Abstract
Circulating tumor cell (CTC) number measured with the CellSearch® assay is prognostic for survival in metastatic castration-resistant prostate cancer (mCRPC) pre- and post-therapy. Using a standard operating protocol for sample collection, processing, and analysis, we compared detection rates of CellSearch® performed using FDA-cleared methodology with a second positive selection assay, AdnaTest®, and a non-selection polymerase chain reaction (PCR)-based (DDPCR) assay in 55 blood samples from 47 men with progressive mCRPC. AdnaTest requires processing within 4 hours of the draw and detects KLK3, PSMA, and EGFR transcripts in cells captured on magnetic beads. The DDPCR assay can be processed up to 7 days after a draw and detects KLK2, KLK3, HOXB13, GRHL2, and FOXA1 genes. AdnaTest and DDPCR were considered positive if at least 1 transcript was detected. AdnaTest detected CTCs in 34 samples (62%, 95% confidence interval [CI] 48%–75%), of which 23 (68%) had unfavorable CTC counts by CellSearch. A positive DDPCR result was seen in 38 cases (69%, 95% CI 55%–81%), including 24 (63%) with unfavorable CellSearch CTC counts. CellSearch found unfavorable CTC counts in 25 samples (45%, 95% CI 33%–58%). Sensitivities were similar between the AdnaTest and DDPCR assays, and both were more sensitive than CellSearch. Concordance probability estimates (possible values 0.5–1.0) associating the biomarker result with survival were similar: 0.77 (standard error [SE] = 0.07) for AdnaTest, 0.72 (SE = 0.08) for DDPCR, and 0.76 (SE = 0.06) for CellSearch. Overall detection rates between the AdnaTest and DDPCR assays were similar and both were superior to CellSearch. The DDPCR assay required the lowest blood volume, least on-site processing and longest stability for batch processing.
Keywords: biomarkers, circulating tumor cells, prostate cancer, prostate-specific markers, AdnaTest, CellSearch
INTRODUCTION
Multiple blood-based assays to detect circulating tumor cells (CTCs) and tumor cell products are in different phases of development, for different contexts of use. These contexts include assessing prognosis, predicting drug sensitivity to therapeutics to guide treatment selection, and as response indicators to assess efficacy. Few assays have undergone the rigorous analytical validation that is required before the evidence generation to establish clinical validity and utility for a specific context of use can proceed.1-3 Presently only 1 CTC assay, CellSearch® (Janssen Diagnostics, Raritan, NJ), has achieved the level of an U.S. Food and Drug Administration (FDA) clearance. This assay uses an immunomagnetic ferrofluid to capture epithelial cell adhesion molecule-1 (EpCAM)-positive cells and reports the number of cells that meet defined criteria.4,5 The enumeration result represents a subset of the CTCs in blood that can be reproducibly identified and scored across laboratories and investigators, and has been shown to be prognostic for survival pre- and post-treatment.6 More recently, the enumeration biomarker in combination with lactate dehydrogenase (LDH) was shown to satisfy the Prentice Criteria for a surrogate endpoint, with respect to survival at the individual patient level in metastatic castration-resistant prostate cancer (mCRPC).7 A limitation of the CellSearch assay is low CTC detection rates at key decision points in management. Needed are assays to detect cells at a higher frequency in a higher percentage of patients.2,5,8-11
AdnaTest® (AdnaGen AG, Langenhagen, Germany) is a positive selection assay that captures cells using immunomagnetic beads coated by antibodies to surface markers. The captured cells are detected using a polymerase chain reaction (PCR)-based method that measures specific transcripts based on tumor type: epidermal growth factor receptor (EGFR), carcinoembryonic antigen (CEA), and EpCAM for colon cancer,12 and MUC-1, human epidermal growth factor receptor 2 (HER2), and GA733-2 for breast cancer, with detection sensitivities similar to that of CellSearch.13,14 The AdnaTest for mCRPC detects CTCs based on KLK3, prostate-specific membrane antigen (PSMA), or EGFR transcripts.15 More recently, a modified platform has been used to detect truncated forms of androgen receptor (AR) as a tumor sensitivity biomarker in mCRPC.16-18
Previously, we developed and analytically validated a non–selection-based direct detection reverse transcription PCR (DDPCR) CTC assay for KLK2, KLK3, HOXB13, GRHL2, and FOXA1, genes that are highly expressed in prostate tissue relative to white blood cells. The assay is performed on blood samples collected in PAXgene tubes that stabilize intracellular RNA, requires minimal on-site processing, and which can be stored and shipped for future analysis at a central laboratory. Relative to CellSearch enumeration, the DDPCR assay was shown to provide a more reliable and robust prediction of overall survival.19 Here, we report the analytical validation and performance characteristics of the AdnaTest Prostate Cancer CTC assay and compare AdnaTest to the CellSearch and DDPCR assays for the context of use of detecting CTCs in mCRPC.
PATIENTS AND METHODS
Patients and Samples
Blood samples were studied from 47 men with progressive mCRPC (both pre- and post-chemotherapy treated) per Prostate Cancer Working Group guidelines,20 treated at Memorial Sloan Kettering Cancer Center (MSKCC) between November 2011 and October 2013. Blood was collected at the time of phlebotomy performed as part of their routine clinical management. Samples for each assay were drawn at the same time and processed in the Department of Laboratory Medicine at MSKCC. Duplicate patient samples were obtained to assess the reproducibility of the AdnaTest assay. Control samples were obtained from 12 volunteers without evidence of prostate cancer. Signed informed consent was obtained from all patients and volunteers on an Institutional Review Board–approved protocol.
Assays
The AdnaTest was performed on 10 mL of whole blood collected into a lavender-top ethylenediaminetetraacetic acid (EDTA) tube that was immediately placed on ice, for processing within 4 hours of blood draw. The test is a two-step procedure to capture CTCs from whole blood using magnetic beads conjugated with antibodies for EpCAM and HER2, and analyzed for expression of prostate-specific genes KLK3, PSMA, and EGFR. The AdnaTest result was considered positive if at least 1 tumor-associated transcript measured >0.1 ng/μl (controls were always performed concurrently with each sample analysis). Analytical validity was established using a previously described method8 (see Supplemental Data).
The DDPCR assay was performed as previously described. In brief, whole blood was collected in 2.5-mL PAXgene tubes and transported at room temperature to the laboratory, and then frozen at −80°C until analyzed.19 This assay was considered positive if at least 1 of the following transcripts was detected: KLK3, KLK2, HOXB13, GRHL2, and FOXA1.
CellSearch CTC enumeration was performed on 7.5 mL of blood collected in CellSave tubes, according to the FDA-cleared manufacturer's method.4 The results are reported as unfavorable (5 or more) or favorable (4 or fewer) number of cells per 7.5 mL of blood.
Data Reporting/Statistical Methods
Fifty-five blood samples were obtained from 47 patients; 40 had all 3 assays performed once, 6 patients twice, and 1 patient 3 times. AdnaTest sensitivity was estimated and compared to that of the DDPCR and CellSearch assays using McNemar's test. The specificity of AdnaTest was computed on 12 healthy volunteers. The concordance probability estimate was used to measure the discriminatory power of each of the assays with respect to survival.21
RESULTS
The clinical characteristics for the 55 samples provided by the 47 patients are presented in Table 1. Forty-five of those samples (82%) were provided by patients who have since died; median survival was 13 months (range 1–42 months).
Table 1.
Baseline Patient Clinical Characteristics
| Characteristic | N=55 Samples* (47 unique patients) |
|---|---|
| Age, years – median (range) | 68 (45–91) |
| Primary Treatment – n (%) | |
| Prostatectomy | 18 (33) |
| Radiation | 20 (36) |
| None | 17 (31) |
| Hormone Therapies – n (%) | |
| ≤2 lines | 16 (29) |
| 3 lines | 18 (33) |
| ≥4 lines | 21 (38) |
| Chemotherapy-naïve – n (%) | 21 (38) |
| Chemotherapy-exposed – n (%) | 34 (62) |
| Metastatic Disease – n (%) | |
| Bone | 52 (95) |
| Lymph Node | 37 (67) |
| Liver | 10 (18) |
| Lung | 12 (22) |
| Other Soft Tissue | 14 (25) |
| Laboratory Measures – median (range) | |
| PSA, ng/mL | 35.84 (<0.05–2481.31) |
| Hgb, g/dl | 11.0 (6.8–14.7) |
| ALK, unit/L | 123 (32–1704) |
| LDH, unit/L | 244 (160–922) |
| ALB, g/dl | 4.1 (3.2–4.6) |
| CTC, cells/7.5mL | 2 (0 – >200) |
| Follow-up, months – median (range) | 13 (1–42) |
| Overall survival, months – median (range) | 13 (1–42) |
| Samples from now-deceased patients – n (%) | 45 (82%) |
The 55 samples were collected from 47 unique patients. All measures in this table were calculated for N=55 (number of samples rather than number of patients).
Abbreviations: ALB, albumin; ALK, alkaline phosphatase; CTC, circulating tumor cells; Hgb, hemoglobin; LDH, lactate dehydrogenase; PSA, prostate-specific antigen.
Sample Collection, Processing and Analysis
A standard operating protocol was generated for sample collection, processing, and analysis for each of the tests, summarized in Table 2.
Table 2.
Sample Collection and Processing Protocol and Detection Frequency of AdnaTest, DDPCR and CellSearch Assays
| Assay | Tube, Volume | Storage/Transport, Time to Processing | Detection Frequency (95% CI) |
|---|---|---|---|
| AdnaTest tumor-specific gene transcript detection | EDTA 10 mL | On ice, <4 hours | 62% (47-75%) |
| DDPCR prostate-specific gene transcript detection in whole blood | PAXgene 2.5 mL | Up to 7 days at room temp, 60 months frozen at −80°C | 69% (55–81%) |
| CellSearch enumeration | CellSave 7.5 mL | Room temp, <96 hours | 45% (33–58%) |
Abbreviations: CI, confidence interval; DDPCR, direct detection polymerase chain reaction; EDTA, ethylenediaminetetraacetic acid.
Detection Rates of AdnaTest, DDPCR, and CellSearch with Cross-test Comparisons
AdnaTest specificity was established using blood from 12 healthy volunteers: it was set at 1.0 as no prostate-expressed genes were detected in any of the volunteer samples tested. The AdnaTest was positive in 34 of the 55 patient samples evaluated (62%, 95% confidence interval [CI] 48%–75%) (Table 2). Overall, AdnaTest detected 3 transcripts in 4 samples, 2 transcripts in 12 samples, and 1 transcript in 18 samples (Table 3). KLK3 was detected in 33 of the positive samples (33/34, 97%); EGFR alone was detected in 1 sample without KLK3 detection. The addition of PSMA transcript (detected in 17 samples, all with KLK3 detection) did not increase the sensitivity of the test.
Table 3.
Concordance Between AdnaTest, DDPCR, and CellSearch Detection of CTC*
| sample # | AdnaTest (No. of genes detected: KLK3, PSMA, EGFR) | Whole blood DDPCR (No. of genes detected: KLK2, KLK3, HOXB13, GRHL2, FOXA1) | CellSearch CTC count/7.5 mL blood |
|---|---|---|---|
| 1 | 3 | 5 | >200 |
| 2 | 3 | 5 | >200 |
| 3 | 3 | 5 | >200 |
| 4 | 2 | 5 | 193 |
| 5 | 2 | 5 | 168 |
| 6 | 2 | 5 | 111 |
| 7 | 0 | 3 | 84 |
| 8 | 2 | 3 | 59 |
| 9 | 2 | 5 | 55 |
| 10 | 1 | 5 | 54 |
| 11 | 1 | 4 | 47 |
| 12 | 3 | 5 | 43 |
| 13 | 2 | 0 | 30 |
| 14 | 1 | 5 | 27 |
| 15 | 2 | 2 | 27 |
| 16 | 2 | 4 | 18 |
| 17 | 2 | 1 | 15 |
| 18 | 1 | 2 | 14 |
| 19 | 1 | 4 | 13 |
| 20 | 1 | 4 | 10 |
| 21 | 0 | 3 | 10 |
| 22 | 1 | 5 | 9 |
| 23 | 1 | 4 | 9 |
| 24 | 1 | 1 | 6 |
| 25 | 2 | 4 | 5 |
| 26 | 2 | 2 | 3 |
| 27 | 0 | 0 | 3 |
| 28 | 1 | 5 | 2 |
| 29 | 0 | 2 | 1 |
| 30 | 0 | 1 | 1 |
| 31 | 1 | 0 | 1 |
| 32 | 1 | 0 | 1 |
| 33 | 1 | 0 | 1 |
| 34 | 0 | 0 | 1 |
| 35 | 0 | 0 | 1 |
| 36 | 2 | 4 | 0 |
| 37 | 1 | 4 | 0 |
| 38 | 1 | 2 | 0 |
| 39 | 0 | 2 | 0 |
| 40 | 0 | 2 | 0 |
| 41 | 0 | 2 | 0 |
| 42 | 1 | 1 | 0 |
| 43 | 1 | 1 | 0 |
| 44 | 0 | 1 | 0 |
| 45 | 0 | 1 | 0 |
| 46 | 1 | 0 | 0 |
| 47 | 0 | 0 | 0 |
| 48 | 0 | 0 | 0 |
| 49 | 0 | 0 | 0 |
| 50 | 0 | 0 | 0 |
| 51 | 0 | 0 | 0 |
| 52 | 0 | 0 | 0 |
| 53 | 0 | 0 | 0 |
| 54 | 0 | 0 | 0 |
| 55 | 0 | 0 | 0 |
For AdnaTest and DDPCR, results ≥1 were considered positive for CTC; CellSearch results are considered unfavorable if ≥5 cells detected per 7.5 mL of blood, and favorable for ≤4 cells. Rows with discordant results are highlighted. (Rows arranged by descending CellSearch results.)
Abbreviations: CTC, circulating tumor cells; DDPCR, direct detection polymerase chain reaction; PSMA, prostate-specific membrane antigen.
The DDPCR assay was positive in 38 samples (69%, 95% CI 55%–81%) based on the detection of one or more genes (Table 2). Overall, the DDPCR assay detected 5 transcripts in 12 samples, 4 transcripts in 8 samples, 3 transcripts in 3 samples, 2 transcripts in 8 samples, and 1 transcript in 7 samples (Table 3). KLK3 was detected in 31 of 55 samples (56%, 95% CI 43%–69%); of these 31 samples, 17 samples also had KLK3 detected by AdnaTest. The DDPCR test detected KLK2 in 27 samples (49%, 95% CI 36%–62%), HOXB13 in 27 samples (49%, 95% CI 36%–62%), FOXA1 in 23 samples (42%, 95% CI 30%–55%), and GRHL2 in 16 samples (29%, 95% CI 19%–42%).
Of the 7 samples positive by DDPCR without KLK3 detection (7/38, 18%), 4 samples had KLK2 plus other gene(s): 1 sample with KLK2+HOXB13+ GRHL2+ FOXA1, 1 sample withKLK2+HOXB13+ FOXA1, 1 sample with KLK2+ HOXB13, and 1 sample with KLK2+ FOXA1. Two samples had only FOXA1 detected and 1 sample had HOXB13 alone detected. Thus, the addition of GRHL2 did not increase the sensitivity of the DDPCR test.
With CellSearch, unfavorable counts (≥5 cells/7.5 mL of blood) were found in 25 of the 55 samples (45%, 95% CI 33%–58%) of which 23 were positive based on the AdnaTest, and 24 were positive by DDPCR (Table 3). Favorable/“negative” counts (≤4 cells/7.5 mL of blood) were found in 30 samples (55%, 95% CI 42%–67%), with 0 cells detected in 20 samples (33%, 95% CI 19%–51%) and 1–4 cells detected in 10 samples (67%, 95% CI 49%–81%). For these 30 samples with favorable CellSearch CTC counts, AdnaTest was positive in 11 (37%, 95% CI 22%–54%), whereas and DDPCR was positive in 14 (47%, 95% CI 29%–65%).
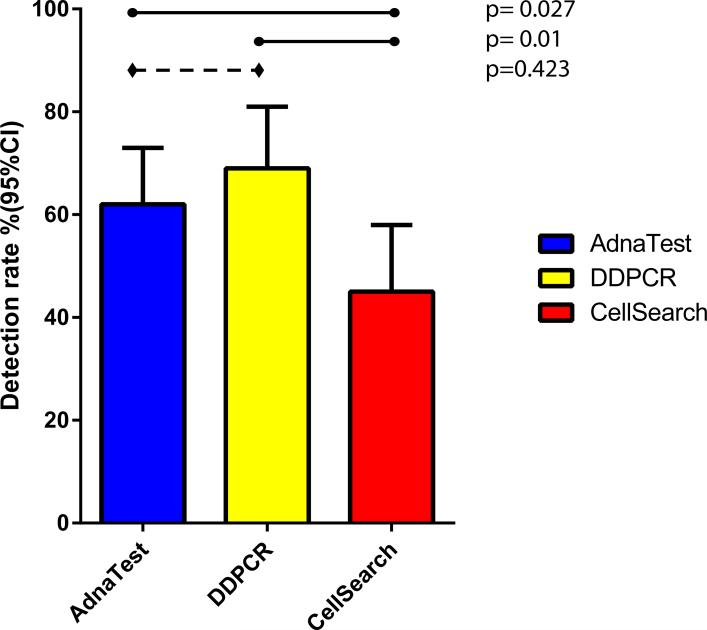
The detection rates of AdnaTest, DDPCR assays and CellSearch unfavorable counts are summarized in Figure 1.
Figure 1.
CTC detection frequencies of the AdnaTest, DDPCR, and CellSearch assays. The level of agreement between the three assays, compared using McNemar's test, showed significant disagreement between DDPCR and CellSearch (p=0.001) and between AdnaTest and CellSearch (p=0.027), but not between AdnaTest and DDPCR (p=0.423).
Concordance Between Assays
The concordance between CTC detection by AdnaTest and DDPCR in whole blood, and CTC enumeration by CellSearch is shown in Table 3, highlighting discordant results for any given sample. There were 5 samples with a positive AdnaTest who had no prostate transcripts detected by the DDPCR in whole blood, while 9 samples with DDPCR-detected prostate transcripts in whole blood had a negative AdnaTest. The AdnaTest and DDPCR were concordant in 41 (75%) samples, of which 29 had detectable cells with both assays and 12 samples had no cells detected with either assay.
The level of agreement between the three assays, based on the positive/negative dichotomization, was compared using McNemar's test (Figure 1). There was significant disagreement between DDPCR and CellSearch (p=0.001) and the AdnaTest and CellSearch (p=0.027). We did not find a disagreement between the AdnaTest and DDPCR (p=0.423).
Estimates of the Concordance Probability of the Assays to Predict Survival
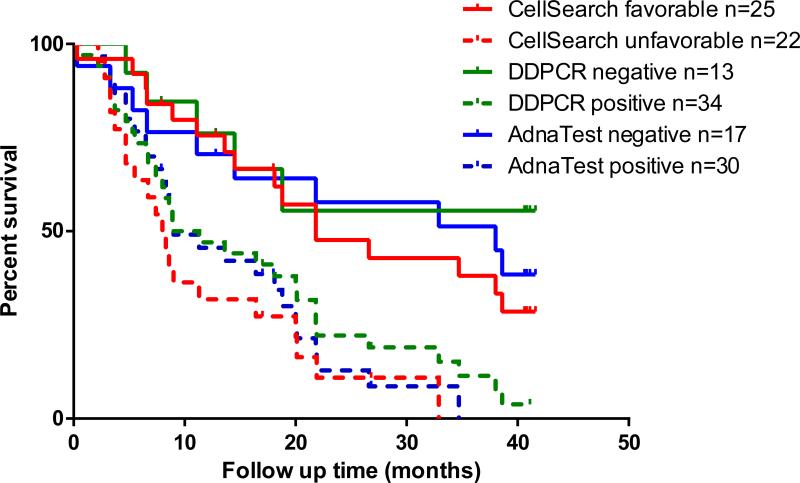
These results are based on the 47 patients. The concordance probability estimates are 0.77 (standard error [SE] = 0.07) for AdnaTest, 0.72 (SE = 0.08) for DDPCR and 0.76 (SE = 0.06) for CellSearch. Figure 2 shows the Kaplan-Meier overall survival estimates for the 3 assays, grouping patients by positive/negative results for each assay. Predictions of survival times were similar between the assays.
Figure 2.
Overall survival estimates by Kaplan-Meier based on each assay. Favorable and unfavorable results were dichotomized based on absent vs present detection of circulating tumor cells (CTC) by AdnaTest and direct detection RT-PCR (DDPCR) and by enumeration of <5 vs ≥5 cells/7.5 mL of blood by CellSearch assay.
DISCUSSION
Generating the evidence to demonstrate the utility of a biomarker for a specific context of use begins by establishing the analytical validity of the assay which includes how the specimen is acquired proceeding through transport to the laboratory and processing, and ending with the method of reporting of the results. CTC detection rates in this population of men with progressive mCRPC were similar between the AdnaTest Prostate assay which identifies CTC based on the presence of KLK3, PSMA, or EGFR transcripts and the DDPCR assay for the presence of prostate-specific KLK2, KLK3, HOXB13, GRHL2, and FOXA1 transcripts. Both were more sensitive than CellSearch, which uses an EpCAM-based CTC capture method. Predictions of survival times, however, were similar between the assays, using the positive/negative and unfavorable/favorable reporting criteria for each test.
All three of the assays — AdnaTest, DDPCR, and CellSearch — have now met the criteria for analytical validation. DDPCR required the lowest blood volume, minimal on-site processing, and the longest stability for batch processing. Both the AdnaTest and DDPCR assays detect prostate-specific transcripts not usually detected in nucleated blood cells. AdnaTest detection in this population was based on KLK3 transcripts in 33 of 34 samples: the addition of PSMA and EGFR detection did not increase sensitivity significantly. With the DDPCR, KLK3 was detected in 31 of 38 positive samples. The additional 7 positive samples were found with KLK2 in 4 samples, FOXA1 alone in 2 samples, and with HOXB13 alone in 1 sample; GRHL2 did not add to detection sensitivity. Importantly, the DDPCR approach is more flexible than AdnaTest, easily enabling inclusion of additional genes.
There are also novel assays currently being evaluated that are designed to detect AR splice variants and ligand binding domain mutations associated with resistance to AR-directed therapies. A phase 3 registration trial was recently initiated comparing galeterone to enzalutamide for patients shown to have ARv7-positive CTCs detected using a modified AdnaTest (NCT02438007). A separate validation of the ability to detect treatment-specific alterations in AR will be required before they can be tested prospectively in the clinic.
Practical issues favor the DDPCR assay. AdnaTest and CellSearch require more blood, which can be an issue when multiple tubes of blood are needed for routine safety assessments and to determine antitumor effects. There are also advantages to the PAXgene tubes used with the DDPCR assay, which require minimal on-site processing and can be transported to a central laboratory at room temperature up to several days after the blood draw for freezing and storage for future analysis. The CellSearch tubes provide consistent results up to 72 hours from the time of blood draw if a stable temperature is maintained. With AdnaTest, the time from blood draw to analysis is critical, as samples must be transported on ice from the phlebotomy site for processing in 4 hours or less. These differences in the time from blood draw to analysis, and the amount of on-site processing required, affect scalability of the tests.
One limitation of our current analysis is the cohort size. While our data showed KLK3 detection to be the primary marker for AdnaTest and DDPCR positive results, replication of these results would be needed before considering removing the less-useful transcripts from those tests.
In conclusion, the DDPCR and CellSearch assays confer easier use in practice than AdnaTest. Importantly, the AdnaTest and DDPCR assays had detection rates similar to each other, and both were superior to the CellSearch detection rate.
Supplementary Material
Acknowledgments
Sources of Funding: DoD Prostate Cancer Research Program Physician Research Award W81XWH-09-1-0307 to DCD. NCI SPORE in Prostate Cancer (P50 CA92629); Prostate Cancer Foundation; DCD, MF and HIS are named on the Memorial Sloan Kettering Cancer Center preliminary patent application for DDPCR assay.
Footnotes
Conflicts of Interest: The authors have no relevant conflicts of interest to declare.
REFERENCES
- 1.McShane LM, Hayes DF. Publication of tumor marker research results: the necessity for complete and transparent reporting. J Clin Oncol. 2012;30:4223–4232. doi: 10.1200/JCO.2012.42.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Danila DC, Pantel K, Fleisher M, et al. Circulating tumors cells as biomarkers: progress toward biomarker qualification. Cancer J. 2011;17:438–450. doi: 10.1097/PPO.0b013e31823e69ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kelloff GJ, Sigman CC, Scher HI. Biomarker development in the context of urologic cancers. Urol Oncol. 2015;33:295–301. doi: 10.1016/j.urolonc.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allard WJ, Matera J, Miller MC, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10:6897–6904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 5.de Bono JS, Scher HI, Montgomery RB, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14:6302–6309. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 6.Shaffer DR, Leversha MA, Danila DC, et al. Circulating tumor cell analysis in patients with progressive castration-resistant prostate cancer. Clin Cancer Res. 2007;13:2023–2029. doi: 10.1158/1078-0432.CCR-06-2701. [DOI] [PubMed] [Google Scholar]
- 7.Scher HI, Heller G, Molina A, et al. Circulating tumor cell biomarker panel as an individual-level surrogate for survival in metastatic castration-resistant prostate cancer. J Clin Oncol. 2015;33:1348–1355. doi: 10.1200/JCO.2014.55.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danila DC, Anand A, Sung CC, et al. TMPRSS2-ERG status in circulating tumor cells as a predictive biomarker of sensitivity in castration-resistant prostate cancer patients treated with abiraterone acetate. Eur Urol. 2011;60:897–904. doi: 10.1016/j.eururo.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danila DC, Heller G, Gignac GA, et al. Circulating tumor cell number and prognosis in progressive castration-resistant prostate cancer. Clin Cancer Res. 2007;13:7053–7058. doi: 10.1158/1078-0432.CCR-07-1506. [DOI] [PubMed] [Google Scholar]
- 10.Scher HI, Jia X, de Bono JS, et al. Circulating tumour cells as prognostic markers in progressive, castration-resistant prostate cancer: a reanalysis of IMMC38 trial data. Lancet Oncol. 2009;10:233–239. doi: 10.1016/S1470-2045(08)70340-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pantel K, Brakenhoff RH, Brandt B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat Rev Cancer. 2008;8:329–340. doi: 10.1038/nrc2375. [DOI] [PubMed] [Google Scholar]
- 12.Raimondi C, Nicolazzo C, Gradilone A, et al. Circulating tumor cells: exploring intratumor heterogeneity of colorectal cancer. Cancer Biol Ther. 2014;15:496–503. doi: 10.4161/cbt.28020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andreopoulou E, Yang LY, Rangel KM, et al. Comparison of assay methods for detection of circulating tumor cells in metastatic breast cancer: AdnaGen AdnaTest BreastCancer Select/Detect versus Veridex CellSearch system. Int J Cancer. 2012;130:1590–1597. doi: 10.1002/ijc.26111. [DOI] [PubMed] [Google Scholar]
- 14.Muller V, Riethdorf S, Rack B, et al. Prognostic impact of circulating tumor cells assessed with the CellSearch System and AdnaTest Breast in metastatic breast cancer patients: the DETECT study. Breast Cancer Res. 2012;14:R118. doi: 10.1186/bcr3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Todenhofer T, Hennenlotter J, Feyerabend S, et al. Preliminary experience on the use of the Adnatest(R) system for detection of circulating tumor cells in prostate cancer patients. Anticancer Res. 2012;32:3507–3513. [PubMed] [Google Scholar]
- 16.Antonarakis ES, Lu C, Luber B, et al. Androgen receptor splice variant 7 and efficacy of taxane chemotherapy in patients with metastatic castration-resistant prostate cancer. JAMA Oncol. 2015;1:582–591. doi: 10.1001/jamaoncol.2015.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakazawa M, Lu C, Chen Y, et al. Serial blood-based analysis of AR-V7 in men with advanced prostate cancer. Ann Oncol. 2015;26:1859–1865. doi: 10.1093/annonc/mdv282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Antonarakis ES, Lu C, Wang H, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371:1028–1038. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Danila DC, Anand A, Schultz N, et al. Analytic and clinical validation of a prostate cancer-enhanced messenger RNA detection assay in whole blood as a prognostic biomarker for survival. Eur Urol. 2014;65:1191–1197. doi: 10.1016/j.eururo.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scher HI, Eisenberger M, D'Amico AV, et al. Eligibility and outcomes reporting guidelines for clinical trials for patients in the state of a rising prostate-specific antigen: recommendations from the Prostate-Specific Antigen Working Group. J Clin Oncol. 2004;22:537–556. doi: 10.1200/JCO.2004.07.099. [DOI] [PubMed] [Google Scholar]
- 21.Gonen M, Heller G. Concordance probability and discriminative power of proportional hazards regression. Biometrika. 2005;92:965–970. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.