Abstract
Caragana microphylla (Leguminosae) is a dominant climax semishrub species in northern China. We evaluated genetic variation within and among populations sampled from three different environmental gradients in Horqin Sandy Land in northern China using intersimple sequence repeats markers and investigated the possible existence of relationships between genetic diversity and environmental factors. The results showed that C. microphylla have high genetic diversity, and environmental gradients affected genetic diversity of C. microphylla populations. Genetic diversity of all populations was affected by many environmental factors and as well correlated with warm index and soil Olsen phosphorus (SOP) concentration. These results have important implications for restoration and management of these degraded ecosystems in arid and semi‐arid areas.
Keywords: Caragana microphylla, environmental factors, genetic diversity, Horqin Sandy Land, intersimple sequence repeat
1. Introduction
The environment plays an important role in evolutionary processes (Scheiner, 1993). A recent surge of studies on plants has explicitly analyzed trait variation among and within species along environmental gradients (Hulshof et al., 2013; Read, Moorhead, Swenson, Bailey, & Sander, 2014). Changes in environmental conditions are predicted to alter diversity within populations (Lovejoy & Hannah, 2005), and genetic diversity is thought to play an essential role in the survival of plant populations in dynamic environments. Environmental factors affect the dynamics of species, even those with high potential for gene flow (Sork et al., 2010; Freeland, Biss, Conrad, & Silvertown, 2010); in turn, genetic diversity of individuals within a population can also affect a range of ecological factors (Vellend & Genber, 2005). The interaction between genetic diversity and ecological factors has been assessed in a few population‐level studies in plants (Hughes, Inouye, Johnson, Underwood, & Vellend, 2008). Genetic variation among plant populations often occurs along different climatic gradients, such as temperature and precipitation gradients (Keller et al., 2011). For example, several studies demonstrated significant effects of simulated climate change on species composition and genetic structure in temperate grassland ecosystems (Fridley, Grime, Askew, Moser, & Stevens, 2011; Harte & Shaw, 1995; Wu, Dijkstra, Koch, & Hungate, 2012).
Genetic variation is nonrandomly distributed among populations and species (Nevo, 1998), with distribution of alleles and genotypes over space and time often affected by numerous factors such as breeding system, seed dormancy and dispersal mechanism, geographic variation and range, life span and other life‐history traits, natural selection, and the history of populations (population genetics, phylogeography, and landscape ecology). (Faye et al., 2016; Hamrick & Godt, 1989; Hamrick, Godt, Murawski, & Loveless, 1991; Hanin, Quaye, Westberg, & Barazani, 2013; Maki, 2003; Manel, Schwartz, Luikart, & Taberlet, 2003; Meloni, Perini, Filigheddu, & Binelli, 2006; Su & Zhang, 2014). Populations in different parts of a species’ range and microhabitats experience and respond to climate change differently (Rehfeldt, Crookston, Warwell, & Evans, 2006; Rehfeldt et al., 2002). This differential response is because of both the genetic composition of local populations and magnitude of climate change, which also vary geographically (Sork et al., 2010). Environmental factors are often responsible for the patterns of genetic structure observed at small spatial scales (Sacks, Brown, & Ernest, 2004). Range expansions characterized by short‐distance dispersal result in reduced genetic diversity in populations at the expanding range front, because these populations suffer from sequential founding events and genetic drift (Austerlitz, Jung, Godelle, & Gouyon, 1997). Moreover, projections of increased temperatures, more frequent droughts, habitat fragmentation, and declining population size indicate that it is questionable whether many plant species will be able persist in their current distributions under such conditions (Richter et al., 2012). Environmental tolerance or local adaptation might allow some species to persist in parts of their current range; other species, even if currently protected, will survive only by colonizing newly suitable areas (Araújo, Cabeza, Thuiller, Hannah, & Williams, 2004).
Horqin Sandy Land is located in the agropastoral transitional zone between the Inner Mongolian Plateau and the Northeast Plains (42°41′–45°45′N, 118°35′–123°30′E) and is one of the four largest sandy areas in northern China; it covers an area of approximately 139,300 km2, which had been desertified sandy land area up to 71,884 km2 of which is desertified sandy land (Wang, 2003; Zhao, Zhao, & Zhang, 2003). Landscape in this area is characterized by sand dunes that alternate with gently undulating lowland areas (Li, Zhang, Duan, & Kang, 2005). This area belongs to the continental semi‐arid monsoon climate and is in the temperate zone, with a mean annual temperature (AMT) of 3–7°C and mean annual rainfall (AP) of 350–500 mm (Zhao et al., 2003). Over recent decades, this region has undergone severe desertification (Li, Zhao, Zhang, Zhang, & Shirato, 2004; Li et al., 2000) and has displayed the northern moving phenomenon of the interlocked agropasturing area of north China in the most recent hundred years (Zhao, Zhao, & Zhang, 2000; Zhao, Zhao, Zhang, & Zhou, 2002).
Caragana microphylla (Leguminosae) is a climax and dominant sand‐fixing shrub species native to northern China; it is an important component of vegetation rehabilitation efforts in the northern China, because it has several highly valuable ecological traits, which include high drought tolerance, antiwind erosion properties, and N2‐fixation capacity (Han, Wang, & Gao, 2011; Zhang et al., 2009; Zhao, 2005), and it is widely planted throughout severely desertified sites to control land desertification in northern China (Su, Zhang, Li, & Wang, 2005; Zhang, Su, Cui, Zhang, & Chang, 2006). Caragana microphylla is distributed in semifixed and fixed sand dunes, and Horqin Sandy Land is the main distribution regions. A unique set of conditions with respect to precipitation and temperature are found in northern China, and Inner Mongolia characterizes the main portion of this species’ distribution (Fu, 1993). Caragana microphylla is long‐lived, perennial, insect‐pollinated, has seed‐based reproduction, and has a broad ecological amplitude (Fu, 1993). Previous studies on C. microphylla have focused on aspects of population distribution patterns and ecological adaptations (Zhao, 2005), morphological characteristics and variations (Li, Jiang, Gu, Ma, & Gao, 2008), physiological adaptations (Li et al., 2008; Ma, Gao, Liu, Wang, & Guo, 2003), nutrient absorption (Li, Chen, Cui, Zhao, & Zhang, 2013), and genetic diversity (Guo et al., 2008; Huang, Zhao, Zhao, Li, & Pan, 2016; Huang et al., 2013). However, the relationship between C. microphylla genetic diversity and environmental gradients has not yet been reported.
The association between genetic and environmental gradients is well‐established evidence of natural selection (Endler, 1986; Manel et al., 2010). In this study, we assessed C. microphylla population genetic variation in different environmental gradients in Horqin Sandy Land using intersimple sequence repeat (ISSR) markers. We used the canonical approach to investigate the potential association between genetic variation and environment, based on correlation analysis between a measure of genetic diversity and environmental variables (Vasemägi & Primmer, 2005).
The aim was to find out how to respond to environmental conditions change in genetic diversity of C. microphylla.
Is there a difference in genetic diversity of C. microphylla along with environmental gradients change (habitat, temperature, and humidity gradients)? If this is the case, what is the change trend and characteristics of genetic diversity within and among C. microphylla populations along environmental gradients across Horqin Sandy Land.
Is there correlation between genetic diversity of C. microphylla and climatic factors in Horqin Sandy Land? If this is the case, whether consistent with previous findings (AP and CI were the major climatic factors that affected genetic diversity indices of C. microphylla populations from northern China; Huang et al., 2016)? If not consistent, which one or several climatic factors affect genetic diversity of C. microphylla populations from Horqin Sandy Land, and how are they correlated?
Is there correlation between genetic diversity of C. microphylla and soil factors in Horqin Sandy Land? If this is the case, which one or several soil factors affect genetic diversity of C. microphylla populations from Horqin Sandy Land, and how are they correlated?
In this article, we explore these hypotheses using ISSR markers and environmental data and provide more information on the genetic diversity of C. microphylla in Horqin Sandy Land, which might be applicable in restoration and management of degraded ecosystems in arid and semi‐arid regions.
2. Materials and Methods
2.1. Sampling
A total of 260 individuals were sampled from 20 natural C. microphylla populations. We sampled 13 individuals from each population (Table 1, Figure 1). Climatic data were obtained from the China Meteorological Administration and are shown in Table 2; climatic means were derived from data from 1971 to 2000. Annual temperature range (ART) was calculated from the formula,
where MTWM = warmest monthly mean temperature, MTCM = coldest monthly mean temperature. Warm index (WI) values were calculated from the formula,
where t = greater than 5°C monthly mean temperature. Cold index (CI) values were calculated from the formula,
where t = lower than 5°C monthly mean temperature. Hydrothermal synthesis index (S) values were calculated from the formula,
where t = month, r t = monthly rainfall, and T t = monthly mean temperature (Bailey, 1979).
Table 1.
Origin of materials and number of samples for 20 populations of Caragana microphylla from Horqin sandy land
| Population | No of plants | Latitude (°N) | Longitude (°E) | Altitude (m) | Habitats |
|---|---|---|---|---|---|
| Pop1 | 13 | 43°10′10″ | 120°37′50″ | 434 | Mobile sand dune |
| Pop2 | 13 | 42°57′35″ | 120°40′46″ | 357 | Lowlands between mobile sand dunes |
| Pop3 | 13 | 42°55′46″ | 120°41′38″ | 367 | Fixed sand dune |
| Pop4 | 13 | 42°55′45″ | 120°41′37″ | 350 | Lowlands between fixed sand dunes |
| Pop5 | 13 | 43°26′48″ | 120°01′26″ | 368 | Semifixed sand dune |
| Pop6 | 13 | 43°04′03″ | 122°17′31″ | 239 | Mobile sand dune |
| Pop7 | 13 | 43°04′03″ | 122°17′31″ | 239 | Lowlands between mobile sand dunes |
| Pop8 | 13 | 43°08′34″ | 122°14′47″ | 220 | Fixed sand dune |
| Pop9 | 13 | 43°08′34″ | 122°14′47″ | 220 | Lowlands between fixed sand dunes |
| Pop10 | 13 | 44°00′09″ | 121°57′15″ | 186 | Mobile sand dune |
| Pop11 | 13 | 44°00′09″ | 121°57′15″ | 186 | Lowlands between mobile sand dunes |
| Pop12 | 13 | 44°25′33″ | 121°16′02″ | 226 | Lowlands between fixed sand dunes |
| Pop13 | 13 | 44°13′21″ | 120°22′48″ | 362 | Mobile sand dune |
| Pop14 | 13 | 44°13′21″ | 120°22′48″ | 362 | Lowlands between mobile sand dunes |
| Pop15 | 13 | 43°51′40″ | 120°13′46″ | 437 | Fixed sand dune |
| Pop16 | 13 | 43°40′34″ | 120°28′26″ | 325 | Semifixed sand dune |
| Pop17 | 13 | 43°22′26″ | 119°33′02″ | 442 | Mobile sand dune |
| Pop18 | 13 | 43°22′26″ | 119°33′02″ | 442 | Lowlands between mobile sand dunes |
| Pop19 | 13 | 43°05′46″ | 119°36′48″ | 485 | Lowlands between fixed sand dunes |
| Pop20 | 13 | 43°09′59″ | 119°34′33″ | 477 | Semifixed sand dune |
Figure 1.
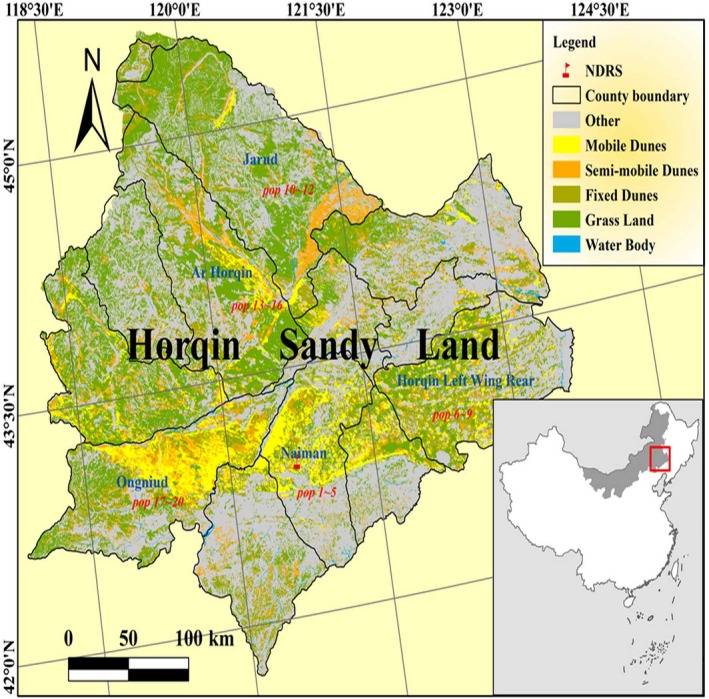
Sampling sites of 20 populations of Caragana microphylla in Horqin Sandy Land
Table 2.
Climatic factors value (average ± SD) for the 20 population sites from the Horqin Sandy Land, obtained from China Meteorological Administration
| Population | Location | AMT (±SD) (°C) | ART (±SD) (°C) | WI (±SD) (°C) | CI (±SD) (°C) | AP (±SD) (mm) | S (±SD) |
|---|---|---|---|---|---|---|---|
| Pop1‐Pop5 | Naiman | 7.5 ± 0.4 | 36.1 ± 1.8 | 56.3 ± 6.7 | 86.2 ± 0.3 | 269.9 ± 40.2 | 21.8 ± 1.9 |
| Pop6‐Pop9 | Horqin Left Wing Rear | 6.9 ± 0.6 | 38.5 ± 2.3 | 60.4 ± 7.2 | 80.4 ± 2.8 | 448.1 ± 39.7 | 36.1 ± 0.5 |
| Pop10‐Pop12 | Jarnd | 6.6 ± 0.5 | 36.5 ± 2.3 | 61.2 ± 4.2 | 80.2 ± 8.5 | 382.5 ± 145.9 | 29.8 ± 9.7 |
| Pop13‐Pop16 | Ar Horqin | 5.9 ± 0.8 | 35.9 ± 2.0 | 74.7 ± 6.4 | 64.0 ± 4.7 | 369.7 ± 121.6 | 30.2 ± 9.9 |
| Pop17‐Pop20 | Ongniud | 6.4 ± 0.5 | 34.7 ± 4.5 | 58.7 ± 3.7 | 75.7 ± 6.6 | 369.9 ± 102.3 | 31.3 ± 9.1 |
AMT, mean annual temperature; ART, annual temperature range; WI, warm index; CI, cold index; AP, mean annual rainfall; S, hydrothermal synthesis index.
This study evaluated three environmental gradients. (1) Habitat gradients: mobile sand dune (vegetation coverage <10%), fixed sand dune (vegetation coverage >50%), semifixed sand dune (vegetation coverage = 30–50%), and lowlands between sand dunes. (2) Thermodynamic gradients: low temperature region (AMT <6.0°C), middle temperature (AMT = 6.0–7.0°C), and high temperature (AMT >7.0°C). (3) Humidity gradients: low humidity (AP <300 mm), middle humidity (AP = 300–400 mm), high humidity region (AP >400 mm).
At each location, a composite soil sample from three depths (0–10, 10–20, and 20–30 cm) was collected from nine sampling points. Soil samples were air‐dried and hand‐sieved through a 2‐mm screen to remove roots and other debris. A portion of each air‐dried soil sample was ground to pass a 0.1‐mm mesh for soil nutrient analyses (Table 3). Soil organic carbon (SOC) concentration was determined using the Walkley–Black dichromate oxidation procedure (Nelson & Sommers, 1982). Soil available nitrogen (SAN) was determined using the alkaline diffusion method, and soil Olsen phosphorus (SOP) was determined using the Olsen method (Olsen, Cole, Watanabe, & Dean, 1954). Young healthy leaves were arbitrarily sampled from plants spaced at least 30 m apart and immediately stored with silica gel in ziplock plastic bags for later DNA extraction.
Table 3.
Soil factors value (average ± SD) for the 20 population sites from the Horqin Sandy Land
| Population | SOC (±SD) (g/kg) | SAN (±SD) (g/kg) | SOP (±SD) (g/kg) | SOC/SAN (±SD) | SOC/SOP (±SD) | SAN/SOP (±SD) |
|---|---|---|---|---|---|---|
| Pop1 | 0.118 ± 0.019 | 0.004 ± 0.001 | 0.006 ± 0.000 | 31.837 ± 8.100 | 18.624 ± 2.833 | 0.603 ± 0.990 |
| Pop2 | 0.113 ± 0.024 | 0.004 ± 0.002 | 0.003 ± 0.000 | 29.021 ± 10.003 | 46.094 ± 12.005 | 1.732 ± 0.413 |
| Pop3 | 1.870 ± 0.822 | 0.008 ± 0.002 | 0.006 ± 0.002 | 248.033 ± 88.055 | 30.805 ± 125.667 | 1.267 ± 0.307 |
| Pop4 | 2.495 ± 0.773 | 0.007 ± 0.003 | 0.006 ± 0.002 | 392.014 ± 101.067 | 419.009 ± 34.012 | 1.115 ± 0.236 |
| Pop5 | 0.360 ± 0.099 | 0.004 ± 0.001 | 0.004 ± 0.001 | 100.002 ± 26.953 | 90.055 ± 45.333 | 0.870 ± 0.303 |
| Pop6 | 0.867 ± 0.180 | 0.018 ± 0.003 | 0.015 ± 0.008 | 48.095 ± 5.701 | 94.160 ± 91.307 | 2.014 ± 1.884 |
| Pop7 | 1.336 ± 0.696 | 0.023 ± 0.006 | 0.010 ± 0.003 | 54.002 ± 15.333 | 16.258 ± 7.126 | 3.049 ± 2.495 |
| Pop8 | 1.930 ± 0.990 | 0.027 ± 0.010 | 0.026 ± 0.014 | 70.043 ± 10.015 | 180.077 ± 72.225 | 2.600 ± 3.310 |
| Pop9 | 1.920 ± 0.840 | 0.028 ± 0.007 | 0.007 ± 0.003 | 70.051 ± 10.005 | 360.089 ± 100.250 | 5.480 ± 3.290 |
| Pop10 | 1.204 ± 0.351 | 0.021 ± 0.004 | 0.006 ± 0.004 | 56.400 ± 13.729 | 288.432 ± 72.025 | 4.953 ± 2.781 |
| Pop11 | 0.300 ± 0.093 | 0.009 ± 0.002 | 0.004 ± 0.001 | 34.067 ± 7.046 | 95.123 ± 35.467 | 2.966 ± 1.736 |
| Pop12 | 1.860 ± 0.790 | 0.022 ± 0.005 | 0.008 ± 0.004 | 80.095 ± 20.028 | 260.065 ± 112.678 | 3.670 ± 3.300 |
| Pop13 | 0.474 ± 0.186 | 0.012 ± 0.002 | 0.007 ± 0.000 | 39.918 ± 15.805 | 73.663 ± 33.111 | 1.828 ± 0.225 |
| Pop14 | 0.692 ± 0.617 | 0.013 ± 0.006 | 0.007 ± 0.001 | 49.006 ± 16.002 | 133.004 ± 45.161 | 2.296 ± 1.682 |
| Pop15 | 1.380 ± 0.420 | 0.019 ± 0.004 | 0.006 ± 0.003 | 70.046 ± 10.051 | 350.089 ± 102.233 | 4.990 ± 3.420 |
| Pop16 | 2.306 ± 1.355 | 0.026 ± 0.008 | 0.025 ± 0.007 | 100.013 ± 30.105 | 100.047 ± 43.915 | 1.100 ± 0.400 |
| Pop17 | 1.176 ± 1.298 | 0.017 ± 0.010 | 0.010 ± 0.004 | 58.291 ± 23.838 | 265.794 ± 57.881 | 3.101 ± 4.876 |
| Pop18 | 0.598 ± 0.330 | 0.014 ± 4.7700 | 0.007 ± 0.002 | 41.037 ± 10.099 | 78.095 ± 30.009 | 1.917 ± 0.410 |
| Pop19 | 2.110 ± 1.610 | 0.025 ± 12.7100 | 0.009 ± 0.005 | 80.095 ± 20.012 | 230.75 ± 80.078 | 2.870 ± 0.770 |
| Pop20 | 3.128 ± 1.403 | 0.040 ± 13.9156 | 0.031 ± 0.001 | 78.205 ± 14.333 | 100.903 ± 45.533 | 1.290 ± 0.473 |
SOC, soil organic carbon; SAN, soil available nitrogen; SOP, soil Olsen phosphorus.
2.2. DNA extraction and ISSR‐PCR amplification
Total DNA was extracted using an AxyPrep genomic DNA mini kit (Axygen, Beijing, China). DNA was quantified spectrophotometrically; samples that yielded high quantities of good‐quality DNA were used in consecutive experiments. After screening 100 ISSR primers from the University of British Columbia (UBC primer set no. 9) for well‐amplified and polymorphic bands among plant populations, we selected 15 primers for use with all individuals.
ISSR amplifications were performed in 25‐μl reaction volumes that contained 40 ng genomic DNA, 1.0 U Taq polymerase, 3 mmol/L MgCl2, 500 μmol/L of each dNTP, 20 mmol/L Tris–HCl (pH 8.3), 100 mmol/L KCl, and 0.3 μmol/L primer. Amplification conditions consisted of an initial step of 3 min at 94°C, followed by 35 cycles of 45 s at 94°C, 45 s at the appropriate annealing temperature (see Table 4 for details), and 2 min at 72°C, and a final 7‐min extension step at 72°C. ISSR reactions were performed at least twice for all individuals and primers to determine the reproducibility of banding patterns. Amplification products along with a 100‐bp DNA ladder were electrophoretically resolved on 1.8% agarose gels that contained ethidium bromide (0.5 μg/ml final concentration) at 100 V for 2 hr and were photographed under ultraviolet light.
Table 4.
Primers sequence, melting temperature and percentage of polymorphism in ISSR analyses of Caragana microphylla
| Primers | Sequences (5′ → 3′) | T m (°C) | No. bands | No. polymorphic loci | Percentage of polymorphic loci | Amplified band size (bp) |
|---|---|---|---|---|---|---|
| UBC 807 | (AG)8T | 54 | 25 | 25 | 100.00 | 150–1750 |
| UBC 810 | (GA)8T | 53 | 19 | 16 | 84.21 | 150–1500 |
| UBC 826 | (AC)8C | 54 | 18 | 17 | 94.44 | 250–1250 |
| UBC 827 | (AC)8G | 54 | 14 | 14 | 100.00 | 250–2000 |
| UBC 835 | (AG)8YC | 53 | 16 | 14 | 87.50 | 150–1250 |
| UBC 836 | (AG)8YA | 52 | 20 | 19 | 95.00 | 150–1350 |
| UBC 840 | (GA)8YT | 58 | 15 | 13 | 86.67 | 200–1500 |
| UBC 842 | (GA)8YG | 58 | 27 | 25 | 92.59 | 150–1500 |
| UBC 848 | (CA)8RG | 48 | 13 | 13 | 100.00 | 225–1250 |
| UBC 857 | (AC)8YG | 58 | 26 | 24 | 92.31 | 150–1750 |
| UBC 864 | (ATG)6 | 52 | 23 | 23 | 100.00 | 250–2000 |
| UBC 880 | (GGAGA)3 | 58 | 25 | 24 | 96.00 | 100–2000 |
| UBC 889 | DBD(AC)7 | 52 | 14 | 13 | 92.86 | 100–900 |
| UBC 892 | TAGATCTGATATCTGAATTCCC | 48 | 18 | 16 | 88.89 | 500–2000 |
| UBC 895 | AGAGTTGGTAGCTCTTGATC | 48 | 15 | 13 | 86.67 | 300–2000 |
y = c/t; b = c/g/t; d = a/g/t.
2.3. Data analysis
During analysis of the resulting gels, only clear and reproducible bands were considered. Amplified fragments were scored for the presence (1) or absence (0) of bands, and the data transformed into a 0/1 binary character matrix. The resulting binary data matrix was analyzed using POPGENE version 1.32 (Yeh & Yang, 1999). Genetic diversity of each population was estimated according to percentage of polymorphic loci (P), observed number of alleles (N a), effective number of alleles (N e), Nei's genetic diversity (h), Shannon's diversity index (I), gene differentiation coefficient (G st), and gene flow (N m). Redundancy analysis (RDA) was used to determine the relative contribution of the measured environmental variables to genetic diversity indices of C. microphylla. Data were first analyzed by detrended correspondence analysis, which indicated that RDA was an appropriate approach (gradient length <3). To avoid overfitting in the regression model due to the large number of explanatory variables, the most discriminating variables were selected using the “forward selection” procedure of the program during analysis. Genetic diversity and environmental data were log (x + 1)‐transformed prior to analysis. RDA was performed using CANOCO version 4.5 (ter Braak & Šmilauer, 2002).
3. Results
3.1. ISSR profiles and genetic diversity
Intersimple sequence repeat band profiles revealed high levels of polymorphism in the surveyed C. microphylla populations. The 15 selected ISSR primers generated a total of 288 clear and distinguishable fragment bands, of which 269 (93.40%) were polymorphic. The size of the amplified fragments ranged from 100 to 2,000 bp, with 19.2 fragments generated on average per primer. The greatest number of bands was generated with the primer UBC842. The least number of bands was generated from the primer UBC848. The percentage of polymorphic bands ranged from 84.21% to 100%. The highest percentage of polymorphic bands was generated with the primers UBC807, UBC827, UBC848, and UBC864. The lowest percentage of polymorphic bands was generated with the primer UBC810 (Table 4).
Calculated genetic diversities within and among the 20 C. microphylla populations are given in Table 5. The number of polymorphic loci (n) ranged from 19 to 121, and P ranged from 6.60% to 42.01%. N a varied from 1.0660 to 1.4201; N e ranged from 1.0385 to 1.3116. h and I values were 0.0233–0.1702 and 0.0352–0.2486, respectively. The highest genetic diversity indices were in population 15, and the lowest genetic diversity values were in population 4. At the species level, h was 0.3365, and I was 0.5041 (Table 5). AMOVA showed that most of the variation (>88.94%) was within populations (Table 6).
Table 5.
Genetic diversity indices of Caragana microphylla populations
| Populations | Sample size | n | P (%) | N a | N e | h | I |
|---|---|---|---|---|---|---|---|
| Pop1 | 13 | 50 | 17.36 | 1.1736 ± 0.3794 | 1.1190 ± 0.2793 | 0.0683 ± 0.1541 | 0.1004 ± 0.2236 |
| Pop2 | 13 | 29 | 10.07 | 1.1007 ± 0.3014 | 1.0661 ± 0.2135 | 0.0385 ± 0.1189 | 0.0570 ± 0.1737 |
| Pop3 | 13 | 27 | 9.38 | 1.0938 ± 0.2920 | 1.0649 ± 0.2156 | 0.0371 ± 0.1191 | 0.0544 ± 0.1726 |
| Pop4 | 13 | 19 | 6.60 | 1.0660 ± 0.2487 | 1.0385 ± 0.1580 | 0.0233 ± 0.0916 | 0.0352 ± 0.1358 |
| Pop5 | 13 | 32 | 11.11 | 1.1111 ± 0.3148 | 1.0720 ± 0.2249 | 0.0417 ± 0.1233 | 0.0619 ± 0.1797 |
| Pop6 | 13 | 40 | 13.89 | 1.1389 ± 0.3464 | 1.0965 ± 0.2579 | 0.0549 ± 0.1420 | 0.0804 ± 0.2052 |
| Pop7 | 13 | 54 | 18.75 | 1.1875 ± 0.3910 | 1.1283 ± 0.2857 | 0.0740 ± 0.1586 | 0.1089 ± 0.2306 |
| Pop8 | 13 | 113 | 39.24 | 1.3924 ± 0.4891 | 1.3116 ± 0.4107 | 0.1700 ± 0.2177 | 0.2441 ± 0.3092 |
| Pop9 | 13 | 55 | 19.10 | 1.1910 ± 0.3938 | 1.1188 ± 0.2608 | 0.0714 ± 0.1510 | 0.1066 ± 0.2230 |
| Pop10 | 13 | 65 | 22.57 | 1.2257 ± 0.4188 | 1.1376 ± 0.2811 | 0.0820 ± 0.1596 | 0.1228 ± 0.2344 |
| Pop11 | 13 | 52 | 18.06 | 1.1806 ± 0.3853 | 1.1227 ± 0.2853 | 0.0701 ± 0.1560 | 0.1032 ± 0.2257 |
| Pop12 | 13 | 37 | 12.85 | 1.1285 ± 0.3352 | 1.0838 ± 0.2395 | 0.0484 ± 0.1323 | 0.0717 ± 0.1924 |
| Pop13 | 13 | 59 | 20.49 | 1.2049 ± 0.4043 | 1.1322 ± 0.2875 | 0.0768 ± 0.1591 | 0.1140 ± 0.2317 |
| Pop14 | 13 | 58 | 20.14 | 1.2014 ± 0.4017 | 1.1346 ± 0.2914 | 0.0777 ± 0.1613 | 0.1146 ± 0.2342 |
| Pop15 | 13 | 121 | 42.01 | 1.4201 ± 0.4944 | 1.2992 ± 0.3802 | 0.1702 ± 0.2076 | 0.2486 ± 0.2988 |
| Pop16 | 13 | 104 | 36.11 | 1.3611 ± 0.4812 | 1.2794 ± 0.3985 | 0.1532 ± 0.2113 | 0.2208 ± 0.3005 |
| Pop17 | 13 | 51 | 17.71 | 1.1771 ± 0.3824 | 1.1135 ± 0.2679 | 0.0664 ± 0.1495 | 0.0987 ± 0.2183 |
| Pop18 | 13 | 32 | 11.11 | 1.1111 ± 0.3148 | 1.0870 ± 0.2505 | 0.0483 ± 0.1382 | 0.0694 ± 0.1980 |
| Pop19 | 13 | 38 | 13.19 | 1.1319 ± 0.3390 | 1.0832 ± 0.2278 | 0.0496 ± 0.1309 | 0.0738 ± 0.1928 |
| Pop20 | 13 | 90 | 31.25 | 1.3125 ± 0.4643 | 1.2193 ± 0.3567 | 0.1242 ± 0.1926 | 0.1819 ± 0.2771 |
| Average | 13 | 56.3 | 19.55 | 1.1955 ± 0.3789 | 1.1354 ± 0.2786 | 0.0773 ± 0.1537 | 0.1134 ± 0.2229 |
| Species level | 260 | 288 | 93.40 | 1.9931 ± 0.0832 | 1.5758 ± 0.3162 | 0.3365 ± 0.1470 | 0.5041 ± 0.1847 |
n, the number of polymorphic loci; P, the percentage of polymorphic loci; N a, observed number of alleles; N e, effective number of alleles; h, Nei's gene diversity; I, Shannon's information index.
Table 6.
Analysis of molecular variance (AMOVA) for Caragana microphylla populations
| Source of variation | df | Percentage of variation | Fixation indices | p |
|---|---|---|---|---|
| Among populationsa | 19 | 11.06 | F ST = 0.1106 | <.001 |
| Within population | 268 | 88.94 | ||
| Total | 287 | |||
| Among groupsb | 2 | 8.71 | F CT = 0.0871 | .005 |
| Among populations with groups | 1 | 1.80 | F SC = 0.0928 | <.001 |
| Total | 268 | 89.50 | F ST = 0.1055 | <.001 |
| Among groupsc | 2 | 8.65 | F CT = 0.0865 | .004 |
| Among populations with groups | 1 | 1.83 | F SC = 0.0937 | <.001 |
| Total | 268 | 89.52 | F ST = 0.1072 | <.001 |
AMOVA from 20 populations as one group.
AMOVA from three groups as represented by WI.
AMOVA from three groups as represented by SOP.
3.2. Pattern of population genetic and its correlation with environmental gradients
Genetic diversity of C. microphylla populations from different environmental gradients is given in Table 7. Along the habitat gradients, N a varied from 1.1294 to 1.3021, N e ranged from 1.0811 to 1.2252, h ranged from 0.0482 to 0.1258, and I values ranged from 0.0718 to 0.1824; G st ranged from 0.5017 to 0.8384, and N m ranged from 0.0964 to 0.4967. Compared with other habitats, lowland populations between fixed sand dunes had the greatest genetic diversity, and those in semifixed sand dunes had the least genetic diversity. Along thermodynamic gradient, N a varied from 1.1090 to 1.2969, N e ranged from 1.0721 to 1.2114, h ranged from 0.0418 to 0.1195, I ranged from 0.0618 to 0.1745, G st ranged from 0.5663 to 0.6864, and N m ranged from 0.2284 to 0.3830. The greatest genetic diversity was in populations in medium‐temperature region populations, and the least genetic diversity was in populations in high‐temperature regions. Along the humidity gradient, C. microphylla populations from high‐humidity regions had the greatest genetic diversity, and low‐humidity regions had the least genetic diversity: N a varied from 1.1090 to 1.2275, N e ranged from 1.0721 to 1.1638, h ranged from 0.0418 to 0.0926, I ranged from 0.0618 to 0.1350; G st ranged from 0.5961 to 0.6864, and N m ranged from 0.2284 to 0.3388.
Table 7.
Genetic diversity indices of Caragana microphylla populations from environmental gradients
| Environmental gradients | N a | N e | h | I | G st | N m | |
|---|---|---|---|---|---|---|---|
| Habitat gradients | Mobile sand dune | 1.1840 ± 0.0330 | 1.1198 ± 0.0162 | 0.0697 ± 0.0104 | 0.1032 ± 0.0162 | 0.7609 | 0.1571 |
| Lowlands between mobile sand dunes | 1.1563 ± 0.0467 | 1.1077 ± 0.0297 | 0.0617 ± 0.0173 | 0.0906 ± 0.0257 | 0.7980 | 0.1266 | |
| Fixed sand dune | 1.2616 ± 0.1326 | 1.1902 ± 0.1067 | 0.1064 ± 0.0579 | 0.1549 ± 0.0828 | 0.5731 | 0.3725 | |
| Lowlands between fixed sand dunes | 1.3021 ± 0.1809 | 1.2252 ± 0.1390 | 0.1258 ± 0.0768 | 0.1824 ± 0.1108 | 0.5017 | 0.4967 | |
| Semifixed sand dune | 1.1294 ± 0.0511 | 1.0811 ± 0.0330 | 0.0482 ± 0.0197 | 0.0718 ± 0.0292 | 0.8384 | 0.0964 | |
| Thermodynamic gradients | Low‐temperature region | 1.1783 ± 0.0486 | 1.1147 ± 0.0278 | 0.0669 ± 0.0170 | 0.0992 ± 0.0258 | 0.6223 | 0.3035 |
| Medium‐temperature region | 1.2969 ± 0.1109 | 1.2114 ± 0.0904 | 0.1195 ± 0.0493 | 0.1745 ± 0.0704 | 0.5663 | 0.3830 | |
| High‐temperature region | 1.1090 ± 0.0397 | 1.0721 ± 0.0292 | 0.0418 ± 0.0164 | 0.0618 ± 0.0239 | 0.6864 | 0.2284 | |
| Humidity gradients | Low‐humidity region | 1.1090 ± 0.0397 | 1.0721 ± 0.0292 | 0.0418 ± 0.0164 | 0.0618 ± 0.0239 | 0.6864 | 0.2284 |
| Medium‐humidity region | 1.1832 ± 0.0905 | 1.1258 ± 0.0638 | 0.0721 ± 0.0357 | 0.1060 ± 0.0523 | 0.6862 | 0.2286 | |
| High‐humidity region | 1.2275 ± 0.1125 | 1.1638 ± 0.0994 | 0.0926 ± 0.0523 | 0.1350 ± 0.0739 | 0.5961 | 0.3388 |
3.3. Correlation between climatic variation and genetic variation
Genetic diversity of the 20 C. microphylla populations from Horqin Sandy Land was influenced by the following climatic factors: AMT, ART, WI, CI, AP, and S (Figure 2). Correlation analyses evidenced that there were positive (WI, AP, and S) and negative (AMT and CI) correlations between genetic diversity indices of C. microphylla populations and climatic factors (Figure 2). RDA showed that the six climate environmental variables (AMT, ART, AP, CI, WI, and S) together explained 78.4% of the total variation in the data, with axes 1 and 2 explaining 69.3% and 9.1% of the total variation, respectively (Figure 2). Of the six environmental variables, only WI was significant according to the Monte Carlo permutation test (p = .001), whereas those of the remaining variables were not significant according to the Monte Carlo permutation test (in all cases p > .05). Of the 78.4% total variation explained by RDA, 57.1% was explained by WI, and the rest (21.3%) by variables that were not significant according to the Monte Carlo permutation test.
Figure 2.
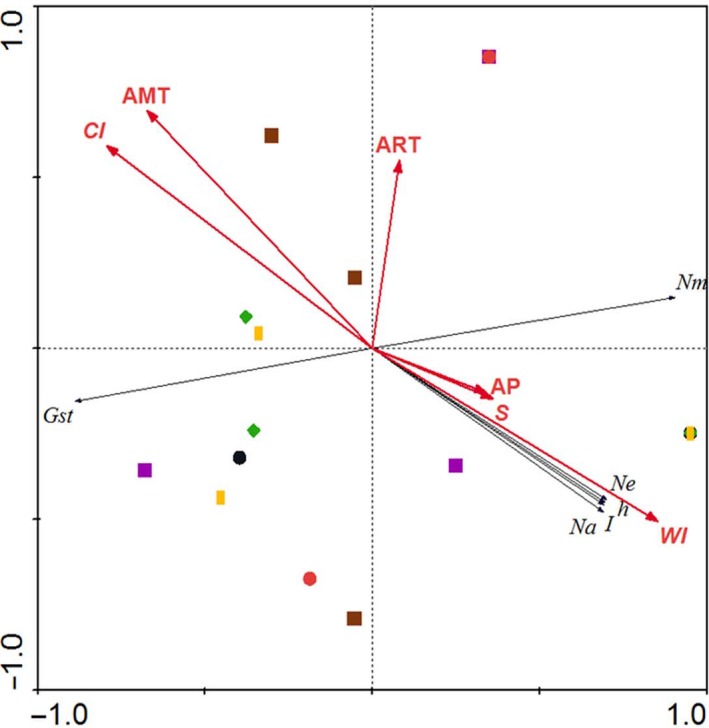
Correlation coefficients between genetic diversity of 20 Caragana microphylla populations and climatic factors
Using RDA, we assessed the relative contribution of the measured climate environmental factors in determining genetic diversity of C. microphylla populations. RDA revealed that the six environmental factors (AMT, ART, AP, CI, WI, and S) together explained 78.4% of the total variation in the genetic diversity indices of C. microphylla populations from Horqin Sandy Land. This result indicates that some other climate factors that were not considered in this study also contribute to the unexplained variation, and WI was the major factor that affected genetic diversity. AMOVA showed that significant genetic differences were detected between the three groups defined as WI conditions, with the variance among groups being 8.71% (p = .005) (Table 6).
3.4. Correlation between soil factor variation and genetic variation
The genetic diversities of the 20 C. microphylla populations from Horqin Sandy Land were influenced by the following soil factors: SOC, SAN, SOP, SOC/SAN, SOC/SOP, and SAN/SOP (Figure 3). Correlation analyses revealed that there were positive correlations between genetic diversity of C. microphylla populations and certain soil factors (SOC, SAN, and SOP) (Figure 3). RDA showed that the six soil environmental variables (SOC, SAN, SOP, SOC/SAN, SOC/SOP, and SAN/SOP) together explained 88.6% of the total variation in the data, with axes 1 and 2 explaining 86.1% and 2.5% of the total variation, respectively (Figure 3). Of the six soil environmental variables, only SOP was significant according to the Monte Carlo permutation test (p = .001), whereas those of the remaining variables were not (in all cases p > .05). Of the total 88.6% variation explained by RDA, 76.1% was explained by SOP and the rest (12.5%) by variables that were not significant according to the Monte Carlo permutation test.
Figure 3.
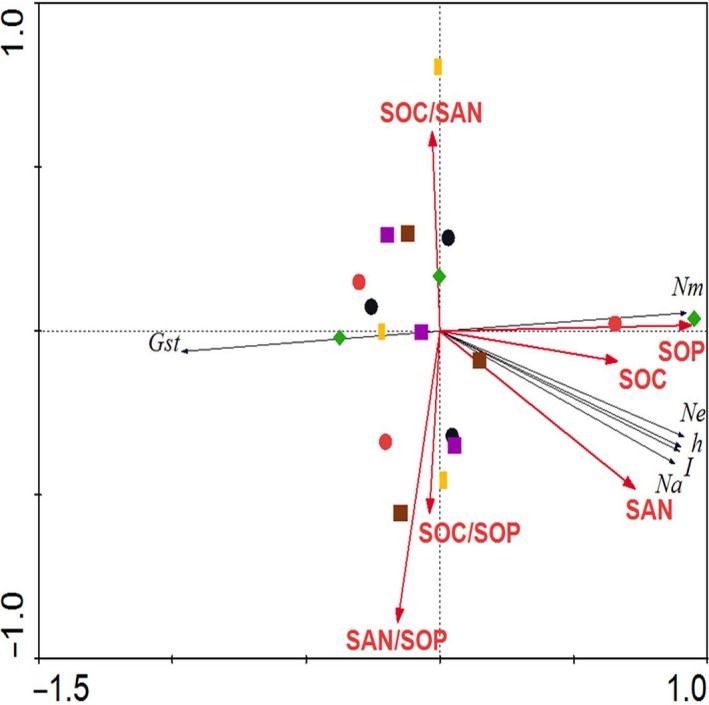
Correlation coefficients between genetic diversity of 20 Caragana microphylla populations and soil factors
Using RDA, we assessed the relative contribution of the measured soil environmental factors in determining genetic diversity indices of C. microphylla populations. RDA revealed that the six environmental factors (SOC, SAN, SOP, SOC/SAN, SOC/SOP, and SAN/SOP) together explained 88.6% of the total variation in the genetic diversity indices of C. microphylla populations from Horqin Sandy Land. This result indicates that some other soil factors that were not considered in this study also contribute to the unexplained variation. SOP was the major factor that affects genetic diversity. AMOVA showed that significant genetic differences were detected between the three groups defined as SOP conditions, with the variance among groups being 8.65% (p = .004) (Table 6).
4. Discussion
4.1. Genetic variation
Genetic diversity of a species is dynamic and shaped by processes that act on widely different spatial and temporal scales. Assessing genetic variation is thus an important component of plant conservation and ecological restoration. In our study, C. microphylla generally exhibited high levels of genetic diversity. At the species level, the value of h estimated in our study for C. microphylla, 0.3365, is higher than that reported from RAPD markers for Stipa grandis from Inner Mongolia (0.2305) (Zhao, Gao, Wang, & Ren, 2008). In C. microphylla, we calculated a value of 0.5041 for I; this is higher than the mean I value for outcrossing species produced by Bussell (1999). Compared with previous study, all values of genetic diversity indices estimated in C. microphylla in Horqin Sandy Land are higher than those reported in northern China based on ISSR analysis (Huang et al., 2016). These differences came from scale difference, because of Horqin Sandy Land was the main natural distribution area of C. Microphylla. On the small scale, the population quantity and the single population size are larger, which makes the gene flow more frequently. C. microphylla is the important sand‐fixation plant in northern China, and there are cultivated populations in natural distribution areas. The genetic diversity indices estimated in this study are higher than plantation populations (Huang et al., 2013). Several reports have indicated that the genetic diversity of natural populations was higher compared with other population type (Chen, Gao, Zhu, & Zhao, 2009; Xue, Liu, & Liu, 1998). But other authors (Hamrick & Godt, 1990; Nybom & Bartish, 2000) noted that levels of genetic variation are strongly dependent on plant life form, geographic range, pollen dispersal mechanisms, and natural selection. Caragana microphylla are long‐lived, perennial, undergo wind pollination, have seed‐based reproduction, and have a broad ecological amplitude. Based on the findings of previous reports, this combination of traits should enable this species to achieve high genetic diversity (Babbel & Selander, 1974; Ge, Wang, Hong, Zhang, & Zu, 1999; Pearse, Crandall, & Beyond, 2004). Caragana microphylla does exhibit high levels of genetic diversity, and this characteristic may have contributed to it being a dominant species in northern China.
4.2. Correlation between genetic variation and environmental gradients
Along the habitat gradient, C. microphylla populations from fixed‐state sand dunes (fixed sand dune and lowlands between fixed sand dunes) had greater genetic diversity than those from mobile‐state sand dunes (mobile sand dunes and lowlands between mobile sand dunes), based on a variety of genetic diversity indices. These results showed that habitat environment change affected genetic diversity of C. microphylla populations, which is consistent with the results about Artemisia halodendron (Huang et al., 2011). These genetic differences between the two types of habitats exhibited that species adapted to the process of desertification land restoration in Horqin Sandy Land, northeast China (Zhao et al., 2000, 2003). This fact might be due to improved soil environments and microclimatic conditions in fixed‐state sand dunes, and individual shrubs were more concentrated than populations from mobile‐state sand dunes, leading to pollen exchange.
In our study, population genetic diversity of C. microphylla was not well correlated with the temperature gradient in Horqin Sandy Land, northeast China. Populations from the high‐temperature region had lower genetic diversity than those from medium‐ and low‐temperature regions. However, these results were not consistent with the results of C. microphylla at relatively smaller geographic distances in Horqin Sandy Land (Huang et al., 2015), which showed that increased temperature, geographical distance, and population size affected genetic diversity of C. microphylla populations. Our data show that C. microphylla population genetic diversity is related to humidity gradients. Populations from the high‐humidity region had higher genetic diversity than those from the medium‐ and low‐humidity regions region. These results showed that genetic diversity of C. microphylla populations was reflected by humidity conditions at a small scale in northern China. These results indicate that desert plants would be able to adapt to an increase in humidity; therefore, genetic differentiation of species would be expected to increase as a result of adaptation (Wang et al., 2009).
4.3. Correlation between environmental and genetic data
Ecological and environmental factors can play roles in shaping genetic diversity patterns (Gaggiotti et al., 2009). With regard to the whole ecosystem, relationships between genomes and environmental factors (e.g., temperature factors, humidity factors, and soil factors) are considered important components of ecological research (Li & Peng, 2001). In our study, correlation analysis revealed a positive association between genetic diversity of C. microphylla and most environmental factors (WI, AP, S, SOC, SAN, and SOP). However, there were negative associations between genetic diversity and a couple of environmental factors (AMT and CI). This result reflected that genetic diversity of C. microphylla was restrained by AMT and CI. This result is not consistent with the previously reported relationship between ISSR diversity and AMT in A. halodendron from Horqin Sandy Land (Huang et al. 2014). Our study revealed that WI and SOP were the major environmental factors that affected genetic diversity indices of the 20 studied C. microphylla populations, which indicates that genetic variation in C. microphylla primarily depends on a greater than 5°C monthly mean temperature and SOP concentration. This result is not consistent with the previously reported relationship between ISSR diversity and environmental factors in C. microphylla from northern China (AP and CI were the major environmental factors that affected genetic diversity indices) (Huang et al., 2016). This suggests that AP and CI were the major environmental factors that affected genetic diversity indices in C. microphylla on the large‐scale field, and WI was the major environmental factors on the small‐scale field. Relevant report found that SAN, SOP, and SAN/SOP contents can exert a chemical control over the evolution of species through changing an organism's rate of growth, or an adaptation to each situation of SAN/SOP (Acquisti, Elser, & Kumar, 2009). However, evaluation of other environmental factors that impact plant population genetic diversity requires further research.
Conflict of Interest
None declared.
Acknowledgments
The authors thank all the members of Naiman Desertification Research Station, and Key Laboratory of Stress physiology and Ecology, Gansu Province, Northwest Institute of Eco‐Environment and Resources, China Academy of Sciences (CAS), for their help in field work and laboratory studies. We acknowledge the China Meteorological Administration (Beijing, China) for his help on the meteorological data information support. This study was financially supported by China national key research and development plan (2016YFC0500907), the national natural science foundation of China (41541003 and 41201561), One hundred person project of the Chinese academy of sciences (Y451H31001) and the natural science foundation of Gansu province (145RJYA269).
Huang, W. , Zhao, X. , Zhao, X. , Li, Y. and Lian, J. (2016), Effects of environmental factors on genetic diversity of Caragana microphylla in Horqin Sandy Land northeast China. Ecology and Evolution, 6: 8256–8266. doi: 10.1002/ece3.2549
References
- Acquisti, C. , Elser, J. J. , & Kumar, S. (2009). Ecological nitrogen limitation shapes the DNA composition of plant genomes. Molecular Biology and Evolution, 26, 953–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo, M. B. , Cabeza, M. , Thuiller, W. , Hannah, L. , & Williams, P. H. (2004). Would climate change drive species out of reserves? An assessment of existing reserve‐selection methods. Global Change Biology, 10, 1618–1626. [Google Scholar]
- Austerlitz, F. , Jung, M. B. , Godelle, B. , & Gouyon, P. H. (1997). Evolution of coalescence times, genetic diversity and structure during colonization. Theoretical Population Biology, 51, 148–164. [Google Scholar]
- Babbel, G. R. , & Selander, R. K. (1974). Genetic variability in edaphically restricted and widespread plant species. Evolution, 28, 619–630. [DOI] [PubMed] [Google Scholar]
- Bailey, H. P. (1979). Semi‐arid climates: their definition and distribution In Hall A. E., Cannell G. H. & Lawton H. W. (Eds.), Agriculture in semiarid environments (Vol. 34, pp. 73–96). New York, NY: Springer‐Verlag. [Google Scholar]
- ter Braak, C. J. F. , & Šmilauer, P. (2002). CANOCO reference manual and CanoDraw for windows user's guide: software for canonical community ordination (version 4.5). Microcomputer Power (Ithaca NY, USA), 500 pp.
- Bussell, J. D. (1999). The distribution of random amplified polymorphic DNA (RAPD) diversity amongst populations of Isotoma petraea (Lobeliaceae). Molecular Ecology, 8, 775–789. [Google Scholar]
- Chen, X. H. , Gao, Y. B. , Zhu, M. J. , & Zhao, T. T. (2009). Genomic DNA extraction and AFLP analysis system establishment of Caragana microphylla . Bulletin of Botanical Research, 29, 529–533. [Google Scholar]
- Endler, J. A. (1986). Natural selection in the wild. Princeton, NJ: Princeton University Press. [Google Scholar]
- Faye, A. , Deblauwe, V. , Mariac, C. , Damien, R. , Sonké, B. , Vigouroux, Y. , & Couvreur, T. L. P. (2016). Phylogeography of the genus Podococcus (Palmae/Arecaceae) in Central African rain forests: climate stability predicts unique genetic diversity. Molecular Phylogenetics and Evolution, 105, 126–138. [DOI] [PubMed] [Google Scholar]
- Freeland, J. R. , Biss, P. , Conrad, K. F. , & Silvertown, J. (2010). Selection pressures have caused genome‐wide population differentiation of Anthoxanthum odoratum despite the potential for high gene flow. Journal of Evolutionary Biology, 23, 776–782. [DOI] [PubMed] [Google Scholar]
- Fridley, J. D. , Grime, P. , Askew, A. P. , Moser, B. , & Stevens, C. J. (2011). Soil heterogeneity buffers community response to climate change in species‐rich grassland. Global Change Biology, 17, 2002–2011. [Google Scholar]
- Fu, X. Q. (1993). The second edition of the flora of Inner Mongolia. Hohhot, China: Inner Mongolia People's Press. [Google Scholar]
- Gaggiotti, O. E. , Bekkevold, D. , Jorgensen, H. B. , Foll, M. , Carvalho, G. R. , Andre, C. , & Ruzzante, D. E. (2009). Disentangling the effects of evolutionary, demographic, and environmental factors influencing genetic structure of natural populations: Atlantic herring as a case study. Evolution, 63, 2939–2951. [DOI] [PubMed] [Google Scholar]
- Ge, S. , Wang, K. Q. , Hong, D. Y. , Zhang, W. H. , & Zu, Y. G. (1999). Comparisons of genetic diversity in the endangered Adenophora lobophylla and its widespread congener, A. potaninii . Conservation Biology, 13, 509–513. [Google Scholar]
- Guo, Q. , Shi, Y. J. , Wei, Z. W. , Yang, Z. H. , Lu, J. , & Jia, Y. Q. (2008). Genetic diversity analysis by SSR marker of fourteen species of Caragana fabr. in He‐xi corridor area of Gansu. Acta Agrestia Sinica, 16, 227–233. [Google Scholar]
- Hamrick, J. L. , & Godt, M. J. W. (1989). Allozyme diversity in plant species In Brown A. H. D., Clegg M. T., Kahler A. L. & Weir B. S. (Eds.), Plant population genetics, breeding and germplasm resources (pp. 43–63). Sunderland, MA: Sinauer. [Google Scholar]
- Hamrick, J. L. , & Godt, M. J. W. (1990). Allozyme diversity in plant species In Brown A. D. H., Clegg M. T., Kahler A. L. & Weir B. S. (Eds.), Plant population genetics, breeding and genetic resources (pp. 43–63). Sunderland, MA: Sinauer Associates. [Google Scholar]
- Hamrick, J. L. , Godt, M. J. W. , Murawski, D. A. , & Loveless, M. D. (1991). Correlation between species traits and allozyme diversity implications for conservation biology In Falk D. A. & Holsinger K. E. (Eds.), Genetics and conservation of rare plants (pp. 75–86). New York, NY: Oxford University Press. [Google Scholar]
- Han, Y. Z. , Wang, Z. , & Gao, H. W. (2011). Optimization of SSR‐PCR system on Caragana mirophylla and its application. Pratacultural Science, 28, 399–413. [Google Scholar]
- Hanin, N. , Quaye, M. , Westberg, E. , & Barazani, O. (2013). Soil seed bank and among‐years genetic diversity in arid populations of Eruca sativa Miller (Brassicaceae). Journal of Arid Environments, 91, 151–154. [Google Scholar]
- Harte, J. , & Shaw, R. (1995). Shifting dominance within a montane vegetation community‐results of a climate‐warming experiment. Science, 267, 876–880. [DOI] [PubMed] [Google Scholar]
- Huang, W. D. , Zhao, X. Y. , Li, Y. L. , Li, Y. Q. , Luo, Y. Y. , Feng, J. , & Su, N. (2015). ISSR analysis of Caragana microphylla (Leguminosae) in different temperature gradients. Sciences in Cold and Arid Regions, 7(1), 0099–0103. [Google Scholar]
- Huang, W. D. , Zhao, X. Y. , Zhao, X. , Li, Y. L., Lian, J. , & Yun, J. Y. (2014). Relationship between the genetic diversity of Artemisia halodendron and climatic factors. Acta Oecologica‐international Journal of Ecology. 55, 97‐103. [Google Scholar]
- Huang, W. D. , Zhao, X. Y. , Zhao, X. , Li, Y. L. , & Pan, C. C. (2016). Environmental determinants of genetic diversity in Caragana microphylla (Fabaceae) in northern China. Botanical Journal of the Linnean Society, 181, 269–278. [Google Scholar]
- Huang, W. D. , Zhao, X. Y. , Zhao, X. , Luo, Y. Q. , Feng, J. , & Su, N. (2013). Genetic variation within the sand‐fixation species Caragana microphylla (Leguminosae) in Horqin sandy land detected by inter‐simple sequence repeats analysis. Biochemical Systematics and Ecology, 51, 343–348. [Google Scholar]
- Huang, W. D. , Zhao, X. Y. , Zhao, X. , Zhao, H. L. , Wang, S. K. , & Lian, J. (2011). A combined approach using ISSR and ITS analysis for the characterization of Artemisia halodendron from Horqin sandy land, northern China. Biochemical Systematics and Ecology, 39, 346–351. [Google Scholar]
- Hughes, A. R. , Inouye, B. D. , Johnson, M. T. J. , Underwood, N. , & Vellend, M. (2008). Ecological consequences of genetic diversity. Ecology Letters, 11, 609–623. [DOI] [PubMed] [Google Scholar]
- Hulshof, C. M. , Violle, C. , Spasojevic, M. J. , Mcgill, B. , Damschen, E. , Harrison, S. , & Enquist, B. J. (2013). Intra‐specific and inter‐specific variation in specific leaf area reveal the importance of abiotic and biotic drivers of species diversity across elevation and altitude. Journal of Vegetation Science, 24, 921–931. [Google Scholar]
- Keller, S. R. , Soolanayakanahally, R. , Guy, R. D. , Silim, S. N. , Olson, M. , & Tiffin, P. (2011). Climate‐driven local adaptation of ecophysiology and phenology in balsam poplar, Populus balsamifera L. (Salicaceae). American Journal of Botany, 98, 99–108. [DOI] [PubMed] [Google Scholar]
- Li, Y. L. , Chen, J. , Cui, J. Y. , Zhao, X. Y. , & Zhang, T. H. (2013). Nutrient resorption in Caragana microphylla along a chronosequence of plantations: implications for desertified land restoration in North China. Ecological Engineering, 53, 299–305. [Google Scholar]
- Li, S. G. , Harazono, Y. , Oikawa, T. , Zhao, H. L. , He, Z. Y. , & Chang, X. L. (2000). Grassland desertification by grazing and the resulting micrometeorological changes in Inner Mongolia. Agricultural and Forest Meteorology, 102, 125–137. [Google Scholar]
- Li, Y. F. , Jiang, S. , Gu, S. , Ma, C. C. , & Gao, Y. B. (2008). Micro‐morphology of leaf epidermis of Caragana in Inner Mongolia. Bulletin of Botanical Research, 28, 668–678. [Google Scholar]
- Li, D. , & Peng, S. L. (2001). Genetic diversity in three Pinnus massoniana populations in different elevations and its relationship with ecological factors. Acta Ecologica Sinica, 21, 415–421. [Google Scholar]
- Li, F. R. , Zhang, A. S. , Duan, S. S. , & Kang, L. F. (2005). Patterns of reproductive allocation in Artemisia halodendron inhabiting two contrasting habitats. Acta Oecologica, 28, 57–64. [Google Scholar]
- Li, F. R. , Zhao, L. Y. , Zhang, H. , Zhang, T. H. , & Shirato, Y. (2004). Wing erosion and airborne dust deposition in farmland during spring in the Horqin Sandy Land of eastern Inner Mongolia, China. Soil and Tillage Research, 75, 121–130. [Google Scholar]
- Lovejoy, T. E. , & Hannah, L. J. (2005). Climate change and biodiversity. New Haven, CT: Yale University Press. [Google Scholar]
- Ma, C. C. , Gao, Y. B. , Liu, H. F. , Wang, J. L. , & Guo, H. Y. (2003). Interspecific transition among Caragana mirophylla, C. davazamcii and C. korshinskii along geographic gradient. I. Ecological and RAPD evidence. Acta Botanica Sinica, 45, 1218–1227. [Google Scholar]
- Maki, M. (2003). Population genetics of threatened wild plants in Japan. Journal of Plant Research, 116, 169–174. [DOI] [PubMed] [Google Scholar]
- Manel, S. , Joost, S. , Epperson, B. K. , Holderegger, R. , Storfer, A. , Rosenberg, M. S. , … Bonin, A. (2010). Perspectives on the use of landscape genetics to detect genetic adaptive variation in the field. Molecular Ecology, 19, 3760–3772. [DOI] [PubMed] [Google Scholar]
- Manel, S. , Schwartz, M. K. , Luikart, G. , & Taberlet, P. (2003). Landscape genetics: combining landscape ecology and population genetics. Trends in Ecology & Evolution, 18(4), 189–197. [Google Scholar]
- Meloni, M. , Perini, D. , Filigheddu, R. , & Binelli, G. (2006). Genetic variation in five Mediterranean populations of Juniperus phoenicea as revealed by inter‐simple sequence repeat (ISSR) markers. Annals of Botany, 97, 299–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, D. W. , & Sommers, L. E. (1982). Total carbon, organic carbon and organic matter In Page A. L., Miller R. H. & Keeney D. R. (Eds.), Methods of soil analysis, part 2 (2nd ed., pp. 539–577). Madison, WI: Agronomy. [Google Scholar]
- Nevo, E. (1998). Genetic diversity in wild cereals: regional and local studies and their bearing on conservation ex situ and in situ. Genetic Resources and Crop Evolution, 45, 355–370. [Google Scholar]
- Nybom, H. , & Bartish, I. V. (2000). Effects of life history traits and sampling strategies on genetic diversity estimates obtained with RAPD markers in plants. Perspectives in Plant Ecology, Evolution and Systematics, 3, 93–114. [Google Scholar]
- Olsen, S. R. , Cole, C. W. , Watanabe, F. S. & Dean, L. A. (1954). Estimation of available phosphorus in soil by extraction with sodium bicarbonate. U.S. Dept. Agric, Washington, Circular. 939.
- Pearse, D. E. , Crandall, K. A. , & Beyond, F. S. T. (2004). Analysis of population genetic data for conservation. Conservation Genetics, 5, 585–602. [Google Scholar]
- Read, Q. D. , Moorhead, L. C. , Swenson, N. G. , Bailey, J. K. , & Sander, N. J. (2014). Convergent effects of elevation on functional leaf traits within and among species. Functional Ecology, 28, 37–45. [Google Scholar]
- Rehfeldt, G. E. , Crookston, N. L. , Warwell, M. V. , & Evans, J. S. (2006). Empirical analyses of plant‐climate relationships for the western United States. International Journal of Plant Sciences, 167, 1123–1150. [Google Scholar]
- Rehfeldt, G. E. , Tchebakova, N. M. , Parfenova, Y. I. , Wykoff, W. R. , Kuzmina, N. A. , & Milyutin, L. I. (2002). Intraspecific responses to climate in Pinus sylvestris . Global Change Biology, 8, 912–929. [Google Scholar]
- Richter, R. , Kipfer, T. , Wohlgemuth, T. , Guerrero, C. C. , Ghazoul, J. , & Moser, B. (2012). Phenotypic plasticity facilitates resistance to climate change in a highly variable environment. Oecologia, 169, 269–279. [DOI] [PubMed] [Google Scholar]
- Sacks, B. N. , Brown, S. K. , & Ernest, H. B. (2004). Population structure of California coyotes corresponds to habitat‐specific breaks and illuminates species history. Molecular Ecology, 13, 1265–1275. [DOI] [PubMed] [Google Scholar]
- Scheiner, S. M. (1993). Genetic and evolution of phenotypic plasticity. Annual Review of Ecology and Systematics, 24, 35–68. [Google Scholar]
- Sork, V. L. , Davis, F. W. , Westfall, R. , Flint, A. , Ikegami, M. , Wang, H. F. , & Grivet, D. (2010). Gene movement and genetic association with regional climate gradients in California valley oak (Quercus lobata Née) in the face of climate change. Molecular Ecology, 19, 3806–3823. [DOI] [PubMed] [Google Scholar]
- Su, Z. H. , & Zhang, M. L. (2014). A range wide geographic pattern of genetic diversity and population structure of Hexinia polydichotoma (Asteraceae) in Tarim Basin and adjacent areas. Biochemical Systematics and Ecology, 56, 49–59. [Google Scholar]
- Su, Y. Z. , Zhang, T. H. , Li, Y. L. , & Wang, F. (2005). Changes in soil properties after establishment of Artemisia halodendron and Caragana microphylla on shifting sand dunes in semiarid Horqin sandy land, Northern China. Environmental Management, 36, 272–281. [DOI] [PubMed] [Google Scholar]
- Vasemägi, A. , & Primmer, C. R. (2005). Challenges for identifying functionally important genetic variation: the promise of combining complementary research strategies. Molecular Ecology, 14, 3623–3642. [DOI] [PubMed] [Google Scholar]
- Vellend, M. , & Genber, M. A. (2005). Connections between species diversity and genetic diversity. Ecology Letters, 8, 767–781. [Google Scholar]
- Wang, T. (2003). Desert and desertification in China. Shijiazhuang, China: Hebei Science and Technology Press. [Google Scholar]
- Wang, C. H. , Li, S. F. , Fu, C. Z. , Gong, X. L. , Huang, L. , Song, X. , & Zhao, Y. (2009). Molecular genetic structure and evolution in native and colonized populations of the Chinese mitten crab, Eriocheir sinensis . Biological Invasions, 11, 389–399. [Google Scholar]
- Wu, Z. , Dijkstra, P. , Koch, G. W. , & Hungate, B. A. (2012). Biogeochemical and ecological feedbacks in grassland responses to warming. Nature Climate Change, 2, 458–461. [Google Scholar]
- Xue, G. X. , Liu, J. , & Liu, J. (1998). RAPD analysis of grass carp population in three‐river waters. Journal of Fishery Sciences of China, 5, 1–5. [Google Scholar]
- Yeh, F. C. , & Yang, R. (1999). POPGENE Version 1.31. Department of renewable resources, University of Alberta, Edmonton. Retrieved from http://www.ualberta.ca/~fyeh.
- Zhang, T. H. , Su, Y. Z. , Cui, J. Y. , Zhang, Z. H. , & Chang, X. X. (2006). A leguminous shrub in semiarid sandy soils of North China. Pedosphere, 16, 319–325. [Google Scholar]
- Zhang, C. J. , Wu, D. X. , Zhang, L. , Zhan, X. Y. , Zhou, S. X. , & Yang, Y. X. (2009). Nodule characteristics of three‐year‐old Caragana microphylla and their responses to environmental changes in an inner Mongolia grassland. Chinese Journal of Plant Ecology, 33, 1165–1176. [Google Scholar]
- Zhao, Y. Z. (2005). The distribution pattern and ecological adaptation of Caragana mirophylla, Caragana davazamcii and Caragana korshinskii . Acta Ecologica Sinica, 25, 3411–3414. [Google Scholar]
- Zhao, N. X. , Gao, Y. B. , Wang, J. L. , & Ren, A. Z. (2008). Population structure and genetic diversity of Stipa grandis P. Smirn, a dominant species in the typical steppe of northern China. Biochemical Systematics and Ecology, 36, 1–10. [Google Scholar]
- Zhao, H. L. , Zhao, X. Y. , & Zhang, T. H. (2000). Causes, processes and countermeasures of desertification in the interlocked agro‐pasturing area of North China. Journal of Desert Research, 20, 22–28. [Google Scholar]
- Zhao, H. L. , Zhao, X. Y. , & Zhang, T. H. (2003). Beijing. Desertification process and its recovery mechanism in Horqin Sandy Land. Beijing, China: Ocean Press. [Google Scholar]
- Zhao, H. L. , Zhao, X. Y. , Zhang, T. H. , & Zhou, R. L. (2002). Boundary line on agro‐pasture zigzag zone in North China and its problems on eco‐environment. Advance in Earth Sciences, 17, 739–747. [Google Scholar]


