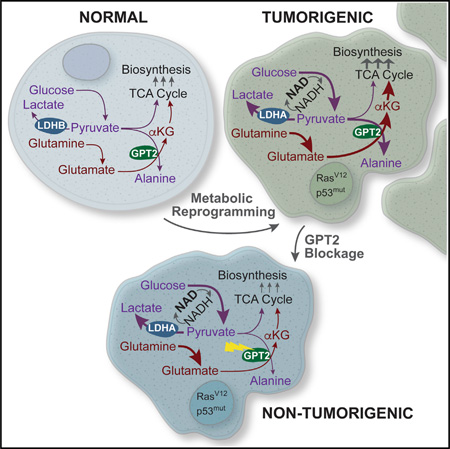
SUMMARY
Metabolic reprogramming is critical to oncogenesis, but the emergence and function of this profound reorganization remain poorly understood. Here we find that cooperating oncogenic mutations drive large-scale metabolic reprogramming, which is both intrinsic to cancer cells and obligatory for the transition to malignancy. This involves synergistic regulation of several genes encoding metabolic enzymes, including the lactate dehydrogenases LDHA and LDHB and mitochondrial glutamic pyruvate transaminase 2 (GPT2). Notably, GPT2 engages activated glycolysis to drive the utilization of glutamine as a carbon source for TCA cycle anaplerosis in colon cancer cells. Our data indicate that the Warburg effect supports oncogenesis via GPT2-mediated coupling of pyruvate production to glutamine catabolism. Although critical to the cancer phenotype, GPT2 activity is dispensable in cells that are not fully transformed, thus pinpointing a metabolic vulnerability specifically associated with cancer cell progression to malignancy.
Graphical Abstract
INTRODUCTION
Many types of cancer cells exhibit pronounced metabolic reprogramming compared with non-transformed cells. The most well documented of these metabolic alterations is the activation of aerobic glycolysis; i.e., the Warburg effect (Warburg, 1956). In addition to glycolytic activation, cancer cells frequently activate fatty acid biosynthesis and glutamine consumption (DeBerardinis et al., 2007; Kuhajda, 2000; Wise et al., 2008). More recently, this metabolic induction has been shown to be an essential feature of the transformed state. A number of metabolic enzymes activated in cancerous cells have been found to be critical for tumorigenesis. These include enzymes involved in glycolysis (Christofk et al., 2008; Fantin et al., 2006; Telang et al., 2006), fatty acid biosynthesis (Bauer et al., 2005; Hatzivassiliou et al., 2005), and glutaminolysis (Gao et al., 2009; Son et al., 2013; Wise et al., 2008; Yuneva et al., 2007). It is also clear that specific oncogenic mutations, for example, those activating the Ras-Akt-mTOR pathways, are critical for activation of common cancer-associated metabolic activities (Deprez et al., 1997; Elstrom et al., 2004; Gaglio et al., 2011; Guo et al., 2011; Kole et al., 1991; Ramanathan et al., 2005; Telang et al., 2007; Vizan et al., 2005; Ying et al., 2012). Little is known, however, about the emergence of metabolic reprogramming and its coordination during the cellular transition to malignancy, due, at least in part, to the presence of multiple causative genetic alterations in cancerous tissues.
Mechanistic insights into the complex structure of cellular regulation underlying malignant cell transformation come from exploration into how distinct oncogenic mutations cooperate to induce such a profound transition (Kinsey et al., 2014; Lloyd et al., 1997; McMurray et al., 2008; Sewing et al., 1997; Smith and Land, 2012; Xia and Land, 2007). In this context, it is notable that numerous genes essential to tumorigenesis can readily be identified by virtue of their synergistic response to cooperating oncogenic mutations. As indicated by genetic perturbation experiments, such genes, termed cooperation response genes (CRGs), contribute to the malignant phenotype at a frequency of >50% (McMurray et al., 2008). CRGs affect diverse cellular mechanisms, including signaling, gene expression, motility, and certain aspects of metabolism, thus pinpointing tangible links by which oncogenic mutations affect metabolic reprogramming, among other effects.
Here we report the emergence of metabolic reprogramming as a function of oncogene cooperation. We utilized a model of oncogenesis in which a constitutively active Ras12V allele and a dominant-negative p53175H allele cooperate to rapidly convert colon crypt cells to malignant cancer cells in vitro (McMurray et al., 2008; Xia and Land, 2007). This enabled direct elucidation of how the expression of individual oncogenic alleles affects metabolic functionality as opposed to dissecting out the multifaceted consequences of inhibiting oncogenic pathways in tumor-derived tissues. We find that cooperation of both p53175H and Ras12V is required and sufficient to induce the majority of cancer cell metabolic phenotypes, including shunting of glucose-derived carbon to lactate, increased glutamine consumption, and fatty acid biosynthesis induction. Furthermore, our results indicate that oncogenic p53 and Ras cooperatively regulate the expression of several metabolic genes we find to be essential for tumorigenesis. These genes include both isoforms of lactate dehydrogenase (LDHA and LDHB), which are induced and repressed, respectively, and GPT2, a mitochondrial glutamate-dependent transaminase that is also oncogenically induced. Reversion of any of these oncogenically driven changes substantially attenuates tumorigenesis. Notably, we show that induction of GPT2 exploits the generation of alanine from the glycolytic end product pyruvate as a means to drive alpha-ketoglutarate formation from glutamate, thus facilitating entry of glutamine carbon into the tricarboxylic acid (TCA) cycle. We also show that this activity is critical to the cancer cell phenotype while being dispensable in cells that are not fully transformed, thus pinpointing a metabolic vulnerability specifically associated with cancer cell proliferation and carcinogenesis. Together, our data provide evidence of a critical link between activated glycolysis and glutamine-dependent TCA cycle anaplerosis, suggesting that production of pyruvate to enable glutamine catabolism is a critical contribution the Warburg effect provides toward oncogenesis.
RESULTS
Oncogenic Ras and Mutant p53 Cooperatively Induce the Cancer Cell Metabolic Program
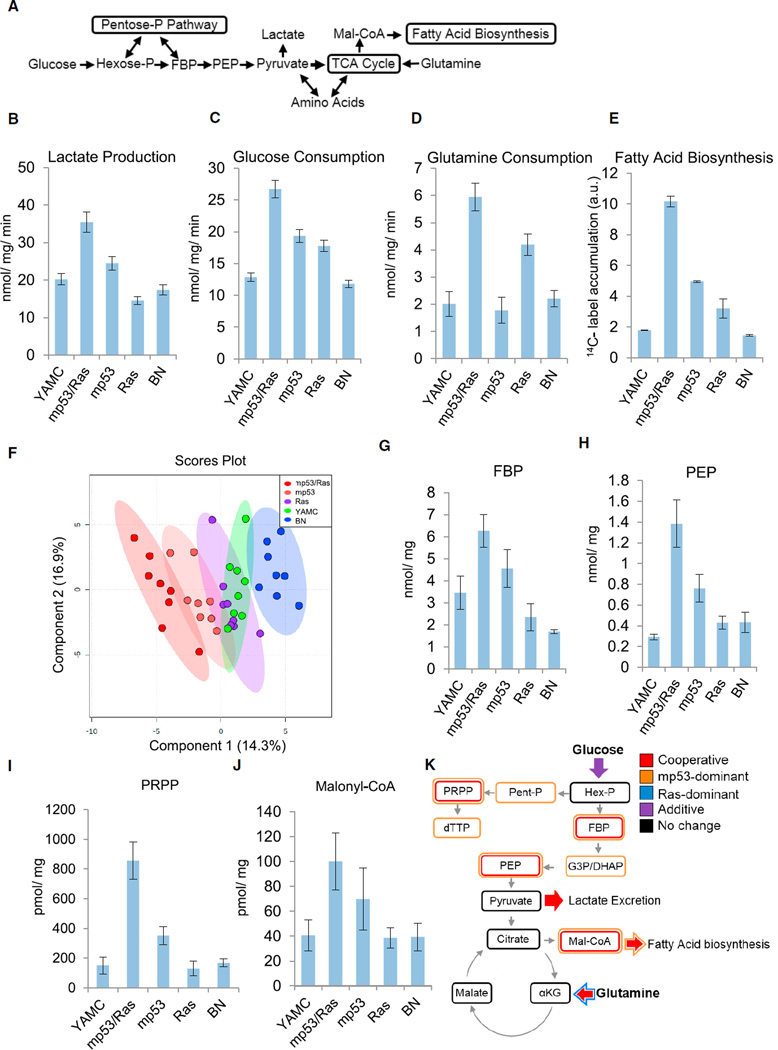
The most widely described metabolic attribute of cancerous cells is the activation of glycolysis with increased secretion of the glycolytic end product lactate; i.e., the Warburg effect (Figure 1A). It remains unclear, however, at what point of the multi-step process of carcinogenesis the glycolytic phenotype emerges and whether this transition is driven by cell-intrinsic mechanisms or by selective forces in the cancer microenvironment (e.g., oxygen limitation). We have extensively used young adult murine colon (YAMC) cells (Whitehead et al., 1993) transduced with activated Ras and mutant p53 to investigate the molecular mechanisms underlying the cooperative nature of malignant cell transformation, which has revealed cancer cell-specific vulnerabilities relevant to human disease (Kinsey et al., 2014; McMurray et al., 2008; Smith and Land, 2012; Xia and Land, 2007; Experimental Procedures). This model is therefore well suited to investigate the metabolic changes associated with the transition to malignancy. It consists of non-transformed parental YAMC cells as well as transduction with either control vectors (bleomycin/neomycin control [BN]) or vectors expressing oncogenic HRas12V (Ras cells), mutant p53175H (mp53 cells), or both oncogenic alleles together (mp53/Ras cells). For all transduced cells, low-passage polyclonal derivatives were utilized to minimize the effects of clonal variation on specific phenotypes. Examination of lactate secretion indicated that transduction of either oncogenic allele individually (i.e., either p53175H or Ras12V) had a negligible effect on lactate excretion compared with parental or control transduced cells (Figure 1B; Table S1). In contrast, cells transduced with both activated Ras and mutant p53 secreted substantially more lactate than control cells or cells transduced with either mutant allele alone (Figure 1B; Table S1). Similarly, expression of both oncogenic alleles increased glucose consumption to a greater extent than expression of either allele in isolation (Figure 1C; Table S1). In addition to glycolytic activation, cancerous cells frequently increase glutamine consumption and fatty acid biosynthesis (Bauer et al., 2005; DeBerardinis et al., 2008; Kuhajda et al., 1994; Rashid et al., 1997; Wise et al., 2008; Figure 1A). Expression of oncogenic RasV12 increased glutamine consumption compared with controls, whereas p53175H expression had little effect (Figure 1D; Table S1). Similar to the observations with glycolysis, co-expression of both oncogenic alleles cooperatively induced glutamine consumption (Figure 1D; Table S1) and also induced fatty acid biosynthesis relative to parental cells or cells expressing either allele in isolation (Figure 1E; Table S1). Combined, these results indicate that individual p53175H or Ras12V mutations in isolation are not sufficient to drive common cancer cell metabolic phenotypes but, rather, that these mutations cooperate to drive core components of the tumor cell metabolic program, including aerobic glycolytic activation, increased glutamine catabolism, and activation of fatty acid biosynthesis. Further, the finding that combined oncogene expression induces the Warburg effect suggests that this central feature of the cancer cell metabolic program can emerge independent of the environmental conditions associated with tumors, such as oxygen limitation.
Figure 1. Oncogenic Mutations Act Synergistically to Induce the Cancer Cell Metabolic Program.
(A) Schematic of central carbon metabolism.
(B–D) Extracellular lactate secretion (B), glucose consumption (C), and glutamine consumption (D) of the indicated derivatives of YAMC: combined mutant (mp53/Ras), mp53 alone, Ras alone, or empty vector (BN) cells.
(E) Measurement of fatty acid biosynthesis.
(B–E) Data are means + SE.
(F) Partial least-squares discriminant analysis of metabolic concentrations and the indicated YAMC derivatives.
(G–J) Absolute intracellular metabolite concentrations determined by LC-MS/MS. Data are means + SE.
(K) Oncogenic cooperativity and allele dominance in metabolic reprogramming of murine colon epithelial cells.
Statistically significant differences were assessed by ANOVA as indicated in the Experimental Procedures (Tables S1 and S4). See also Figures S1 and S2 and Tables S1-S4.
The observed oncogene-induced metabolic changes do not arise from simple differences in cell proliferation rates because metabolic induction does not correlate with proliferation rates. For example, mp53/Ras cells produce 2-fold more lactate than Ras cells despite proliferating at similar rates (Figure 1B; Figure S1A). Further, Ras cells produce less lactate than mp53 cells even though Ras cells proliferate faster (Figures 1A and 1D; Figure S1A). Moreover, although Ras overexpression can induce senescence in specific contexts (Serrano et al., 1997), the robust proliferation of Ras cells (Figure S1A) coupled with a lack of senescence-associated β-galactosidase (β-gal) staining (Figures S1B and S1C) indicates that the Ras cells were not undergoing senescence at the time of metabolic measurements. Their metabolic status thus cannot be attributed to a senescent cell state. Taken together, these results demonstrate that the metabolic alterations associated with these alleles are not simply a function of either increased proliferation or senescence.
To examine how p53 and Ras mutations affect cell metabolism more broadly, we employed liquid chromatography-tandem mass spectrometry (LC-MS/MS) to globally profile how metabolite concentrations are affected by p53175H and Ras12V expression. The pool sizes of 69 different metabolites are reported in Tables S2 and S3. Using the levels of these metabolite pools, a partial least-squares discriminant analysis (PLS-DA) model was constructed and subsequently validated through cross-validation and permutation testing (Figures S1C and S2D). In the resulting PLS-DA scores plot, the mp53/Ras cells segregated from the non-transformed control cells (Figure 1F). The mp53 cells exhibited an intermediate separation between the mp53/Ras cells and the controls, whereas the metabolite concentrations of the Ras cells largely overlapped with the control cells (Figure 1F). The metabolites whose levels were significantly modulated by oncogene expression (Table S4), and that contributed the most to the principal component separation of the different cell types largely belonged to key oncogenically activated pathways such as glycolysis, nucleotide biosynthesis, and lipid biosynthesis (Figure S1E). For glycolysis, these oncogene-induced metabolites include fructose bisphosphate (FBP) (Figure 1G) and phosphoenolpyruvate (PEP) (Figure 1H), the production of which is heavily regulated and two of the rate-controlling steps of glycolysis (Mazurek et al., 2002; Voet and Voet, 2004). Various pentose phosphate pathway metabolites were also induced, including ribose phosphate (Table S4) and 5-phosphoribosyl-α-pyrophosphate (PRPP) (Figure 1I), whose production is a rate-controlling step of de novo nucleotide biosynthesis (Voet and Voet, 2004). Although these pentose phosphate pathway intermediates were elevated in the transformed cells, the rate of oxidative pentose phosphate flux, which generates ribose and nicotinamide adenine dinucleotide phosphate (NADPH) for biosynthesis and protection from oxidative stress, was only modestly increased in mp53/Ras cells (Figures S2A and S2B). Consistent with the observed oncogenic increases in fatty acid biosynthesis, joint expression of p53175H and RasV12 substantially increased malonyl-coenzyme A (CoA) levels (Figure 1J), whose production is the tightly regulated initial reaction of fatty acid biosynthesis (Figure 1A; Brownsey et al., 2006).
Oncogenic p53 and Ras mutations cooperate to induce many transformation-specific phenotypes (Kinsey et al., 2014; McMurray et al., 2008; Smith and Land, 2012; Xia and Land, 2007), and, accordingly, many of the metabolic phenotypes observed here were cooperatively induced (Figure 1K). However, a number of these metabolic activities were induced predominately by expression of either mp53 or oncogenic Ras. For example, expression of oncogenic Ras predominantly drove the increases in glutamine consumption (Figures 1D and 1K) but did not affect lactate secretion (Figure 1B). Alternatively, expression of mp53 was the major factor responsible for increasing the various glycolytic and pentose phosphate metabolites (Figure 1K). These mp53-induced concentrations were the major factors that led to the intermediate segregation of mp53 cells relative to the fully transformed and control cells observed in the PLS-DA model (Figure 1F; Figures S1C–S1E). Notably, the most widely defining feature of cancer metabolism (i.e., induction of lactate secretion) was the most cooperatively induced phenotype observed (Figures 1B and 1K). It has been reported previously that oncogenic Ras expression in non-transformed fibroblasts drives increased lactate secretion (Gaglio et al., 2011). Our data indicate that these Ras-induced changes are context-dependent and strongly suggest that, although expression of different oncogenic alleles activates specific metabolic activities, complete oncogenic reprogramming involves cooperative action between both mp53 and oncogenic Ras.
Cooperative De-regulation of Lactate Dehydrogenase Activity Is Critical for Tumorigenesis
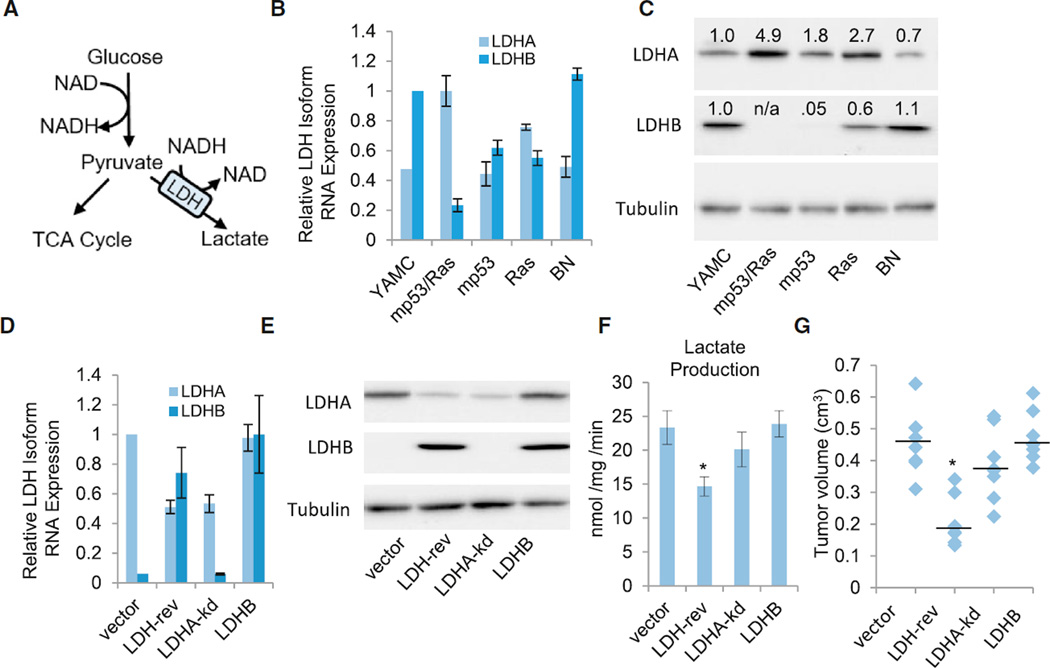
The highly cooperative induction of lactate production in transformed cells suggests that a coordinated deregulation of LDH isoform expression underlies emergence of the glycolytic phenotype during oncogenesis. LDH mediates conversion of the glycolytic end product pyruvate to lactate, utilizing the cofactor NADH (Figure 2A). The enzyme comprises a tetramer that can be composed of any ratio of two isoforms, A and B, encoded by distinct genes, LDHA and LDHB. Tetramers containing more LDHA isoforms exhibit increased activity compared with those with more LDHB (Read et al., 2001). Notably, LDHA expression has been found to be both induced during and important for tumorigenesis in some experimental models (Fantin et al., 2006; Le et al., 2010). However, the coordination of LDHA and LDHB isoform expression in controlling the glycolytic phenotype of cancer cells is poorly understood. Analysis of LDH isoform mRNA and protein levels in our model indicated that oncogenic Ras is dominant in upregulating LDHA expression, whereas mp53 is sufficient to suppress LDHB (Figures 2B and 2C). However, cooperation of both oncogenic alleles was required for the LDHA:LDHB ratio observed in mp53/Ras transformed cells (Figures 2B and 2C), consistent with the pattern of lactate production (Figure 1B). A similar switch in LDH isoform expression from LDHB to LDHA was observed in human pancreatic (Figures S3A and S3B) and prostate (Figures S3D and S3E) cancers compared with respective normal tissues, indicating relevance to human disease. In colon cancer, both LDH isoforms were elevated (Figures S3G and S3H), suggesting that alternative paths to LDH deregulation may exist. Further, although expression of LDHA and LDHB is significantly different in pancreatic cancer specimens with both p53 and Ras mutations versus cancers with neither of these mutations, LDH isoform expression did not correlate with the p53/Ras mutational status of prostate or colon tumor samples (Figures S3C, S3F, and S3H). Overall, this suggests that LDH deregulation frequently emerges with the cancer phenotype in general rather than being limited to mutant p53- and Ras-driven cancers.
Figure 2. The LDH Isoform Switch Is Critical to Tumorigenesis.
(A) Glycolysis schematic.
(B and C) LDHA or LDHB RNA expression (B) and protein expression (C) in parental (YAMC) cells or the indicated derivatives of YAMC cells: empty vector (BN), mp53, Ras, and combined mutant cells (mp53/Ras).
(D and E) LDHA or LDHB RNA expression (D) and protein expression (E) in the indicated derivatives of PR cells: vector control (vector), LDHA knockdown plus LDHB overexpression (LDH-rev), LDHA-kd, or LDHB overexpression (LDHB).
(B and E) RNA expression of LDHA or LDHB was measured via real-time PCR using gene-specific primers.
(D and C) Protein expression values of LDHA and LDHB were normalized to tubulin expression.
(F) Lactate secretion from the indicated derivatives of mp53/Ras cells.
(B–F) Data are means + SE. *p < 0.01 versus all other cell lines by Student’s t test.
(G) Tumor volumes 4 weeks after eight subcutaneous implantations of the indicated derivatives of mp53/Ras cells in the flanks of nude mice. Median tumor volume is indicated by the horizontal line. *p < 0.01 versus all other cell lines by Student’s t test.
See also Figure S3.
To test the effect of coordinated deregulation of both LDHA and LDHB on oncogenic phenotypes, we reset their expression in mp53/Ras cells to non-transformed levels using LDHB expression and LDHA short hairpin RNA (shRNA) constructs to increase LDHB and decrease LDHA levels, respectively (Figures 2D and 2E). Analysis of lactate excretion upon targeting of LDH expression indicated that, individually, neither knockdown of LDHA expression nor overexpression of LDHB were sufficient to significantly reduce lactate secretion (Figure 2F). However, when both LDHA expression was knocked down and LDHB expression was induced, the levels of lactate excretion were significantly reduced (Figure 2F). Similarly, individual knockdown of LDHA or overexpression of LDHB did not affect tumor formation (Figure 2G). However, the tumor growth potential was substantially reduced upon simultaneous reversion of both LDHA and LDHB in mp53/Ras cells (also known as LDH-reversed cells) (Figure 2G). Notably, tumors that did arise following xeno-implantation of LDH-reversed mp53/Ras cell populations exhibited signs of escape; i.e., LDHA expression increased and LDHB expression decreased in the tumors (Figures S4A and S4B), further indicating that LDH reversion is incompatible with tumor growth. Together, these results suggest that cooperative oncogene-driven expression changes to specific LDH isoforms are critical for both LDH enzyme activity (i.e., lactate production) and tumorigenesis.
Oncogenically Induced LDH Isoform Switching Is Critical for Maintaining TCA Anaplerosis during Hypoxia
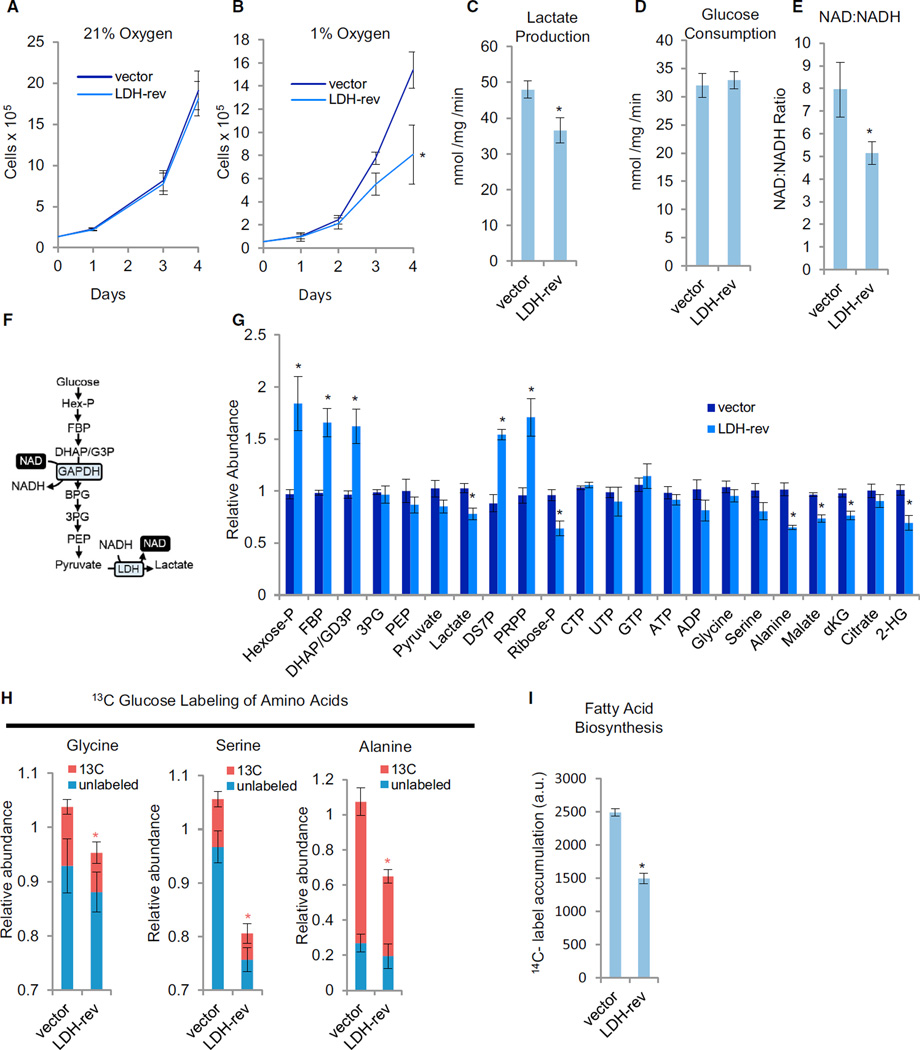
LDH activity is critical for both tumorigenesis and proliferation under hypoxia but not in the presence of atmospheric oxygen (Fantin et al., 2006; Xie et al., 2014), a notion confirmed in LDH-reversed mp53/Ras cells (Figures 2G and 3A and 3B). Prolonged exposure to hypoxia has been reported to reduce both mitochondrial membrane potential and ATP concentrations (Fantin et al., 2006; Figure S4C). The LDH-mediated effects that initiate the collapse of the malignant state, however, remain unclear. Here we show that the earliest detectable metabolic changes following oxygen limitation in LDH-reverted cells not only include substantial changes to glycolysis and glucose-mediated biosynthesis but also to glutamine-driven TCA anaplerosis (detailed below).
Figure 3. The Effect of LDH Isoform Reversal under Hypoxia.
(A and B) Adherent cell proliferation of vector control (vector) and LDH isoform-reversed (LDH-rev) mp53/Ras cells under 21 % oxygen (A) or 1 % oxygen (B). Data are means + SD. *p < 0.01 versus vector control by Student’s t test.
(C and D) Extracellular lactate secretion (C) and glucose consumption (D) at 1 % oxygen.
(E) Total intracellular NAD:NADH ratio at 1 % oxygen determined by LC-MS/MS.
(F) Schematic of the glycolytic pathway.
(G) Relative intracellular metabolite levels at 1 % oxygen determined by LC-MS/MS.
(B–E) Data are means + SE. *p < 0.01 versus vector control by Student’s t test.
(H) Cells were labeled with 13C-glucose for 90 min at 1% oxygen, and the relative abundance of 13C and 12C isoforms was determined by LC-MS/MS. Columns represent total relative abundance, and the 13C portion represents the sum of all detectable isotopologues of a given metabolite. *p < 0.01 for 13C isoform abundance of the indicated metabolite versus that metabolite’s 13C isoform abundance in vector control cells by Student’s t test. Data are means + SE.
(I) Measurement of fatty acid biosynthesis at 1 % oxygen. Data are means + SE. *p < 0.01 versus vector control by Student’s t test.
(C–I) All measurements were made following 8- to 10-hr cell culture in 1 % oxygen.
See also Figures S4 and S5 and Table S5.
LDH reversion substantially reduced lactate secretion under hypoxia (Figure 3C). In contrast, glucose consumption rates were not affected (Figure 3D), indicative of glucose-derived carbon being shunted away from lactate because of reduced LDH activity. Notably, under limiting oxygen, LDH serves to regenerate nicotinamide adenine dinucleotide (NAD+), which is typically supplied by respiration, thus providing a rationale for the hypoxia-specific proliferative dependency of cancer cells on high LDH activity. Consistent with this scenario, the LDH-reverted cells exhibited a lower NAD:NADH ratio under hypoxia (Figure 3E) but not under non-limiting oxygen (data not shown). Reduced NAD+ pools can create a glycolytic bottleneck because NAD+ is required for the reaction catalyzed by GAPDH (Figure 3F). This bottleneck at GAPDH was observed, with LDH reversion inducing substantial increases in the glycolytic metabolites upstream of GAPDH, including Hexose-P, FBP, and dihydroxyacetone phosphate (DHAP)/glyceraldehyde 3-phosphate (G3P), while reducing the glycolytic metabolites downstream of GAPDH, such as PEP, pyruvate, and lactate (Figure 3G). Pentose phosphate pathway (PPP) intermediates were also affected, with an induction of sedoheptulose-7P and PRPP (Figure 3G), possibly reflecting increased mass action flow through PPP resulting from a GAPDH bottleneck. Moreover, at early time points after oxygen limitation, the levels of several nucleotides, including ATP, as well as the ATP:ADP ratio, were similar between control and LDH-reversed cells (Figure 3G), whereas the levels of alanine, serine, and the TCA cycle metabolites malate and α-ketoglutarate (αKG) were substantially reduced (Figure 3G). The levels of 2-hydroxyglurate (2-HG) were also reduced in LDH-reversed cells (Figure 3G), consistent with recent results indicating that LDHA is important for the induction of 2-HG under hypoxic conditions (Intlekofer et al., 2015), although changes to specific 2-HG enantiomers could not be resolved from our analysis.
Glucose is a major carbon source for alanine, serine, and glycine, and the decreased levels of these metabolites could reflect a decrease in their glucose-derived biosynthesis upon oxygen limitation. Glucose labeling patterns supported this idea, with LDH reversion decreasing the amount of glucose-derived alanine, serine, and glycine (Figure 3H; Figure S4D; see Figure S6 for labeling kinetics). Consistent with a compensatory response to a blockage in serine biosynthesis, the LDH-reverted cells increased their medium consumption of serine (Figure S4E). Glucose, via citrate, is also a major source of acetyl-CoA for fatty acid biosynthesis, which could depend on LDH activity upon oxygen limitation. Compatible with this hypothesis, LDH-reverted cells exhibited decreased fatty acid biosynthesis relative to vector controls (Figure 3I). Combined, our data indicate that, in LDH-reversed cancer cells, oxygen limitation produces multiple defects in glucose-fueled biosynthesis.
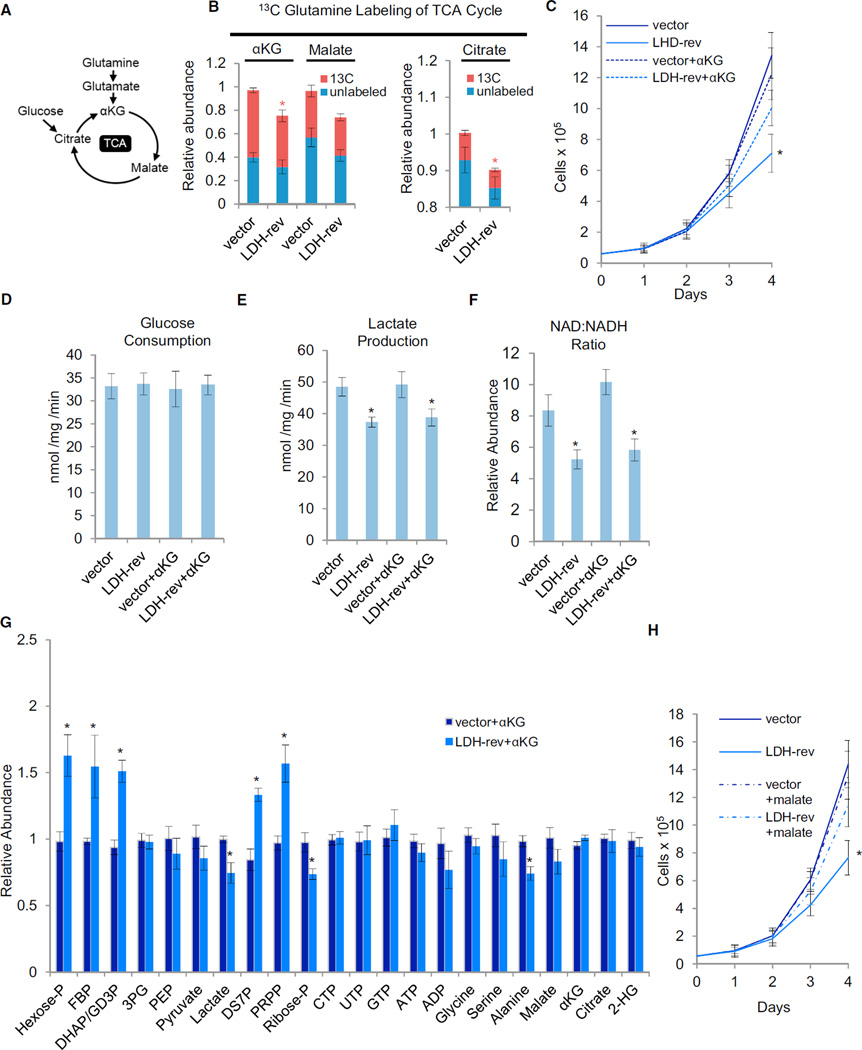
Glutamine is a major TCA cycle carbon source in cancerous cells, a process known as glutamine-driven TCA anaplerosis (DeBerardinis et al., 2007; Wise et al., 2008; Figure 4A). In addition to glutaminolysis, pyruvate carboxylation can also anaplerotically replenish TCA cycle pools, an activity that has been found to be increased in certain tumors (Fan et al., 2009). However, in contrast to glutamine consumption, expression of oncogenic Ras and/or mp53 had little effect on the levels of pyruvate carboxylation (Figures S2C and S2D) suggesting that, in this context, oncogene expression preferentially drives glutaminolytic over pyruvate carboxylation-based TCA anaplerosis. Given that LDH-reversed cells had reduced malate and αKG levels under hypoxic conditions (Figure 3G), we analyzed how LDH reversion affected glutamine-derived TCA cycle anaplerosis. Oxygen limitation reduced levels of glutamine-derived αKG, citrate, and malate in LDH-reversed cells (Figure 4B; Figure S4F), consistent with the observed decreases in the concentrations of these metabolites (Figure 3G). LDH reversion thus negatively affects the ability of glutamine to supply TCA cycle pools, suggesting that glutamine flux into the TCA cycle is partially dependent on LDH activity under hypoxic conditions.
Figure 4. LDH Supports Glutamine-Driven TCA Anaplerosis under Hypoxia.
(A) Schematic of the tricarboxylic acid (TCA) cycle
(B) Vector control (vector) or LDH-rev mp53/Ras cells were labeled with 13C-glutamine for 90 min, and the relative abundance of 13C and 12C isoforms was determined by LC-MS/MS. Columns represent total relative abundance, and the 13C portion represents the sum of all detectable isotopologues of a given metabolite. Measurements were made following 8- to 10-hr cell culture in 1 % oxygen. Data are means + SE. *p < 0.01 for 13C isoform abundance of the indicated metabolite versus that metabolite’s 13C isoform abundance in vector control cells by Student’s t test.
(C) Adherent cell proliferation in 1 % oxygen and in the presence of 4 mM αKG. Data are means + SD. *p < 0.01 versus all other cell lines by Student’s t test.
(D and E) Extracellular lactate secretion (D) and glucose consumption (E) at 1 % oxygen.
(F) Total intracellular NAD:NADH ratio at 1 % oxygen determined by LC-MS/MS.
(G) Relative intracellular metabolite levels at 1 % oxygen determined by LC-MS/MS.
(D–G) Measurements were made following 8- to 10-hr cell culture in 1% oxygen and in the presence of 4 mM αKG where indicated. Data are means + SE. *p< 0.01 versus vector control by Student’s t test.
(H) Adherent cell proliferation in 1 % oxygen and in the presence of 4 mM dimethyl malate (mal) where indicated. Data are means + SD. *p < 0.01 versus all other cell lines by Student’s t test.
See also Figures S4-S6 and Table S5.
To explore which metabolic pathways are relevant for cell proliferation under hypoxia, we assessed whether medium supplementation with specific metabolites whose levels were significantly down in LDH-reversed cells could rescue cell proliferation under limiting oxygen. As shown in Figure 4C, medium supplementation with αKG significantly increased the ability of LDH-reversed cells to proliferate under hypoxic conditions, thus indicating its functional relevance to the hypoxic proliferation defect exhibited by the LDH-reversed cells. Notably, supplementation with αKG did not affect glucose consumption, lactate secretion, or the NAD:NADH ratio of LDH-reversed cells (Figures 4D-4F), suggesting that the functional effect of αKG supplementation was independent of LDH’s glycolytic role. Further supporting this view, medium supplementation with αKG did not affect the impact of low oxygen on glycolytic metabolites; i.e., those pools upstream of GAPDH were still increased and those downstream of GAPDH were still decreased (Figure 4G). Alanine pools were also not affected by αKG supplementation, whereas the decrease in cellular αKG was rescued (Figure 4G). Similarly to αKG, medium supplementation with malate also substantially rescued the proliferation of LDH-reversed cells at low oxygen concentrations (Figure 4H), further highlighting the importance of TCA anaplerosis for this phenotype. In contrast, medium supplementation with alanine had no effect on cellular proliferation (Figure S4G), indicating that decreases in alanine do not play a critical role in the proliferation defect displayed by the LDH-reversed cells at low oxygen concentrations. Our data thus point toward a critical role for LDH in glutamine-driven TCA cycle anaplerosis under hypoxic conditions, suggesting this is an essential function of activated glycolysis in facilitating cancer cell proliferation.
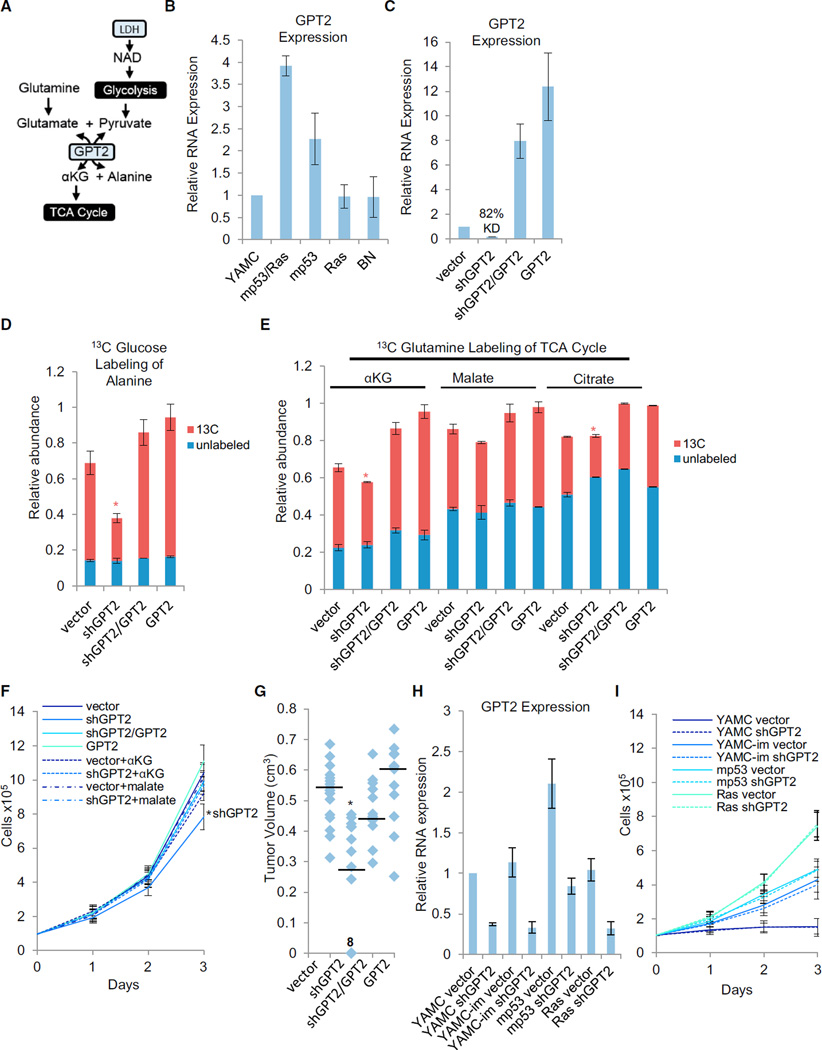
GPT2 Is Critical for TCA Anaplerosis and Tumorigenesis
Glutamine-driven TCA anaplerosis requires the transfer of nitrogen from glutamate to an acceptor molecule to form αKG. GPT2 transfers glutamate nitrogen to pyruvate, thus forming alanine and αKG (Figure 5A), making GPT2 a strong candidate for coupling glycolytic flux to TCA anaplerosis. This reaction readily occurs in mp53/Ras cells because we observed substantial nitrogen transfer from glutamine to alanine after labeling with α-15N-glutamine (Figure S6C). Moreover, in contrast with LDH isoform switching, whose TCA contributions only emerge under limiting oxygen concentrations (see above), the GPT2-catalyzed transamination would be predicted to contribute to TCA anaplerosis irrespective of oxygen availability. Consistent with this prediction, GPT2 expression was not only cooperatively induced in mp53/Ras cells (Figure 5B), but reversal of this induction via shRNA-mediated knockdown (Figure 5C) substantially attenuated both glucose-derived alanine labeling (Figure 5D; Figure S6A) and 13C-glutamine labeling of αKG and citrate under atmospheric oxygen (Figure 5E; Figure S6B). We thus conclude that GPT2 plays an essential role in both glutamine-mediated TCA anaplerosis and the utilization of glucose/glycolysis-derived pyruvate to drive this activity. GPT2 knockdown also inhibited cell proliferation at atmospheric oxygen in vitro, an effect that was restored by medium supplementation either with αKG or malate (Figure 5F) but not by alanine (Figure S6D), thus linking the requirement of GPT2 for maximal cell proliferation to its role in TCA anaplerosis. Pyruvate supplementation did not rescue proliferation (Figure S6E), consistent with GPT2 knockdown (kd) blocking the means for pyruvate to drive TCA anaplerosis. Pyruvate, on the other hand, does rescue proliferation of LDH-reversed cells in hypoxia, where pyruvate supply becomes limiting (Figure S6F). Reduction of GPT2 expression in mp53/Ras cells also significantly inhibited tumor formation in xenograft assays (Figure 5G). Moreover, tumors that did arise following xeno-implantation of GPT2-kd cells were comprised of cells that had lost knockdown (Figure S6G), further supporting the idea that low GPT2 expression is incompatible with tumor growth. In addition, the effect of GPT2 knockdown on tumor formation and cell proliferation as well as 13C-glucose and 13C-glutmaine labeling could be rescued by expression of an shRNA-resistant GPT2 allele (Figures 5D-5F), ruling out shRNA off-target effects as an underlying cause of these phenotypes. Notably, the GPT2 proliferative requirement emerged only in the context of malignant cell transformation because GPT2 knockdown did not affect the proliferation of the parental, immortalized parental, or single oncogene transduced cells (Figures 5H and 5I). Taken together, our data suggest that GPT2-mediated TCA cycle anaplerosis is critical and specific to the cancer phenotype.
Figure 5. Cooperative Induction of GPT2 Expression Is Important for Glutamine-Driven TCA Anaplerosis and Tumorigenesis.
(A) Schematic of GPT2-mediated coupling of glutamine-driven TCA anaplerosis to glycolytic production of pyruvate.
(B) GPT2 RNA expression in parental (YAMC) cells or the indicated derivatives of YAMC cells: combined mutant (mp53/Ras), mp53 alone, Ras alone, or empty vector (BN) cells.
(C) GPT2 RNA expression in the indicated derivatives of mp53/Ras cells: vector control (vector), GPT2 knockdown (shGPT2), GPT2 knockdown plus shRNA-resistant GPT2 cDNA expression (shGPT2/GPT2), or GPT2 overexpression (GPT2).
(B and C) Expression of GPT2 was measured via real-time PCR using gene-specific primers.
(D and E) Cells were labeled with 13C-glucose (D) or 13C-glutamine (E) for 90 min, and the relative abundance of 13C and 12C isoforms was determined by LC-MS/MS. Columns represent total relative abundance, and the 13C portion represents the sum of all detectable isotopologues of a given metabolite. *p < 0.01 for 13C isoform abundance of the indicated metabolite versus that metabolite’s 13C isoform abundance in all other cell lines by Student’s t test. Data are means + SE.
(F) Adherent cell proliferation in the presence of 4 mM αKG or 4 mM dimethyl malate where indicated. Data are means + SD. *p < 0.01 versus all other cell lines by Student’s t test.
(G) Tumor volumes 4 weeks after 12–18 subcutaneous implantations of the indicated derivatives of mp53/Ras cells in the flanks of nude mice. Median tumor volume is indicated by the horizontal line. *p<0.01 versus all other cell lines by Student’s t test. “8” on the graph indicates the number of data points represented by the indicated marker.
(H) GPT2 RNA expression in the indicated derivatives of YAMC, immortalized YAMC (YAMC-im), mp53, or Ras cells: vector control (vector) or GPT2 knockdown (shGPT2). Expression of GPT2 was measured via real-time PCR using gene-specific primers.
(I) Adherent cell proliferation of the indicated YAMC derivatives.
Data are means + SD. See also Figures S5 and S6 and Table S5.
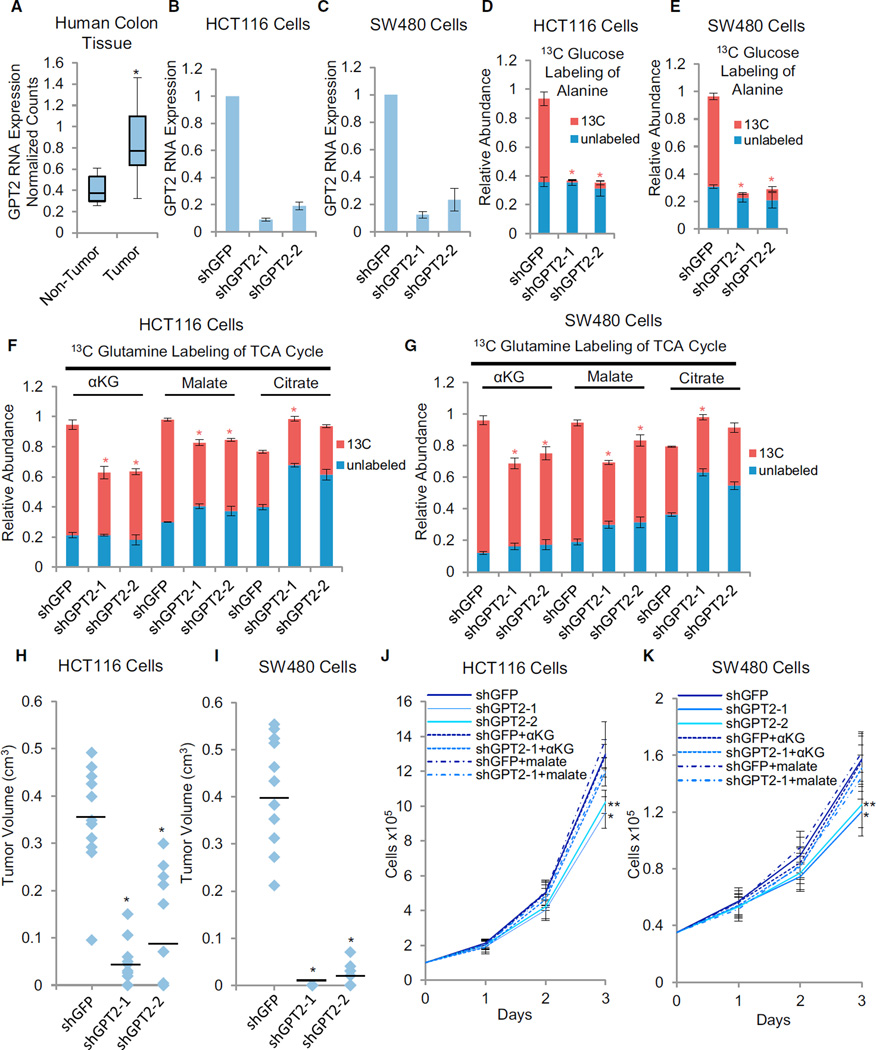
GPT2 expression is elevated in human colon cancer tissue compared with non-tumor control tissue (Figure 6A; Figure S7A). This increase appears to be independent of the p53 and Ras mutational status of these human tumors (Figure S7B), suggesting that GPT2 induction is a general feature of colon cancers that emerges as a consequence of malignant transformation rather than being driven specifically by oncogenic p53 and/or Ras mutations. To test the importance of GPT2 in a human colon cancer context, we targeted GPT2 expression with shRNA in two separate human colon cancer cell lines. GPT2 knockdown substantially reduced GPT2 expression in both HCT116 and SW480 cells and induced strong reductions in 13C-glucose-derived alanine labeling (Figures 6B-6E; Figure S7C). Similar to our observations in the murine system, knockdown of GPT2 in both human cancer lines reduced 13C-glutamine-derived labeling of TCA constituents, including reduced labeling of αKG, malate, and citrate (Figures 6F and 6G; Figures S7D and S7E). GPT2 knockdown also substantially inhibited the ability of either cell line to form tumors in mice (Figures 6H & I), as well as induced an in vitro proliferation defect. The proliferation defect of GPT2-kd cells could be rescued through malate or αKG medium supplementation (Figures 6J and 6K) but not alanine or pyruvate supplementation (Figures S7F–S7I). Notably, the in vivo tumor inhibitory effect of GPT2 knockdown was phenocopied by knocking down expression of glutaminase (Figures S7J–S7L), an enzyme that converts glutamine to glutamate. This further supports the functional importance of glutamine catabolism in GPT2-dependent tumorigenesis. Together, our results indicate that GPT2 is a critical link between glycolysis and the TCA cycle that harnesses glycolytically produced pyruvate to accept glutamine-derived nitrogen, thereby exploiting glycolytic activation to enable both TCA cycle anaplerosis and the malignant state of colon cancer cells.
Figure 6. GPT2 Is Critical for Human Colon Cancer Cell Proliferation.
(A) GPT2 RNA expression in human colon tumor versus non-tumor samples. *p < 0.01 versus non-tumor by Student’s t test. The y axis values represent linear scale.
(B and C) GPT2 RNA expression in the indicated derivatives of HCT116 (B) or SW480 (C) cells: shGFP control (shGFP), GPT2 knockdown-1 (shGPT2-1), or GPT2 knockdown-2 (shGPT2-2). Expression of GPT2 was measured via real-time PCR using gene-specific primers.
(D–G). Cells were labeled with 13C-glucose (D and E) or 13C-glutamine (F and G) for 90 min, and the relative abundance of 13C and 12C isoforms was determined by LC-MS/MS. Columns represent total relative abundance, and the 13C portion represents the sum of all detectable isotopologues of a given metabolite. *p<0.01 for 13C isoform abundance of the indicated metabolite versus that metabolite’s 13C isoform abundance in shGFP control by Student’s t test.
(B–E) Data are means + SE.
(H and I) Tumor volumes 4 weeks after 12 subcutaneous implantations of the indicated cell lines in the flanks of nude mice. Median tumor volume is indicated by the horizontal line. *p < 0.01 versus shGFP by Student’s t test.
(J and K) Adherent cell proliferation in the presence of 4 mM αKG or 4 mM dimethyl malate where indicated. Data are means + SD. *p < 0.01 versus all other cell lines, except shGPT2-2, by Student’s t test. **p < 0.01 versus shGFP by Student’s t test.
See also Figures S5 and S7 and Tables S6 and S7.
DISCUSSION
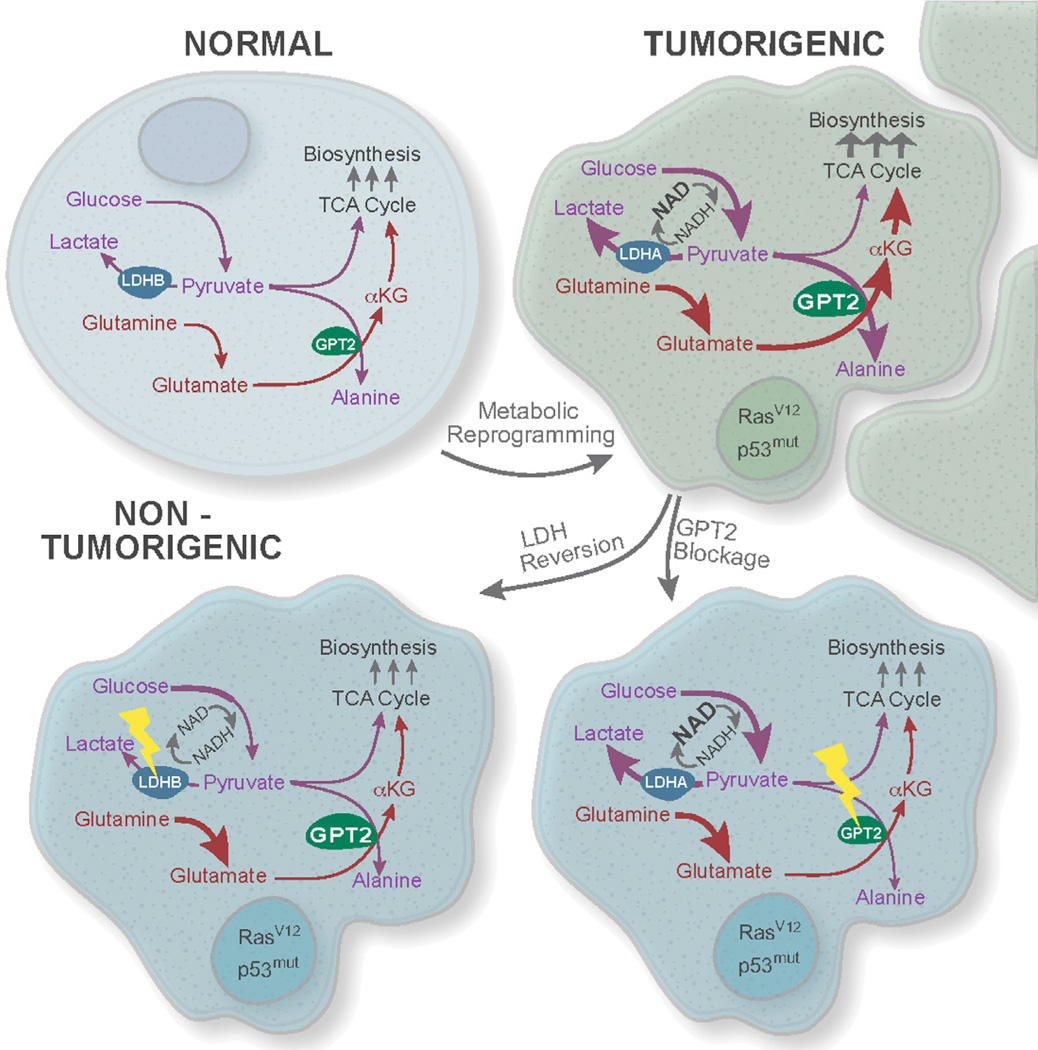
Oncogenic transformation of diverse tissue types frequently results in a global metabolic shift, with activation of both glycolysis and glutamine-driven TCA cycle anaplerosis as hallmarks critical to many cancers. Here we show that these two key features of cancer cell metabolic reprogramming are causally linked via coupling of pyruvate production to glutamine catabolism, thus revealing a critical contribution the Warburg effect makes toward oncogenesis (Figure 7). Coupling of glycolysis and TCA cycle anaplerosis occurs at the level of the mitochondrial enzyme GPT2, which plays an essential role in cancer cell proliferation. We also show that large-scale metabolic reprogramming by oncogenic p53 and Ras apparently is not dependent on environmental selective pressures such as oxygen limitation and acidosis, as suggested previously (Gatenby and Gillies, 2008). Oncogene-mediated metabolic reprogramming thus not only preconditions cancer cells to proliferate under adverse conditions but also creates vulnerabilities that may be exploited by intervention strategies targeting nodes in the metabolic network critical and specific to cancer cells.
Figure 7. Oncogene-lnduced Coupling of Aerobic Glycolysis and Glutamine-Driven TCA Cycle Anaplerosis Is Mediated by GPT2 and Critical to the Cancer Phenotype.
Oncogenic mutations induce metabolic reprogramming in part through switching expression of LDHB to LDHA and inducing GPT2 expression, resulting in both activation and coupling of aerobic glycolysis and TCA cycle anaplerosis to support cancer cell proliferation. We suggest that this link between glycolytically derived pyruvate and glutamine metabolism reflects a critical feature of the Warburg effect.
In general, oncogenic p53 and Ras are both required for driving full cancer cell metabolic reprogramming. In part, the individual mutations affect metabolic function in distinct ways. For example, expression of p53175H alone partially induces the levels of most glycolytic metabolites (Figure 1). These results are consistent with many reports that p53 inhibits the expression of glucose transporters while activating expression of negative regulators of glycolysis (Bensaad et al., 2006; Kawauchi et al., 2008; Schwartzenberg-Bar-Yoseph et al., 2004; Zhang et al., 2011). In contrast, although oncogenic Ras12V expression alone increases glucose consumption slightly, it had little effect on glycolytic pools, and it actually reduces lactate secretion (Figure 1). Activated Ras signaling is well known to stimulate glycolysis (reviewed in Bryant et al., 2014); however, its effects are strongly context-dependent (Baer et al., 2014; Ridley et al., 1988; To et al., 2013; Vartanian et al., 2013), and the observed muted effect of oncogenic Ras on glycolysis likely reflects a requirement for cooperative oncogenic signaling to induce lactate secretion. In contrast to glycolysis, oncogenic Ras expression is capable of inducing glutamine consumption (Figure 1), consistent with the notion that activated Ras is required for enhanced glutamine metabolism (Gaglio et al., 2011; Son et al., 2013; Telang et al., 2007). Nevertheless, the full extent of these changes only emerged with combined mp53 and oncogenic Ras expression, with synergistic induction of pathways such as glycolysis, lactate secretion, glutamine consumption, and fatty acid biosynthesis (Figure 1). Collectively, our data indicate that cancer cell metabolic reprogramming is a complex process and emerges in response to multiple oncogenic mutations.
Tumor-derived tissues exhibit pronounced genetic diversity and instability, thus complicating the detection of causal relationships relevant to the process of malignant transformation. In contrast, our work indicates that cooperating oncogenic mutations can induce a coordinated cancer cell metabolic program with intrinsic and thus cancer cell-specific metabolic vulnerabilities. In this context, synergistic regulation of LDH isoforms and GPT2 in response to cooperating oncogenic mutations served as a powerful identifier of downstream effectors critical to both carcinogenesis and metabolic reprogramming, although their regulation, at least in part, may reflect the nature of the malignant state rather than the identity of oncogenic driver mutations (Figure S3).
The two LDH isoforms LDHA and LDHB are regulated in opposite directions. Both induction of LDHA and a decrease in LDHB levels are required for full glycolytic activation and lactate secretion (Figure 2). Conversely, an inhibition of tumorigenesis only occurred when both oncogene-induced changes to LDH mRNAs were reversed (Figure 2), consistent with the notions that LDHA exhibits increased activity over LDHB (Read et al., 2001) and that LDHA expression is critical to tumorigenesis (Fantin et al., 2006). LDH activity can maintain glycolysis under limiting oxygen concentrations by providing NAD+ to the reaction catalyzed by GAPDH, which is thought to be the enzyme’s major contribution to tumorigenesis (reviewed in Augoff et al., 2015). Notably, here we report that LDH dependent glycolytic activity is critical for generating pyruvate to serve as a nitrogen acceptor for glutamine-driven TCA cycle anaplerosis under hypoxia (Figure 4). This highlights a link between glycolytically derived pyruvate and glutamine metabolism that is critical to cancer cell proliferation.
The coupling between glycolysis and TCA anaplerosis described here may shed further light on the unexpected finding that 2HG pools in cancer cells are increased during hypoxia and correlate with LDH activity (Intlekofer et al., 2015; Oldham et al., 2015). Given that 2-HG is produced from αKG, our findings indicating that αKG levels are dependent on LDH activity suggest that LDH activity could be important to supply αKG for 2-HG biosynthesis, a possibility that warrants further investigation.
Our results indicate that malignant cell transformation induces dependence on the aminotransferase GPT2. Aminotransferases facilitate amino acid-mediated TCA anaplerosis and are emerging as critical determinants of oncogenesis. Aspartate amino transaminase (GOT1) (Son et al., 2013), the branched-chain amino acid transferase (BCAT1) (Tönjes et al., 2013), and phosphoserine aminotransferase (PSAT1) (Possemato et al., 2011) have been found to be essential for tumorigenesis of pancreatic adenocarcinoma, glioblastoma, and breast cancer cells, respectively. A requirement of aminotransferase activity for glutamine-driven anaplerosis in colon cancer has been suggested previously through in vitro work inhibiting GPT2 (Weinberg et al., 2010). However, direct evidence for a critical role of GPT2 activity in glutamine-driven TCA anaplerosis and in vivo tumorigenesis has been lacking. Here we provide genetic evidence for in vivo cancer cell GPT2 dependency coupled with the non-essential nature of GPT2 in non-transformed cells. GPT2 catalyzes the transfer of glutamate nitrogen to pyruvate, forming alanine and αKG in the process. Our findings indicate that GPT2 is critical for glutamine-driven TCA anaplerosis and couples glycolytic pyruvate output to driving glutamine catabolism in murine and human colon cancer cells. We also show that the critical role of GPT2 for cancer cell proliferation is specific to tumorigenic cells expressing multiple oncogenic mutations, whereas proliferation rates of non-tumorigenic cells expressing single oncogenic mutations or normal parental cells remain unaltered in response to GPT2 knockdown.
Our work demonstrates in vivo growth dependency of colon cancer cells on glutamine catabolism. Recent studies on lung cancer cells have questioned whether the phenomenon of glutamine addiction observed in vitro is relevant in vivo (Davidson et al., 2016; Hensley et al., 2016). Here we show that knockdown of both GPT2 and glutaminase (GLS), which converts glutamine to glutamate, inhibits tumor formation by human colon cancer cells in xenograft assays. Our results, along with recent work targeting in vivo glutamine metabolism in breast cancer cells (Gross et al., 2014; van Geldermalsen et al., 2016), suggest that the in vivo role of glutamine metabolism in tumors may be context-dependent. Further studies are thus needed to elucidate whether certain contexts, such as tissue of origin, might determine the relevance of cancer cell glutamine dependency in vivo.
In summary, our results identify a mechanism through which the Warburg effect (i.e., cancer-associated glycolytic activation) contributes to oncogenesis. Although the observation that cancerous cells frequently activate glycolysis is decades old, the mechanisms through which it contributes to oncogenesis have been unclear. Some have postulated that the Warburg effect contributes through the increased speed of energy generation associated with glycolysis, although direct evidence for this hypothesis is limited (reviewed in Liberti and Locasale, 2016). Others have suggested that activated glycolysis contributes through the production of biosynthetic intermediates (reviewed in DeBerardinis et al., 2008). On the other hand, the bulk of glycolytic carbon is excreted as lactate and alanine rather than funneled into biosynthesis (Liberti and Locasale, 2016). Although contributions of the above-mentioned pathways are possible, our results indicate that the Warburg effect critically supports oncogenesis through the production of pyruvate to accept nitrogen from glutamine catabolism. Notably, this oncogenic coupling of glucose and glutamine metabolism is induced by common oncogenic mutations that drive LDH switching, the induction of GPT2, and the emergence of specific metabolic vulnerabilities. Many of these metabolic phenotypes are shared by diverse types of cancer, which suggests the exciting possibility that these metabolic drivers and vulnerabilities may provide attractive molecular targets for novel broad-spectrum cancer interventions.
EXPERIMENTAL PROCEDURES
Cells
YAMC cells (Whitehead et al., 1993), YAMC cells with empty vectors (BN), and YAMC cells expressing p53175H (mp53), HRas12v (Ras), and both p53175H and HRas12v (mp53/Ras) were derived and maintained as described previously (McMurray et al., 2008; Xia and Land, 2007). HCT-116 and SW-480 human colon cancer cells were purchased from the ATCC. Both human cell lines were cultured at 37°C in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Hyclone) and 2.5 µg/mL gentamycin (Invitrogen). Genetically perturbed cells were generated by retroviral infection with a virus containing appropriate shRNA or cDNA expression constructs. The retrovirus used to infect murine cells and human colon cancer cells was produced by transient transfection of ΦNX-eco cells or 293TN cells (ATCC). Infections were carried out in media supplemented with 8 µg/mL Polybrene to increase infection efficiency. After infection, polyclonal cell populations with stable expression of the indicated shRNAs and/or cDNAs were generated by selection with 5 µg/mL puromycin, 5 µg/mL blasticidin, and/or 75 µg/mL hygromycin B.
Adherent Cell Proliferation Assays
Prior to the assay, cells were grown without selective drugs for 48 hr under standard culture conditions. Cells were then plated under standard culture conditions and at the indicated densities. Twenty-four hours after plating, cells were treated with the indicated concentrations of dimethyl alpha-ketoglutarate (Sigma), dimethyl malate (Sigma), or L-alanine (Sigma) and cultured under the indicated oxygen conditions. Cells were counted every 24 hr, and counts were normalized to cell number at initial time of treatment; i.e., 24 hr after plating the cells.
LC-MS/MS Methodology
Metabolites were extracted with −78°C methanol:water (80:20) prior to analysis using reverse-phase chromatography with an ion-paring reagent in a Shimadzu high-performance liquid chromatography (HPLC) coupled to a Thermo Quantum triple quadrupole mass spectrometer. Mass spectrometry was performed in negative mode with selected reaction monitoring (SRM)-specific scans as described in DeVito et al. (2014) and Munger et al. (2008). Further specifics regarding labeling, extraction, chromatography, and mass spectrometry data analysis can be found in the Supplemental Experimental Procedures.
Extracellular Lactate, Glucose, and Glutamine Measurements
Prior to measurement, cells were grown without selective drugs for 48 hr at 39°C under standard medium conditions and then cultured for an additional 20 hr without serum. After 20 hr of serum starvation, cells were given fresh serum free media, and measurements were made 6 hr later using a NOVA bio-profile basic 4 according to the manufacturer’s instructions (http://www.novabiomedical.com). For measurements under hypoxia, cells were moved to 1% oxygen for the indicated time prior to sample collection and analysis. Results were normalized for protein content measurement by Bradford assay.
Xenograft Assay
Tumor formation was assessed as described previously (McMurray et al., 2008). Briefly, 5 × 105 mp53/Ras cell derivatives or 2.5 × 106 human colon cancer cell derivatives were implanted subcutaneously into the flanks of CD-1 nude mice (Crl:CD-1-Foxn1nu, Charles River Laboratories). Prior to injection, all cells were grown without selective drugs for a minimum of 48 hr in standard culture medium. Tumor diameters along the x, y, and z axes were measured by caliper at indicated time points, and the averages of these three measurements were used to calculate volume, utilizing the formula volume = (4/3)Πr3. The statistical significance of tumor volume differences was determined by Student’s t test. All animal procedures conform to the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC) regulations and were approved by the local University Committee on Animal Resources.
Statistics
PLS-DA and ANOVA of the metabolite pool size data were performed using the publicly available software MetaboAnalyst 3.0 (http://www.metaboanalyst.ca) (Xia et al., 2015). Prior to analysis, protein-normalized concentration data were corrected for batch effects using the Combat method (Johnson et al., 2007). Batch-corrected data were subsequently auto-scaled. For PLS-DA, PLS regression was performed using the plsr function provided by the R pls package (Mevik and Wehrens, 2007). Classification and cross-validation were performed using the corresponding wrapper function offered in the R caret package (Kuhn, 2008). Permutation testing was performed to assess the significance of class discrimination using the optimal number of components as determined by leave-one-out cross-validation (LOOCV), which was four components (Figures S1C and S1D). Permutation testing of the PLS-DA model, with 2,000 permutations, yielded a p value that was less than 5e4 (Figure S1).
For all experiments in which more than two samples types were compared (i.e., more than two cell lines), significant features were identified through a one-way ANOVA of the batch-corrected data, followed by Fisher’s least significant difference (LSD) post hoc testing. For comparisons between two cell types, a Student’s t test was employed. In either case, where multiple tests were performed, p values were corrected using the Benjamini-Hochberg false discovery rate (FDR) correction.
Metabolic activities were deemed to be cooperative when the activities/pool size increases for the murine colon cells satisfied the following criteria:
Metabolic activities or pool size increases were deemed mp53- or Ras allele-dominant when their fold changes relative to the YAMC cells and the opposite single mutation were both >2-fold.
Supplementary Material
In Brief.
Metabolic reprogramming is critical to cancer, but its emergence and contribution to malignancy are poorly understood. Smith et al. find that cancer cells depend on glutamic pyruvate transaminase to exploit activated glycolysis and drive glutamine-based TCA cycle anaplerosis. Their work reveals a cancer-specific metabolic vulnerability of potential therapeutic interest.
Highlights.
Oncogene cooperation drives large-scale metabolic reprogramming
The Warburg effect is critical to glutamine-based TCA anaplerosis
Oncogenic transformation induces dependence on glutamic pyruvate transaminase 2
GPT2 exploits activated glycolysis to facilitate glutamine-driven TCA anaplerosis
Acknowledgments
We thank Hannah S. Land for graphics. This work was supported by NIH grants CA120317 and CA138249 (to H.L.) and AI081773 (to J.M.). J.M. is a Damon Runyon-Rachleff Innovator supported by the Damon Runyon Cancer Research Foundation (DRR-09-10) and by a Research Scholar Grant from the American Cancer Society (RSG-15-049-01-MPC).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, six figures, and seven tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2016.09.045.
AUTHOR CONTRIBUTIONS
Conceptualization, B.S., H.L., and J.M.; Formal Analysis, B.S., A.A., H.L., and J.M.; Investigation, B.S., X.L.S., A.A., and C.M.S.; Writing, B.S., H.L., and J.M.; Visualization, B.S., H.L., and J.M.; Project Administration, B.S., H.L., and J.M.; Funding Acquisition, H.L. and J.M.
REFERENCES
- Augoff K, Hryniewicz-Jankowska A, Tabola R. Lactate dehydrogenase 5: an old friend and anew hope in the war on cancer. Cancer Lett. 2015;358:1–7. doi: 10.1016/j.canlet.2014.12.035. [DOI] [PubMed] [Google Scholar]
- Baer R, Cintas C, Dufresne M, Cassant-Sourdy S, Schönhuber N, Planque L, Lulka H, Couderc B, Bousquet C, Garmy-Susini B, et al. Pancreatic cell plasticity and cancer initiation induced by oncogenic Kras is completely dependent on wild-type PI 3-kinase p110α. Genes Dev. 2014;28:2621–2635. doi: 10.1101/gad.249409.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer DE, Hatzivassiliou G, Zhao F, Andreadis C, Thompson CB. ATP citrate lyase is an important component of cell growth and transformation. Oncogene. 2005;24:6314–6322. doi: 10.1038/sj.onc.1208773. [DOI] [PubMed] [Google Scholar]
- Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Brownsey RW, Boone AN, Elliott JE, Kulpa JE, Lee WM. Regulation of acetyl-CoA carboxylase. Biochem. Soc. Trans. 2006;34:223–227. doi: 10.1042/BST20060223. [DOI] [PubMed] [Google Scholar]
- Bryant KL, Mancias JD, Kimmelman AC, Der CJ. KRAS: feeding pancreatic cancer proliferation. Trends Biochem. Sci. 2014;39:91–100. doi: 10.1016/j.tibs.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- Davidson SM, Papagiannakopoulos T, Olenchock BA, Heyman JE, Keibler MA, Luengo A, Bauer MR, Jha AK, O’Brien JP, Pierce KA, et al. Environment Impacts the Metabolic Dependencies of Ras-Driven Non-Small Cell Lung Cancer. Cell Metab. 2016;23:517–528. doi: 10.1016/j.cmet.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. USA. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Deprez J, Vertommen D, Alessi DR, Hue L, Rider MH. Phosphorylation and activation of heart 6-phosphofructo-2-kinase by protein kinase B and other protein kinases of the insulin signaling cascades. J. Biol. Chem. 1997;272:17269–17275. doi: 10.1074/jbc.272.28.17269. [DOI] [PubMed] [Google Scholar]
- DeVito SR, Ortiz-Riaño E, Martínez-Sobrido L, Munger J. Cytomegalovirus-mediated activation of pyrimidine biosynthesis drives UDP-sugar synthesis to support viral protein glycosylation. Proc. Natl. Acad. Sci. USA. 2014;111:18019–18024. doi: 10.1073/pnas.1415864111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elstrom RL, Bauer DE, Buzzai M, Karnauskas R, Harris MH, Plas DR, Zhuang H, Cinalli RM, Alavi A, Rudin CM, Thompson CB. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 2004;64:3892–3899. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- Fan TW, Lane AN, Higashi RM, Farag MA, Gao H, Bousamra M, Miller DM. Altered regulation of metabolic pathways in human lung cancer discerned by (13)C stable isotope-resolved metabolomics (SIRM) Mol. Cancer. 2009;8:41. doi: 10.1186/1476-4598-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425–434. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- Gaglio D, Metallo CM, Gameiro PA, Hiller K, Danna LS, Balestrieri C, Alberghina L, Stephanopoulos G, Chiaradonna F. Oncogenic K-Ras decouples glucose and glutamine metabolism to support cancer cell growth. Mol. Syst. Biol. 2011;7:523. doi: 10.1038/msb.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, Dang CV. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatenby RA, Gillies RJ. A microenvironmental model of carcinogenesis. Nat. Rev. Cancer. 2008;8:56–61. doi: 10.1038/nrc2255. [DOI] [PubMed] [Google Scholar]
- Gross MI, Demo SD, Dennison JB, Chen L, Chernov-Rogan T, Goyal B, Janes JR, Laidig GJ, Lewis ER, Li J, et al. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol. Cancer Ther. 2014;13:890–901. doi: 10.1158/1535-7163.MCT-13-0870. [DOI] [PubMed] [Google Scholar]
- Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JM, Karantza V, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzivassiliou G, Zhao F, Bauer DE, Andreadis C, Shaw AN, Dhanak D, Hingorani SR, Tuveson DA, Thompson CB. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 2005;8:311–321. doi: 10.1016/j.ccr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Hensley CT, Faubert B, Yuan Q, Lev-Cohain N, Jin E, Kim J, Jiang L, Ko B, Skelton R, Loudat L, et al. Metabolic Heterogeneity in Human Lung Tumors. Cell. 2016;164:681–694. doi: 10.1016/j.cell.2015.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer AM, Dematteo RG, Venneti S, Finley LW, Lu C, Judkins AR, Rustenburg AS, Grinaway PB, Chodera JD, Cross JR, et al. Hypoxia induces production of L-2-hydroxyglutarate. Cell Metab. 2015;22:304–311. doi: 10.1016/j.cmet.2015.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Kawauchi K, Araki K, Tobiume K, Tanaka N. p53 regulates glucose metabolism through an IKK-NF-kappaB pathway and inhibits cell transformation. Nat. Cell Biol. 2008;10:611–618. doi: 10.1038/ncb1724. [DOI] [PubMed] [Google Scholar]
- Kinsey C, Balakrishnan V, O’Dell MR, Huang JL, Newman L, Whitney-Miller CL, Hezel AF, Land H. Plac8 links oncogenic mutations to regulation of autophagy and is critical to pancreatic cancer progression. Cell Rep. 2014;7:1143–1155. doi: 10.1016/j.celrep.2014.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kole HK, Resnick RJ, Van Doren M, Racker E. Regulation of 6-phosphofructo-1-kinase activity in ras-transformed rat-1 fibroblasts. Arch. Biochem. Biophys. 1991;286:586–590. doi: 10.1016/0003-9861(91)90084-v. [DOI] [PubMed] [Google Scholar]
- Kuhajda FP. Fatty-acid synthase and human cancer: new perspectives on its role in tumor biology. Nutrition. 2000;16:202–208. doi: 10.1016/s0899-9007(99)00266-x. [DOI] [PubMed] [Google Scholar]
- Kuhajda FP, Jenner K, Wood FD, Hennigar RA, Jacobs LB, Dick JD, Pasternack GR. Fatty acid synthesis: a potential selective target for antineoplastic therapy. Proc. Natl. Acad. Sci. USA. 1994;91:6379–6383. doi: 10.1073/pnas.91.14.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn M. Building predictive models in R using the caret package. J Stat Softw. 2008;28:1–26. [Google Scholar]
- Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, Deck LM, Royer RE, Vander Jagt DL, Semenza GL, Dang CV. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc. Natl. Acad. Sci. USA. 2010;107:2037–2042. doi: 10.1073/pnas.0914433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberti MV, Locasale JW. The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem. Sci. 2016;41:211–218. doi: 10.1016/j.tibs.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd AC, Obermüller F, Staddon S, Barth CF, McMahon M, Land H. Cooperating oncogenes converge to regulate cyclin/cdk complexes. Genes Dev. 1997;11:663–677. doi: 10.1101/gad.11.5.663. [DOI] [PubMed] [Google Scholar]
- Mazurek S, Grimm H, Boschek CB, Vaupel P, Eigenbrodt E. Pyruvate kinase type M2: a crossroad in the tumor metabolome. Br. J. Nutr. 2002;87(Suppl 1):S23–S29. [PubMed] [Google Scholar]
- McMurray HR, Sampson ER, Compitello G, Kinsey C, Newman L, Smith B, Chen SR, Klebanov L, Salzman P, Yakovlev A, Land H. Synergistic response to oncogenic mutations defines gene class critical to cancer phenotype. Nature. 2008;453:1112–1116. doi: 10.1038/nature06973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevik B-H, Wehrens R. The pls Package. Principal Component and Partial Least Squares Regression in R. J. Stat. Soft. 2007;18:23. [Google Scholar]
- Munger J, Bennett BD, Parikh A, Feng XJ, McArdle J, Rabitz HA, Shenk T, Rabinowitz JD. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat. Biotechnol. 2008;26:1179–1186. doi: 10.1038/nbt.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham WM, Clish CB, Yang Y, Loscalzo J. Hypoxia-Mediated Increases in L-2-hydroxyglutarate Coordinate the Metabolic Response to Reductive Stress. Cell Metab. 2015;22:291–303. doi: 10.1016/j.cmet.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan A, Wang C, Schreiber SL. Perturbational profiling of a cell-line model of tumorigenesis by using metabolic measurements. Proc. Natl. Acad. Sci. USA. 2005;102:5992–5997. doi: 10.1073/pnas.0502267102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid A, Pizer ES, Moga M, Milgraum LZ, Zahurak M, Pasternack GR, Kuhajda FP, Hamilton SR. Elevated expression of fatty acid synthase and fatty acid synthetic activity in colorectal neoplasia. Am. J. Pathol. 1997;150:201–208. [PMC free article] [PubMed] [Google Scholar]
- Read JA, Winter VJ, Eszes CM, Sessions RB, Brady RL. Structural basis for altered activity of M- and H-isozyme forms of human lactate dehydrogenase. Proteins. 2001;43:175–185. doi: 10.1002/1097-0134(20010501)43:2<175::aid-prot1029>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Paterson HF, Noble M, Land H. Ras-mediated cell cycle arrest is altered by nuclear oncogenes to induce Schwann cell transformation. EMBO J. 1988;7:1635–1645. doi: 10.1002/j.1460-2075.1988.tb02990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res. 2004;64:2627–2633. doi: 10.1158/0008-5472.can-03-0846. [DOI] [PubMed] [Google Scholar]
- Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- Sewing A, Wiseman B, Lloyd AC, Land H. High-intensity Raf signal causes cell cycle arrest mediated by p21Cip1. Mol. Cell. Biol. 1997;17:5588–5597. doi: 10.1128/mcb.17.9.5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B, Land H. Anticancer activity of the cholesterol exporter ABCA1 gene. Cell Rep. 2012;2:580–590. doi: 10.1016/j.celrep.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son J, Lyssiotis CA, Ying H, Wang X, Hua S, Ligorio M, Perera RM, Ferrone CR, Mullarky E, Shyh-Chang N, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telang S, Yalcin A, Clem AL, Bucala R, Lane AN, Eaton JW, Chesney J. Ras transformation requires metabolic control by 6-phosphofructo-2-kinase. Oncogene. 2006;25:7225–7234. doi: 10.1038/sj.onc.1209709. [DOI] [PubMed] [Google Scholar]
- Telang S, Lane AN, Nelson KK, Arumugam S, Chesney J. The oncoprotein H-RasV12 increases mitochondrial metabolism. Mol. Cancer. 2007;6:77. doi: 10.1186/1476-4598-6-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To MD, Rosario RD, Westcott PM, Banta KL, Balmain A. Interactions between wild-type and mutant Ras genes in lung and skin carcinogenesis. Oncogene. 2013;32:4028–4033. doi: 10.1038/onc.2012.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tönjes M, Barbus S, Park YJ, Wang W, Schlotter M, Lindroth AM, Pleier SV, Bai AH, Karra D, Piro RM, et al. BCAT1 promotes cell proliferation through amino acid catabolism in gliomas carrying wild-type IDH1. Nat. Med. 2013;19:901–908. doi: 10.1038/nm.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Geldermalsen M, Wang Q, Nagarajah R, Marshall AD, Thoeng A, Gao D, Ritchie W, Feng Y, Bailey CG, Deng N, et al. ASCT2/SLC1A5 controls glutamine uptake and tumour growth in triple-negative basal-like breast cancer. Oncogene. 2016;35:3201–3208. doi: 10.1038/onc.2015.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartanian S, Bentley C, Brauer MJ, Li L, Shirasawa S, Sasazuki T, Kim JS, Haverty P, Stawiski E, Modrusan Z, et al. Identification of mutant K-Ras-dependent phenotypes using a panel of isogenic cell lines. J. Biol. Chem. 2013;288:2403–2413. doi: 10.1074/jbc.M112.394130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizan P, Boros LG, Figueras A, Capella G, Mangues R, Bassilian S, Lim S, Lee WN, Cascante M. K-ras codon-specific mutations produce distinctive metabolic phenotypes in NIH3T3 mice [corrected] fibroblasts. Cancer Res. 2005;65:5512–5515. doi: 10.1158/0008-5472.CAN-05-0074. [DOI] [PubMed] [Google Scholar]
- Voet D, Voet JG. Biochemistry. Third. John Wiley & Sons; 2004. [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Weinberg F, Hamanaka R, Wheaton WW, Weinberg S, Joseph J, Lopez M, Kalyanaraman B, Mutlu GM, Budinger GR, Chandel NS. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc. Natl. Acad. Sci. USA. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead RH, VanEeden PE, Noble MD, Ataliotis P, Jat PS. Establishment of conditionally immortalized epithelial cell lines from both colon and small intestine of adult H-2Kb-tsA58 transgenic mice. Proc. Natl. Acad. Sci. USA. 1993;90:587–591. doi: 10.1073/pnas.90.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, Thompson CB. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl. Acad. Sci. USA. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia M, Land H. Tumor suppressor p53 restricts Ras stimulation of RhoA and cancer cell motility. Nat. Struct. Mol. Biol. 2007;14:215–223. doi: 10.1038/nsmb1208. [DOI] [PubMed] [Google Scholar]
- Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0-making metabolomics more meaningful. Nucleic Acids Res. 2015;43:W251–W257. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Hanai J, Ren JG, Kats L, Burgess K, Bhargava P, Signoretti S, Billiard J, Duffy KJ, Grant A, et al. Targeting lactate dehydrogenase-a inhibits tumorigenesis and tumor progression in mouse models of lung cancer and impacts tumor-initiating cells. Cell Metab. 2014;19:795–809. doi: 10.1016/j.cmet.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-San-anikone E, Locasale JW, Son J, Zhang H, Coloff JL, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J. Cell Biol. 2007;178:93–105. doi: 10.1083/jcb.200703099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Lin M, Wu R, Wang X, Yang B, Levine AJ, Hu W, Feng Z. Parkin, a p53 target gene, mediates the role of p53 in glucose metabolism and the Warburg effect. Proc. Natl. Acad. Sci. USA. 2011;108:16259–16264. doi: 10.1073/pnas.1113884108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.