Abstract
Quantifying the effects of cis-regulatory DNA on gene expression is a major challenge. Here, we present the multiplexed editing regulatory assay (MERA); a high-throughput CRISPR/Cas9-based approach that analyzes the functional impact of the regulatory genome in its native context. MERA tiles thousands of mutations across ~40 kb of cis-regulatory genomic space and uses knock-in green fluorescent protein (GFP) reporters to read out gene activity. Using this approach, we obtain quantitative information on the contribution of cis-regulatory regions to gene expression. We identify proximal and distal regulatory elements necessary for expression of four embryonic stem cell-specific genes. We show a consistent contribution of neighboring gene promoters to gene expression and identify unmarked regulatory elements (UREs) that control gene expression but do not have typical enhancer epigenetic or chromatin features. We compare thousands of functional and non-functional genotypes at a genomic location and identify the basepair-resolution functional motifs of regulatory elements.
Gene regulation provides the basis for cell type-specific function. Although differences in cis-regulatory DNA are known to underlie human variation and disease, predicting the effects of cis-regulatory variants on gene expression remains challenging.
Important strides have been made over the past decade to catalog gene regulatory elements. A histone modification code has been found to correlate with cis-regulatory elements, such as enhancers and promoters, and states, such as active and poised1–5. Gene expression reporter assays, which can now be done in high-throughput formats6–8, have confirmed elements that are sufficient to activate gene expression in heterologous contexts. Additionally, techniques to identify distal DNA interactions have begun to associate enhancers with their cognate promoters9–12, which are often not in close proximity and can at times be megabases apart.
However, existing gene regulatory techniques have several shortcomings. Reporter assays focus on elements that are sufficient to activate gene expression in a heterologous context and therefore cannot characterize elements that are necessary but not sufficient for gene expression or elements whose activity does not transfer to a non-native context. Additionally, genes can have many regulatory elements, and there is no high-throughput approach capable of determining the relative importance of each gene regulatory element on native gene expression levels. Efforts to systematically test enhancers, predicted using histone modification data from reporter assays, have found that the majority of predicted enhancers do not activate gene expression as expected13. This suggests that additional assays are required to decipher native gene regulation.
CRISPR/Cas9 has been used in genome-wide mutation screens to identify genes required for survival, drug resistance and tumor metastasis24–28 In these screens, guide RNAs (gRNAs) targeting tens of thousands of sites within genes are cloned into lentiviral vectors and delivered as a pool into target cells along with Cas9. By identifying gRNAs that are enriched or depleted in the cells after selection for the desired phenotype, genes that are required for this phenotype can be systematically identified.
Here we develop CRISPR/Cas9-based MERA to analyze the regulatory genome at single base resolution in its native context. MERA employs Cas9, which has been shown to cleave DNA when paired with a single-gRNA14–17. In MERA, Cas9-induced double-strand breaks (DSBs) are repaired in an error-prone fashion by cellular non-homologous end joining (NHEJ), inducing a wide range of mutations initiated at the cleavage site which are typically small (<10 bp) insertion/deletions (indels) but can include larger (>100 bp) indels15, 16, 23 and altered individual bases.
The MERA assay first carries out a high-throughput screen that maps the effects of genomic variation on gene expression. Selected elements can then be characterized by functional motif discovery and validated. We map elements that are required for gene expression by expressing gRNAs that tile a gene’s cis-regulatory region and measuring how likely each gRNA is to diminish gene expression. We then perform deep sequencing of the gRNA-induced mutations in targeted regions to reveal thousands of genotypes that either did or did not lose gene expression. This enables us to characterize the functional importance of each base. Finally, we validate the results of the MERA screen by the replacement of selected genomic elements by homologous recombination.
Results
Developing the MERA assay
There are two distinctions between MERA and previous gene mutation screening approaches that spurred us to alter the CRISPR/Cas9-based mutation screening technique. First, the targeted sites in our screen are often close together, so cells receiving more than one gRNA may delete a region instead of mutate that region, which would complicate downstream analysis. While this issue can be addressed for lentiviral libraries by lowering the multiplicity of infection (MOI), we sought a more elegant approach to limit cells to a single gRNA. Second, each gene for which we perform MERA requires a different gRNA library.
All high-throughput CRISPR/Cas9-based approaches to date have required cloning gRNA libraries into a lentiviral vector and producing a batch of virus, a time-consuming process that would have to be done separately for each library. We sought an approach that would allow a library to be used on the day it arrives.
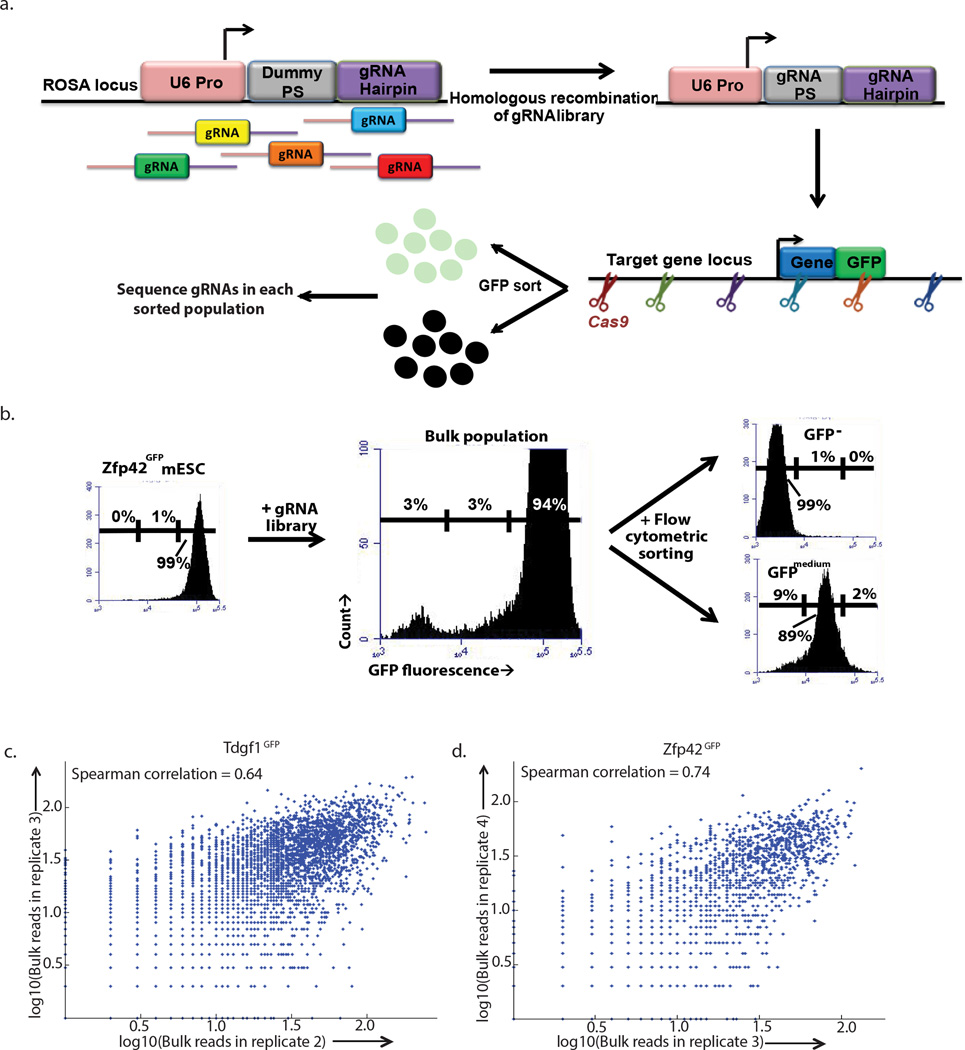
To enable the efficient targeting of precisely one regulatory element per cell, we devised a strategy that ensures that only one gRNA can be expressed per cell and allows gRNA libraries to be used without any molecular cloning into a delivery vector. We integrated a single copy of the gRNA expression construct (a U6 promoter driving expression of a dummy gRNA hairpin) into the universally accessible ROSA locus of mouse embryonic stem cells (mESCs) using CRISPR/Cas9-mediated homologous recombination (Fig. 1a). We then use CRISPR/Cas9-mediated homologous recombination to replace the dummy gRNA with a gRNA from our library. We use PCR to add 79–90 bp homology arms to our gRNA library, as we found that longer homology arms increase background cutting of gRNAs transcribed from unintegrated PCR fragments (Supplementary Fig. 1). We then introduce the pool of gRNA homology fragments into cells along with Cas9 and a gRNA plasmid that induces a DSB at the dummy gRNA site. In a substantial fraction of cells (~30%), the dummy gRNA is repaired by homologous recombination, creating a functional gRNA expression construct targeting a single genomic site from the library (Fig. 1a, Supplementary Fig. 2). It is random chance which gRNA is integrated in each cell, allowing a pooled screen in which each cell expresses only one gRNA.
Figure 1. Multiplexed editing regulatory assay (MERA).
(a) In MERA, a genomically integrated dummy gRNA is replaced with a pooled library of gRNAs through CRISPR/Cas9-based homologous recombination such that each cell receives a single gRNA. Guide RNAs are tiled across the cis-regulatory regions of a GFP-tagged gene locus, and cells are flow cytometrically sorted according to their GFP expression levels. Deep sequencing on each population is used to identify gRNAs preferentially associated with partial or complete loss of gene expression. (b) Zfp42GFP mESCs express uniformly strong GFP. After bulk gRNA integration, a subpopulation of cells lose partial or complete GFP expression. These cells are flow cytometrically isolated for deep sequencing. (c,d) Bulk reads for gRNAs are highly correlated between replicates of (c) Tdfg1 or (d) Zfp42, indicating consistent and replicable integration rates.
Of note, the genomic integration-based gRNA screening platform used in MERA could also be applied to other CRISPR-based high-throughput screens as long as the cell line undergoes homologous recombination at appreciable frequency, and it could be modified to achieve expression of any set number of gRNAs per cell for combinatorial screening. While the integration-based approach is thus ill-suited to in vivo screens or screens in cells with limited homologous recombination, it provides an alternative to lentiviral screening that substantially reduces the time, effort, and cost involved in CRISPR library screening for applicable cell lines such as ESCs.
We generated GFP knock-in lines for four mESC-specific genes, Nanog, Rpp25, Tdgf1, and Zfp4229, and synthesized corresponding gRNA libraries, each with 3908 gRNAs tiling cis-regulatory regions. In case of Tdgf1, the library targeted the 40 kb region proximal to the gene in an unbiased manner. In other cases, we selected proximal regions to the gene most likely to be involved in regulation based on enhancer-like features that are a maximum of ~150kb away from the gene as well as distal regions up to 92 mB away from the gene when ChIA-PET distal interaction data2 suggested a possible interaction with the target gene promoter3 (Supplementary Information). In Figures 2 and 3, the bulk density panel shows the distribution of integrated gRNAs along the region probed. Among the 3621 gRNAs found to be integrated in at least 1 replicate of Tdgf1, the mean distance between adjacent gRNAs was 11bp. Of note, repetitive and unmappable genomic regions cannot be tiled with gRNAs, and gRNAs targeting regions whose sequence differs from that of the reference genome cannot be appropriately tiled without genome sequence data of the cell line. Each library also contained 10 positive control gRNAs targeting the GFP open reading frame that we expected would cause GFP loss.
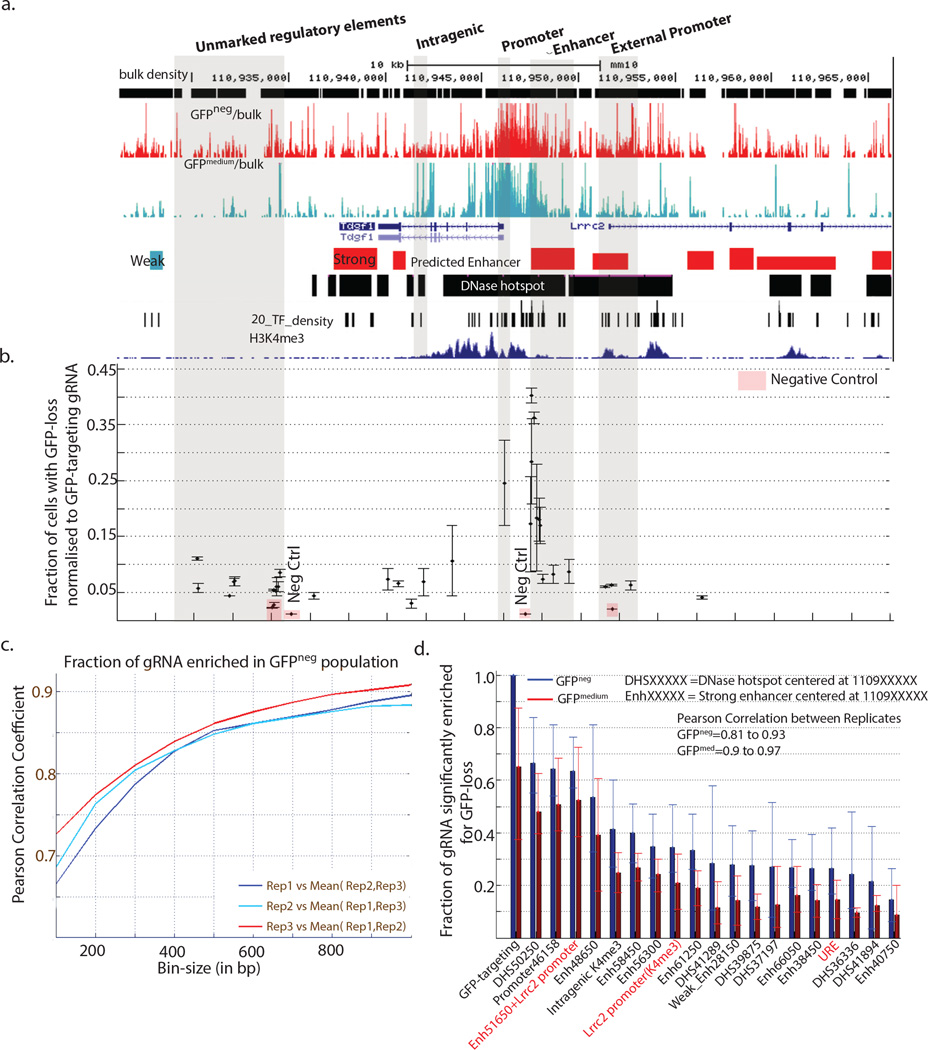
Figure 2. MERA enables systematic identification of required cis-regulatory elements for Tdgf1.
(a) A genomic view the Tdgf1 proximal regulatory region showing in track order (i) the location of gRNAs that did not result in GFP loss, (ii) enriched gRNAs in GFPneg cells (red), (iii) enriched gRNAs in GFPmedium cells (green), (iv) annotated genes, (v) predicted enhancers (green=weak, red=strong), (vi) DNase-I hotspot regions, (vii) transcription factor binding density based on ChIP-seq data, (vii) H3K4me3 ChIP-seq data. Several active regulatory elements coincide with dense clusters of overlapping gRNAs. A large number of gRNA significantly enriched in GFPneg population are also observed in regions devoid of regulatory element features (UREs). Genomic regions of interest are shaded, annotated above the plot, and described in further detail in the text. (b) Individual validation of specific gRNAs detected as enriched in the GFPneg population in the MERA assay using the self-cloning CRISPR system. The proportion of cells undergoing GFP loss upon incorporation of a particular gRNA divided by the proportion of cells undergoing GFP loss upon incorporation of GFP-targeting positive control gRNA are plotted against the actual genomic location of the gRNA. Negative controls or gRNA showing no reads in either GFPneg and GFPmedium populations are highlighted in red. (c) Correlation of gRNAs significantly enriched in the GFPneg population in fixed size bins varying from 100bp to 1kb for biological replicates in Tdgf1 (d) Fraction of GFPneg enriched gRNA among the different functional genomic categories surrounding the Tdgf1 gene.
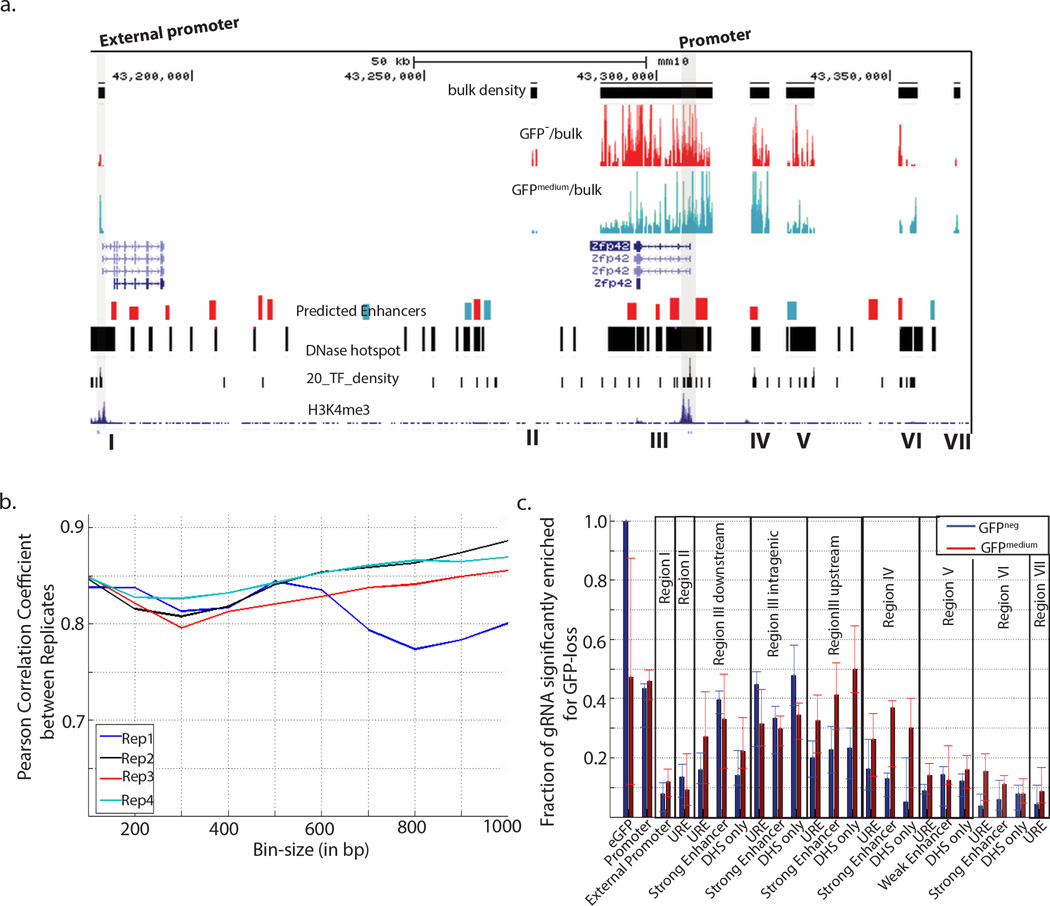
Figure 3. MERA enables systematic identification of required cis-regulatory elements for Zfp42.
(a) A genomic view the Zfp42 proximal regulatory region showing in track order (i) the location of gRNAs that did not result in GFP loss, (ii) enriched gRNAs in GFPneg cells (red), (iii) enriched gRNAs in GFPmedium cells (green), (iv) annotated genes, (v) predicted enhancers (green=weak, red=strong), (vi) DNase-I hotspot regions, (vii) transcription factor binding density based on ChIP-seq data, (vii) H3K4me3 ChIP-seq data. Several active regulatory elements coincide with dense clusters of overlapping gRNAs. Genomic regions of interest are shaded, annotated above the plot, and described in further detail in the text. (b) Correlation of gRNAs significantly enriched in the GFPneg population in fixed size bins varying from 100bp to 1kb for biological replicates in Tdgf1. c.) Fraction of GFPneg enriched gRNA among the different functional genomic categories surrounding the Tdgf1 gene.
MERA screens identify required regulatory regions
We performed four biological replicate screens for Zfp42 and Tdgf1, two replicates for Nanog and a single replicate for Rpp25. Selected screen hits were independently confirmed as described below. Starting one week after electroporation, we collected genomic DNA of the unsorted library-integrated cells to examine differences in gRNA integration. Over 90% of correctly synthesized gRNAs were detected in the genomic DNA for both Tdgf1 and Zfp42 libraries (Supplementary Methods). In addition, gRNA integration rates in biological replicates showed concordance (Fig. 1c,d; Supplementary Fig. 4a). All of the regulatory regions that we surveyed had adequate coverage of gRNAs to assay their detailed function (Bulk density track, Fig. 2,3; Supplementary Figure 5–6).
Library-integrated mESCs were then flow cytometrically sorted to identify gRNAs inducing loss of GFP expression. Separate GFPneg and GFPmedium populations were sorted in the Tdgf1GFP and Zfp42GFP experiments, whereas GFPneg and GFPmedium populations were combined in the NanogGFP and Rpp25GFP experiments because of incomplete population separation (Fig. 1b; Supplementary Fig. 3).
The distribution of gRNA abundance in GFPneg and GFPmedium populations in all screens clearly indicates that a subset of cis-regulatory genomic space is required for gene expression at all four gene loci (Fig. 2a,b, Fig. 3). We detected significant overrepresentation of nearly all integrated positive control GFP coding region targeting gRNAs in all replicates (Fig. 2d, Fig. 3c, Supplementary Fig. 4b) suggesting that MERA robustly identifies gRNAs inducing loss of gene expression. Using the relative abundances of GFP open reading frame (ORF)-targeting positive control gRNAs and the dummy gRNA as a negative control, we devised a method to detect gRNAs with statistically significant overrepresentation in GFPneg and GFPmedium populations (Supplementary Fig. 4b,c, Supplementary Methods).
In our MERA screen of Tdgf1 we observed differential enrichment of gRNAs in established functional categories of genomic elements associated with gene regulation33, 34,35,36,37 (Fig. 2a,d, Supplementary Fig. 5). The highest density of significant gRNAs in the genomic regions were observed at the promoter region for Tdgf1, the strong proximal enhancer 4kb upstream of Tdgf1, and the strong enhancer overlapping the Lrrc2 promoter (Fig. 2a,d).
Surprisingly we observed a novel class of genomic elements downstream of Tdgf1 (Figure 2a, highlighted in grey) which did not coincide with any known markers of regulatory activity such as H3K27ac, H3K4me1, H3K4me3, known transcription factor (TF) binding sites, DNase-I hypersensitivity, predicted DNase-I hotspots, or enhancers predicted from chromatin modifications. We designated such elements that do not contain these markers as unmarked regulatory elements (UREs). Unmarked regulatory regions were often over 1 kb in length and produced comparable loss of GFP as some distant enhancers (Fig. 2d).
In our MERA screen of Zfp42, we also observed the strongest enrichment for GFP loss in the promoter and proximal enhancer regions (Fig. 3a,c). We observed enrichment of gRNAs in the GFPneg and GFPmedium population at UREs in regions II, III, VI and VII (Fig. 3a, Supplementary Fig. 6a) and observed the participation of the neighboring Trim12 promoter in regulating Zfp42 (Figure 3a, Supplementary Figure 6b). We also note that regulatory regions upstream of Zfp42 tended to cause intermediate loss of GFP as compared to a complete loss of GFP (GFPmedium in red versus GFPneg in blue; Fig. 3c), suggesting that these enhancers are each responsible for only part of the overall Zfp42 expression level in cells.
Validation of MERA hits
To determine the accuracy of the MERA screen in systematically determining required cis-regulatory regions, we first examined replicate consistency among our Tdgf1, Zfp42, and Nanog MERA data. Spatial patterns of GFPneg gRNA enrichment were largely conserved between replicates with Pearson correlation values of 0.8 at 300 bp bin size (Fig. 2c, Fig. 3b, Supplementary Fig. 6c). At an individual level, the overlap between gRNAs enriched in GFPneg populations between replicates was significant for all replicates (hyper geometric p-value <0.001); however, it was not as high as for binned regions, likely because a single gRNA can cause thousands of distinct mutant genotypes with varying phenotypes.
To analyze false positives caused by off-target effects, we examined how putative off-target effects affect MERA results using a model based on GUIDE-Seq38 (Supplementary Methods). We found that when we eliminate gRNAs with potential off-target effects from our analysis, the global distribution of significantly enriched gRNAs along the regulatory landscape of the gene is unaltered and relative contributions of different functional categories are unaffected (Supplementary Fig. 5a and 6a,c). Furthermore, several gRNAs with no predicted off-target effects support the regulation of Tdgf1 by the promoter of Lrrc2 (Supplementary Fig. 5b), the promoter of Trim12, and a URE region (Supplementary Fig. 6a–c), and none of these regions are more likely to contain off-target effects than other screened regions.
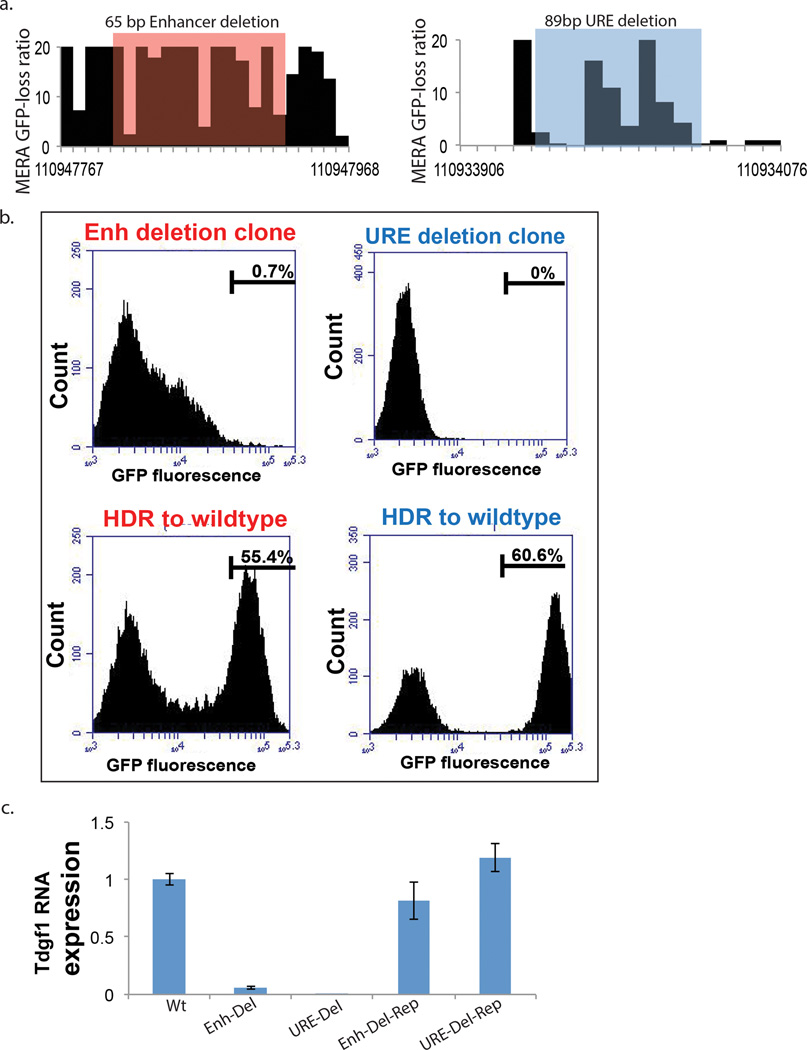
Figure 6. Local genotypes at an enhancer and a URE dictate Tdgf1 expression phenotype.
(a.) Tdgf1 MERA screen ratio of GFPmedium/neg/bulk reads for each gRNA at an upstream enhancer (left) and a downstream URE (right) region. (b) Flow cytometric measurement of Tdgf1-GFP expression in clonal cell lines following CRISPR-induced deletion of the shaded regions from (a) show loss of GFP (1st and 3rd plots from left). CRISPR-mediated homology-directed repair (HDR) back to the wildtype genotype induced robust GFP recovery at both loci (2nd and 4th plots from left). c.) Tdgf1 RNA expression in wild-type mESCs (left), clonal mESC lines with deletions of the enhancer and URE shaded in (a) (2nd and 3rd from left), and bulk mESC lines following HDR back to the wildtype genotype (4th and 5th from left), all normalized to wildtype expression level.
To analyze potential off-target effects with an independent method, we asked whether any gRNAs from the Tdgf1 library would extinguish Zfp42-GFP activity and vice versa. We found that a much smaller percentage of cells lose GFP upon targeting by a mismatched gRNA library than by the matched library (Supplementary Fig. 8). Sequencing revealed that the gRNAs enriched in GFPneg mismatched library-targeted cells were predominantly GFP control gRNAs with a small number of non-clustered gRNAs displaying off-target activity (Supplementary Figs 5,6). Thus, the clustered enrichment of GFP loss at enhancers, neighboring promoters, and UREs in MERA is not replicated by computationally predicted or experimentally determined off-target effects, leading us to conclude that GFP loss in these regions is a result off on-target gRNA effects (Supplementary Fig. 5a–c, Supplementary Fig. 6a,b).
To determine the false-positive rate at the level of individual gRNAs, we introduced individual gRNAs to determine whether their rate of GFP loss correlated with their activity in the pooled MERA screen. These gRNAs fell within several of the functional categories including UREs and neighboring promoters (Fig. 2a highlighted in grey, Fig. 2b). We confirmed significantly increased GFP loss in 29/30 gRNAs from these screens as compared to five similarly located control gRNAs (Fig. 2b). Altogether, we conclude that MERA has a low false positive rate.
We next sought to determine the false negative rate of MERA. As opposed to ORF-targeting screens in which all gRNAs are assumed to be equivalently likely to induce frameshift mutations that inactivate gene function, we find that regulatory mutations induce more variable phenotypes with regards to gene expression (see Supplementary Discussion). In our individual follow-up assays, we find that gRNAs targeting the GFP ORF induce GFP loss in >90% of cells, those targeting promoter regions induce GFP loss in 20–40% of cells, those targeting distal regulatory elements induce GFP loss in 5–40% of cells, while negative controls induce GFP loss in <2% of cells (Fig. 2b). We assert that this phenotypic diversity results from the wide spectrum of mutations at target sites, which are differentially likely to disrupt functional regulatory elements such as transcription factor binding sites. We confirm this hypothesis in several cases by performing functional motif discovery, described later in the text.
To assess the false-negative rate of MERA gRNAs, we examined regions in our data with strong likelihood to induce GFP loss. We found 10/10 GFP-targeting gRNAs in all 4 GFP lines are highly enriched in GFPneg cells (Fig. 2d, Fig. 3c). Additionally, 67/83 (81%) gRNAs that target the first 700bp of the Rpp25 open-reading frame are highly enriched in GFPneg cells. In a 500bp around the Tdgf1 promoter region, 48/59 (81%) of gRNAs induce GFP loss in multiple replicates (Supplementary Fig. 4f). Thus, we find that a high percentage of gRNAs expected to have an effect on gene expression are enriched in GFPneg cells. It is unclear whether the 20% of gRNAs in these regions that do not induce GFP loss are false negatives or true negatives, as their mechanism of inducing GFP loss is not as direct as when the GFP ORF itself is targeted. However, even if this appreciable percentage of individual gRNAs are false negatives, it does not impair the ability of MERA to determine required regulatory regions, as the high density of gRNAs in a region (~1 per 8 bp) allows highly reproducible resolution at the level of 100–1000 bp (Fig. 2c, 3b). We then asked if annotated regulatory regions are necessary for gene function. An appreciable percentage of gRNAs induce significant GFP loss at 9/9 of Tdgf1 predicted enhancers (+/− 20kb around Tdgf1) and 6/7 of predicted Zfp42 enhancers (−21 to +45kb around Zfp42). (Supplementary Tables 2,3). However, there is substantial heterogeneity in the percentage of gRNAs within an enhancer that induce GFP loss, and some DNase-hypersensitive sites without enhancer histone modifications contain a high fraction of GFP loss-inducing gRNAs (Supplementary Tables 2,3), indicating that enhancer histone modifications do not entirely predict required regulatory regions. We cannot rule out that certain regions may suffer from systematic inefficiencies in gRNA targeting.
Gene regulatory trends emerging from MERA screens
Our MERA results revealed that Tdgf1, Nanog, Rpp25 and Zfp42 have different regulatory architectures (Fig. 2, Fig. 3, Supplementary Figs 5,6,10). All regulatory regions within 20kb of the Nanog promoter were associated with clusters of highly enriched gRNAs, and 20% to 40% of the tested gRNAs in predicted enhancers and DNase-I hotspots proximal to Nanog resulted in GFPneg cells (Supplementary Fig. 10c). In contrast, the Rpp25 gene shows a dense concentration of significant gRNAs at its promoter and short ORF region. Other proximal regulatory regions of Rpp25 had 12% of tested gRNAs resulting in GFPneg cells (Supplementary Fig. 10d). Tdgf1 shows a similar trend to Nanog with dense clusters of significant gRNA in the proximal regulatory regions (Fig. 2a,d). UREs were also seen in cis-regulatory regions near Rpp25 (Supplementary Fig. 10b). In Nanog, a distal ChIA-Pet region >92 mB away showed several strongly enriched gRNAs whereas three other distal ChIA-Pet regions showed no strongly enriched gRNAs (Supplementary Fig. 10a), indicating that MERA is capable of measuring the functionality of long-distance chromatin interactions.
One observation common to all genes is the participation of the promoters of other genes in regulation. In some cases these gene promoters are several million bases away. Examples of foreign promoter involvement can be seen in the case of Lrrc2 promoter in Tdgf1 (Fig. 2a,d), Mirc35hg in Nanog (Supplementary Fig. 10a), Scamp5 and Cox5a in Rpp25 (Supplementary Fig. 10b). Previous studies have documented the existence of dual property elements40 that can act as either promoter or enhancer in different cellular contexts. Additionally, it is known that neighboring promoters often interact with each other41 and that neighboring gene expression is often coordinated42. Here we observe that active promoters may coordinate gene expression patterns of neighboring genes by functioning as enhancers within the same cellular context.
Functional motif discovery to examine MERA-predicted regulatory regions
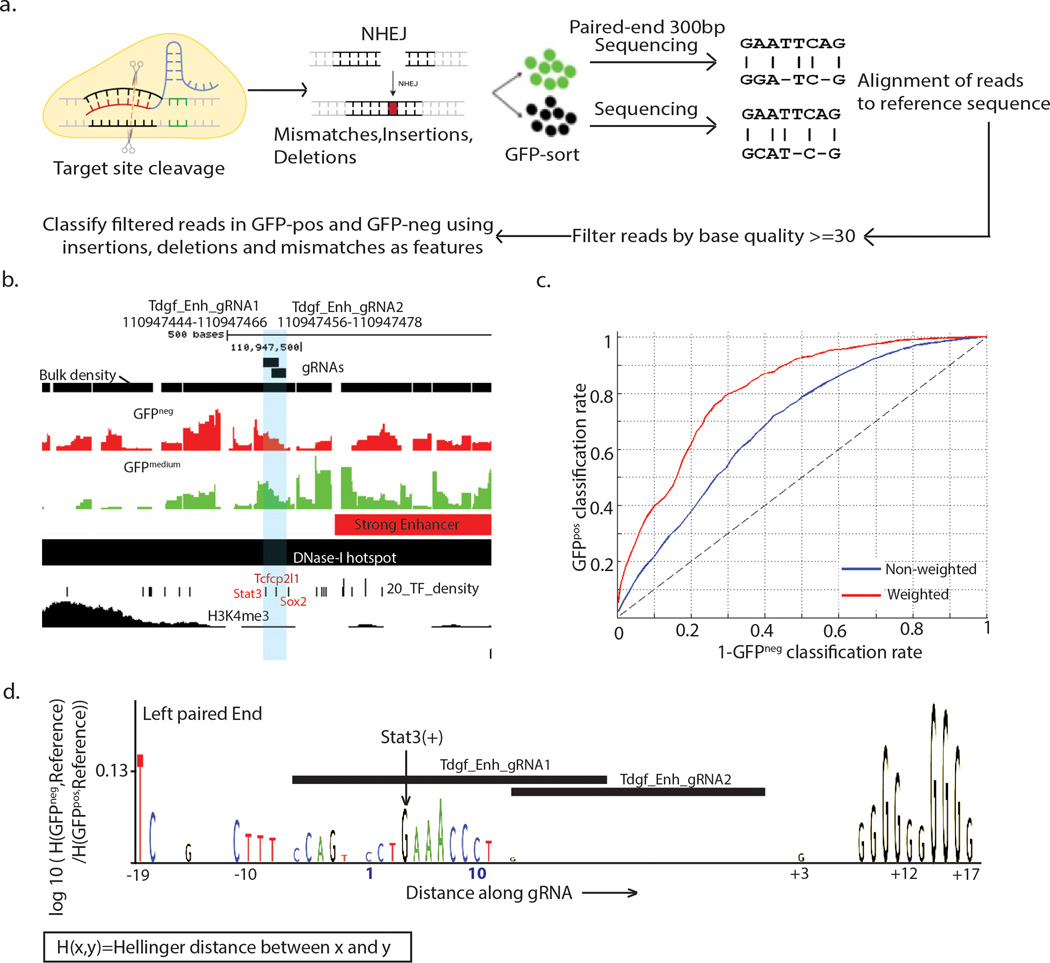
The second phase of MERA uses functional motif discovery to identify the causal elements governing expression at MERA screen hits. Because Cas9 induces random mutations, a pool of mESCs treated with Cas9 and a single gRNA will contain thousands of distinct mutant genotypes centered on the gRNA cleavage site. Recently, TAL effector nucleases have been used to derive functional footprints of regulatory DNA43. We hypothesized that we could pinpoint DNA sequence motif(s) that cause GFP loss by identifying sequence features that consistently differ between thousands of GFPpos and GFPneg genotypes at a given site (Fig. 4a). Functional motif discovery proceeds by performing individual Cas9-mediated mutation by a selected gRNA, obtaining thousands of genotypes from both GFPpos and GFPmedium/neg cells by high-throughput sequencing, and then summarizing the observed genotypes as motifs that reveal which bases are important for gene expression (Fig. 4a, Supplementary Methods). Using the differences in fractions of genotypes at positions along the gRNA, we defined a base-level importance score that was independent of the cutting biases of the gRNA and built a Random Forest44 classifier to gauge the accuracy of distinguishing GFPneg or GFPpos genotypes using base-level features (Supplementary Methods).
Figure 4. Functional motif discovery analysis of region-specific mutant genotypes at enhancers reveals required regulatory motifs.
(a) A schematic of the procedure involved in finding mutations induced by a particular gRNA (b) Plot showing the genomic regions surrounding two gRNAs at a proximal Tdgf1 enhancer region (gRNAs are shaded) showing overlap with DNase-I hotspot and predicted enhancer regions, and transcription factor binding sites Stat3, Tcfcp2l1 and Sox2. (c) ROC curve for fivefold classification of GFPneg and GFPpos genotypes using mutations within −20 to +20bp of the gRNA along left and right paired end reads as features. (d) Motif logo for region mutated by gRNAs with base scores computed as log-ratios of the hellinger distance of the GFPneg genotypes at a base to the reference base to the hellinger distance of the GFPpos genotypes at a base to the reference base, caused by Tdgf_gRNA_1 andTdgf_gRNA_2 along the left paired end read.
We first tested to see if functional motif discovery in Tdgf1 and Zfp42 enhancer regions would permit us to classify genotypes held-out of initial algorithmic training as GFPneg or GFPpos. We selected two overlapping gRNAs for functional motif discovery in a Tdgf1 proximal enhancer that overlapped binding sites for the key mESC transcription factors Stat3, Sox2 and Tcfcp2lI, of which Stat3 is the only factor with a direct binding site. We were able to classify held out genotypes with an area under the curve (AUC) of 0.81 (Fig. 4c), and observed an enrichment of the bases for the Stat3 motif30 in both the left and right paired end reads (Fig. 4d, Supplementary Fig. 12e). We achieved similar success at Zfp42 enhancer sites, identifying required bases around Nrf1 and p300 binding sites (Supplementary Fig. 13).
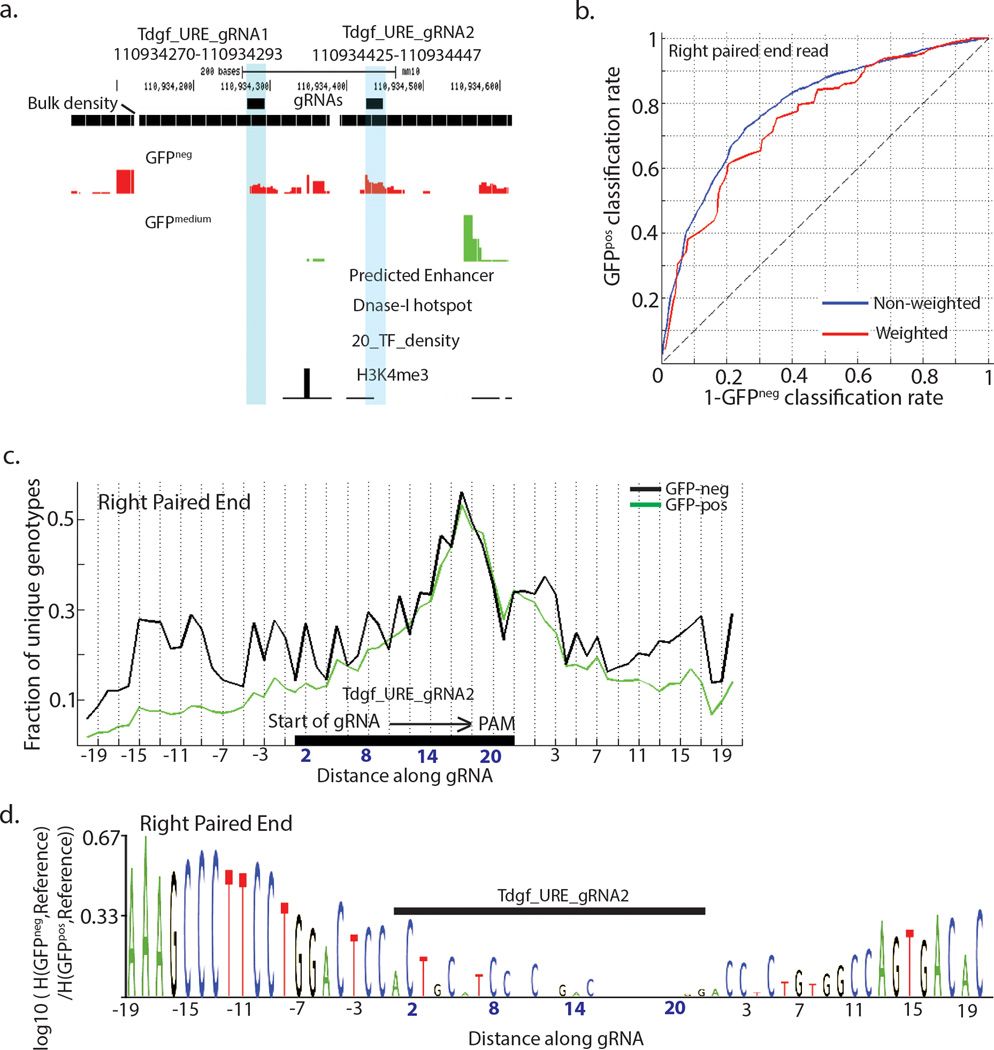
We next applied functional motif discovery to two gRNAs in a URE ~12 kb downstream of the Tdgf1 transcript (Fig. 5a). We obtained high classification accuracy for held out genotypes from both gRNAs (AUC 0.81 and 0.76, Fig. 5b, Supplementary Fig. 15c), and we observed blocks of consecutive bases whose deletion correlated with GFP loss (Fig. 5c-d, Supplementary Fig. 15d-e), suggesting focal regions of the genome that are required for URE function. Altogether, we conclude that functional motif discovery is a valuable method for ascertaining which bases at MERA-identified regulatory regions are required for gene expression. In enhancer regions, these bases correspond to known binding motifs, and in UREs, we identify blocks of bases which are required for gene expression.
Figure 5. Functional motif discovery analysis of a URE reveals critical base positions involved in gene regulation.
(a) Plot showing the genomic regions surrounding two gRNAs (gRNAs are shaded) showing their absence of active histone modifications, known transcription factor binding, predicted enhancers or DNase-I hotspots. (b) Receiver-operating characteristic (ROC) curve for fivefold classification of GFPneg and GFPpos genotypes using mutations on the right paired end read within –20 to +20bp of Tdgf_URE_gRNA2. Unweighted classification (in blue) counts each unique genotype in the test-set only once while weighted classification (red) counts each unique genotype in the test-set as many times as the number of reads assigned to it, for calculating sensitivity and specificity. (c) Fraction of unique genotypes in GFPneg and GFPpos populations with mutations at bases along the right paired end read reveals pattern of cleavage around Tdgf_URE_gRNA2. (d) Motif logo for the region mutated by Tdgf_URE_gRNA2 along the right paired end read with base scores computed as log-ratios of the hellinger distance of the GFPneg genotypes at a base to the reference base to the hellinger distance of the GFPpos genotypes at a base to the reference base.
We then used homologous recombination to validate that the Tdgf1 enhancer and URE regulatory elements are truly required for gene expression in the third phase of MERA. We used flanking gRNAs to induce short (>100 bp) deletions in two regions predicted to induce GFP loss by our MERA screen, one in the Tdgf1 enhancer and one at a URE. As expected, a subset of cells lost GFP expression, and we obtained clonal GFPneg lines containing the deletion genotype (Fig. 6a,b). We then used homology-directed repair to restore the wild-type genotype in these cells, finding at each site that a large percentage of cells reverted to a GFPpos state (Fig. 6b). We replicated this experiment in wildtype cells without a Tdgf1–GFP allele, finding that clonal deletion cells lost Tdgf1 RNA expression, and clonal repaired lines restored Tdgf1 expression (Fig. 6c). This robust and straightforward relationship between local genotype and GFP expression provides compelling evidence that the local DNA sequence at a URE is required for Tdgf1 expression.
Discussion
MERA offers an unbiased, high-resolution approach to directly interrogate the function of the regulatory genome. It not only provides a survey of required cis-regulatory elements, but also enables functional motif discovery to dissect the precise nature of identified regulatory elements. We find evidence that neighboring gene promoters as well as unmarked regulatory elements (UREs) that are not associated with conventionally expected DNase hypersensitivity and histone mark features play unexpectedly large roles in controlling gene expression. This observation reinforces the importance of direct perturbation analysis to definitively characterize genome function, as we observe that correlative genome annotation does not fully predict regulatory requirement.
While we do not yet have definitive data as to the function of UREs, we find that a URE downstream of the Tdgf1 gene is highly sensitive to base substitution at a string of consecutive bases, suggesting that its DNA sequence is crucial to its regulatory activity. Furthermore, we find the first half of this URE to be highly conserved (phastcons score>0.85, Supplementary Fig. 14e) indicating potential functional significance of the genomic region. Consistent with these data, UREs may be RNA templates, elements bound by uncharacterized protein factors, or spacers where their precise base sequence is of secondary importance. We cannot exclude the possibility that UREs are only active in a cellular subpopulation and thus conventionally expected DNase hypersensitivity and histone mark features are not detected when the entire cellular population is assayed.
We designed our gRNA libraries to target a mix of previously annotated and unannotated cis-regulatory regions, and thus we did not uniformly tile the proximal regions of any of these genes. Therefore, we cannot estimate the frequency of UREs, and we expect that future MERA screens with even more extensive coverage at more loci will elucidate how pervasive UREs and neighboring gene promoters are in the regulatory architecture of the genome.
MERA is complementary to high-throughput reporter assays, and future experiments performing both approaches should provide insight into the degree of concordance between necessary and sufficient gene regulatory elements. MERA also enables quantitative assessment of the relative effects of distinct cis-regulatory elements on gene expression and could potentially provide insights into how regulatory regions combine to achieve desired levels of expression. Extending MERA to explore how changes in individual cis-regulatory elements alter gene networks will aid our understanding of how cis-regulatory variants lead to human disease. We expect that the direct interrogation of variant locations discovered in genome-wide association studies by MERA will provide a rapid way to screen such variants for function in relevant cell types.
Supplementary Material
References
- 1.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 3.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melnikov A, et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nature biotechnology. 2012;30:271–277. doi: 10.1038/nbt.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arnold CD, et al. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013;339:1074–1077. doi: 10.1126/science.1232542. [DOI] [PubMed] [Google Scholar]
- 8.Patwardhan RP, et al. Massively parallel functional dissection of mammalian enhancers in vivo. Nature biotechnology. 2012;30:265–270. doi: 10.1038/nbt.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fullwood MJ, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simonis M, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nature genetics. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 12.Dostie J, et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome research. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwasnieski JC, Fiore C, Chaudhari HG, Cohen BA. High-throughput functional testing of ENCODE segmentation predictions. Genome research. 2014;24:1595–1602. doi: 10.1101/gr.173518.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jinek M, et al. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nature biotechnology. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho SW, et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome research. 2014;24:132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilbert LA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nature biotechnology. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kleinstiver BP, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cradick TJ, Fine EJ, Antico CJ, Bao G. CRISPR/Cas9 systems targeting beta-globin and CCR5 genes have substantial off-target activity. Nucleic acids research. 2013;41:9584–9592. doi: 10.1093/nar/gkt714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou Y, et al. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509:487–491. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
- 27.Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera Mdel C, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nature biotechnology. 2014;32:267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- 28.Chen S, et al. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell. 2015;160:1246–1260. doi: 10.1016/j.cell.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arbab M, Srinivasan S, Hashimoto T, Geijsen N, Sherwood Richard I. Cloning-free CRISPR. Stem Cell Reports. doi: 10.1016/j.stemcr.2015.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yue F, et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh AM, Hamazaki T, Hankowski KE, Terada N. A heterogeneous expression pattern for Nanog in embryonic stem cells. Stem cells. 2007;25:2534–2542. doi: 10.1634/stemcells.2007-0126. [DOI] [PubMed] [Google Scholar]
- 33.Rajagopal N, et al. RFECS: a random-forest based algorithm for enhancer identification from chromatin state. PLoS Comput Biol. 2013;9:e1002968. doi: 10.1371/journal.pcbi.1002968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.John S, et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nature genetics. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sherwood RI, et al. Discovery of directional and nondirectional pioneer transcription factors by modeling DNase profile magnitude and shape. Nature biotechnology. 2014;32:171–178. doi: 10.1038/nbt.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo Y, Mahony S, Gifford DK. High resolution genome wide binding event finding and motif discovery reveals transcription factor spatial binding constraints. PLoS computational biology. 2012;8:e1002638. doi: 10.1371/journal.pcbi.1002638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsai SQ, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nature biotechnology. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leung D, et al. Integrative analysis of haplotype-resolved epigenomes across human tissues. Nature. 2015;518:350–354. doi: 10.1038/nature14217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li G, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woo YH, Walker M, Churchill GA. Coordinated expression domains in mammalian genomes. PloS one. 2010;5:e12158. doi: 10.1371/journal.pone.0012158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vierstra J, et al. Functional footprinting of regulatory DNA. Nature methods. 2015 doi: 10.1038/nmeth.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Breiman L. Random Forests. Machine Learning. 2001;45:5–32. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.