Abstract
Background
Variation at TP63 has recently been shown to be associated with lung adenocarcinoma in the Asian population.
Methods
To investigate how this finding translates to the European population we compared the genotypes of SNPs annotating the TP63 locus at 3q28 in 4,462 lung cancer patients, including 911 with adenocarcinoma, and 8,235 controls from the United Kingdom (UK).
Results
A statistically significant association between adenocarcinoma risk and SNP genotype was shown: rs10937405, odds ratio (OR) =1.21, P=1.82x10-4; rs17429138, OR=1.23, P=7.49x10-5; and rs4396880, OR=1.21, P=2.03x10-4. Haplotype analysis was consistent with a single TP63 risk locus defined by SNPs rs10937405, rs17429138 and rs4396880. While no association between SNPs and small cell lung cancer was shown, the rs10937405 and rs439680 associations were significant for squamous cancer (respective P-values, 0.0022 and 0.02).
Conclusions
These findings show TP63 variation is a risk factor for the development of lung adenocarcinoma in the UK population. Furthermore, they provide additional insight into the subtype-specificity of the 3q28 lung cancer association.
Impact
Our data confirm the association of 3q28 with lung adenocarcinoma and that this association is not confined to the Asian population. Elucidating the functional basis of this association will be contingent on future fine mapping of the TP63 loci.
Keywords: 3q28, risk, cancer, lung polymorphism, association study
Introduction
Primary lung cancer is a major cause of cancer death worldwide causing over 1 million deaths each year(1). The various histological forms of lung cancer are typically divided into small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC) comprising adenocarcinoma and squamous tumours. Each of the lung cancer types has different clinico-pathological characteristics reflective of differences in carcinogenesis(2).
While lung cancer is largely caused by tobacco smoking, previous studies have implicated inherited genetic factors in disease etiology. Notably, genome-wide association (GWA) studies of lung cancer have robustly demonstrated that polymorphic variation at 5p15.33 (TERT-CLPTM1), 6p21.33 (BAT3-MSH5), and 15q25.1 (CHRNA5-CHRNA3-CHRNA4) influences lung cancer risk in European populations(3–7). Given the biological differences between the different types of lung cancer, searches for histology-specific associations have been conducted. Analysis of European GWA datasets has shown that the single nucleotide polymorphism (SNP) rs2736100 (TERT) is principally associated with adenocarcinoma risk(8–9). A recent GWA study has replicated the rs2736100 adenocarcinoma risk association in Japanese and Korean populations and additionally also showed an association between rs10937405, annotating TP63 at 3q28 and adenocarcinoma risk(10).
Understanding the effects of these risk variants in different populations is important in terms of inferring disease causality as well as for the translation of these results to risk prediction in different populations. The risk variants may confer different magnitudes of increased risk in different populations for a variety of reasons, including differences in allele frequency and linkage disequilibrium (LD) structure, and difference in genetic and environmental backgrounds that interact with the variants.
To provide further insights into the relationship between 3q28 variation and adenocarcinoma of the lung we have analyzed a large series of cases and controls from the UK.
Materials and Methods
Study participants
This analysis is based on data previously generated from a two-stage GWA study of lung cancer (6, 8). Briefly, Phase 1 comprised 1,978 cases with pathologically confirmed lung cancer ascertained through the Genetic Lung Cancer Predisposition Study (GELCAPS)(11). 472 of the 1,978 cases (24%) had a diagnosis of adenocarcinoma. 5,199 Individuals from the 1958 Birth cohort and National Blood Service served as source of Phase 1 controls (12, 13). Phase 2 consisted of an additional 2,484 lung cancer cases ascertained through GELCAPS(11). 439 of the 2,484 cases (18%) had a diagnosis of adenocarcinoma. Control blood samples were obtained from 3,036 healthy individuals recruited to the National Cancer Research Network genetic epidemiologic studies, the National Study of Colorectal Cancer (1999–2006; n=541)(14), GELCAPS (1999–2004; n=1,520), and the Royal Marsden Hospital Trust/Institute of Cancer Research Family History and DNA Registry (1999–2004; n=975). All of the cases and controls were British residents and had self-reported European Ancestry. Table 1 provides details of the cases and controls. In Phase 1, demographic information for the public accessible controls is not available. In phase 2, cases tended to be older than controls and higher proportion were male. Furthermore, the proportion of cases which were smokers was higher than in controls and cigarette consumption was greater (Table 1). Collection of blood samples and clinico-pathologiocal information from patients and controls was undertaken with informed consent and ethical review board approval in accordance with the tenets of the Declaration of Helsinki.
Table 1. Details of the lung cancer patients and controls studied.
| Phase 1 | Phase 2¥ | ||||
|---|---|---|---|---|---|
| Cases | Control subjects | Cases | Control subjects | ||
| Number (Male; %) | 1,978 (1,203, 60%) | 5,999 (-,-) | 2,484 (1,690, 68%) | 3,036 (1,497, 49%) | |
| Mean age (SD) years | 57 (6) | - | 72 (7) | 61 (11) | |
| Family history of lung cancer* | 287 (15%) | - | 348 (14%) | - | |
| Lung cancer histology | - | ||||
| NSCLC | 1,441 (73%) | - | 1,938 (78%) | - | |
| Adenocarcinoma | 472 (24%) | - | 439 (18%) | - | |
| Squamous | 620 (31%) | - | 1,072 (43%) | - | |
| Other | 349 (18%) | - | 427 (17%) | - | |
| SCLC | 535 (27%) | - | 512 (21%) | - | |
| Other | 2 (< 1%) | - | 34 (1%) | - | |
| Smoking Status | |||||
| Former smokers | 1,188 (60%) | - | 1,726 (69%) | 585 (19%) | |
| Ever Smokers | 680 (34%) | - | 549 (22%) | 341 (11%) | |
| Never smokers | 110 (6%) | - | 132 (5%) | 553 (18%) | |
| Unknown | 0 (0%) | - | 77 (3%) | 1,557 (51%) | |
| Mean CPD (SD) | 23 (12) | - | 22 (13) | 18 (11) | |
defined as having at least one first-degree relative affected with lung cancer
Chi-square test is used in the test of gender difference between cases and controls; and student-t test is used for test of age and mean CPD differences; P-values are all <0.05.
SD, standard deviation
NSCLC, non-small cell lung cancer
SCLC, small cell lung cancer
CPD, cigarettes per day
SNP selection and genotyping
DNA was extracted from samples using conventional methodologies and quantified using PicoGreen (Invitrogen, Carlsbad, USA). Genotyping of Phase 1 and Phase 2 was conducted using Illumina Human550 BeadChips and Illumina Infinium custom arrays, respectively, according to the manufacturer's protocols as previously described(6, 8). Our selection of SNPs for analysis was largely dictated by previously published data. The study reported by Miki et al reported an association between rs10937405 and adenocarcinoma risk in the Asian population (10). In addition they provided evidence for a weak association between rs4396880 and lung cancer risk in the Central European population using data previously generated by IARC researchers (10).
For Phase 1 in addition to analysing these two SNPs we derived the genotypes for 35 SNPs which map to a 169 Kb region of LD encompassing rs10937405 (190,865,877bps) at 3q28 (190,707,812bps-190,876,439bps; Supplementary Table 1). For Phase 2 analysis we derived rs4396880 (190,838,915bps) genotypes from Illumina Phase 2 data but genotyped rs10937405 and rs17429138 (190,728,287bps) directly using allele-specific PCR (KBiosciences). In all assays a DNA sample was deemed to have failed if it generated genotypes at <95% of loci. A SNP was deemed to have failed if fewer than 95% of DNA samples generated a genotype at the locus. To monitor QC genotyping, a series of duplicate samples were genotyped in the same batches.
Statistical and bioinformatic analysis
In all analyses a two-sided P-value of 0.05 of less was considered statistically significant. Statistical analyses were undertaken in R (v2.8) software. Deviation of the genotype frequencies in the controls from those expected under Hardy-Weinberg Equilibrium (HWE) was assessed by χ2 test. Odds ratios (OR) and associated 95% confidence intervals (CI) were calculated by unconditional logistic regression. Because of the unavailability of demographic information on the Phase 1 controls, adjustment of ORs for age and gender was only undertaken for Phase 2 data. To investigate the relationship between genotype with age, sex and family history we conducted a case-only analysis using both Phase 1 and Phase 2 case data. To examine the impact of genotype on smoking quantity, we tested the equality of medium cigarette consumption of the three genotype strata using Kruskal-Wallis test. The contributing population attributable risk (PAR) from TP63 variants was derived the formulae: where Pi is the prevalence in controls of the lung cancer risk allele at the ith locus, and is the ORi is the OR of the risk allele at the ith locus.
Haplotype analysis was performed in PLINK (v.1.07) software (15) whereby a standard E-M algorithm is used to compare the distribution of probabilistically-inferred set of haplotypes for each individual. LD metrics between HapMap SNPs were based on HapMapIII Release27, viewed using Haploview (v4.2) (16) and plotted using SNAP. LD blocks were defined on the basis of HapMap recombination rate (cM/Mb) as defined using Oxford recombination hotspots(17) and on the basis of distribution of confidence intervals previously defined(18). Prediction of the untyped SNPs was carried out using IMPUTEv2, based on HapMapIII Release27 (Feb2009, NCBI B36, dbSNP26) and the 1000 Genomes Project. Imputed data were analyzed using SNPTESTv2 to account for uncertainties in SNP prediction, using a threshold for maximum posterior probability of calling of ≥95%.
Results
While this study was primarily a study of the relationship between TP63 variation and risk of adenocarcinoma, we also investigated the relationship between genotype and other lung cancer subtypes. Genotypes were obtained for >95% of cases and controls for all SNPs irrespective of genotyping platform; hence there was no evidence of any systematic bias in genotyping. There was complete concordance between duplicate samples. The SNP allele frequencies in each of the control series in our study were similar to previously published data on the Northern European population. Furthermore, there was no evidence of population stratification as the genotype distribution in controls for each SNP satisfied HWE (i.e. P>0.05; Supplementary Table 1).
Confining our analysis to the relationship between 3q28 variation and adenocarcinoma risk, in Phase 1, 17 of the 37 SNPs provided evidence for an association at P<0.05 (Figure 1). The strongest association was provided by rs17429138 (per allele P=2.51x10-3; Table 2). Evidence for an association was also provided by rs10937405 (per allele P=9.24x10-3) and rs4396880 (per allele P=1.00x10-2; Table 2).
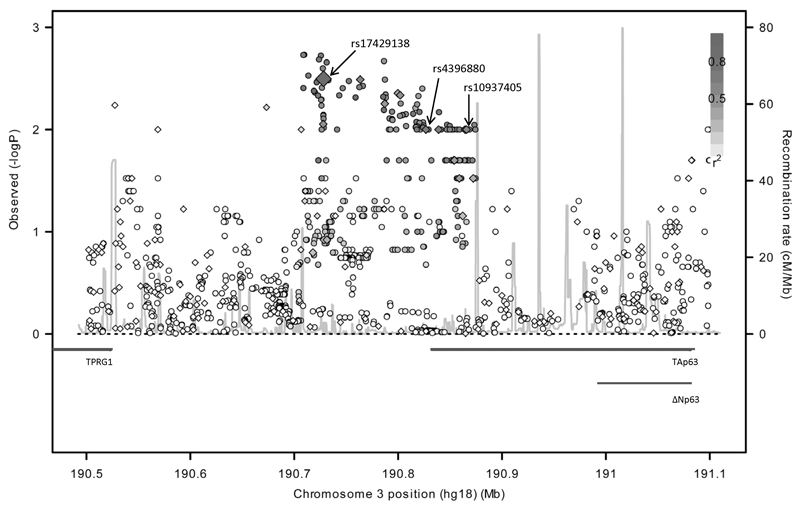
Figure 1. Case-control association plot for lung adenocarcinoma, LD map and regional plot of the genomic structure of the TP63 region in chromosome 3q28.
−log10 P values (y-axis) of the SNPs are shown according to their chromosomal position (x-axis). The genotyped and imputed SNPs are labelled by diamonds and circles respectively. The colour intensity of each symbol reflects the extent of LD with rs17429138: black (r2>0.8) through to white (r2<0.2). Genetic recombination rates (cM/Mb), estimated using HapMap CEU samples, are shown with a light grey line. Physical positions are based on NCBI build 36 of the human genome. Also shown are the relative position of transcripts mapping to the region.
Table 2. Lung cancer risk associated with TP63 variants rs17429138, rs4396880 and rs10937405.
| Phase 1 |
Phase 2 |
Combined |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases (%) | Controls (%) | OR (95% CI) | P-value | Cases (%) | Controls (%) | OR (95% CI) | P-value | OR (95% CI) | P-value | P-value§ | ||
| TP63 | ||||||||||||
| Adenocarcinoma | ||||||||||||
| rs17429138 | ||||||||||||
| AA† | 213 (45.9) | 2025 (39) | 1 (Ref) | 179 (41.6) | 1106 (37.3) | 1 (Ref) | ||||||
| AG | 197 (42.5) | 2400 (46.2) | 0.78 (0.64-0.96) | 0.02 | 206 (47.9) | 1411 (47.5) | 0.9 (0.73-1.12) | 0.35 | 0.84 (0.72-0.97) | 0.02 | 0.34 | |
| GG | 54 (11.6) | 772 (14.9) | 0.67 (0.49-0.91) | 9.88×10-03 | 45 (10.5) | 451 (15.2) | 0.62 (0.44-0.87) | 5.95×10-03 | 0.64 (0.51-0.81) | 1.71×10-04 | 0.75 | |
| HWE | 0.41 | 0.16 | 0.21 | 0.98 | ||||||||
| RAF, per allele OR | 0.67 | 0.62 | 1.24 (1.08-1.43) | 2.51×10-03 | 0.66 | 0.61 | 1.22 (1.05-1.42) | 0.01 | 1.23 (1.11-1.37) | 7.49×10-05 | 0.84 | |
| rs10937405 | ||||||||||||
| GG† | 178 (38.4) | 1699 (32.7) | 1 (Ref) | 153 (35.3) | 924 (31.1) | 1 (Ref) | ||||||
| AG | 214 (46.1) | 2527 (48.6) | 0.81 (0.66-1) | 0.05 | 219 (50.5) | 1452 (48.9) | 0.91 (0.73-1.14) | 0.41 | 0.85 (0.73-0.99) | 0.04 | 0.44 | |
| AA | 72 (15.5) | 973 (18.7) | 0.71 (0.53-0.94) | 0.02 | 62 (14.3) | 591 (19.9) | 0.63 (0.46-0.87) | 4.21×10-03 | 0.67 (0.54-0.83) | 2.19×10-04 | 0.61 | |
| HWE | 0.57 | 0.54 | 0.25 | 0.63 | ||||||||
| RAF, per allele OR | 0.61 | 0.57 | 1.20 (1.05-1.38) | 9.24×10-03 | 0.6 | 0.56 | 1.22 (1.06-1.41) | 7.03×10-03 | 1.21 (1.10-1.34) | 1.82×10-04 | 0.86 | |
| rs4396880 | ||||||||||||
| GG† | 201 (43.2) | 1939 (37.3) | 1 (Ref) | 177 (40.4) | 1080 (35.9) | 1 (Ref) | ||||||
| AG | 203 (43.7) | 2437 (46.9) | 0.8 (0.66-0.99) | 0.04 | 211 (48.2) | 1427 (47.5) | 0.9 (0.73-1.12) | 0.35 | 0.85 (0.73-0.98) | 0.03 | 0.44 | |
| AA | 61 (13.1) | 822 (15.8) | 0.72 (0.53-0.96) | 0.03 | 50 (11.4) | 498 (16.6) | 0.61 (0.44-0.85) | 3.75×10-03 | 0.67 (0.54-0.83) | 3.52×10-04 | 0.49 | |
| HWE | 0.39 | 0.22 | 0.27 | 0.47 | ||||||||
| RAF, per allele OR | 0.65 | 0.61 | 1.20 (1.04-1.38) | 0.01 | 0.64 | 0.6 | 1.23 (1.06-1.42) | 6.79×10-03 | 1.21 (1.09-1.34) | 2.03×10-04 | 0.83 | |
| Squamous | ||||||||||||
| rs17429138 | ||||||||||||
| AA+ | 247 (40.5) | 2025 (39) | 1 (Ref) | 412 (38.9) | 1106 (37.3) | 1 (Ref) | ||||||
| AG | 281 (46.1) | 2400 (46.2) | 0.96 (0.8-1.15) | 0.66 | 502 (47.4) | 1411 (47.5) | 0.96 (0.82-1.11) | 0.55 | 0.96 (0.85-1.08) | 0.46 | 0.97 | |
| GG | 82 (13.4) | 772 (14.9) | 0.87 (0.67-1.13) | 0.3 | 144 (13.6) | 451 (15.2) | 0.86 (0.69-1.07) | 0.17 | 0.86 (0.73-1.02) | 0.09 | 0.93 | |
| HWE | 0.88 | 0.16 | 0.65 | 0.98 | ||||||||
| RAF, per allele OR | 0.64 | 0.62 | 1.06 (0.94-1.2) | 0.32 | 0.63 | 0.61 | 1.07 (0.97-1.19) | 0.19 | 1.07 (0.99-1.16) | 0.1 | 0.93 | |
| rs10937405 | ||||||||||||
| GG† | 216 (35.4) | 1699 (32.7) | 1 (Ref) | 369 (34.8) | 924 (31.1) | 1 (Ref) | ||||||
| AG | 290 (47.5) | 2527 (48.6) | 0.9(0.75-1.09) | 0.28 | 514 (48.5) | 1452 (48.9) | 0.89 (0.76-1.04) | 0.13 | 0.89 (0.79-1.01) | 0.07 | 0.88 | |
| AA | 104 (17) | 973 (18.7) | 0.84 (0.66-1.08) | 0.17 | 176 (16.6) | 591 (19.9) | 0.75 (0.61-0.92) | 5.49×10-03 | 0.78 (0.67-0.92) | 2.59×10-03 | 0.47 | |
| HWE | 0.69 | 0.54 | 0.89 | 0.63 | ||||||||
| RAF, per allele OR | 0.59 | 0.57 | 1.09 (0.97-1.23) | 0.14 | 0.59 | 0.56 | 1.15 (1.04-1.27) | 5.46×10-03 | 1.13 (1.04-1.22) | 2.15×10-03 | 0.51 | |
| rs4396880 | ||||||||||||
| GG+ | 238 (39.1) | 1939 (37.3) | 1(Ref) | 412 (38.6) | 1080 (35.9) | 1 (Ref) | ||||||
| AG | 282 (46.3) | 2437 (46.9) | 0.94 (0.79-1.13) | 0.53 | 507 (47.5) | 1427 (47.5) | 0.93 (0.8-1.08) | 0.36 | 0.94 (0.83-1.05) | 0.27 | 0.92 | |
| AA | 89 (14.6) | 822 (15.8) | 0.88 (0.68-1.14) | 0.34 | 148 (13.9) | 498 (16.6) | 0.78 (0.63-0.97) | 0.02 | 0.82 (0.7-0.97) | 0.02 | 0.47 | |
| HWE | 0.71 | 0.22 | 0.69 | 0.47 | ||||||||
| RAF, per allele OR | 0.62 | 0.61 | 1.06 (0.94-1.2) | 0.32 | 0.62 | 0.6 | 1.12 (1.01-1.24) | 0.03 | 1.10 (1.01-1.18) | 0.02 | 0.53 | |
| SCLC | ||||||||||||
| rs17429138 | ||||||||||||
| AA+ | 206 (38.9) | 2025 (39) | 1 (Ref) | 182 (36.3) | 1106 (37.3) | 1 (Ref) | ||||||
| AG | 256 (48.3) | 2400 (46.2) | 1.05 (0.86-1.27) | 0.63 | 246 (49) | 1411 (47.5) | 1.06 (0.86-1.3) | 0.58 | 1.05 (0.91-1.21) | 0.47 | 0.94 | |
| GG | 68 (12.8) | 772 (14.9) | 0.87 (0.65-1.15) | 0.32 | 74 (14.7) | 451 (15.2) | 1 (0.74-1.33) | 0.98 | 0.93 (0.76-1.14) | 0.47 | 0.5 | |
| HWE | 0.4 | 0.16 | 0.54 | 0.98 | ||||||||
| RAF, per allele OR | 0.63 | 0.62 | 1.04 (0.91-1.19) | 0.54 | 0.39 | 0.39 | 1.01 (0.88-1.16) | 0.87 | 1.03 (0.93-1.13) | 0.58 | 0.76 | |
| rs10937405 | ||||||||||||
| GG† | 170 (32.1) | 1699 (32.7) | 1 (Ref) | 161 (32.1) | 924 (31.1) | 1 (Ref) | ||||||
| AG | 265 (50) | 2527 (48.6) | 1.05 (0.86-1.28) | 0.65 | 250 (49.9) | 1452 (48.9) | 0.99 (0.8-1.22) | 0.91 | 1.02 (0.88-1.18) | 0.8 | 0.7 | |
| AA | 95 (17.9) | 973 (18.7) | 0.98 (0.75-1.27) | 0.86 | 90 (18) | 591 (19.9) | 0.87 (0.66-1.15) | 0.34 | 0.93 (0.77-1.12) | 0.43 | 0.57 | |
| HWE | 0.64 | 0.54 | 0.68 | 0.63 | ||||||||
| RAF, per allele OR | 0.57 | 0.57 | 1 (0.88-1.14) | 0.95 | 0.57 | 0.56 | 1.06 (0.93-1.21) | 0.39 | 1.03 (0.94-1.13) | 0.52 | 0.55 | |
| rs4396880 | ||||||||||||
| GG+ | 193 (36.5) | 1939 (37.3) | 1 (Ref) | 182 (36) | 1080 (35.9) | 1 (Ref) | ||||||
| AG | 261 (49.3) | 2437 (46.9) | 1.08 (0.89-1.31) | 0.46 | 243 (48.1) | 1427 (47.5) | 1.01 (0.82-1.24) | 0.92 | 1.04 (0.91-1.2) | 0.55 | 0.67 | |
| AA | 75 (14.2) | 822 (15.8) | 0.92 (0.69-1.21) | 0.54 | 80 (15.8) | 498 (16.6) | 0.95 (0.72-1.27) | 0.74 | 0.93 (0.77-1.14) | 0.5 | 0.85 | |
| HWE | 0.38 | 0.22 | 0.94 | 0.47 | ||||||||
| RAF, per allele OR | 0.61 | 0.61 | 1.02 (0.89-1.16) | 0.8 | 0.6 | 0.6 | 1.02 (0.89-1.16) | 0.8 | 1.02 (0.93-1.12) | 0.72 | 1 | |
| NSCLC | ||||||||||||
| rs17429138 | ||||||||||||
| AA+ | 605 (42.7) | 2025 (39) | 1 (Ref) | 754 (39.7) | 1106 (37.3) | 1 (Ref) | ||||||
| AG | 629 (44.4) | 2400 (46.2) | 0.88 (0.77-1) | 0.04 | 895 (47.1) | 1411 (47.5) | 0.93 (0.82-1.05) | 0.26 | 0.9 (0.83-0.99) | 0.03 | 0.52 | |
| GG | 183 (12.9) | 772 (14.9) | 0.79 (0.66-0.95) | 1.00×10-02 | 251 (13.2) | 451 (15.2) | 0.82 (0.68-0.98) | 0.03 | 0.81 (0.71-0.92) | 9.89×10-04 | 0.83 | |
| HWE | 0.33 | 0.16 | 0.57 | 0.98 | ||||||||
| RAF, per allele OR | 0.65 | 0.62 | 1.13 (1.03-1.23) | 6.22×10-03 | 0.63 | 0.61 | 1.1 (1.01-1.2) | 0.03 | 1.11 (1.05-1.18) | 5.07×10-04 | 0.67 | |
| rs10937405 | ||||||||||||
| GG† | 512 (36.1) | 1699 (32.7) | 1 (Ref) | 663 (34.7) | 924 (31.1) | 1 (Ref) | ||||||
| AG | 675 (47.6) | 2527 (48.6) | 0.89 (0.78-1.01) | 0.07 | 942 (49.3) | 1452 (48.9) | 0.9 (0.79-1.03) | 0.13 | 0.9 (0.82-0.98) | 0.02 | 0.83 | |
| AA | 231 (16.3) | 973 (18.7) | 0.79 (0.66-0.94) | 7.28×10-03 | 304 (15.9) | 591 (19.9) | 0.72 (0.6-0.85) | 1.31×10-04 | 0.75 (0.66-0.85) | 4.02×10-06 | 0.45 | |
| HWE | 0.73 | 0.54 | 0.32 | 0.63 | ||||||||
| RAF, per allele OR | 0.6 | 0.57 | 1.13 (1.04-1.23) | 5.43×10-03 | 0.59 | 0.56 | 1.17 (1.08-1.27) | 2.22×10-04 | 1.15 (1.08-1.22) | 4.59×10-06 | 0.55 | |
| rs4396880 | ||||||||||||
| GG+ | 583 (41.1) | 1939 (37.3) | 1 (Ref) | 750 (39) | 1080 (35.9) | 1 (Ref) | ||||||
| AG | 640 (45.2) | 2437 (46.9) | 0.87 (0.77-0.99) | 0.04 | 916 (47.6) | 1427 (47.5) | 0.92 (0.82-1.05) | 0.22 | 0.9 (0.82-0.98) | 0.02 | 0.53 | |
| AA | 194 (13.7) | 822 (15.8) | 0.78 (0.65-0.94) | 9.03×10-03 | 259 (13.5) | 498 (16.6) | 0.75 (0.63-0.89) | 1.34×10-03 | 0.77 (0.68-0.87) | 3.80×10-05 | 0.72 | |
| HWE | 0.39 | 0.22 | 0.43 | 0.47 | ||||||||
| RAF, per allele OR | 0.64 | 0.61 | 1.13 (1.04-1.23) | 4.22×10-03 | 0.63 | 0.6 | 1.14 (1.05-1.24) | 2.36×10-03 | 1.14 (1.07-1.21) | 2.98×10-05 | 0.94 | |
HWE: Hardy-Weinberg Equilibrium
RAF: risk allele frequency
risk allele
test of heterogeneity P-value
Each of these three SNPs provided support for a relationship between TP63 variation and adenocarcinoma risk in Phase 2 data (Table 2). Odds ratios were unaffected, adjusting for age and sex (Supplementary Table 2). Moreover, pooling data from the two case-control series provided statistically significant evidence for an association between rs10937405, (per allele P=1.82x10-4), rs17429138 (per allele P=7.49x10-5) and rs4396880 (per allele P=2.03x10-4) even with adjustment for multiple testing ascribable to evaluation of 37 SNPs. For all three SNPs the association with lung adenocarcinoma risk was dose-dependent, with the highest risks being conferred by homozygosity for risk genotype (Table 2).
Following these analyses we investigated the relationship between rs10937405, rs17429138 and rs4396880 genotype and the other lung cancer histologies (Table 2). None of the three SNPs provided evidence for an association between 3q28 variation and risk of SCLC (Table 2). In contrast a strong relationship with NSCLC was shown; respective combined per allele P-values were 4.49x10-6, 5.07x10-4 and 2.98x10-5 (Table 2). In addition to this association being driven by an association for adenocarcinoma support was also provided by an association with squamous cancer, notably with rs10937405 and rs4396880 for which respective per allele P-values in the combined analysis were 2.15x10-3 and 2.00x10-2 (Table 2).
To explore for age- and sex-specific differences we conducted a case-only analysis of rs10937405, rs17429138 and rs4396880, using age 65 to stratify age at diagnosis of adenocarcinoma. This analysis provided no evidence that the risk associated with TP63 genotype is modified by age or gender (Table 3). We also found no evidence to support a relationship between TP63 genotype and a family history of lung cancer (based on the definition of having at least one first-degree relative affected with lung cancer; Table 3). Using either all cases or controls we found no evidence that TP63 genotype defined by either rs10937405, rs17429138 or rs4396880 influences cigarette consumption (Table 4).
Table 3. Relationship between sex, age at diagnosis, family history of lung cancer and TP63 genotype in adenocarcinoma cases.
| TP63 | |||||||
|---|---|---|---|---|---|---|---|
| rs17429138 | |||||||
| n (%) | n (%) | OR (95% CI) | P-value | ||||
| Sex | |||||||
| Male | Female | ||||||
| AA† | 205 (42.2) | 187 (45.8) | 1 (Ref) | ||||
| AG | 230 (47.3) | 173 (42.4) | 1.21 (0.92-1.6) | 0.18 | |||
| GG | 51 (10.5) | 48 (11.8) | 0.97 (0.62-1.51) | 0.89 | |||
| RAF, per allele OR | 0.34 | 0.33 | 1.06 (0.87-1.29) | 0.59 | |||
| Age | |||||||
| ≤ 65 | > 65 | ||||||
| AA† | 224 (45.3) | 168 (42.1) | 1 (Ref) | ||||
| AG | 216 (43.6) | 187 (46.9) | 0.87 (0.65-1.15) | 0.31 | |||
| GG | 55 (11.1) | 44 (11) | 0.94 (0.6-1.46) | 0.78 | |||
| RAF, per allele OR | 0.67 | 0.66 | 1.07 (0.88-1.31) | 0.49 | |||
| Family history | |||||||
| Yes | No | ||||||
| AA† | 47 (47.5) | 345 (43.4) | 1 (Ref) | ||||
| AG | 39 (39.4) | 364 (45.8) | 0.79 (0.5-1.23) | 0.29 | |||
| GG | 13 (13.1) | 86 (10.8) | 1.11 (0.57-2.14) | 0.76 | |||
| RAF, per allele OR | 0.67 | 0.66 | 1.04 (0.76-1.43) | 0.8 | |||
| rs10937405 | |||||||
| Sex | |||||||
| Male | Female | ||||||
| GG† | 182 (37.1) | 149 (36.6) | 1 (Ref) | ||||
| AG | 239 (48.7) | 194 (47.7) | 1.01 (0.76-1.34) | 0.95 | |||
| AA | 70 (14.3) | 64 (15.7) | 0.9 (0.6-1.34) | 0.59 | |||
| RAF, per allele OR | 0.61 | 0.6 | 1.04 (0.86-1.26) | 0.68 | |||
| Age | |||||||
| ≤ 65 | > 65 | ||||||
| GG† | 190 (38.4) | 141 (35) | 1 (Ref) | ||||
| AG | 230 (46.5) | 203 (50.4) | 0.84 (0.63-1.12) | 0.24 | |||
| AA | 75 (15.2) | 59 (14.6) | 0.94 (0.63-1.41) | 0.78 | |||
| RAF, per allele OR | 0.62 | 0.6 | 1.06 (0.88-1.29) | 0.53 | |||
| Family history | |||||||
| Yes | No | ||||||
| GG† | 39 (39.8) | 292 (36.5) | 1 (Ref) | ||||
| AG | 39 (39.8) | 394 (49.2) | 0.74 (0.46-1.18) | 0.21 | |||
| AA | 20 (20.4) | 114 (14.2) | 1.31 (0.73-2.35) | 0.36 | |||
| RAF, per allele OR | 0.4 | 0.39 | 1.06 (0.78-1.44) | 0.7 | |||
| rs4396880 | |||||||
| Sex | |||||||
| Male | Female | ||||||
| GG† | 203 (41.2) | 175 (42.7) | 1 (Ref) | ||||
| AG | 231 (46.9) | 183 (44.6) | 1.09 (0.82-1.44) | 0.55 | |||
| AA | 59 (12) | 52 (12.7) | 0.98 (0.64-1.49) | 0.92 | |||
| RAF, per allele OR | 0.35 | 0.35 | 1.02 (0.84-1.24) | 0.86 | |||
| Age | |||||||
| ≤ 65 | > 65 | ||||||
| GG† | 215 (43.3) | 163 (40) | 1 (Ref) | ||||
| AG | 217 (43.8) | 197 (48.4) | 0.84 (0.63-1.11) | 0.21 | |||
| AA | 64 (12.9) | 47 (11.5) | 1.03 (0.67-1.58) | 0.88 | |||
| RAF, per allele OR | 0.65 | 0.64 | 1.04 (0.86-1.27) | 0.67 | |||
| Family history | |||||||
| Yes | No | ||||||
| GG† | 44 (44.4) | 334 (41.5) | 1 (Ref) | ||||
| AG | 38 (38.4) | 376 (46.8) | 0.77 (0.49-1.21) | 0.26 | |||
| AA | 17 (17.2) | 94 (11.7) | 1.37 (0.75-2.51) | 0.3 | |||
| RAF, per allele OR | 0.36 | 0.35 | 1.06 (0.78-1.44) | 0.72 | |||
RAF: risk allele frequency
risk allele
Table 4. Smoking intensity and dependence by TP63 genotype.
| Lung adenocarcinoma | Control subjects | |||||
|---|---|---|---|---|---|---|
| Genotype | n | Mean CPD | P-value* | n | Mean CPD | P-value* |
| rs17429138 | ||||||
| AA† | 358 | 19.4 | 347 | 17.7 | ||
| AG | 377 | 19.0 | 425 | 18.8 | ||
| GG | 94 | 19.5 | 121 | 18.6 | ||
| 0.99 | 0.62 | |||||
| rs10937405 | ||||||
| GG† | 301 | 19.9 | 288 | 17.5 | ||
| AG | 405 | 18.9 | 441 | 19.1 | ||
| AA | 127 | 18.4 | 165 | 17.6 | ||
| 0.55 | 0.07 | |||||
| rs4396880 | ||||||
| GG† | 345 | 19.7 | 333 | 18.3 | ||
| AG | 387 | 18.8 | 435 | 18.7 | ||
| AA | 106 | 18.8 | 139 | 17.2 | ||
| 0.44 | 0.13 | |||||
CPD: cigarette per day
From Kruskal-Wallis test
Risk genotype
Figure 1 shows the position of the SNPs rs10937405, rs17429138 and rs4396880 mapping to 3q28 and the relative positions of the two isoforms of TP63; the TA and N-terminal-truncated (ΔN) TP63. Also shown is the LD structure across the region. The SNPs rs10937405, rs17429138 and rs4396880 are highly correlated within the CEU population; rs10937405-rs17429138 (r2=0.60, D’=0.86), rs17429138-rs4396880 (r2=0.66, D’=0.83), rs10937405-rs4396880 (r2=0.82, D’=0.98), thus defining a single risk haplotype (Supplementary Table 3).
Using Phase 1 data we sought to establish whether we could identify SNPs better correlated with risk of adenocarcinoma at 3q28 (190.5-191.1 Mb, encompassing TP63; Figure 1) through imputation of untyped SNPs referencing HapMap. In total 1,497 additional HapMap SNPs mapping to the remainder of the interval were successfully imputed. Nine SNPs provided slightly superior evidence for an association with adenocarcinoma risk to that provided by rs17429138, all mapping 5’ to TP63 (rs190726018, rs34000992, rs16864458, rs35218873, rs1597774, rs2378502, rs6787097, rs9290894, rs6444380; Figure 1).
Discussion
Our findings provide evidence that polymorphic variation annotating TP63 plays a role in determining the risk of developing lung adenocarcinoma; thereby confirming the recent observation made by Miki et al (10) in an analysis of Japanese and Korean populations. In addition our analysis provides evidence that the association while not extending to SCLC appears to also influences other forms of NSCLC.
A major strength of our study is that these data have been systematically ascertained in a consistent fashion and by making use of GWA data bias from population stratification confounding has been avoided. Population stratification is a concern in all association studies as a source of bias, as the genotype frequencies for many polymorphic variants differ markedly between ethnic groups. We have sought to further minimize this form of bias by excluding subjects with self-reported non-European ethnicity and the use of GWA SNP data to identify non-CEU individuals. Moreover the frequency of SNP genotypes in controls were directly comparable to those seen in previously published data on the UK population. It is entirely conceivable that polymorphic variation, for example in TP63, may contribute to the differing rates of adenocarcinoma shown between ethnic groups. The risk of adenocarcinoma associated with rs10937405 reported by Miki et al (10) in Asians was higher than that seen in the UK (per allele ORs of 1.31 and 1.20 respectively), however the risk allele is more common (0.43 vs 0.33), suggesting the variant contributes to ~8% of the PAR for adenocarcinoma in both populations.
While it will be challenging to identify the precise mechanism by which 3q28 variation affects lung adenocarcinoma development, accumulation of DNA damage and lack of response to genotoxic stress is recognised to contribute to lung carcinogenesis. TP63 is a member of the tumour suppressor TP53 gene family, which is pivotal to cellular differentiation and responsiveness to cellular stress(19). Exposure of cells to DNA damage leads to induction of TP63 and both isoforms have the ability to transactivate TP53 target genes, hence impacting on cellular responsiveness to DNA damage(20–21). The TAp63 isoforms are transcribed using a promoter located upstream of exon 1 of the gene, whereas expression of the ΔNp63 isoforms are regulated by a promoter within intron 3 of TP63(22). rs10937405, rs17429138 and rs4396880 appear to define a single risk haplotype to which a functional variant maps. While it is probable that the association annotated by this haplotype reflects a single risk variant it does preclude the possibility that the haplotype may capture multiple functional risk alleles. Although elucidating a functional basis for the SNP associations will be contingent on fine mapping it is entirely plausible that they may impact either directly or through LD on TP63 expression, especially as our imputed data implies a functional association 5’ to the coding region of TP63.
In summary, our data confirm TP63 as a susceptibility gene for lung adenocarcinoma and that the association is not confined to the Asian population. Furthermore, our data provides evidence that the association may extend to other forms of NSCLC.
Supplementary Material
Acknowledgements
This work was supported by Cancer Research UK (C1298/A8780 and C1298/A8362- Bobby Moore Fund for Cancer Research UK) who provided principal funding for this study. Athena Matakidou was the recipient of a clinical research fellowship from the Allan J Lerner Fund. We are also grateful to National Cancer Research Network, Helen Rollason Heal Cancer Charity and Sanofi-Aventis. We acknowledge NHS funding for the Royal Marsden Biomedical Research Centre. We would like to thank all individuals that participated in this study and the clinicians who took part in the GELCAPS consortium. This study made use of genotyping data on the 1958 Birth Cohort, this data was generated and generously supplied to us by Panagiotis Deloukas of the Wellcome Trust Sanger Institute. A full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk.
Footnotes
Competing Interests Statement
The authors declare no competing financial interests.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Daigo Y, Nakamura Y. From cancer genomics to thoracic oncology: discovery of new biomarkers and therapeutic targets for lung and esophageal carcinoma. Gen Thorac Cardiovasc Surg. 2008;56:43–53. doi: 10.1007/s11748-007-0211-x. [DOI] [PubMed] [Google Scholar]
- 3.Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–7. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 4.McKay JD, Hung RJ, Gaborieau V, Boffetta P, Chabrier A, Byrnes G, et al. Lung cancer susceptibility locus at 5p15.33. Nat Genet. 2008;40:1404–6. doi: 10.1038/ng.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–22. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Broderick P, Webb E, Wu X, Vijayakrishnan J, Matakidou A, et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat Genet. 2008;40:1407–9. doi: 10.1038/ng.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–42. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broderick P, Wang Y, Vijayakrishnan J, Matakidou A, Spitz MR, Eisen T, et al. Deciphering the impact of common genetic variation on lung cancer risk: a genome-wide association study. Cancer Res. 2009;69:6633–41. doi: 10.1158/0008-5472.CAN-09-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landi MT, Chatterjee N, Yu K, Goldin LR, Goldstein AM, Rotunno M, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 2009;85:679–91. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miki D, Kubo M, Takahashi A, Yoon KA, Kim J, Lee GK, et al. Variation in TP63 is associated with lung adenocarcinoma susceptibility in Japanese and Korean populations. Nat Genet. 42:893–6. doi: 10.1038/ng.667. [DOI] [PubMed] [Google Scholar]
- 11.Eisen T, Matakidou A, Houlston R. Identification of low penetrance alleles for lung cancer: the GEnetic Lung CAncer Predisposition Study (GELCAPS) BMC Cancer. 2008;8:244. doi: 10.1186/1471-2407-8-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Power C, Elliott J. Cohort profile: 1958 British birth cohort (National Child Development Study) Int J Epidemiol. 2006;35:34–41. doi: 10.1093/ije/dyi183. [DOI] [PubMed] [Google Scholar]
- 14.Penegar S, Wood W, Lubbe S, Chandler I, Broderick P, Papaemmanuil E, et al. National study of colorectal cancer genetics. Br J Cancer. 2007;97:1305–9. doi: 10.1038/sj.bjc.6603997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 17.Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–4. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- 18.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–9. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 19.Flores ER. The roles of p63 in cancer. Cell Cycle. 2007;6:300–4. doi: 10.4161/cc.6.3.3793. [DOI] [PubMed] [Google Scholar]
- 20.Petitjean A, Ruptier C, Tribollet V, Hautefeuille A, Chardon F, Cavard C, et al. Properties of the six isoforms of p63: p53-like regulation in response to genotoxic stress and cross talk with DeltaNp73. Carcinogenesis. 2008;29:273–81. doi: 10.1093/carcin/bgm258. [DOI] [PubMed] [Google Scholar]
- 21.Katoh I, Aisaki KI, Kurata SI, Ikawa S, Ikawa Y. p51A (TAp63gamma), a p53 homolog, accumulates in response to DNA damage for cell regulation. Oncogene. 2000;19:3126–30. doi: 10.1038/sj.onc.1203644. [DOI] [PubMed] [Google Scholar]
- 22.Moll UM, Slade N. p63 and p73: roles in development and tumor formation. Mol Cancer Res. 2004;2:371–86. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.