Abstract
White-blooded Antarctic crocodile icefish are the only vertebrates known to lack functional hemoglobin genes and red blood cells throughout their lives. We do not yet know, however, whether extinction of hemoglobin genes preceded loss of red blood cells or vice versa, nor whether erythropoiesis regulators disappeared along with hemoglobin genes in this erythrocyte-null clade. Several microRNAs, which we here call erythromiRs, are expressed primarily in developing red blood cells in zebrafish, mouse, and humans. Abrogating some erythromiRs, like Mir144 and Mir451a, leads to profound anemia, demonstrating a functional role in erythropoiesis. Here, we tested two not mutually exclusive hypotheses: 1) that the loss of one or more erythromiR genes extinguished the erythropoietic program of icefish and/or led to the loss of globin gene expression through pseudogenization; and 2) that some erythromiR genes were secondarily lost after the loss of functional hemoglobin and red blood cells in icefish. We explored smallRNA transcriptomes generated from the hematopoietic kidney marrow of four Antarctic notothenioids: two red-blooded species (bullhead notothen Notothenia coriiceps and emerald notothen Trematomus bernacchii) and two white-blooded icefish (blackfin icefish Chaenocephalus aceratus and hooknose icefish Chionodraco hamatus). The N. coriiceps genome assembly anchored analyses. Results showed that, like the two red-blooded species, the blackfin icefish genome possessed and the marrow expressed all known erythromiRs. This result indicates that loss of hemoglobin and red blood cells in icefish was not caused by loss of known erythromiR genes. Furthermore, expression of only one erythromiR, mir96, appears to have been lost after the loss of red blood cells and hemoglobin – expression was not detected in the erythropoietic organ of hooknose icefish but was present in blackfin icefish. All other erythromiRs investigated, including mir144 and mir451a, were expressed by all four species and thus are present in the genomes of at least the two white-blooded icefish. Our results rule out the hypothesis that genomic loss of any known erythromiRs extinguished erythropoiesis in icefish, and suggest that after the loss of red blood cells, few erythromiRs experienced secondary loss. Results suggest that functions independent of erythropoiesis maintained erythromiRs, thereby highlighting the evolutionary resilience of miRNA genes in vertebrate genomes.
Keywords: miRNA, Notothenioidei, hematopoiesis, Channichthyidae, mirc144
1. Introduction
Since the first report by the Norwegian biologist Ditlef Rustad of a fish with “colorless blood” that he caught near the sub-Antarctic Bouvet Island (Bouvetøya) in December 1927 and the confirmation in 1954 by Johan Ruud that this species, the blackfin icefish Chaenocephalus aceratus, lacks both red blood cells and hemoglobin (Hb) (Ruud, 1954), the “crocodile icefish” of the Southern Ocean have puzzled physiologists. Icefish comprise the only vertebrate clade whose members lack red blood cells and the oxygen transport protein hemoglobin throughout their life cycles. Spurred by these seminal observations, contemporary molecular biologists and physiologists have sought to understand the evolutionary mechanism(s) that led to the loss of mature red blood cells and hemoglobin in icefish and the physiological traits that enable these unique vertebrates to survive without oxygen-binding proteins in their blood (Braasch et al., 2015; Cheng and Detrich, 2007; Holeton, 1970; Kock, 2005a, 2005b; Near et al., 2006; Sidell and O’Brien, 2006).
Among the approximately 130 notothenioid species currently recognized, only the 16 species of the notothenioid crown group, the icefish family Channichthyidae, have lost functional hemoglobin genes (Cheng and Detrich, 2007; Giordano et al., 2015; Near et al., 2006; Sidell and O’Brien, 2006). The loss of functional hemoglobin genes is a shared, derived feature (synapomorphy) that left 15 of the 16 icefish species with a pseudogenized α-globin gene and no β-globin gene, and one species, Jonah’s icefish (Neopagetopsis ionah), with an inactive αβ-globin pseudogene complex, a “genomic fossil” derived by gene introgression (Cocca et al., 1995; di Prisco et al., 2002; Near et al., 2006). Furthermore, all notothenioids lost myoglobin (Mb) expression in their skeletal muscles (Sidell et al., 1997), and six species of icefish have lost expression of myoglobin in addition in their heart muscle by multiple independent mutational events (Borley and Sidell, 2010; Grove et al., 2004; Sidell et al., 1997; Sidell and O’Brien, 2006; Small et al., 2003). Several other teleost lineages have also lost cardiac myoglobin expression, including the circum-Arctic three-spined stickleback (Gasterosteus aculeatus), which possesses a pseudogenized myoglobin gene (Hoffmann et al., 2011; Macqueen et al., 2014).
Whether the loss of functional hemoglobin genes in icefish evolution preceded the loss of red blood cells or extinction of erythropoiesis came first is as yet an unanswered question. To address this issue, we must understand the cascade of events that led to the disruption of globin genes in icefish genomes and the disappearance of red blood cells, which are produced in the pronephric (head) kidney marrow of teleost fish (Fänge, 1994; Witeska, 2013). Did the fixation of deleterious mutations in hemoglobin genes lead to the loss of red blood cells, despite their near-universal additional role in carbon dioxide/bicarbonate metabolism and transport (Maffia et al., 2001; Tufts et al., 2002)? Alternatively, did red blood cells disappear first, followed by mutations that rendered hemoglobin genes nonfunctional? If erythropoiesis disappeared first, what mechanism led to the suppression of red blood cell development? Did positive regulators of erythropoiesis become non-functional? Or did ancestral icefish evolve mechanisms that actively suppress erythroid development?
Although many protein-coding genes regulate erythropoiesis, such as gata1 (Galloway et al., 2005; Welch et al., 2004), myb (Vegiopoulos et al., 2006), spi1b (also known as pu.1 (Rhodes et al., 2005)) and bty (discovered by subtractive hybridization using the hematopoietic transcriptomes of red-blooded and white-blooded notothenioids (Yergeau et al., 2005)), microRNAs also play a key role. miRNAs are small endogenous non-coding RNAs that regulate gene expression post-transcriptionally by binding to specific mRNAs and, together with the RNA-Induced Silencing Complex, either mediate transcript decay or repress transcript translation (See Carthew and Sontheimer, 2009; Christodoulou et al., 2010; Desvignes et al., 2015; Kosik, 2010 for reviews on biogenesis and function). In vertebrates, including teleost fish, several miRNAs are necessary for the formation of red blood cells and functional hemoglobin, and we refer to them here as “erythromiRs” in analogy to the well-described muscle-specific microRNAs referred to as “myomiRs” (McCarthy, 2008). For example, the erythromiR MIR155 is expressed strongly in early stages of erythropoiesis but dramatically weaker at later stages. In contrast, MIR451A displays the opposite expression pattern, increasing in expression more than 200-fold between progenitor stages and late stages of erythropoiesis (Masaki et al., 2007). Furthermore, knockdown and knockout experiments showed that mir451a and its clustered companion mir144 produce microRNAs that are major erythropoietic factors required for erythroid precursor maturation in zebrafish (Dore et al., 2008; Du et al., 2009; Fu et al., 2009; Pase et al., 2009; Yu et al., 2010), mouse (Patrick et al., 2010; Rasmussen et al., 2010; Yu et al., 2010; Zhan et al., 2007) and human (Kim et al., 2015, 2013). In addition, the mature products of both Mir144 and Mir451a protect red blood cells from oxidative stress in zebrafish, mouse, and human (Sangokoya et al., 2010; Yu et al., 2010). Other miRNA genes are also important erythropoietic factors, including mir23a, mir126, and mir223 (see Azzouzi et al., 2012; Bhagavathi and Czader, 2010; Havelange and Garzon, 2010; Lawrie, 2010; Listowski et al., 2012; Mohammdai-asl et al., 2015; Sayed and Abdellatif, 2011; Undi et al., 2013; Zhang et al., 2012 for reviews).
In addition to a role in erythropoiesis, miRNAs participate more generally in hematopoiesis. Some miRNAs, such as mir150, mir155, and mir223, also play roles in the development of megakaryocytes/platelets and the T cell and B cell lineages (see Bhagavathi and Czader, 2010; Havelange and Garzon, 2010; Sayed and Abdellatif, 2011; Undi et al., 2013; Zhang et al., 2012 for review), while some other miRNAs, such as mir142 or mir181, regulate development of blood cell lineages other than erythrocytes (Chen et al., 2004; Fan et al., 2014; Kramer et al., 2015). These data show that miRNAs are key regulators of hematopoiesis, including erythropoiesis, and suggest the hypothesis that loss of one or more miRNAs could have been important in the evolution of the erythrocyte-null, hemoglobin-null phenotypes of white-blooded icefish.
Recently, Xu et al. (2015) looked for potential erythropoietic suppressor miRNAs in white-blooded icefish and reported that mir16b, mir152, and mir1388 were over-expressed in the pronephric kidney of hooknose icefish Chionodraco hamatus (See Appendix for etymology) compared to their expression in the red-blooded emerald notothen Trematomus bernacchii. Injection of each of these three miRNAs into zebrafish embryos reduced the production of red blood cells (Xu et al., 2015), but other developmental defects also occurred that can appear due to non-specific deleterious effects of over-expression experiments (Jin et al., 2015; Zhang et al., 2013). Despite their interesting findings and important dataset, these authors did not examine the global conservation of erythromiRs in icefish, leaving open the possibility that icefish genomes may have lost some key erythromiR genes that are required for normal erythropoiesis or erythroid maturation.
We took advantage of the recently published reference genome of the red-blooded bullhead notothen, Notothenia coriiceps (Shin et al., 2014), smallRNA sequencing data from the red-blooded emerald notothen T. bernacchii and the white-blooded hooknose icefish C. hamatus (Xu et al., 2015), and smallRNA sequencing data that we generated from N. coriiceps and the white-blooded blackfin icefish Chaenocephalus aceratus, to test two hypotheses regarding the role of erythromiRs in the novel erythroid phenotypes of icefish. In Hypothesis 1), the loss of known erythromiR genes in Antarctic icefish correlates with the loss of functional red blood cells and/or the disruption of their globin genes; and in Hypothesis 2), the loss of hemoglobin and red blood cells by icefish secondarily allowed the loss of some or all erythromiR genes in some icefish lineages.
2. Materials and Methods
2.1. Fish samples
Specimens of C. aceratus and N. coriiceps were collected by bottom trawls or baited fish traps deployed from the ARSV Laurence M. Gould south of Low Island or west of Brabant Island in the Palmer Archipelago (April–May, 2014). Fish were transported alive to Palmer Station, Antarctica, where they were maintained in flow-through seawater aquaria at −1.5 to 1°C. Samples of pronephric (head) kidney, the major site of erythropoiesis in teleost fish (Fänge, 1994; Witeska, 2013), were dissected and stored in RNAlater until further use at the University of Oregon. Procedures were performed according to protocols approved by the Institutional Animal Care and Use Committees (IACUC) of the University of Oregon (#10-26) and of Northeastern University (#12-0306 R).
2.2. SmallRNA-sequencing and analysis
Total RNAs were extracted from the pronephric kidney of one male C. aceratus and one male N. coriiceps using the Zymo Research Direct-zolTM RNA MiniPrep kit according to the manufacturer’s instructions. Two species-specific smallRNA libraries were then prepared and barcoded using the BiooScientififc NEXTflexTM small RNA sequencing kit with 15 PCR cycles and sequenced by Illumina HiSeq2500 at the University of Oregon Genomics Core Facility. Raw single-end 50-nt long reads were deposited in the NCBI Short Read Archive under accession numbers SRP069031 and SRP069032 for C. aceratus and N. coriiceps respectively. Pronephric kidney smallRNA reads from T. bernacchii and C. hamatus were retrieved from (Xu et al., 2015). The method used to generate the libraries for the latter two species was not reported in detail (Xu et al., 2015).
Reads from the four pronephric kidney libraries were processed identically using a new bioinformatic tool, Prost!, which is available online at https://github.com/uoregon-postlethwait/prost (Batzel et al., 2015). Briefly, raw reads were trimmed from adapter sequences, filtered for quality using the FASTX-Toolkit, size-selected for lengths between 17 and 25 nucleotides, filtered for a minimum of five identical reads, and grouped by genomic location using the published N. coriiceps genome assembly as a reference genome (Shin et al., 2014). Groups of sequences were then annotated against mature and hairpin sequences present in miRBase Release 21 (Kozomara and Griffiths-Jones, 2013), the extended zebrafish miRNA annotation (Desvignes et al., 2014), and the spotted gar annotation (Braasch et al., 2016). Gene nomenclature follows recent conventions (Desvignes et al., 2015), including those for zebrafish (Bradford et al., 2011).
3. Results and Discussion
3.1. Sequencing statistics
After sequence filtering and read grouping as described in section 2.2, the four Antarctic species N. coriiceps, T. bernacchii, C. hamatus and C. aceratus provided about 200,000, 6.3 million, 7.8 million, and 4.8 million reads, respectively. Because variations in library preparation protocols precluded statistically sound differential expression analysis among samples (e.g. see Baran-Gale et al., 2013; Hafner et al., 2011; Pritchard et al., 2012; Raabe et al., 2014; Tian et al., 2010), we were not able to perform reliable differential expression analysis with this dataset. The data are, however, robust for qualitative analysis, such as identifying specific miRNAs in each species’ smallRNA transcriptome and, therefore providing positive proof for the presence of the encoding gene in each species’ genome.
3.2. Antarctic fish genomes possess erythropoietic miRNAs
The hypothesis that the loss of erythropoietic miRNAs led to the loss of red blood cells and hemoglobin in white-blooded icefish predicts that red-blooded notothenioids would possess erythromiRs known from other vertebrates but white-blooded icefish would lack one or more of them. To test this prediction, we first examined whether miRNAs currently known to be important in erythropoiesis in vertebrates were expressed in erythropoietic organs of red-blooded and white-blooded notothenioids.
Output of smallRNA transcriptomic reads using the software Prost! for the N. coriiceps pronephric kidney identified sequences for all miRNAs known to be involved in erythropoiesis in vertebrates (Table 1) and we mapped them onto the N. coriiceps genome assembly (Additional File 1). Similar analyses identified all known erythromiRs in the T. bernacchii smallRNA dataset (Table 1). The presence of these miRNAs in these two red-blooded Antarctic notothenioids and their expression in the pronephric kidney is consistent with the hypothesis that these miRNAs play a conserved role in erythropoiesis in Antarctic red-blooded notothenioids as they do in other fish and in tetrapods. While some species had sequencing reads from both arms of nearly all erythromiR hairpins, in other species, reads appeared from only one arm, the 5’ or 3’ arm; for example, mir23a in N. coriiceps and C. hamatus, or mir155 in N. coriiceps, T. bernacchii, and C. hamatus. This situation can occur due to either unequal sequencing levels or asymmetries in arm degradation. The identification of sequencing reads from only one of the two strands, especially when they have perfect sequence conservation, nevertheless definitively demonstrates that the gene is 1) present in the genome, 2) expressed, and 3) has at least one strand processed into a mature form at a significant level. For example, the most highly expressed strand for mir155 is the 5p strand (MiR155-5p), which is present in libraries from all four species; the complementary strand, however, was only found in C. aceratus, likely due to deeper sequencing in this species.
Table 1.
Presence of erythromiR genes in Antarctic notothenioid genomes deduced from smallRNA-sequencing data. The list of erythromiRs was compiled from several reviews on miRNA function in erythropoiesis and hematopoiesis (Azzouzi et al., 2012; Bhagavathi and Czader, 2010; Havelange and Garzon, 2010; Lawrie, 2010; Listowski et al., 2012; Mohammdai-asl et al., 2015; Sayed and Abdellatif, 2011; Undi et al., 2013; Zhang et al., 2012). Gene names in boldface indicate those shown to be involved in erythropoiesis in teleost fish, and the supporting reference is given. Superscript “P” and “W” identify miRNAs known to be involved in platelet and white blood cell formation respectively.
| miRNA gene | Trematomus bernacchii | Notothenia coriiceps | Chaenocephalus aceratus | Chionodraco hamatus | References in teleosts |
|---|---|---|---|---|---|
| mir15a | □ | □ | □ | □ | |
| mir16b | □ | □ | □ | □ | (Xu et al., 2015) |
| mir23a | □ | □ | □ | □ | (Zhu et al., 2013) |
| mir24 W | □ | □ | □ | □ | |
| mir96 | □ | □ | □ | □ | |
| mir103 W | □ | □ | □ | □ | |
| mir126 P | □ | □ | □ | □ | (Grabher et al., 2011) |
| mir145 | □ | □ | □ | □ | (Muhseen and Abbood, 2014) |
| mir144/451a | □ | □ | □ | □ | (Du et al., 2009; Fu et al., 2009; Pase et al., 2009; Yu et al., 2010) |
| mir150 P,W | □ | □ | □ | □ | |
| mir152 | □ | □ | □ | □ | (Xu et al., 2015) |
| mir155 P,W | □ | □ | □ | □ | |
| mir462 (aka | □ | □ | □ | □ | |
| mir191) | □ | □ | □ | □ | |
| mir210 | □ | □ | □ | □ | |
| mir221/222 W | □ | □ | □ | □ | |
| mir223 P,W | □ | □ | □ | □ | (Roberto et al., 2015) |
| mir1388 | □ | □ | □ | □ | (Xu et al., 2015) |
Analysis of smallRNA sequencing data from the white-blooded C. aceratus pronephric kidney also revealed expression of all known erythropoietic miRNAs. This finding demonstrates that the blackfin icefish genome possesses the known set of erythomiR genes (Table 1). The C. hamatus pronephric kidney sequencing data, in contrast, contained reads for all known erythromiRs with the exception of mir96, which was undetected (Table 1). Because MiR96-5p was present in the smallRNA transcriptome of C. aceratus hematopoietic marrow, and its sequence is identical to the MiR96-5p of red-blooded notothenioids (Additional File 1), we reject the possibility that the loss of detectable expression of the mir96 erythromiR gene in an ancestor of all extant icefish caused the loss of red blood cells and hemoglobin.
In sum, the presence and expression of known erythromiRs in the hematopoietic marrow of at least one white-blooded icefish rules out the hypothesis that the loss of one or more known erythromiRs by the most recent common ancestor of icefish triggered the loss of red blood cells and/or functional hemoglobin because such gene losses should be shared by the clade.
3.3. Evolution of erythropoietic miRNAs following the loss of erythropoiesis
Given datasets for two white and two red-blooded notothenioids, we then asked whether icefish lost any miRNA genes implicated in vertebrate erythropoiesis secondarily after the clade lost red blood cells and hemoglobin. Because expression of neither the 5’ nor the 3’ strand of mir96 was detected in C. hamatus but was readily detected in C. aceratus, mir96 expression loss in C. hamatus is likely a secondary event that followed red blood cell and globin gene losses.
Assuming that the function of mir96 in humans – the regulation of embryonic globin expression (Azzouzi et al., 2011) – is conserved in red-blooded teleost fish, then the absence of mir96 expression in the pronephric kidney of C. hamatus may be due to prior evolutionary loss of globin genes, which relaxed selective pressures for conservation of this erythromiR and its pronephric kidney expression. Alternatively, failure to detect mir96 expression in C. hamatus kidney marrow could reflect either: 1) insufficient sequencing depth of its pronephric kidney transcriptome; or 2) loss of expression of mir96 in pronephric kidney but maybe not in other tissues. These possibilities can be evaluated by deeper sequencing of pronephric kidney libraries, sequencing of a larger variety of tissues, generation of whole genome sequences, and wider phylogenetic sampling with species related to hooknose icefish, such as the ocellated icefish Chionodraco rastrospinosus, Myer’s icefish Chionodraco myersi, or spiny icefish Chaenodraco wilsoni (Near et al., 2012).
3.4. A case study: the conservation of erythropoietic mir144 and mir451a genes in white-blooded icefish
In vertebrates, the mir144/451a cluster (alias mirc144), plays a central role in the developmental progression of erythroid precursors to mature red blood cells (Dore et al., 2008; Du et al., 2009; Fu et al., 2009; Kim et al., 2015, 2013; Pase et al., 2009; Patrick et al., 2010; Rasmussen et al., 2010; Yu et al., 2010; Zhan et al., 2007). In mammals, in addition to the erythropoietic function of mirc144, recent work suggests a role of Mir144 and Mir451a in cardiomyocyte function and development (Kuwabara et al., 2015; Song et al., 2014; Wang et al., 2012). In teleosts, some erythropoietic miRNAs (e.g., mir150, mir155 and mir223, Table1) are known to participate in the formation of other blood cell types in addition to erythrocytes as well as in the development of other tissues. In contrast, in teleosts, mir144 and mir451a are the only erythromiRs we are aware of that have been shown by functional experiments to play a role exclusively in erythropoiesis and not in the development of other blood cell lineages or other tissue and organs. When these genes are knocked-down or knocked-out in zebrafish (Dore et al., 2008; Du et al., 2009; Fu et al., 2009; Pase et al., 2009; Yu et al., 2010), erythropoiesis is impaired but no other embryonic defects, including heart defects, appear. Thus, disruption of the mirc144 cluster would be an attractive candidate for causing loss of erythropoiesis in icefish, without disrupting the development of other blood cell types, including myeloid and lymphoid lineages, because if the mirc144 cluster was indeed erythropoiesis-specific in teleosts, then its loss could have hypothetically impaired red blood cell maturation in icefish ancestors as loss of the cluster loss does in zebrafish, mouse and human today. And because the function of the cluster in teleost fish appears to relate only to erythropoiesis, the loss of red blood cells might have occurred first, followed by the loss of this mirc144 cluster in disuse analogous to pseudogenization of hemoglobin genes. The detection of mir144 and mir451a expression in the erythropoietic tissues of two species of erythrocyte-null icefish, however, shows that icefish genomes conserve the clustered mir144 and mir451a genes.
Conservation of the mir144 and mir451a genes in icefish genomes and their expression in the icefish hematopoietic organ, however, doesn’t necessarily mean that the function of these miRNAs is also conserved. Indeed, rearrangements of genes within genomes are sometimes associated with changes in gene regulation. Therefore, we analyzed whether syntenies around mirc144 were conserved between the bullhead notothen and other ray-finned fish and humans. Conservation of synteny and expression for the mirc144 cluster in N. coriiceps would suggest a conserved function of the cluster at least in red-blooded notothens. Our results showed that the genomic environment of the mirc144 cluster was well conserved between N. coriiceps and several vertebrates including human (Figure 1A–B). Conserved synteny analyses in the white-blooded icefish C. aceratus and C. hamatus are not yet possible due to the lack of a reference genome assembly for any icefish species. Consequently, whether the icefish mirc144 cluster experienced rearrangements that may have altered mir144 and mir451a function remains an open question.
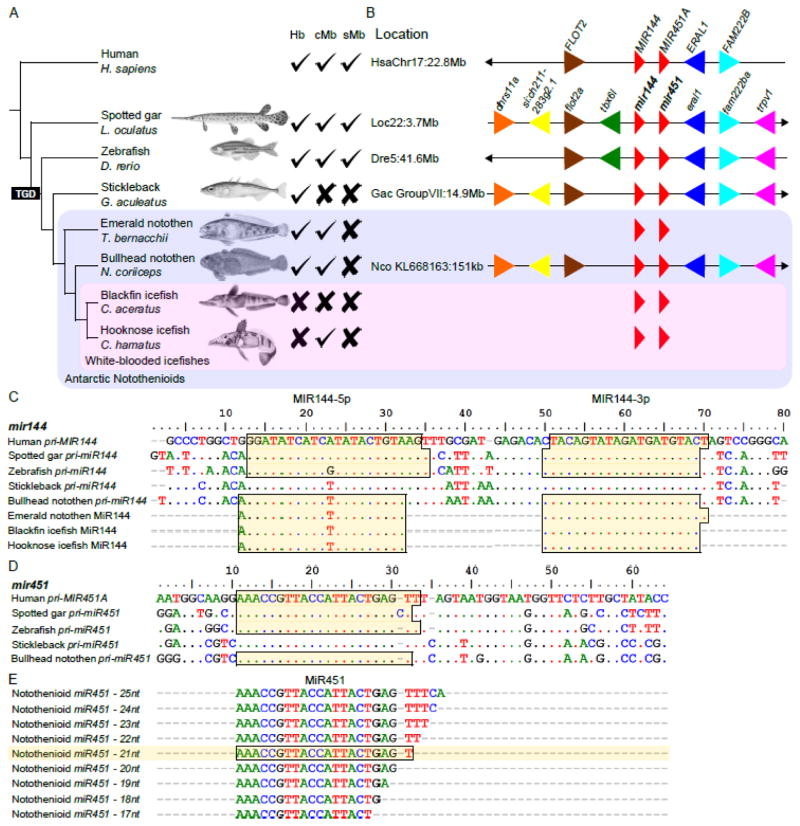
Figure 1. Evolutionary conservation of the miR144/451a miRNA gene cluster.
A) Phylogenetic relationships among vertebrates with human (Homo sapiens), spotted gar (Lepisosteus oculatus), zebrafish (Danio rerio), three-spined stickleback (Gasterosteus aculeatus), and four Antarctic notothenioids. The table records for each species the presence/absence of genes encoding hemoglobin (Hb) and cardiac and skeletal muscle myoglobin (cMb and sMb, respectively). B) Synteny conservation of the mir144/451a cluster (mirc144) among vertebrates. The figure lists the approximate location of the cluster for each species. Small black arrows at the ends of chromosome segments indicate the direction of the chromosome/scaffold in the corresponding genome assemblies deposited in Ensembl as of November 2015. Note that synteny relationships for the mir144 and mir451 genes of the emerald notothen and the two icefish species are unknown due to the absence of reference genomes. C) Alignment of cDNA sequences for human, spotted gar, zebrafish, stickleback, and bullhead notothen partial primary miR144 RNAs (pri-miR144). Dots denote conserved nucleotides, and dashes indicate indels. The most highly expressed isomiRs in the smallRNA sequencing data are highlighted in pale yellow for emerald notothen, blackfin icefish and hooknose icefish; for these mature MiR144 sequences, dashes are introduced to align the isomiRs with respect to the five pri-miR144 sequences. Mature MiR144 sequences for stickleback are unknown. Genomic pri-miR144 sequences are not available for T. bernacchii, C. aceratus, and C. hamatus. D) Sequence alignments for human, spotted gar, zebrafish, stickleback, and bullhead notothen partial primary miR451a RNAs (pri-miR451a). Dots denote conserved nucleotides; dashes denote indels (insertions or deletions). The most highly expressed isomiRs for MiR451a are highlighted in pale yellow. E) In all four Antarctic notothenioids, isomiRs both smaller and larger than the most highly expressed isomiRs were also found in the sequencing data and reflect post-transcriptional enzymatic trimming. The mature MiR451a sequence of stickleback is unknown.
Another hypothesis for a role of the mirc144 cluster in the white-blood phenotype would be that icefish process these miRNAs differently from red-blooded vertebrates, so that these miRNAs are non-functional in the erythropoietic context. Our analysis of the miRNA-seq data showed that the nucleotide sequences of MiR144-3p, the mature miRNA originating from the 3’ side of the precursor hairpin, was perfectly conserved across all investigated ray-finned fish (Figure 1C), consistent with the observation that MiR144-3p is the active mature product originating from the mir144 gene in erythropoiesis (Dore et al., 2008; Fu et al., 2009; Kim et al., 2013; Rasmussen et al., 2010). In contrast, the nucleotide sequence of MiR144-5p, the mature miRNA originating from the 5’ side of the precursor hairpin, showed evolved nucleotide differences in vertebrates, including a lineage-specific one-nucleotide change (A to T) in perciformes, represented here by the three-spined stickleback and notothenioids (Near et al., 2015, 2013) (Figure 1C). MiR144-5p additionally displays a seed-shift of one nucleotide at the 5’ end in Antarctic fish, which is a change in the position of the seed region of the miRNA by one nucleotide (Figure 1C), and which can potentially have major functional repercussions (Desvignes et al., 2015). Given that the reference MiR144-5p sequences for human, spotted gar and zebrafish were obtained from tissues other than pronephric kidney (Braasch et al., 2016; Desvignes et al., 2014) and/or miRBase (Kozomara and Griffiths-Jones, 2013), we cannot, however, rule out the possibility that the most expressed MiR144-5p isomiR in some tissue other than pronephric kidney has a 5’ start similar to the one observed in human, gar and zebrafish, and that, in the pronephric-kidney specifically, the most expressed MiR144-5p isomiR has a 5’ start similar to the one observed in notothenioid fish. SmallRNA sequencing of pronephric-kidney from other teleost species, especially those closely related to notothenioids should provide answers to this issue.
Sequencing data originating from the mir451a gene in all four species clearly demonstrate that the primary miRNA pri-miR451a and the mature MiR451a are well conserved across evolution (Figure 1D–E). MiR451a is the only known miRNA to be processed in a non-canonical, Dicer-independent pathway involving Argonaute2 cleavage followed by exonuclease nibbling of the 3’ tail of the miRNA, which results in the formation of multiple isomiRs displaying a tiling pattern (Cheloufi et al., 2010; Cifuentes et al., 2010; Yang et al., 2010). Our finding that all four notothenioid species exhibit this characteristic tiling pattern (Figure 1E) demonstrates that the maturation process for MiR451a is also conserved in Antarctic notothenioids.
Together, the conservation of the mir144 and mir451 genes in the genomes of the two icefish species studied here and the shared pattern of isomiR processing between red-blooded and white-blooded notothenioids in hematopoietic tissues demonstrate that neither the loss of these miRNA genes nor the modification of their processing are directly responsible for the erythrocyte-null phenotype of icefish. A role for the cluster in the white-blooded phenotype, however, can’t be entirely ruled out because the paucity of samples available for this study prevented us from conducting a statistically robust differential expression analysis among species and because reference genome assemblies for icefish species are not yet in hand.
Nevertheless, the conservation of the mirc144 cluster, its expression in kidney marrow, and its conserved processing in two species of icefish despite the lack of erythropoiesis suggest that the cluster may perform functions other than erythropoiesis in the pronephric kidney and/or in other tissues (e.g. heart ventricle) of notothenioids and perhaps of other teleosts.
4. Conclusions
Study of erythromiRs in erythropoietic organs of red-blooded and white-blooded Antarctic fish revealed that the loss of red blood cells and hemoglobin was caused neither by the loss of miRNA genes known to be necessary for erythropoiesis in red-blooded vertebrates nor by the loss of their expression in hematopoietic marrow. Furthermore, our results showed that expression of only one erythromiR (mir96) appears to have reduced below detection in one of the two icefish species, the hooknose icefish, consistent with secondary loss after the extinction of hemoglobin genes and erythropoiesis. The conservation of erythromiRs in sequence and expression despite the loss of erythropoiesis could be explained if these non-coding RNAs play roles in the development of non-erythropoietic blood cell lineages, perhaps other myeloid lineages, like megakaryocytes, mast cells, or myeloblasts, or in the lymphoid lineage leading to B-cells and T-cells, lineages that apparently develop normally in icefish (Bhagavathi and Czader, 2010; Havelange and Garzon, 2010; Sayed and Abdellatif, 2011; Undi et al., 2013; Zhang et al., 2012). In addition, erythromiRs may persist in icefish because they perform additional functions in non-hematopoietic tissues or functions other than erythropoiesis in blood-producing organs. For example they might help protect cells from oxidative stress (Sangokoya et al., 2010; Yu et al., 2010), which is harsher in Antarctic fish due to the 1.6 fold increase in oxygen content of the frigid Southern Ocean (Giordano et al., 2015). The persistence of these miRNAs in icefish genomes may also highlight the evolutionary resilience of miRNA genes once they have become embedded as fine regulators in several genetic pathways (Lee et al., 2007; Peterson et al., 2009; Wheeler et al., 2009).,
Our finding of erythromiRs in white-blooded icefish does not, however, rule out the possibility that evolution of the miRNA system participated in the evolution of the white-blooded icefish phenotype. One hypothesis is that erythromiR binding sites on targeted messenger RNAs were lost or greatly modified or that new targets evolved. Precise annotation of potential targets of erythromiRs in the 3’UTRs of Antarctic fish mRNAs and the sequencing of both the miRNAs and their mRNA target sites will be necessary to unravel the full role of miRNAs in the loss of red blood cells and hemoglobin and the dramatic adaptive evolution that followed in the Channichthyidae family.
Supplementary Material
Highlights.
Loss of hemoglobin and red blood cells in white-blooded icefish is not due to the extinction of microRNA genes currently known to regulate vertebrate erythropoiesis (erythromiRs).
Known vertebrate erythromiR genes are conserved and expressed in blackfin icefish, but mir96 expression in hematopoietic marrow appears to have been secondarily lost in hooknose icefish.
The major erythromiR genes mir144 and mir451a are conserved in white-blooded Antarctic icefishes despite the lack of red-blood cells.
Results highlight the resilience of miRNA genes in animal genomes and suggest that some erythromiRs may have functions beyond their roles in red blood cell formation.
Acknowledgments
The authors would like to thank the captain and crew of the ARSV Laurence M. Gould and the personnel of the US Antarctic Program support contractors for assistance in Chile, at sea, and at Palmer Station, Antarctica. The authors also thank Jason Sydes and Pete Batzel for running Prost! software on the four miRNA libraries, and Brian Eames for helping to develop Prost!. The authors also thank Qianghua Xu and colleagues at Shanghai Ocean University for making their important sequencing resource publicly available. This work was supported by NIH grants R01 OD011116 and NIH R01AG31922 (JHP), and by NSF grants ANT-1247510 and PLR-1444167 (HWD) and PLR- 1543383 (JHP & HWD).
Appendix. The hooknose icefish Chionodraco hamatus
The Antarctic icefish Chionodraco hamatus (Lönnberg, 1905) lacks a common name. Because the use of common names sometimes facilitates communication, we propose to apply “hooknose icefish” to the Antarctic icefish Chionodraco hamatus (Lönnberg, 1905). The qualifier “hooknose” derives from the etymology of hamatus, from the Latin “h mus,” meaning “hook”, referring to the prominent spine on the nose. The genus Chionodraco contains two other species: the ocellated icefish Chionodraco rastrospinosus, DeWitt & Hureau, 1979, and Myer’s icefish Chionodraco myersi, DeWitt & Tyler, 1960. C. rastrospinosus also has a well-developed rostral spine as its scientific name suggests, whereas C. myersi has only a small rostral knob.
Footnotes
Competing interests:
Authors have no competing interests to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Azzouzi I, Moest H, Winkler J, Fauchère JC, Gerber AP, Wollscheid B, Stoffel M, Schmugge M, Speer O. MicroRNA-96 Directly Inhibits γ-Globin Expression in Human Erythropoiesis. PLoS ONE. 2011;6:e22838. doi: 10.1371/journal.pone.0022838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzouzi I, Schmugge M, Speer O. MicroRNAs as components of regulatory networks controlling erythropoiesis. Eur J Haematol. 2012;89:1–9. doi: 10.1111/j.1600-0609.2012.01774.x. [DOI] [PubMed] [Google Scholar]
- Baran-Gale J, Erdos MR, Sison C, Young A, Fannin EE, Chines PS, Sethupathy P. Massively differential bias between two widely used Illumina library preparation methods for small RNA sequencing. bioRxiv. 2013;001479 doi: 10.1101/001479. [DOI] [Google Scholar]
- Batzel P, Desvignes T, Postlethwait JH, Eames BF, Sydes J. Prost!, a tool for miRNA annotation and next generation smallRNA sequencing experiment analysis. Zenodo. 2015 doi: 10.5281/zenodo.35422. [DOI] [Google Scholar]
- Bhagavathi S, Czader M. MicroRNAs in Benign and Malignant Hematopoiesis. Arch Pathol Lab Med. 2010;134:1276–1281. doi: 10.1043/2009-0178-RS.1. [DOI] [PubMed] [Google Scholar]
- Borley KA, Sidell BD. Evolution of the myoglobin gene in Antarctic Icefishes (Channichthyidae) Polar Biol. 2010;34:659–665. doi: 10.1007/s00300-010-0921-x. [DOI] [Google Scholar]
- Braasch I, Gehrke AR, Smith JJ, Kawasaki K, Manousaki T, Pasquier J, Amores A, Desvignes T, Batzel P, Catchen J, Berlin AM, Campbell MS, Barrell D, Martin KJ, Mulley JF, Ravi V, Lee AP, Nakamura T, Chalopin D, Fan S, Wcisel D, Cañestro C, Sydes J, Beaudry FEG, Sun Y, Hertel J, Beam MJ, Fasold M, Ishiyama M, Johnson J, Kehr S, Lara M, Letaw JH, Litman GW, Litman RT, Mikami M, Ota T, Saha NR, Williams L, Stadler PF, Wang H, Taylor JS, Fontenot Q, Ferrara A, Searle SMJ, Aken B, Yandell M, Schneider I, Yoder JA, Volff J-N, Meyer A, Amemiya CT, Venkatesh B, Holland PWH, Guiguen Y, Bobe J, Shubin NH, Di Palma F, Alföldi J, Lindblad-Toh K, Postlethwait JH. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat Genet. 2016;48:427–437. doi: 10.1038/ng.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braasch I, Peterson SM, Desvignes T, McCluskey BM, Batzel P, Postlethwait JH. A new model army: Emerging fish models to study the genomics of vertebrate Evo-Devo. J Exp Zoolog B Mol Dev Evol. 2015;324:316–341. doi: 10.1002/jez.b.22589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford Y, Conlin T, Dunn N, Fashena D, Frazer K, Howe DG, Knight J, Mani P, Martin R, Moxon SAT, Paddock H, Pich C, Ramachandran S, Ruef BJ, Ruzicka L, Schaper HB, Schaper K, Shao X, Singer A, Sprague J, Sprunger B, Slyke CV, Westerfield M. ZFIN: enhancements and updates to the zebrafish model organism database. Nucleic Acids Res. 2011;39:D822–D829. doi: 10.1093/nar/gkq1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S, Dos Santos CO, Chong MMW, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs Modulate Hematopoietic Lineage Differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Cheng C-HC, Detrich HW. Molecular ecophysiology of Antarctic notothenioid fishes. Philos Trans R Soc B Biol Sci. 2007;362:2215–2232. doi: 10.1098/rstb.2006.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christodoulou F, Raible F, Tomer R, Simakov O, Trachana K, Klaus S, Snyman H, Hannon GJ, Bork P, Arendt D. Ancient animal microRNAs and the evolution of tissue identity. Nature. 2010;463:1084–1088. doi: 10.1038/nature08744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, Wolfe SA, Giraldez AJ. A Novel miRNA Processing Pathway Independent of Dicer Requires Argonaute2 Catalytic Activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocca E, Ratnayake-Lecamwasam M, Parker SK, Camardella L, Ciaramella M, Prisco G, di Detrich HW. Genomic remnants of alpha-globin genes in the hemoglobinless antarctic icefishes. Proc Natl Acad Sci. 1995;92:1817–1821. doi: 10.1073/pnas.92.6.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvignes T, Batzel P, Berezikov E, Eilbeck K, Eppig JT, McAndrews MS, Singer A, Postlethwait JH. miRNA Nomenclature: A View Incorporating Genetic Origins, Biosynthetic Pathways, and Sequence Variants. Trends Genet. 2015;31:613–626. doi: 10.1016/j.tig.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvignes T, Beam MJ, Batzel P, Sydes J, Postlethwait JH. Expanding the annotation of zebrafish microRNAs based on small RNA sequencing. Gene. 2014;546:386–389. doi: 10.1016/j.gene.2014.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Prisco G, Cocca E, Parker SK, Detrich HW., III Tracking the evolutionary loss of hemoglobin expression by the white-blooded Antarctic icefishes. Gene, Papers presented at the 3rd Anton Dohrn Workshop “Fish Genomics: Structural and Functional Aspects”; Ischia. 1-2 June 2001; Giorgio Bernardi, Giacomo Bernardi (Organizers); 2002. pp. 185–191. [DOI] [PubMed] [Google Scholar]
- Dore LC, Amigo JD, Santos CO, dos Zhang Z, Gai X, Tobias JW, Yu D, Klein AM, Dorman C, Wu W, Hardison RC, Paw BH, Weiss MJ. A GATA-1-regulated microRNA locus essential for erythropoiesis. Proc Natl Acad Sci. 2008;105:3333–3338. doi: 10.1073/pnas.0712312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du T-T, Fu Y-F, Dong M, Wang L, Fan H-B, Chen Y, Jin Y, Chen S-J, Chen Z, Deng M, Huang Q-H, Liu TX. Experimental validation and complexity of miRNA–mRNA target interaction during zebrafish primitive erythropoiesis. Biochem Biophys Res Commun. 2009;381:688–693. doi: 10.1016/j.bbrc.2009.02.122. [DOI] [PubMed] [Google Scholar]
- Fänge R. Blood cells, haemopoiesis and lymphomyeloid tissues in fish. Fish Shellfish Immunol. 1994;4:405–411. doi: 10.1006/fsim.1994.1036. [DOI] [Google Scholar]
- Fan HB, Liu YJ, Wang L, Du TT, Dong M, Gao L, Meng ZZ, Jin Y, Chen Y, Deng M, Yang HT, Jing Q, Gu AH, Liu TX, Zhou Y. miR-142-3p acts as an essential modulator of neutrophil development in zebrafish. Blood. 2014;124:1320–1330. doi: 10.1182/blood-2013-12-545012. [DOI] [PubMed] [Google Scholar]
- Fu YF, Du TT, Dong M, Zhu KY, Jing CB, Zhang Y, Wang L, Fan HB, Chen Y, Jin Y, Yue GP, Chen SJ, Chen Z, Huang QH, Jing Q, Deng M, Liu TX. Mir-144 selectively regulates embryonic α-hemoglobin synthesis during primitive erythropoiesis. Blood. 2009;113:1340–1349. doi: 10.1182/blood-2008-08-174854. [DOI] [PubMed] [Google Scholar]
- Galloway JL, Wingert RA, Thisse C, Thisse B, Zon LI. Loss of Gata1 but Not Gata2 Converts Erythropoiesis to Myelopoiesis in Zebrafish Embryos. Dev Cell. 2005;8:109–116. doi: 10.1016/j.devcel.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Giordano D, Russo R, Coppola D, Altomonte G, di Prisco G, Bruno S, Verde C. “Cool” adaptations to cold environments: globins in Notothenioidei (Actynopterygii, Perciformes) Hydrobiologia. 2015;761:293–312. doi: 10.1007/s10750-015-2306-1. [DOI] [Google Scholar]
- Grabher C, Payne EM, Johnston AB, Bolli N, Lechman E, Dick JE, Kanki JP, Look AT. Zebrafish microRNA-126 determines hematopoietic cell fate through c-Myb. Leukemia. 2011;25:506–514. doi: 10.1038/leu.2010.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove TJ, Hendrickson JW, Sidell BD. Two species of antarctic icefishes (genus Champsocephalus) share a common genetic lesion leading to the loss of myoglobin expression. Polar Biol. 2004;27:579–585. doi: 10.1007/s00300-004-0634-0. [DOI] [Google Scholar]
- Hafner M, Renwick N, Brown M, Mihailović A, Holoch D, Lin C, Pena JTG, Nusbaum JD, Morozov P, Ludwig J, Ojo T, Luo S, Schroth G, Tuschl T. RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. RNA. 2011;17:1697–1712. doi: 10.1261/rna.2799511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havelange V, Garzon R. Micrornas: Emerging key regulators of hematopoiesis. Am J Hematol. 2010;85:935–942. doi: 10.1002/ajh.21863. [DOI] [PubMed] [Google Scholar]
- Holeton GF. Oxygen uptake and circulation by a hemoglobinless antarctic fish (Chaenocephalus aceratus Lonnberg) compared with three red-blooded antartic fish. Comp Biochem Physiol. 1970;34:457–471. doi: 10.1016/0010-406X(70)90185-4. [DOI] [PubMed] [Google Scholar]
- Jin HY, Gonzalez-Martin A, Miletic AV, Lai M, Knight S, Sabouri-Ghomi M, Head SR, Macauley MS, Rickert RC, Xiao C. Transfection of microRNA Mimics Should Be Used with Caution. Front Genet. 2015;6 doi: 10.3389/fgene.2015.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Tan YS, Cheng WC, Civin CI. MiR-144 and MiR-451 Regulate Human Erythropoiesis By Targeting RAB14. Blood. 2013;122:942–942. [Google Scholar]
- Kim M, Tan YS, Cheng W-C, Kingsbury TJ, Heimfeld S, Civin CI. MIR144 and MIR451 regulate human erythropoiesis via RAB14. Br J Haematol. 2015;168:583–597. doi: 10.1111/bjh.13164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kock K-H. Antarctic icefishes (Channichthyidae): a unique family of fishes. A review, Part I. Polar Biol. 2005a;28:862–895. doi: 10.1007/s00300-005-0019-z. [DOI] [Google Scholar]
- Kock K-H. Antarctic icefishes (Channichthyidae): a unique family of fishes. A review, Part II. Polar Biol. 2005b;28:897–909. doi: 10.1007/s00300-005-0020-6. [DOI] [Google Scholar]
- Kosik KS. MicroRNAs and Cellular Phenotypy. Cell. 2010;143:21–26. doi: 10.1016/j.cell.2010.09.008. [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2013;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer NJ, Wang WL, Reyes EY, Kumar B, Chen CC, Ramakrishna C, Cantin EM, Vonderfecht SL, Taganov KD, Chau N, Boldin MP. Altered lymphopoiesis and immunodeficiency in miR-142 null mice. Blood. 2015;125:3720–3730. doi: 10.1182/blood-2014-10-603951. [DOI] [PubMed] [Google Scholar]
- Kuwabara Y, Horie T, Baba O, Watanabe S, Nishiga M, Usami S, Izuhara M, Nakao T, Nishino T, Otsu K, Kita T, Kimura T, Ono K. MicroRNA-451 Exacerbates Lipotoxicity in Cardiac Myocytes and High-Fat Diet-Induced Cardiac Hypertrophy in Mice Through Suppression of the LKB1/AMPK Pathway. Circ Res. 2015;116:279–288. doi: 10.1161/CIRCRESAHA.116.304707. [DOI] [PubMed] [Google Scholar]
- Lawrie CH. microRNA expression in erythropoiesis and erythroid disorders. Br J Haematol. 2010;150:144–151. doi: 10.1111/j.1365-2141.2009.07978.x. [DOI] [PubMed] [Google Scholar]
- Lee C-T, Risom T, Strauss WM. Evolutionary Conservation of MicroRNA Regulatory Circuits: An Examination of MicroRNA Gene Complexity and Conserved MicroRNA-Target Interactions through Metazoan Phylogeny. DNA Cell Biol. 2007;26:209–218. doi: 10.1089/dna.2006.0545. [DOI] [PubMed] [Google Scholar]
- Listowski M, Heger E, Bogusławska D, Machnicka B, Kuliczkowski K, Leluk J, Sikorski A. microRNAs: fine tuning of erythropoiesis. Cell Mol Biol Lett. 2012;18:34–46. doi: 10.2478/s11658-012-0038-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macqueen DJ, Serrana DG, de la Johnston IA. Cardiac myoglobin deficit has evolved repeatedly in teleost fishes. Biol Lett. 2014;10:20140225. doi: 10.1098/rsbl.2014.0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffia M, Rizzello A, Acierno R, Rollo M, Chiloiro R, Storelli C. Carbonic anhydrase activity in tissues of the icefish Chionodraco hamatus and of the red-blooded teleosts Trematomus bernacchii and Anguilla anguilla. J Exp Biol. 2001;204:3983–3992. doi: 10.1242/jeb.204.22.3983. [DOI] [PubMed] [Google Scholar]
- Masaki S, Ohtsuka R, Abe Y, Muta K, Umemura T. Expression patterns of microRNAs 155 and 451 during normal human erythropoiesis. Biochem Biophys Res Commun. 2007;364:509–514. doi: 10.1016/j.bbrc.2007.10.077. [DOI] [PubMed] [Google Scholar]
- McCarthy JJ. MicroRNA-206: The skeletal muscle-specific myomiR. Biochim Biophys Acta BBA - Gene Regul Mech MicroRNA. 2008;1779:682–691. doi: 10.1016/j.bbagrm.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammdai-asl J, Ramezani A, Norozi F, Malehi AS, Asnafi AA, Far MAJ, Mousavi SH, Saki N. MicroRNAs in erythropoiesis and red blood cell disorders. Front Biol. 2015;10:321–332. doi: 10.1007/s11515-015-1365-z. [DOI] [Google Scholar]
- Muhseen ZT, Abbood NN. MiR-145 Regulates Hematopoiesis during Zebrafish Development. Am J Biosci Bioeng. 2014;2:44. doi: 10.11648/j.bio.20140203.12. [DOI] [Google Scholar]
- Near TJ, Dornburg A, Eytan RI, Keck BP, Smith WL, Kuhn KL, Moore JA, Price SA, Burbrink FT, Friedman M, Wainwright PC. Phylogeny and tempo of diversification in the superradiation of spiny-rayed fishes. Proc Natl Acad Sci. 2013;110:12738–12743. doi: 10.1073/pnas.1304661110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Near TJ, Dornburg A, Harrington RC, Oliveira C, Pietsch TW, Thacker CE, Satoh TP, Katayama E, Wainwright PC, Eastman JT, Beaulieu JM. Identification of the notothenioid sister lineage illuminates the biogeographic history of an Antarctic adaptive radiation. BMC Evol Biol. 2015;15:109. doi: 10.1186/s12862-015-0362-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Near TJ, Dornburg A, Kuhn KL, Eastman JT, Pennington JN, Patarnello T, Zane L, Fernández DA, Jones CD. Ancient climate change, antifreeze, and the evolutionary diversification of Antarctic fishes. Proc Natl Acad Sci. 2012;201115169 doi: 10.1073/pnas.1115169109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Near TJ, Parker SK, Detrich HW. A Genomic Fossil Reveals Key Steps in Hemoglobin Loss by the Antarctic Icefishes. Mol Biol Evol. 2006;23:2008–2016. doi: 10.1093/molbev/msl071. [DOI] [PubMed] [Google Scholar]
- Pase L, Layton JE, Kloosterman WP, Carradice D, Waterhouse PM, Lieschke GJ. miR-451 regulates zebrafish erythroid maturation in vivo via its target gata2. Blood. 2009;113:1794–1804. doi: 10.1182/blood-2008-05-155812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick DM, Zhang CC, Tao Y, Yao H, Qi X, Schwartz RJ, Huang LJ-S, Olson EN. Defective erythroid differentiation in miR-451 mutant mice mediated by 14- 3-3ζ. Genes Dev. 2010;24:1614–1619. doi: 10.1101/gad.1942810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson KJ, Dietrich MR, McPeek MA. MicroRNAs and metazoan macroevolution: insights into canalization, complexity, and the Cambrian explosion. BioEssays News Rev Mol Cell Dev Biol. 2009;31:736–747. doi: 10.1002/bies.200900033. [DOI] [PubMed] [Google Scholar]
- Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet. 2012;13:358–369. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raabe CA, Tang T-H, Brosius J, Rozhdestvensky TS. Biases in small RNA deep sequencing data. Nucleic Acids Res. 2014;42:1414–1426. doi: 10.1093/nar/gkt1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen KD, Simmini S, Abreu-Goodger C, Bartonicek N, Giacomo MD, Bilbao-Cortes D, Horos R, Lindern MV, Enright AJ, O’Carroll D. The miR-144/451 locus is required for erythroid homeostasis. J Exp Med. 2010;207:1351–1358. doi: 10.1084/jem.20100458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes J, Hagen A, Hsu K, Deng M, Liu TX, Look AT, Kanki JP. Interplay of Pu.1 and Gata1 Determines Myelo-Erythroid Progenitor Cell Fate in Zebrafish. Dev Cell. 2005;8:97–108. doi: 10.1016/j.devcel.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Roberto VP, Tiago DM, Gautvik K, Cancela ML. Evidence for the conservation of miR-223 in zebrafish (Danio rerio): Implications for function. Gene. 2015;566:54–62. doi: 10.1016/j.gene.2015.04.022. [DOI] [PubMed] [Google Scholar]
- Ruud JT. Vertebrates without Erythrocytes and Blood Pigment. Nature. 1954;173:848–850. doi: 10.1038/173848a0. [DOI] [PubMed] [Google Scholar]
- Sangokoya C, Telen MJ, Chi JT. microRNA miR-144 modulates oxidative stress tolerance and associates with anemia severity in sickle cell disease. Blood. 2010;116:4338–4348. doi: 10.1182/blood-2009-04-214817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed D, Abdellatif M. MicroRNAs in Development and Disease. Physiol Rev. 2011;91:827–887. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- Shin SC, Ahn DH, Kim SJ, Pyo CW, Lee H, Kim M-K, Lee J, Lee JE, Detrich HW, Postlethwait JH, Edwards D, Lee SG, Lee JH, Park H. The genome sequence of the Antarctic bullhead notothen reveals evolutionary adaptations to a cold environment. Genome Biol. 2014;15:468. doi: 10.1186/s13059-014-0468-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidell BD, O’Brien KM. When bad things happen to good fish: the loss of hemoglobin and myoglobin expression in Antarctic icefishes. J Exp Biol. 2006;209:1791–1802. doi: 10.1242/jeb.02091. [DOI] [PubMed] [Google Scholar]
- Sidell BD, Vayda ME, Small DJ, Moylan TJ, Londraville RL, Yuan M-L, Rodnick KJ, Eppley ZA, Costello L. Variable expression of myoglobin among the hemoglobinless Antarctic icefishes. Proc Natl Acad Sci. 1997;94:3420–3424. doi: 10.1073/pnas.94.7.3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small DJ, Moylan T, Vayda ME, Sidell BD. The myoglobin gene of the Antarctic icefish, Chaenocephalus aceratus, contains a duplicated TATAAAA sequence that interferes with transcription. J Exp Biol. 2003;206:131–139. doi: 10.1242/jeb.00067. [DOI] [PubMed] [Google Scholar]
- Song L, Su M, Wang S, Zou Y, Wang X, Wang Y, Cui H, Zhao P, Hui R, Wang J. MiR-451 is decreased in hypertrophic cardiomyopathy and regulates autophagy by targeting TSC1. J Cell Mol Med. 2014;18:2266–2274. doi: 10.1111/jcmm.12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G, Yin X, Luo H, Xu X, Bolund L, Zhang X. Sequencing bias: comparison of different protocols of MicroRNA library construction. BMC Biotechnol. 2010;10:64. doi: 10.1186/1472-6750-10-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufts B, Gervais M, Staebler M, Weaver J. Subcellular distribution and characterization of gill carbonic anhydrase and evidence for a plasma carbonic anhydrase inhibitor in Antarctic fish. J Comp Physiol B. 2002;172:287–295. doi: 10.1007/s00360-002-0253-4. [DOI] [PubMed] [Google Scholar]
- Undi RB, Kandi R, Gutti RK, Undi RB, Kandi R, Gutti RK. MicroRNAs as Haematopoiesis Regulators, MicroRNAs as Haematopoiesis Regulators. Adv Hematol Adv Hematol. 2013;2013:e695754. doi: 10.1155/2013/695754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vegiopoulos A, García P, Emambokus N, Frampton J. Coordination of erythropoiesis by the transcription factor c-Myb. Blood. 2006;107:4703–4710. doi: 10.1182/blood-2005-07-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhu H, Zhang X, Liu Y, Chen J, Medvedovic M, Li H, Weiss MJ, Ren X, Fan G-C. Loss of the miR-144/451 cluster impairs ischaemic preconditioning-mediated cardioprotection by targeting Rac-1. Cardiovasc Res. 2012;94:379–390. doi: 10.1093/cvr/cvs096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JJ, Watts JA, Vakoc CR, Yao Y, Wang H, Hardison RC, Blobel GA, Chodosh LA, Weiss MJ. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood. 2004;104:3136–3147. doi: 10.1182/blood-2004-04-1603. [DOI] [PubMed] [Google Scholar]
- Wheeler BM, Heimberg AM, Moy VN, Sperling EA, Holstein TW, Heber S, Peterson KJ. The deep evolution of metazoan microRNAs. Evol Dev. 2009;11:50–68. doi: 10.1111/j.1525-142X.2008.00302.x. [DOI] [PubMed] [Google Scholar]
- Witeska M. Erythrocytes in teleost fishes: a review. Zool Ecol. 2013;23:275–281. doi: 10.1080/21658005.2013.846963. [DOI] [Google Scholar]
- Xu Q, Cai C, Hu X, Liu Y, Guo Y, Hu P, Chen Z, Peng S, Zhang D, Jiang S, Wu Z, Chan J, Chen L. Evolutionary suppression of erythropoiesis via the modulation of TGF-β signalling in an Antarctic icefish. Mol Ecol. 2015;24:4664–4678. doi: 10.1111/mec.13344. [DOI] [PubMed] [Google Scholar]
- Yang J-S, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, Papapetrou EP, Sadelain M, O’Carroll D, Lai EC. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc Natl Acad Sci. 2010;107:15163–15168. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yergeau DA, Cornell CN, Parker SK, Zhou Y, Detrich HW., III bloodthirsty, an RBCC/TRIM gene required for erythropoiesis in zebrafish. Dev Biol. 2005;283:97–112. doi: 10.1016/j.ydbio.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Yu D, Santos CO, dos Zhao G, Jiang J, Amigo JD, Khandros E, Dore LC, Yao Y, D’Souza J, Zhang Z, Ghaffari S, Choi J, Friend S, Tong W, Orange JS, Paw BH, Weiss MJ. miR-451 protects against erythroid oxidant stress by repressing 14-3-3ζ. Genes Dev. 2010;24:1620–1633. doi: 10.1101/gad.1942110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Shykind B, Sun T. Approaches to manipulating microRNAs in neurogenesis. Front Neurosci. 2013;6:196. doi: 10.3389/fnins.2012.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Sankaran VG, Lodish HF. MicroRNAs in erythroid and megakaryocytic differentiation and megakaryocyte–erythroid progenitor lineage commitment. Leukemia. 2012;26:2310–2316. doi: 10.1038/leu.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan M, Miller CP, Papayannopoulou T, Stamatoyannopoulos G, Song C-Z. MicroRNA expression dynamics during murine and human erythroid differentiation. Exp Hematol. 2007;35:1015–1025. doi: 10.1016/j.exphem.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Wang D, Wang F, Li T, Dong L, Liu H, Ma Y, Jiang F, Yin H, Yan W, Luo M, Tang Z, Zhang G, Wang Q, Zhang J, Zhou J, Yu J. A comprehensive analysis of GATA-1-regulated miRNAs reveals miR-23a to be a positive modulator of erythropoiesis. Nucleic Acids Res. 2013;gkt093 doi: 10.1093/nar/gkt093. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.