Abstract
Advances in biomedical research allow for the capture of an unprecedented level of genetic, molecular, and clinical information from large patient cohorts, where the quest for precision medicine can be pursued. An overarching goal of precision medicine is to integrate the large–scale genetic and molecular data with deep phenotypic information to identify a new mechanistic disease classification. This classification can ideally be used to meet the clinical goal of the right medication for the right patient at the right time. Glomerular disease presents a formidable challenge for precision medicine. Patients present with similar signs and symptoms, which cross the current disease categories. The diseases are grouped by shared histopathologic features, but individual patients have dramatic variability in presentation, progression, and response to therapy, reflecting the underlying biologic heterogeneity within each glomerular disease category. Despite the clinical challenge, glomerular disease has several unique advantages to building multilayered datasets connecting genetic, molecular, and structural information needed to address the goals of precision medicine in this population. Kidney biopsy tissue, obtained during routine clinical care, provides a direct window into the molecular mechanisms active in the affected organ. In addition, urine is a biofluid ideally suited for repeated measurement from the diseased organ as a liquid biopsy with potential to reflect the dynamic state of renal tissue. In our review, current approaches for large–scale data generation and integration along the genotype-phenotype continuum in glomerular disease will be summarized. Several successful examples of this integrative biology approach within glomerular disease will be highlighted along with an outlook on how achieving a mechanistic disease classification could help to shape glomerular disease research and care in the future.
Keywords: genetics and development, glomerular disease, glomerulonephritis, glomerulopathy, molecular genetics, transcriptional profiling, Biological Products, Biology, Biomedical Research, Biopsy, Genotype, Goals, Humans, Kidney Glomerulus, nephrology, Phenotype, Precision Medicine
Introduction
The last decade has seen rapid advances in biomedical research, now allowing the capture of genetic, molecular, and clinical information from large patient cohorts. These advances motivated the strategy proposed in the 2011 Institute of Medicine (IOM) report Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease (1). This strategy aims to use prospective cohorts of patients under routine clinical care to map genetic and molecular profiles with clinical disease course. The overarching goal is to use large–scale data integration paired with deep phenotypic clinical data to establish a disease classification (i.e., a new taxonomy) rooted in molecular causes as well as clinical presentation. The approach of precision medicine, endorsed by a major initiative from the National Institutes of Health (NIH) (2), strives to achieve the goal of targeted treatment assignment. Precision medicine in this context takes into account individual disease presentation and variability in mechanistic terms to realize the concept of the right medication for the right patient at the right time.
This concept of precision nephrology certainly resonates with patients affected by glomerular disease and the health providers caring for them. Patients present with multiple signs and symptoms (e.g., edema, proteinuria, hematuria, and reduced renal function), which cross the current disease categories. At the same time, patients grouped into a common diagnosis by histopathologic features often have dramatic variability in presentation, progression, and response to therapy. The mainstay of therapy includes immunosuppressive drugs, which have an unpredictable response at the individual patient level and frequent toxicity (3–5). These clinical observations are a direct consequence of the underlying biologic heterogeneity within each glomerular disease currently lumped together by our descriptive classifications. The application of precision medicine to glomerular disease provides an opportunity to identify molecular predictors of underlying disease mechanisms, response to therapy, and progression of disease. Combined with currently available clinical and pathologic features as well as validating the findings in model systems, this approach ultimately strives to inform treatment decisions and improve patient outcomes.
The field of oncology is commonly looked to as a model of success in leveraging this approach to not only better understand disease heterogeneity but also, translate such insights into novel therapies (6). The successful use of whole-exome sequencing to identify the genetic basis for response to cytotoxic T-lymphocyte antigen 4 blockade in melanoma is one such example (7). In this landmark study, a discovery cohort of only 25 patients (11 who responded to therapy and 14 who did not) was analyzed through a bioinformatic pipeline, which included sequencing for mutational load in the tumor tissue, translating mutations into altered peptides, and simulating MHC class 1 binding, leading to the identification of a neoepitope signature that correlated with clinical response. Furthermore, the Cancer Genome Atlas (TCGA) aims to establish a core resource to facilitate this approach in other cancer diagnoses. TCGA is funded by the National Cancer Institute and the National Human Genome Research Institute and has collected tumor and normal tissue from 11,000 patients with 33 types of cancer to generate multiple large–scale databases of DNA, RNA, and proteomics. Among other advances, this database was used to identify biologically based subtypes of glioblastoma, setting up improved trial design, and enabling novel therapeutic developments (8,9).
This article will highlight definitions, important limitations, and approaches to large–scale dataset generation in glomerular disease (excluding genetics, which will be discussed separately in this series). Examples of the successful application of an integrative approach in glomerular disease will be provided, with the anticipation that more success stories are expected in the future. The power of analyzing the human disease as a unified whole has independent value, but combining with parallel work in model systems, cell cultures, and observational and interventional clinical studies allows each approach to inform the other and expedite discovery and clinical translation.
Generation of Large-Scale Data in Glomerular Disease
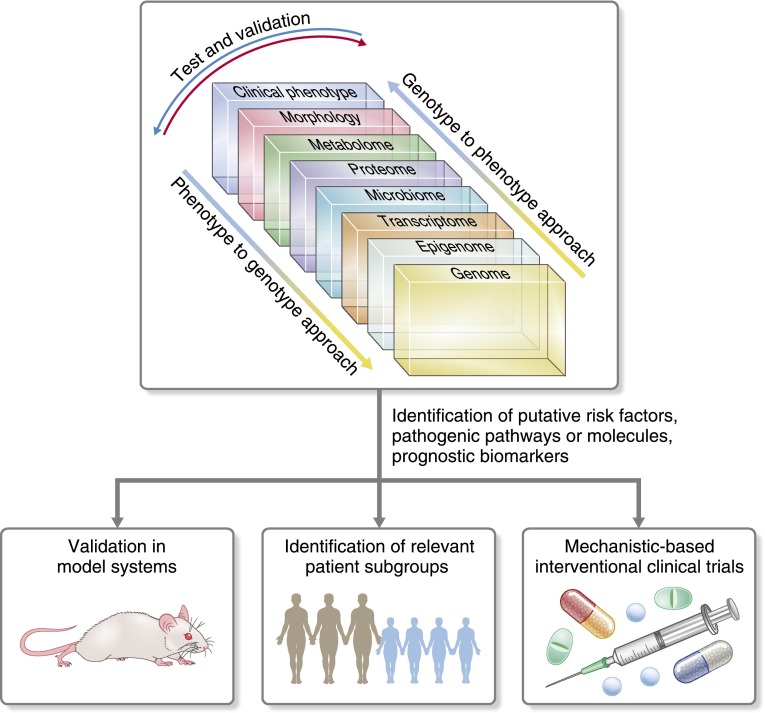
Generation of multilayered large–scale datasets for glomerular disease begins to address the goals of precision medicine for nephrology (Figure 1). Not unlike oncology, there are unique advantages of applying this approach to kidney disease, where the biospecimens needed to generate the datasets highlighted in Figure 1 are frequently obtained during routine clinical care. For example, biopsy tissue (morphology) not only provides valuable structural information currently in clinical use, but also offers a window into the molecular mechanisms active in the tissue of the affected organ (transcriptome). Another advantage is the easy access to urine, which can be tested repeatedly and serves as a potential liquid biopsy to capture changes in the renal tissue with high precision (proteome and metabolome).
Figure 1.
Large–scale dataset integration across the genotype-phenotype continuum. Integrative biology approaches can be initiated with any dataset across the genotype-phenotype continuum and then, evaluated against the other domains to help identify novel risk factors, pathogenic pathways, or biomarkers of progression and therapeutic response. As a result, novel mechanisms can be further investigated in model systems to confirm findings and test therapies. Mechanistic–based patient groups can be identified and targeted for enrollment in proof of concept clinical trials.
Epigenetics
Epigenetics generally refers to changes in nucleic acids of a given cell or cell type and its descendants that do not involve alteration in the underlying DNA sequence. Epigenetic modifications include DNA methylation, histone modification, and noncoding RNA (e.g., microRNAs), which can activate or silence genes. These modifications help to explain why a given transcription factor may have different effects in different cell types (10). These changes may be induced by aging and environmental exposures and have been linked to many chronic diseases (10,11). There are multiple methods to generate epigenetic datasets that capture the variety of modifications, but in general, they can be assessed in a genome-wide fashion using sequencing (e.g., bisulfite-seq) or microarrays. As with other large–scale datasets, the generation method has advantages and disadvantages, including coverage of the genome, quantitative information availability, and resolution (11). Of note, epigenetic changes in the kidney vary by cell type and may change over time, and should be accounted for in the analysis of this type of data.
Transcriptomics
Transcriptomics refers to the set of mRNA produced by the genome in a cell or tissue. Also referred to as gene expression, it is the integrated response of a cellular system to its environment and can vary widely by cell type, by tissue type, by disease state, and in response to treatment. As a result, it is far more dynamic than the epigenome. These data can be obtained from prospectively procured or archival tissue samples and analyzed by quantitative PCR for transcripts of interest, microarrays that can detect thousands of transcripts and transcript-wide sequencing (e.g., RNA-seq). Expression profiles can be analyzed at the specific transcript level, or prior knowledge can be used to aggregate transcripts known to interact in functional groups for association with disease groups or clinical disease progression (12). Of note, the mRNA levels may not necessarily correlate with actual protein levels given the additional regulation of protein production, modification, and degradation. Also, mRNA analysis at the tissue level will be affected by the cell composition. Newer techniques, such as Drop-seq, can generate single–cell transcription profiles (13).
Proteomics
Proteomics, including phospho- and glycoproteomics, aims to capture the complexity of the human protein machinery, which further exceeds that of the genome by several orders of magnitude. Proteomics refers to the complete set of proteins expressed within a cell, tissue, or organism. Detection approaches have diversified substantially ranging from antibody–based, targeted strategies (identification of a previously known protein) to unbiased mass spectrometry profiling (screening proteins that may or may not have previously been identified). Microarrays and suspension-bead assays can detect multiple proteins in a single analysis. Detection depth is increasing steadily to capture several thousand molecules per analysis run but has not reached the same comprehensive level as for nucleic acids. Proteomic analysis of serum, plasma, urine, and kidney tissue is readily possible and may ultimately prove fruitful in numerous forms of glomerular disease with regard to unbiased biomarker discovery (14). However, each approach has its set of limitations, including that more abundant proteins are more easily identified, that mapping of peptides to proteins remains imperfect, and that results may be influenced by various methods of protein extraction and purification before analysis. That said, proteomic analyses were critical to identifying phospholipase A2 receptor (PLA2R) as the key antigen in idiopathic membranous nephropathy (MN) (15), and a comparison of the proteome of urinary microvesicles from patients with idiopathic MN, patients with FSGS, and healthy controls revealed a potential association of increased lysosome membrane protein-2 as a biomarker in idiopathic MN (16).
Metabolomics
Metabolites (i.e., amino acids, bile acids, organic acids, and sugars) are the downstream products of enzymatic activity and considered the “currency of the cell,” reflecting the metabolic state of an organism. Most currently used biochemical markers of renal function fall into this category (e.g., creatinine, urea, uric acid, and cystatin C). They represent not only the active biologic processes, but they are also thought to have possible functional roles in health and disease (17). Metabolomics datasets can be generated using high–field nuclear magnetic resonance spectroscopy or mass spectrometry to separate and detect metabolites followed by data processing and bioinformatics analyses for identification (18,19). Targeted approaches can be used to identify a smaller number of metabolites with high accuracy. Finally, separate approaches, using heavy isotope–labeled substrates, allow for analysis of the amount of a metabolite derived from a particular substance or cellular pathway (20). Of note, metabolites are very diverse biochemical entities, and no single platform is able to capture the full spectrum given their chemical diversity, huge range of observed concentrations, and variations by cell and tissue type. Additionally, metabolites can be alternated by environmental exposures (e.g., food, medication, or microbes), which vary by person (20).
Microbiomics
Microbiomics captures the commensal flora present on and in the human body, most often by identification of known microbe DNA or RNA. There has been limited application of microbiomics to glomerular disease with regard to analysis of flora from different areas of the body, including but not limited to saliva and stool (21). However, with an increased understanding of the tight interaction of the resident human flora with a multitude of physiologic functions ranging from immune response to metabolism, discoveries could be made in glomerular diseases that have strong pathogenetic associations with intestinal flora and the immune system. This connection is starting to be made with regard to IgA nephropathy, and a recent study provides the first evidence in support of an association of stool microbiota in patients with and without progressive IgA nephropathy (22).
Large–Scale Data Capture for Structural, Clinical, and Environmental Measures
Structural information from tissue biopsies has been the prime source of disease classification of glomerular diseases. The rich visual information can now be captured by digital whole–slide imaging and made accessible to data mining tools used for the genetic and molecular information described above (23). These images can be accessed remotely for analysis by pathologists for systematic scoring, morphometric measurements, and potentially, computer–assisted image analysis.
With the mandated implementation of electronic health records in routine care, data sources for clinical information are starting to match the scale and complexity of the genetic and molecular datasets described above. A detailed discussion of research using electronic health records, its opportunities, and its challenges is beyond the scope of this review. However, it is noted that simplifying the ability to easily extract, annotate, and understand unstructured clinical data from electronic health records through natural language processing has been developed and is starting to be deployed for renal diseases (24).
Comprehensive capture of glomerular disease etiologies should also include the definition of potential environmental determinants. Approaches that link environmental exposures to defined neighborhoods on a national scale, referred to as “geomapping,” can be used to effectively evaluate the interaction of the environment with genetic and molecular disease determinants (25).
Team Science and Data Integration
One of the main opportunities and challenges of an integrative biology approach is the wide range of research approaches deployed and data types generated as noted above. This mandates investigative teams with the ability to integrate diverse research strategies and expertise. This team science approach maximizes the potential of any individual multilayered dataset by analyses conducted across the different data domains, facilitating discovery of novel associations and disease mechanisms (26).
Making the large-scale data accessible to the research teams and in particular, researchers collaborating outside of their domain expertise is an essential step of the precision medicine approach, but it requires unique tools to facilitate the engagement of the scientific community. Indeed, the 2015 IOM report on team science highlighted diversity of expertise in team membership and deep knowledge integration as two of the key challenges for team science in precision medicine (27).
Several approaches have been developed for exploration of large-scale datasets from renal disease and are currently used by the community. One example is Nephroseq, a web-based tool designed to make gene expression datasets readily searchable by researchers with a wide range of expertise. Renal gene expression datasets are obtained from the public domain, such as with Gene Expression Omnibus (National Center for Biotechnology Information; www.ncbi.nlm.nih.gov/geo), or directly from investigators, annotated against a standardized ontology, and loaded into Nephroseq. The datasets are linked and dynamically analyzed with a wide variety of systems biology tools. Users can search the datasets by a single gene, a list of genes, or a predefined gene list to conduct a variety of analyses with visualization, including differential expression between groups, coexpression, and outlier sample identification.
Another example is data sharing and analytic platforms designed to allow a group of investigators with diverse methodologic expertise to collaborate around a cohort of patients with several large–scale datasets. These platforms for joint data analysis and data mining by research consortia, for example, are a critical prerequisite for successful collaboration. One of the several currently available platforms is tranSMART, which was initially developed by Johnson and Johnson and subsequently released as open source software in 2012. It is now supported by the tranSMART Foundation (transmartfoundation.org) and its open user community. A research consortium can load their independently governed tranSMART instance with a wide variety of data types, including clinical data and genomic, proteomics, metabolomics, and expression datasets. Loaded data are then available for a wide range of analyses (e.g., descriptive statistics, correlations, survival analysis, clustering, and heat maps). Furthermore, using the same data–sharing platform across diseases and consortia allows for more efficient collaboration by scientists with different expertise and tools inside research teams, and most excitingly, across different research efforts in a globally distributed manner, while maintaining scientific independence and control by each participating network.
Examples of Successful Integrative Biology Approaches in Nephrology
Identification of Disease Etiology: M-Type PLA2R
In the pivotal work to identify M-type PLA2R as an autoantigen in idiopathic MN, proteomic work played a critical role (15). After serum from patients with MN was used to identify a previously unseen band in extracts of normal human glomeruli, mass spectrometry proteomics allowed for identification of candidate peptide sequences contained in the human protein band extract without prior knowledge. These peptides were then compared with existing databases of human proteins to narrow to a list of 18 candidate proteins. Candidate proteins, in turn, could then be evaluated against available antibodies and recombinant proteins, which led to the identification of PLA2R. The ability to map peptide sequences from this unknown Western blot band against known proteins is an example of the value of large-scale data available to inform and enhance mechanistic laboratory approaches. However, the MN story of integrating additional genome–scale datasets goes even farther. Independent of the immunology efforts, a genome–wide association study performed on European patients with MN showed two significant associations. One was located, as expected for an autoimmune disease, in the HLA region, but the second signals tagged PLA2R as a genetic risk factor for MN, linking genetic risk with immunopathology in this disease and implicating PLA2R into the pathophysiology (28). Efforts are underway to combine genetics, transcriptomics, proteomic, and immunology to define the exact mechanism linking the genetic polymorphism with the immunopathology (29).
Identification of Progression Risk Factors: Urinary EGF
A data–driven, unbiased approach implemented across several large–scale data domains recently identified a urinary marker that predicts progression of kidney disease in three independent cohorts, including two glomerular disease cohorts (30). The pipeline began with identifying renal biopsy mRNA transcripts that correlated with eGFR and also were differentially regulated compared with live kidney donor tissue. Next, the gene list was narrowed by including those transcripts that underwent validation in a separate cohort, had kidney-specific expression, and had compelling biology for a mechanistic role in CKD. EGF was the top gene transcript meeting these criteria and is a protein that modulates the response to tubulointerstitial damage. EGF was then evaluated as a potential noninvasive urine marker in three independent glomerular disease cohorts. Urine EGF protein level not only correlated with tissue gene expression level but also added predictive value in predicting progression of CKD above baseline eGFR, urine albumin, and demographics. It is proposed that EGF may capture the functional reserve of the tubulointerstitial compartment not currently captured by eGFR and urine protein. These results were highlighted in the NIH Director’s blog as an example of a precision medicine approach to identifying novel prognostic markers as well as identifying biologic mechanisms, which can then be further studied in a more targeted fashion (31).
Identification of Novel Therapeutic Targets: Baricitinib for Diabetic Nephropathy
A systems-level analysis was to undertaken to better understand the molecular commonalities and differences in underlying disease mechanisms between human diabetic nephropathy and diabetic mouse models, which do not completely recapitulate the human disease. In two studies, analysis of kidney tissue mRNA expression in human diabetic kidney biopsies compared with normal human samples and diabetic mouse models revealed an association of the JAK-STAT pathway expression with human disease progression (32,33). Baricitinib is a small molecule inhibitor of JAK-1 and -2, and it was previously approved for the treatment of rheumatoid arthritis. On the basis of the human systems–level analysis and prior tissue and animal studies, a phase 2 clinical trial, sponsored by Eli Lilly (Indianapolis, IN), was conducted in 129 patients. Lower urinary albumin-to-creatinine ratio was seen at 3 and 6 months in the treatment groups compared with placebo groups, supporting additional study of baricitinib as a new therapy for diabetic nephropathy (34). Using approaches such as these to better understand disease drivers in both human disease and animal models holds promise for novel treatment design and drug repurposing in glomerular disease.
Identification of Novel Diagnostic Markers: Anti-Myeloperoxidase Autoantibodies in Patients with ANCA-Negative Vasculitis
Although many technologies described here are unbiased in their approach, there are new immunologic technologies emerging that allow for clarification of unanswered questions in a targeted manner. One such example is a technology termed autoantigen excision (35). There is a small subset of patients with pauci–immune necrotizing and crescentic GN who have signs and symptoms consistent with ANCA-associated GN but whose ANCA serology is negative. For years, this was a perplexing clinical and serologic discrepancy that resulted in diagnostic and therapeutic delays until the advent of autoantigen excision. Briefly, this methodology involves incubation of autoantigen with autoantibody bound to a protein column, which then is exposed to trypsin, thereby leaving only the complementarity-determining region of the autoantigen bound to the antibody followed by mass spectrometry for protein identification. The application of autoantigen excision to serum from patients with ANCA-negative GN resulted in the identification of anti-myeloperoxidase autoantibodies. Additional studies determined that ceruloplasmin, the endogenous inhibitor of myeloperoxidase, was removed during purification of IgG, which explained why the standard clinical serum–based ANCA test was negative (35). Such a methodology and test will not only expedite diagnosis and treatment of patients with ANCA-negative GN but also, allow for unbiased identification of autoantigens associated with other purported humoral autoimmune glomerular diseases, such as minimal change glomerulopathy and FSGS, which have autoantigens that remain unknown.
The Road Ahead
Glomerular disease research has made significant progress in understanding underlying disease mechanisms. In many patients with inherited glomerular diseases, single-gene defects with pathologic molecules and pathways have been defined (36). These specific molecular signals are now integrated into the larger functional context using comprehensive datasets linking genotype to molecular pathways, structural lesions, and clinical outcomes. For example, the interaction of genetic risk loci with steady–state gene expression profiles and clinical outcomes can be defined using expression quantitative trait loci, where genetic variants can be tested for their effect on gene expression variation. This has been successfully applied for APOL1–associated glomerular disease, lupus nephritis, and CKD (37–40). Linking genome–wide association study, gene expression, and clinical outcomes allows inferences beyond pure associations toward identification of potential causal disease mechanisms as therapeutic targets. An intriguing strategy in this context is the Mendelian randomization approach (i.e., stratifying a disease population on the basis of a genetic marker closely linked with a specific molecular function). The subgroups exhibiting the genetically determined molecular difference are then evaluated for differences in clinical outcomes using the genetic variability as the intervention in an in silico clinical trial.
The approaches above still constrain the analyses toward a predefined set of molecules. A series of computational strategies has been developed in the field of artificial intelligence to identify patterns and interdependencies in multidimensional datasets referred to as machine-learning approaches. In a variety of biomedical fields, these approaches are being tested for their utility on large–scale clinical and genomic datasets. For example, the NIH-International Business Machines (IBM)–funded Dialogue for Reverse Engineering Assessments and Methods Challenges provide information from a study with genome-scale datasets and outcome data in an open competition for investigators to develop outcome or treatment predictor algorithms for the given dataset (www.dreamchallenges.org). The algorithms from the participating scientists compete on a test dataset, and the best performing algorithms are published and in some instances, directly used in clinical trials for patient selection (41).
In summary, we are seeing glomerular research transition to an information-rich science, offering intriguing opportunities and new challenges. Extracting disease–relevant, actionable knowledge from the novel big data sources will be a critical task ahead. Efforts are ongoing to generate and share high-quality samples and datasets in globally distributed research networks to define key drivers of glomerular disease. With access to biopsy tissue and noninvasive biofluids for molecular profiling strategies, nephrology offers unique opportunities to advance our molecular understanding of not only glomerular diseases but also chronic diseases in general. With these advantages in our field, we have to attract the best and brightest trainees to move precision medicine from laboratory benches and computer hard drives to our nephrology clinics.
Disclosures
L.H.M. and W.F.P. have no financial conflicts of interest in relationship to the data presented. M.K. leads a precompetitive consortium partially supported by a grant from Eli Lilly (Indianapolis, IN) and has provided consultations to Eli Lilly as a University of Michigan employee. M.K. also has a patent pending covering the use of urinary EGF as a prognostic biomarker in CKD.
Acknowledgments
L.H.M. is supported by University of Michigan Institute for Clinical and Health Research Career Development grant KL2-TR000434. W.F.P is supported by National Institute of Diabetes and Digestive and Kidney Diseases grants P01DK058335-15 and 1UM1DK100845-01 and receives translational research funding from The Broad Institute (Cambridge, MA). M.K. is supported by National Institutes of Health grants U54DK083912 Nephrotic Syndrome Rare Disease Clinical Research Network II, UM1DK100845 CureGN, and P30DK081943 George M. O'Brien Kidney Research Core Center at the University of Michigan.
Footnotes
Published online ahead of print. Publication date available at www.cjasn.org.
References
- 1.Institute of Medicine: Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease, Washington, DC, The National Academies Press, 2011 [PubMed]
- 2.Collins FS, Varmus H: A new initiative on precision medicine. N Engl J Med 372: 793–795, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hingorani SR, Weiss NS, Watkins SL: Predictors of peritonitis in children with nephrotic syndrome. Pediatr Nephrol 17: 678–682, 2002 [DOI] [PubMed] [Google Scholar]
- 4.Kerlin BA, Ayoob R, Smoyer WE: Epidemiology and pathophysiology of nephrotic syndrome-associated thromboembolic disease. Clin J Am Soc Nephrol 7: 513–520, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monach PA, Arnold LM, Merkel PA: Incidence and prevention of bladder toxicity from cyclophosphamide in the treatment of rheumatic diseases: A data-driven review. Arthritis Rheum 62: 9–21, 2010 [DOI] [PubMed] [Google Scholar]
- 6.de Bono JS, Ashworth A: Translating cancer research into targeted therapeutics. Nature 467: 543–549, 2010 [DOI] [PubMed] [Google Scholar]
- 7.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, Hollmann TJ, Bruggeman C, Kannan K, Li Y, Elipenahli C, Liu C, Harbison CT, Wang L, Ribas A, Wolchok JD, Chan TA: Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 371: 2189–2199, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chin L, Andersen JN, Futreal PA: Cancer genomics: From discovery science to personalized medicine. Nat Med 17: 297–303, 2011 [DOI] [PubMed] [Google Scholar]
- 9.Tomczak K, Czerwińska P, Wiznerowicz M: The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp Oncol (Pozn) 19[1A]: A68–A77, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Susztak K: Understanding the epigenetic syntax for the genetic alphabet in the kidney. J Am Soc Nephrol 25: 10–17, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reddy MA, Natarajan R: Recent developments in epigenetics of acute and chronic kidney diseases. Kidney Int 88: 250–261, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan Y, Martini S, Mariani LH: Genes caught in flagranti: Integrating renal transcriptional profiles with genotypes and phenotypes. Semin Nephrol 35: 237–244, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA: Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161: 1202–1214, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rovin BH, Klein JB: Proteomics and autoimmune kidney disease. Clin Immunol 161: 23–30, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beck LH Jr., Bonegio RG, Lambeau G, Beck DM, Powell DW, Cummins TD, Klein JB, Salant DJ: M-type phospholipase A2 receptor as target antigen in idiopathic membranous nephropathy. N Engl J Med 361: 11–21, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rood IM, Merchant ML, Wilkey DW, Zhang T, Zabrouskov V, van der Vlag J, Dijkman HB, Willemsen BK, Wetzels JF, Klein JB, Deegens JK: Increased expression of lysosome membrane protein 2 in glomeruli of patients with idiopathic membranous nephropathy. Proteomics 15: 3722–3730, 2015 [DOI] [PubMed] [Google Scholar]
- 17.Sharma K, Karl B, Mathew AV, Gangoiti JA, Wassel CL, Saito R, Pu M, Sharma S, You YH, Wang L, Diamond-Stanic M, Lindenmeyer MT, Forsblom C, Wu W, Ix JH, Ideker T, Kopp JB, Nigam SK, Cohen CD, Groop PH, Barshop BA, Natarajan L, Nyhan WL, Naviaux RK: Metabolomics reveals signature of mitochondrial dysfunction in diabetic kidney disease. J Am Soc Nephrol 24: 1901–1912, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhee EP: Metabolomics and renal disease. Curr Opin Nephrol Hypertens 24: 371–379, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao YY: Metabolomics in chronic kidney disease. Clin Chim Acta 422: 59–69, 2013 [DOI] [PubMed] [Google Scholar]
- 20.Sas KM, Karnovsky A, Michailidis G, Pennathur S: Metabolomics and diabetes: Analytical and computational approaches. Diabetes 64: 718–732, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Couser WG, Johnson RJ: The etiology of glomerulonephritis: Roles of infection and autoimmunity. Kidney Int 86: 905–914, 2014 [DOI] [PubMed] [Google Scholar]
- 22.De Angelis M, Montemurno E, Piccolo M, Vannini L, Lauriero G, Maranzano V, Gozzi G, Serrazanetti D, Dalfino G, Gobbetti M, Gesualdo L: Microbiota and metabolome associated with immunoglobulin A nephropathy (IgAN). PLoS One 9: e99006, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barisoni L, Nast CC, Jennette JC, Hodgin JB, Herzenberg AM, Lemley KV, Conway CM, Kopp JB, Kretzler M, Lienczewski C, Avila-Casado C, Bagnasco S, Sethi S, Tomaszewski J, Gasim AH, Hewitt SM: Digital pathology evaluation in the multicenter Nephrotic Syndrome Study Network (NEPTUNE). Clin J Am Soc Nephrol 8: 1449–1459, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L, Cheng WY, Glicksberg BS, Gottesman O, Tamler R, Chen R, Bottinger EP, Dudley JT: Identification of type 2 diabetes subgroups through topological analysis of patient similarity. Sci Transl Med 7: 311ra174, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hicken MT, Gipson DS: Matching the genotype in resolution: Innovative ways of phenotype capture. Semin Nephrol 35: 279–290, 2015 [DOI] [PubMed] [Google Scholar]
- 26.Mirel B, Luo A, Harris M: Research infrastructure for collaborative team science: Challenges in technology-supported workflows in and across laboratories, institutions, and geographies. Semin Nephrol 35: 291–302, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cooke NJ, Hilton ML, editors: Enhancing the Effectiveness of Team Science, Washington, DC, The National Academies Press, 2015 [PubMed] [Google Scholar]
- 28.Stanescu HC, Arcos-Burgos M, Medlar A, Bockenhauer D, Kottgen A, Dragomirescu L, Voinescu C, Patel N, Pearce K, Hubank M, Stephens HA, Laundy V, Padmanabhan S, Zawadzka A, Hofstra JM, Coenen MJ, den Heijer M, Kiemeney LA, Bacq-Daian D, Stengel B, Powis SH, Brenchley P, Feehally J, Rees AJ, Debiec H, Wetzels JF, Ronco P, Mathieson PW, Kleta R: Risk HLA-DQA1 and PLA(2)R1 alleles in idiopathic membranous nephropathy. N Engl J Med 364: 616–626, 2011 [DOI] [PubMed] [Google Scholar]
- 29.Francis JM, Beck LH Jr., Salant DJ: Membranous nephropathy: A journey from bench to bedside. Am J Kidney Dis 68: 138–147, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ju W, Nair V, Smith S, Zhu L, Shedden K, Song PX, Mariani LH, Eichinger FH, Berthier CC, Randolph A, Lai JY, Zhou Y, Hawkins JJ, Bitzer M, Sampson MG, Thier M, Solier C, Duran-Pacheco GC, Duchateau-Nguyen G, Essioux L, Schott B, Formentini I, Magnone MC, Bobadilla M, Cohen CD, Bagnasco SM, Barisoni L, Lv J, Zhang H, Wang HY, Brosius FC, Gadegbeku CA, Kretzler M; ERCB, C-PROBE, NEPTUNE, and PKU-IgAN Consortium: Tissue transcriptome-driven identification of epidermal growth factor as a chronic kidney disease biomarker. Sci Transl Med 7: 316ra193, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collins FS: Pursuing Precision Medicine for Chronic Kidney Disease, NIH Director’s Blog, 2015. Available at: https://directorsblog.nih.gov/2015/12/15/pursuing-precision-medicine-for-chronic-kidney-disease/. Accessed July 28, 2016 [Google Scholar]
- 32.Berthier CC, Zhang H, Schin M, Henger A, Nelson RG, Yee B, Boucherot A, Neusser MA, Cohen CD, Carter-Su C, Argetsinger LS, Rastaldi MP, Brosius FC, Kretzler M: Enhanced expression of Janus kinase-signal transducer and activator of transcription pathway members in human diabetic nephropathy. Diabetes 58: 469–477, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hodgin JB, Nair V, Zhang H, Randolph A, Harris RC, Nelson RG, Weil EJ, Cavalcoli JD, Patel JM, Brosius FC 3rd, Kretzler M: Identification of cross-species shared transcriptional networks of diabetic nephropathy in human and mouse glomeruli. Diabetes 62: 299–308, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tuttle KB, Brosius FC, Adler SG, Kretzler M, Mehta RL, Tumlin JA, Liu J, Silk ME, Cardillo TE, Duffin KL, Haas JV, Macias WL, Janes JM: Baricitinib in Diabetic Kidney Disease: Results from a Phase 2 Multicenter, Randomized Double-Blind, Placebo-Controlled Study, In: 75th Scientific Sessions of the American Diabetes Association, Boston, MA, June 5–9, 2015. Abstract 114-LB [Google Scholar]
- 35.Roth AJ, Ooi JD, Hess JJ, van Timmeren MM, Berg EA, Poulton CE, McGregor J, Burkart M, Hogan SL, Hu Y, Winnik W, Nachman PH, Stegeman CA, Niles J, Heeringa P, Kitching AR, Holdsworth S, Jennette JC, Preston GA, Falk RJ: Epitope specificity determines pathogenicity and detectability in ANCA-associated vasculitis. J Clin Invest 123: 1773–1783, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hildebrandt F: Genetic kidney diseases. Lancet 375: 1287–1295, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chung SA, Brown EE, Williams AH, Ramos PS, Berthier CC, Bhangale T, Alarcon-Riquelme ME, Behrens TW, Criswell LA, Graham DC, Demirci FY, Edberg JC, Gaffney PM, Harley JB, Jacob CO, Kamboh MI, Kelly JA, Manzi S, Moser-Sivils KL, Russell LP, Petri M, Tsao BP, Vyse TJ, Zidovetzki R, Kretzler M, Kimberly RP, Freedman BI, Graham RR, Langefeld CD; International Consortium for Systemic Lupus Erythematosus Genetics: Lupus nephritis susceptibility loci in women with systemic lupus erythematosus. J Am Soc Nephrol 25: 2859–2870, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ledo N, Ko YA, Park AS, Kang HM, Han SY, Choi P, Susztak K: Functional genomic annotation of genetic risk loci highlights inflammation and epithelial biology networks in CKD. J Am Soc Nephrol 26: 692–714, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sampson MG, Robertson CC, Martini S, Mariani LH, Lemley KV, Gillies CE, Otto EA, Kopp JB, Randolph A, Vega-Warner V, Eichinger F, Nair V, Gipson DS, Cattran DC, Johnstone DB, O’Toole JF, Bagnasco SM, Song PX, Barisoni L, Troost JP, Kretzler M, Sedor JR; Nephrotic Syndrome Study Network: Integrative genomics identifies novel associations with APOL1 risk genotypes in black NEPTUNE subjects. J Am Soc Nephrol 27: 814–823, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trudu M, Janas S, Lanzani C, Debaix H, Schaeffer C, Ikehata M, Citterio L, Demaretz S, Trevisani F, Ristagno G, Glaudemans B, Laghmani K, Dell’Antonio G, Loffing J, Rastaldi MP, Manunta P, Devuyst O, Rampoldi L; Swiss Kidney Project on Genes in Hypertension (SKIPOGH) team: Common noncoding UMOD gene variants induce salt-sensitive hypertension and kidney damage by increasing uromodulin expression. Nat Med 19: 1655–1660, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Küffner R, Zach N, Norel R, Hawe J, Schoenfeld D, Wang L, Li G, Fang L, Mackey L, Hardiman O, Cudkowicz M, Sherman A, Ertaylan G, Grosse-Wentrup M, Hothorn T, van Ligtenberg J, Macke JH, Meyer T, Schölkopf B, Tran L, Vaughan R, Stolovitzky G, Leitner ML: Crowdsourced analysis of clinical trial data to predict amyotrophic lateral sclerosis progression. Nat Biotechnol 33: 51–57, 2015 [DOI] [PubMed] [Google Scholar]