestin, an early neuronal precursor marker, starts off uniformly expressed in all cells on day 2 (Fig. 10B). By day 7 it has increased in intensity, processes have begun to extend and C9orf72 can be seen in these processes. By day 8, nestin‐positive processes have been replaced by neurofilament and nucleo‐cytoplasmic staining is seen in the nestin‐negative cells with neuronal morphology. Underlying these cells, nestin‐positive but C9orf72‐negative epithelial‐like cells can be seen (Fig. 10B, asterisk), mirroring the pattern of C9orf72 seen with neurons and glia in the brain.
Expression of neurofilament indicating mature neurons and the frequency of C9orf72‐positive neurites, increases between day 6 and 8 and cell bodies associated with neurofilament‐positive neurites have nucleo‐cytoplasmic C9orf72 staining rather than strongly cytoplasmic alone (Fig. 10C, asterisks). A later marker of neuronal maturity, the synaptic vesicle marker SV2, is weakly expressed on day 4 but can be seen expressed robustly on day 8, distributed throughout neurites and neuronal cell bodies (Fig. 10D). Again, SV2 is present in neuronal cells which stain positive for the speckled nucleo‐cytoplasmic C9orf72 distribution. The C9orf72 staining in the neurites appears stronger where SV2 is being expressed (Fig. 10D, asterisk). Around 20% of the neurons generated by day 8 are motor neurons and stain positive for the transcription factors Islet1/2 (Fig. 10E). P19‐derived motor neurons display the same intracellular distribution of C9orf72 as other P19‐derived neurons (Fig. 10F).
Quantitation of C9orf72 transcript‐specific expression during development and in vitro differentiation
We also quantified the levels of the different mouse C9orf72 transcripts during embryonic and postnatal mouse developments as well as in adult brain to support the immunohistochemistry data. C9orf72 transcript 1 shows a steady increase in levels throughout the embryonic stages to E18.5, when a clear change in expression levels can be seen with lower levels in mid/hind brain (Fig. 11A). During postnatal stages expression of C9orf72, transcript 1 appears to reach steady levels between P5 and P15 in the cerebrum, whereas a rapid increase in expression levels can be seen in the cerebellum from P5 onwards. C90rf72 transcript 2 also appears to steadily increase during development. During postnatal stages, C9orf72 transcript 2 appears to increase in expression in both the cerebrum and cerebellum concurrently to similar levels by P15. C9orf72 transcript 2 is initially present at levels below that of C9orf72 transcript 1 and 7 but reaches similar levels at least in the cerebrum by P15. The expression levels of C9orf72 transcript 7 expression at all stages appears to be between that of transcript 1 and 2. Its expression follows a pattern more similar to C9orf72 transcript 2, with a concurrent increase in expression in both cerebrum and cerebellar tissue from P5.
Figure 11.
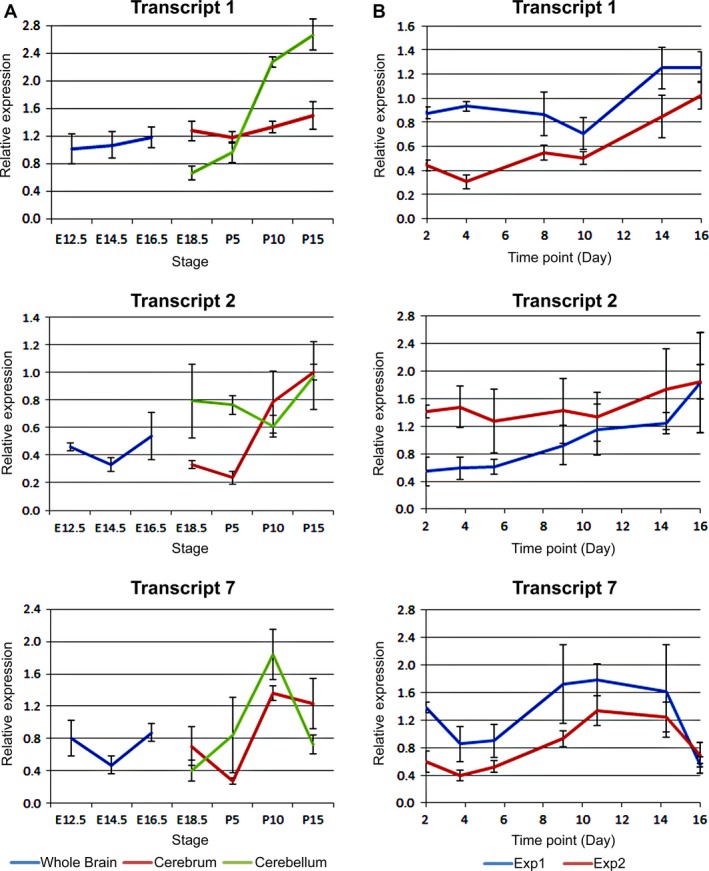
C9orf72 expression levels increase throughout development and differentiation in the CNS. Expression levels of mC9ORF79 transcripts relative to Actb determined by qRT‐PCR. (A) C9orf72 expression in a developmental series of mouse brains; E12.5 head; E14.5 and E16.5 brains; E18.5 fore and mid/hind brain; P5, 10 and 15 cerebrum and cerebellum. RNA from four siblings pooled in duplicate. Levels of all three transcripts increase over development, particularly transcript 1 in the cerebellum. (B) A time course of C9orf72 expression in P19 embryonal carcinoma cells differentiated on matrigel as described in Materials and Methods at the indicated time points. As with transcripts in brain tissue, all three C9orf72 transcripts appear to increase steadily during differentiation. RNA from two independent experiments and all reactions performed in triplicate.
Analysis of expression of the three C9orf72 transcripts during neuronal differentiation of pluripotent P19 cells showed a steady increase in expression over the time course of the differentiation similar to the steady increase in levels seen by immunostaining (Fig. 11B). There is no sudden peak during the time course as seen during the developmental series from post‐natal stages. By day 16, expression of C9orf72 transcript 1 and 2 peaked at levels below that of post‐natal mouse brains. C9orf72 transcript 2 levels appear higher than in embryonic brains. C9orf72 transcript 7 appears to peak midway through the differentiation and maintain a steady level until day 16 where it appears to have dropped dramatically.
Discussion
C9orf72 is linked to neurodegenerative diseases, and the hexanucleotide repeat expansions in the first intron of this gene have been shown to cause neuronal cell death but very little is known about the distribution, cellular function or normal role for the C9orf72 gene product in the adult brain or during neuronal differentiation. We have observed substantial region specific differences in expression domains in the developing and adult brain, such as increased levels in particular cortical layers. We, like Atkinson et al. (2015), show that C9orf72 protein is detected throughout the mouse brain in a punctate speckled manner. In addition, we have analysed the distribution of C9orf72 in early embryonic stages. We also show hitherto unreported notable expression in the olfactory bulb, hippocampus and cerebellum in addition to the cortex, which is traditionally associated with FTD. We show in addition, that novel distinct changes occur both in the domains of expression of C9orf72 and in the intracellular localisation. This is not an artefact of tissue processing, as we also demonstrate this switch in distribution using an in vitro model of neuronal differentiation.
In contrast to the lacZ knock‐in mouse used by Suzuki et al. (2013), where lacZ appeared more frequently in the projection neurons of layers V and II/III, we found that in the adult cortex increased C9orf72 staining in layers I, IV and VI. This may be due to the lacZ expression highlighting the cell body of the C9orf72‐positive neuron, while the immunostaining we observed highlights synapses clustered around dendritic processes. This is particularly apparent in the staining seen in layer I, a molecular layer consisting of large numbers of apical dendrites from pyramidal cells in lower layers. During development, C9orf72 can be seen as a distinct layer in the intermediate zone where neuronal precursors are migrating from the proliferating progenitors in the ventricular zone through to the nascent cortical plate.
The involvement of the cerebellum in FTD is unclear and patients do not show classic signs of cerebellar defects such as ataxia. Despite this, atrophy in the cerebellum has been identified in C9orf72 ALS‐FTD patients (Hornberger, 2012; Tan et al. 2014; Bocchetta et al. 2016) and p62‐positive inclusion bodies have been identified in the cerebellum as well as the cortex in FTD (King et al. 2009). These inclusions are present in patients with C9orf72 HREs and those found in the cortex, hippocampus and cerebellum have been shown to contain dipeptide repeat proteins (Mann et al. 2013; Davidson et al. 2014; May et al. 2014). However, in contrast to the strong localisation of C9orf72 seen by us in the molecular layer of the cerebellum, DPR inclusions are typically found in the cell bodies on the internal granule layer which are the source of the projections into the molecular layer.
In the hippocampus, the strongest expression of C9orf72 is within the granule cells of the dentate gyrus and pyramidal cells of CA2/3, where DPR deposits are found in human patients but not in the CA1. (Mann et al. 2013; Davidson et al. 2014; May et al. 2014). Therefore expression of the DPR from the HRE may be linked to overall expression levels of the C9orf72 gene or expression of a particular transcript. In agreement with our observation that transcript 1 is dramatically upregulated during post‐natal development of the cerebellum and increased levels of C9orf72 protein in the expanding cerebellar molecular layer, Atkinson et al. (2015) show that the 550kDa isoform of transcript 1 is highly enriched in synaptosome preparations.
Olfactory defects and impaired sense of smell have been identified in some cases of FTD, although this is more commonly associated with Alzheimer's (AD) and Parkinson's disease (PD) (Luzzi et al. 2007; Pardini et al. 2009; Heyanka et al. 2014; Orasji et al. 2016). The olfactory glomeruli is a structure dense with synapses and is the first level of synaptic processing in the olfactory system (Hamilton et al. 2005). As with the molecular layers of the cortex and cerebellum, C9orf72 staining is particularly intense in this region. C9orf72 can be also be seen in the cells of the internal and external plexiform areas.
Suzuki et al. (2013) did not observe C9orf72 expression in astrocytes and microglia. In mixed cultures of cortical neurons and astrocytes we found that C9orf72 expression was virtually undetectable in astrocytes but strongly expressed in the neurons. Similarly, in the spinal cord we observed expression in spinal motor neurons as well as the DRGs.
C9orf72 is predicted to be a DENN‐domain protein (differentially expressed in normal and neoplastic cells) (Zhang et al. 2012; Levine et al. 2013). This family of proteins comprises Rab guanine nucleotide exchange factors (RabGEFs) which together with Rab family GTPases are involved in membrane and vesicular trafficking (Stenmark, 2009; Marat et al. 2011). In the mouse brain and in P19‐derived neurons, we observed that C9orf72 expression and localisation were closely linked to the synaptic vesicle marker SV2 and dense pre‐synaptic areas. Consistent with this, the densest C9orf72 staining and SV2 overlap was seen in the molecular layer of the adult mouse cerebellum where granule cell axons form synapses with Purkinje dendrites.
Xiao et al. (2015) using isoform specific antibody, described the intracellular localisation of the long C9orf72 isoform in Purkinje cells as diffuse in the cytoplasm with distinct speckles within the cell body. As of now there is no description of the distribution of the long human C9orf72 isoform for other areas of the human brain. Interestingly, Xiao et al. (2015) show that the short human isoform has a very distinct association with the nuclear membrane in Purkinje cells. While the human short isoform is the first 222 amino acid of the human long isoform, the mouse short isoform comprises the last 317 amino acids of transcript 1. Because of this it is not possible to predict without further characterisation what the role of the shorter mouse isoform may be in comparison with human.
We also show a distinct switch in intracellular localisation of C9orf72 from a predominantly cytoplasmic distribution predominantly in early developmental stages, to a marked increase in nuclear C9orf72 in addition to the cytoplasmic expression as neurons mature in the brain. We see this switch distinctly in the developing cortex and the external plexiform area of the olfactory bulb, and it is recapitulated in vitro during P19 differentiation to mature neurons. A few previous studies have shown C9orf72 distribution in mature neurons; however, we are the first to demonstrate the early cytoplasmic distribution followed by a switch to nuclear and cytoplasmic distribution in mature neurons. Nuclear import of C9orf72 is not well characterised and the protein is not predicted to contain an NLS; however, Xiao et al. (2015) have shown association of both human C9orf72 isoforms with importin‐β1, lamin B and Ran‐GTPase by immunostaining and immunoprecipitation. What triggers this shift in distribution is unclear. Whether this difference in C9orf72 distribution has an effect on the role of the protein during neurogenesis is unknown. The proteins presenilin and ApoE associated with Alzheimer's disease have been shown to have a role in adult hippocampal neurogenesis (Chevallier et al. 2005; Mu & Gage, 2011; Kiyota et al. 2015) as does the FTD‐associated progranulin (Arrant et al. 2015).
It would be informative to better characterise the sub‐cellular localisation in vitro and in vivo epitope tag knock‐in approach. In this approach, normal C9orf72 expression would be unaffected, potentially yielding clearer results over the heterozygous knock‐out required in LacZ gene‐trap methods (Suzuki et al. 2013).
Our study is the first to provide a detailed characterisation of the expression domains and localisation of C9orf72 protein within the tissues of the mouse brain, from early embryonic stages through to mature adult. We also confirm others’ descriptions of intracellular distribution in mature cells and show a novel C9orf72 distribution within neuronal precursors. The correlation between C9orf72 expression domains and FTD‐affected areas highlights that understanding the role of C9orf72 during normal development and cellular function will yield insight into FTD‐ALS pathology.
Author contributions
R.F. and E.S.P. carried out the experimental work and data analysis, R.F. wrote the manuscript and participated in the design of the study. V.S. conceived and designed the study, carried out the data analysis and wrote the manuscript.
Supporting information
Fig. S1. Comparison of two C9orf72 antibodies by Western blot. (A) ClustalO alignment of mouse and human C9orf72 protein isoforms showing antibody epitopes. (B) Western blot for purified recombinant human C9orf72 using either ProteinTech (PT) or SantaCruz (SC) primary antibody showing a strong band at 55 kDa for His‐tagged C9orf72 isoform 1. (C) Western blot of P10 mouse cortex and cerebellum using SantaCruz primary antibody shows isoform 1 is the predominant form. Each lane represents tissue pooled from two mice. GAPDH used as loading control. Unpurified bacterial lysate of recombinant human his‐tagged C9orf72 isoform 1 used as positive control (rC9).
Fig. S2. Comparison of two C9orf72 antibodies by immunostaining on brain sections. Comparison of adjacent sections of mouse brain immunostained with either ProteinTech or SantaCruz anti C9orf72 antibody and counterstained with DAPI in the cerebellum (C), hind brain (D) and cortex (E).
Fig. S3. C9orf72 expression in the spinal cord is associated strongly with neurofilament. Transverse sections of mid‐thoracic spinal cord from the indicated embryonic stages. Stained with C9orf72 and neurofilament (NF), counterstained with DAPI. C9orf72 expression in the ventral spinal cord is strongest in the transverse tracts of the lateral (lf) and ventral funiculus (vf) at E12.5 (A). C9orf72 expression is also found with associated neurofilament in the dorsal root ganglia with expression increasing from E12.5 to E16.5. Scale bars: 100 μm.
Acknowledgements
We would like to acknowledge the Developmental Studies Hybridoma Bank for the 2H3, Rat401 and 40.2D6 antibodies. We thank Ravi Acharya and Shalini Iyer for providing the recombinant purified C9orf72 protein and the crude bacterial lysates with C9orf72 used to verify the antibodies to C9orf72. The authors declare no conflict of interests.
References
- Arrant AE, Patel AR, Roberson ED (2015) Effects of exercise on progranulin levels and gliosis in progranulin‐insufficient mice. eNeuro 2, pii: 0061‐14,1‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson RAK, Fernandez‐Martos CM, Atkin JD, et al. (2015) C9ORF72 expression and cellular localization over mouse development. Acta Neuropathol Commun 3, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bede P, Bokde ALW, Byrne S, et al. (2013) Multiparametric MRI study of ALS stratified for the C9orf72 genotype. Neurology 81, 361–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bede P, Elamin M, Byrne S, et al. (2015) Patterns of cerebral and cerebellar white matter degeneration in ALS. J Neurol Neurosurg Psychiatry 86, 468–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Blitterswijk M, DeJesus‐Hernandez M, Rademakers R (2012) How do C9ORF72 repeat expansions cause amyotrophic lateral sclerosis and frontotemporal dementia: can we learn from other noncoding repeat expansion disorders? Curr Opin Neurol 25, 689–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocchetta M, Cardoso MJ, Cash DM, et al. (2016) Patterns of regional cerebellar atrophy in genetic frontotemporal dementia. Neuroimage Clin 11, 287–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevallier NL, Soriano S, Kang DE, et al. (2005) Perturbed neurogenesis in the adult hippocampus associated with presenilin‐1 A246E mutation. Am J Pathol 167, 151–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper‐Knock J, Higginbottom A, Stopford MJ, et al. (2015) Antisense RNA foci in the motor neurons of C9ORF72‐ALS patients are associated with TDP‐43 proteinopathy. Acta Neuropathol (Berl) 130, 63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson YS, Barker H, Robinson AC, et al. (2014) Brain distribution of dipeptide repeat proteins in frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9ORF72. Acta Neuropathol Commun 2, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehay C, Kennedy H (2007) Cell‐cycle control and cortical development. Nat Rev Neurosci 8, 438–450. [DOI] [PubMed] [Google Scholar]
- DeJesus‐Hernandez M, Mackenzie IR, Boeve BF, et al. (2011) Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p‐linked FTD and ALS. Neuron 72, 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H‐X, Chen W, Hong S‐T, et al. (2011) Mutations in UBQLN2 cause dominant X‐linked juvenile and adult‐onset ALS and ALS/dementia. Nature 477, 211–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratta P, Mizielinska S, Nicoll AJ, et al. (2012) C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G‐quadruplexes. Sci Rep 2, 1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gijselinck I, Van Langenhove T, van der Zee J, et al. (2012) A C9orf72 promoter repeat expansion in a Flanders‐Belgian cohort with disorders of the frontotemporal lobar degeneration‐amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet Neurol 11, 54–65. [DOI] [PubMed] [Google Scholar]
- Gomez‐Deza J, Lee Y, Troakes C, et al. (2015) Dipeptide repeat protein inclusions are rare in the spinal cord and almost absent from motor neurons in C9ORF72 mutant amyotrophic lateral sclerosis and are unlikely to cause their degeneration. Acta Neuropathol Commun 3, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton KA, Heinbockel T, Ennis M, et al. (2005) Properties of external plexiform layer interneurons in mouse olfactory bulb slices. Neuroscience 133, 819–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartikainen P, Räsänen J, Julkunen V, et al. (2012) Cortical thickness in frontotemporal dementia, mild cognitive impairment, and Alzheimer's disease. J Alzheimers Dis JAD 30, 857–874. [DOI] [PubMed] [Google Scholar]
- Heyanka DJ, Golden CJ, McCue RB, et al. (2014) Olfactory deficits in frontotemporal dementia as measured by the Alberta Smell Test. Appl Neuropsychol Adult 21, 176–182. [DOI] [PubMed] [Google Scholar]
- Hornberger M (2012) Assessment of psychiatric changes in C9ORF72 frontotemporal dementia. Alzheimers Res Ther 4, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiung G‐YR, DeJesus‐Hernandez M, Feldman HH, et al. (2012) Clinical and pathological features of familial frontotemporal dementia caused by C9ORF72 mutation on chromosome 9p. Brain 135, 709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashi E, Valdmanis PN, Dion P, et al. (2008) TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet 40, 572–574. [DOI] [PubMed] [Google Scholar]
- Khalaf‐Nazzal R, Francis F (2013) Hippocampal development – old and new findings. Neuroscience 248, 225–242. [DOI] [PubMed] [Google Scholar]
- King A, Al‐Sarraj S, Shaw C (2009) Frontotemporal lobar degeneration with ubiquitinated tau‐negative inclusions and additional alpha‐synuclein pathology but also unusual cerebellar ubiquitinated p62‐positive, TDP‐43‐negative inclusions. Neuropathol Off J Jpn Soc Neuropathol 29, 466–471. [DOI] [PubMed] [Google Scholar]
- Kiyota T, Morrison CM, Tu G, et al. (2015) Presenilin‐1 familial Alzheimer's disease mutation alters hippocampal neurogenesis and memory function in CCL2 null mice. Brain Behav Immun 49, 311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppers M, Blokhuis AM, Westeneng H‐J, et al. (2015) C9orf72 ablation in mice does not cause motor neuron degeneration or motor deficits. Ann Neurol 78, 426–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski TJ, Bosco DA, Leclerc AL, et al. (2009) Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323, 1205–1208. [DOI] [PubMed] [Google Scholar]
- Laakso MP, Frisoni GB, Könönen M, et al. (2000) Hippocampus and entorhinal cortex in frontotemporal dementia and Alzheimer's disease: a morphometric MRI study. Biol Psychiatry 47, 1056–1063. [DOI] [PubMed] [Google Scholar]
- Lee Y‐B, Chen H‐J, Peres JN, et al. (2013) Hexanucleotide repeats in ALS/FTD form length‐dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep 5, 1178–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine TP, Daniels RD, Gatta AT, et al. (2013) The product of C9orf72, a gene strongly implicated in neurodegeneration, is structurally related to DENN Rab‐GEFs. Bioinformatics 29, 499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling S‐C, Polymenidou M, Cleveland DW (2013) Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron 79, 416–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu DC, Niu T, Alaynick WA (2015) Molecular and cellular development of spinal cord locomotor circuitry. Front Mol Neurosci 8, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzzi S, Snowden JS, Neary D, et al. (2007) Distinct patterns of olfactory impairment in Alzheimer's disease, semantic dementia, frontotemporal dementia, and corticobasal degeneration. Neuropsychologia 45, 1823–1831. [DOI] [PubMed] [Google Scholar]
- Mackenzie IRA, Frick P, Grässer FA, et al. (2015) Quantitative analysis and clinico‐pathological correlations of different dipeptide repeat protein pathologies in C9ORF72. Acta Neuropathol (Berl) 130, 845–861. [DOI] [PubMed] [Google Scholar]
- Mann DM, Rollinson S, Robinson A, et al. (2013) Dipeptide repeat proteins are present in the p62 positive inclusions in patients with frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9ORF72. Acta Neuropathol Commun 1, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marat AL, Dokainish H, McPherson PS (2011) DENN domain proteins: regulators of Rab GTPases. J Biol Chem 286, 13791–13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmigère F, Ernfors P (2007) Specification and connectivity of neuronal subtypes in the sensory lineage. Nat Rev Neurosci 8, 114–127. [DOI] [PubMed] [Google Scholar]
- Martinez S, Andreu A, Mecklenburg N, et al. (2013) Cellular and molecular basis of cerebellar development. Front Neuroanat 7, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May S, Hornburg D, Schludi MH, et al. (2014) C9orf72 FTLD/ALS‐associated Gly‐Ala dipeptide repeat proteins cause neuronal toxicity and Unc119 sequestration. Acta Neuropathol (Berl) 128, 485–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu Y, Gage FH (2011) Adult hippocampal neurogenesis and its role in Alzheimer's disease. Mol Neurodegener 6, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orasji SSS, Mulder JL, de Bruijn SFTM, et al. (2016) Olfactory dysfunction in behavioral variant frontotemporal dementia. Clin Neurol Neurosurg 141, 106–110. [DOI] [PubMed] [Google Scholar]
- Pardini M, Huey ED, Cavanagh AL, et al. (2009) Olfactory function in corticobasal syndrome and frontotemporal dementia. Arch Neurol 66, 92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, et al., ITALSGEN Consortium (2011) A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21‐linked ALS‐FTD. Neuron 72, 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringholz GM, Appel SH, Bradshaw M, et al. (2005) Prevalence and patterns of cognitive impairment in sporadic ALS. Neurology 65, 586–590. [DOI] [PubMed] [Google Scholar]
- Schludi MH, May S, Grässer FA, et al. , German Consortium for Frontotemporal Lobar Degeneration, Bavarian Brain Banking Alliance (2015) Distribution of dipeptide repeat proteins in cellular models and C9orf72 mutation cases suggests link to transcriptional silencing. Acta Neuropathol (Berl) 130, 537–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H (2009) Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol 10, 513–525. [DOI] [PubMed] [Google Scholar]
- Stewart H, Rutherford NJ, Briemberg H, et al. (2012) Clinical and pathological features of amyotrophic lateral sclerosis caused by mutation in the C9ORF72 gene on chromosome 9p. Acta Neuropathol (Berl) 123, 409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Dykes IM, Liang X, et al. (2008) A central role for Islet1 in sensory neuron development linking sensory and spinal gene regulatory programs. Nat Neurosci 11, 1283–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Maroof AM, Merkle FT, et al. (2013) The mouse C9ORF72 ortholog is enriched in neurons known to degenerate in ALS and FTD. Nat Neurosci 16, 1725–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan RH, Devenney E, Dobson‐Stone C, et al. (2014) Cerebellar integrity in the amyotrophic lateral sclerosis – frontotemporal dementia continuum. PLoS ONE 9, e105632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treloar HB, Miller AM, Ray A, et al. (2010). Development of the olfactory system In: The Neurobiology of Olfaction, Frontiers in Neuroscience. (ed. Menini A.), Chapter 5. Boca Raton: CRC Press/Taylor & Francis. [Google Scholar]
- Van Langenhove T, van der Zee J, Van Broeckhoven C (2012) The molecular basis of the frontotemporal lobar degeneration‐amyotrophic lateral sclerosis spectrum. Ann Med 44, 817–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waite AJ, Bäumer D, East S, et al. (2014) Reduced C9orf72 protein levels in frontal cortex of amyotrophic lateral sclerosis and frontotemporal degeneration brain with the C9ORF72 hexanucleotide repeat expansion. Neurobiol Aging 35, 1779.e5–1779.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheaton MW, Salamone AR, Mosnik DM, et al. (2007) Cognitive impairment in familial ALS. Neurology 69, 1411–1417. [DOI] [PubMed] [Google Scholar]
- Whitwell JL, Weigand SD, Boeve BF, et al. (2012) Neuroimaging signatures of frontotemporal dementia genetics: C9ORF72, tau, progranulin and sporadics. Brain 135, 794–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, MacNair L, McGoldrick P, et al. (2015) Isoform‐specific antibodies reveal distinct subcellular localizations of C9orf72 in amyotrophic lateral sclerosis. Ann Neurol 78, 568–583. [DOI] [PubMed] [Google Scholar]
- Zhang D, Iyer LM, He F, et al. (2012) Discovery of novel DENN proteins: implications for the evolution of eukaryotic intracellular membrane structures and human disease. Front Genet 3, 283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Liu Y, Bañez‐Coronel M, et al. (2013) RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci U S A 110, E4968–4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Comparison of two C9orf72 antibodies by Western blot. (A) ClustalO alignment of mouse and human C9orf72 protein isoforms showing antibody epitopes. (B) Western blot for purified recombinant human C9orf72 using either ProteinTech (PT) or SantaCruz (SC) primary antibody showing a strong band at 55 kDa for His‐tagged C9orf72 isoform 1. (C) Western blot of P10 mouse cortex and cerebellum using SantaCruz primary antibody shows isoform 1 is the predominant form. Each lane represents tissue pooled from two mice. GAPDH used as loading control. Unpurified bacterial lysate of recombinant human his‐tagged C9orf72 isoform 1 used as positive control (rC9).
Fig. S2. Comparison of two C9orf72 antibodies by immunostaining on brain sections. Comparison of adjacent sections of mouse brain immunostained with either ProteinTech or SantaCruz anti C9orf72 antibody and counterstained with DAPI in the cerebellum (C), hind brain (D) and cortex (E).
Fig. S3. C9orf72 expression in the spinal cord is associated strongly with neurofilament. Transverse sections of mid‐thoracic spinal cord from the indicated embryonic stages. Stained with C9orf72 and neurofilament (NF), counterstained with DAPI. C9orf72 expression in the ventral spinal cord is strongest in the transverse tracts of the lateral (lf) and ventral funiculus (vf) at E12.5 (A). C9orf72 expression is also found with associated neurofilament in the dorsal root ganglia with expression increasing from E12.5 to E16.5. Scale bars: 100 μm.


