Abstract
Skeletal muscle is a highly ordered yet complex tissue containing several cell types that interact with each other in order to maintain structure and homeostasis. It is also a highly regenerative tissue that responds to damage in a highly intricate but stereotypic manner, with distinct spatial and temporal kinetics. Proper examination of this process requires one to look at the three-dimensional orientation of the cellular and subcellular components, which can be accomplished through tissue clearing. While there has been a recent surge of protocols to study biology in whole tissue, it has primarily focused on the nervous system. This chapter describes the workflow for whole mount analysis of murine skeletal muscle for LacZ reporters, fluorescent reporters and immunofluorescence staining. Using this technique, we are able to visualize LacZ reporters more effectively in deep tissue samples, and to perform fluorescent imaging with a depth greater than 1700 μm.
Keywords: Skeletal muscle, Tissue imaging, tdTomato, Green fluorescence protein (GFP), Satellite cell, Tissue clearing, lacZ reporter
1 Introduction
Skeletal muscle is the largest tissue in the body and possesses incredible regenerative capacity. It is composed of several different types of cells including muscle fibers, satellite cells, blood vessels, fibroblasts, and immune cells [1]. These diverse cell types have a complicated interplay and changes in anatomy from the basal state during injury and disease. Understanding the structure–function relationships of the cells within the tissue is important for modeling the dynamic changes during muscle development and regeneration. Imaging is one of the most powerful tools to study this interplay of different cell types. Typically, cross sections or longitudinal sections, obtained from frozen or paraffin embedded tissue blocks, are immunolabeled and a single slice is examined under the microscope. While this is informative in the lateral plane, the information in the axial direction is lost leading to underrepresentation or misrepresentation of the actual data. As such, it is important to obtain information from all three dimensions in the tissue to study the anatomical and cellular interactions in the tissue. An example of such interaction is of that between satellite cells and blood vessel endothelial cells. It has been implicated that satellite cells are located preferentially close to blood vessels [2]. However, this interaction may be underestimated in typical two-dimensional analysis (Fig. 1). This scenario highlights the importance of analyzing cellular interactions in muscle tissue in its native three-dimensional state. The techniques in this chapter would lead to whole-tissue quantification, at a single cell resolution, to allow for the understanding of spatial interactions between cell types in the muscle during homeostasis, injury and repair.
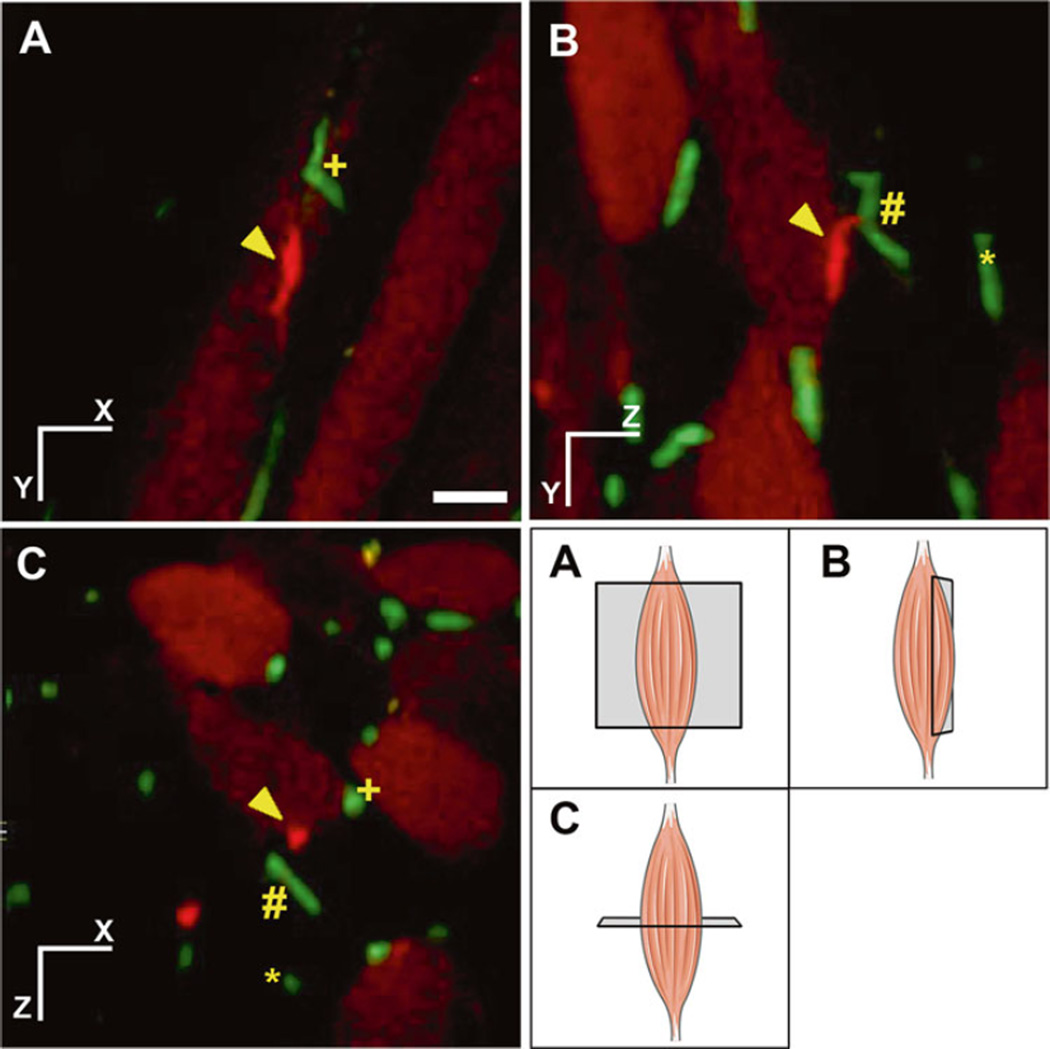
Fig. 1.
Risk of under sampling when looking at cellular proximity in sections in muscle tissue. Spatial relationship is lost or misrepresented in thin section analysis. Orthogonal view of tdTomato labeled satellite cell (red) and green fluorescence protein (GFP) labeled blood vessels (green) from Pax7CreERT2:R26R/tdTomato:Flk-1+/GFP mice. Schematic representation of the orthogonal view is presented in the bottom right panel. Three different blood vessels are marked as (plus), (hash) and (asterisk). One satellite cell is marked as (arrowhead). Cross sectional analysis in panel (A) incorrectly shows the closest proximity between satellite cell (arrowhead) and blood vessels to be (plus). In addition, panel (A) and (B) greatly under-sample the amount of blood vessels in the proximity of the satellite cell, while panel (C) shows a better representation of the information. In traditional sectional analysis, only one section would be available for analysis thereby greatly underestimating the interactions between satellite cells and blood vessel. Tissue imaging sampling in three dimensions allows for more accurate quantification of cell proximity. Scale bar denotes 25 μm
One approach to studying the spatial interaction of cells in their three dimensional (3D) anatomical state is by obtaining serial sections, imaging each section and reconstructing the 3D representation through manual or automated methods [3]. However, this process is technically challenging, error prone and labor intensive. Optical sectioning using software deconvolution and confocal microscopy also allows for imaging in 3D, but is severely limited in muscle tissue due to the presence of abundant light-scattering molecules such as lipids, extracellular matrix, and endogenously fluorescent molecules such as myoglobin and NADH. In addition, due to poor and unreliable labeling reagents for populations such as satellite cells, genetic reporter mouse lines encoding fluorescent molecules [4–7] or β-galactosidase (LacZ) [6, 8, 9] have been developed that allow for reliable and consistent assessment of cellular populations with high fidelity. LacZ reporters have been extensively utilized in studies of skeletal muscle regeneration and transplantations. X-gal staining for LacZ reporter has the benefit of maintaining stability over time and has high signal to noise ratio. One limitation of LacZ staining is the penetration depth of the staining and opacity of the muscle tissue hinders the ability to detect the signal far from the surface of the tissue.
An ideal optical tissue clearing protocol for muscle would reduce light-scattering via refractive index mismatching, signal from pigmented molecules, preserve fluorescence reporter molecule and be amenable to immunolabeling. In this chapter we adapt several tissue clearing and staining protocols to optimize them for the study of murine adult skeletal muscle [10–15]. In addition, we expand the utility of tissue clearing protocols by utilizing them for clearing LacZ stained samples. To the best of our knowledge, this is the first report to show the dramatic improvement in visualization of LacZ signal post-tissue clearing. Using the current protocol we can image completely through thinner muscles such as the cremaster and diaphragm muscles using a confocal microscope and through an extensor digitorum longus (EDL) or soleus muscle using a two-photon microscope (see Note 2).
2 Materials
Nutator mixer at 4 °C.
Bacterial shaker at 37 °C.
Rotating mixer at 37 °C.
15 ml and 50 ml conical tubes.
Dissection tools.
Glass coverslips.
Silicone imaging spacers (Electron Microscopy Science).
2 % paraformaldehyde (PFA) in phosphate buffered saline (PBS), pH 7.4.
PBST: phosphate buffered saline (PBS), pH 7.4, 0.01 % Triton.
X-gal wash buffer. 0.1 M sodium phosphate buffer, pH 7.2, 2 mM MgCl2, 0.01 % sodium deoxycholate, and 0.02 % Nonidet-P40.
X-gal reaction buffer. 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 10 mg/ml X-gal reagent (5-bromo-4-chloro-3-indolyl-β-D-galactoside) in X-gal wash buffer.
Polymerization solution. Dissolve 50 g of acrylamide and 1.25 g of VA-044 (Wako Pure Chemical) in 500 ml of PBS. Aliquot and freeze. Thaw solution overnight at 4 °C and degas the solution using nitrogen gas purge for 5 min on ice before use.
Clearing solution. 50 % N, N, N′, N′-Tetrakis(2-hydroxypropyl) ethylenediamine (87640, Sigma-Aldrich), 4 M urea in PBS.
Fluorescence wash buffer. 0.1 % Triton X-100, 0.1 % Tween 20, 0.1 % sodium deoxycholate, 10 % DMSO in PBS, pH 7.4.
Fluorescence staining buffer. 5 % donkey/goat/sheep serum, 0.1 % Triton X-100, 0.1 % Tween 20, 0.1 % sodium deoxycholate, 10 % DMSO in PBS, pH 7.4.
Refractory index matching solution (RIMS). 10 g of Histodenz (D2158 Sigma-Aldrich) in 7.5 ml of PBS, 0.1 % Tween 20, 0.1 % sodium azide (or alternative antibacterial agent).
Immunolabeling reagents (Table 1).
Image acquisition. Confocal or two-photon microscope with long working distance objective.
Deconvolution. Fiji (www.Fiji.sc) [16] (ImageJ) with DeconvolutionLab plugin [17]. Huygens Professional (Scientific Volume Imaging. www.svi.nl).
Image processing and visualization. Fiji (www.Fiji.sc) [16] with 3D viewer plugin [18], Icy (www.icy.bioimageanalysis.org) [19], Imaris (Bitplane www.bitplane.com), and Amira 3D (FEI www.fei.com).
Table 1.
Reagents for fluroscent labeling of skeletal muscle
| Antigen | Company | Cat # | Concentration | Comment |
|---|---|---|---|---|
| CD31-Alexa 488 | Biolegend | 102514 | 5–10 µg for intravital |
Marks blood vessels. Does not work on fixed tissue |
| DyLight 488 or 649 labeled tomato lectin |
Vector staining |
DL-1174, DL-1177 |
70 µg for intravital 1:50 for whole mount |
Marks blood vessels. Fluorescently labelled fixable dextran or Isolectin B4 may also be used |
| Laminin | Sigma- Aldrich |
L-9393, L-0663 |
1:100 for whole mount |
Marks muscle membrane and blood vessels |
| Dystrophin | Abcam | Ab15277 | 1:100 for whole mount |
Marks skeletal muscle membranes |
| Alexa Fluor® conjugated secondary antibodies |
Thermo Fisher Scientific |
1:200 | Classical fluorescence dyes are not recommended |
|
| Donkey/goat/sheep serum |
Sigma- Aldrich |
5 % | Block based on the species the secondary antibody is from (see Note 1) |
3 Methods
3.1 Preparation of Tissue for Clearing
For proper quantification analysis, it is important to have homogenous signal throughout the sample. For this purpose, reporter lines are far superior to immunolabeling. For labeling satellite cells, we utilized the Pax7CreErT2 mice [20] crossed with (loxP-stoploxP-tdTomato) reporter mice [21], which are currently the strongest conditional reporter mice available for satellite cells (see Note 2). For assessing LacZ signals, we utilized the Myf5+/nLacZ mice [22] for satellite cells and Flt-1+/LacZ [23] mice for blood vessels.
Sedate mice according to the institutionally approval protocol.
Administer fluorescent-labeled antibody or lectin through either tail vein or retro-orbital sinus for intravital staining of the vasculature or circulating cells. Allow the reagents to circulate and bind for 10 min.
Prepare cold 2 % PFA in PBS for perfusion fixation of the mouse (see Notes 3 and 4).
Perfuse the animal with 50 ml of fixative at a rate of 2.5 ml/min, as previously described [24] (see Note 5).
Dissect the tissue required for the study. For initial screening, we recommend the male cremaster muscle, which is thin, readily cleared and easily imaged. Steps for proper dissections are detailed in Fig. 2. Other suitable muscle includes the EDL and the soleus muscle. Their dissections have been previously described [25].
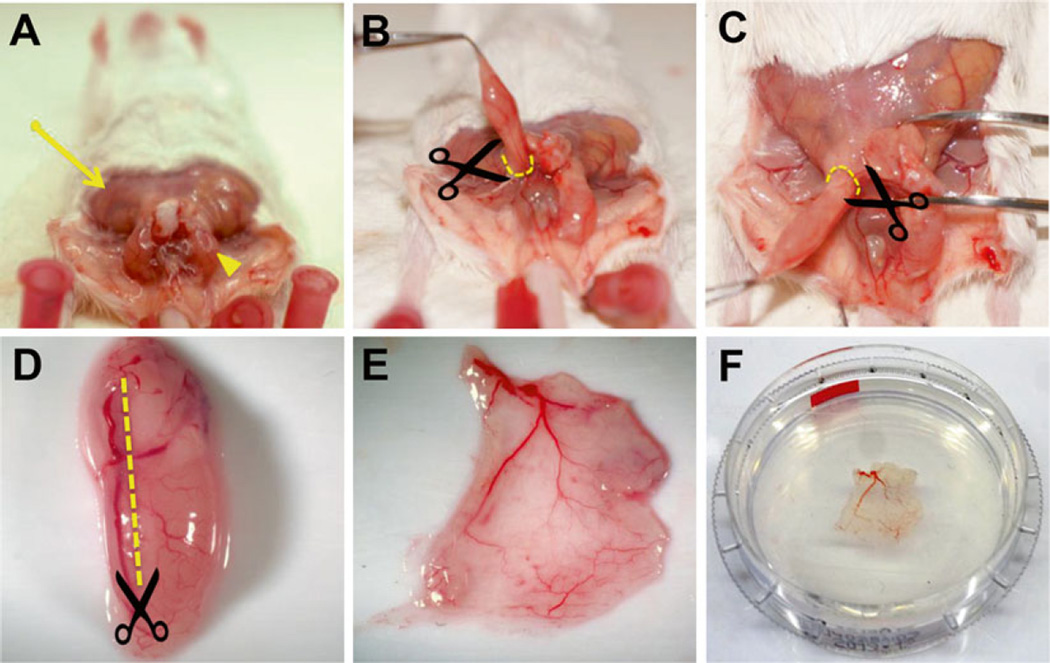
Fig. 2.
Preparation of mouse cremaster muscle. (A) Remove the overlying skin (arrow) over the cremaster muscle (arrowhead). Gently apply pressure on the abdomen to allow the testicles to descend into the cremaster sac and grasp the muscle at the distal tip using blunt tweezers. (B, C) Retract the cremaster sac up and away from the body, gently removing the connective tissue as the muscle is freed. Make an incision around the cremaster sac (broken lines) and obtain the muscle along with the gonads. (D) Make an incision longitudinally along the muscle (broken line) taking care to avoid the primary artery feeding the muscle. (E) Flatten the muscle on a soft surface and remove the internal soft tissue. (F) Post-fix the muscle flat by submerging it underneath a coverslip to maintain the flat shape
3.2 LacZ Staining
Tissue clearing allows for greater transparency compared to traditional preparations following LacZ staining. Cleared tissue can be visualized all the way through to allow for proper assessment of signal deep within the tissue that would be traditionally missed (Fig. 3).
Wash the tissue samples using X-gal wash buffer three times at 37 °C for 2 h (see Note 6).
Light shield with aluminum foil and incubate the tissue in X-gal reaction buffer overnight at 37 °C in a bacterial shaker. Large pieces of tissue such as the adult gastrocnemius muscle should be cut into smaller pieces to allow to sufficient penetration of the substrate.
Wash the tissue three times in ice-cold PBST for 30 min each with rotation at 4 °C. 4.
Incubate the tissue in polymerization solution for 6 h with rotation at 4 °C.
Transfer the tissue into fresh nitrogen degassed polymerization solution. Overfill a 15 ml conical with the polymerization solution and place a plastic wrap over the tube before capping it to avoid introducing any air into the filled conical tube. Place the tube on a rotation mixer at 37 °C for 3 h. Discard the acrylamide waste appropriately.
Wash the tissue in clearing solution for 30 min and discard the residual acrylamide waste appropriately.
Incubate the tissue in clearing solution for 1–3 day at 37 °C in a bacterial shaker. Replace clearing solution each day.
Wash the tissue in PBST three times for 30 min.
Incubate the tissue in RIMS solution overnight.
Image the tissue using a stereoscope while it is immersed in RIMS. Flat muscle such as the cremaster muscle may be submerged in RIMS and flattened by a glass coverslip. Thicker muscle may be imaged by placing them into silicone isolator surrounded by coverslips (see Note 7).
The samples can remain in RIMS indefinitely and stored at room temperature.
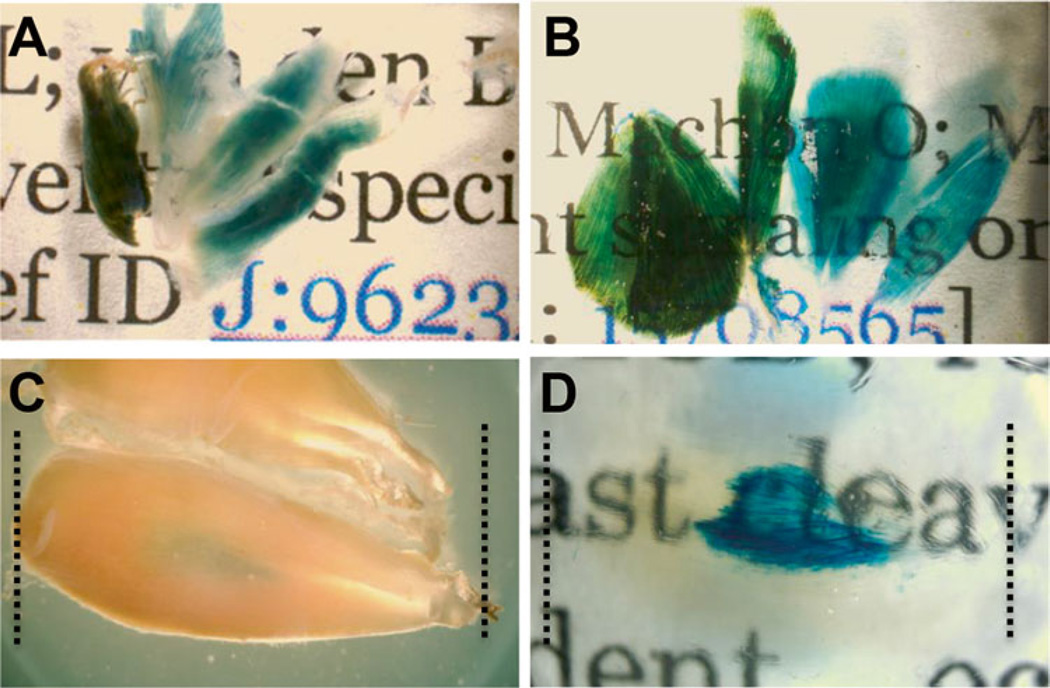
Fig. 3.
Tissue clearing following LacZ staining allows for greater transparency in muscle allowing for visualization of deep tissue staining. Skeletal muscle from Flt-1+/LacZ mice marking blood vessels lacZ staining before (A) and after (B) tissue clearing. Tibialis anterior (TA) muscle before tissue clearing (C) shows moderate lacZ staining. TA muscle after tissue clearing (D) shows extensive signal in the middle of the tissue
3.3 Tissue Clearing for Fluorescent Reporter and Immunofluorescence
For deep tissue fluorescent imaging, it is recommended that the red or far-red fluorescent probes be used to avoid interference from endogenously fluorescent molecules. Skeletal muscle presents an additional challenge in selecting fluorophores due to the presence of myoglobin, NADH and FAD and non-homogenous concentration of these molecules. These molecules are especially problematic in the same spectral region as GFP [26]. As such, weak markers or myofiber markers should not be visualized in the green spectrum. GFP reporters may also be red-shifted using anti-GFP antibody. If multi-color-imaging spanning the spectra is desired, the green spectra should be reserved for non-myogenic markers.
Wash the tissue in fluorescence wash buffer three times for 60 min at 37 °C to permeabilize the tissue (see Note 8).
Block the tissue in fluorescence staining buffer for 4 h at 37 °C.
Incubate the tissue with primary antibody in fluorescence staining buffer for 24–48 h depending on the thickness of the tissue at 37 °C (see Note 9).
Transfer the tissue to 4 °C overnight to allow the antibody to bind.
Wash the tissue in fluorescence wash buffer three times at 37 °C in a bacterial shaker for 8 h total.
Incubate the tissue in 1:200 dilution of fluorescently conjugated secondary antibody (see Note 10) for 24–48 h at 37 °C.
Wash the tissue in fluorescence wash buffer three times at 37 °C in a bacterial shaker for 8 h total.
Incubate the tissue in RIMS overnight.
Mount the tissue in fresh RIMS between coverslips in silicone gaskets or 3D printed slide holders.
For long term storage, transfer the tissue into PBS and store at 4 °C for a few months. Samples should be incubated in RIMS again prior to imaging.
3.4 Image Acquisition
Image acquisition is highly dependent on the type of instrument used and the manufacturer or core-facility should be consulted for the best use case for a particular instrument. In this section, several tips will be discussed to allow deep tissue imaging. Typically, depth of 450 μm can be achieved using a confocal microscope system. Depth of more than 1700 μm can be obtained using a two-photon microscope system on cleared tissue. Two-photon excitation can be particularly challenging to predict and therefore proper planning is required. The spectra database hosted at the University of Arizona (http://www.spectra.arizona.edu) is a great resource for obtaining the proper excitation of various florescent dyes and molecules and the two-photon spectra is labeled as “-2P”. The regular periodic organization of the sarcomeres also allows for label free second harmonic generation (SHG) detection of the myofibers [27] at an excitation wavelength of 870 nm. This can be particularly useful to delineate between intact and injured muscle fibers (Fig. 4). However, the emission will be in the same range as common DNA binding dyes such as DAPI or Hoechst 33342 and thus require separate acquisitions.
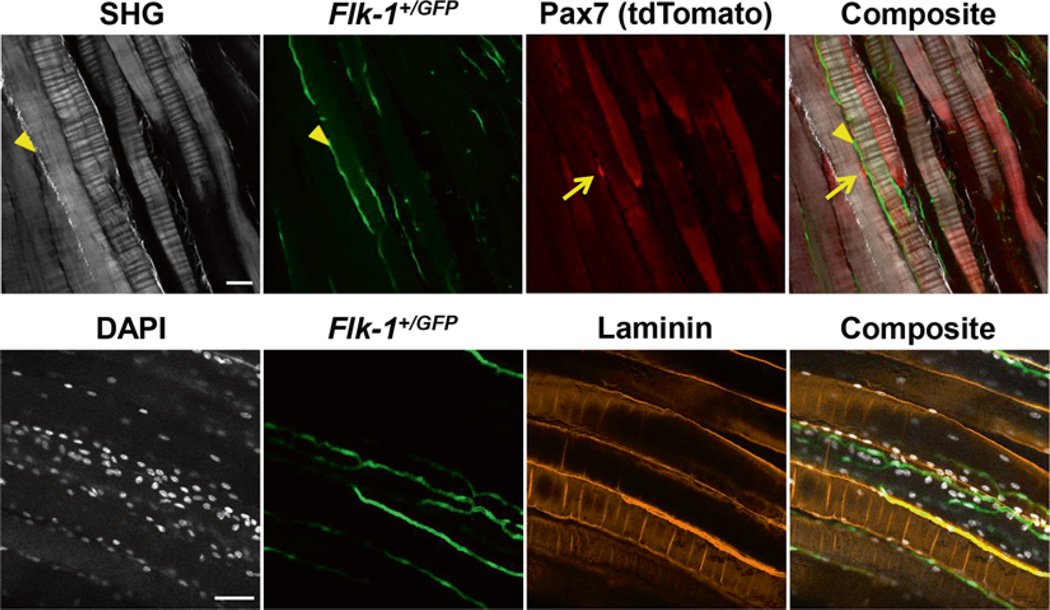
Fig. 4.
Examples of cleared tissue with fluorescent reporter and immunostaining. (Top) Maximum intensity projection of 3 × 1 μm slices of cleared tissue with fluorescent reporters acquired using a two-photon microscope system. Second harmonic generation (SHG) for label free detection of collagen and muscle fiber sarcomere. tdTomato positive satellite cells (arrows) and Flk-1+/GFP positive blood vessels. Collagen fibers are also marked by SHG signal (arrowheads). (Bottom) Maximum intensity projection of 15 × 1 μm slices of cleared tissue with immunofluorescence and fluorescent reporter acquired using a confocal microscope system. DAPI labels all nuclei, Flk-1+/GFP marks the blood vessels and laminin immunofluorescence marks the myofiber and blood vessel membrane. Scale bar denotes 50 μm
3.5 Image Processing
Some image processing is required to obtain good quality images suitable for analysis. While optical sectioning through confocal allows for excitation at a single point or layer, there is still some inherent scattering of light that causes noise and scattering of the signal, particularly in the axial direction. Software deconvolution can mathematically correct for this by redistributing the scattered light signal to its original state. A detailed discussion of deconvolution is out of the scope of this chapter. Some practical considerations have been addressed in previous reports [17, 28]. We utilize Huygens Professional, which has a user-friendly interface and allows for theoretical calculation of a point spread function (PSF). Detailed tutorial on utilizing the software can be found on the manufacturers website and elsewhere [28].
3.6 Visualization of Data
Deconvoluted images can be visualized in several manner including intensity projection, orthogonal view, and 3D render. This can be accomplished in several program including Imaris, Amira 3D, Icy, and FIJI. FIJI [16] and Icy [19] are free open source software. For this chapter instructions for obtaining these views in FIJI are provided. Open the image file using the Bio-Formats Import plugin (File → Import → Bio Formats) as a Hyperstack.
Intensity projection is a convenient method to visualize data from many different focal planes in a 2D format. It is more suited for visualizing rare populations over multiple planes or complex vascular anatomy. To view an intensity projection, open the Z-Projection dialog from Image → Stacks → Z Project. Select the range for the projection and select the desired projection type. Maximum Intensity Projection (MIP) is the most popular projection algorithm for visualizing 3D data. However it is very susceptible to imaging artifacts and saturated hotspots. Standard Deviation Intensity Projection (SDIP) is an alternative to MIP that takes into account the contrast variability across the entire data set. SDIP is suited for samples with good signal to noise ratios (SNR) and membrane staining. Additional projections such as minimum intensity, average intensity, sum slices and median are less useful for use in projection of fluorescent samples.
Orthogonal view is a simple method to visualize 3D information. To view the hyperstack in orthogonal view go to Image → Stack → Orthogonal view. It displays the XY plane along with the XZ and YZ plane. In Orthogonal Views, the two alternative planes will dock along the respective dimension of the XY plane. Two intersecting yellow lines will live update in the alternative views automatically.
3D Render. 3D rendering is possible but limited in FIJI. It requires the 3D viewer plugin [18]. Images can be 3D rendered by Plugin → 3D viewer. Detailed explanation of the 3D viewer is out of the scope of this chapter but can be found at the Virtual Insect Brain Project website (www.3dviewer.neurofly.de).
Footnotes
Animal serum may be substituted by 1 % bovine serum albumin (BSA). However, IgG free BSA should be used if antigoat secondary antibodies are used due to similarities between bovine and goat IgG.
As with any Cre based system, it is important to validate the specificity of the Cre using a strong reporter system. For labeling satellite cells, we tested two popular tamoxifen inducible Cre mouse lines, Pax7tm2.1(cre/ERT2)Fan/J(Fan) and Pax7tm1(cre/ERT2)Gaka(Gaka), available in Jackson Laboratory.
Fresh PFA is important for imaging of fluorescently labeled tissue. Commercially available PFA solutions may contain methanol as preservative. Methanol can bleach fluorescent proteins and dampen β-gal activity.
If immunostaining is not required, 0.05 % or 1.0 % glutaraldehyde may be added the fixative solution for fluorescently labeled and β-gal labeled tissue, respectively.
Perfusion fixation followed by overnight immersion fixation in best for preserving fluorescent molecules. However, if immunostaining is susceptible to PFA fixation, tissue may be immersion fixed for only 30 min (with perfusion) or 4 h (without perfusion) 4 °C.
All washes are done in a bacterial shaker in 15 ml or 50 ml conical tubes filled 2/3 of the way and laying at a 45°. The vigorous agitation is required for proper penetration of the labeling regents. Prolong fixation time if the samples tend to break apart during the wash steps.
Silicone spacers may be washed, dried and reused. Use adhesive tape to remove dust particles prior to adhering the spacer onto a glass coverslip.
Light shield all samples using aluminum foil during the wash and incubation period to prevent loss of fluorescence.
Highly abundant epitope will require higher amounts of antibody as compared to immunostaining in cross sections. For example, anti-laminin staining requires tenfold higher antibody concentration as compared to immunostaining in cross sections.
It is important to use Alexa Fluor or DyLight-conjugated antibodies versus classical dyes such as FITC, rhodamine, Cy5, or other such dyes as they are much more susceptible to photobleaching and changes in pH.
References
- 1.Bentzinger CF, Wang YX, Dumont NA, Rudnicki MA. Cellular dynamics in the muscle satellite cell niche. EMBO Rep. 2013;14:1062–1072. doi: 10.1038/embor.2013.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Christov C, Chretien F, Abou-Khalil R, et al. Muscle satellite cells and endothelial cells: close neighbors and privileged partners. Mol Biol Cell. 2007;18:1397–1409. doi: 10.1091/mbc.E06-08-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoshida S, Sukeno M, Nabeshima Y-I. A vasculature-associated niche for undifferentiated spermatogonia in the mouse testis. Science. 2007;317:1722–1726. doi: 10.1126/science.1144885. [DOI] [PubMed] [Google Scholar]
- 4.Keefe AC, Lawson JA, Flygare SD, et al. Muscle stem cells contribute to myofibres in sedentary adult mice. Nat Commun. 2015;6:7087. doi: 10.1038/ncomms8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosnakovski D, Xu Z, Li W, et al. Prospective isolation of skeletal muscle stem cells with a Pax7 reporter. Stem Cells. 2008;26:3194–3204. doi: 10.1634/stemcells.2007-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sambasivan R, Gayraud-Morel B, Dumas G, et al. Distinct regulatory cascades govern extraocular and pharyngeal arch muscle progenitor cell fates. Dev Cell. 2009;16:810–821. doi: 10.1016/j.devcel.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 7.Relaix F, Rocancourt D, Mansouri A, Buckingham M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 2005;435:948–953. doi: 10.1038/nature03594. [DOI] [PubMed] [Google Scholar]
- 8.Asakura A, Seale P, Girgis-Gabardo A, Rudnicki MA. Myogenic specification of side population cells in skeletal muscle. J Cell Biol. 2002;159:123–134. doi: 10.1083/jcb.200202092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tajbakhsh S, Bober E, Babinet C, et al. Gene targeting the myf-5 locus with nlacZ reveals expression of this myogenic factor in mature skeletal muscle fibres as well as early embryonic muscle. Dev Dyn. 1996;206:291–300. doi: 10.1002/(SICI)1097-0177(199607)206:3<291::AID-AJA6>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 10.Renier N, Wu Z, Simon D, Yang J, et al. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell. 2014;159(4):896–910. doi: 10.1016/j.cell.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 11.Tainaka K, Kubota SI, Suyama TQ, et al. Whole-body imaging with single-cell resolution by tissue decolorization. Cell. 2014;159:911–924. doi: 10.1016/j.cell.2014.10.034. [DOI] [PubMed] [Google Scholar]
- 12.Susaki EA, Tainaka K, Perrin D, et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell. 2014;157:726–739. doi: 10.1016/j.cell.2014.03.042. [DOI] [PubMed] [Google Scholar]
- 13.Yang B, Treweek JB, Kulkarni RP, et al. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell. 2014;158:945–958. doi: 10.1016/j.cell.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomer R, Ye L, Hsueh B, Deisseroth K. Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat Protoc. 2014;9:1682–1697. doi: 10.1038/nprot.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hama H, Hioki H, Namiki K, et al. ScaleS: an optical clearing palette for biological imaging. Nat Neurosci. 2015;18:1518–1529. doi: 10.1038/nn.4107. [DOI] [PubMed] [Google Scholar]
- 16.Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vonesch C, Unser M. A fast thresholded Landweber algorithm for wavelet-regularized multidimensional deconvolution. IEEE Trans Image Process. 2008;17:539–549. doi: 10.1109/TIP.2008.917103. [DOI] [PubMed] [Google Scholar]
- 18.Schmid B, Schindelin J, Cardona A, et al. A high-level 3D visualization API for Java and ImageJ. BMC Bioinformatics. 2010;11:274. doi: 10.1186/1471-2105-11-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Chaumont F, Dallongeville S, Chenouard N, et al. Icy: an open bioimage informatics platform for extended reproducible research. Nat Methods. 2012;9:690–696. doi: 10.1038/nmeth.2075. [DOI] [PubMed] [Google Scholar]
- 20.Murphy MM, Lawson JA, Mathew SJ, et al. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development. 2011;138:3625–3637. doi: 10.1242/dev.064162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madisen L, Zwingman TA, Sunkin SM, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tajbakhsh S, Rocancourt D, Cossu G, Buckingham M. Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act upstream of MyoD. Cell. 1997;89:127–138. doi: 10.1016/s0092-8674(00)80189-0. [DOI] [PubMed] [Google Scholar]
- 23.Kearney JB, Ambler CA, Monaco KA, et al. Vascular endothelial growth factor receptor Flt-1 negatively regulates developmental blood vessel formation by modulating endothelial cell division. Blood. 2002;99:2397–2407. doi: 10.1182/blood.v99.7.2397. [DOI] [PubMed] [Google Scholar]
- 24.Brazelton TR, Blau HM. Optimizing techniques for tracking transplanted stem cells in vivo. Stem Cells. 2005;23:1251–1265. doi: 10.1634/stemcells.2005-0149. [DOI] [PubMed] [Google Scholar]
- 25.Collins CA, Zammit PS. Isolation and grafting of single muscle fibres. Methods Mol Biol. 2009;482:319–330. doi: 10.1007/978-1-59745-060-7_20. [DOI] [PubMed] [Google Scholar]
- 26.Jackson KA, Snyder DS, Goodell MA. Skeletal muscle fiber-specific green autofluorescence: potential for stem cell engraftment artifacts. Stem Cells. 2004;22:180–187. doi: 10.1634/stemcells.22-2-180. [DOI] [PubMed] [Google Scholar]
- 27.Liu W, Raben N, Ralston E. Quantitative evaluation of skeletal muscle defects in second harmonic generation images. J Biomed Opt. 2013;18:26005. doi: 10.1117/1.JBO.18.2.026005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wimmer VC, Möller A. High-resolution confocal imaging in tissue. Methods Mol Biol. 2010;611:183–191. doi: 10.1007/978-1-60327-345-9_15. [DOI] [PubMed] [Google Scholar]