Abstract
Deletion of chromosome 17p with a loss of p53 is an unfavorable cytogenetic change in chronic lymphocytic leukemia (CLL) with poor clinical outcome. Since p53 affects mitochondrial function and integrity, we examined possible mitochondrial changes in CLL mice with TCL1-Tg/p53−/− and TCL1-Tg/p53+/+ genotypes and in primary leukemia cells from CLL patients with or without 17p-deletion. Although the expression of mitochondrial COX1, ND2, and ND6 decreased in p53−/−CLL cells, there was an increase in mitochondrial biogenesis as evidenced by higher mitochondrial mass and mtDNA copy number associated with an elevated expression of TFAM and PGC-1α. Surprisingly, the overall mitochondrial respiratory activity and maximum reserved capacity increased in p53−/− CLL cells. Our study suggests that leukemia cells lacking p53 seem able to maintain respiratory function by compensatory increase in mitochondrial biogenesis.
Keywords: Chronic Lymphocytic Leukemia, p53, mitochondrial biogenesis
1. Introduction
Chronic lymphocytic leukemia (CLL) is the most common form of adult leukemia and is characterized by B-cell expansion of CD5+ lymphocytes along with expression of cell surface antigens including CD19, CD20, and CD23 (Hallek, 2013; Rozman and Montserrat, 1995). Cytogenetic studies reveal that nearly 50% of patients have genetic abnormalities, which can influence and predict disease aggressiveness, response to therapy and overall survival (Juliusson and Gahrton, 1993). A recent study further identified certain non-coding recurrent mutations (driver mutations) in CLL that may affect gene expression and splicing, contributing to CLL disease development (Puente et al., 2015). Approximately 5–7% of the CLL cases have a deletion on the short arm of chromosome 17 (del17p). Importantly, del17p in CLL patients is consistently associated with a significant decrease in the median treatment-free interval and a shorter median survival time compared to CLL with other genetic abnormalities (Döhner et al., 2000). A contributing factor to this dismal statistics is that the TP53 tumor suppressor gene is located in chromosome 17p. Enumerable research has shown that loss of p53 gene, or its mutation, results in altered response to DNA damage, cellular metabolism and mitochondria function. Previous studies support the occurrence of mitochondria alterations in CLL. Increased number of mitochondria was observed in CLL patients compared to normal lymphocytes and correlated to the level of endogenous nitric oxide (Carew et al., 2004). However, the impact of loss of p53 on mitochondrial function in CLL leukemia cells has not been directly examined. Previous studies suggest that p53 plays an important role in maintaining mitochondrial genetic integrity and function (Achanta et al., 2005; Matoba et al., 2006). It is unclear if loss of p53 would lead to a decrease in mitochondrial respiration in CLL cells, or if the leukemia cells might be able to compensate the effect of p53 loss by up-regulating mitochondrial function. To investigate this important question and its physiological relevance, it is essential to use proper experimental models.
The TCL1 transgenic mouse model has been shown to mimic human CLL disease, and it is a useful model for in vivo study to test potential therapeutic agents to investigate the biology of this disease (Bichi et al., 2002). Since TCL1 transgenic mice retain functional wild-type TP53, it is necessary to compare with TCL1 transgenic mice with p53 deletion in order to evaluate the effect of p53 on CLL disease development and other biological consequences in vivo. To this end, we recently developed a TCL1/p53−/− mouse colony by crossing of TCL1 (p53+/+) transgenic mice with p53−/− mice (Liu et al., 2014). Similar to the TCL1/p53+/+ transgenic mice, TCL1/p53−/− mice showed expansion of the CD5+/IgM+ B-cells but with an earlier disease onset. In addition, TCL1/p53−/− mice had significantly shorter survival time, were more resistant to DNA-damaging drugs used in clinic, and had increased expression of anti-apoptotic proteins including Mcl-1, Bcl-XL, and Bcl-2 (Liu et al., 2014). Significantly, these alterations were comparable to what was observed in CLL patient samples with del17p. P53 has been shown to be actively involved in mitochondrial biogenesis and function either through direct regulation of mitochondrial genes by transcriptional activation/repression or through influencing other signaling pathways such as NFκB (Ak and Levine, 2010; Johnson and Perkins, 2012). Mitochondria are unique in that it possesses its own mtDNA that encode for certain components of the electron transport chain and replication machinery. Aerobic respiration is executed by five multi-subunit enzymes including complex I, II, III, IV and F1-FO-ATP synthase located within the mitochondria. Loss of p53 has been shown to result in decreased aerobic respiratory capacity in part through affecting SCO2 (synthesis of cytochrome c oxidase 2), which aids in assembly of cytochrome c oxidase (Matoba et al., 2006). In addition, p53 may directly interact with the mitochondria polymerase gamma (DNA Pol γ) upon mtDNA damage, leading to an enhancement of replication activity of DNA Pol γ (Achanta et al., 2005).
We have previously shown that a loss of p53 increased the expression of anti-apoptotic proteins in TCL1/p53−/− mice, providing a possible explanation for why these cells responsed poorly to therapy. Likewise, conventional chemotherapeutic agents, which often activate the pro-apoptotic function of p53 as a mechanism to induce cell death, are not highly effective in treating del17p patients. Since mitochondria play a major role in cellular metabolism, apoptosis, and cellular response to anticancer agents, it is important to exam if loss of p53 function might affect mitochondrial function in vivo. In this study, we used the TCL1/p53−/−mouse model as an experimental system to evaluate the in vivo effect of p53 on mitochondrial function, and further tested if the findings in these mice could be observed in primary leukemia samples from CLL patients.
2. Materials and Methods
Mouse models
The generation of Eu-TCL1 and TCL1/p53 −/− mice and their maintenance were described previously (Bichi et al., 2002; Liu et al., 2014). Both TCL1/p53−/− and TCL1/p53+/+ mice were housed in the pathogen-free animal facility at the University of Texas MD Anderson Cancer Center, and the animal study was carried out under a research protocol approved by the Institutional Animal Care and Use Committee (IACUC).
Reagents
Cells were maintained in RPMI-1640 (SigmaAldrich, St. Louis, MO) supplemented with 10% fetal bovine serum (SigmaAldrich, St. Louis, MO), penicillin (100 units/ml) + streptomycin (100 μg/ml) (P4333, SigmaAldrich, St. Louis, MO) at 37 °C in a humidified 5% CO2 incubator. Rabbit anti-PGC1α (SC-13067) was from Santa Cruz Biotechnology(Dallas, TX), rabbit anti-TFAM (ARP31400_P050) was from Aviva Systems Biology(San Diego, CA), mouse anti-MitoProfile Total OXPHOS Rodent WB Antibody Cocktail (ab110413) and mouse anti-MitoProfile® Total OXPHOS Human WB Antibody Cocktail (ab110411), were purchased from AbCam (Cambridge, MA), mouse anti-β-actin (A5441) was from Sigma_Aldrich (St. Louis, MO).
Isolation of CLL cells
Primary CLL cells were obtained from the peripheral blood samples of B-CLL patients, who were diagnosed according to National Cancer Institute criteria (Zwiebel and Cheson, 1998). Prior to blood collection, informed consents under a research protocol approved by the Institutional Review Board of MD Anderson Cancer Center were obtained from all patients in accordance with the proper policy and regulation. In all experiments, CLL cells were isolated from blood samples by density gradient centrifugation (Huang et al., 2000) and incubated in RPMI-1640 medium containing 10% FBS and penicillin (100 U/ml1) + streptomycin (100 μg/ml) overnight before experiments. Mouse splenocytes were isolated as previously described (Liu et al., 2014).
Assessment of mitochondria mass
Mitochondria mass was measured by incubating 1×106 cells for 15 min with 0.1μM MitoTracker Green FM (M7514, Life Technologies, Grand Island, NY), followed by washing and re-suspension in PBS. Cellular fluorescence was then measured using a FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA) equipped with CellQuest Pro software. Flow cytometric data were analyzed using FlowJo (Tree Star, Ashland, OR).
Real time PCR
RNA was isolated using the RNEasy mini kit (74104, Qiagen, Valencia, CA) with addition of the DNase clean-up step. cDNA was prepared using 1μg of RNA for reverse transcriptase using RevertAid First Strand cDNA Synthesis Kit (K1621, Thermo Scientific, Rockford, IL). mRNA transcript levels were measured by running the reaction mix of cDNA template, primers and SYBR Green PCR Master Mix (4344463, Applied Biosystems, Grand Island, NY) on the Viia7 Real-Time PCR System (Life Technologies, Grand Island, NY). Fold changes were determined by normalizing to β–actin as an internal control and presented as 2−ΔCT. The primers used in this study are listed in Table 1.
Table 1.
| Gene | Sequence | Reference |
|---|---|---|
| Mouse | ||
| HuActinB For393 | CATGTACGTTGCTATCCAGGC | (Wang and Seed, 2003) |
| Hu ActinB Rev642 | CTCCTTAATGTCACGCACGAT | (Wang and Seed, 2003) |
| MsActinB mouse For-16 | CACCCGCGAGCACAGCTTCTT | |
| MsActinB Rev-133 | TTTGCACATGCCGGAGCCGTT | |
| Ms β2M For-21582 | TTGAGACTGTGATTGGCAATGCCT | (Santos et al., 2002) |
| Ms β2M Rev-30345 | CCTTTAATGCCCATCCCGGA CT | (Santos et al., 2002) |
| Ms COX1-For | TGGAGGCTTTGGAAACTGAC | (Yatsuga and Suomalainen, 2012) |
| Ms COX1-Rev | TCCTGCATGGGCT AGATTTC | (Yatsuga and Suomalainen, 2012) |
| Ms CytB-For | AGACAAAGCCACCTTGACCC GAT | (Tewari et al., 2012) |
| Ms CytB-Rev | ACGATTGCTAGGGCCGCGAT | (Tewari et al., 2012) |
| Ms NRF1 For-696 | TCTCACCCTCCAAACCCAAC | (Thundathil et al., 2005) |
| Ms NRF1 Rev-931 | CCCGACCTGTGGAATACTTG | (Thundathil et al., 2005) |
| Hu PGC1a-For | GTCACCACCCAAATCCTTAT | (Wan et al., 2012) |
| Hu PGC1a-Rev | ATCTACTGCCTGGAGACCTT | (Wan et al., 2012) |
| Ms PolgA For-2025 | CCTAAGCTCATGGCACTGAC | (Thundathil et al., 2005) |
| Ms PolgA Rev-2208 | TGCTGCTTCCCCTGTTCAAG | (Thundathil et al., 2005) |
| Ms Polrmt For-3051 | GTCTACAGGAGATGTTCACC | (Thundathil et al., 2005) |
| Ms Polrmt Rev-3295 | CAGGGAGTGGATGAAGTTG | (Thundathil et al., 2005) |
| Ms PolG2-For | TCTGGGAAACTACGGGCGACTCT | (Tewari et al., 2012) |
| Ms PolG2-Rev | TGCTGGGTGTCTGATTGCTGTTC | (Tewari et al., 2012) |
| Ms TFAM For-590 | CATTTATGTATCTGAAAGCTT CC | (Thundathil et al., 2005) |
| Ms TFAM Rev-761 | CTCTTCCCAAGACTTCATTTC | (Thundathil et al., 2005) |
| Ms Tfb1m For-665 | AAGTTGATGTAGGAGTGGTG | (Thundathil et al., 2005) |
| Ms Tfb1m Rev-838 | ATGTCTGCCAACTGTAACAG | (Thundathil et al., 2005) |
| Ms Twinkle-For | CATCCCCCGGCCTGG TGTCT | (Tewari et al., 2012) |
| Ms Twinkle-Rev | GCCCCG TCGACTGGCTCAAG | (Tewari et al., 2012) |
Mitochondria DNA copy number
DNA was extracted using the DNeasy Blood & Tissue Kit (69504, Qiagen, Valencia, CA). 100 ng of DNA was used to measure mitochondria DNA (mtDNA) copy number using nuclear DNA beta-2 microglobulin (β2M) primers and mitochondrial DNA primers for cytochrome b (Cytb) (Table 1). The reaction mix of DNA, primers and SYBR Green was run under the conditions as previously described (Venegas and Halberg, 2012), using a 7900HT Fast Real-time PCR system (Applied Biosystems, Grand Island, NY).
Western blot analysis
Cells (1×10^6) were lysed by addition of ice-cold RIPA buffer (50mM Tris-HCl pH 8, 150mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) supplemented with 1x cOmplete ULTRA protease inhibitor cocktail (05892970001, Roche, Indianapolis, IN) and 1x PhosSTOP phosphatase inhibitor cocktail (04906837001, Roche, Indianapolis, IN) on ice for 30 minutes. Samples were centrifuged at 20,000 x g for 15 minutes and protein concentration measured with the BCA protein assay (23225, Thermo Scientific, Rockford, lL). 20μg of protein was separated on a SDS-PAGE gel, transferred onto a nitrocellulose membrane and analyzed with various antibodies.
Extracellular Flux (XF) Measurement
Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) was measured using the Seahorse XFe24 Extracellular Flux Analyzer (Seahorse Bioscience, North Billerica, MA) according to manufacturer’s instructions and further detailed by Wu et al. (Wu et al., 2007). Briefly, 1×106 TCL1/p53 cells isolated from splenocytes were seeded in a 24-well cell culture plate (100777-0044, Seahorse Bioscience) coated with Cell-Tak (354240, BD Bioscience, Bedford, MA) in DMEM supplemented with 2g/L glucose, 1mM sodium pyruvate and 2mM glutamine, titrated to pH 7.4 with sodium hydroxide. Plates were centrifuged on a swing-bucket rotor for 5 min at 150x g (accel 1, brake 1) followed by rotating plate and repeating centrifugation for 5 min at 150x g (accel 1, brake 0). Cells were then incubated in a CO2 free incubator 1 h before the assay. OCR and ECAR was measured every 8 min followed by sequential injection of assay media, oligomycin, FCCP and rotenone up to 2.5h. Rates for OCR and ECAR were normalized to the cell number in each well and presented as relative units: OCR (pmol/min/1×106 cells) and ECAR (mpH/min/1×106 cells).
ATP Measurement
ATP levels were measured using the ATPlite 1Step luminescence assay kit (Perkin Elmer, Waltham, MA). Cells were seeded at 5×105 cells in a black 96-well microplate and left to equilibrate to room temperature. Following the addition of the substrate solution, the microplate was left to shake for 10 minutes in the Fluoroskan Ascent™ Microplate Fluorometer prior to measuring luminescence.
Statistical analysis
All experiments were done in CLL cells from at least three different mice and patient samples. Statistical significance was analyzed by Student’s t-test, and P values of <0.05 were considered statistically significant. Graph bars and plots were generated using Prizm software (GraphPad, La Jolla, CA)
3. Results
3.1. Altered expression of mitochondria-encoded genes in CLL cells with loss of p53
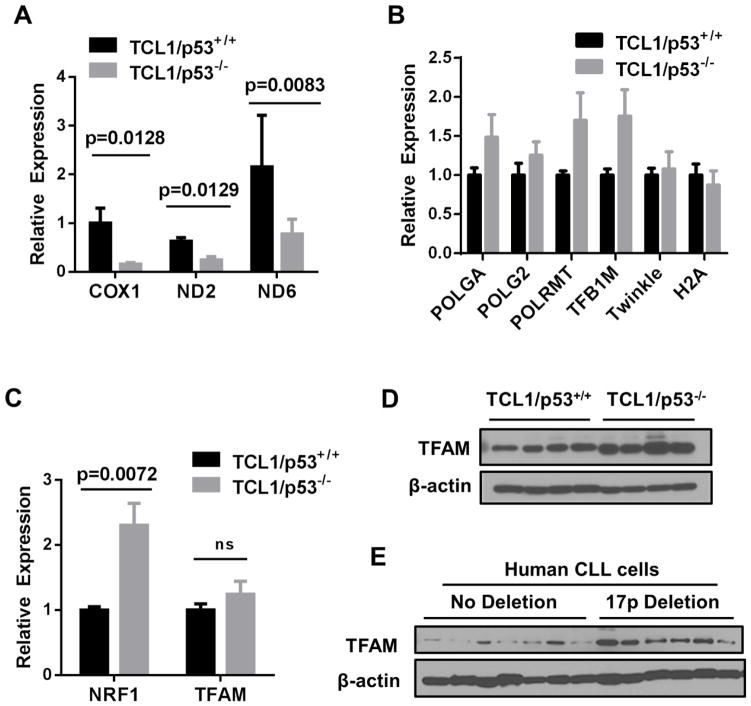
We first examined the expression of mitochondria-encoded respiratory chain components to assess potential mitochondrial changes in CLL cells with loss of p53. The expression of NADH dehydrogenases 2 and 6 (ND2 and ND6, two components of mitochondrial respiratory complex I) and cytochrome c oxidase I (COX1, a component of complex IV) were measured in the leukemic splenocytes isolated from TCL1-Tg/p53−/− CLL mice in comparison with the leukemic splenocytes from TCL1-Tg/p53+/+ CLL mice. Quantitative real-time PCR revealed a significant decrease of ND2, ND6, and COX1 transcripts in p53−/− leukemia cells (Fig 1A), suggesting that a loss of p53 might cause a reduction of mitochondria-encoded gene expression.
Figure 1. Effect of loss of p53 on expression of mitochondria-related genes in TCL1/p53−/− mice and CLL patient samples.
A, Real time RT-PCR was used to determine differences in RNA expression of mitochondria and nuclear encoded genes. Mitochondria-encoded gene expression was compared between mouse groups by measuring COXI, ND2 and ND6. B, Nuclear genes involved in mitochondrial DNA transcription and replication: POLGA, POLG2, POLRMT, TFB1M, and Twinkle, along with H2A were measured between the indicated mouse groups. C, Comparison of mRNA transcript levels for two nuclear encoded transcriptional regulators of mitochondria biogenesis NRF-1 and TFAM are shown for TCL1/p53+/+ mice (n= 5 mice) and TCL1/p53−/− mice (n=6). D, TFAM protein expression was measured in TCL1/p53+/+ and TCL1/p53−/− mouse samples by western blot. E, Primary CLL cells isolated from patient blood samples with and without 17p deletion were analyzed by western blotting for TFAM; β-actin was probed as a loading control. Student’s t test was used to determine p values. ns, not significant.
To further test if any key molecules that regulate mitochondria biogenesis and gene expression might have altered in p53−/− leukemia cells, expression of genes involved in mitochondria replication was measured. DNA polymerase γ is a nuclear DNA-encoded, mitochondria-specific DNA polymerase. POLGA encodes for the catalytic subunit of the enzyme while POLG2 encodes for an accessory subunit. Interestingly, the expression of both POLGA and POLG2 genes seemed to show an increasing trend in p53−/− mice, but did not reach statistical significance (Fig. 1B). Similarly, the expression of POLRMT, the mitochondrial DNA-directed RNA polymerase that regulates mitochondria gene expression, and TFB1M, a mitochondrial transcription factor B1 that functions as a methyltransferase to affect mitochondrial 12S rRNA and thus regulating the assembly and stability the mitochondrial ribosome, also showed an increasing trend without reaching statistical significance (Fig. 1B). No changes were observed for Twinkle, the mitochondria DNA helicase, and histone 2A (H2A), a nuclear protein associated with chromatin (Fig. 1B). These data together suggest that the decrease in expression of the mitochondrial-encoded molecules (ND2, ND6, and COX1) was not due to any decrease in POLGA, POLG2, POLRMT, or TFB1M.
As mitochondrial transcription is also regulated by the nuclear-encoded NRF-1 and TFAM, we next sought to measure the expression patterns of these genes between the two mouse groups with different p53 genotypes. Surprisingly, TCL1/p53−/− mice had significantly increased the expression for NRF-1 (p=0.0072, n=5), while TFAM seemed also elevated but did not reach significance (n= 5) (Fig. 1C). Western blot analysis confirmed the elevated TFAM protein expression in TCL1/p53−/− mice compared to TCL1/p53+/+ mice (Fig. 1D). Consistently, the increase in TFAM expression as was observed in primary leukemia cells isolated from CLL patients with chromosome 17p-deletion (thus loss of p53 gene) compared to CLL cells without del17p (Fig. 1E).
3.2. Elevated expression of PGC1a in CLL cells with 17p deletion
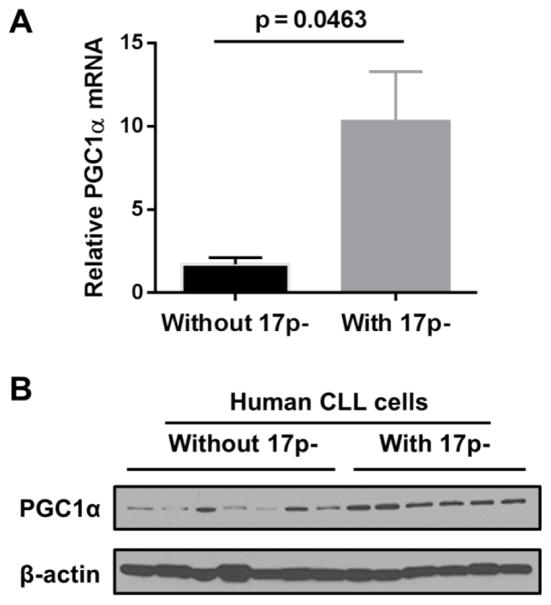
In addition to TFAM, Peroxisome proliferator-activated receptor gamma coactivator 1-α (PGC-1α) is another major regulator of mitochondria biogenesis. We next asked if PGC-1α might also be up-regulated in CLL cells with loss of p53. RT-PCR analysis revealed an elevated PGC-1α mRNA in CLL patients with 17p deletion compared to the non-17p deletion group (p=0.0463, n=3) (Fig. 2A). To determine if the increased expression of PGC-1α transcripts would lead to elevated PGC-1α protein, we performed western blot analysis using CLL cells from patients with and without 17p deletion. Akin to the RT-PCR results, the level of PGC-1α protein in CLL cells with 17p-deletion was higher than that in CLL cells without 17p deletion (Fig. 2B).
Figure 2. Elevated PGC-1α expression in CLL cells with 17-deletion.
A. RT-PCR analysis was used to compare the expression of PGC-1α RNA transcripts in primary CLL cells isolated from patients with and without 17p deletion (n=3 each group). B. Primary CLL cells from patient samples with and without 17p deletion were probed for PGC-1α. β-actin was probed as a loading control.
3.3. Increased mitochondrial mass and mitochondria DNA copy number in leukemic splenocytes of TCL1/p53−/− mice
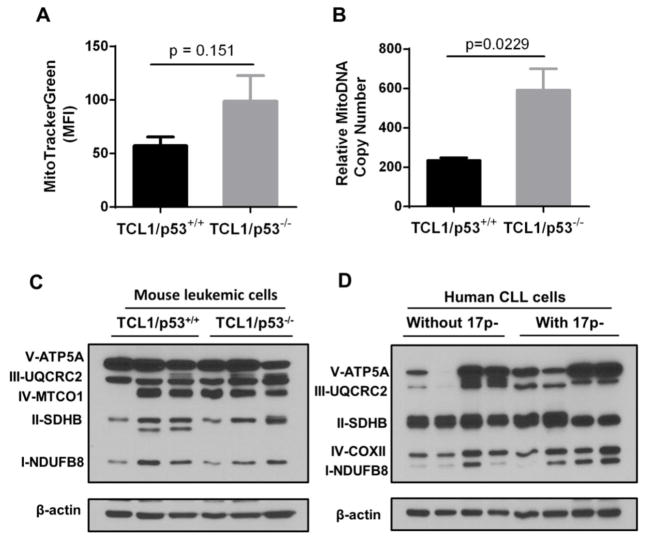
As the expression of mitochondrial biogenesis regulators NRF, TFAM and PGC1α was elevated in leukemia cells with loss of p53, we next asked whether these changes might lead to an increase in mitochondrial mass. Total mitochondrial mass in leukemia cells was measured by staining with MitoTracker Green, which stains mitochondria in a transmembrane potential-independent manner, and quantitatively analyzed by flow cytometry. TCL1/p53−/− mice exhibited a trend of elevated fluorescence but did not reach significance (p=0.151, Fig. 3A). As TFAM has been shown to regulate mtDNA copy number, the ratios of mitochondrial to nuclear DNA were compared between TCL1/p53+/+ and TCL1/p53−/− mice by qPCR, which revealed a significant increase in relative mtDNA copy number in leukemia cells from TCL1/p53−/− mice (p=0.0229, n=7) (Fig. 3B). This data suggests that the decrease in expression of the ND2, ND6, and COX1 observed in p53−/− leukemia cells was not due to a decrease in mitochondrial mass or mtDNA copy number. Instead, it appears that there was likely a compensatory increase in mitochondrial biogenesis.
Figure 3. Comparison of mitochondrial mass, mitochondrial DNA copy number, and respiratory chain components in leukemic cells with or without loss of p53.
A, Mitochondrial mass of splenocytes was determined by staining with MitoTracker Green and analyzed by FACS in TCL1/p53+/+ mice (n=4) and TCL1/p53−/− mice (n=4). MFI, mean fluorescence intensity; Student’s t test was used to determine p values. B, Relative mitochondria DNA (mtDNA) copy number was determined by qPCR using DNA isolated from the leukemic splenocytes of TCL1/p53+/+ mice (n=5) and TCL1/p53−/− mice (n=7). The ratios of mitochondria to nuclear DNA (β2M) of the two mouse groups were compared (p=0.0229). C–D, Expression of mitochondrial electron transport complexes I–V in TCL1/p53+/+ and TCL1/p53−/− mice and in primary CLL patient samples with and without 17p deletion was measured by western blot analysis. β-actin was used as a loading control.
Western blot analysis was then used to measure the expression of the mitochondrial electron transport chain components in leukemia cells with or without loss of p53. As shown in Fig 3C, although there was some heterogeneity in expression of the respiratory chain components among the individual mice, the overall expression patterns of the five respiratory complexes were similar in leukemia cells from TCL1/p53+/+ and TCL1/p53−/− mice. Consistently, similar patterns were also observed in primary human CLL cells with or without 17p-deletion (Fig. 3D).
3.4. Increased basal oxygen consumption rate in leukemic splenocytes of TCL1/p53−/− mice
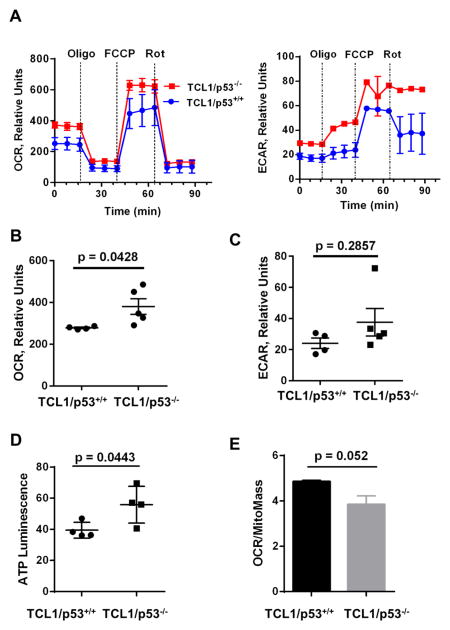
Loss of p53 has been shown to reduce optimal mitochondrial respiration via suppressing the expression of synthesis of cytochrome c oxidase 2 (SCO2), which is required for proper assembling of mitochondrial respiratory chain complex IV (cytochrome c oxidase complex) (Matoba et. al, 2006). To determine whether there were inherent changes in mitochondrial function in TCL1 mice upon loss of p53, a mitochondria stress test was performed using a Seahorse XF24 extracellular flux analyzer. Figure 4A shows a representative graph of the oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) between TCL1/p53+/+ and TCL1/p53−/− mice at the basal culture conditions and in the presence of various metabolic inhibitors such as oligomycin and rotenone. The basal OCR was significantly elevated in TCL1/p53−/− mice (p=0.0428, n=5 mice, Fig. 4B), while the ECAR also showed an increasing trend but did not reach statistical significance (p=0.2857, Fig. 4C).
Figure 4. Effect of loss of p53 on mitochondria activity in leukemic splenocytes of TCL1 mice.
A, Basal oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) was measured inleukemic splenocytes isolated from TCL1/p53+/+ mice (n=4) and TCL1/p53−/−mice (n=5). Three measurement cycles in duplicate was taken for each mouse sample. Representative graphs for OCR and ECAR are shown. Dotted lines indicate the time points of injection of 1 μM oligomycin (Oligo), 4 μM FCCP, or 2μM Rotenone (Rot). B, Comparison of the mean basal OCR between TCL1/p53+/+ mice (n=4) and TCL1/p53−/− mice (n=5). C, Comparison of the mean ECAR between TCL1/p53+/+ mice (n=4) and TCL1/p53−/− mice (n=5). D, Cellular ATP levels in the splenocytes from TCL1/p53+/+ and TCL1/p53−/− mice (n=4). E, Comparison between TCL1/p53+/+ mice (n=4) and TCL1/p53−/− mice (n=5) for their relative OCR normalized by mitochondrial mass measured by MitoTracker Green staining.
Further analyses of mitochondria function were performed by sequential addition of complex V inhibitor oligomycin, the mitochondrial uncoupler FCCP, and complex I inhibitor rotenone. Cells from TCL1/p53+/+ and p53−/− mice were similarly sensitive to oligomycin, as evidenced by a similar drop in OCR from the basal levels (Fig. 4A). The maximum respiratory capacity, measured by the increase in oxygen consumption upon addition of FCCP, appeared substantially increased in TCL1/p53−/− mice. Inhibition of mitochondrial-respiration by rotenone revealed that the residual oxygen consumption rates (non-mitochondrial OCR) were similar in both groups. Measurement of cellular ATP revealed that the leukemia cells from TCL1/p53−/− mice contained higher ATP compared to TCL1/p53+/+ mice (Fig. 4D, p=0.0443, n=4). Since OCR and mitochondria mass were both elevated in p53−/− cells, we normalized the OCR values of p53+/+ and p53−/− cells by their respective mitochondrial mass to evaluate the relative respiratory activity per mitochondrion. Interestingly, after such normalization, the respiratory activity per mitochondrion was apparently lower in TCL1/p53−/− mice (Fig 4E), although it did not reach statistical significance (p=0.052).
4. Discussion
Previous studies suggest that p53 is an important regulator of apoptosis and metabolism due in part to its significant effect on mitochondria. Alterations in mitochondrial function and mutations in mitochondria DNA have been observed in CLL patients who received chemotherapy, suggesting a potential role of mitochondria in CLL disease development and drug sensitivity (Carew et al., 2003). Furthermore, mitochondria respiration was decreased in p53-null colon cancer cells attributed to its direct involvement in activating synthesis of cytochrome c oxidase (SCO2) (Matoba et al., 2006). In this study, we aimed to determine if alterations in the mitochondria are present in CLL leukemia cells with loss of p53 and how this could functionally affect CLL cells.
Our findings suggest that an initial decrease in expression of certain mitochondrial encoded genes upon loss of p53, and a possible compensatory response in mitochondrial biogenesis. The decrease in mRNA expression of the mitochondria-encoded COX1, ND2, and ND6 indicates that either transcriptional regulation of these mitochondria encoded genes may be attenuated or/and certain components of mtDNA might be compromised. In an early study, Achanta et al demonstrated that a binding of wild-type p53 protein with POLγ increased its function and enhanced the stability of mtDNA against oxidative damage, therefore maintaining mtDNA integrity (Achanta et al., 2005). In the current study, the increase in expression of the mitochondria biogenesis regulators TFAM and NRF-1 observed in p53-null leukemia cells suggests a possible compensatory response to the initial reduction in mitochondria-encoded gene expression. Higher expression of TFAM and NRF-1 has been observed in CLL patients relative to normal lymphocytes (Carew et. al, 2004). TFAM not only regulates mtDNA copy number and protects its stability, but can also interact directly with p53, thus aiding in maintenance of mtDNA (Yoshida et al., 2003). Importantly, the increase in TFAM transcript levels also translated to elevated protein expression leading to an increase in mtDNA copy number. It is known that TFAM elevation can overcome mitochondria deficiencies induced by physiological damage such as myocardial infarction (Ikeuchi et al., 2005). Similarly, overexpression of NRF-1 was shown to minimize chemically induced mitochondria damage. For instance, neuronal cells treated with MPTP, which metabolizes to the complex I inhibitor MPP+, triggered mitochondria damage and elevated ROS production. Transfection of NRF-1 or TFAM could minimize the inhibitory effects of MPP+ and significantly reduce the amount of ROS production, mitochondrial membrane potential change and ATP loss (Piao et al., 2012).
The molecular mechanism that regulates the compensatory response in mitochondrial biogenesis upon the loss of p53 is still unclear at present time. Transcription of TFAM can be directly regulated by NRF-1 and NRF-2; while NRF-1 contains transcriptional response elements for NFkB in its promoter region. It has been observed that activation of NFκB by lipopolysaccharide (LPS) induction could lead to an increase in expression of NRF-1 and the downstream molecules such as TFAM (Suliman et al., 2010). Interestingly, aberrant activation of NFκB has been reported in CLL (Furman et al., 2000). It is also important to note that the increased protein expression of TFAM as well PGC1α was also observed in primary leukemia cells from CLL patients with del17p, suggesting that the findings in TCL1/p53−/− mice are likely of clinical relevance.
Functional assays by the XF metabolic analysis suggest that the compensatory response of mitochondrial biogenesis was functionally competent, as the overall cellular OCR was significantly elevated in the splenocytes of TCL1/p53−/− mice (Fig 4A–4B). However, since the mitochondria mass seemed substantially elevated in TCL1/p53−/− mice, the observed increase in OCR might not necessarily suggest that each individual mitochondrion in p53−/− cells has elevated respiration. Indeed, when the OCR was normalized with the mitochondria mass, the respiratory activity per mitochondrion was apparently lower in TCL1/p53−/− mice (Fig 4E). These data together suggest that each mitochondrion in p53−/− cells might has compromised mitochondrial respiration, but the overall cellular OCR was not decreased or even increased due to a compensatory increase in mitochondrial biogenesis. It is also worth noting that the apparent increase of mitochondrial mass in p53−/− cells did not reach statistical significance (p=0.151). This could due to the small sample sizes (4 mice per group). It is also possible that the loss of p53 might affect the mitochondrial fission process, restraining the expansion of mitochondrial mass in the p53−/− cells. This potential possibility requires further study.
Determining the oligomycin-sensitive and insensitive proportions of OCR can reveal whether there were differences in proton leakage and ATP synthesis via complex V between TCL1/p53+/+ and TCL1/p53−/− mice, as the oligomycin-sensitive portion of OCR represents ATP synthesis while the portion of oligomycin-insensitive OCR mainly reflects proton leakage. Since no significant differences were observed between the TCL1/p53−/− and TCL1/p53+/+ mice in their sensitivity to oligomycin, the mitochondrial coupling efficiency seems similar between the two mouse groups. Similarly, the portion of non-mitochondrial respiration was similar between the two mouse groups, as evidenced by similar residual respiratory rates following addition of rotenone.
In addition to mitochondria dysfunction observed in various cancers, increase in glycolysis is another common metabolic alteration. Interestingly, the glycolytic activity in leukemia cells from TCL1/p53−/− mice seems unchanged or slightly increased, as indicated by ECAR measurement (Fig 4A & 4C). The significant increase in cellular ATP observed in TCL1/p53−/− mice was likely due to the increase in both mitochondrial metabolism and cytosolic glycolysis.
In summary, our study showed that although the expression of mitochondrial COX1, ND2, and ND6 decreased in p53−/− CLL cells, there was a compensatory increase in mitochondrial biogenesis. As TCL1/p53−/− mice have a significantly shorter life span compared to TCL1/p53+/+ mice, it is possible that having a high functioning mitochondria might contributes to CLL cell viability and the aggressiveness of the leukemia cells. Further studies are needed to determine whether mitochondria-targeting agents would be useful in treating CLL with loss of p53.
Highlights.
Loss of p53 in chronic lymphocytic leukemia (CLL) cells leads to decreased expression of mitochondrial COX1, ND2, and ND6.
CLL cells lacking p53 exhibit a compensatory increase in mitochondrial biogenesis and elevated respiration.
TFAM and PGC-1α seem to play a role in the mitochondrial compensatory response to loss of p53.
Acknowledgments
The authors thank M.R. Nelson, B.A. Hayes and R. LaPushin for their assistance in handling CLL samples. This work was supported in part by grants CA085563 and CA172724 from the National Institutes of Health, a grant from the CLL Global Research Foundation, and the generous philanthropic contributions to The University of Texas MD Anderson CLL Moon Shot Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Achanta G, Sasaki R, Feng L, Carew JS, Lu W, Pelicano H, Keating MJ, Huang P. Novel role of p53 in maintaining mitochondrial genetic stability through interaction with DNA Pol gamma. EMBO J. 2005;24:348–492. doi: 10.1038/sj.emboj.7600819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ak P, Levine AJ. p53 and NF-κB: different strategies for responding to stress lead to a functional antagonism. The FASEB Journal. 2010;24:364–652. doi: 10.1096/fj.10-160549. [DOI] [PubMed] [Google Scholar]
- Bichi R, Shinton SA, Martin ES, Koval A, Calin GA, Cesari R, Russo G, Hardy RR, Croce CM. Human chronic lymphocytic leukemia modeled in mouse by targeted TCL1 expression. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:695–960. doi: 10.1073/pnas.102181599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carew JS, Nawrocki ST, Xu RH, Dunner K, McConkey DJ, Wierda WG, Keating MJ, Huang P. Increased mitochondrial biogenesis in primary leukemia cells: the role of endogenous nitric oxide and impact on sensitivity to fludarabine. Leukemia. 2004;18:193–940. doi: 10.1038/sj.leu.2403545. [DOI] [PubMed] [Google Scholar]
- Carew JS, Zhou Y, Albitar M, Carew JD, Keating MJ, Huang P. Mitochondrial DNA mutations in primary leukemia cells after chemotherapy: clinical significance and therapeutic implications. Leukemia. 2003;17:143–447. doi: 10.1038/sj.leu.2403043. [DOI] [PubMed] [Google Scholar]
- Döhner H, Stilgenbauer S, Benner A, Leupolt E, Kröber A, Bullinger L, Döhner K, Bentz M, Lichter P. Genomic Aberrations and Survival in Chronic Lymphocytic Leukemia. New England Journal of Medicine. 2000;343:191–916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- Furman RR, Asgary Z, Mascarenhas JO, Liou2 HC, Schattner EJ. Modulation of NF-κB Activity and Apoptosis in Chronic Lymphocytic Leukemia B Cells. The Journal of Immunology. 2000;164:2200–2206. doi: 10.4049/jimmunol.164.4.2200. [DOI] [PubMed] [Google Scholar]
- Hallek M. Chronic lymphocytic leukemia: 2013 update on diagnosis, risk stratification and treatment. American Journal of Hematology. 2013;88:803–816. doi: 10.1002/ajh.23491. [DOI] [PubMed] [Google Scholar]
- Huang P, Feng L, Oldham EA, Keating MJ, Plunkett W. Superoxide dismutase as a target for the selective killing of cancer cells. Nature. 2000;407:390–395. doi: 10.1038/35030140. [DOI] [PubMed] [Google Scholar]
- Ikeuchi M, Matsusaka H, Kang D, Matsushima S, Ide T, Kubota T, Fujiwara T, Hamasaki N, Takeshita A, Sunagawa K, Tsutsui H. Overexpression of Mitochondrial Transcription Factor A Ameliorates Mitochondrial Deficiencies and Cardiac Failure After Myocardial Infarction. Circulation. 2005;112:683–690. doi: 10.1161/CIRCULATIONAHA.104.524835. [DOI] [PubMed] [Google Scholar]
- Johnson RF, Perkins ND. Nuclear factor-kappaB, p53, and mitochondria: regulation of cellular metabolism and the Warburg effect. Trends in biochemical sciences. 2012;37:317–324. doi: 10.1016/j.tibs.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Juliusson G, Gahrton G. Cytogenetics in CLL and related disorders. Bailliere’s clinical haematology. 1993;6:821–848. doi: 10.1016/s0950-3536(05)80178-7. [DOI] [PubMed] [Google Scholar]
- Liu J, Chen G, Feng L, Zhang W, Pelicano H, Wang F, Ogasawara MA, Lu W, Amin HM, Croce CM, Keating MJ, Huang P. Loss of p53 and altered miR15-a/16–1short right arrowMCL-1 pathway in CLL: insights from TCL1-Tg:p53(−/−) mouse model and primary human leukemia cells. Leukemia. 2014;28:118–128. doi: 10.1038/leu.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- Piao Y, Kim HG, Oh MS, Pak YK. Overexpression of TFAM, NRF-1 and myr-AKT protects the MPP(+)-induced mitochondrial dysfunctions in neuronal cells. Biochimica et biophysica acta. 2012;1820:577–585. doi: 10.1016/j.bbagen.2011.08.007. [DOI] [PubMed] [Google Scholar]
- Puente XS, Bea S, Valdes-Mas R, Villamor N, Gutierrez-Abril J, Martin-Subero JI, Munar M, Rubio-Perez C, Jares P, Aymerich M, Baumann T, Beekman R, Belver L, Carrio A, Castellano G, Clot G, Colado E, Colomer D, Costa D, Delgado J, Enjuanes A, Estivill X, Ferrando AA, Gelpi JL, Gonzalez B, Gonzalez S, Gonzalez M, Gut M, Hernandez-Rivas JM, Lopez-Guerra M, Martin-Garcia D, Navarro A, Nicolas P, Orozco M, Payer AR, Pinyol M, Pisano DG, Puente DA, Queiros AC, Quesada V, Romeo-Casabona CM, Royo C, Royo R, Rozman M, Russinol N, Salaverria I, Stamatopoulos K, Stunnenberg HG, Tamborero D, Terol MJ, Valencia A, Lopez-Bigas N, Torrents D, Gut I, Lopez-Guillermo A, Lopez-Otin C, Campo E. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015 doi: 10.1038/nature14666. [DOI] [PubMed] [Google Scholar]
- Rozman C, Montserrat E. Chronic Lymphocytic Leukemia. New England Journal of Medicine. 1995;333:1052–1057. doi: 10.1056/NEJM199510193331606. [DOI] [PubMed] [Google Scholar]
- Santos J, Mandavilli B, Van Houten B. Measuring Oxidative mtDNA Damage and Repair Using Quantitative PCR. In: Copeland W, editor. Mitochondrial DNA. Humana Press; 2002. pp. 159–176. [DOI] [PubMed] [Google Scholar]
- Suliman HB, Sweeney TE, Withers CM, Piantadosi CA. Co-regulation of nuclear respiratory factor-1 by NFκB and CREB links LPS-induced inflammation to mitochondrial biogenesis. Journal of Cell Science. 2010;123:2565–2575. doi: 10.1242/jcs.064089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewari S, Santos JM, Kowluru RA. Damaged mitochondrial DNA replication system and the development of diabetic retinopathy. Antioxid Redox Signal. 2012;17:492–504. doi: 10.1089/ars.2011.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thundathil J, Filion F, Smith LC. Molecular control of mitochondrial function in preimplantation mouse embryos. Molecular Reproduction and Development. 2005;71:405–413. doi: 10.1002/mrd.20260. [DOI] [PubMed] [Google Scholar]
- Venegas V, Halberg MC. Measurement of mitochondrial DNA copy number. Methods in molecular biology. 2012;837:327–335. doi: 10.1007/978-1-61779-504-6_22. [DOI] [PubMed] [Google Scholar]
- Wan X, Gupta S, Zago MP, Davidson MM, Dousset P, Amoroso A, Garg NJ. Defects of mtDNA replication impaired mitochondrial biogenesis during Trypanosoma cruzi infection in human cardiomyocytes and chagasic patients: the role of Nrf1/2 and antioxidant response. Journal of the American Heart Association. 2012;1:e003855. doi: 10.1161/JAHA.112.003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Seed B. A PCR primer bank for quantitative gene expression analysis. Nucleic acids research. 2003;31:e154. doi: 10.1093/nar/gng154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Neilson A, Swift AL, Moran R, Tamagnine J, Parslow D, Armistead S, Lemire K, Orrell J, Teich J, Chomicz S, Ferrick DA. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. American journal of physiology. Cell physiology. 2007;292:C125–136. doi: 10.1152/ajpcell.00247.2006. [DOI] [PubMed] [Google Scholar]
- Yatsuga S, Suomalainen A. Effect of bezafibrate treatment on late-onset mitochondrial myopathy in mice. Hum Mol Genet. 2012;21:526–535. doi: 10.1093/hmg/ddr482. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Izumi H, Torigoe T, Ishiguchi H, Itoh H, Kang D, Kohno K. P53 physically interacts with mitochondrial transcription factor A and differentially regulates binding to damaged DNA. Cancer research. 2003;63:3729–3734. [PubMed] [Google Scholar]
- Zwiebel JA, Cheson BD. Chronic lymphocytic leukemia: staging and prognostic factors. Seminars in oncology. 1998;25:42–59. [PubMed] [Google Scholar]