Abstract
Innate immunity refers to the body’s initial response to curb infection upon exposure to invading organisms. While the detection of pathogen-associated molecules is an ancient form of host defense, if dysfunctional, autoimmune disease may result. The innate immune response during pathogenic infection is initiated through the activation of receptors recognizing conserved molecular patterns, such as nucleic acids from a virus’ genome or replicative cycle. Additionally, the host’s own nucleic acids are capable of activating an immune response. Therefore, it follows that the nucleic acid-sensing pathways must be tightly controlled to avoid an autoimmune response from recognition of self, yet still be unimpeded to respond to viral infections. In this review, we will describe the nucleic acid sensing pathways and how they respond to virus infection. Moreover, we will discuss autoimmune diseases that develop when these pathways fail to signal properly and identify knowledge gaps that are prime for interrogation.
Keywords: autoimmunity, interferon, RIG-I, MDA5, STING, cGAS
Introduction
The innate immune response is the first line of defense to microbial infection, and it is initiated through the activation of receptors recognizing molecules that are signature of pathogenic infection. However, when the innate immune response is misregulated, autoimmunity can result. While innate immune pathways are found as far back as early branching metazoans, these signaling pathways have evolved extensively and become increasingly complex in higher organisms. The innate immune response provides the first line of defense against pathogens by responding to foreign molecules within the cell that are a signature of pathogenic infection, such as cytosolic DNA or double-stranded RNA, which are by-products of bacterial and viral infections. Specifically, the innate immune response is initiated through the activation of pattern recognition receptors (PRRs) that recognize conserved pathogen motifs called pathogen-associated molecular patterns (PAMPs) [1,2]. Intracellular mammalian PRRs consist of the Toll-like receptor (TLR), nucleotide-binding oligomerization domain (NOD)-like receptor (NLR), the family of RIG-I-like receptors (RLR), and the cytosolic DNA receptors, among other families of PRRs [1,3,4]. The activation of these PRRs with their respective PAMPs results in the recruitment of a number of intermediate adaptor molecules, such as IFN regulatory factor 3 (IRF3), and IRF7, which results in the nuclear translocation of NFκB, [5]. This cascade culminates with the induction of IRF-responsive genes, including IFNβ, a cytokine produced during the early stages of infection that binds to its receptor and induces IFN-stimulated gene (ISG) expression through the activation of the JAK-STAT pathway [6–8]. IFNβ is also able to activate NFκB, thus amplifying the IFN response via a positive feedback loop due to increased NFκB activity leading to increased IFNβ and pro-inflammatory cytokine induction [9,10]. Pro-inflammatory cytokines are important for the recruitment of specialized immune cells to the site of infection to curb pathogen levels. However, if pro-inflammatory cytokines are activated in the absence of infection, autoimmune disease may occur. As such, one can imagine that while the innate immune response is important to control pathogenic infection, if the response is left unchecked, detrimental autoimmunity, characterized by high levels of inflammation in the absence of infection, may result.
Innate immune signaling must be finely tuned so that it can provide a sufficient response against pathogenic infection, yet not be constitutively or hyperactive in such a way that autoinflammatory or autoimmune disease results [11]. More than 20 million Americans suffer from forms of autoimmune disease, such as rheumatoid and juvenile arthritis, Crohn’s disease (CD), systemic lupus erythematosus (SLE), Acardi Goutières Syndrome (AGS), and Sjogren’s Syndrome. Moreover, autoimmune diseases can affect children. Approximately one in every thousand children is affected by juvenile arthritis, an autoimmune disease that causes persistent joint pain, swelling, and stiffness. Other autoimmune diseases occurring in children include celiac disease, type 1 diabetes (T1D), SLE, and scleroderma. Autoimmune diseases that occur in childhood often have significant, life-long health consequences (reviewed in [12]). While the exact etiology of many of these diseases still remains unknown, it is reasonable that chronic microbial infection or the presence of nucleic acids in the cytosol, such as DNA or RNA from apoptotic or necrotic cells, results in prolonged activation of the innate immune response, potentially causing inflammation-mediated autoimmune disease [13]. Understanding how innate immune pathways function may explain the mechanisms for autoimmune disease and lead to therapies to treat the disorders.
In this review, we will delineate the major nucleic acid sensing pathways in mammals, with a focus on the RLRs and cytosolic DNA sensing pathways. Upon describing these pathways, we will discuss how defects in the pathways manifests into disease. Specifically, we will describe autoimmune diseases that result from mutations in innate immune response genes as well as how the protein products from these genes are essential for responses to virus infections. We will conclude by discussing therapeutic measures that are being developed to target these pathways to protect against viral infection and autoimmunity as well as animal models that can be used to interrogate these pathways.
The RNA Sensing Pathways
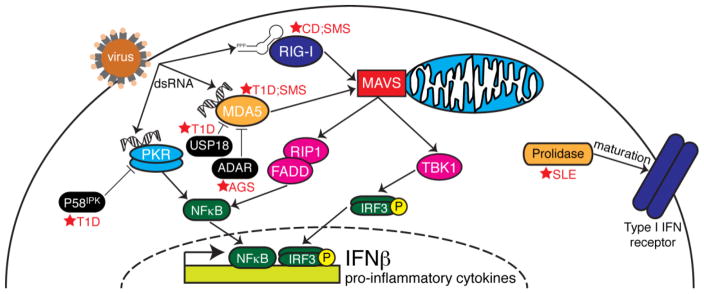
RNA viruses and cytosolic dsRNA are recognized by the RIG-I-like Receptors (RLRs) [14]. There are three RLRs, which are involved in the innate immune response: Retinoic acid-inducible gene I (RIG-I) [3], Melanoma differentiation-associated gene 5 (MDA5) [15], and Laboratory of genetics and physiology 2 (LGP2) [16]. RIG-I and MDA5 have been demonstrated to sense viral RNA and signal for an innate immune response, and LGP2 is likely a component of negative feedback for IFNβ induction [3,17]. RIG-I and MDA5 both contain a DExD/H-box helicase domain that recognizes foreign RNA and two caspase-recruitment domains (CARDs) to interact with mitochondrial antiviral signaling (MAVS) [18]. Upon activation, RIG-I and MDA5 signal to the adaptor protein MAVS, which leads to the IRF3 and NFκB activation and induction of the cytokine IFNβ [19] (Fig. 1).
Figure 1. The RNA sensing pathways.
Following virus infection, dsRNAs are produced in the cell cytosol as a byproduct of the virus life cycle. While dsRNAs activate PKR and MDA5, short (<200 bp) RNAs with a 5′ triphoosphate group activate RIG-I. MDA5 and RIG-I signal through the adapter MAVS which is located on the mitochondrial membrane. MAVS then signals though TBK1 and RIP1/FADD to activate the transcription factors IRF3 and NFκB, respectively. PKR is capable to activating NFκB. When activated, these transcription factors localize to the nucleus where they induce IFNβ and pro-inflammatory cytokines, which are secreted from the cell and bind to the type I IFN receptor and recruit specialized immune cells to the site of infection. Dysfunction in certain nodes of pathway can result in autoimmune diseases (marked by red stars). Loss of the PKR inhibitor, P58IPK results in late onset of Type I diabetes (T1D), as does downregulation of the MDA5 inhibitor, USP18, or mutations in the gene encoding MDA5. Mutations in the MDA5 inhibitor ADAR may lead to Aicardi-Goutières syndrome (AGS), while mutations in RIG-I and MDA5 may lead to Singleton-Merten syndrome (SMS). Crohn’s disease (CD) has been linked to RIG-I dysfunction. Finally, loss of prolidase, which aids in the proper maturation and surface expression of the type I IFN receptor, is linked with systemic lupus erythematosus (SLE).
RIG-I and MDA5 both recognize non-self RNA and have varying functions in RNA sensing. RIG-I can recognize the 5′-triphosphate of single-stranded RNA [20,21]. RIG-I recognizes shorter double-stranded RNA sequences, and MDA5 recognizes longer double-stranded RNA sequences [14]. This length-dependence makes these two cytosolic sensors complementary to each other for viral nucleic acid sensing in the same pathway. These differences in RNA recognition allow each sensor to detect different types of viruses. For example, MDA5 senses picornaviruses and RIG-I senses many paramyxoviruses [20]. However, there is some overlap in viral recognition, as both sensors can respond to Dengue virus, for example [22].
Dysfunction in RLR pathways can lead to immune deficiencies and increased susceptibility to viruses such as Hepatitis C virus, West Nile virus (WNV), influenza virus, and Dengue virus (DENV). The adaptor MAVS is required to initiate an immune response to both WNV and DENV [23,24], indicating the importance of the RLRs that detect viral RNA and initiate an interferon response via MAVS. Interestingly, mutations in RFC1, the gene which codes for a subunit to activate DNA polymerase, are associated with patient susceptibility to neuroinvasive West Nile virus infection [25].
RNA sensing dysfunction and autoimmunity
RIG-I and MDA5 are both implicated in autoimmune disease. Specifically, MDA5 has been linked to Type I diabetes (T1D). T1D can be triggered in an individual by a combination of viral infection in pancreatic beta cells and genetic predisposition [26]. For example, enterovirus and coxsackie virus infections localize to β-cells [27], and neutralizing antibodies to these infections have been found in TID patients [28]. Additionally, cohort studies have indicated an association between enterovirus infection and TID [29]. A Genome Wide Association Study identified that certain single-nucleotide polymorphisms (SNPs) cause higher expression of the gene that codes for MDA5, IFIH1 (Interferon induced with helicase domain I), leading to an increase in risk for T1D [30]. Additionally, a missense mutation in the IFIH1 gene allowing MDA5 to be constitutively active in a mouse model induces lupus-like nephritis and autoimmunity, indicating a link between RNA sensing and autoimmunity [31]. The downstream adaptor MAVS is critical for the development of autoimmunity via MDA5, as mice carrying a mutation in MDA5 leading to hyperactivity rescues the autoimmune phenotype [31]. Moreover, additional components of the innate immune response activated by MDA5 may be implicated in T1D. Santin et al. hypothesize that inhibition of Ubiquitin-specific peptidase 18 (USP18) leads to an increase in interferon due to an increased MDA5 expression to signal through MAVS. The increase in interferon results in an increase in STAT signaling, leading to inflammation and apoptosis in pancreatic β-cells [32]. This study proposes USP18 as a “master regulator” of the interferon response. More recently, it was shown that mutations in the RNA-editing enzyme ADAR1 were associated with the autoimmune disease Aicardi-Goutières syndrome (AGS) [33]. Pestal et al. clarified the mechanism of this pathology by showing that ADAR1 is necessary to prevent AGS through its inhibition of MDA5-mediated MAVS activation [34]. Together, these studies show how MDA5 dsRNA sensing must be tightly regulated in order for normal innate immune signaling and to avoid inflammatory autoimmune disease.
While MDA5 hyperactivity leads to autoimmunity, reduced MDA5 expression has been correlated with resistance to T1D. Mice heterozygous for MDA5 are protected from T1D when infected with coxsackievirus serotype B4 (CB4) [35]. However, the tradeoff for this protection to T1D from MDA5-deficiciency leaves organisms susceptible to encephalomyocarditis virus strain D (EMCV-D). McCartney et al. determined that MDA5 and TLR3 are both required to prevent T1D in EMCV-D-infected mice [36]. This requirement for median levels of MDA5 illustrates the immune system’s balancing act between immune deficiency and autoimmunity: high levels of active MDA5 can lead to autoimmunity and T1D, but a dearth of MDA5 leads to susceptibility to viral infections that can cause chronic disease from cell death.
Dnajc3, the gene that codes for P58IPK, has also been implicated in the innate immune response to viral infection and diabetes. P58IPK was initially discovered in experiments where influenza virus superinfection in cells infected with an adenovirus mutant lacking the gene to inhibit PKR, the interferon-induced dsRNA-activated eIF2α kinase, restored PKR inhibition [37,38]. Subsequent studies delineated that P58IPK is proviral during influenza virus infection [39], vaccinia virus infection [40], and coxsackievirus infection [41].
Alternatively, the loss of P58IPK rendered mice more susceptible to influenza virus infection [42] due to a hyperactive innate immune response and loss of P58IPK-mediated PKR regulation. P58IPK also inhibits PERK, an eIF2a kinase that is activated during ER stress [43]. Due to the inhibitory role that P58IPK has on PERK, mice lacking P58IPK display T1D and late-stage type 2 diabetes [44], although the disease phenotype is less severe than in mice lacking PERK [45]. In both models, the mice display pancreatic β-cell apoptosis due to disruptions in ER stress homeostasis. However, only recently has it been shown that in humans, the deletion of Dnajc3 results in juvenile-onset diabetes and multisystemic neurodegenerative disorders [46]. Together, these studies show how P58IPK has diverse roles in both viral infection and autoimmune disease through its ability to interact with two different eIF2α kinases.
RIG-I, which is encoded by the DDX58 gene and is the RLR complementary to MDA5, has been implicated in immune deficiencies that cause inflammatory bowel disease and Crohn’s disease (CD). Mice lacking RIG-I have fewer and smaller Peyer’s patches, as well as an increase in apoptotic B220+ cells within the patches [47]. Additionally, CD patients have decreased levels of RIG-I in the intestinal epithelium [48]. In a healthy individual, the interferon response is negatively regulated via autophagy through the association with RIG-I and MAVS [49] or with NOD2 [50], and the development of autoimmunity is avoided. When autophagy is reduced, there are more reactive oxygen species within the cell, and more RLR activity to produce inflammation [51]. This indicates that the processes of autophagy and interferon-mediated immunity are in balance with each other. Autophagy is inhibited in CD epithelial cells, leading to an increase in inflammation during E. coli infection [52], and autophagy is increased when NFκB, the link to interferon activation, is inhibited [53]. Additionally, CD has been linked to mutations in the IRGM gene [54], which encodes a GTP-binding protein that induces autophagy by signaling through RIG-I.
It has also been demonstrated that RIG-I and NOD2 negatively regulate each other [50]. Particularly, knockdown of RIG-I results in increased NFκB activation through NOD2, which is corroborated by the finding that the main NOD2 mutations in CD patients are associated with increased negative regulation of RIG-I [50]. These interferon-producing pathways must be kept in balance in a healthy individual. In CD patients with reduced levels of RIG-I, the characteristic inflammation may be caused by an increase in NOD2 function.
Mutations in DDX58 and IFIH1 also converge on another autosomal disorder, called Singleton-Merten syndrome (SMS) [55,56]. While the originally identified syndrome in IFIH1 patients was characterized by dental abnormalities, aortic calcification, glaucoma, and skeletal abnormalities, DDX58 patients exhibited all of the above traits except for dental abnormalities. These patients were this diagnosed with atypical SMS. Nevertheless, in both studies, elevated levels of IFNβ were observed, thus identifying a common pathogenic autoimmune mechanism.
Finally, mutations in the PEPD gene causes prolidase deficiency (PD) and are associated with SLE, among other severe symptoms [57]. PD is a rare autosomal disorder, affecting 1 in 1–2 million newborns. A recent report has revealed that prolidase is required for normal expression of the type I IFN receptor and an IFN response during RNA virus infection [58]. Interestingly, Lubick et al. show that while prolidase deficiency results in a decreased type I IFN response, the mechanism of increased ISG production in PD patients may be due to an alternate mechanism via RSAD2/viperin activation. Taken together, these studies show how tight regulation of type I IFN and ISG induction must be controlled to defend against RNA virus infection and to avoid autoimmunity (Fig. 1).
The DNA Sensing Pathways
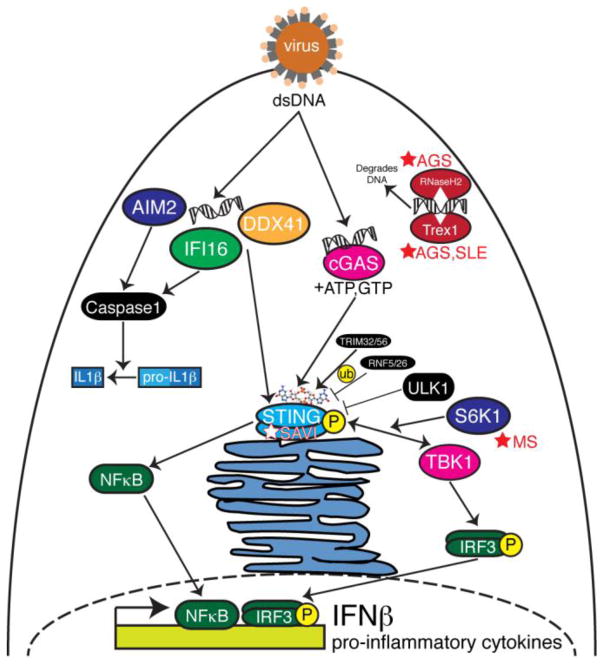
While the innate immune signaling pathways downstream from the TLRs, NLRs, and RLRs converge on IFNβ and pro-inflammatory cytokine induction, so do signaling pathways from receptors that recognize cytosolic DNA. In 2008, four independent groups identified STING (stimulator of interferon genes, also known as TMEM173, MITA, ERIS, MYPS) as a critical component of the innate immune response to cytosolic DNA and acting upstream of TANK binding kinase 1 (TBK1) and IRF3 phosphorylation [59–62]. However, at the time, it was not know if DNA bound directly to STING to activate the innate immune response. One of the first innate immune DNA sensors to be identified was DNA-dependent activator of IFN regulatory factors (DAI). Takaoka et al. demonstrated that DAI is critical for DNA-mediated IRF3 activation and subsequent IFNβ induction in response to HSV-1 infection [63]. In dendritic cells, DDX41 was shown to recognize exogenous dsDNA as well as genomic DNA from HSV-1 infection to activate IRF3 and NFκB to lead to IFNβ induction in a STING-dependent manner [64]. IFI16, another DNA sensor, has been shown to bind DNA from vaccinia virus and signal through STING and TBK1 [65]. IFI16 also responds to the herpesviruses herpes simples virus 1 (HSV-1) and human cytomegalovirus, activating the inflammasome [66,67]. Further, while IFI16 is able to recognize HSV-1 viral DNA in the nucleus, the virus has established a mechanism to degrade IFI16-mediated DNA sensing via the viral protein ICP0 [68]. Finally, AIM2 has been identified as a DNA sensor whose activation leads to inflammasome activation [69,70], and AIM2 deficient mice fail to produce IL1β in response to viral and bacterial infection [71]. In fact, a number of AIM2-like receptors (ALRs) that contain PYHIN domains have been shown to colocalize with STING to induce IFNβ via a STING-mediated pathway [72].
Upon showing that STING binds directly to cyclic dinucleotides (CDNs) [73–77], the search for the metabolic source for these cytosolic CDNs began in earnest. It has been shown that c-di-AMP is secreted during Listeria monocytogenes infection, inducing IFNβ in a STING-dependent manner [78,79]. In 2013, the Chen lab discovered cyclic GMP-AMP synthase (cGAS), and when activated by cytosolic DNA ligands, cGAS metabolizes ATP and GTP into non-canonical cyclic GMP-AMP (cGAMP) containing 2′-5′ phosphodiester linkages which then bind to and activate STING [80–84]. When activated by cGAMP, STING dimerizes and translocates to perinuclear regions where TBK1 is recruited, leading to STING and IRF3 phosphorylation [85]. The addition of cGAS to the family of DNA sensors provides the missing link between STING’s natural ligand, CDNs, and the recognition of cytosolic DNA (Fig. 2).
Figure 2. The DNA sensing pathways.
Following DNA virus infection, genomic dsDNA is sensed in the cytosol by AIM2, IFI16, DDX41, and cGAS. AIM2 and IFI16 signal though caspase 1 to activate IL1β and the inflammasome. IFI16 and DDX41 have been shown to activate STING. When activated by dsDNA, cGAS metabolizes cyclic GMP-AMP (cGAMP) which activates STING. A number of proteins regulate STING such as ubiquitin (ub) ligases, TRIM32, TRIM56, RNF5, and RNF26, and ULK1. The STING/TBK1/S6K1 complex activates IRF3, a transcription factor that localizes to the nucleus to induce IFNβ. STING also activates NFκB that localizes to the nucleus to induce pro-inflammatory cytokines. The DNA nucleases Trex1 and RNaseH2 act as checkpoints to degrade cytosolic DNA so that the DNA sensing pathways are not hyperactive. Mutations in these DNA nucleases lead to autoimmune inflammatory diseases such as Aicardi-Goutières syndrome (AGS) and systemic lupus erythematosus (SLE). Mutations in STING lead to STING-associated vasculopathy with onset in infancy (SAVI). Chemical inhibition of S6K1 protects mice from experimental autoimmune encephalomyelitis, a mouse model for studying multiple sclerosis (MS).
These studies exemplify how the sensing of cytosolic DNA by cGAS initiates the metabolism of cGAMP and thus the amplification of the STING-mediated IFNβ response. It is thus no surprise that STING-mediated signaling must be kept under tight control to effectively respond to virus infection, but not exhibit hyperactivity and lead to an autoimmune response. A number of mechanisms have been shown to regulate STING activity. For example, K63-linked ubiquitination of STING by TRIM56 or TRIM32 leads to STING dimerization and interaction withTBK1 [86,87]. Alternatively, K48-linked ubiquitination of STING by RNF5 and RNF26 leads to decreased anti-viral activity due to STING degradation [88,89]. Additionally, ULK1-mediated phosphorylation of STING at S366 upon cGAMP stimulation has been shown to inhibit STING activity [90]. However, a subsequent report shows that TBK1 phosphorylates STING at S366 to activate IRF3 [85]. Taken together, positive and negative regulation of STING occurs through a number of mechanisms, each of which may play different roles in terms of STING-mediated microbial pathogenesis or autoimmune disease.
A number of viral proteins, including those from RNA virus infection, are able to inhibit the cGAS/STING pathway to inhibit the innate immune response. One of the first viruses to be utilized in studying the STING-mediated innate immune response was HSV-1, and it was shown both in vitro and in vivo that the loss of STING resulted in increased viral replication and mortality in animals [59,91]. Interesting, the HSV-1 protein ICP0 was shown to interact with and stabilize STING in certain cell lines to achieve maximal viral replication [92]. However, cGAS is responsible for initiating the innate immune response during HSV-1 infection since cGAS-deficient mice succumb more rapidly to HSV-1 infection [93]. Members of the gammaherpesviridae, such as Kaposi’s sarcoma herpesvirus or Epstein-Barr virus, encode inhibitors of STING and cGAS such as vIRF1, LANA, and ORF52 [94,95]. The function of these proteins is to inhibit either the binding of DNA to cGAS or STING activation. Human papillomavirus and adenovirus encode oncogenes that inhibit STING activity, namely E7 and E1A [96]. These studies provide mechanistic evidence as to how viral oncogenes inhibit STING, whose signaling is important to defend against tumorigenesis [97,98].
DNA sensing implicated in autoimmune diseases
While the cGAS/STING-mediated innate immune response is necessary to defend against DNA virus infection, DNA-mediated signaling must also be controlled to prevent against certain autoimmune diseases. TREX1 encodes a 3′-5′ DNA exonuclease, and a loss of function mutation in this gene results in AGS, SLE, and other autoinflammatory diseases [99,100]. Interestingly, crossing TREX1-deficient mice with mice lacking either STING or the IFNα/β receptor resulted in a rescue of the autoinflammatory phenotypes [101,102], placing STING signaling central to these interferonopathies. Supporting this centrality, children bearing gain-of-function mutations in exon 5 of STING display a hyperactive interferon response that results in neonatal-onset systemic inflammation. The clinical syndrome is called STING-associated vasculopathy with onset in infancy (SAVI) [103]. An independent study also showed that the V155M mutation in STING resulted in spontaneous activation of STING and familial lupus-like disease [104].
Mutations in RNASEH2A, RNASEH2B, and RNASEH2C have also been linked to AGS [33,105]. The proteins encoded by these genes form the RNase H2 complex and function to degrade cellular RNA:DNA hybrids. In a mouse model of the disease due to a homozygous A174T knock-in mutation, the cGAS/STING pathway was central to the autoinflammatory pathology, since the loss of STING in these mice reduced levels of inflammatory cytokines.
Recently, it was shown that the kinase domain of the ribosomal protein S6 kinase 1 (S6K1) interacts with STING to mediate the formation of the S6K1-STING-TBK1 complex necessary for phosphorylation of IRF3 [106]. Considering that the pan-ribosomal S6 kinase (RSK) inhibitor BI-D1870 is able to protect mice from experimental autoimmune encephalomyelitis (EAE), a mouse model for studying multiple sclerosis, this may be evidence for the role of the STING signalosome in multiple sclerosis autoimmune disease [107]. Lemos et al. provide further evidence to link STING and multiple sclerosis in their study showing that the activation of cGAS/STING in an EAE mouse model suppresses autoimmunity [108]. Taken together, there is increasing evidence that STING plays a central role in autoimmune interferonopathies (Fig. 2). Seeing that cGAS is hyperactive in Trex1- and DNaseII-deficient mice, and that deletion of cGAS in these backgrounds rescues the lethal autoimmune phenotypes [109], it is possible that there are human genetic variants in cGAS that also result in autoimmune hyperinflammatory diseases.
Conclusions
Defining how innate immune pathways function is central to understanding the mechanisms underlying the development of autoimmune disease and is a necessary prerequisite to devising targeted therapies for such disorders. Central to these autoimmune pathologies is elevated levels of type I interferon, a powerful molecule that is critical to combat virus infection and bridge the innate and adaptive immune responses. For some microbial diseases and cancers, interferon therapy is still the current standard of care, although this treatment is associated with severe side effects. The development of methods to inhibit nucleic acid sensors will be important for therapeutic intervention to treat autoimmune interferonopathies. Seeing as viruses already encode a number of inhibitors for different nodes of these nucleic acid sensing pathways, is it possible that we can turn to these mechanisms of viral inhibition to develop novel therapies?
Acknowledgments
Research in the Goodman Lab is funded by NIH Grant R00 AI106963 and funds from Washington State University. L.R.H. Ahlers is supported by NIH Training Grant T32 GM008336.
Footnotes
Conflict of Interest
Laura Ahlers and Alan Goodman declare that they have no conflict of interest.
Compliance with Ethics Guidelines
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Contributor Information
Laura R. H. Ahlers, Email: laurahylden@vetmed.wsu.edu.
Alan G. Goodman, Email: agoodman@vetmed.wsu.edu.
References
- 1.Kawai T, Akira S. Toll-like Receptors and Their Crosstalk with Other Innate Receptors in Infection and Immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 2.Sabin LR, Hanna SL, Cherry S. Innate antiviral immunity in Drosophila. Current Opinion in Immunology. 2010;22:4–9. doi: 10.1016/j.coi.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 4.Paludan SR, Bowie AG. Immune sensing of DNA. Immunity. 2013;38:870–880. doi: 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma S, tenOever BR, Grandvaux N, Zhou G-P, Lin R, Hiscott J. Triggering the Interferon Antiviral Response Through an IKK-Related Pathway. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- 6.Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, Nakaya T, Katsuki M, Noguchi S, Tanaka N, et al. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity. 2000;13:539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- 7.Goodman AG, Zeng H, Proll SC, Peng X, Cilloniz C, Carter VS, Korth MJ, Tumpey TM, Katze MG. The alpha/beta interferon receptor provides protection against influenza virus replication but is dispensable for inflammatory response signaling. J Virol. 2010;84:2027–2037. doi: 10.1128/JVI.01595-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 9.Levy DE, Marie I, Smith E, Prakash A. Enhancement and Diversification of IFN Induction by IRF-7-Mediated Positive Feedback. Journal of Interferon & Cytokine Research. 2002;22:87–93. doi: 10.1089/107999002753452692. [DOI] [PubMed] [Google Scholar]
- 10.Pfeffer LM, Kim JG, Pfeffer SR, Carrigan DJ, Baker DP, Wei L, Homayouni R. Role of Nuclear Factor-{kappa}B in the Antiviral Action of Interferon and Interferon-regulated Gene Expression. Journal Of Biological Chemistry. 2004;279:31304–31311. doi: 10.1074/jbc.M308975200. [DOI] [PubMed] [Google Scholar]
- 11.Ahn J, Barber GN. Self-DNA, STING-dependent signaling and the origins of autoinflammatory disease. Current Opinion in Immunology. 2014;31:121–126. doi: 10.1016/j.coi.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 12.Crow YJ, Manel N. Aicardi-Goutieres syndrome and the type I interferonopathies. Nat Rev Immunol. 2015;15:429–440. doi: 10.1038/nri3850. [DOI] [PubMed] [Google Scholar]
- 13.Yoshida H, Okabe Y, Kawane K, Fukuyama H, Nagata S. Lethal anemia caused by interferon-beta produced in mouse embryos carrying undigested DNA. Nat Immunol. 2005;6:49–56. doi: 10.1038/ni1146. [DOI] [PubMed] [Google Scholar]
- 14.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid–inducible gene-I and melanoma differentiation–associated gene 5. The Journal of Experimental Medicine. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang DC, Gopalkrishnan RV, Wu Q, Jankowsky E, Pyle AM, Fisher PB. mda-5: An interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc Natl Acad Sci U S A. 2002;99:637–642. doi: 10.1073/pnas.022637199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui Y, Li M, Walton KD, Sun K, Hanover JA, Furth PA, Hennighausen L. The Stat3/5 locus encodes novel endoplasmic reticulum and helicase-like proteins that are preferentially expressed in normal and neoplastic mammary tissue. Genomics. 2001;78:129–134. doi: 10.1006/geno.2001.6661. [DOI] [PubMed] [Google Scholar]
- 17.Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo Y-M, Gale M, Akira S, et al. Shared and Unique Functions of the DExD/H-Box Helicases RIG-I, MDA5, and LGP2 in Antiviral Innate Immunity. The Journal of Immunology. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 18.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nature Immunology. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 19.Seth RB, Sun L, Ea C-K, Chen ZJ. Identification and Characterization of MAVS, a Mitochondrial Antiviral Signaling Protein that Activates NF-κB and IRF3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 20.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 21.Hornung V, Ellegast J, Kim S, Brzózka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann K-K, Schlee M, et al. 5′-Triphosphate RNA Is the Ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 22.Loo Y-M, Fornek J, Crochet N, Bajwa G, Perwitasari O, Martinez-Sobrido L, Akira S, Gill MA, García-Sastre A, Katze MG, et al. Distinct RIG-I and MDA5 Signaling by RNA Viruses in Innate Immunity. Journal of Virology. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang X, Yue Y, Li D, Zhao Y, Qiu L, Chen J, Pan Y, Xi J, Wang X, Sun Q, et al. Antibody-dependent enhancement of dengue virus infection inhibits RLR-mediated Type-I IFN-independent signalling through upregulation of cellular autophagy. Scientific Reports. 2016;6:22303. doi: 10.1038/srep22303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suthar MS, Ma DY, Thomas S, Lund JM, Zhang N, Daffis S, Rudensky AY, Bevan MJ, Clark EA, Kaja MK, et al. IPS-1 is essential for the control of West Nile virus infection and immunity. PLoS Pathog. 2010;6:e1000757. doi: 10.1371/journal.ppat.1000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loeb M, Eskandarian S, Rupp M, Fishman N, Gasink L, Patterson J, Bramson J, Hudson TJ, Lemire M. Genetic Variants and Susceptibility to Neurological Complications Following West Nile Virus Infection. Journal of Infectious Diseases. 2011;204:1031–1037. doi: 10.1093/infdis/jir493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knip M, Veijola R, Virtanen SM, Hyöty H, Vaarala O, Åkerblom HK. Environmental Triggers and Determinants of Type 1 Diabetes. Diabetes. 2005;54:S125–S136. doi: 10.2337/diabetes.54.suppl_2.s125. [DOI] [PubMed] [Google Scholar]
- 27.Ylipaasto P, Klingel K, Lindberg AM, Otonkoski T, Kandolf R, Hovi T, Roivainen M. Enterovirus infection in human pancreatic islet cells, islet tropism in vivo and receptor involvement in cultured islet beta cells. Diabetologia. 2004;47:225–239. doi: 10.1007/s00125-003-1297-z. [DOI] [PubMed] [Google Scholar]
- 28.Clements GB, Galbraith DN, Taylor KW. Coxsackie B virus infection and onset of childhood diabetes. Lancet. 1995;346:221–223. doi: 10.1016/s0140-6736(95)91270-3. [DOI] [PubMed] [Google Scholar]
- 29.Yeung WC, Rawlinson WD, Craig ME. Enterovirus infection and type 1 diabetes mellitus: systematic review and meta-analysis of observational molecular studies. BMJ. 2011;342:d35. doi: 10.1136/bmj.d35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu S, Wang H, Jin Y, Podolsky R, Reddy MVPL, Pedersen J, Bode B, Reed J, Steed D, Anderson S, et al. IFIH1 polymorphisms are significantly associated with type 1 diabetes and IFIH1 gene expression in peripheral blood mononuclear cells. Human Molecular Genetics. 2009;18:358–365. doi: 10.1093/hmg/ddn342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Funabiki M, Kato H, Miyachi Y, Toki H, Motegi H, Inoue M, Minowa O, Yoshida A, Deguchi K, Sato H, et al. Autoimmune Disorders Associated with Gain of Function of the Intracellular Sensor MDA5. Immunity. 2014;40:199–212. doi: 10.1016/j.immuni.2013.12.014. Of importance: This paper describes the generation of a mouse carrying a mutation in MDA5 through ENU mutagenesis and the mechanisms by which lupus-like autoimmune disease develops. [DOI] [PubMed] [Google Scholar]
- 32.Santin I, Moore F, Grieco FA, Marchetti P, Brancolini C, Eizirik DL. USP18 is a key regulator of the interferon-driven gene network modulating pancreatic beta cell inflammation and apoptosis. Cell Death & Disease. 2012;3:e419. doi: 10.1038/cddis.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crow YJ, Chase DS, Lowenstein Schmidt J, Szynkiewicz M, Forte GM, Gornall HL, Oojageer A, Anderson B, Pizzino A, Helman G, et al. Characterization of human disease phenotypes associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR, and IFIH1. Am J Med Genet A. 2015;167A:296–312. doi: 10.1002/ajmg.a.36887. Of importance: This paper describes a number of interferonopathies due to familial mutations in nucleic acid sensing proteins. They also describe potential therapeutic windows for autoimmune disease treatment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pestal K, Funk CC, Snyder JM, Price ND, Treuting PM, Stetson DB. Isoforms of RNA-Editing Enzyme ADAR1 Independently Control Nucleic Acid Sensor MDA5-Driven Autoimmunity and Multi-organ Development. Immunity. 2015;43:933–944. doi: 10.1016/j.immuni.2015.11.001. Of importance: This paper provides mechanistic data as to how mutations in ADAR control MDA5-MAVS signaling and organ development, leading to human diseases caused by ADAR mutations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lincez PJ, Shanina I, Horwitz MS. Reduced Expression of the MDA5 Gene IFIH1 Prevents Autoimmune Diabetes. Diabetes. 2015;64:2184–2193. doi: 10.2337/db14-1223. [DOI] [PubMed] [Google Scholar]
- 36.McCartney SA, Vermi W, Lonardi S, Rossini C, Otero K, Calderon B, Gilfillan S, Diamond MS, Unanue ER, Colonna M. RNA sensor-induced type I IFN prevents diabetes caused by a β cell-tropic virus in mice. The Journal of Clinical Investigation. 2011;121:1497–1507. doi: 10.1172/JCI44005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katze MG, Tomita J, Black T, Krug RM, Safer B, Hovanessian AG. Influenza virus regulates protein synthesis during infection by repressing the autophosphorylation and activity of the cellular 68,000- M r protein kinase. Journal Of Virology. 1988;62:3710–3717. doi: 10.1128/jvi.62.10.3710-3717.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee TG, Tomita J, Hovanessian AG, Katze MG. Purification and partial characterization of a cellular inhibitor of the interferon-induced protein kinase of M r 68,000 from influenza virus-infected cells. Proc Natl Acad Sci U S A. 1990;87:6208–6212. doi: 10.1073/pnas.87.16.6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goodman AG, Smith JA, Balachandran S, Perwitasari O, Proll SC, Thomas MJ, Korth MJ, Barber GN, Schiff LA, Katze MG. The cellular protein P58IPK regulates influenza virus mRNA translation and replication through a PKR-mediated mechanism. J Virol. 2007;81:2221–2230. doi: 10.1128/JVI.02151-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goodman AG, Tanner BCW, Chang ST, Esteban M, Katze MG. Virus infection rapidly activates the P58IPK pathway, delaying peak kinase activation to enhance viral replication. Virology. 2011;417:27–36. doi: 10.1016/j.virol.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang HM, Qiu Y, Ye X, Hemida MG, Hanson P, Yang D. P58(IPK) inhibits coxsackievirus-induced apoptosis via the PI3K/Akt pathway requiring activation of ATF6a and subsequent upregulation of mitofusin 2. Cell Microbiol. 2014;16:411–424. doi: 10.1111/cmi.12229. [DOI] [PubMed] [Google Scholar]
- 42.Goodman AG, Fornek JL, Medigeshi GR, Perrone LA, Peng X, Dyer MD, Proll SC, Knoblaugh SE, Carter VS, Korth MJ, et al. P58(IPK): A Novel “CIHD” Member of the Host Innate Defense Response against Pathogenic Virus Infection. PLoS Pathog. 2009;5:e1000438. doi: 10.1371/journal.ppat.1000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan W, Frank CL, Korth MJ, Sopher BL, Novoa I, Ron D, Katze MG. Control of PERK eIF2a kinase activity by the endoplasmic reticulum stress-induced molecular chaperone P58 IPK. Proc Natl Acad Sci U S A. 2002;99:15920–15925. doi: 10.1073/pnas.252341799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ladiges WC, Knoblaugh SE, Morton JF, Korth MJ, Sopher BL, Baskin CR, MacAuley A, Goodman AG, LeBoeuf RC, Katze MG. Pancreatic beta-cell failure and diabetes in mice with a deletion mutation of the endoplasmic reticulum molecular chaperone gene P58IPK. Diabetes. 2005;54:1074–1081. doi: 10.2337/diabetes.54.4.1074. [DOI] [PubMed] [Google Scholar]
- 45.Harding HP, Zeng H, Zhang Y, Jungries R, Chung P, Plesken H, Sabatini DD, Ron D. Diabetes mellitus and exocrine pancreatic dysfunction in Perk −/− mice reveals a role for translational control in secretory cell survival. Mol Cell. 2001;7:1153–1163. doi: 10.1016/s1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- 46.Synofzik M, Haack TB, Kopajtich R, Gorza M, Rapaport D, Greiner M, Schonfeld C, Freiberg C, Schorr S, Holl RW, et al. Absence of BiP co-chaperone DNAJC3 causes diabetes mellitus and multisystemic neurodegeneration. Am J Hum Genet. 2014;95:689–697. doi: 10.1016/j.ajhg.2014.10.013. Of major importance: This study screened 226,194 individuals for mutations in Dnajc3 and found 8 individuals who all had mutations in Dnajc3. Individuals lacking Dnajc3 had a recessive form of diabetes mellitus and widespread neurodegeneration. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Zhang H-X, Sun Y-P, Liu Z-X, Liu X-S, Wang L, Lu S-Y, Kong H, Liu Q-L, Li X-H, et al. Rig-I −/− mice develop colitis associated with downregulation of Gαi2. Cell Research. 2007;17:858–868. doi: 10.1038/cr.2007.81. [DOI] [PubMed] [Google Scholar]
- 48.Funke B, Lasitschka F, Roth W, Penzel R, Meuer S, Saile M, Gretz N, Sido B, Schirmacher P, Autschbach F. Selective downregulation of retinoic acid-inducible gene I within the intestinal epithelial compartment in Crohn’s disease. Inflammatory Bowel Diseases. 2011;17:1943–1954. doi: 10.1002/ibd.21572. [DOI] [PubMed] [Google Scholar]
- 49.Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin K-Q, Ishii KJ, Kawai T, Akira S, Suzuki K, et al. The Atg5–Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci U S A. 2007;104:14050–14055. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morosky SA, Zhu J, Mukherjee A, Sarkar SN, Coyne CB. Retinoic Acid-induced Gene-I (RIG-I) Associates with Nucleotide-binding Oligomerization Domain-2 (NOD2) to Negatively Regulate Inflammatory Signaling. Journal of Biological Chemistry. 2011;286:28574–28583. doi: 10.1074/jbc.M111.227942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc Natl Acad Sci U S A. 2009;106:2770–2775. doi: 10.1073/pnas.0807694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bretin A, Carrière J, Dalmasso G, Bergougnoux A, B’Chir W, Maurin A-C, Müller S, Seibold F, Barnich N, Bruhat A, et al. Activation of the EIF2AK4-EIF2A/eIF2α-ATF4 pathway triggers autophagy response to Crohn disease-associated adherent-invasive Escherichia coli infection. Autophagy. 2016;12:1–14. doi: 10.1080/15548627.2016.1156823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao H, Lin L, Haq IU, Zeng SM. Inhibition of NF-κB promotes autophagy via JNK signaling pathway in porcine granulosa cells. Biochemical and Biophysical Research Communications. 2016;473:311–316. doi: 10.1016/j.bbrc.2016.03.101. [DOI] [PubMed] [Google Scholar]
- 54.Palomino-Morales RJ, Oliver J, Gómez-García M, López-Nevot MA, Rodrigo L, Nieto A, Alizadeh BZ, Martín J. Association of ATG16L1 and IRGM genes polymorphisms with inflammatory bowel disease: a meta-analysis approach. Genes and Immunity. 2009;10:356–364. doi: 10.1038/gene.2009.25. [DOI] [PubMed] [Google Scholar]
- 55.Rutsch F, MacDougall M, Lu C, Buers I, Mamaeva O, Nitschke Y, Rice GI, Erlandsen H, Kehl HG, Thiele H, et al. A specific IFIH1 gain-of-function mutation causes Singleton-Merten syndrome. Am J Hum Genet. 2015;96:275–282. doi: 10.1016/j.ajhg.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jang MA, Kim EK, Now H, Nguyen NT, Kim WJ, Yoo JY, Lee J, Jeong YM, Kim CH, Kim OH, et al. Mutations in DDX58, which encodes RIG-I, cause atypical Singleton-Merten syndrome. Am J Hum Genet. 2015;96:266–274. doi: 10.1016/j.ajhg.2014.11.019. Of importance: This study describes a family with atypical SMS carries a genetic variant in the gene encoding RIG-I. This variant results in constitutive activation of RIG-I and increased IFN levels. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Butbul Aviel Y, Mandel H, Avitan Hersh E, Bergman R, Adiv OE, Luder A, Brik R. Prolidase deficiency associated with systemic lupus erythematosus (SLE): single site experience and literature review. Pediatr Rheumatol Online J. 2012;10:18. doi: 10.1186/1546-0096-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lubick KJ, Robertson SJ, McNally KL, Freedman BA, Rasmussen AL, Taylor RT, Walts AD, Tsuruda S, Sakai M, Ishizuka M, et al. Flavivirus Antagonism of Type I Interferon Signaling Reveals Prolidase as a Regulator of IFNAR1 Surface Expression. Cell Host Microbe. 2015;18:61–74. doi: 10.1016/j.chom.2015.06.007. Of major importance: This study describes the use of West Nile virus infection to uncover that prolidase is required for normal type I IFN receptor expression and provides mechanistic data as to why mutations in prolidase lead to autoimmune disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jin L, Hill KK, Filak H, Mogan J, Knowles H, Zhang B, Perraud A-L, Cambier JC, Lenz LL. MPYS Is Required for IFN Response Factor 3 Activation and Type I IFN Production in the Response of Cultured Phagocytes to Bacterial Second Messengers Cyclic-di-AMP and Cyclic-di-GMP. The Journal of Immunology. 2011;187:2595–2601. doi: 10.4049/jimmunol.1100088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun W, Li Y, Chen L, Chen H, You F, Zhou X, Zhou Y, Zhai Z, Chen D, Jiang Z. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci U S A. 2009;106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 63.Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 64.Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Johnson KE, Bottero V, Flaherty S, Dutta S, Singh VV, Chandran B. IFI16 restricts HSV-1 replication by accumulating on the hsv-1 genome, repressing HSV-1 gene expression, and directly or indirectly modulating histone modifications. PLoS Pathog. 2014;10:e1004503. doi: 10.1371/journal.ppat.1004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li T, Chen J, Cristea IM. Human cytomegalovirus tegument protein pUL83 inhibits IFI16-mediated DNA sensing for immune evasion. Cell Host Microbe. 2013;14:591–599. doi: 10.1016/j.chom.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Orzalli MH, Conwell SE, Berrios C, DeCaprio JA, Knipe DM. Nuclear interferon-inducible protein 16 promotes silencing of herpesviral and transfected DNA. Proc Natl Acad Sci U S A. 2013;110:E4492–4501. doi: 10.1073/pnas.1316194110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brunette RL, Young JM, Whitley DG, Brodsky IE, Malik HS, Stetson DB. Extensive evolutionary and functional diversity among mammalian AIM2-like receptors. J Exp Med. 2012;209:1969–1983. doi: 10.1084/jem.20121960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ouyang S, Song X, Wang Y, Ru H, Shaw N, Jiang Y, Niu F, Zhu Y, Qiu W, Parvatiyar K, et al. Structural Analysis of the STING Adaptor Protein Reveals a Hydrophobic Dimer Interface and Mode of Cyclic di-GMP Binding. Immunity. 2012;36:1073–1086. doi: 10.1016/j.immuni.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shang G, Zhu D, Li N, Zhang J, Zhu C, Lu D, Liu C, Yu Q, Zhao Y, Xu S, et al. Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP. Nat Struct Mol Biol. 2012;19:725–727. doi: 10.1038/nsmb.2332. [DOI] [PubMed] [Google Scholar]
- 76.Shu C, Yi G, Watts T, Kao CC, Li P. Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nat Struct Mol Biol. 2012;19:722–724. doi: 10.1038/nsmb.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yin Q, Tian Y, Kabaleeswaran V, Jiang X, Tu D, Eck MJ, Chen ZJ, Wu H. Cyclic di-GMP sensing via the innate immune signaling protein STING. Mol Cell. 2012;46:735–745. doi: 10.1016/j.molcel.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP Secreted by Intracellular Listeria monocytogenes Activates a Host Type I Interferon Response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sauer JD, Sotelo-Troha K, von Moltke J, Monroe KM, Rae CS, Brubaker SW, Hyodo M, Hayakawa Y, Woodward JJ, Portnoy DA, et al. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect Immun. 2011;79:688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, Hopfner KP, Ludwig J, Hornung V. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Diner EJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, Hyodo M, Hayakawa Y, Hammond MC, Vance RE. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep. 2013;3:1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP Synthase Is a Cytosolic DNA Sensor That Activates the Type I Interferon Pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. Of major importance: This is the first paper to identify cGAS and show that it binds cytosolic DNA to generate cGAMP and activate type I IFN. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gao D, Wu J, Wu YT, Du F, Aroh C, Yan N, Sun L, Chen ZJ. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 2013;341:903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, Du F, Ren J, Wu YT, Grishin NV, et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015;347:aaa2630. doi: 10.1126/science.aaa2630. Of importance: This study describes a common mechanism that both MAVS and STING use to activate IRF3 through TBK1 and TRIF. [DOI] [PubMed] [Google Scholar]
- 86.Tsuchida T, Zou J, Saitoh T, Kumar H, Abe T, Matsuura Y, Kawai T, Akira S. The Ubiquitin Ligase TRIM56 Regulates Innate Immune Responses to Intracellular Double-Stranded DNA. Immunity. 2010;33:765–776. doi: 10.1016/j.immuni.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 87.Zhang J, Hu MM, Wang YY, Shu HB. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J Biol Chem. 2012;287:28646–28655. doi: 10.1074/jbc.M112.362608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Qin Y, Zhou MT, Hu MM, Hu YH, Zhang J, Guo L, Zhong B, Shu HB. RNF26 temporally regulates virus-triggered type I interferon induction by two distinct mechanisms. PLoS Pathog. 2014;10:e1004358. doi: 10.1371/journal.ppat.1004358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhong B, Zhang L, Lei C, Li Y, Mao A-P, Yang Y, Wang Y-Y, Zhang X-L, Shu H-B. The Ubiquitin Ligase RNF5 Regulates Antiviral Responses by Mediating Degradation of the Adaptor Protein MITA. Immunity. 2009;30:397–407. doi: 10.1016/j.immuni.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 90.Konno H, Konno K, Barber GN. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell. 2013;155:688–698. doi: 10.1016/j.cell.2013.09.049. Of importance: This study shows how STING is negatively regulated through ULK1, which is also activated by cGAMP, to prevent STING hyperactivity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kalamvoki M, Roizman B. HSV-1 degrades, stabilizes, requires, or is stung by STING depending on ICP0, the US3 protein kinase, and cell derivation. Proc Natl Acad Sci U S A. 2014;111:E611–617. doi: 10.1073/pnas.1323414111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ma Z, Jacobs SR, West JA, Stopford C, Zhang Z, Davis Z, Barber GN, Glaunsinger BA, Dittmer DP, Damania B. Modulation of the cGAS-STING DNA sensing pathway by gammaherpesviruses. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1503831112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wu JJ, Li W, Shao Y, Avey D, Fu B, Gillen J, Hand T, Ma S, Liu X, Miley W, et al. Inhibition of cGAS DNA Sensing by a Herpesvirus Virion Protein. Cell Host Microbe. 2015;18:333–344. doi: 10.1016/j.chom.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lau L, Gray EE, Brunette RL, Stetson DB. DNA tumor virus oncogenes antagonize the cGAS-STING DNA-sensing pathway. Science. 2015;350:568–571. doi: 10.1126/science.aab3291. Of major importance: This study identifies that transformed cell lines harbor viral oncogenes that inhibit the cGAS/STING pathway. [DOI] [PubMed] [Google Scholar]
- 97.Xia T, Konno H, Ahn J, Barber GN. Deregulation of STING Signaling in Colorectal Carcinoma Constrains DNA Damage Responses and Correlates With Tumorigenesis. Cell Rep. 2016;14:282–297. doi: 10.1016/j.celrep.2015.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ahn J, Konno H, Barber GN. Diverse roles of STING-dependent signaling on the development of cancer. Oncogene. 2015;34:5302–5308. doi: 10.1038/onc.2014.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Crow YJ, Hayward BE, Parmar R, Robins P, Leitch A, Ali M, Black DN, van Bokhoven H, Brunner HG, Hamel BC, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat Genet. 2006;38:917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- 100.Lee-Kirsch MA, Gong M, Chowdhury D, Senenko L, Engel K, Lee YA, de Silva U, Bailey SL, Witte T, Vyse TJ, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat Genet. 2007;39:1065–1067. doi: 10.1038/ng2091. [DOI] [PubMed] [Google Scholar]
- 101.Gall A, Treuting P, Elkon KB, Loo Y-M, Gale M, Jr, Barber GN, Stetson DB. Autoimmunity Initiates in Nonhematopoietic Cells and Progresses via Lymphocytes in an Interferon-Dependent Autoimmune Disease. Immunity. 2012;36:120–131. doi: 10.1016/j.immuni.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ahn J, Ruiz P, Barber GN. Intrinsic self-DNA triggers inflammatory disease dependent on STING. J Immunol. 2014;193:4634–4642. doi: 10.4049/jimmunol.1401337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu Y, Jesus AA, Marrero B, Yang D, Ramsey SE, Sanchez GA, Tenbrock K, Wittkowski H, Jones OY, Kuehn HS, et al. Activated STING in a Vascular and Pulmonary Syndrome. N Engl J Med. 2014 doi: 10.1056/NEJMoa1312625. Of major importance: This paper shows that children carrying conserved mutations in the gene encoding STING display a form of autoimmune disease through STING hyperactivation and constituitive Stat1 signaling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jeremiah N, Neven B, Gentili M, Callebaut I, Maschalidi S, Stolzenberg MC, Goudin N, Fremond ML, Nitschke P, Molina TJ, et al. Inherited STING-activating mutation underlies a familial inflammatory syndrome with lupus-like manifestations. J Clin Invest. 2014;124:5516–5520. doi: 10.1172/JCI79100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Crow YJ, Leitch A, Hayward BE, Garner A, Parmar R, Griffith E, Ali M, Semple C, Aicardi J, Babul-Hirji R, et al. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection. Nat Genet. 2006;38:910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- 106.Wang F, Alain T, Szretter KJ, Stephenson K, Pol JG, Atherton MJ, Hoang HD, Fonseca BD, Zakaria C, Chen L, et al. S6K-STING interaction regulates cytosolic DNA-mediated activation of the transcription factor IRF3. Nat Immunol. 2016 doi: 10.1038/ni.3433. Of importance: This paper uncovers the role of the ribosomal protein S6K1 in STING/TBK1-mediated IRF3 activation, implicating S6K1 as a target to treat autoimmune disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Takada I, Yogiashi Y, Makishima M. The ribosomal S6 kinase inhibitor BI-D1870 ameliorated experimental autoimmune encephalomyelitis in mice. Immunobiology. 2016;221:188–192. doi: 10.1016/j.imbio.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 108.Lemos H, Huang L, Chandler PR, Mohamed E, Souza GR, Li L, Pacholczyk G, Barber GN, Hayakawa Y, Munn DH, et al. Activation of the STING adaptor attenuates experimental autoimmune encephalitis. J Immunol. 2014;192:5571–5578. doi: 10.4049/jimmunol.1303258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gao D, Li T, Li XD, Chen X, Li QZ, Wight-Carter M, Chen ZJ. Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proc Natl Acad Sci U S A. 2015;112:E5699–5705. doi: 10.1073/pnas.1516465112. Of major importance: This study provides mechanistic data as to how cGAS hyperactivation may lead to a hyperinflammatory response, implicating it as a major target to treat autoimmune diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]