Abstract
The design of cancer-targeting particles with precisely-tuned physiocochemical properties may enhance delivery of therapeutics and access to pharmacological targets. However, molecular level understanding of the interactions driving the fate of nanomedicine in biological systems remains elusive. Here, we show that ultrasmall (< 10 nm in diameter) poly(ethylene glycol) (PEG)-coated silica nanoparticles, functionalized with melanoma-targeting peptides, can induce a form of programmed cell death known as ferroptosis in starved cancer cells and cancer-bearing mice. Tumor xenografts in mice intravenously injected with nanoparticles using a high-dose multiple injection scheme exhibit reduced growth or regression, in a manner that is reversed by the pharmacological inhibitor of ferroptosis, liproxstatin-1. These data demonstrate that ferroptosis can be targeted by ultrasmall silica nanoparticles and may have therapeutic potential.
Nanomaterials designed to target cancer may hold therapeutic potential by enhancing the delivery of therapeutics1–4, but their interactions with cancer in animal models, and long-term effects on cells, remain poorly understood5–9. To examine the effects of nanoparticle ingestion on cells, we utilized ~6 nm surface-functionalized poly(ethylene glycol)-coated (PEGylated) near-infrared (NIR) fluorescent silica nanoparticles, referred to as Cornell dots (C′ dots), with diameters controllable down to the sub-10 nm range, as reported in detail elsewhere10. This FDA Investigational New Drug (IND)-approved hybrid organo-silica particle was previously shown to be a promising cancer molecular imaging agent in metastatic melanoma patients after functionalizing its surface with αvβ3 integrin-targeting peptides and radiolabels2,11. Preferential accumulation was observed within integrin-expressing primary and/or metastatic melanomatous lesions in human subjects and animal melanoma models11,12, while, at the same time, demonstrating rapid renal clearance.
Given its potential clinical utility and its early adaptation for drug delivery applications13, we initially performed detailed cell biological studies using the latest generation of C dots, synthesized in water, referred to as C′ dots10, surface-functionalized with a 14-mer peptide analog, alpha-melanocyte stimulating hormone (αMSH)14, which targets a different surface receptor expressed on malignant melanoma cells (i.e., melanocortin-1 receptor, MC1-R). Using a multiple high-dose injection scheme, with and without pharmacologic inhibition of ferroptosis, the resulting αMSH-PEG-C′ dots (Fig. 1a) were used herein to determine whether alterations in cell survival occurred in cancer cell lines and tumor xenografts over a wide dose range relative to that seen under control conditions, and whether cellular pathways were modulated by particle ingestion.
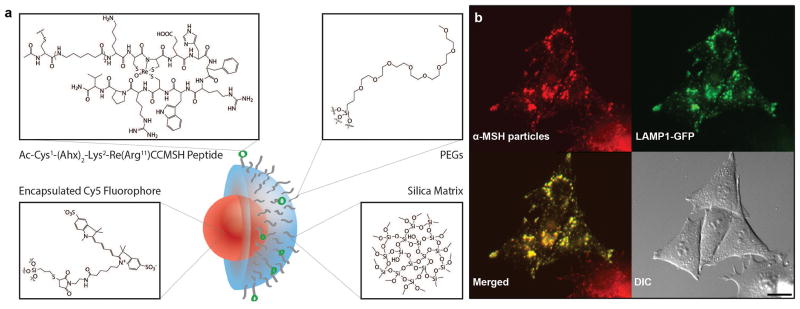
Figure 1. αMSH-PEG-C′ dot particles and their localization to lysosomal networks.
a, Silica-based and ultrasmall αMSH-PEG-C′ dots of 6 nm diameter silica-based particles with a fluorescent (Cy5 encapsulated) core and polyethylene glycol (PEG) coating and alpha melanocyte-stimulating hormone (αMSH)-modified exterior. b, αMSH-PEG-C′ dots localize to lysosomal networks in cells. M21 melanoma cells expressing LAMP1-GFP (green) were treated with αMSH-PEG-C′ dots (15μM) for 24 hours. Note colocalization between nanoparticles (Cy5 fluorescence, pseudo-colored red) and LAMP1-GFP in merged image. Scale bar=10μm.
αMSH-PEG-C′ dots reside in lysosomes and are well-tolerated
Live imaging of MC1-R expressing human melanoma cells (M21) treated with particles for 24 hours revealed colocalization of fluorescent αMSH-PEG-C′ dots with lysosomes, the latter visualized by expression of a GFP-tagged lysosomal-associated membrane protein 1 (LAMP1), indicating that ingested particles reside in lysosomal or late endosomal networks (Fig. 1b). M21 cells treated with increasing concentrations of αMSH-PEG-C′ dots up to 15 μM showed similar survival and proliferation rates to those of control cells (Fig. 2a), demonstrating that incubation at even high particle concentrations is well-tolerated. We next examined if lysosomes were functioning properly within cells treated with αMSH-PEG-C′ dots. To determine this, we examined the autophagy pathway, which targets intracellular substrates for lysosomal degradation. Autophagy was examined by quantifying the basal levels and turnover rates of the autophagy protein microtubule-associated protein 1 light chain 3 (LC3), which is lipidated onto autophagosomal membranes and becomes degraded following the fusion of autophagosomes with lysosomes15. The accumulation of the autophagosome-associated, lipidated form of LC3, or LC3-II, identified by its faster electrophoretic mobility in SDS-PAGE gels relative to the non-lipidated form, LC3-I, can be quantified as a measure of autophagy induction or flux through lysosomes15. Cells treated with increasing concentrations of αMSH-PEG-C′ dots for 24 hours, from 0.15 to 15 μM, had similar relative LC3-II levels compared to control cells, suggesting that autophagy is not induced or perturbed by nanoparticle treatment (Fig. 2b). Importantly, the treatment of cells with a lysosomal inhibitor, concanamycin A (ConA), which inhibits lysosome function by raising pH, and blocks autophagosome degradation, resulted in a similar accumulation of LC3-II in particle-treated cells as compared to controls, demonstrating that lysosomes are functioning properly even when cells are loaded with high concentrations of αMSH-PEG-C′ dots (Fig. 2b).
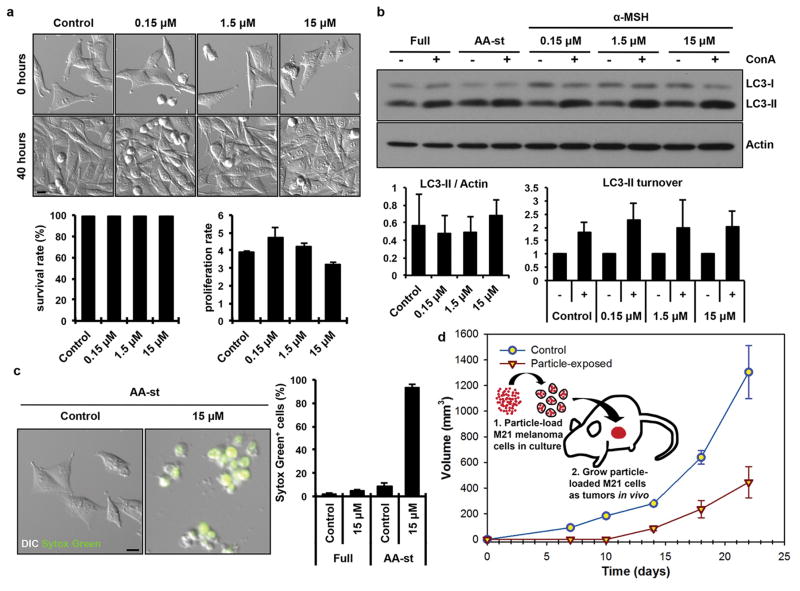
Figure 2. αMSH-PEG-C′ dot particles induce cell death in amino acid-deprived conditions.
a, Nanoparticles are well-tolerated in nutrient-replete media. Images show M21 cells treated with the indicated αMSH-PEG-C′ dot concentrations and cultured for 40 hours. Nanoparticles had no significant effect on cell survival (left graph) or cell proliferation (right graph), as quantified by time-lapse microscopy-based tracking of individual cell fates. Error bars indicate mean+/− standard error of the mean. N=3 biological experiments, with five independent fields of view for each. See Supp. Figure 6a,b for individual experimental values. Scale bar=10μm. b, Autophagy and lysosome function in nanoparticle-treated cells are unperturbed. Western blot shows LC3-I and –II in cells treated with increasing doses of αMSH-PEG-C′ dots for 24 hours compared to untreated (Full media) and amino acid-starved (AA-st) cells, in the presence (+) and absence (−) of the lysosome inhibitor concanamycin A (ConA, 1 hour at 100nM). Levels of LC3-II (left graph) are unaltered by nanoparticle treatment, and ConA-inducible LC3-II accumulation (right graph), a measure of autophagy flux, is similar between treated and untreated cells. Error bars indicate mean+/− standard error of the mean. N=3 biological replicates for each group. See Supp. Figure 6c,d for individual experimental values. c, Nanoparticle treatment induces cell death of M21 cells cultured in amino acid-free media. Images show live control cells and dead (Sytox green-positive) nanoparticle-treated cells in AA-st conditions. Scale bar=10μm. Graph shows percent Sytox green-positive cells in full media (Full) or AA-st conditions after 50 hours, as determined by time-lapse microscopy. Error bars indicate mean+/− standard deviation. N=4 for each group. Each replicate is from one biological experiment, quantified with five independent fields of view. d, M21 cells treated with 15μM αMSH-PEG-C′ dots in full media for 72 hours prior to create xenografts in immunodeficient (SCID/Beige) mice demonstrate growth inhibition (open inverted triangles) relative to untreated control cells (open circles). Schematic shows workflow, consisting of (1) particle-loading M21 melanoma cells, by treatment at 15uM for 48 hours in culture under full media conditions, and (2) injecting 5×106 particle-loaded M21 cells into mice to assay xenograft tumor growth versus control untreated cells. Data show mean tumor volume over 22 days of growth from three tumors per group. Error bars indicate mean+/− standard error of the mean. Particle-treated M21 cells showed statistically significant (p<0.001) growth inhibition compared with untreated control cells over the study interval. P-value is from a Wald test in a regression model estimated by generalized estimating equations to take into account the longitudinal nature of the data.
αMSH-PEG-C′ dots induce the death of starved cells
Although αMSH-PEG-C′ dots were well-tolerated by cells cultured under nutrient-replete conditions, and the autophagy pathway and lysosome function appeared to be unperturbed, we further examined if particle treatment might affect cells cultured under nutrient-deprived conditions where autophagy is induced. Cells cultured in amino acid-free media were treated with αMSH-PEG-C′ dots and examined by time-lapse imaging. While amino acid deprivation was well-tolerated by M21 cells in the absence of particles, the treatment of amino acid-deprived cells with 15 μM αMSH-PEG-C′ dots, which had no effect on cells in nutrient-replete media, surprisingly led to cell death at high rates, detected by the uptake of Sytox Green, a membrane-impermeable nucleic acid dye that labels cells with ruptured plasma membranes (Fig. 2c). This demonstrates that while αMSH-PEG-C′ dots are generally well-tolerated, nutrient-deprived cancer cells are sensitive to treatment. To determine whether this finding could have consequences for tumor growth in vivo, we incubated M21 cells with αMSH-PEG-C′ dots in culture under nutrient-replete conditions, which had no effect on cell viability (Fig. 2a and Supp. Fig. 1a), and then injected particle-exposed cells, as well as particle-unexposed cells, into mice as flank tumor xenografts, to promote a nutrient-deprived state that is known to result from a lack of vascularization in early xenograft tumors16. M21 cells loaded with αMSH-PEG-C′ dots demonstrated statistically significant growth-inhibition (p<0.001) relative to non-particle exposed cells (Fig. 2d). In fact, no measurable tumor growth occurred from particle-exposed cells up to 10 days following cell injection. These findings suggested that treatment with αMSH-PEG-C′ dots at high concentrations may induce cell death under conditions of nutrient deprivation in culture and in vivo.
αMSH-PEG-C′ dot-induced death occurs by ferroptosis
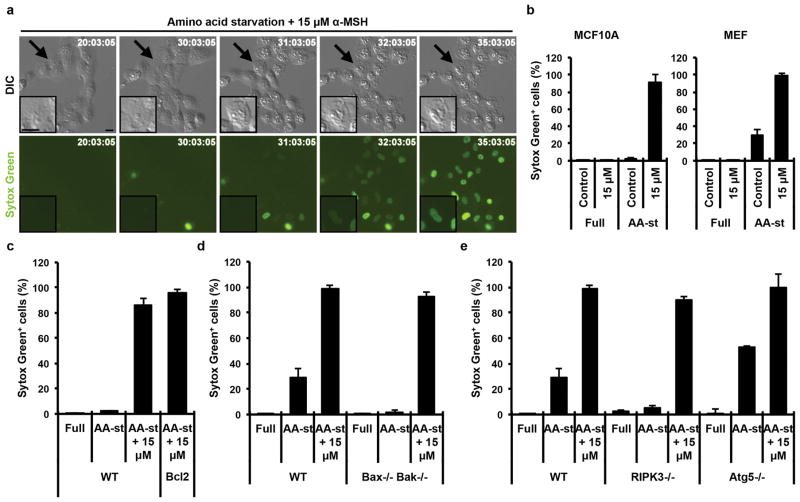
We sought to identify the mechanism of how cells treated with αMSH-PEG-C′ dots undergo cell death under nutrient-deprived conditions. Careful inspection of the morphology of dying cells suggested a form of necrosis, involving cell swelling and plasma membrane rupture, in the absence of cell blebbing and fragmentation that is typically observed during apoptosis (Fig. 3a, Supp. Fig. 1b). In order to more definitively identify the mechanism of cell death, we utilized two non-tumor cell lines, MCF10A human mammary epithelial cells and mouse embryo fibroblasts (MEF), that were also observed to die at high rates when cultured in amino acid-free media in the presence of αMSH-PEG-C′ dots (Fig. 3b). Cells rendered resistant to apoptosis, by overexpression of the anti-apoptotic protein Bcl-2 (MCF10A-Bcl2) (Supp. Fig. 1c) or by genetic deletion of Bax and Bak (Bax/Bak−/− MEFs)17,18, underwent cell death at rates similar to control cells, suggesting that αMSH-PEG-C′ dot-induced cell death does not occur by apoptosis (Fig. 3c, d). Next, we examined if cell death was occurring by necroptosis, a programmed form of necrosis that requires the RIPK3 kinase19. Ripk3−/− knockout MEFs, which are resistant to necroptosis (Supp. Fig. 1d), also underwent cell death at rates similar to controls, suggesting that nanoparticle treatment does not induce necroptosis (Fig. 3e). We further determined if a recently-described form of cell death involving the autophagy pathway, autosis20, could be involved, by treating autophagy-related gene 5 knockout MEFs (Atg5 −/− MEFs), completely deficient for autophagy, with α-MSH-PEG-C′ dots in the absence of amino acids. Atg5 −/− MEFs underwent cell death at rates similar to those of controls, demonstrating that αMSH-PEG-C′ dot-induced cell death does not involve autophagy, and is not autosis (Fig. 3e). Collectively, the foregoing data demonstrate that cell death, induced by a combination of particle treatment and amino acid deprivation, occurs independently of apoptosis, necroptosis, and autosis.
Figure 3. αMSH-PEG-C′ dot particle-induced cell death is not apoptosis, necroptosis or autosis.
a, MCF10A human mammary epithelial cells cultured in the absence of amino acids with 15μM αMSH-PEG-C′ dots undergo cell death after 30 hours with necrotic features. Insets show a dying cell indicated by an arrow. Fluorescence images show Sytox green-labeling of dead cell nuclei. Scale bar=10μm. b, Quantification of cell death (Sytox green +) in MCF10A and mouse embryo fibroblast (MEF) cultures in full media or amino acid-starved (AA-st) conditions in the presence or absence of 15μM αMSH-PEG-C′ dots, and after 40 hours (MCF10A) or 45 hours (MEF), as determined by time-lapse microscopy. Error bars indicate mean+/− standard deviation. N=5 per group. Each replicate is from one biological experiment, quantified with five independent fields of view. c–e, Cell death assays, as in (b), indicate that inhibition of apoptosis by Bcl2 overexpression in MCF10A (c), quantified after a 38 hour time-lapse experiment, or deletion of Bax and Bak in MEF (d), quantified after 45 hours, or inhibition of necroptosis by deletion of ripk3 in in MEF (e), quantified after 45 hours, or inhibition of autophagy by knockout of Atg5 in MEF after 45 hours (e) does not inhibit cell death induced by amino acid starvation and treatment with 15μM αMSH-PEG-C′ dots. Error bars indicate mean+/-standard deviation. N=5 per group. Each replicate is from one biological experiment, quantified with five independent fields of view.
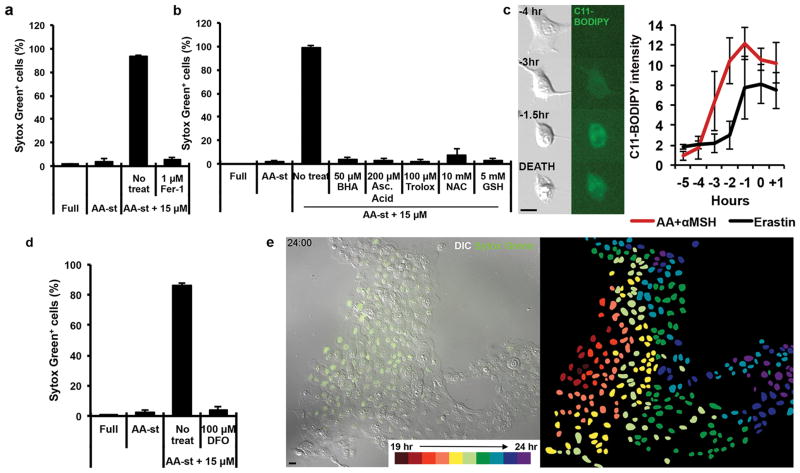
We next examined if ferroptosis, a recently described cell death mechanism that occurs via an iron- and lipid reactive oxygen species (ROS)-dependent process, and is induced by glutathione depletion21, could be involved in αMSH-PEG-C′ dot-induced cell death. We first tested whether ferrostatin-1 and liproxstatin-1, pharmacological inhibitors of ferroptosis, that are scavengers of lipid ROS, could block cell death in this context. Treatment with either inhibitor rescued cell viability, reducing cell death to levels occurring under amino acid-deprived conditions in the absence of nanoparticles (Fig. 4a and Supp. Fig. 1e). Nanoparticle-induced cell death was also inhibited by treatment with other antioxidants, including butylated hydroxyanisole (BHA), ascorbic acid (Asc Acid) and trolox, or, alternatively, by glutathione repletion through the addition of glutathione or N-acetylcysteine (NAC), a precursor of glutathione (Fig. 4b). To examine if lipid ROS accumulate during nanoparticle-induced cell death, we imaged particle-exposed cells in the presence of the lipid oxidation indicator C11-BODIPY. Increased staining prior to cell death was seen to occur in response to treatment with the known ferroptosis-inducing agent erastin, in a liproxstatin-1-inhibitable manner (Fig. 4c, Supp. Fig. 1f). Like cell death induced by erastin treatment, lipid ROS detected by C11-BODIPY staining accumulated several hours prior to the induction of cell death by nanoparticle treatment under amino acid-free conditions (Fig. 4c). To further examine if nanoparticle-induced death is dependent on iron, a known requirement for ferroptosis, we found that cells treated with deferoxamine (DFO), an iron chelator used for treating iron overload and an agent reported to block ferroptosis21, almost completely inhibited cell death (Fig. 4d). Together these data demonstrate that treatment of amino acid-starved cells with high αMSH-PEG-C′ dot concentrations induces ferroptosis. Interestingly, ferroptosis in this context was also observed to propagate from cell to cell in a wave-like manner (Fig. 4e, Supplementary Movie 1), unlike that found for cells undergoing other types of death, such as apoptosis (Supp. Fig. 1g, Supplementary Movie 2). These findings suggested cell-cell communication of a death-inducing signal, similar to a recent report of ferroptosis occurring in renal tubules in response to treatment with erastin22.
Figure 4. Ferroptosis is the underlying mechanisms of αMSH particle-induced cell death.
a, Quantification of cell death (Sytox green +) in MCF10A cells cultured in full media (Full) or amino acid-starved (AA-st) conditions in the presence or absence of 15μM αMSH-PEG-C′ dots and (a) 1μM Ferrostatin-1 (Fer-1) after 40 hours. Error bars indicate mean +/− standard deviation. N=3 per group. Each replicate is from one biological experiment, quantified with five independent fields of view. b, Quantification of cell death (Sytox green +) in MCF10A cells cultured in full media (Full) or amino acid-starved (AA-st) conditions in the presence or absence of 15μM αMSH-PEG -C′ dots and 50νM butylated hydroxyanisole (BHA), 200νM ascorbic acid (Asc Acid), 100νM Trolox, 10mM N-acetylcusteine (NAC), or 5mM glutathione (GSH) after 40 hours. Error bars indicate mean +/− standard deviation. N=5 per group. Each replicate is from one biological experiment, quantified with five independent fields of view. c, Lipid ROS accumulate prior to death in cells treated with 15μM αMSH-PEG-C′dots and amino acid withdrawal. Images show treated cells cultured in the presence of C11-BODIPY that detects lipid ROS. Note that the fluorescence intensity of C11-BODIPY increases several hours before cell death (times indicated on each image prior to cell death in bottom image). Scale bar=10μm. Graph: quantification of C11-BODIPY fluorescence in particle-treated and amino acid-starved cells (red line), or erastin-treated cells (black line). Mean intensities out of five cells +/− standard deviation are shown from one biological experiment. Time zero indicates the time of cell death determined by DIC microscopy. Note that C11-BODIPY staining increases in intensity between three and four hours prior to cell death. d, Quantification of cell death (Sytox green +) in MCF10A cells cultured in full media (Full) or amino acid-starved (AA-st) conditions in the presence or absence of 15μM αMSH-PEG-C′ dots and 100μM deferoxamine (DFO). Error bars indicate mean +/− standard deviation. N=3 per group. Each replicate is from one biological experiment, quantified with five independent fields of view. e, Images from time-lapse analysis of MCF10A undergoing ferroptosis in amino acid-starved conditions with 15μM αMSH-PEG-C′ dots. Note that death (Sytox green positivity) spreads cell-to-cell from the left side of the image to the right. Right image shows the position of each dead cell nucleus pseudocolored to represent the relative timing of cell death. Scale bar=10μm.
αMSH-PEG-C′ dots inhibit tumor growth
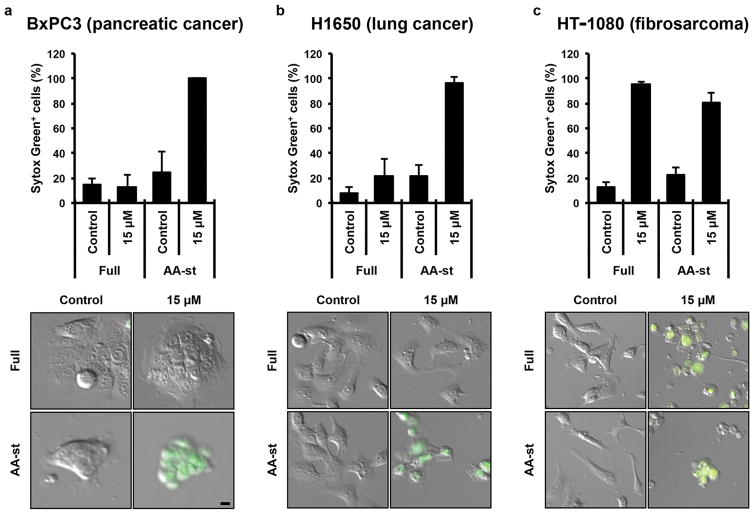
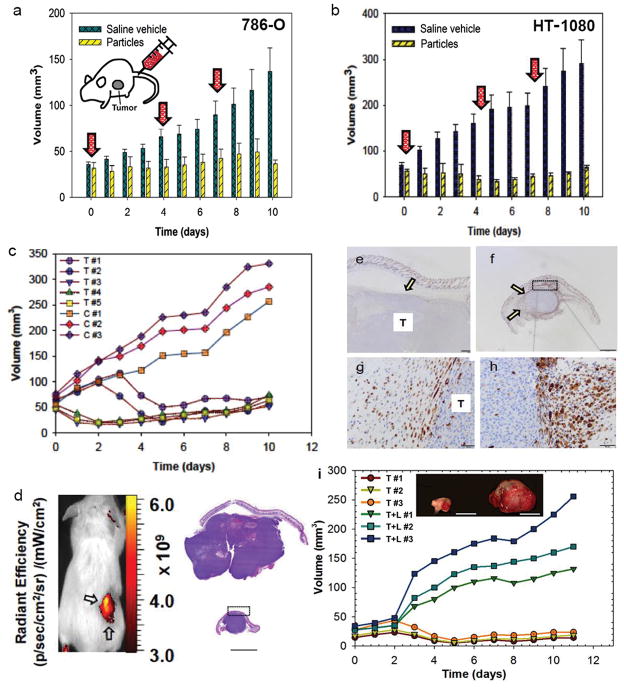
We next examined if ferroptosis could be induced by nanoparticle treatment in a wider panel of cancer cells. Like M21 cells, BxPC3 pancreatic carcinoma cells, H1650 lung carcinoma cells, HT-1080 fibrosarcoma cells, and 786-O renal carcinoma cells underwent high rates of necrosis when treated under amino acid free conditions with αMSH-PEG-C′ dots, indicating that cell death can be induced by a combination of particle treatment and nutrient deprivation in a variety of different cancer cell types (Fig. 5a–c, Supp. Fig. 2a). Interestingly, HT-1080 cells underwent necrosis in response to nanoparticle treatment even when cultured in full media (Fig. 5c), as well as in starvation media at 10-fold lower particle concentrations (Supp. Fig. 2b), suggesting that these cells are particularly sensitive to particle-induced ferroptosis. We further investigated whether particle-induced treatment responses could be generated in 786-O renal carcinoma and HT-1080 fibrosarcoma xenograft models. Using a multi-dosing delivery scheme, tumor growth was assessed over a 10-day period after three high-dose intravenous (i.v.) treatments of either targeted particle probe (n = 5) or 0.9% saline solution (n = 3) in immunosuppressed mice bearing flank 786-O or HT-1080 tumors. Relative to the rapidly increasing tumor volumes measured after injection with saline vehicle, statistically significant inhibition of tumor growth was observed with multiple-dose particle treatments for both tumor types (Fig. 6a,b), greater for HT-1080 xenografts. Surprisingly, this was accompanied by partial tumor regression exceeding 50% for all particle-treated HT-1080 tumors (range: 57%-78%; mean 64%) within a 4–5 day interval after initial injection (Fig. 6c). At study termination, statistically significant reductions in treated tumor volumes (HT-1080: mean ~85%; p<0.001; 786-O: mean ~73%; p<0.01) were found relative to control volumes (Figs. 6a,b). Intense fluorescence signal overlying the xenograft by whole-body optical imaging (Fig. 6d) suggested particle localization. Immunohistochemical staining for the macrophage marker, Mac-2, revealed large numbers of recruited macrophages surrounding treated tumors relative to that seen around control tumors, both at low and high magnification (Figs. 6e–h).
Figure 5. αMSH-PEG-C′ dots induce cell death in different types of cancer cells.
a–c, Quantification of cell death (Sytox green +) in (a) BxPC3 pancreatic carcinoma cells after 40 hours, (b) H1650 lung carcinoma cells after 45 hours and (c) HT-1080 fibrosarcoma cells after 65 hours, in full media (Full) or amino acid-free media (AA-st), in the presence or absence of 15μM αMSH-PEG-C′ dots. Error bars indicate mean+/-standard deviation. For parts a,b, N=5 per group. For part c, N=3 per group. Each replicate is from one biological experiment, quantified with five independent fields of view. Scale bars=10′m.
Figure 6. αMSH-PEG-C′ dots inhibit tumor growth in 786-O and HT-1080 xenograft models.
a,b, Graphical summary of 786-O (a) and HT-1080 (b) average tumor volume measurements in αMSH-PEG-C′ dot-treated (‘particles’; n=5) and control (‘saline-vehicle; n=3) mice; error bars indicate standard deviation. Three, high-dose (12 nanomoles (nmol)/dose) αMSH-PEG-C′ dot or saline vehicle control treatments (30 micromoles/dose) were i.v.-injected (arrows) over a 10-day period, with particle-treated tumours, on average, demonstrating growth inhibition, greater for HT-1080, relative to saline-treated controls. c, Individual HT-1080 tumor volume measurements from part (b) are shown for particle-treated (‘T’) versus saline-treated (‘C’) mice. Relative to control tumor volumes, data show marked inhibition of tumor growth and partial tumor regression after particle treatments (HT-1080: p<0.001; 786-OL P<0.01). P-values are from a Wald test in a regression model estimated by generalized estimating equations to take into account the longitudinal nature of the data. d, Left image: whole body Cy5 fluorescent imaging of a representative H1080 tumor xenograft (arrows). Right image: low-power view of H&E-stained tissue sections from representative control (top) and treated (bottom) tumors reveal a densely cellular and invasive neoplasm exhibiting multifocal necrosis. Control specimens are noted to be disproportionately larger in size than the corresponding treated ones without evident morphological differences. e–h, Immunohistochemical staining of tumor sections with macrophage marker, Mac-2, shows scattered macrophages (arrow) surrounding control tumor sections (T) on (e) low and (g) high power views, while corresponding (f) low and (h) high power views of Mac-2 stained treated sections show large numbers of Mac-2 positive cells circumscribing the tumor at similar locations (boxes, d, f; arrows). Small numbers of intratumoral Mac-2 positive cells are also noted. i, Graphical summary of individual HT-1080 tumor volume measurements in mice undergoing combined inhibitor (Liproxstatin-1) and particle treatment (T+L; n=3) versus particle treatment alone (T; n=3). Three high-dose (12 nmol/dose) αMSH-PEG-C′ dot treatments (with and without i.p.-injected Liproxstatin-1) were given over a 10-day period. Relative to particle treatment alone, marked progression of tumor growth is seen following combined inhibitor and particle treatment (p<0.001). Image: Representative particle-exposed tumors reveal specimens to be disproportionately larger in size when additionally treated with Liproxstatin-1 (right tumor). Scale bars: 1 mm (d, e, and f); 50 μm (g, h); 1 cm (j).
To further examine if the inhibition of tumor growth resulting from nanoparticle treatment could be related to ferroptosis, we treated tumor-bearing mice with daily intraperitoneal (i.p.) doses of liproxstatin-1 for 10 days to determine the effects on particle-induced tumor shrinkage. Notably, in HT-1080 xenografted mice (n = 3) subsequently administered three high-dose particle treatments, liproxstatin-1 administration significantly reduced growth inhibition to levels nearly equivalent to those seen in non particle-exposed tumors (Fig. 6i). Using a generalized estimating equations (GEE) model with a logit link, average daily growth in particle-exposed tumors treated with liproxstatin-1 was 14.6 mm3 (95% CI: 10.1–18.9), as compared with −0.87 mm3 (95% CI: −1.06 to −0.69) for particle treatment alone, a difference of 15.3 mm3 (CI: 13.1 to 17.6; p <0.001, Wald test from GEE). Corresponding particle-exposed tumor specimens were significantly smaller on average than particle-treated tumors receiving daily liproxstatin-1 (Fig. 6i, inset).
Conclusions
Here we demonstrate that the combined treatment of cells with αMSH-PEG-C′ dots and amino acid starvation can synergize to induce the cell death program, ferroptosis, and that high-dose particle delivery can inhibit tumor growth and cause tumor regression; these effects are reversible with liproxstatin-1. Although their role in wound repair (through engulfment of cellular debris) is well established, the significance of increased numbers of macrophages around particle-treated, as opposed to control, tumours remains unclear23. It is known that a high degree of macrophage plasticity can occur in response to local cues from the tumor microenvironment24, and that, upon activation, macrophages can assume a spectrum of roles needed to maintain tissue homeostasis25, GPX including shifts in function associated with tumor shrinkage24.
These data thus define a potential therapeutic application for molecularly-targeted C′ dots, already in clinical trials for cancer imaging and detection2, but without the need for surface-attached cytotoxic agents. An important question raised by these findings is how these particles induce ferroptosis. Surface-modification of particles with αMSH peptides for targeting cancers enhances cellular internalization (data not shown). Interestingly, particle surface modification with αMSH is not required for ferroptosis in the cell lines tested. We have observed its induction, albeit more slowly, with unmodified PEGylated C′ dots. The slower rate of ferroptosis induction may reflect a slower rate of internalization of unmodified particles relative to αMSH-modified platforms (Supp. Fig. 3a).
We therefore have considered that the native silica particle itself has ferroptosis-inducing activity, as deprotonated surface silanol groups and/or fractal internal structure may lead to iron adsorption and/or incorporation (i.e., loading) within its structure. We have indeed found iron loading of αMSH-PEG-C′ dots incubated with culture media, as compared with doubly de-ionized water preparations (Supp. Fig. 3b, Supp. S1). Increasing amounts of iron were loaded into particles in a concentration-dependent manner upon exposure to ferric oxide solutions. These findings were accompanied by a decrease in iron-loading capacity as they became saturated at high iron concentration (Supp. S1). Further, increased intracellular iron levels were found for αMSH-PEG-C′ dot nanoparticle-treated cells compared to non-treated cells (Supp. Fig. 3b, Table S1). Consistent with a model in which particles deliver iron into cells, particle-treated cells upregulated expression of the heavy chain of ferritin (FTH1) that binds cytosolic iron (Supp. Fig. 3c). We further found that iron loading into cells by treatment with ferric ammonium citrate (FAC) is sufficient to mimic particle treatment and induce ferroptosis in amino acid-starved cells (Supp. Fig. 3d), suggesting that nanoparticles may engage ferroptosis by loading iron into cells.
Increased iron uptake could lead to the depletion of glutathione, conceivably due to increased ROS generation. We do find suppression of glutathione levels in particle-treated cells (Supp. Fig. 3e). Pre-treatment with erastin, which inhibits glutathione production by blocking cystine uptake, sensitizes cells to particle-induced ferroptosis, suggesting that glutathione depletion is rate-limiting for particle-induced death (Supp. Fig. 3f). Taken together, our data support a model whereby particle-induced ferroptosis is executed following iron uptake into cells, suppression of glutathione, and accumulation of lipid ROS (Supp. Fig. 4). Lipid ROS may accumulate in glutathione-suppressed cells due to lowered activity of the glutathione peroxidase 4 (GPX4) enzyme that protects cells against lipid peroxidation and inhibits ferroptosis26,27. We have not found that particle treatment inhibits GPX4 activity in an enzymatic assay from treated cell lysates (Supp. Fig. 3g), consistent with the model that particle treatment does not lead to lipid peroxidation by direct inhibition of GPX4. We additionally note that particle-induced ferroptosis does not require the presence of glutamine, unlike ferroptosis occurring in starved cells28, as our studies also utilize amino acid-free conditions in which enhanced iron uptake may bypass requirements for glutamine.
While the concentration of nanoparticles used here to either induce in vitro cell death or inhibit in vivo tumor growth is at least four orders of magnitude higher than what is used currently in human subjects for single-dose imaging-based studies2, local concentrations could be driven to much higher levels at tumor sites as part of a multi-dosing strategy, combinatorial treatment regimen, and/or by direct catheter infusion at the target site. Such a dosing schedule would be designed to yield maximum tumor-to-background ratios while reducing off-target toxicities and promoting efficient renal clearance. It is notable that the leakiness of tumor vasculature is thought to allow the accumulation of systemically injected nanoparticles in tumor tissues29, while contributing to nutrient deprivation within tumors, suggesting that the synergism between nanoparticles and nutrient deprivation may be restricted to tumor sites in vivo. Some cancers may also be particularly sensitive to this mechanism of cell death, which could lower the threshold of particle concentration needed to achieve an anti-tumor effect. Indeed, we have found that HT-1080 cancer cells undergo αMSH-PEG-C′ dot-induced ferroptosis even under nutrient-replete conditions (Fig. 5c), and these cells are also killed in amino acid-deprived conditions with 10-fold lower nanoparticle concentration (Supp. Fig. 2b). It is notable that HT-1080 tumors are well-vascularized (Supp. Fig. 5a), suggesting that the sensitivity of these cancer cells to particle-induced death, even under nutrient-replete conditions (Fig. 5c), may contribute to the strong anti-tumor effect that is observed following intravenous particle delivery.
While conceivably the sensitivity of some cancers could lower the particle concentrations needed to exert an anti-tumor effect, we have also found that multiple high-dose αMSH-PEG-C′ dot treatments are well tolerated. Evaluation of tumor, hepatic, renal, and hematologic specimens from particle- and vehicle-treated HT-1080- and 786-O-bearing mice, performed at the termination of the study, showed no significant group differences in complete blood counts, serum chemistries, and hepatic and renal histopathology, with the exception of indirect and total bilirubin serum concentrations, which were moderately elevated in particle-treated mice as against controls (Supp. Tables S2, S3). However, no mechanism could be ascribed to this finding given the lack of hemolysis on complete blood counts and absence of liver injury on histopathology and serum chemistries. The extent of macrophage staining among particle-treated and vehicle-treated tumor specimens is also summarized (Supp. Table S4).
The discovery of nanoparticle-induced ferroptosis as a redox modulator of cell fate, as well as a mediator of tumor regression and growth inhibition, suggests that it may be possible to exploit this process therapeutically to synchronously and selectively kill those cancers most susceptible to this mechanism. Elucidating the basis for such differential tumor sensitivity is essential. It will be important, for instance, to assess the extent of tumor vascularity, as this parameter will critically influence particle delivery, nutrient and oxygenation status, and treatment response. As such, its assessment across a variety of pre-clinical tumor types may enable better screening of models suitable for induction of cell death by this mechanism, as well as for stratifying patients in the future to appropriate combinatorial treatment paradigms.
It will also be critical to determine whether controlled variations in C′ dot properties –structure, composition, or surface chemistry– enhance, or even abrogate, induction of ferroptosis. We found, for instance, that C′ dot size plays a critical role in the magnitude of the effect observed under amino acid-deprived conditions (Supp. Fig. 5b). Smaller diameter (~6 nm) PEGylated C′ dots led to significantly higher percentages of Sytox Green labeled HT-1080 cells (i.e., 80%) cells compared to that found for larger diameter (~10 nm) C′ dots (~25%).
Finally, we note that while the response to liproxstatin-1 provides evidence for ferroptosis as an anti-tumor mechanism in vivo, other mechanisms may be contributory, including modulation of the tumor microenvironment, given marked macrophage recruitment to particle-treated tumors (Fig. 6g,h, HT-1080; Supp. Fig. 5c, 786-O)24, an area under active investigation. Altogether, our findings lay the groundwork for exploiting these ultrasmall silica nanoparticles as effective ferroptosis-inducing agents in sensitive tumors.
Materials and Methods
Cell culture and constructs
MEF and HT-1080 cells (American Type Culture Collection (ATCC)) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (MSKCC Media Preparation Facility) supplemented with 10% fetal bovine serum (FBS, Sigma, St. Louis, MO) with penicillin/streptomycin (Corning, Corning, NY). MCF10A cells (ATCC) were cultured in DMEM/F12 (Gibco, Grand Island, NY) supplemented with 5% horse serum (Atlanta Biologicals, Flowery Branch, GA), 20 ng/ml EGF (Peprotech, Rocky Hill, NJ), 10 μg/ml insulin (Sigma), 0.5 μg/ml hydrocortisone (Sigma), and 100 ng/ml cholera toxin (Sigma) with penicillin/streptomycin. M21 (ATCC), BxPC3 (ATCC), H1650 (ATCC) and 786-O (ATCC) cells were cultured in RPMI-1640 (Gibco) supplemented with 10% FBS with penicillin/streptomycin. Cell lines have not been authenticated, as these were purchased from ATCC, except for MEFs, which were validated with functional studies as being the correct knockouts. Cells were routinely verified as mycoplasma-free by DAPI imaging. Amino acid-free medium was prepared by dialyzing heat-inactivated FBS (for MEF, HT-1080, M21, BxPC3, and H1650 cells) or horse serum (for MCF10A cells) for 4 h, followed by overnight incubation at 4°C in phosphate-buffered saline (PBS) in MWCO 3500 dialysis tubing (21-152-9; Fisherbrand, Pittsburgh, PA) and addition to base media prepared without amino acids. pRetro–Lamp1–GFP was introduced into M21 cells by retroviral transduction, and stable cell lines were selected with puromycin (2 μg/ml). Sample sizes for all cell studies were chosen to comply with logistical and financial constraints, and they are all commensurate with the traditions of the field.
Reagents
The following reagents were used at the indicated concentrations: Concanamycin A (ConA) (Sigma) 100 nM; SYTOX Green Nucleic Acid Stain (S7020; Invitrogen, Carlsbad, CA) 5 nM; Ferrostatin-1 (Fer-1) (EMD Millipore, Billerica, MA) 1 μM; Liproxstatin-1 (Selleckchem) 1 μM and 125 mg/kg for in vitro and in vivo, respectively; Deferoxamine (DFO) (Sigma) 100 μM; Butylated hydroxyanisole (BHA) (Sigma) 50 μM; Ascorbic acid (Asc Acid) (Sigma) 200 μM; Trolox (Sigma) 100 μM; N-acetylcysteine (NAC) (Sigma) 10 mM; Glutathione (GSH) (Sigma) 5 mM; TNFα (Sigma) 100 ng/ml; Cycloheximide (CHX) (Sigma) 1 μg/ml and 50 μg/ml to induce necroptosis and apoptosis, respectively; zVAD (Sigma) 20 μM; Necrostatin-1 (Sigma) 30 μM; Buthionine sulphoximine (BSO) (Sigma) 400 μM; Ferric ammonium citrate (FAC) (Sigma, F5879) 400 μM; Erastin (Sigma), 5 μM, C11-BODIPY(581/591) 2 μM (Invitrogen). Reagents were added to cultures at the start of biological assays with the exception of ConA which was added 1 hour prior to lysis for western blotting.
Peptide synthesis
A modified melanocortin-1 receptor targeting peptide Re(Arg11)CCMSH14 with a double aminohexanoic acid (Ahx2) aliphatic linker and N-Ac-Cys was synthesized using standard solid phase Fmoc peptide chemistry. The rhenium-cyclized αMSH peptide analog, Ac-Cys1-(Ahx)2-dLys2-Re[Cys-Cys-Glu-His-dPhe-Arg-Trp-Cys]-Arg-Pro-Val-NH2, was analyzed and purified on a Beckman Coulter High Performance Liquid Chromatography (HPLC) system coupled with an LCQ FLEET Ion Trap Mass spectrometer (Thermo Fisher Scientific) and finally recovered by lyophilization.
Synthesis and characterization of αMSH-PEG-C′ dots
Fluorescent silica nanoparticles (C′ dots) of different size, encapsulating the organic dye, Cy5, were synthesized in water as previously described30. αMSH peptides were conjugated to maleimido-terminated heterobifunctional polyethylene glycol silane (mal-PEG-silane) via its N-terminal acetylated cysteine thiol to form αMSH-PEG-silane. Conjugates were attached to the particle surface in the PEGylation step as described previously10,30 to generate αMSH functionalized C′ dots, or αMSH-PEG-C′ dots. Synthesized particle samples were dialyzed in water and purified by gel permeation chromatography (GPC, Bio-Rad Laboratories, Inc, Hercules, California) prior to further characterization10. Absorption and emission spectral profiles for the encapsulated and native Cy5 dye were obtained using a Varian Cary 5000 spectrophotometer (Varian, Palo Alto, CA) and a fluorescence spectrofluorometer (Photon Technology International, Inc, Birmington, NJ). Hydrodynamic radius, brightness, and concentration of αMSH-PEG-C′ dots, as against free Cy5 dye, were determined using a homebuilt fluorescence correlation spectroscopy (FCS) set up configured with a solid-state 633-nm excitation10.
Western blotting
Cells were scraped into ice-cold RIPA buffer (50 mM Tris at pH 7.4, 150 mM NaCl, 2 mM EDTA, 1% NP40, 0.1% SDS with protease inhibitor cocktail) and lysed for 10 min on ice. Lysates were then centrifuged at 15,870g for 20 min at 4 °C and protein was quantified by BCA assay (Pierce, Waltham, MA). Samples were separated on 15% polyacrylamide SDS–PAGE gels and transferred to a polyvinyldifluoride membrane which was blocked with TBST plus 5% BSA and incubated overnight at 4°C with primary antibodies (anti-LC3A/B (4108; Cell Signaling, Danvers, MA), anti-FTH1 (3998; Cell Signaling), and anti-Actin (A1978; Sigma)), diluted in blocking buffer. Blots were incubated with horseradish peroxidase conjugated to secondary antibodies and protein was detected using enhanced chemiluminescence detection (Invitrogen). Densitometry analysis was carried out using ImageJ software (NIH).
Time-lapse microscopy
Cells were plated onto glass-bottom dishes (MatTek, Ashland, MA) overnight and fluorescence and differential interference contrast (DIC) images were acquired every 30 min for indicated times using a Nikon TI-E inverted microscope, a CoolSNAP HQ2 CCD (charge-coupled device) camera (Photometrics, Tucson, AZ), a live-cell incubation chamber to maintain cells at 37 °C and 5% CO2, and NIS Elements software (Nikon, Melville, NY). Cell fates, including cell survival, death, and proliferation, were manually quantified and processed using NIS Elements software and Image J.
Glutathione quantification
For glutathione measurements, HT-1080 cells were plated on 6cm cell culture dishes, incubated with 15 μM αMSH-tagged nanoparticles in amino acid-free DMEM + 10% dialyzed FBS and harvested approximately 2 hours before the expected time of death. As controls, cells were treated with a mix of 3 parts full or amino acid-free DMEM and 1 part H2O, or 100 μM BSO in full media. Cells were washed 3× with cold PBS and harvested through cell scraping in 50 μL cold lysis buffer. Protein concentrations were measured using the BCA assay and sample volumes were adjusted so that each had the same final protein concentration. Total glutathione was measured using the Glutathione Assay Kit (Cayman Chemicals, 703002) and reduced glutathione was measured using the QuantiChromTM Glutathione Assay Kit (BioAssay Systems, DIGT-250) according to manufacturer’s instructions.
Iron measurements
For iron loading capacity measurements of C′ dots, 50 μL αMSH-PEG-C′ dots (60 μM) were added to either 150 μL iron-containing media (8.6μM) or to 150 μL FeCl3 solutions prepared over a range of iron (Fe3+) concentrations (2 μM to 2 mM, Table S1). Solutions were spun at room temperature (160 rpm) for 48 hours, followed by separation of free iron from C′ dots using a PD-10 column, and eluting in water. Cellular samples were incubated with and without particles in media for 48 hours prior to centrifugation, pelleted, and washed three times prior to re-suspending in phosphate buffered saline solution. Iron measurements (parts per billion, ppb) were determined using Microwave Plasma - Atomic Emission Spectroscopy, along with iron-loading capacities. Iron loading capacity was computed as the ratio of the amount of iron in purified particle-exposed C′ dots (or cells), divided by the total amount of initial iron measured prior to purification, and multiplied by 100. All experiments were performed in triplicate.
GPX4 Activity Assay
The Gpx4 specific activity assay was performed according to Roveri and colleagues31. In brief, frozen cell pellets were resuspended in 100 μL lysis buffer (100 mM KH2PO4/K2HPO4, pH 7.4, 1 mM EDTA, 150 mM KCl, 0.1 % CHAPS, 3 mM β-mercaptoethanol and protease inhibitor cocktail) and homogenized by 50 pestle strokes. Samples were incubated for 15 min on ice and cell debris was removed by centrifugation (20000 g and 10 min, 4°C). 50 μL of the supernatant from homogenized cells were used to measure the enzymatic activity in 1 ml of assay buffer (100 mM TrisHCl pH 7.8 containing 5 mM EDTA, 0.1 % Triton X, 3 mM GSH, 200 μM NADPH and 0.6 U/ml glutathione reductase) in the presence of 20 μM phosphatidylcholine hydroperoxide (PCOOH). Gpx4 activity is determined by the glutathione reductase-dependent consumption of NADPH detectable by a decrease in absorbance at 340 nm in a SpectraMax plate reader (Molecular Device GmbH). Protein content in the samples was determined by the colorimetric 660 nm Pierce Protein Assay method (Pierce, Waltham, MA).
Animal models and tumor inoculation
All animal experiments were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee of Memorial Sloan-Kettering Cancer Center and followed NIH guidelines for animal welfare. Human melanoma (M21) xenografts were generated on the shaved flanks of immunodeficient male SCID/Beige C.B-17/IcrHsd-PrkdcscidLystbg-J mice (6–8 weeks old, Harlan Laboratories, South Easton, MA). Human sarcoma HT-1080 and 786-O flank xenografts (~2.0 × 106 cells/100 μl) were additionally generated in the same model. Average initial tumor volumes of 45 – 75 mm3 were used for all studies. For all in vivo studies, three animals per group were used. No randomization was used and no blinding was performed.
In vivo dosing strategy and examination
Five million M21 cells, cultured in serum-supplemented media, were subcutaneously implanted into the right flank of mice using a 23-gauge trocar needle to establish melanoma xenografts 2 days after particle exposure (n=3) or without exposure (n=3). In a subsequent study, mice were assigned to one of two different treatment groups to evaluate the response of HT-1080 and 786-O tumors to high-concentrations (60 μM) of i.v.-injected αMSH-PEG-C′ dots (n=5 mice; 200 μl) administered three times over a 10-day period (i.e., days 0, 4, 7). Control HT-1080 and 786-O mice (n=3) were administered 0.9% saline vehicle at the same time points. In a third treatment study, HT-1080 mice were assigned to one of two groups to evaluate response to three, high-concentration doses (60 μM) of i.v.-injected αMSH-PEG-C′ dots (n=3 mice; 200 μl) alone or following intraperitoneal administration of liproxstatin-1 (n=3 mice, 125 mg/kg) over a 10-day period. Tumor sizes were measured using calipers over the treatment interval. All mice were examined by palpation at the site of tumor cell inoculation, and were observed daily over until the termination of tumor growth studies for signs of morbidity or mortality. Two perpendicular diameters (d1 ≤ d2) of the tumor used to calculate the tumor volume (V; 4/3·π·d12·d2/8) were measured with calipers daily following the injection of cells.
In vivo fluorescence imaging
Animals were anesthetized using isoflurane and whole body optical fluorescence imaging was acquired to identify nanoparticle fluorescence at the tumor site. Mice were scanned for 0.1 to 1 seconds using the IVIS Spectrum photon-counting device optical imaging system (Xenogen, Alameda, CA) with the blocks and filters for Cy5 fluorescence (excitation 650 nm, emission 680 nm), and for background fluorescence (excitation 465 nm, emission 600 nm), selected according to the manufacturer’s recommendations. Fluorescence background was likewise subtracted according to the manufacturers’ instructions. Fluorescence signal was reported as radiant efficiency ((photons/s/cm2/sr)/mW/cm2).
Histopathologic analysis
Immediately after terminating the in vivo imaging study, HT-1080 and 786-O male and female mice were euthanized by CO2 inhalation, and representative particle-exposed (n=2) and control (n=1) tumors, as well as hepatic and renal specimens, were excised at necropsy. Hematologic specimens were additionally obtained for complete blood counts and serum chemistries. Excised tumors, livers, and kidneys were fixed in 10% neutral buffered formalin for 24 hours, processed in alcohol and xylene, embedded in paraffin, sectioned at 5-micron thickness, and stained with hematoxylin and eosin (H&E). Additional tumor sections were stained by immunohistochemistry for Mac-2 (primary antibody Cedarlane CL8942B applied at concentration of 1:100 following heat-induced epitope retrieval [HIER] in a pH 6.0 buffer), myeloperoxidase (Dako A0398, 1:1000, HIER pH 6.0), cleaved caspase-3 (Cell Signaling Technology 9661, 1:250, HIER pH6.0), and Ki-67 (Abcam ab16667, 1:100, HIER pH 9.0). Mac-2 staining was performed manually with an avidin-biotin detection system (Vectastain ABC Elite Kit, Vector Laboratories, PK-6100). Other stains were performed on a Leica Bond RX automated stainer using the Bond Polymer Refine detection kit (Leica Biosystem DS9800). Tumor sections were also stained by the TUNEL method as previously described. All slides were examined by a board-certified veterinary pathologist.
Tumor Vascularity Assessment
Tumor vascularity was assessed by staining HT-1080 tumors by immunohistochemistry for CD31 on a Leica Bond RX automated staining platform (Leica Biosystems). Following heat-induced epitope retrieval at pH 9.0, the primary antibody (rat monoclonal, catalog #DIA-310; Dianova) was applied at a concentration of 1:250 and was followed by application of a polymer detection system (Novocastra Bond Polymer Refine Detection, Leica Biosystems).
Statistics
Volume-time profiles were compared between the two treatment groups using robust standard errors calculated by a generalized estimating equations approach32. Particle-treated tumor growth profiles, with and without a pharmacological inhibitor to ferroptosis, liproxstatin-1, were compared using a linear model. The longitudinal aspect of the data was taken into account by using generalized estimating equations. We assigned statistical significance at P < 0.05.
Supplementary Material
Supplementary Figure 1. Control assays for apoptosis and necroptosis-inhibited cells, and Liproxstatin-1. a, Image shows M21 cells cultured in full media in the absence (left) and presence (right) of 15μM αMSH-PEG-C′ dots for 72 hours (particle fluorescence is shown in cyan). Graph shows percent cell death (Sytox green + cells) for control and particle-treated cells, at 3 days and 10 days after 72 hours of particle treatment in full media. Note that particle treatment in full media conditions does not inhibit cell viability. Error bars indicate mean +/− standard deviation. N=5 per group. Each replicate is from one biological experiment, quantified with five independent fields of view. b, MCF10A cells cultured in amino acid-free media and treated with 100 nM concanamycin A (ConA) undergo cell death with morphologic features of apoptosis involving cell blebbing and fragmentation. c, Quantification of cell death (Sytox green +) in MCF10A cells cultured in full media (Full) or amino acid-starved (AA-st) conditions, in the presence or absence of Bcl2 overexpression and treatment with ConA. Note Bcl2 inhibits apoptosis induced by ConA treatment in AA-st conditions. Error bars indicate mean+/− standard deviation. N=5 per group. Each replicate is from one biological experiment, quantified with five independent fields of view. d, Knockout of RIPK3 inhibits necroptosis. Graph shows percent cell death (Sytox green +) of wild-type (wt) and RIPK3−/− MEFs treated with a combination of 100 ng/ml TNFα, 1 μg/ml cycloheximide (CHX) and 20 μM zVAD to induce necroptosis. Note RIPK3 −/− MEFs do not undergo cell death, similar to wild-type cells treated with 30 μM necrostatin-1 (Nec-1), a necroptosis inhibitor. Error bars indicate mean+/− standard deviation. N=5 per group. Each replicate is from one biological experiment, quantified with five independent fields of view. e, Liproxstatin-1 treatment inhibits particle-induced cell death. Quantification of cell death (Sytox green +) in MCF10A cells cultured in amino acid-starved (AA-st) conditions in the presence or absence of 15μM α-MSH-PEG-C′ dots and 1μM Liproxstatin-1 after 40 hours. Error bars indicate mean+/− standard deviation. N=5 per group. Each replicate is from one biological experiment, quantified with five independent fields of view. f, Liproxstatin-1 treatment inhibits C11-BODIPY staining induced by erastin treatment. Image shows C11-BODIPY-incubated cells treated with erastin (5μM), 6 hours after control cells underwent cell death as in Fig. 4c. Note the absence of C11-BODIPY staining in erastin+liproxtstatin-1-treated cells. g, MCF10A cells undergoing apoptosis in response to treatment with 50 μg/ml cycloheximide exhibit an asynchronous pattern of cell death. Left image shows image from time-lapse microscopy of cells that have undergone apoptosis (green = Sytox green). Right image shows Sytox green-positive nuclei from left image, pseudocolored to represent the timing of individual cell deaths. The temporal pattern of cell death here is asynchronous compared to Fig 4d. All scale bars = 10μm.
Supplementary Figure 2. Nanoparticle-induced cell death in 786-O and HT-1080 cells, and vascularization of HT-1080 tumors. a, Quantification of cell death (Sytox green +) in amino acid-starved 786-O renal cancer cells in the presence or absence of 15μM αMSH-PEG-C′ dots. Error bars indicate mean+/− standard deviation. N=5 per group. Each replicate is from one biological experiment, quantified with five independent fields of view. Scale bar = 10μm. Necrotic cell deaths were determined by morphology and rapid Sytox green positivity (top images), as compared to apoptotic deaths (bottom images). b, Quantification of cell death (Sytox green +) in HT-1080 fibrosarcoma cells cultured under AA-st conditions in the presence of 1.5 or 15μM αMSH-PEG-C′ dots. Error bars indicate mean+/− standard deviation. N=5 per group. Each replicate is from one biological experiment, quantified with five independent fields of view.
Supplementary Figure 3. Nanoparticles affect iron uptake and glutathione levels. a, Quantification of cell death (Sytox green +) of MCF10A cells cultured in AA-st conditions in the presence of 15μM αMSH-PEG-C′ dots (black bars) or PEG-C′ dots lacking the αMSH peptide (gray bars). Error bars indicate mean+/− standard deviation. N=5 per group. Each replicate is from one biological experiment, quantified with five independent fields of view. b, αMSH-PEG-C′ dots bind iron and treated cells have increased iron levels. Graph shows iron concentrations determined from αMSH nanoparticle stock samples and untreated cells (both are “Controls”, below the limit of detection), and αMSH-PEG-C′ dots purified from culture media (NP+ Media, black), as well as cells treated with nanoparticles (MP+ Cells, red). Iron concentration standards are shown as red data points.
All measurements are means from one biological experiment performed in triplicate. See Materials and Methods section for methodology. Raw data are supplied as Table S1. c, Ferritin heavy chain expression is increased in amino acid-starved (AA-st) cells by treatment with 15μM αMSH-PEG-C′ dots, or by treatment with ferric ammonium citrate (FAC), and is decreased by treatment with DFO. Western blot shows ferritin heavy chain (FTH1) expression compared to actin loading control, 24 hours after treatment with the indicated reagents. d, Treatment of cells with iron (FAC) is sufficient to mimic αMSH-PEG-C′ dot-induced death in amino acid-starvation (AA-st) conditions. Graph shows quantification of percent cell death (Sytox green +) for cells treated as indicated. e, Upper graph: quantification of reduced glutathione levels in MCF10A cells cultured in full media (Full) or amino acid-free media (AA-st) for 24 hours, in the presence or absence of 15μM αMSH-PEG-C′ dots or 400 μM buthionine sulphoximine (BSO), an inhibitor of γ-glutamylcysteine synthetase, the rate-limiting enzyme for glutathione. Note that nanoparticle treatment and amino acid deprivation reduce glutathione levels alone and in combination, similar to treatment with BSO. Error bars indicate mean+/− standard error of the mean, out of three independent experiments. See Supp. Figure 8d for individual experimental values. Lower graph: total glutathione levels are reduced in nanoparticle-treated cells. Graph shows relative levels of total glutathione in amino acid-starved cells, nanoparticle-treated cells (αMSH), and cells treated with the inhibitor of glutathione production BSO, after 24 hours. f, Pre-treatment with erastin sensitizes to nanoparticle-induced ferroptosis. Graph shows percent HT-1080 cell death (Sytox green+ cells) 18 hours (black bars) or 24 hours (red bars) after the indicated treatments. Pre-treatment with erastin for 4 hours is indicated as ‘−’ or ‘+’. Note that erastin-pre-treatment does not induce cell death on its own (‘No treat’), but sensitizes to both continued erastin treatment (‘Erastin’), and the combination of αMSH-PEG-C′ dots (15μM) and amino acid starvation (‘AA-st + αMSH′), as pre-treated cultures (‘+’) undergo more cell death at 18 and 24 hours. Error bars indicate mean+/− standard deviation. N=5 per group. Each replicate is from one biological experiment, quantified with five independent fields of view. g, GPX4 activity is not inhibited by particle treatment. Graph shows specific activity of GPX4 (see Materials and Methods) from cell lysates collected after the indicated treatments. Data are from three independent biological experiments; error bars represent mean+/− standard error of the mean.
Supplementary Figure 4. Model for αMSH-PEG-C′ dot-induced ferroptosis. Amino acid deprivation (I) and nanoparticle ingestion by lysosomes (II) lead to depletion of glutathione (see Supp. Figure 2c) and, when combined, induction of ferroptosis that is dependent upon lysosomal iron, a known inducer of reactive oxygen species (ROS), and lipid peroxidation, the latter blocked by Ferrostatin-1 and Liproxstatin-1.
Supplementary Figure 5. Images of particle-treated tumors. a, HT-1080 tumors are well vascularized. Images show a representative HT-1080 xenograft specimen, harvested 10 days after particle injection, and stained by immunohistochemistry for the endothelial marker CD31. Low magnification (4x) microscopic image of whole tumor (left image) and high magnification (40x) image of tumor region (right image). Note the presence of blood vessels throughout the tumor. Scale bars = 500μm (low power); 50μm (high power). b, Cell death measurements in HT-1080 cells treated with differently sized PEG-C′ dots. Quantification of HT-1080 cell death (Sytox green +) cultured in full media or AA-st conditions for 48 hours in the absence or presence of 15μM PEG-C′ dots, 6nm or 10nm, lacking the αMSH peptide. Error bars indicate mean+/− standard deviation. N=5 per group. c, αMSH-PEG-C′ dot-treated 786-O tumor shows large number of recruited macrophages. Low (top) and high (bottom) power views of Mac-2 stained treated sections show marked Mac-2 positive cells circumscribing the tumor at similar locations (box, arrows).
Supplementary Figure 6. Individual data points for graphs from Figures 2 and 3. a, Data from the cell survival graph (on left) in Figure 2a are shown. N=3 biological experiments, each data point represents the mean from five independent fields of view. b, Data from the cell proliferation graph (on right) in Figure 2a are shown. N=3 biological experiments, each data point represents the mean from five independent fields of view. c, Individual LC3-II/actin values are shown from Figure 2b, left graph. N=3 western blots for each group. Note that the 15μm condition has two values plotted at 0.79. d, Individual LC3-II turnover values are shown from Figure 2b, right graph. N=3 western blots for each group.e, Data from the cell death graph in Figure 2c are shown. N=4 biological experiments, each data point represents the mean from five independent fields of view. f, Data from the cell death graph on the left in Figure 3b are shown for N=5 for each group. Each replicate is an independent field of view of the same biological experiment. g, Data from the cell death graph on the right in Figure 3b are shown for N=5 for each group. Each replicate is an independent field of view of the same biological experiment. h, Data from the cell death graph in Figure 3c are shown for N=5 for each group. Each replicate is an independent field of view of the same biological experiment. i, Data from the cell death graph in Figure 3d are shown for N=5 for each group. Each replicate is an independent field of view of the same biological experiment. j, Data from the cell death graph in Figure 3e are shown for N=5 for each group. Each replicate is an independent field of view of the same biological experiment.
Supplementary Figure 7. Individual data points for graphs from Figures 4, 5 and Supp. Figure 1. a, Data from the cell death graph in Figure 4a are shown. N=3 biological experiments, each data point represents the mean from five independent fields of view. b, Data from the cell death graph in Figure 4b are shown. N=5 for each group. Each replicate is an independent field of view of the same biological experiment. c, Data from the cell death graph in Figure 4d are shown. N=3 biological experiments, each data point represents the mean from five independent fields of view. d, Data from the cell death graph in Figure 5a are shown. N=5 for each group. Each replicate is an independent field of view of the same biological experiment. e, Data from the cell death graph in Figure 5b are shown. N=5 for each group. Each replicate is an independent field of view of the same biological experiment. f, Data from the cell death graph in Figure 5c are shown. N=3 biological experiments, each data point represents the mean from five independent fields of view. g, Data from the cell death graph in Supp. Figure 1c are shown. N=5 for each group. Each replicate is an independent field of view of the same biological experiment. h, Data from the cell death graph in Supp. Figure 1d are shown. N=5 for each group. Each replicate is an independent field of view of the same biological experiment. i, Data from the cell death graph in Supp. Figure 1e are shown. N=5 for each group. Each replicate is an independent field of view of the same biological experiment.
Supplementary Figure 8. Individual data points for graphs from Supp. Figures 2 and 3. a, Data from the cell death graph in Supp. Figure 2b are shown. N=5 for each group. Each replicate is an independent field of view of the same biological experiment. b, Data from the cell death graph in Supp. Figure 3a are shown. N=5 for each group. Each replicate is an independent field of view of the same biological experiment. c, Data from the cell death graph in Supp. Figure 3d are shown. N=5 for each group. Each replicate is an independent field of view of the same biological experiment. d, Data for the glutathione quantifications from Supp. Figure 3e (top graph) are shown. N=3 independent biological experiments. e, Data from the cell death graph in Supp. Figure 3f are shown. N=5 for each group. Each replicate is an independent field of view of the same biological experiment.
Table S1. Iron Concentrations measured by microwave plasma atomic emission spectroscopy (MP-AES).
Table S2. Metabolic concentration proflles in tumor-bearing mice.
Table S3. Hematologic profiles in tumor-bearing mice.
Table S4. Histopathologic profiles in tumor-bearing mice.
Supplementary Movie 1. Time-lapse analysis of MCF10A cells undergoing ferroptosis in amino acid-starved conditions with 15μM aMSH-PEG-C′ dots. Left image panel shows Sytox Green fluorescence (green) merged with DIC. Right image panel shows Sytox green-positive nuclei from left image, pseudocolored to represent the timings of individual cell deaths. Note that death (Sytox green positivity) spreads cell-to-cell from the left side of the image to the right. Time is shown as hours:minutes on left image. Pseudocolored nuclei represent time scale from Fig. 4d.
Supplementary Movie 2. Time-lapse analysis of MCF10A cells undergoing apoptosis in response to cycloheximide treatment. Left image panel shows Sytox Green fluorescence (green) merged with DIC. Right image panel shows Sytox green-positive nuclei from left image, pseudocolored to represent the timings of individual cell deaths. Note that cell death (Sytox green positivity) occurs asynchronously compared to Supplementary Movie 1. Time is shown as hours:minutes on left image. Pseudocolored nuclei represent time scale from Supplementary Fig. 1d.
Acknowledgments
This study was funded by grants from the NIH (R01GM111350, to M.O.; 1R01CA161280-01A1, to M.B. and U.W.; 1U54 CA199081-01 to M.B. and U.W.; R01GM113013; R01CA166413 to X.J.; Sloan Kettering Institute Core Grant P30 CA008748CCSG; and the Benjamin Friedman Research Fund (M.O.). Peptide synthesis was conducted by the University of Missouri Structural Biology Core.
Footnotes
Author contributions. Product preparation (K.M., T.Q., and M.T.); experimental design (S.E.K., M.O., M.B., K.M., L.Z., M.Go, X.J., P.Z., S.M., F.C., T.Q., and U.W.); data acquisition (S.E.K., L.Z., K.M., M.T., M.Go., X.J., P.M., F.C., S.M., M.B., M.O., I.I, M.C. and M.R.); data analysis/interpretation (S.E.K., M.O., M.B., K.M., P.M., F.C., P.Z., M.G., S.M., L.Z., U.W., I.I, M.C. and M.R.); and preparing the manuscript (S.E.K., M.O., M.B., M.Go., P.Z., K.M., S.M., T.Q., U.W.). All authors discussed the results and implications and commented on the manuscript.
Additional Information: Supplementary information is available in the online version of the paper.
Competing financial interests. The authors declare an intellectual property interest in a provisional patent 62/280,960.
References
- 1.Davis ME, Chen ZG, Shin DM. Nanoparticle therapeutics: an emerging treatment modality for cancer. Nat Rev Drug Discov. 2008;7:771–782. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 2.Phillips E, et al. Clinical translation of an ultrasmall inorganic optical-PET imaging nanoparticle probe. Sci Transl Med. 2014;6:260ra149. doi: 10.1126/scitranslmed.3009524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duncan R, Richardson SC. Endocytosis and intracellular trafficking as gateways for nanomedicine delivery: opportunities and challenges. Mol Pharm. 2012;9:2380–2402. doi: 10.1021/mp300293n. [DOI] [PubMed] [Google Scholar]
- 4.Duncan R, Gaspar R. Nanomedicine(s) under the microscope. Mol Pharm. 2011;8:2101–2141. doi: 10.1021/mp200394t. [DOI] [PubMed] [Google Scholar]
- 5.Ma X, et al. Gold nanoparticles induce autophagosome accumulation through size-dependent nanoparticle uptake and lysosome impairment. ACS Nano. 2011;5:8629–8639. doi: 10.1021/nn202155y. [DOI] [PubMed] [Google Scholar]
- 6.Li JJ, Hartono D, Ong CN, Bay BH, Yung LY. Autophagy and oxidative stress associated with gold nanoparticles. Biomaterials. 2010;31:5996–6003. doi: 10.1016/j.biomaterials.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 7.Chen N, et al. Long-term effects of nanoparticles on nutrition and metabolism. Small. 2014;10:3603–3611. doi: 10.1002/smll.201303635. [DOI] [PubMed] [Google Scholar]
- 8.Li C, et al. PAMAM nanoparticles promote acute lung injury by inducing autophagic cell death through the Akt-TSC2-mTOR signaling pathway. J Mol Cell Biol. 2009;1:37–45. doi: 10.1093/jmcb/mjp002. [DOI] [PubMed] [Google Scholar]
- 9.Stern ST, Adiseshaiah PP, Crist RM. Autophagy and lysosomal dysfunction as emerging mechanisms of nanomaterial toxicity. Part Fibre Toxicol. 2012;9:20. doi: 10.1186/1743-8977-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma K, et al. Control of Ultrasmall Sub-10 nm Ligand-Functionalized Fluorescent Core–Shell Silica Nanoparticle Growth in Water. Chem Mater. 2015;27:4119–4133. doi: 10.1021/acs.chemmater.5b01222. [DOI] [Google Scholar]
- 11.Bradbury MS, et al. Clinically-translated silica nanoparticles as dual-modality cancer-targeted probes for image-guided surgery and interventions. Integr Biol (Camb) 2013;5:74–86. doi: 10.1039/c2ib20174g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benezra M, et al. Multimodal silica nanoparticles are effective cancer-targeted probes in a model of human melanoma. J Clin Invest. 2011;121:2768–2780. doi: 10.1172/JCI45600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoo B, et al. Ultrasmall dual-modality silica nanoparticle drug conjugates: Design, synthesis, and characterization. Bioorg Med Chem. 2015;23:7119–7130. doi: 10.1016/j.bmc.2015.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miao Y, Benwell K, Quinn TP. 99mTc- and 111In-labeled alpha-melanocyte-stimulating hormone peptides as imaging probes for primary and pulmonary metastatic melanoma detection. J Nucl Med. 2007;48:73–80. [PubMed] [Google Scholar]
- 15.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nelson DA, et al. Hypoxia and defective apoptosis drive genomic instability and tumorigenesis. Genes Dev. 2004;18:2095–2107. doi: 10.1101/gad.1204904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kandasamy K, et al. Involvement of proapoptotic molecules Bax and Bak in tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced mitochondrial disruption and apoptosis: differential regulation of cytochrome c and Smac/DIABLO release. Cancer Res. 2003;63:1712–1721. [PubMed] [Google Scholar]
- 18.Jacobson MD, et al. Bcl-2 blocks apoptosis in cells lacking mitochondrial DNA. Nature. 1993;361:365–369. doi: 10.1038/361365a0. [DOI] [PubMed] [Google Scholar]
- 19.He S, et al. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell. 2009;137:1100–1111. doi: 10.1016/j.cell.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y, et al. Autosis is a Na+, K+-ATPase-regulated form of cell death triggered by autophagy-inducing peptides, starvation, and hypoxia-ischemia. Proc Natl Acad Sci U S A. 2013;110:20364–20371. doi: 10.1073/pnas.1319661110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dixon SJ, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linkermann A, et al. Synchronized renal tubular cell death involves ferroptosis. Proc Natl Acad Sci U S A. 2014;111:16836–16841. doi: 10.1073/pnas.1415518111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruhrberg C, De Palma M. A double agent in cancer: deciphering macrophage roles in human tumors. Nat Med. 2010;16:861–862. doi: 10.1038/nm0810-861. [DOI] [PubMed] [Google Scholar]
- 24.Guiducci C, Vicari AP, Sangaletti S, Trinchieri G, Colombo MP. Redirecting in vivo elicited tumor infiltrating macrophages and dendritic cells towards tumor rejection. Cancer Res. 2005;65:3437–3446. doi: 10.1158/0008-5472.can-04-4262. [DOI] [PubMed] [Google Scholar]
- 25.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang WS, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friedmann Angeli JP, et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat Cell Biol. 2014;16:1180–1191. doi: 10.1038/ncb3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao M, Monian P, Quadri N, Ramasamy R, Jiang X. Glutaminolysis and Transferrin Regulate Ferroptosis. Mol Cell. 2015;59:298–308. doi: 10.1016/j.molcel.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gabizon A, et al. Cancer nanomedicines: closing the translational gap. Lancet. 2014;384:2175–2176. doi: 10.1016/s0140-6736(14)61457-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma K, Zhang D, Cong Y, Wiesner U. Elucidating the Mechanism of Silica Nanoparticle PEGylation Processes Using Fluorescence Correlation Spectroscopies. Chem Mater. 2016;28:1537–1545. doi: 10.1021/acs.chemmater.6b00030. [DOI] [Google Scholar]
- 31.Roveri A, Maiorino M, Ursini F. Enzymatic and immunological measurements of soluble and membrane-bound phospholipid-hydroperoxide glutathione peroxidase. Methods Enzymol. 1994;233:202–212. doi: 10.1016/s0076-6879(94)33023-9. [DOI] [PubMed] [Google Scholar]
- 32.Zeger SL, Liang KY, Albert PS. Models for longitudinal data: a generalized estimating equation approach. Biometrics. 1988;44:1049–1060. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Control assays for apoptosis and necroptosis-inhibited cells, and Liproxstatin-1. a, Image shows M21 cells cultured in full media in the absence (left) and presence (right) of 15μM αMSH-PEG-C′ dots for 72 hours (particle fluorescence is shown in cyan). Graph shows percent cell death (Sytox green + cells) for control and particle-treated cells, at 3 days and 10 days after 72 hours of particle treatment in full media. Note that particle treatment in full media conditions does not inhibit cell viability. Error bars indicate mean +/− standard deviation. N=5 per group. Each replicate is from one biological experiment, quantified with five independent fields of view. b, MCF10A cells cultured in amino acid-free media and treated with 100 nM concanamycin A (ConA) undergo cell death with morphologic features of apoptosis involving cell blebbing and fragmentation. c, Quantification of cell death (Sytox green +) in MCF10A cells cultured in full media (Full) or amino acid-starved (AA-st) conditions, in the presence or absence of Bcl2 overexpression and treatment with ConA. Note Bcl2 inhibits apoptosis induced by ConA treatment in AA-st conditions. Error bars indicate mean+/− standard deviation. N=5 per group. Each replicate is from one biological experiment, quantified with five independent fields of view. d, Knockout of RIPK3 inhibits necroptosis. Graph shows percent cell death (Sytox green +) of wild-type (wt) and RIPK3−/− MEFs treated with a combination of 100 ng/ml TNFα, 1 μg/ml cycloheximide (CHX) and 20 μM zVAD to induce necroptosis. Note RIPK3 −/− MEFs do not undergo cell death, similar to wild-type cells treated with 30 μM necrostatin-1 (Nec-1), a necroptosis inhibitor. Error bars indicate mean+/− standard deviation. N=5 per group. Each replicate is from one biological experiment, quantified with five independent fields of view. e, Liproxstatin-1 treatment inhibits particle-induced cell death. Quantification of cell death (Sytox green +) in MCF10A cells cultured in amino acid-starved (AA-st) conditions in the presence or absence of 15μM α-MSH-PEG-C′ dots and 1μM Liproxstatin-1 after 40 hours. Error bars indicate mean+/− standard deviation. N=5 per group. Each replicate is from one biological experiment, quantified with five independent fields of view. f, Liproxstatin-1 treatment inhibits C11-BODIPY staining induced by erastin treatment. Image shows C11-BODIPY-incubated cells treated with erastin (5μM), 6 hours after control cells underwent cell death as in Fig. 4c. Note the absence of C11-BODIPY staining in erastin+liproxtstatin-1-treated cells. g, MCF10A cells undergoing apoptosis in response to treatment with 50 μg/ml cycloheximide exhibit an asynchronous pattern of cell death. Left image shows image from time-lapse microscopy of cells that have undergone apoptosis (green = Sytox green). Right image shows Sytox green-positive nuclei from left image, pseudocolored to represent the timing of individual cell deaths. The temporal pattern of cell death here is asynchronous compared to Fig 4d. All scale bars = 10μm.
Supplementary Figure 2. Nanoparticle-induced cell death in 786-O and HT-1080 cells, and vascularization of HT-1080 tumors. a, Quantification of cell death (Sytox green +) in amino acid-starved 786-O renal cancer cells in the presence or absence of 15μM αMSH-PEG-C′ dots. Error bars indicate mean+/− standard deviation. N=5 per group. Each replicate is from one biological experiment, quantified with five independent fields of view. Scale bar = 10μm. Necrotic cell deaths were determined by morphology and rapid Sytox green positivity (top images), as compared to apoptotic deaths (bottom images). b, Quantification of cell death (Sytox green +) in HT-1080 fibrosarcoma cells cultured under AA-st conditions in the presence of 1.5 or 15μM αMSH-PEG-C′ dots. Error bars indicate mean+/− standard deviation. N=5 per group. Each replicate is from one biological experiment, quantified with five independent fields of view.
Supplementary Figure 3. Nanoparticles affect iron uptake and glutathione levels. a, Quantification of cell death (Sytox green +) of MCF10A cells cultured in AA-st conditions in the presence of 15μM αMSH-PEG-C′ dots (black bars) or PEG-C′ dots lacking the αMSH peptide (gray bars). Error bars indicate mean+/− standard deviation. N=5 per group. Each replicate is from one biological experiment, quantified with five independent fields of view. b, αMSH-PEG-C′ dots bind iron and treated cells have increased iron levels. Graph shows iron concentrations determined from αMSH nanoparticle stock samples and untreated cells (both are “Controls”, below the limit of detection), and αMSH-PEG-C′ dots purified from culture media (NP+ Media, black), as well as cells treated with nanoparticles (MP+ Cells, red). Iron concentration standards are shown as red data points.
All measurements are means from one biological experiment performed in triplicate. See Materials and Methods section for methodology. Raw data are supplied as Table S1. c, Ferritin heavy chain expression is increased in amino acid-starved (AA-st) cells by treatment with 15μM αMSH-PEG-C′ dots, or by treatment with ferric ammonium citrate (FAC), and is decreased by treatment with DFO. Western blot shows ferritin heavy chain (FTH1) expression compared to actin loading control, 24 hours after treatment with the indicated reagents. d, Treatment of cells with iron (FAC) is sufficient to mimic αMSH-PEG-C′ dot-induced death in amino acid-starvation (AA-st) conditions. Graph shows quantification of percent cell death (Sytox green +) for cells treated as indicated. e, Upper graph: quantification of reduced glutathione levels in MCF10A cells cultured in full media (Full) or amino acid-free media (AA-st) for 24 hours, in the presence or absence of 15μM αMSH-PEG-C′ dots or 400 μM buthionine sulphoximine (BSO), an inhibitor of γ-glutamylcysteine synthetase, the rate-limiting enzyme for glutathione. Note that nanoparticle treatment and amino acid deprivation reduce glutathione levels alone and in combination, similar to treatment with BSO. Error bars indicate mean+/− standard error of the mean, out of three independent experiments. See Supp. Figure 8d for individual experimental values. Lower graph: total glutathione levels are reduced in nanoparticle-treated cells. Graph shows relative levels of total glutathione in amino acid-starved cells, nanoparticle-treated cells (αMSH), and cells treated with the inhibitor of glutathione production BSO, after 24 hours. f, Pre-treatment with erastin sensitizes to nanoparticle-induced ferroptosis. Graph shows percent HT-1080 cell death (Sytox green+ cells) 18 hours (black bars) or 24 hours (red bars) after the indicated treatments. Pre-treatment with erastin for 4 hours is indicated as ‘−’ or ‘+’. Note that erastin-pre-treatment does not induce cell death on its own (‘No treat’), but sensitizes to both continued erastin treatment (‘Erastin’), and the combination of αMSH-PEG-C′ dots (15μM) and amino acid starvation (‘AA-st + αMSH′), as pre-treated cultures (‘+’) undergo more cell death at 18 and 24 hours. Error bars indicate mean+/− standard deviation. N=5 per group. Each replicate is from one biological experiment, quantified with five independent fields of view. g, GPX4 activity is not inhibited by particle treatment. Graph shows specific activity of GPX4 (see Materials and Methods) from cell lysates collected after the indicated treatments. Data are from three independent biological experiments; error bars represent mean+/− standard error of the mean.
Supplementary Figure 4. Model for αMSH-PEG-C′ dot-induced ferroptosis. Amino acid deprivation (I) and nanoparticle ingestion by lysosomes (II) lead to depletion of glutathione (see Supp. Figure 2c) and, when combined, induction of ferroptosis that is dependent upon lysosomal iron, a known inducer of reactive oxygen species (ROS), and lipid peroxidation, the latter blocked by Ferrostatin-1 and Liproxstatin-1.
Supplementary Figure 5. Images of particle-treated tumors. a, HT-1080 tumors are well vascularized. Images show a representative HT-1080 xenograft specimen, harvested 10 days after particle injection, and stained by immunohistochemistry for the endothelial marker CD31. Low magnification (4x) microscopic image of whole tumor (left image) and high magnification (40x) image of tumor region (right image). Note the presence of blood vessels throughout the tumor. Scale bars = 500μm (low power); 50μm (high power). b, Cell death measurements in HT-1080 cells treated with differently sized PEG-C′ dots. Quantification of HT-1080 cell death (Sytox green +) cultured in full media or AA-st conditions for 48 hours in the absence or presence of 15μM PEG-C′ dots, 6nm or 10nm, lacking the αMSH peptide. Error bars indicate mean+/− standard deviation. N=5 per group. c, αMSH-PEG-C′ dot-treated 786-O tumor shows large number of recruited macrophages. Low (top) and high (bottom) power views of Mac-2 stained treated sections show marked Mac-2 positive cells circumscribing the tumor at similar locations (box, arrows).
Supplementary Figure 6. Individual data points for graphs from Figures 2 and 3. a, Data from the cell survival graph (on left) in Figure 2a are shown. N=3 biological experiments, each data point represents the mean from five independent fields of view. b, Data from the cell proliferation graph (on right) in Figure 2a are shown. N=3 biological experiments, each data point represents the mean from five independent fields of view. c, Individual LC3-II/actin values are shown from Figure 2b, left graph. N=3 western blots for each group. Note that the 15μm condition has two values plotted at 0.79. d, Individual LC3-II turnover values are shown from Figure 2b, right graph. N=3 western blots for each group.e, Data from the cell death graph in Figure 2c are shown. N=4 biological experiments, each data point represents the mean from five independent fields of view. f, Data from the cell death graph on the left in Figure 3b are shown for N=5 for each group. Each replicate is an independent field of view of the same biological experiment. g, Data from the cell death graph on the right in Figure 3b are shown for N=5 for each group. Each replicate is an independent field of view of the same biological experiment. h, Data from the cell death graph in Figure 3c are shown for N=5 for each group. Each replicate is an independent field of view of the same biological experiment. i, Data from the cell death graph in Figure 3d are shown for N=5 for each group. Each replicate is an independent field of view of the same biological experiment. j, Data from the cell death graph in Figure 3e are shown for N=5 for each group. Each replicate is an independent field of view of the same biological experiment.
Supplementary Figure 7. Individual data points for graphs from Figures 4, 5 and Supp. Figure 1. a, Data from the cell death graph in Figure 4a are shown. N=3 biological experiments, each data point represents the mean from five independent fields of view. b, Data from the cell death graph in Figure 4b are shown. N=5 for each group. Each replicate is an independent field of view of the same biological experiment. c, Data from the cell death graph in Figure 4d are shown. N=3 biological experiments, each data point represents the mean from five independent fields of view. d, Data from the cell death graph in Figure 5a are shown. N=5 for each group. Each replicate is an independent field of view of the same biological experiment. e, Data from the cell death graph in Figure 5b are shown. N=5 for each group. Each replicate is an independent field of view of the same biological experiment. f, Data from the cell death graph in Figure 5c are shown. N=3 biological experiments, each data point represents the mean from five independent fields of view. g, Data from the cell death graph in Supp. Figure 1c are shown. N=5 for each group. Each replicate is an independent field of view of the same biological experiment. h, Data from the cell death graph in Supp. Figure 1d are shown. N=5 for each group. Each replicate is an independent field of view of the same biological experiment. i, Data from the cell death graph in Supp. Figure 1e are shown. N=5 for each group. Each replicate is an independent field of view of the same biological experiment.
Supplementary Figure 8. Individual data points for graphs from Supp. Figures 2 and 3. a, Data from the cell death graph in Supp. Figure 2b are shown. N=5 for each group. Each replicate is an independent field of view of the same biological experiment. b, Data from the cell death graph in Supp. Figure 3a are shown. N=5 for each group. Each replicate is an independent field of view of the same biological experiment. c, Data from the cell death graph in Supp. Figure 3d are shown. N=5 for each group. Each replicate is an independent field of view of the same biological experiment. d, Data for the glutathione quantifications from Supp. Figure 3e (top graph) are shown. N=3 independent biological experiments. e, Data from the cell death graph in Supp. Figure 3f are shown. N=5 for each group. Each replicate is an independent field of view of the same biological experiment.
Table S1. Iron Concentrations measured by microwave plasma atomic emission spectroscopy (MP-AES).
Table S2. Metabolic concentration proflles in tumor-bearing mice.
Table S3. Hematologic profiles in tumor-bearing mice.
Table S4. Histopathologic profiles in tumor-bearing mice.
Supplementary Movie 1. Time-lapse analysis of MCF10A cells undergoing ferroptosis in amino acid-starved conditions with 15μM aMSH-PEG-C′ dots. Left image panel shows Sytox Green fluorescence (green) merged with DIC. Right image panel shows Sytox green-positive nuclei from left image, pseudocolored to represent the timings of individual cell deaths. Note that death (Sytox green positivity) spreads cell-to-cell from the left side of the image to the right. Time is shown as hours:minutes on left image. Pseudocolored nuclei represent time scale from Fig. 4d.
Supplementary Movie 2. Time-lapse analysis of MCF10A cells undergoing apoptosis in response to cycloheximide treatment. Left image panel shows Sytox Green fluorescence (green) merged with DIC. Right image panel shows Sytox green-positive nuclei from left image, pseudocolored to represent the timings of individual cell deaths. Note that cell death (Sytox green positivity) occurs asynchronously compared to Supplementary Movie 1. Time is shown as hours:minutes on left image. Pseudocolored nuclei represent time scale from Supplementary Fig. 1d.