Abstract
Aims
Our aim is to explore the associations between mitochondrial DNA (mtDNA) content and basal plasma glucose, plasma glucose after oral glucose administration and oxidative stress in a Chinese population with different levels of glucose tolerance. We also aimed to investigate the effect of mtDNA content on basal and oral glucose‐stimulated insulin secretion.
Methods
Five hundred and fifty‐six Chinese subjects underwent a 75‐g, 2‐h oral glucose tolerance test. Subjects with diabetes (n = 159), pre‐diabetes (n = 197) and normal glucose tolerance (n = 200) were screened. Blood lipid profile was assessed, and levels of the oxidative stress indicators superoxide dismutase, glutathione reductase (GR) and 8‐oxo‐2′‐deoxyguanosine (8‐oxo‐dG) were measured. Levels of HbA1c, plasma glucose, insulin and C‐peptide were also determined. Measurements were taken at 0, 30, 60 and 120 min after 75 g oral glucose tolerance test. Peripheral blood mtDNA content was assessed using a real‐time polymerase chain reaction assay. Insulin sensitivity was evaluated by homeostatic model assessment of insulin resistance and Matsuda index (ISIM). Basal insulin secretion index (HOMA‐β), early phase disposition index (DI30) and total phase disposition index (DI120) indicate insulin levels at different phases of insulin secretion.
Results
Peripheral blood mtDNA content was positively associated with DI30 and DI120 and was negatively associated with plasma glucose measured 30, 60 and 120 min after oral glucose administration. However, there was no correlation between mtDNA content and basal insulin secretion (HOMA‐β), serum lipid or oxidative stress indicators (8‐oxo‐dG, superoxide dismutase, GR). HbA1c was negatively associated with GR (r = −0.136, p = 0.001). Multiple linear regression analysis showed that reduced peripheral blood mtDNA content increased the risk of impaired glucose‐stimulated β cell function (DI30: β = 0.104, p = 0.019; DI120: β = 0.116, p = 0.009).
Conclusions
Decreased peripheral blood mtDNA content was more closely associated with glucose‐stimulated insulin secretion than with basal secretion. Reduction in glucose‐stimulated insulin secretion causes postprandial hyperglycaemia. The oxidative stress was probably largely influenced by hyperglycaemia; it was probably that the decreased mt DNA content led to hyperglycaemia, which caused elevated oxidative stress. © 2016 The Authors. Diabetes/Metabolism Research and Reviews Published by John Wiley & Sons Ltd.
Keywords: mitochondrial DNA content, oxidative stress, glucose tolerance, islet β cell function
Introduction
Mitochondria are unique organelles that are present in all eukaryotic cells. They contain multiple copies of their own genome and are responsible for cellular processes such as adenosine triphosphate (ATP) synthesis, β‐oxidation of fatty acids and apoptosis 1, 2, 3. Accumulating evidence suggests that alterations in mitochondrial DNA (mtDNA) are associated with type 2 diabetes mellitus (T2DM) 4, 5. The mtDNA content is reduced in the islets of diabetes‐prone Goto–Kakizaki rats and in mitochondrial transcriptional factor A (Tfam) – defective mice 6, 7. In addition, in patients with T2DM, mtDNA content is reduced in the skeletal muscle and peripheral blood 8. It has been shown that decreased mtDNA content precedes T2DM 9. However, the role of mtDNA content in the onset of T2DM is not fully understood.
The relationship of mtDNA content to hyperglycaemia and oxidative stress is complex. In islet β cells, glucose‐stimulated insulin release requires ATP. Under conditions of decreased mtDNA content, a variety of cell types, including β cells, have reduced bioenergetic functions and ATP synthesis, resulting in closing of K‐ATP channels and decreased insulin release 10, 11. In addition, the mitochondrial network is no longer maintained, and there is a concomitant increase in reactive oxygen species (ROS). The collapse of mitochondrial homeostasis has a profound effect on nuclear–mitochondrial crosstalk, causing increased insulin resistance, islet β cell apoptosis and hyperglycaemia 12, 13, 14, 15. The relationship between peripheral blood mtDNA content, hyperglycaemia and oxidative stress has been characterized primarily in vitro, and there have been only few large population‐based studies. While previous studies have shown that reduced mtDNA content is associated with fasting hyperglycaemia 9, 16, the levels of fasting plasma glucose (FPG) alone do not comprehensively reflect islet β cell function. The relationship between peripheral blood mtDNA content and glucose‐stimulated insulin secretion has not been explored.
In the present study, we evaluated peripheral blood mtDNA content and oxidative stress in a Chinese population with different glucose tolerance statuses, ranging from normoglycaemia to pre‐diabetes and diabetes. We explored the correlation between mtDNA content and basal plasma glucose, plasma glucose after oral glucose and oxidative stress, and also to investigate the influence of mtDNA content on basal and glucose‐stimulated insulin secretion.
Methods
Study population
All subjects were recruited from a type 2 diabetes project in a Beijing suburb in China between March 2014 and January 2015. Five hundred and ninety‐nine subjects underwent a 75 g oral glucose tolerance test (OGTT). The 75 g OGTT was conducted after an overnight fast (>10 h). Blood samples were collected at 0, 30, 60 and 120 min following the OGTT. The glucose tolerance status of each subject was classified based on the 1999 criteria of the World Health Organization. Normal glucose tolerance (NGT) was indicated by FPG <6.1 mmol/L and 2‐h postprandial glucose (2‐h PG) <7.8 mmol/L. Pre‐diabetes was indicated by impaired fasting glucose measured as 6.1 mmol/L ≤ FPG < 7.0 mmol/L and 2‐h PG < 7.8 mmol/L, by impaired glucose tolerance (IGT) measured as FPG < 6.1 mmol/L and 7.8 ≤ 2 h PG < 11.1 mmol/L, or by both impaired fasting glucose and IGT. Diabetes was indicated by FPG ≥ 7.0 mmol/L or 2 h PG ≥ 11.1 mmol/L.
Subjects were excluded if they tested positive for type 1 diabetes mellitus‐related antibodies, were taking hypoglycaemic or steroid drugs or were taking drugs interfering with lipid metabolism. Subjects who had cardiovascular diseases, cerebrovascular diseases or nephropathy were also excluded. According to these criteria, subjects with NGT (n = 200), pre‐diabetes (n = 197) and diabetes (n = 159) were selected for this study. The study protocol was approved by the Ethics Committee of Peking Union Medical College Hospital. The subjects voluntarily signed informed consent forms.
Clinical measurement
A standardized medical history and accurate physical examination were undertaken in all of the subjects before a 75‐g OGTT was administered. Measurements of waist circumference (midway between the iliac crest and the costal margin) and hip circumference (HC) (at the level of the trochanters) were performed twice by the same observer, and the mean value was recorded. Weight and height were measured without shoes in light clothing, and body mass index (BMI) was calculated by dividing the body weight in kilogrammes by the square of the height in metres. Blood pressure measurements from subjects at rest were obtained twice with a standard mercury sphygmomanometer, and the mean value was calculated.
Biochemical measurements
Plasma glucose was measured in glucose oxidase assay. Cholesterol (TC), triglyceride (TG), high‐density lipoprotein‐cholesterol (HDL‐C) and low‐density lipoprotein‐cholesterol (LDL‐C) were determined using an automated analyser. Serum insulin and C‐peptide were measured by chemiluminescent enzyme immunoassay. HbA1c analysis was performed by high‐performance liquid chromatography (intra‐assay coefficient of variation (CV) <3%, inter‐assay CV <10%).
Assessment of insulin resistance
Homeostatic model assessment of insulin resistance (HOMA‐IR) and Matsuda insulin sensitivity index (ISIM) were calculated to evaluate the insulin resistance 17.
Assessment of β cell function
The homeostasis model assessment of insulin secretion (HOMA‐β) was calculated as basal insulin release 17. Early phase insulin release was calculated as the total insulin area under the curve divided by the total glucose area under the curve during the first 30 min of the OGTT (InsAUC30/GluAUC30), which was shown to have a strong correlation with first‐phase insulin secretion. Insulin secretion relative to insulin sensitivity (ISIM) was expressed as the disposition index (DI), calculated as follows: early phase DI30 = [InsAUC30/GluACU30] × ISIM, and total‐phase DI120 = [InsAUC120/GluACU120] × ISI. The values calculated from the formulas earlier are highly correlated with first‐phase insulin secretion in intravenous glucose tolerance tests 18.
Measurement of mtDNA content
Genomic DNA in leukocytes was extracted from peripheral blood samples using the QIAamp DNA blood mid kit (Qiagen, Hilden, Germany). Purified DNA samples were diluted and quantified using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA). The relative mtDNA copy numbers were measured by quantitative real‐time polymerase chain reaction and corrected by simultaneous measurement of nuclear DNA. The forward and reverse primers for the single‐copy nuclear gene human β‐actin were 5′‐TGGGTACAATGAGGAGTAGG‐3′ and 5′‐GGAGTAATCCAGGTCGGT‐3′. The forward and reverse primers for the mtDNA gene NADH dehydrogenase, subunit 1 (ND1) were 5′‐CCCTAAAACCCG CCACA TCT‐3′ and 5′‐GAGCG ATGGT GAGAGCTAAGGT‐3′.
The quantitative real‐time polymerase chain reaction procedures for ND1 and human β‐actin were performed in separate 96‐well plates with the same samples in the same well positions to avoid possible position effects. A negative control (water), a positive control (calibrator DNA) and a standard curve were included in each run. The calibrator DNA was a genomic DNA sample from a healthy control subject that was used to compare results from different independent assays. Each plate contained randomly selected samples, thus ensuring equal representation of all cases and controls. The lab personnel were blind to case or control status. As reference DNA, we used pooled DNA from 26 participants who had been randomly selected as controls in this study (200 ng of genomic DNA for each sample). The reference DNA sample was serially diluted twofold to generate a 5‐point standard curve with between 40 and 2.5 ng of DNA in each reaction. The r 2 correlation for each standard curve was ≥0.99, with the acceptable standard deviation for the Ct values set at 0.25. The ratio of the mtDNA copy number to human β‐actin copy number was determined for each sample using standard curves. This ratio was proportional to the mtDNA copy number in each sample. The ratio for each sample was then normalized to the calibrator DNA sample to standardize between different runs. Finally, in order to assess any intra‐assay variation, we assayed ten blood DNA samples from healthy control subjects at three times. We then repeated the assay with the same blood DNA samples on three different days to evaluate inter‐assay variation. Average coefficients of intra‐assay and inter‐assay variance were 4.2% (range = 1.6–9.8%) and 4.6% (range = 0.9–7.8%), respectively.
Measurement of superoxide dismutase, glutathione reductase and 8‐oxo‐dG activity
Serum was collected from fasting blood samples. The levels of superoxide dismutase (SOD), glutathione reductase (GR) and 8‐oxo‐2′‐deoxyguanosine (8‐oxo‐dG) were determined as per the manufacturer's instructions (Cloud‐Clone Corp., Houston, TX, USA). Absorbance kinetics was measured through an ELISA reader.
Statistical analysis
All statistical analyses were performed using spss software, version 17.0 (IBM Corp., Chicago, IL, USA). The data are presented as the mean ± standard deviation. Parameters not normally distributed were transformed. Categorical data were analysed using the χ2 test. The significance of the mean differences was tested by analysis of variance (followed by Bonferroni's post hoc pairwise comparisons). The mtDNA content was adjusted by age and sex. Pearson correlation was assessed between variables and risk factors. Stepwise multiple linear regression analysis was performed to exclude the influences of potential confounding variables between mtDNA and β cell function. All p‐values were two sided, and p < 0.05 was considered statistically significant.
Results
Clinical characteristics of subgroups divided by plasma glucose profiles
Subjects were divided into three groups according to their plasma glucose levels: NGT, pre‐diabetes or diabetes. The characteristics of the three groups are presented in Table 1. The pre‐diabetes and diabetes subjects were older and had larger BMIs, larger WCs, larger HCs and higher systolic blood pressure than the NGT subjects. In both pre‐diabetes and diabetes subjects, HbA1c, FPG and plasma glucose measured 30, 60 and 120 min after taking 75 g glucose were significantly higher than those in the NGT subjects. The diabetes subjects had higher fasting serum insulin levels but lower serum insulin and C‐peptide levels 30, 60 and 120 min after taking 75 g glucose than the pre‐diabetes and the NGT subjects.
Table 1.
Characteristics of different glucose tolerance statuses
| Parameter | NGT n = 200 | Pre‐diabetes n = 197 | Diabetes n = 159 | p value |
|---|---|---|---|---|
| Age, years | 48.73 ± 12.06 | 55.34 ± 10.24 | 57.80 ± 10.42 | 0.000** |
| Sex (male : female) | 61 : 141 | 77 : 127 | 72 : 128 | 0.000** |
| BMI, kg/m2 | 25.09 ± 3.44 | 26.66 ± 3.78 | 26.10 ± 3.80 | 0.000** |
| Waist circumference, cm | 84.69 ± 9.70 | 88.73 ± 9.29 | 88.36 ± 9.39 | 0.000** |
| Hip circumference, cm | 90.23 ± 9.88 | 93.77 ± 9.09 | 95.00 ± 11.22 | 0.000** |
| SBP, mm Hg | 123.38 ± 18.73 | 129.06 ± 16.16 | 131.55 ± 19.29 | 0.000** |
| DBP, mm Hg | 75.66 ± 9.82 | 76.73 ± 9.96 | 76.40 ± 10.29 | 0.553 |
| HbA1c (%) | 5.27 ± 0.29 | 5.71 ± 0.33 | 7.19 ± 1.55 | 0.000** |
| FPG, mmol/L | 5.45 ± 0.36 | 6.10 ± 0.48 | 8.89 ± 3.30 | 0.000** |
| PG 30′, mmol/L | 8.97 ± 1.91 | 10.71 ± 2.02 | 14.16 ± 4.44 | 0.000** |
| PG 60′,mmol/L | 7.60 ± 1.98 | 10.09 ± 2.78 | 16.11 ± 5.13 | 0.000** |
| PG 120′, mmol/L | 5.82 ± 1.21 | 7.61 ± 1.75 | 14.68 ± 5.68 | 0.000** |
| Ln (insulin 0′, mU/L) | 2.20 ± 0.49 | 2.31 ± 0.51 | 2.40 ± 0.74 | 0.003** |
| Ln (insulin 30′, mU/L) | 4.28 ± 0.68 | 4.20 ± 0.68 | 3.53 ± 0.84 | 0.000** |
| Ln (insulin 60′, mU/L) | 4.09 ± 0.66 | 4.28 ± 0.68 | 3.89 ± 0.89 | 0.000** |
| Ln (insulin 120′, mU/L) | 3.43 ± 0.72 | 3.87 ± 0.77 | 3.84 ± 0.90 | 0.000** |
| Ln (C‐peptide 0′, ng/mL) | 0.18 ± 0.36 | 0.36 ± 0.39 | 0.29 ± 0.53 | 0.000** |
| Ln (C‐peptide 30′, ng/mL) | 1.63 ± 0.42 | 1.57 ± 0.42 | 0.96 ± 0.63 | 0.000** |
| Ln (C‐peptide 60′, ng/mL) | 1.74 ± 0.44 | 1.83 ± 0.39 | 1.32 ± 0.69 | 0.000** |
| Ln (C‐peptide 120′, ng/mL) | 1.46 ± 0.43 | 1.76 ± 0.43 | 1.51 ± 0.65 | 0.000** |
| Ln HOMA‐IR | 1.18 ± 0.34 | 1.34 ± 0.38 | 1.68 ± 0.66 | 0.000** |
| Sqrt Matsuda index (ISIM) | 10.44 ± 2.53 | 9.11 ± 2.44 | 8.21 ± 2.91 | 0.000** |
| Sqrt HOMA‐β | 10.01 ± 2.54 | 9.18 ± 2.44 | 7.58 ± 3.43 | 0.000** |
| Sqrt DI30 | 24.94 ± 4.94 | 19.57 ± 4.63 | 12.11 ± 4.57 | 0.000** |
| Sqrt DI120 | 27.93 ± 3.87 | 23.63 ± 6.29 | 14.86 ± 6.15 | 0.000** |
| TC, mmol/L | 5.27 ± 1.04 | 5.63 ± 1.01 | 5.36 ± 1.16 | 0.002** |
| Log (TG), mmol/L | 0.31 ± 0.89 | 0.64 ± 0.81 | 0.70 ± 0.92 | 0.000** |
| Log (HDL‐C), mmol/L | 0.40 ± 0.03 | 0.34 ± 0.31 | 0.30 ± 0.34 | 0.016* |
| LDL‐C, mmol/L | 2.67 ± 0.75 | 2.96 ± 0.69 | 2.84 ± 0.74 | 0.000** |
| Log (TG/HDL‐C) | −0.08 ± 1.05 | 0.30 ± 0.97 | 0.40 ± 1.02 | 0.000** |
p < 0.05.
p < 0.01.
DI30 = [InsAUC30/GluACU30] × ISIM, DI120 = [InsAUC120/GluACU120] × ISIM. Insulin, C‐peptide, HOMA‐IR, TG, HDL‐C and TG/HDL‐C have been natural logarithm transformed, and ISIM, HOMA‐β, DI30 and DI120 have been square root transformed.
NGT, normal glucose tolerance; BMI, body mass index; SBP, systolic blood pressure; DBP, diastolic blood pressure; FPG, fasting plasma glucose; PG, postprandial plasma glucose; HOMA‐IR, homeostatic model assessment of insulin resistance; DI, disposition index; HDL‐C, high‐density lipoprotein‐cholesterol.
Homeostatic model assessment of insulin resistance was significantly higher, and ISIM was lower in both the pre‐diabetes and diabetes subjects than in the NGT subjects. In addition, HOMA‐β, DI30, DI120 and AIR/HOMA‐IR were significantly lower in both the pre‐diabetes and diabetes subjects than in the NGT subjects. The pre‐diabetes and diabetes subjects also had higher lipid profiles (TC, TG, LDL‐C and TG/HDL‐C) and lower HDL‐C than the NGT subjects.
Peripheral blood mtDNA content and oxidative stress of patients with different glucose tolerance statuses
Peripheral blood mtDNA content (adjusted by age and sex) was significantly lower in the diabetes subjects compared with the NGT subjects (Figure 1). 8‐oxo‐dG is a modified nucleoside base detected as a by‐product of DNA damage. 8‐oxo‐dG levels were higher in the diabetes subjects than in the NGT subjects, but there was no difference between the diabetes subjects and the pre‐diabetes subjects. Levels of the antioxidant enzymes SOD and GR were significantly lower in the diabetes subjects compared with the NGT subjects, but there was no difference between the diabetes subjects and the pre‐diabetes subjects (Figure 2).
Figure 1.
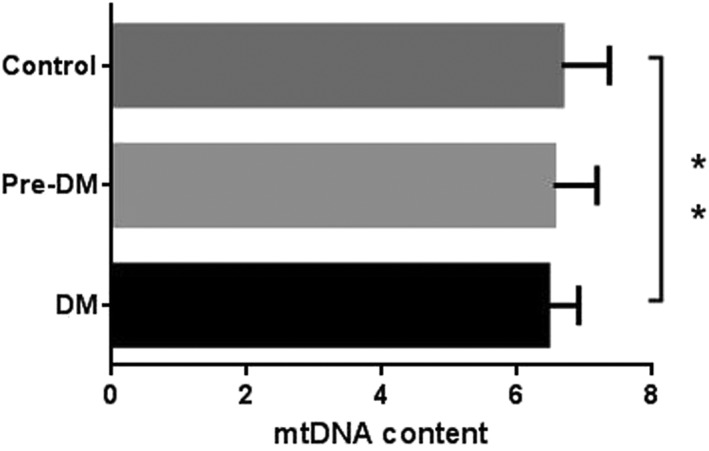
Peripheral blood mitochondrial DNA (mtDNA) content in groups with different glucose tolerance statuses (** p < 0.01). DM, diabetes mellitus.
Figure 2.
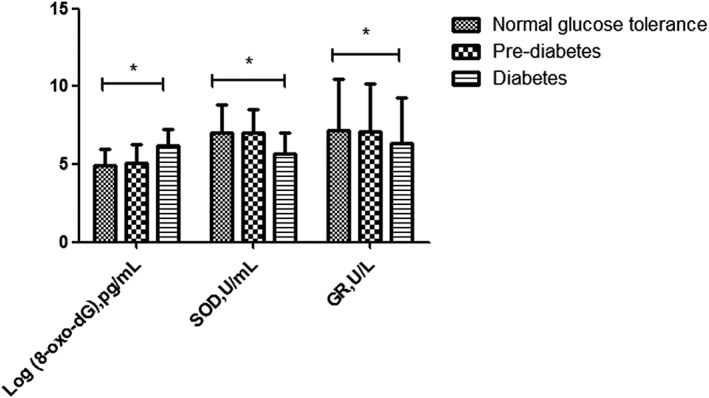
Serum oxidative stress indicators [8‐oxo‐2′‐deoxyguanosine (8‐oxo‐dG), superoxide dismutase (SOD) and glutathione reductase (GR)] of groups with different glucose tolerance statuses.
Association between peripheral blood mtDNA content and plasma glucose, insulin resistance, islet β cell function and oxidative stress
Peripheral blood mtDNA content was negatively associated with age. In addition, peripheral blood mtDNA content adjusted by age was not correlated with BMI, WC, HC, blood pressure, FPG, insulin, C‐peptide and insulin, or C‐peptide measured 30, 60 or 120 min after taking 75 g glucose. mtDNA content was significantly negatively associated with HbA1c and plasma glucose levels measured 30, 60 and 120 min after taking 75 g glucose. mtDNA content was significantly positively correlated with indices reflecting early phase insulin secretion (DI30) and total insulin secretion (DI120). In contrast, mtDNA content was not correlated with the basal insulin secretion index HOMA‐β, serum lipid or the oxidative stress indicators 8‐oxo‐dG, SOD and GR (Table 2). HbA1c was negatively associated with GR (r = –0.136, p = 0.001).
Table 2.
Correlation of peripheral blood mtDNA with metabolic risk factors
| Parameter | mtDNA | p value |
|---|---|---|
| r value | ||
| Age | −0.124 | 0.005** |
| BMI | −0.037 | 0.390 |
| WC | −0.066 | 0.128 |
| HC | −0.074 | 0.090 |
| Systolic blood pressure | −0.051 | 0.241 |
| Diastolic blood pressure | −0.045 | 0.303 |
| HbA1c% | −0.135 | 0.003** |
| FPG | −0.054 | 0.217 |
| PG 30′ | −0.114 | 0.009** |
| PG 60′ | −0.094 | 0.033* |
| PG 120′ | −0.109 | 0.013* |
| Ln (insulin 0′) | −0.013 | 0.606 |
| Ln (insulin 30′) | −0.025 | 0.562 |
| Ln (insulin 60′) | −0.470 | 0.285 |
| Ln (insulin 120′) | −0.014 | 0.751 |
| Ln (C‐peptide 0′) | −0.091 | 0.056 |
| Ln (C‐peptide 30′) | −0.043 | 0.332 |
| Ln (C‐peptide 60′) | −0.078 | 0.075 |
| Ln (C‐peptide 120′) | −0.037 | 0.399 |
| Ln HOMA‐IR | −0.052 | 0.235 |
| Sqrt ISIM | 0.083 | 0.062 |
| Sqrt HOMA‐β | 0.045 | 0.306 |
| Sqrt DI30 | 0.104 | 0.025* |
| Sqrt DI120 | 0.120 | 0.009** |
| TC | −0.032 | 0.480 |
| Log (TG) | −0.068 | 0.122 |
| Log (HDL‐C) | −0.020 | 0.647 |
| LDL‐C | −0.008 | 0.860 |
| Log (TG/HDL‐C) | −0.050 | 0.251 |
| Log (8‐oxo‐dG) | −0.012 | 0.781 |
| SOD | 0.053 | 0.232 |
| GR | 0.030 | 0.498 |
p < 0.05.
p < 0.01.
mtDNA, mitochondrial DNA; BMI, body mass index; WC, waist circumference; HC, hip circumference; FPG, fasting plasma glucose; PG, postprandial plasma glucose; HOMA‐IR, homeostatic model assessment of insulin resistance; TG, triglyceride; LDL‐C, low‐density lipoprotein‐cholesterol; ISIM, Matsuda insulin sensitivity index; DI, disposition index; TC, Cholesterol; HDL‐C, high‐density lipoprotein‐cholesterol; SOD, superoxide dismutase; GR, glutathione reductase.
Reduced peripheral blood mtDNA content increased the risk of impaired glucose‐stimulated β cell function
Multiple linear regression analysis was used to determine the association between peripheral blood mtDNA content and glucose‐stimulated β cell function. mtDNA content had a significant positive association with DI30 and DI120 even after adjustment by age, sex, BMI, WC, HC, HOMA‐IR, lipid profiles and oxidative stress (Table 3).
Table 3.
Multiple linear regression analysis using β cell function (DI30 and DI120) as the dependent variable
| β coefficient | SE | F‐value | p value | ||
|---|---|---|---|---|---|
| DI30 as dependent variable | Independent variable: mtDNA adjusted for age, sex, BMI, WC, HC, HOMA‐IR, lipid profiles and oxidative stress | 0.104 | 0.011 | 5.56 | 0.019* |
| DI120 as dependent variable | Independent variable: mtDNA adjusted for age, sex, BMI, WC, HC, HOMA‐IR, lipid profiles and oxidative stress | 0.116 | 0.013 | 6.923 | 0.009** |
p < 0.05.
p < 0.01.
SE, standard error; DI, disposition index; mtDNA, mitochondrial DNA; BMI, body mass index; WC, waist circumference; HC, hip circumference; HOMA‐IR, homeostatic model assessment of insulin resistance.
Discussion
Accumulating evidence suggests that mtDNA content is associated with T2DM 4, 5, 9, 10. Previous studies focused on the relationship between peripheral blood mtDNA content and FPG 5, 16. However, the results from these studies were inconsistent with one another because of subjects with the different and narrow ranges of FPG 5, 19. The range of FPG in the present study was 3.49 to 24.76 mmol/L, which was wider than ranges used in previous studies 5, 19; however, our results showed that peripheral blood mtDNA content was not associated with FPG and HOMA‐β. Study that comprehensively explored the effect of mtDNA content and glucose‐stimulated insulin secretion was still lacking. In this study, we evaluated β cell function by OGTT; the results indicated that peripheral blood mtDNA content was positively associated with β cell function at early phase insulin release and upon total‐phases insulin release and was negatively associated with plasma glucose at 30 and 60 min after oral glucose administration. Moreover, multiple linear regression analysis indicated that decreased peripheral blood mtDNA is associated with impaired insulin secretion at early phase insulin release and upon total‐phase insulin release. Glucose‐stimulated insulin release is ATP‐dependent. In β cells, increased ATP production in mitochondria induces Ca2+ influx, which stimulates insulin secretion 20. In addition, β cell mitochondria with decreased mtDNA content synthesize less ATP upon glucose stimulation. This results in K‐ATP channel closing and decreased postprandial insulin release, which leads to hyperglycaemia 10, 21. In the present study, we demonstrated a positive relationship between mtDNA content and glucose‐stimulated insulin secretion. This relationship was present among a population with a continuous and wide range of plasma glucose levels. We found that decreased peripheral blood mtDNA content was more closely associated with glucose‐stimulated insulin secretion than with basal insulin secretion, suggesting a role in causing postprandial hyperglycaemia.
In the present study, reduction in peripheral blood mtDNA content led to decreased β cell function and hyperglycaemia. In addition, levels of the DNA damage byproduct 8‐oxo‐dG increased, whereas levels of the antioxidants SOD and GR decreased. These changes were indicative of elevated oxidative stress and could in part reflect the effect of mitochondrial function on plasma glucose and oxidative stress. According to a previous study, loss of mtDNA leads to decreased bioenergetic functions, mitochondrial fission and increased ROS 12. In this study, peripheral blood mtDNA content was not associated with indicators of oxidative stress. However, oxidative stress was mildly elevated in pre‐diabetic subjects and was significantly elevated in diabetic subjects. Furthermore, in diabetic subjects, HbA1c was negatively associated with GR. According to previous studies, hyperglycaemia induces increased ROS production in mitochondria, causing an imbalance between excessive ROS and antioxidant defences 22. This study indicated that in populations with IGT, oxidative stress was probably largely influenced by hyperglycaemia rather than decreased mtDNA content. Our findings suggest a possible scenario in which decreased mtDNA content leads to hyperglycaemia, which causes elevated oxidative stress; increased oxidative stress further damages mtDNA, leading to a vicious cycle of damage.
There is a potential interaction between medication and mtDNA content. To limit this bias, we excluded subjects taking oral hypoglycaemic drugs, lipid‐lowering drugs and insulin in this study. We concluded that there was no association between mtDNA content and insulin resistance, lipid profiles adjusted for age and sex, onset of diabetes or disease duration.
This study has several limitations. The sample size was not large enough. Subjects who had cardiovascular disease were excluded from this study; however, cardiovascular disease is the major killer in T2DM, and it shows loss of mtDNA as well 23, excluding these patients may actually remove ‘worst‐case’ scenario patients. Although we investigated the existence of an association between peripheral blood mtDNA content and plasma glucose in this cross‐sectional study, it remains unclear whether mitochondrial dysfunction is primary or secondary to T2DM; this question may be better addressed by longitudinal studies.
In summary, the present study suggests that in a population with a wide range of plasma glucose levels, decreased peripheral blood mtDNA content was probably more closely associated with glucose‐stimulated insulin secretion than with basal secretion. Peripheral blood mtDNA content was positively associated with β cell function at early phase insulin release and upon total‐phases insulin release, which is associated with postprandial hyperglycaemia, and consequently induces increased oxidative stress. Our findings provide increased insight into the role of mitochondrial function in diabetes.
Conflicts of interest
The authors have no conflicts of interest.
Acknowledgements
We thank all of the participants who participated in the study.
This project was supported by the National Natural Science Foundation of China (grant no. 81270878) and the National Key Program of Clinical Science of China (WBYZ2011‐873).
This article has been thoroughly edited by a native English speaker from an editing company. Editing Certificate will be provided upon request.
Zhou, M. , Zhu, L. , Cui, X. , Feng, L. , Zhao, X. , He, S. , Ping, F. , Li, W. , and Li, Y. (2016) Reduced peripheral blood mtDNA content is associated with impaired glucose‐stimulated islet β cell function in a Chinese population with different degrees of glucose tolerance. Diabetes Metab Res Rev, 32: 768–774. doi: 10.1002/dmrr.2814.
References
- 1. DiMauro S, Schon EA. Mitochondrial respiratory‐chain diseases. New Engl J Med 2003; 348: 2656–2668. [DOI] [PubMed] [Google Scholar]
- 2. Legros F, Malka F, Frachon P, et al. Organization and dynamics of human mitochondrial DNA. J Cell Sci 2004; 117(Pt 13): 2653–2662. [DOI] [PubMed] [Google Scholar]
- 3. Rebelo AP, Dillon LM, Moraes CT, et al. Mitochondrial DNA transcription regulation and nucleoid organization. J Inherit Metab Dis 2011; 34: 941–951. [DOI] [PubMed] [Google Scholar]
- 4. Wong J, McLennan SV, Molyneaux L, et al. Mitochondrial DNA content in peripheral blood monocytes: relationship with age of diabetes onsetand diabetic complications. Diabetologia 2009; 52: 1953–1961. [DOI] [PubMed] [Google Scholar]
- 5. Weng SW, Lin TK, Liou CW, et al. Peripheral blood mitochondrial DNA content and dysregulation of glucose metabolism. Diabetes Res Clin Pract 2009; 83: 94–99. [DOI] [PubMed] [Google Scholar]
- 6. Silva JP, Köhler M, Graff C, et al. Impaired insulin secretion and beta‐cell loss in tissue‐specific knockout mice with mitochondrial diabetes. Nat Genet 2000; 26: 336–340. [DOI] [PubMed] [Google Scholar]
- 7. Serradas P, Giroix MH, Saulnier C, et al. Mitochondrial deoxyribonucleic acid content is specially decreased in adult, but not fetal, pancreatic islets of the Goto–Kakizaki rat, a genetic model of noninsulin‐dependent diabetes. Endocrinology 1995; 136: 5623–5631. [DOI] [PubMed] [Google Scholar]
- 8. Antonetti DA, Reynet C, Kahn CR. Increased expression of mitochondrial encoded genes in skeletal muscles of humans with diabetes mellitus. J Clin Invest 1995; 95: 1383–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee HK, Song JH, Shin CS, et al. Decreased mitochondrial DNA content in peripheral blood precedes the development of non‐insulin‐dependent diabetes mellitus. Diabetes Res Clin Pract 1998; 42: 161–167. [DOI] [PubMed] [Google Scholar]
- 10. Nile DL, Brown AE, Kumaheri MA, et al. Age‐related mitochondrial DNA depletion and the impact on pancreatic Beta cell function. PLoS One 2014; 9: e115433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Reiling E, Ling C, Uitterlinden AG, et al. The association of mitochondrial content with prevalent and incident type 2 diabetes. J Clin Endocrinol Metab 2010; 95: 1909–1915. [DOI] [PubMed] [Google Scholar]
- 12. Herrera A, Garcia I, Gaytan N, et al. Endangered species: mitochondrial DNA loss as a mechanism of human disease. Front Biosci (Schol Ed) 2015; 7: 109–124. [DOI] [PubMed] [Google Scholar]
- 13. Marella M, Seo BB, Matsuno‐Yagi A, Yagi T. Mechanism of cell death caused by complex I defects in a rat dopaminergic cell line. J Biol Chem 2007; 282: 24146–24156. [DOI] [PubMed] [Google Scholar]
- 14. Kroemer G, Zamzami N, Susin SA. Mitochondrial control of apoptosis. Immunol Today 1997; 18: 44–51. [DOI] [PubMed] [Google Scholar]
- 15. Wang J, Silva JP, Gustafsson CM, et al. Increased in vivo apoptosis in cells lacking mitochondrial DNA gene expression. Proc Natl Acad Sci U S A 2001; 98: 4038–4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xu FX, Zhou X, Shen F, et al. Decreased peripheral blood mitochondrial DNA content is related to HbA1c, fasting plasma glucose level and age of onset in type 2 diabetes mellitus. Diabet Med 2012; 29: e47–e54. [DOI] [PubMed] [Google Scholar]
- 17. Matthews DR, Hosker JP, Rudenski AS, et al. Homeostasis model assessment: insulin resistance and beta‐cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985; 28: 412–419. [DOI] [PubMed] [Google Scholar]
- 18. Stancakova A, Javorsky M, Kuulasmaa T, et al. Changes in insulin sensitivity and insulin release in relation to glycemia and glucose tolerance in 6,414 Finnish men. Diabetes 2009; 58: 1212–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Singh R, Hattersley AT, Harries LW. Reduced peripheral blood mitochondrial DNA content is not a risk factor for type 2 diabetes. Diabet Med 2007; 24: 784–787. [DOI] [PubMed] [Google Scholar]
- 20. Gembal M, Detimary P, Gilon P, et al. Mechanisms by which glucose can control insulin release independently from its action on adenosine triphosphate‐sensitive K+ channels in mouse B cells. J Clin Invest 1999; 91: 871–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kadowaki T, Kadowaki H, Mori Y, et al. A subtype of diabetes mellitus associated with a mutation of mitochondrial DNA. New Engl J Med 1994; 330: 962–968. [DOI] [PubMed] [Google Scholar]
- 22. Nishikawa T, Araki E. Impact of mitochondrial ROS production in the pathogenesis of diabetes mellitus and its complications. Antioxid Redox Signal 2007; 9: 343–353. [DOI] [PubMed] [Google Scholar]
- 23. Chen S, Xie X, Wang Y, et al. Association between leukocyte mitochondrial DNA content and risk of coronary heart disease: a case–control study. Atherosclerosis 2014; 237: 220–226. [DOI] [PubMed] [Google Scholar]


