Abstract
To identify risk variants for multiple myeloma (MM), we conducted a genome-wide association study totaling of 1,675 MM cases and 5,903 controls. We identified risk loci for MM at 3p22.1 (rs1052501, ULK4; odds ratio [OR]=1.32; P=7.47x10-9) and 7p15.3 (rs4487645, OR=1.38; P=3.33x10-15). In addition, we observed a promising association at 2p23.3 (rs6746082, OR=1.29; P=1.22x10-7). Our study reports previously unidentified genomic regions associated with MM risk that may lead to new etiological insights.
Multiple myeloma (MM) is a malignancy of plasma cells primarily located within the bone marrow1. In the US ~16,000 individuals are diagnosed each year with MM and ~11,000 die of the disease2. Monoclonal gammopathy of undetermined significance (MGUS; a pre-malignant clone of plasma cells producing a monoclonal paraprotein) is present in ~2% of individuals older than 50 years, and the risk of progressing to multiple myeloma is 1% each year3. The increased risk of MM in the relatives of MGUS cases is consistent with MGUS being a marker of genetic susceptibility4. To date no lifestyle or environmental exposures have been consistently linked to an increased risk of MM or MGUS. Predicated on the hypothesis that part of the 2- to 4-fold elevated risk of MM in relatives of individuals with MM5 is a consequence of the co-inheritance of multiple low-risk variants, we conducted two genome-wide association studies (GWAS).
The two GWAS were conducted in the UK and Germany (UK-GWAS and German-GWAS). Genotyping of both case series was conducted using Illumina Omni Express BeadChips. Cases for the UK-GWAS comprised 1,371 patients ascertained through the UK Medical Research Council (MRC) Myeloma-IX trial6. Genotype frequencies were compared with publicly accessible genotype data generated by the UK Wellcome Trust Case-Control Consortium 2 (WTCCC2) study of 2,699 individuals from the 1958 British Birth Cohort (58C)7 and 2,501 individuals from the UK Blood Service collections (UKBS), that had been genotyped using Illumina Human1.2M-Duo Custom_v1 Array BeadChips (Supplementary Methods). The German-GWAS was based on genotyping 384 MM cases ascertained through Heidelberg University Clinic. Genotype frequencies were compared with publicly accessible genotype data generated by the Heinz-Nixdorf Recall (HNR) study of 704 individuals8 from the German population that had been genotyped using Illumina HumaOmni-Quad BeadChips (Supplementary Methods).
Genotype data from the two GWAS were filtered on the basis of pre-specified quality-control measures (Supplementary Methods). Individual SNPs were excluded from further analysis if they showed deviation from the Hardy–Weinberg equilibrium with a P<1.0x10-6 in controls, an individual SNP genotype yield <95%, or a minor allele frequency <1%. This filtering resulted in the use of 422,839 autosomal SNPs, common to both case-control series. A total of 80 case samples were removed during quality control steps for reasons including a failure to genotype, unknown duplicates and closely related individuals or non-CEU ancestry (Supplementary Figure 1; Supplementary Figure 2).
Prior to undertaking the meta-analysis of the two GWAS, we searched for potential errors and biases in the datasets. Quantile-quantile plots of genome-wide chi-square values showed there was minimal inflation of the test statistics rendering substantial cryptic population substructure or differential genotype calling between cases and controls unlikely in either GWAS (genomic control inflation factor9, λgc=1.033 and 1.059 in UK and German GWAS respectively; Supplementary Figure 3). For completeness principal components analysis was performed using the Eigenstrat10 software to determine the effects of population substructure on our findings (λcorrected=1.010 and 1.005 in UK and German GWAS respectively; Supplementary Figure 3).
Using data on all cases and controls from both series, we derived joint odds ratios (ORs) and confidence intervals (CIs) under a fixed effects model for each SNP, and associated P-values11. In the combined analysis 19 SNPs, which annotate three genomic regions, showed evidence for an association with MM at Pcombined<5.0x10-7 with evidence of an association at P<0.05 in both datasets (Supplementary Table 1). We successfully genotyped the most highly associated SNPs mapping to the three regions in 169 MM cases ascertained through the UK MRC Myeloma-VII trial (Supplementary Methods). For controls we made use of Illumina Hap550K BeadChip genotype data generated on 927 healthy individuals from the UK as part of a study of colorectal cancer we had previously conducted12.
In a combined analysis the rs1052501 association at 3p22.1 and the rs4636103 association at 7p15.3 attained genome-wide significance (P-values 7.47x10-9 and 3.33x10-15 respectively; Table 1)
Table 1. Summary results for the rs4487645 (7p15.3), rs1052501 (3p22.1) and rs6746082 (2p23.3) SNPs associated with multiple myeloma risk.
| RAFa | Case genotypes | RAFa | Control genotypes | ORb | 95% CIc | P-value | Padjustedd | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs4487645 (7p15.3) | CC | AC | AA | CC | AC | AA | ||||||
| UK-GWAS | 0.71 | 669 | 548 | 103 | 0.65 | 2216 | 2333 | 646 | 1.34 | 1.22-1.47 | 1.07x10-9 | 3.23x10-9 |
| German-GWAS | 0.76 | 206 | 126 | 22 | 0.67 | 320 | 300 | 83 | 1.55 | 1.26-1.90 | 2.66x10-5 | 6.40x10-5 |
| UK replication | 0.73 | 92 | 64 | 13 | 0.65 | 383 | 421 | 113 | 1.50 | 1.16-1.95 | 0.002 | - |
| Combined | 1.38 | 1.28-1.50 | 3.33x10-15 | 2.62x10-14 | ||||||||
| Phet=0.36, I2=1% | ||||||||||||
| rs1052501 (3p22.1) | GG | AG | AA | GG | AG | AA | ||||||
| UK-GWAS | 0.20 | 54 | 422 | 845 | 0.16 | 137 | 1391 | 3668 | 1.31 | 1.18-1.46 | 8.65x10-7 | 6.04x10-7 |
| German-GWAS | 0.19 | 10 | 118 | 226 | 0.16 | 17 | 185 | 502 | 1.32 | 1.04-1.68 | 0.021 | 0.085 |
| UK replication | 0.21 | 9 | 52 | 108 | 0.16 | 25 | 248 | 643 | 1.34 | 1.00-1.79 | 0.049 | - |
| Combined | 1.32 | 1.20-1.45 | 7.47x10-9 | 1.81x10-8 | ||||||||
| Phet=0.99, I2=0% | ||||||||||||
| rs6746082 (2p23.3) | AA | AC | CC | AA | AC | CC | ||||||
| UK-GWAS | 0.82 | 889 | 392 | 40 | 0.79 | 3202 | 1758 | 238 | 1.26 | 1.13-1.41 | 4.17x10-5 | 3.79x10-5 |
| German-GWAS | 0.84 | 247 | 97 | 10 | 0.77 | 415 | 258 | 31 | 1.50 | 1.18-1.90 | 8.21x10-4 | 3.63x10-3 |
| UK replication | 0.82 | 115 | 47 | 7 | 0.79 | 574 | 294 | 49 | 1.22 | 0.91-1.64 | 0.177 | - |
| Combined | 1.29 | 1.17-1.42 | 1.22X10-7 | 4.02x10-7 | ||||||||
| Phet=,0.39 I2=0% | ||||||||||||
Risk allele frequency (RAF).
Odds ratio.
95% Confidence Interval.
Eigenstrat adjusted P-values.
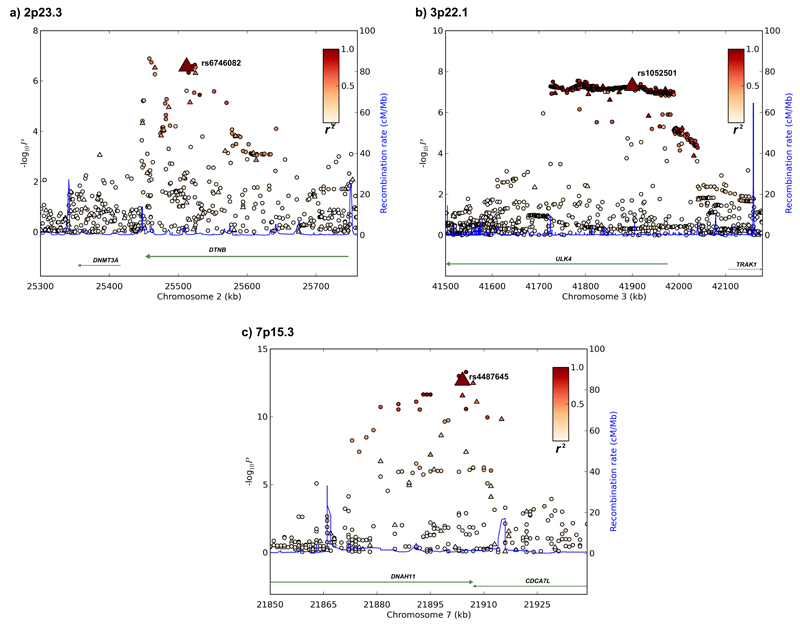
rs1052501 localizes to exon 17 of the serine/threonine-protein kinase unc-51-like kinase 4 (ULK4) gene (41,900,402bps) within a 516kb region of linkage disequilibrium (LD) on 3p22.1 (Figure 1). The G to A substitution at rs1052501 results in an alanine to threonine change at amino acid 542 which is predicted to be tolerated / benign by the in silico algorithms SIFT and Polyphen respectively. The Atg1/ULK complex is a key regulator of mTOR-mediated autophagy13 but have thus far not been directly implicated in MM pathogenesis. However, autophagy genes are increasingly being considered as tumor suppressors14 and intriguingly mTOR regulation represents an important therapeutic target in myelomatous plasma cells15. In addition to ULK4 the region of extensive LD encompasses the 5’ part of TRAK1 (trafficking protein, kinesin binding 1; MIM 608112) which has a crucial role in regulating the endocytic trafficking of GABA(A) receptors.
Figure 1. Regional plots of association results and recombination rates for the 2p23.3, 3p22.1 and 7p15.3 susceptibility loci.
(a-c) Association results of both genotyped (triangles) and imputed (circles) SNPs in the GWAS samples and recombination rates within the loci: (a) 2p23.3, (b) 3p22.1, (b) 7p15.3. For each plot, −log10 P values (y axis) of the SNPs are shown according to their chromosomal positions (x axis). The top genotyped SNP in each combined analysis is a large triangle and is labeled by its rsID. The color intensity of each symbol reflects the extent of LD with the top genotyped SNP: white (r2=0) through to dark red (r2=1.0). Genetic recombination rates (cM/Mb), estimated using HapMap CEU samples, are shown with a light blue line. Physical positions are based on NCBI build 36 of the human genome. Also shown are the relative positions of genes and transcripts mapping to each region of association. Genes have been redrawn to show the relative positions; therefore, maps are not to physical scale.
rs4487645 maps to intron 80 of the DNAH11 (dynein, axonemal, heavy chain 11 MIM 603339) gene at 7p15.3 (21,904,765bps; Figure 1). DNAH11 encodes a dynein heavy chain microtubule-dependent motor ATPase, which is involved in respiratory cilia movement; germline DNAH11 mutations causing Kartagener Syndrome (situs inversus totalis and primary ciliary dyskinesia). The 88kb region of LD annotated by rs4487645 also encompasses the 3’ part of the CDCA7L (cell division cycle associated 7-like; MIM 609685) gene (Figure 1). Deregulation of MYC typifies plasma cell neoplasms16,17. CDCA7L therefore represents an attractive candidate for the functional basis of the rs4487645 association since CDCA7L is a MYC-interacting protein, acts as a binding partner of p75 and potentiates MYC-transformational activity18,19.
In addition to the 3p22.1 and 7p15.3 loci the rs6746082 association at 2p23.3 was promising but did not attain genome-wide significance (P=1.22x10-7). rs6746082 localizes to intron 12 of DTNB (25,512,748 bps; Figure 1) within a 256kb region of LD on 2p23.3. DTNB (MIM 602415) encodes dystrobrevin beta, a component of the dystrophin-associated protein complex which is abundantly expressed in brain and other tissues but not in muscle. Alternatively spliced transcript variants encoding different DTNB isoforms exist but none have thus far been implicated in the biology of MM.
Elucidation of the basis of the 2p23.3, 3p22.1, and 7p15.3 associations will require fine–mapping and functional analyses, however to explore the region further we imputed unobserved genotypes in UK and German GWA cases and controls using HapMap Phase III and 1000genomes data (Supplementary Methods). This analysis did not provide for substantive evidence of a stronger association at each of the three loci to that provided by rs4487645, rs1052501, or rs6746082 (Supplementary Methods, Figure 1; Supplementary Table 2). To examine if any directly typed or imputed SNPs annotate a putative transcription factor binding/enhancer element, we conducted a bioinformatic search of the region of association using Transfac Matrix Database20, and PReMod21 software. These analyses did not provide evidence that rs4487645, rs1052501, or rs6746082, or closely correlated SNP maps with a known or predicted transcription regulatory region (Supplementary Table 2).
To explore whether the rs4487645, rs1052501, rs6746082 associations reflects cis-acting regulatory effects on DNAH11 or CDCA7L, ULK4 or TRAK1, and DTNB respectively, we studied mRNA expression in the plasma cells of 191 MM patients using Human Genome U133 Plus 2.0 arrays (Affymetrix, Santa Clara, USA)22 and 90 EBV–lymphoblastoid cell lines using Sentrix Human-6 Expression BeadChips (Illumina, San Diego, USA)23,24. There was no consistent statistically significant relationship between rs4487645, rs1052501, rs6746082 and expression after adjustment for multiple testing (Supplementary Figure 4). This does not preclude the possibility of subtle effects of genotype with a cumulative long-term impact since we could only detect >5% difference in expression by genotype with 80% power and levels of RNA at a single time point may not adequately capture the impact of differential expression in tumor development.
Multiple myeloma is characterized by male predominance. We assessed the relationship between sex, age at diagnosis and rs4487645, rs1052501, rs6746082 by case-only analysis using data from all series (Supplementary Table 3). The association with MM was not related to age or sex (Supplementary Table 3). Several subtypes of MM are recognized which have unique clinico-pathological phenotypes25. Hierarchically MM can be divided into hyperdiploid and non-hyperdiploid subtypes26,27. The latter is primarily composed of cases harboring IGH translocations (t(11;14)(q13;q32), t(4;14)(p16;q32) and t(14;16)(q32;q23), and is typified by more aggressive disease. Trisomies and a more indolent form of the disease characterize hyperdiploid MM25,28. Case-only analysis provided no evidence for a subtype specific association after adjustment for multiple testing, consistent with the risk variant having a generic effect on MM (Supplementary Table 3).
The risks of MM associated with rs4487645, rs1052501 and rs6746082 are modest, collectively accounting for only ~4% of the familial risk of MM. However, the carrier frequency of risk alleles are high in the European population therefore the loci make a significant contribution to the development of MM in terms of population attributable fraction, underlying ~37% of cases. It will be intriguing to explore how our findings translate to non-European populations, which have a lower prevalence of MGUS and MM29. As the frequencies of rs4487645, rs1052501 and rs6746082 genotypes in the CHB/JPT and YRI populations are significantly different to the CEU population it is possible that 2p23.3, 3p22.1 and 7p15.3 variation in part underscores differences in disease incidence.
Our findings provide evidence that common genetic variation influences MM risk. Furthermore, these findings in conjunction with recent observations from Hodgkin’s lymphoma30 and chronic lymphocytic leukemia31 GWAS raise the possibility that genetic determined dysregulation of MYC may be a common mechanism of predisposition to hematological malignancies of B-cell lineage. Given the modest size of our study and as evidenced by an over-representation of association signals after exclusion of SNPs mapping to regions of LD at the 2p23.3, 3p22.1 and 7p15.3 associations (Supplementary Figure 3), it is likely that further risk variants for MM will be identified through additional studies.
Methods
Subjects
Genome-wide association study
UK-GWAS: The UK study was based on MM cases (ICD-10 C90.0; 819 male; mean age at diagnosis 64.1 years, SD 10.3) ascertained through the UK Medical Research Council (MRC) Myeloma-IX trial6. All cases were UK residents. For controls, we used publicly accessible data generated by the Wellcome Trust Case Control Consortium from the 1958 Birth Cohort (58C; also known as the National Child Development Study)7 and UK Blood Service (UKBS). Genotyping of both sets of controls was conducted using Illumina Human 1.2M-Duo Custon_v1 Array chips. SNP calling was performed using Illuminus Software. Full details of genotyping, SNP calling and QC have been previously reported (www.wtccc.org.uk). Concordant with previous findings comparison of the two control series showed little evidence for systematic bias (inflation factor λ=1.019; Supplementary Figure 1)30.
German-GWAS: The German study was based on 384 MM patients (ICD-10 C90.0; 229 male; mean age at diagnosis 54.5 years, SD 8.0) ascertained through Heidelberg University Centre. For controls, we used publicly accessible genotype data on 704 healthy individuals, with no past history of malignancy enrolled into the Heinz Nixdorf Recall (HNR) study8.
Replication
For replication we genotyped 169 additional cases of MM (93 male) collected through the UK Medical Research Council (MRC) Myeloma-VII trial. For controls we made use of previously generated data from a UK GWAS of colorectal cancer (420 males, 507 females, aged 18-69 years)12.
Ethics
Collection of blood samples and clinico-pathological information from subjects was undertaken with informed consent and relevant ethical review board approval in accordance with the tenets of the Declaration of Helsinki.
Karotyping
Conventional cytogenetic studies of MM cells were conducted using standard karotyping methodologies and standard criteria to define a clone were applied. Where possible meta-phase FISH was used to confirm iFISH abnormalities were present in the same cells as the abnormalities detected by conventional cytogenetics. iFISH and ploidy classification of UK samples was conducted using the methodology described by Chiecchio et al32. iFISH and ploidy classification of German samples was performed as previously described33,34.
Genotyping
DNA was extracted from EDTA-venous blood samples using Qiagen (Crawley, UK) Flexigene or QIAamp methodologies and quantified using PicoGreen (Invitrogen, Carlsbad, USA). Genotyping of cases in the GWAS was conducted using Illumina OmniExpress BeadChips according to the manufacturer's protocols (Illumina, San Diego, USA). To ensure quality of genotyping, duplicates were included on each sample plate (showing a concordance >99.99%). DNA samples with GenCall scores <0.25 at any locus were considered “no calls”. In each sample series a SNP was deemed to have failed if <95% of DNA samples generated a genotype at the locus. Cluster plots were manually inspected for all SNPs considered for replication.
Evaluating and editing cluster positions
Intensity data from arrays were imported into Illumina's BeadStudio clustering and calling software application. For the small subset of loci that were not clustered properly by the automated algorithm, the data were reviewed to identify loci that needed to be removed, manually edited or left unchanged. Clustered SNPs were evaluated using the metrics listed in the SNP Table of the BeadStudio software. These metrics are based on all samples for each locus and thus provide overall performance information for each locus. To identify loci potentially needing to be edited or removed, each quality metric column in the SNP table was sequentially sorted. Metrics used for identifying poorly or incorrectly clustered data included intensity, cluster separation, position of each cluster (AA, AB, BB), Hardy-Weinberg equilibrium, call frequency and variation of cluster width. The reproducibility of control samples on each plate, as well as replicates were also used to identify missclustered loci. Although not all cluster plots were assessed, ~10% of the lowest-performing loci were examined. Of these 10%, ~20% were edited or annotated (to indicate loci with nearby polymorphisms or hemizygous deletions) and ~2% were excluded. Review of data was conducted by a second individual to determine if any metrics were missed or if further editing was required. Overall, this process provides for substantially increased genotyping accuracy.
Validation of Illumina SNP genotypes
To confirm genotyping accuracy for rs4487645, rs1052501, rs6746082 SNPs we confirmed genotypes by ABI 3730xl Sanger sequencing in >184 randomly selected samples from each of the UK case, 58C and HNR control series (concordance >99.5%); PCR primers available on request.
Replication genotyping
Replication of rs4487645, rs1052501, rs6746082 associations were performed by ABI 3730xl Sanger sequencing of all MRC Myeloma-VII trial samples.
Statistical and bioinformatic analysis
We applied pre-determined quality control metrics to the GWAS data. We restricted analyses to samples for whom >95% of SNPs were successfully genotyped, thus eliminating 28 cases. We computed identity-by-state (IBS) probabilities for all pairs (cases and controls) to search for duplicates and closely related individuals amongst samples (defined as IBS ≥0.80, thereby excluding first-degree relatives). For all identical pairs the sample having the highest call rate was retained, eliminating 17 MM cases. To identify individuals who might have non-Western European ancestry, we merged our case and control data with phase II HapMap samples (60 western European [CEU], 60 Nigerian [YRI], 90 Japanese [JPT] and 90 Han Chinese [CHB]). For each pair of individuals we calculated genome-wide IBS distances on markers shared between HapMap and our SNP panel, and used these as dissimilarity measures upon which to perform principal component analysis. The first two principal components for each individual were plotted and any individual not present in the main CEU cluster was excluded from analyses. We removed 35 cases with non-CEU ancestry and one 58C control which had previously been identified as being diagnosed with Hodgkin Lymphoma (HL)30. In each sample series we filtered out SNPs having a minor allele frequency [MAF] <1%, and a call rate <95%. We also excluded SNPs showing departure from Hardy-Weinberg equilibrium (HWE) at P<10-6 in controls. For replication and validation analysis call rates were >95% per 384-well plate for each SNP; cluster plots were visually examined by two researchers.
Main analyses were undertaken using R (v2.6), Stata v.10 (State College, Texas, US) and PLINK (v1.06)35 software. The association between each SNP and risk of MM was assessed by the Cochran-Armitage trend test. The adequacy of the case-control matching and possibility of differential genotyping of cases and controls were formally evaluated using quantile-quantile (Q-Q) plots of test statistics. The inflation factor λ was based on the 90% least significant SNPs9. We undertook adjustment for possible population substructure using Eigenstrat software. Odds ratios (ORs) and associated 95% confidence intervals (CIs) were calculated by unconditional logistic regression. Meta-analysis was conducted using standard methods11. Cochran’s Q statistic to test for heterogeneity11 and the I2 statistic to quantify the proportion of the total variation due to heterogeneity were calculated36.I2 values ≥75% are considered characteristic of large heterogeneity36,37. To conduct a pooled analysis incorporating Eigenstrat adjusted P-values from the GWAS we used the weighted Z-method implemented in the program METAL38. Associations by sex, age and clinic-pathological phenotypes were examined by logistic regression in case-only analyses.
The familial relative risk of MM attributable to any locus is given by the formula39:
where p is the population frequency of the minor allele, q=1-p, and r1 and r2 are the relative risks (approximated by the odds ratios) for heterozygotes and the rarer homozygotes, relative to the more common homozygotes. From λ* it is possible to quantify the impact the locus makes to the overall familial risk of MM seen in first-degree relatives. Assuming a multiplicative interaction between risk alleles the proportion of the overall familial risk attributable to the locus is given by log(λ*)/log(λ0), where λ0, the overall familial risk of MM is assumed on the basis of epidemiological studies to be 2.455.
The population attributable fraction was estimated from 1 - Πi1 - xi, where xi = p.(ORpa-1)/(p.(ORpa-1)+1), p is the frequency of the risk allele in the population and ORpa is the per allele ORs.
Prediction of the untyped SNPs was carried out using IMPUTEv2, based on HapMap Phase III haplotypes release 2 (HapMap Data Release 27/phase III Feb 2009 on NCBI B36 assembly, dbSNP26) and 1000genomes. Imputed data were analysed using SNPTEST v2 to account for uncertainties in SNP prediction. LD metrics between HapMap SNPs were based on Data Release 27/phase III (Feb 2009) on NCBI B36 assembly, dbSNP26, viewed using Haploview software (v4.2) and plotted using SNAP. LD blocks were defined on the basis of HapMap recombination rate (cM/Mb) as defined using the Oxford recombination hotspots40 and on the basis of distribution of confidence intervals defined by Gabriel et al41. To annotate potential regulatory sequences within disease loci we implemented in silico searches using Transfac Matrix Database v7.2920, and PReMod1021 software. We used the in silico algorithms SIFT and PolyPhen to predict the impact of amino acid substitutions.
Relationship between SNP genotype and mRNA expression
To examine for a relationship between SNP genotype and expression levels of CDCA7L, DNAH11, ULK4, TRAK1 and DTNB in MM, we made use of Affymetrix Human Genome U133 Plus 2.0 array data we previously generated on the plasma cells from 192 MM patients from the MRC Myeloma IX trial22. To examine for a relationship between SNP genotype and expression levels in lymphocytes we made use of publicly available expression data generated from analysis of 90 Caucasian derived Epstein-Barr virus–transformed lymphoblastoid cell lines using Sentrix Human-6 Expression BeadChips (Illumina, San Diego, USA)23,24. Online recovery of data was performed using WGAViewer Version 1.25 Software. Differences in the distribution of levels of mRNA expression between SNP genotypes were compared using a Wilcoxon-type test for trend42. Power of assays to establish a relationship between genotype and expression we made using STATA software assuming allele-based test of difference in normalized logRNA expression (imposing a Bonferroni corrected P-value of 0.005 to address multiple testing).
Supplementary Material
Note: Supplementary information is available on the Nature Genetics website
Acknowledgements
Myeloma UK provided principal funding for the study. Additional funding was provided by Cancer Research UK (C1298/A8362 supported by the Bobby Moore Fund) and Leukaemia Lymphoma Research Fund and the NHS via the Biological Research Centre of the National Institute for Health Research at the Royal Marsden Hospital NHS Trust. This study made use of genotyping data on the 1958 Birth Cohort. Genotyping data on controls was generated by the Wellcome Trust Sanger Institute. A full list of the investigators who contributed to the generation of the data are available from http://www.wtccc.org.uk. We are grateful to all investigators who contributed to the Colorectal Tumour Gene Identification (CORGI) consortium, from which controls in the replication were drawn. In Germany funding was provided to Dietmar-Hopp-Stiftung Walldorf, the University Hospital Heidelberg and EU Health-F4-2007-200767 (APO-SYS). The GWAS made use of genotyping data from the population based HNR study. The HNR study is supported by the Heinz Nixdorf Foundation (Germany). Additionally, the study is funded by the German Ministry of Education and Science and the German Research Council (DFG; Project SI 236/8-1, SI236/9-1, ER 155/6-1). Funding was provided to LE by the Medical Faculty of the University Hospital of Essen (IFORES). The genotyping of the Illumina HumanOmni-1 Quad BeadChips of the HNR subjects was financed by the German Centre for Neurodegenerative Disorders (DZNE), Bonn. We are extremely grateful to all investigators who contributed to the generation of this dataset. Finally we are grateful to all the patients and investigators at the individual centers for their participation. We also thank the staff of the CTRU University of Leeds and the NCRI haematology Clinical Studies Group.
Footnotes
Author Contributions
RSH designed the study. RSH and GM obtained financial support in the UK and KH and HG in Germany. RSH drafted the manuscript. DC performed principal statistical and bioinformatic analyses; YM and SED performed additional statistical and bioinformatic analyses; PB coordinated laboratory analyses; AL and BO performed genotyping. DCJ managed and prepared Myeloma IX and Myeloma XI Case Study DNA samples. HG, KN and NW coordinated and managed German DNA samples, KH and AF coordinated genotyping. BAW performed UK expression analyses. FMR performed UK and AJ German FISH analyses. GJM, FED, WAG, GHJ and JAC performed ascertainment and collection of Case Study samples. PH, TWN, MMN performed and coordinated genotyping of German cases and controls. SM ascertained and managed the HNR sample. IT acquired CRC control samples. All authors contributed to the final paper.
Competing Interests Statement
The authors declare no competing financial interests.
URLs
The R suite can be found at http://www.r-project.org/
Detailed information on the tag SNP panel can be found at http://www.illumina.com/
dbSNP: http://www.ncbi.nlm.nih.gov/projects/SNP/
HapMap: http://www.hapmap.org/
1000Genomes: http://www.1000genomes.org/
SNAP http://www.broadinstitute.org/mpg/snap/
IMPUTE: https://mathgen.stats.ox.ac.uk/impute/impute.html
SNPTEST: http://www.stats.ox.ac.uk/~marchini/software/gwas/snptest.html
PReMod: http://genomequebec.mcgill.ca/PReMod/welcome.do
Transfac Matrix Database: http://www.biobase-international.com/pages/index.php?id=transfac
JASPAR2 database: http://jaspar.cgb.ki.se/
EIGENSTRAT: http://genepath.med.harvard.edu/~reich/Software.htm
Wellcome Trust Case Control Consortium: www.wtccc.org.uk
METAL: www.sph.umich.edu/csg/abecasis/metal
Mendelian Inheritance In Man: http://www.ncbi.nlm.nih.gov/omim
Medical Research Council (MRC) Myeloma-IX trial: http://public.ukcrn.org.uk
Medical Research Council (MRC) Myeloma-VII trial: http://ctru.leeds.ac.uk/myelomaVII
WGAViewer: http://www.genome.duke.edu/centers/pg2/downloads/wgaviewer.php
SIFT: http://sift.jcvi.org/
PolyPhen: http://genetics.bwh.harvard.edu/pph/
References
- 1.Kyle RA, Rajkumar SV. Multiple myeloma. N Engl J Med. 2004;351:1860–73. doi: 10.1056/NEJMra041875. [DOI] [PubMed] [Google Scholar]
- 2.Palumbo A, Anderson K. Multiple myeloma. N Engl J Med. 2011;364:1046–60. doi: 10.1056/NEJMra1011442. [DOI] [PubMed] [Google Scholar]
- 3.Kyle RA, et al. A long-term study of prognosis in monoclonal gammopathy of undetermined significance. N Engl J Med. 2002;346:564–9. doi: 10.1056/NEJMoa01133202. [DOI] [PubMed] [Google Scholar]
- 4.Landgren O, et al. Risk of plasma cell and lymphoproliferative disorders among 14621 first-degree relatives of 4458 patients with monoclonal gammopathy of undetermined significance in Sweden. Blood. 2009;114:791–5. doi: 10.1182/blood-2008-12-191676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altieri A, Chen B, Bermejo JL, Castro F, Hemminki K. Familial risks and temporal incidence trends of multiple myeloma. Eur J Cancer. 2006;42:1661–70. doi: 10.1016/j.ejca.2005.11.033. [DOI] [PubMed] [Google Scholar]
- 6.Morgan GJ, et al. First-line treatment with zoledronic acid as compared with clodronic acid in multiple myeloma (MRC Myeloma IX): a randomised controlled trial. Lancet. 2010;376:1989–99. doi: 10.1016/S0140-6736(10)62051-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Power C, Elliott J. Cohort profile: 1958 British birth cohort (National Child Development Study) Int J Epidemiol. 2006;35:34–41. doi: 10.1093/ije/dyi183. [DOI] [PubMed] [Google Scholar]
- 8.Schmermund A, et al. Assessment of clinically silent atherosclerotic disease and established and novel risk factors for predicting myocardial infarction and cardiac death in healthy middle-aged subjects: rationale and design of the Heinz Nixdorf RECALL Study. Risk Factors, Evaluation of Coronary Calcium and Lifestyle. Am Heart J. 2002;144:212–8. doi: 10.1067/mhj.2002.123579. [DOI] [PubMed] [Google Scholar]
- 9.Clayton DG, et al. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nat Genet. 2005;37:1243–6. doi: 10.1038/ng1653. [DOI] [PubMed] [Google Scholar]
- 10.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 11.Pettiti D. Meta-analysis decision analysis and cost-effectivness analysis. Oxford University Press; 1994. [Google Scholar]
- 12.Tomlinson IP, et al. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet. 2008;40:623–30. doi: 10.1038/ng.111. [DOI] [PubMed] [Google Scholar]
- 13.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–95. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang C, Jung JU. Autophagy genes as tumor suppressors. Curr Opin Cell Biol. 2010;22:226–33. doi: 10.1016/j.ceb.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ocio EM, Mateos MV, Maiso P, Pandiella A, San-Miguel JF. New drugs in multiple myeloma: mechanisms of action and phase I/II clinical findings. Lancet Oncol. 2008;9:1157–65. doi: 10.1016/S1470-2045(08)70304-8. [DOI] [PubMed] [Google Scholar]
- 16.Taddesse-Heath L, Meloni-Ehrig A, Scheerle J, Kelly JC, Jaffe ES. Plasmablastic lymphoma with MYC translocation: evidence for a common pathway in the generation of plasmablastic features. Mod Pathol. 2010;23:991–9. doi: 10.1038/modpathol.2010.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dib A, Gabrea A, Glebov OK, Bergsagel PL, Kuehl WM. Characterization of MYC translocations in multiple myeloma cell lines. J Natl Cancer Inst Monogr. 2008:25–31. doi: 10.1093/jncimonographs/lgn011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang A, et al. Identification of a novel c-Myc protein interactor, JPO2, with transforming activity in medulloblastoma cells. Cancer Res. 2005;65:5607–19. doi: 10.1158/0008-5472.CAN-05-0500. [DOI] [PubMed] [Google Scholar]
- 19.Maertens GN, Cherepanov P, Engelman A. Transcriptional co-activator p75 binds and tethers the Myc-interacting protein JPO2 to chromatin. J Cell Sci. 2006;119:2563–71. doi: 10.1242/jcs.02995. [DOI] [PubMed] [Google Scholar]
- 20.Matys V, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–10. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferretti V, et al. PReMod: a database of genome-wide mammalian cis-regulatory module predictions. Nucleic Acids Res. 2007;35:D122–6. doi: 10.1093/nar/gkl879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walker BA, et al. Integration of global SNP-based mapping and expression arrays reveals key regions, mechanisms, and genes important in the pathogenesis of multiple myeloma. Blood. 2006;108:1733–43. doi: 10.1182/blood-2006-02-005496. [DOI] [PubMed] [Google Scholar]
- 23.Stranger BE, et al. Genome-wide associations of gene expression variation in humans. PLoS Genet. 2005;1:e78. doi: 10.1371/journal.pgen.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stranger BE, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–53. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fonseca R, et al. International Myeloma Working Group molecular classification of multiple myeloma: spotlight review. Leukemia. 2009;23:2210–21. doi: 10.1038/leu.2009.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dewald GW, Kyle RA, Hicks GA, Greipp PR. The clinical significance of cytogenetic studies in 100 patients with multiple myeloma, plasma cell leukemia, or amyloidosis. Blood. 1985;66:380–90. [PubMed] [Google Scholar]
- 27.Debes-Marun CS, et al. Chromosome abnormalities clustering and its implications for pathogenesis and prognosis in myeloma. Leukemia. 2003;17:427–36. doi: 10.1038/sj.leu.2402797. [DOI] [PubMed] [Google Scholar]
- 28.Walker BA, et al. A compendium of myeloma-associated chromosomal copy number abnormalities and their prognostic value. Blood. 2010;116:e56–65. doi: 10.1182/blood-2010-04-279596. [DOI] [PubMed] [Google Scholar]
- 29.Landgren O, Weiss BM. Patterns of monoclonal gammopathy of undetermined significance and multiple myeloma in various ethnic/racial groups: support for genetic factors in pathogenesis. Leukemia. 2009;23:1691–7. doi: 10.1038/leu.2009.134. [DOI] [PubMed] [Google Scholar]
- 30.Enciso-Mora V, et al. A genome-wide association study of Hodgkin's lymphoma identifies new susceptibility loci at 2p16.1 (REL), 8q24.21 and 10p14 (GATA3) Nat Genet. 2010;42:1126–30. doi: 10.1038/ng.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crowther-Swanepoel D, et al. Common variants at 2q37.3, 8q24.21, 15q21.3 and 16q24.1 influence chronic lymphocytic leukemia risk. Nat Genet. 42:132–6. doi: 10.1038/ng.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiecchio L, et al. Deletion of chromosome 13 detected by conventional cytogenetics is a critical prognostic factor in myeloma. Leukemia. 2006;20:1610–7. doi: 10.1038/sj.leu.2404304. [DOI] [PubMed] [Google Scholar]
- 33.Neben K, et al. Combining information regarding chromosomal aberrations t(4;14) and del(17p13) with the International Staging System classification allows stratification of myeloma patients undergoing autologous stem cell transplantation. Haematologica. 2010;95:1150–7. doi: 10.3324/haematol.2009.016436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wuilleme S, et al. Ploidy, as detected by fluorescence in situ hybridization, defines different subgroups in multiple myeloma. Leukemia. 2005;19:275–8. doi: 10.1038/sj.leu.2403586. [DOI] [PubMed] [Google Scholar]
- 35.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–58. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 37.Ioannidis JP, Ntzani EE, Trikalinos TA. 'Racial' differences in genetic effects for complex diseases. Nat Genet. 2004;36:1312–8. doi: 10.1038/ng1474. [DOI] [PubMed] [Google Scholar]
- 38.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Houlston RS, Ford D. Genetics of coeliac disease. QJM. 1996;89:737–43. doi: 10.1093/qjmed/89.10.737. [DOI] [PubMed] [Google Scholar]
- 40.Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–4. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- 41.Gabriel SB, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–9. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 42.Cuzick J. A Wilcoxon-type test for trend. Stat Med. 1985;4:87–90. doi: 10.1002/sim.4780040112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.