Abstract
BACKGROUND
Genetic factors predispose to attention deficit/hyperactivity disorder (ADHD). Previous studies have reported linkage and association to ADHD of gene variants within ADGRL3. In this study, we functionally analyzed non-coding variants in this gene as likely pathological contributors.
METHODS
In silico, in vitro and in vivo approaches were used to identify and characterize evolutionary conserved elements within the ADGRL3 linkage region (~207 Kb). Family-based genetic analyses on 838 individuals (372 affected and 466 unaffected) identified ADHD-associated SNPs harbored in some of these conserved elements. Luciferase assays and zebrafish GFP transgenesis tested conserved elements for transcriptional enhancer activity. Electromobility shift assays were used to verify transcription factor binding disruption by ADHD risk alleles.
RESULTS
An ultraconserved element was discovered (ECR47) that functions as a transcriptional enhancer. A three-variant ADHD risk haplotype in ECR47, formed by rs17226398, rs56038622 and rs2271338, reduced enhancer activity by 40% in neuroblastoma and astrocytoma cells (PBonferroni<0.0001). This enhancer also drove GFP expression in the zebrafish brain in a tissue-specific manner, sharing aspects of endogenous ADGRL3 expression. The rs2271338 risk allele disrupts binding of YY1, an important factor in the development and function of the central nervous system. Expression quantitative trait loci analysis of post-mortem human brain tissues revealed an association between rs2271338 and reduced ADGRL3 expression in the thalamus.
CONCLUSIONS
These results uncover the first functional evidence of common non-coding variants with potential implications for the pathology of ADHD.
Keywords: ADHD, Genetics, ADGRL3, LPHN3, Latrophilin, Cis-acting regulatory element, Enhancer, Evolutionary conserved regions, Zebrafish
INTRODUCTION
Attention deficit/hyperactivity disorder (ADHD) is a complex heritable trait that affects more children and adolescents than any other psychiatric disorder: approximately 5.3% of the world population is estimated to be affected (1, 2). ADHD increases the risk of disruptive symptoms, substance use, legal problems, and underemployment, which reduces the quality of life of ADHD sufferers and their families (3-6). ADHD heritability has been estimated at 76%, suggesting a strong genetic component (7). Candidate gene and genome-wide studies of SNP and copy number variants have identified a number of susceptibility loci (8), but very few molecular studies have attempted to elucidate the functional effects of ADHD risk variants.
Common genetic variants within the adhesion G protein-coupled receptor L3 gene (ADGRL3, also known as latrophilin 3 or LPHN3) in 4q13.2 are strongly linked and associated with ADHD (9-19). The ADHD-linked region in ADGRL3 (hereinafter referred to as “minimal critical region”, or MCR) spans exons 4 through 19 of the gene (approximately 207 Kb). Searching for variants that might affect ADGRL3 protein function, Domene et al. (20) sequenced the entire coding region of the gene in a small cohort of subjects with ADHD, but no missense coding changes or canonical splice site alterations were found to be associated with the disorder. This suggests that intronic non-coding variants are the likely pathological contributors.
In the present study, we interrogated the ADGRL3 MCR aiming to identify transcriptional enhancer elements with potential functional implications for ADHD.
METHODS AND MATERIALS
Subjects
Individuals with and without ADHD were ascertained from the Medellin metropolitan area (Antioquia, Colombia). The Paisa community is considered a genetic isolate of Caucasian descent with low admixture with Amerindian and Negroid ethnicities (21). The cohort consisted of 14 multi-generational families and 125 nuclear families for a total of 838 individuals: 372 affected and 466 unaffected, 335 children and adolescents (3-16 years old), and 503 adults (17 years and older). The multi-generational families had an average size of 28 members (range 9-57) and an average of 2.93 generations (range 2-4). Full details of the clinical, demographic, and genetic ascertainment features, as well as the methodology of neuropsychological evaluation have been published elsewhere (22-24). The study was reviewed and approved by the Institutional Review Board of the National Human Genome Research Institute (Protocol 00-HG-0058) and the University of Antioquia Ethics Committee (Protocol 11-13-342).
Genetic statistical analyses
SNP genotyping methods are presented in Supplement 1. Genotype data were imported into Golden Helix® SVS 8.3.1 (Golden Helix, Bozeman, MT) for family-based association testing (FBAT). Genotype and allelic frequencies were estimated by maximum likelihood. FBAT analyses using ADHD status as a categorical variable were applied to the whole set of markers that passed quality control. We also performed family-based haplotype analyses to compare with the results at the marker level. Co-morbid conditions (substance use and disruptive symptoms) and neuropsychological endophenotypes (Wechsler Intelligence Scale for Children [WISC] Block Design, WISC Performance Intelligence Quotient [PIQ], WISC Full-scale Intelligence Quotient [FSIQ], “A”-cancelation and Vigilance Test Correct Responses [ACVTCR] and Omissions [ACVTO], and Rey-Osterrieth Complex Figure Test [ROCFT Copy]) were used as ADHD-interacting variables. We used endophenotype data only for 255 children and adolescents (170 affected and 85 unaffected) with an FSIQ≥81 and regular school grades adequate for their age, in order exclude participants with potential learning disabilities.
Because the ADGRL3 genomic region under study is known to be linked to ADHD (11), the null hypothesis of linkage and no association was tested. Individual genotypes inconsistent with Mendelian transmission were excluded from the analyses. All markers were tested for deviation from Hardy-Weinberg equilibrium. Allelic tests of association were applied using a dominant model of inheritance.
Identification of evolutionary conserved elements within the ADGRL3 MCR and prediction of regulatory function
We used the ECR Browser (25) to identify highly conserved elements within the ADGRL3 MCR (Accession no. NC_000004.11, GRCh37.p13/hg19, coordinates chr4:62,688,419-62,895,842). We looked at DNA sequence conservation across several vertebrate species including human, mouse, chicken and zebrafish, using a sliding window of >150 bp long and >70% identity. To predict transcriptional regulatory function, we gathered annotation data from three Regulation tracks of the UCSC Table Browser: Chromatin State Segmentation by a Hidden Markov Model (Broad ChromHMM) (26, 27), EP300 Transcription Factor ChIP-seq (28), and Uniform DNAse I Hypersensitivity Sites (29). Additional information on these annotation tracks can be found in Supplement 1.
Luciferase assays
The generation of ECR-luciferase constructs, cell culture conditions and transient transfections are described in Supplement 1. After 24h of transfection, culture supernatants containing secreted luciferases were collected. Luciferase assays were performed using the Pierce™ Luciferase Flash Assay kits (Thermo Fisher Scientific, Waltham, MA). Each ECR-luciferase construct was assayed in triplicate and each transfection experiment was repeated at least three times (n=9). Luciferase activity data were analyzed using a one-way ANOVA with Bonferroni correction, as implemented in Prism 6 (GraphPad Software, La Jolla, CA).
Electromobility shift assays (EMSA)
EMSAs were performed using standard procedures. ECR47 DNA probes were incubated with Myc-DDK-tagged recombinant transcription factors or whole-cell expression lysates (Origene, Rockville, MD). For supershift reactions, an anti-DDK (FLAG) monoclonal antibody (Origene) was added.
Zebrafish bioassays
Zebrafish stocks and manipulations followed the Animal Care and Use protocols used in our Zebrafish Core facility (NIH/NHGRI). Zebrafish transgenesis and in situ hybridization were performed following standards procedures (see Supplement 1 for details).
RESULTS
Identification of potential regulatory elements within ADGRL3
Using the ECR Browser we identified highly conserved elements harbored in the ADGRL3 MCR. Although it is currently well established that the inter-genome comparisons of distant species (such as humans and fish) are very powerful in identifying critical distant regulatory elements, only 5% of the genes in the human genome contain a human/fugu non-coding ECR in their genomic neighborhood (30-32). For that reason, the analysis with species more closely related than fish is required for many human genes to identify regulatory elements. We therefore used the human/chicken alignment to select candidate sequences. The human/opossum alignment (an evolutionarily closer species) was arbitrarily used to give an identity to the conserved elements, thus naming 51 regions ECR1 through ECR51 (Figure S1 in Supplement 1 and Table S1 in Supplement 2).
Given that enhancer activity has been correlated with certain properties of chromatin (33), we gathered p300 binding sites, histone marks and DNase I hypersensitivity sites annotation data to predict ECR enhancer function (Table S1 in Supplement 2). From the list of elements conserved in chicken we excluded those ECRs not predicted to be functional by any of the annotation tracks above and that did not contain SNPs with a minor allele frequency (MAF) >1%, as genetic studies predominantly suggest that ADHD risk is mainly explained by common variants (1, 7, 8). Following this approach, the number of candidate ECRs was reduced to 10: ECR1, ECR2, ECR4, ECR9, ECR20, ECR22, ECR26, ECR37, ECR46, and ECR47. Because coding regions can overlap enhancer sequences (34, 35), we did not exclude those elements containing ADGRL3 exons (ECR20, ECR26, ECR37, and ECR47).
Genetic statistical analyses of ECR variants
Table S2 in Supplement 1 shows a list of the common variants harbored in ECR sequences predicted to be functional. We performed family-based association tests in 838 individuals from the same Colombian multigenerational and nuclear families that previously showed linkage and association of ADHD to 4q13.2 (11, 22). Genotype proportions did not significantly depart from Hardy-Weinberg equilibrium. SNPs in 6 of the 10 ECRs showed significant association with ADHD, comorbid disorders and/or endophenotypes after correction by false discovery rate (Table 1). ECR46, ECR47 and ECR4 showed the highest association with ADHD (all five variants, PFDR = 0.00219), followed by ECR26 (both PFDR = 0.00342), ECR2 (variant rs1868790, PFDR = 0.00988), and ECR37 (variant rs1397548, PFDR = 0.03173). Interestingly, only variants in ECR46 (rs11131352) and ECR47 (rs17226398, rs56038622 and rs2271338) consistently showed association with disruptive symptoms (ODD [all PFDR = 0.00005] and CD [all PFDR = 0.00076]), substance use (alcohol [all PFDR =0.00199] and nicotine [all PFDR = 0.00203]) and endophenotypes (ACVTCR [all PFDR = 0.00098], ACVTO [all PFDR = 0.00098], and ROCFT [all PFDR = 0.02775]). Variants in ECR1, ECR9 and ECR22 did not show association with ADHD, comorbid conditions or endophenotypes, and, therefore, were not considered for functional analysis. None of the variants showed association with WISC Block Design, WISC PIQ or WISC FSIQ. Haplotype analyses for ADHD affection status are presented in Table S3 in Supplement 1.
Table 1.
Association between ECR common variants and ADHD, disruptive symptoms, substance use and neuropsychological endophenotypes in the Paisa dataset.
| ECR ID |
Variant | Allele (Freq.) |
ADHD p- value |
ODD p-value | CD p-value | Nicotine p- value |
Alcohol p- value |
ACVTCR p- value |
ACVTO p- value |
ROCFT p- value |
Function | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Raw | FDR | Raw | FDR | Raw | FDR | Raw | FDR | Raw | FDR | Raw | FDR | Raw | FDR | Raw | FDR | ||||
| ECR46 | rs11131352 | T (0.132) | 0.00073 | 0.00219 | 0.00002 | 0.00005 | 0.00020 | 0.00076 | 0.00054 | 0.00203 | 0.00066 | 0.00199 | 0.00046 | 0.00098 | 0.00046 | 0.00098 | 0.01295 | 0.02775 | intronic |
| ECR47 | rs17226398 | C (0.132) | 0.00073 | 0.00219 | 0.00002 | 0.00005 | 0.00020 | 0.00076 | 0.00054 | 0.00203 | 0.00066 | 0.00199 | 0.00046 | 0.00098 | 0.00046 | 0.00098 | 0.01295 | 0.02775 | intronic |
| rs56038622 | T (0.132) | 0.00073 | 0.00219 | 0.00002 | 0.00005 | 0.00020 | 0.00076 | 0.00054 | 0.00203 | 0.00066 | 0.00199 | 0.00046 | 0.00098 | 0.00046 | 0.00098 | 0.01295 | 0.02775 | intronic | |
| rs2271338 | A (0.132) | 0.00073 | 0.00219 | 0.00002 | 0.00005 | 0.00020 | 0.00076 | 0.00054 | 0.00203 | 0.00066 | 0.00199 | 0.00046 | 0.00098 | 0.00046 | 0.00098 | 0.01295 | 0.02775 | intronic | |
| ECR4 | rs10021694 | T (0.313) | 0.00063 | 0.00219 | 0.03921 | ns | 0.03274 | ns | 0.03740 | ns | 0.03092 | ns | 0.00003 | 0.00016 | 0.00003 | 0.00017 | 0.00015 | 0.00228 | intronic |
| ECR26 | rs734644 | T (0.281) | 0.00155 | 0.00342 | 0.000003 | 0.00003 | 0.02510 | ns | 0.02262 | 0.04241 | 0.00119 | 0.00255 | 0.00201 | 0.00376 | 0.00199 | 0.00373 | ns | ns | coding-syn |
| rs2305339 | G (0.278) | 0.00160 | 0.00342 | 0.000003 | 0.00003 | 0.02641 | ns | 0.02223 | 0.04241 | 0.00082 | 0.00204 | 0.00552 | 0.00920 | 0.00548 | 0.00913 | ns | ns | intronic | |
| ECR2 | rs1868790 | A (0.427) | 0.00527 | 0.00988 | 0.02291 | 0.03819 | 0.01442 | 0.04327 | 0.02062 | 0.04241 | 0.02062 | 0.03866 | ns | ns | ns | ns | ns | ns | intronic |
| rs73823249 | G (0.064) | ns | ns | ns | ns | ns | ns | 0.02191 | 0.04241 | 0.000001 | 0.00001 | ns | ns | ns | ns | 0.00186 | 0.00928 | intronic | |
| ECR37 | rs1397548 | A (0.308) | 0.01904 | 0.03173 | 0.00272 | 0.00582 | ns | ns | ns | ns | ns | ns | 0.00002 | 0.00013 | 0.00002 | 0.00013 | 0.02180 | 0.04088 | coding-syn |
| rs1397547 | G (0.059) | ns | ns | 0.01799 | 0.03372 | 0.02892 | ns | 0.04169 | ns | 0.04748 | ns | 0.000004 | 0.00006 | 0.000004 | 0.00006 | 0.00035 | 0.00261 | coding-syn | |
FDR: Benjamini-Hochberg false discovery rate. ns: not significant. MAF: Minor allele frequency. syn: synonymous
Given the identical P values for some single-variant associations and the physical proximity of variants, we calculated linkage disequilibrium (LD) between SNPs. We used phased genotype data from the 1000 Genomes Project for the CEU population (Utah residents with Northern and Western European ancestry). For our dataset, we performed an LD pairwise analysis using the Golden Helix® SVS software. Variants rs2305339 and rs734644 in ECR26 were in complete LD (r2 = 0.99, D’ = 1.00, haplotypes AC/GT [protective/risk]), as well as variants rs11131352 (ECR46), rs17226398, rs56038622 and rs2271338 (ECR47) (r2 = 1.00, D’ = 1.00, haplotypes AGAG/TCTA [protective/risk]) (Table S4 in Supplement 1). For initial evaluation in this study, risk alleles in complete LD were tested in luciferase assays as a haplotype rather than individually, and compared to the protective haplotype.
ECR46 and ECR47 appear as two independent conserved elements in chicken, but they are part of a single, “core” ECR (≥350 bp long, ≥77% identity) (31) in species evolutionarily closer to humans such as opossum, mouse and chimpanzee. For that reason, these two sequences were evaluated in luciferase assays independently and together.
ECR47 functions as a tissue-specific enhancer
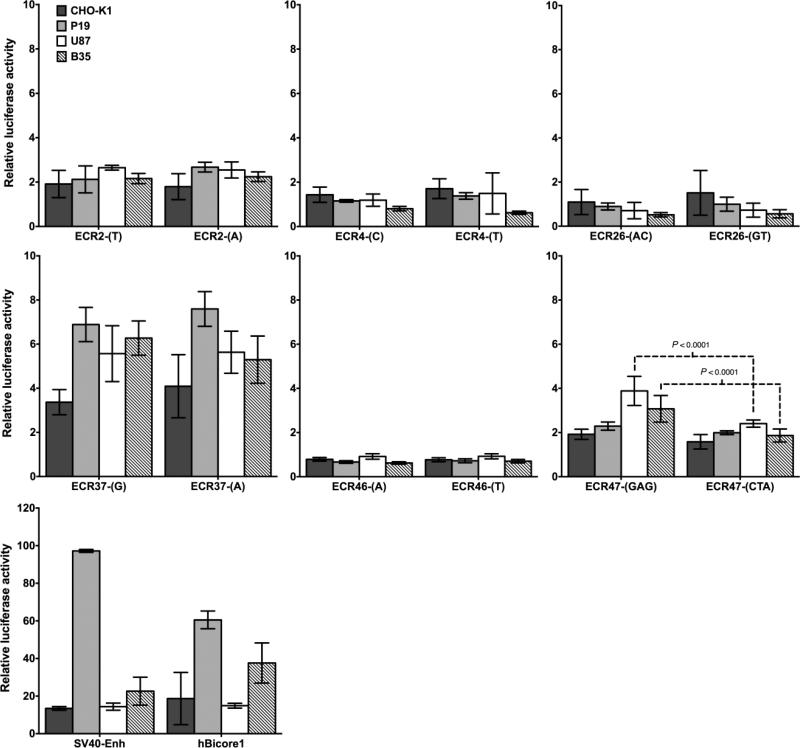
We tested the ADHD-associated ECRs for enhancer activity using a dual secreted luciferase reporter assay in four different cell lines (Figure 1). We compared luciferase activity of ECRs containing the protective versus the risk allele/haplotype. As described above, ECR26 and ECR47 contain markers in complete LD, thus, we compared haplotypes rather than pairwise combinations of alleles. ECR37 and ECR47 were the only elements to stimulate luciferase expression. ECR37 showed weak enhancer activity in all cell lines; however, its ADHD-associated variant rs1397548 did not affect enhancer function. ECR47 also showed weak enhancer activity, but only in B35 neuroblastoma and U87 astrocytoma cells, suggesting tissue-specific activity. Unlike ECR37, ECR47 risk haplotype CTA reduced luciferase activity by approximately 40% in both cells lines (PBonferroni < 0.0001). ECR2, ECR4, ECR26 and ECR46 showed no stimulation of luciferase activity in any of the cell lines. The activity of core element ECR46/47 was similar to that of ECR47 alone (Figure S2 in Supplement 1), indicating that regulatory activity resides on the ECR47 moiety.
Figure 1.
Secreted luciferase assays testing ADHD-associated ECR sequences for enhancer activity. Four different cell lines were transfected with the ECR-luciferase constructs for 24 h. Luciferase activity was normalized against a constitutive Gaussia luciferase plasmid and expressed as relative luciferase activity (Cypridina/Gaussia ratio). Results are presented as the stimulation of luciferase activity above the basal, enhancer-less vector containing only the minimal promoter. Letters in parenthesis indicate the alleles or haplotypes being tested. ECR37 and ECR47 showed weak enhancer activity compared to the strong control enhancers SV40 and human Bicore1. ECR47 risk haplotype CTA decreased enhancer activity in B35 neuroblastoma and U87 astrocytoma cells (PBonferroni < 0.0001). Statistical differences were defined using one-way ANOVA with correction for multiple comparisons.
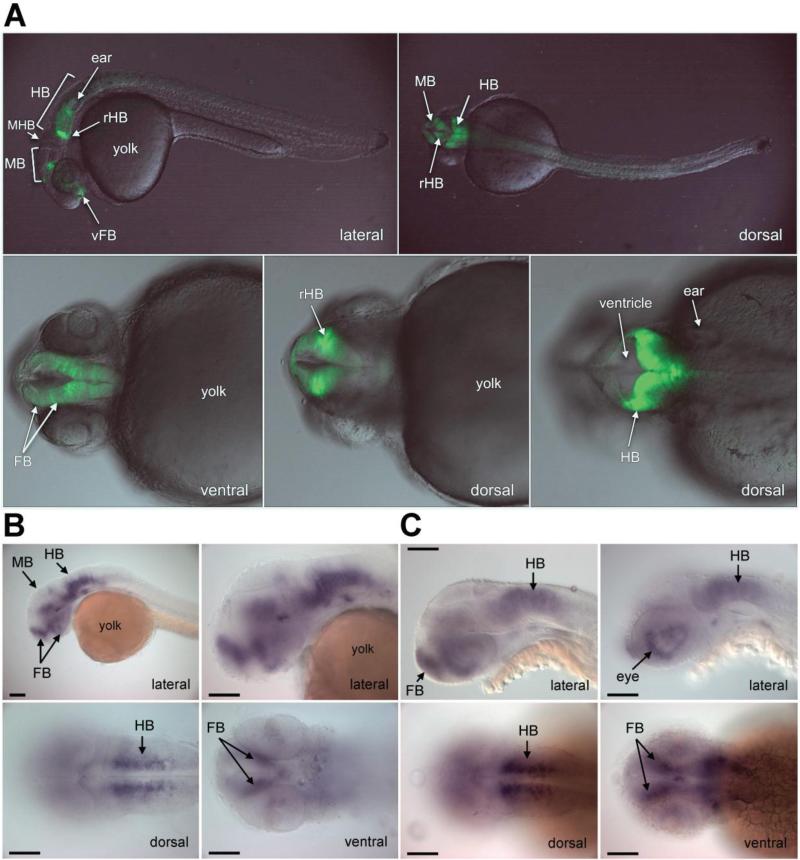
Subsequently, we evaluated the ability of the human ECR47 sequence to drive GFP reporter expression in stable transgenic zebrafish lines. A clear GFP signal was detected in the zebrafish brain that was restricted to the ventral forebrain, the midbrain and the hindbrain of embryos 28-30 hpf (Figure 2A). In contrast with the weak enhancer activity detected in luciferase assays, a strong GFP signal was observed in the zebrafish. This may be explained by the fact that multi-copy integration events can occur during transgenesis or that ECR47 may function as a developmental enhancer, thus behaving differently during embryogenesis compared to differentiated cells in culture. Interestingly, the ECR47-driven GFP expression pattern shared specific aspects of endogenous adgrl3.1 expression in forebrain, midbrain and hindbrain, but not in telencephalon and retina, as evaluated by in situ hybridization (Figure 2B,C). Because the ZED vector system is not robust enough to allow quantitative measurement of enhancer activity in vivo, we did not investigate the effect of ECR47 risk haplotype (CTA) on enhancer function in the zebrafish. A number of variables make it challenging to compare ECR47 risk and protective haplotypes quantitatively using the ZED system, for example: 1) different copy number integration during zebrafish transgenesis; 2) structural complexity of transgene integration loci in the genome (heterochromatin vs. euchromatin); 3) DNA methylation effects; 4) zebrafish tissue autofluorescence and non-specific, background GFP expression.
Figure 2.
In vivo enhancer testing and correlation with adgrl3.1 expression in the zebrafish. ECR47-driven GFP expression was monitored in the brain of transgenic embryos at 28-30 hours post fertilization (hpf). (A) Stable transgenic F2 embryo showing GFP expression restricted to the central nervous system (i.e. forebrain, midbrain and hindbrain). (B and C) Expression of adgrl3.1 was detected by in situ hybridization of embryos at (B) 36 hpf and (C) 48 hpf (top row left mid-sagittal and right retina in focus). ECR47-driven GFP expression represents several specific aspects of endogenous adgrl3.1 consistent with the location of neuronal tissue in the developing brain. Endogenous adgrl3.1 mRNA expression is detected in the telencephalon, the ventral forebrain, the midbrain, the hindbrain, and the retina throughout the developmental stages analyzed. Abbreviations: FB, forebrain; HB, hindbrain; MB, midbrain; MHB, midbrain-hindbrain boundary; r, rostral; v, ventral. Views as indicated. Orientation: anterior to left in all images, dorsal up in lateral views. Scale bar is 100 μm, in C all images are in the same scale.
It is important to highlight that ECR47 is also conserved in the zebrafish (Figure S3 in Supplement 1), which makes it an ultraconserved element likely to have an important biological function.
Transcription factors preferentially associated with brain function are overrepresented in ECR47
Analysis of the TF binding profiles of ECR47 and 144 in vivo tested brain enhancers from the VISTA Enhancer Browser (36) (Table S5 in Supplement 3) revealed a significant overrepresentation of developmental TF families preferentially associated with brain tissue, such as distal-less homeodomain (V$DLXF), NK6 homeobox (V$NKX6), PAX homeodomain (V$PAXH), and Brn-5 POU (V$BRN5) transcription factors, as suggested by the high Z-scores (Table S6 in Supplement 1). Homeobox transcription factors (V$HBOX), Brn POU domain (V$BRNF), Cart-1 (V$CART), and Lim homeodomain (V$LHXF) factors were also overrepresented, but while they may participate in neural development and function, they are not preferentially associated with the central nervous system (CNS).
The potential effects of ECR47 SNP allele substitutions on TF binding were examined using SNPInspector. All three ADHD risk alleles (CTA) were predicted to produce loss or gain of binding sites for important neurodevelopmental TFs, such as GRHL1, PAX2, PAX3, YY1, HRE, CREB, LHX3, BRN2, BRN3, POU6F2, PIT1, and CRX (Table 2).
Table 2.
Transcription factor binding sites predicted to be affected by the ADHD-associated haplotype in ECR47. Search was performed using SNPInspector from the Genomatix® Software Suite. The analysis is based on MatInspector and Genomatix® library of matrix descriptions for transcription factor (TF) binding sites (MatBase).
| SNP rsID |
Major allele |
Minor allele |
Allele change |
Predicted effect |
TF family/matrix |
TF name | Optimized threshold* |
Strand | Core similarity** |
Matrix similarity*** |
|---|---|---|---|---|---|---|---|---|---|---|
| rs17226398 | G | C | G > C | Gained | V$PAX3/PAX3.02 | Pax-3 paired domain protein | 0.85 | + | 1 | 0.893 |
| Gained | V$HIFF/HRE.02 | Hypoxia-response elements | 0.97 | − | 1 | 0.978 | ||||
| Lost | V$GRHL/GRHL1.01 | Grainyhead-like 1 | 0.86 | + | 1 | 0.864 | ||||
| Lost | V$CP2F/TCFCP2L1.01 | Transcription factor CP2-like 1 | 0.87 | + | 0.815 | 0.894 | ||||
| rs56038622 | A | T | A > T | Gained | V$CREB/TAXCREB.02 | Tax/CREB complex | 0.71 | + | 0.75 | 0.739 |
| rs2271338 | G | A | G > A | Gained | V$LHXF/LHX3.02 | LIM-homeodomain 3 | 0.82 | + | 1 | 0.84 |
| Gained | V$BRNF/BRN3.01 | Brn-3, POU-IV protein class | 0.78 | − | 0.75 | 0.815 | ||||
| Gained | V$BRN5/POU6F2.01 | Retina-derived POU-domain factor 1, dimeric | 0.76 | + | 0.81 | 0.779 | ||||
| Gained | V$BRNF/BRN2.04 | POU class 3 homeobox 2 (POU3F2) | 0.82 | + | 1 | 0.827 | ||||
| Gained | V$OCT1/OCT1.03 | Octamer-binding protein 1 (POU2F1) | 0.85 | + | 0.767 | 0.854 | ||||
| Gained | V$PIT1/PIT1.02 | Pituitary transcription factor 1 (POU1F1) | 0.81 | + | 1 | 0.821 | ||||
| Gained | V$HNF6/OC2.01 | One CUT-homeodomain protein | 0.82 | + | 1 | 0.876 | ||||
| Lost | V$PAX2/PAX2.01 | Zebrafish PAX2 paired domain protein | 0.78 | + | 0.789 | 0.784 | ||||
| Lost | V$YY1F/YY1.02 | Yin and Yang 1 repressor | 0.94 | − | 1 | 0.979 | ||||
| Lost | V$BCDF/CRX.01 | Cone-rod homeobox protein | 0.94 | + | 1 | 0.946 | ||||
At the matrix similarity optimized threshold a minimum number of matches is found in non-regulatory test sequences; i.e., with this matrix similarity the number of false positive matches is minimized.
Core similarity refers to the degree of matching to the “core sequence” of a matrix (highest conserved positions of the matrix, usually 4 bases). The maximum core similarity of 1.0 is only reached when the highest conserved bases of a matrix match exactly in the sequence.
Matrix similarity refers to the degree of matching to each sequence position in the matrix. A perfect match to the highest conserved nucleotide at each position gets a score of 1.00. A “good” match usually has a similarity of > 0.80. Mismatches in highly conserved positions of the matrix decrease the matrix similarity more than mismatches in less conserved regions.
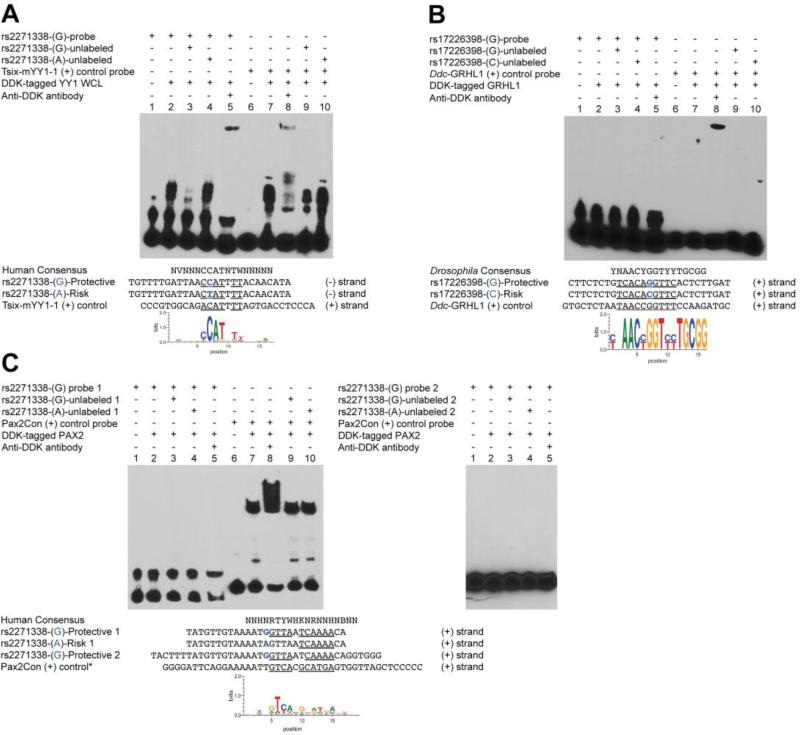
YY1 binding to ECR47 is disrupted by rs2271338 ADHD risk allele
The bindings sites of three important neurodevelopmental TFs were predicted to be disrupted by ECR47 risk allele substitutions. PAX2 and YY1 were predicted to be disrupted by rs2271338 G>A and GRHL1 was predicted to be disrupted by rs17226398 G>C (Table 2). While GRHL1 and PAX2 did not bind to their predicted sites (Figure 3B,C), YY1 produced a clear gel shift when added to a 27-bp fragment containing rs2271338 protective allele. YY1 binding was completely abrogated by addition of excess unlabeled protective sequence, but not by the sequence containing the risk allele (Figure 3A).
Figure 3.
YY1 binding to ECR47 enhancer is disrupted by rs2271338 risk allele.
(A) A biotin-labeled DNA fragment containing rs2271338 ADHD protective allele was incubated with a HEK293 whole cell lysate (WCL) expressing DDK (FLAG)-tagged human YY1 (lanes 2-5 from left to right). Mobility shift in lane 2 indicates protein binding to the probe, which was abrogated by incubation with molar excess of the unlabeled fragment (lane 3), but not of an unlabeled fragment containing the rs2271338 risk allele (lane 4). Higher molecular weight shift with the addition of anti-DDK antibody identifies YY1 as the binding factor (lane 5). Lanes 6-10 correspond to a known YY1 binding sequence used as positive control (87). Lanes 7-10 show protein binding to the control probe, with the anti-DDK antibody producing a similar supershift thus confirming YY1 identity (lane 8). Molar excess of the unlabeled ECR47 protective fragment was capable of reducing protein binding (lane 9), but the risk fragment was not (lane 10).
No binding was detected for GRHL1 (B) or PAX2 (C) transcription factors. (B) Lanes 1-5, biotin-labeled fragment containing the rs17226398 protective allele; lanes 6-10, GRHL1 positive control sequence (89). Interestingly, no shift was observed in the positive control except when the antibody was added (lane 8). Apparently, the addition of the antibody stabilizes GRHL1-DNA interaction, which otherwise is labile under the experimental conditions used. (C) Left panel, lanes 1-5, same DNA fragment used for YY1 in A; lanes 6-10, PAX2 positive control sequence (90). Nether the protective nor the risk ECR47 fragments affected PAX2 binding to the positive control probe (lanes 9 and 10). The right panel shows the results for a longer ECR47 probe (Protective 2) extending to the 3’ end of the sequence. PAX2 still did not bind to DNA.
Position weight matrices were taken from MotifMap for human hg19 (YY1, PAX2) and Drosophila dm3 (GRHL1) assemblies (91).
We next examined whether YY1 affected endogenous ADGRL3 expression. Real-time PCR analysis revealed a strong expression of endogenous ADGRL3 mRNA in SH-SY5Y neuroblastoma but not in U87 astrocytoma cells (Figure S4A in Supplement 1), suggesting that expression of this gene may be specific to the neuronal lineage in the CNS. After YY1 siRNA transfection we could not detect any significant changes in ADGRL3 mRNA expression in these cell lines (Figure S4B in Supplement 1). This result might suggest that the ECR47-YY1 pair plays a developmental role or that its spatiotemporal regulation of ADGRL3 expression is complex and cannot be modeled properly outside its native regulatory circuit in the brain.
rs2271338 is associated with decreased ADGRL3 mRNA expression in the thalamus
Unlike other behavioral disorders, such as schizophrenia and major depression, brain tissue from ADHD patients is not readily available. However, since SNPs associated with complex diseases are likely to function as expression quantitative trait loci (eQTLs), the tissues of unaffected individuals can be used for gene expression association analyses. eQTL analysis of brain tissue from 137 neuropathologically confirmed controls (age 16-102) revealed a significant association between the rs2271338 AA risk genotype and reduced ADGRL3 expression in the thalamus (P < 0.01) (Figure S5 and Table S7 in Supplement 1). Rs2271338 was either absent or not associated with ADGRL3 expression in GTEx, SNPExpress and GRASP databases (see Supplement 1).
DISCUSSION
Very few molecular studies have attempted to explain the molecular effects of ADHD associated genetic variants. Studies on dopamine transporter (DAT1) (37-40), tryptophan hydroxylase 2 (TPH2) (41, 42) and T-cadherin (CDH13) (43) have examined the functional properties of rare missense mutations of moderate and large effects, but these findings fail to explain the higher incidence of ADHD and larger phenotypic variance observed in populations. Instead, the common disease-common variant hypothesis is better supported by a substantial number of genetic epidemiological studies, with common variants accounting for ~40% of ADHD heritability (7, 8).
ADGRL3 is a strong ADHD candidate gene. ADGRL3 common variants predispose to ADHD, modulate brain metabolism, and predict ADHD severity and comorbidity with disruptive symptoms and substance use disorder (11, 13, 15, 16), When combined with other risk factors, ADGRL3 risk variants improve the prediction of ADHD severity, dysfunctional comorbidity, long-term outcome, and response to treatment with stimulant medication (10-16). Recent work from our group also showed genetic linkage and association of ADGRL3 variants to neuropsychological endophenotypes, providing a more powerful framework for ADHD clinical classification and for the identification of causative genetic variation (44).
ADGRL3 encodes a member of the latrophilin subfamily of adhesion G protein-coupled receptors that is highly expressed in brain regions implicated in the dopaminergic systems (11, 45). ADGRL3 endogenous ligand has been identified as FLRT3, a postsynaptic membrane protein involved in axon guidance and neuronal cell migration during embryonic development (46). More importantly, the ADGRL3-FLRT3 synaptic pair regulates excitatory transmission in vitro and in vivo (47). Animal models also support ADGRL3 implication in ADHD pathophysiology (48, 49).
Using a combination of evolutionary sequence conservation and regulatory annotation data we identified candidate sequences within the ADGRL3 MCR with potential regulatory function. Several variants revealed significant association with ADHD, comorbid disorders and/or neuropsychological endophenotypes, with a four-marker haplotype in the ECR46/ECR47 core element showing the highest level of association across the board. This result may suggest that ECR46/47 participates in a common neurobiological pathway to ADHD. Given the complexity of the ADHD phenotype, we must expect complex genetic interactions within the ADGRL3 locus and with other genomic regions, which may explain the different levels of association observed across ECRs (Table 1).
The genetic association of ADGRL3 with disruptive behaviors and substance use supports previous observations. Families with ADHD cluster ODD, CD and SUD (23); children diagnosed with ADHD monitored during the transition into adolescence exhibit higher rates of alcohol, tobacco, and psychoactive drug use than unaffected children (50, 51); and the lifetime risk for substance use is approximately 50% in subjects with childhood ADHD persisting into adulthood (52, 53).
Linkage and association of ADGRL3 with neuropsychological endophenotypes was recently reported by Mastronardi et al. (44). In agreement with their findings, we show a significant association of ECR variants with ACVT Correct Responses lower scores and ACVT Omissions higher scores. Impaired response inhibition and poor sustained attention, as measured by these two tests, are fundamental components of the executive dysfunction present in ADHD (54-57). In the same vein, the inattentive and hyperactive/impulsive motor phenotype associated with ADHD is expected to affect the ability to perform the ROCFT Copy test, hence the significant association with ECR risk variants. However, the complexity of the multiple cognitive domains assessed by ROCFT (including visuospatial constructional ability, visual memory and several components of executive function) can confound the interpretation of scores, which might explain the lower level of significance (58). We did not detect any association with the WISC intellectual measures (Block Design, PIQ or FSIQ), which might indicate that ADGRL3 variation does not contribute to the cognitive deficits evaluated by these particular tests.
Functional testing in vitro identified ECR47 as a transcriptional enhancer. ECR47 was active in cultured neurons and astrocytes in a tissue-specific manner and its function was disrupted by the ADHD risk haplotype defined by variants rs17226398/rs56038622/rs2271338 (C/T/A) in complete LD. The non-random association between these risk alleles may suggest evolutionary selective pressure to conserve an important biological function. Although neurons and astrocytes have distinct transcriptome profiles, they both share a common neuroepithelial origin and over the past two decades it has become clear that astrocytes participate in a wide variety of complex functions in the CNS, including crucial roles in synaptic transmission and information processing in neural circuits (59-61). Evidence also suggests that glial cells may play a role in the pathogenesis of neuropsychiatric disorders including ADHD (62, 63). However, further studies are required to determine whether ECR47 function in astrocytes is functionally relevant for the pathology of ADHD, as ADGRL3 expression in astrocytoma cells was very low compared to human neuroblastoma cells (Figure S4 in Supplement 1).
Strikingly, we also found that ECR47 functions as a brain enhancer in vivo. ECR47 was able to drive GFP expression in the zebrafish brain consistent with various aspects of endogenous adgrl3.1 expression. These results are in concert with a previous report by Lange et al. (49) showing wide adgrl3.1 expression in the zebrafish brain at 24, 48 and 72 hpf, which suggests that ECR47 activation may share brain-specific factors with the adgrl3.1 transcriptional machinery during zebrafish development.
Enhancer elements have signatures that define tissue specificity (64, 65). Analysis of ECR47 and a large set of in vivo tested brain enhancers revealed a significant overlapping of binding sites for homeobox TF families preferentially associated with neurodevelopmental processes. DLX (DLX-1, DLX-2 and DLX-5), BRN5/POU6F1, PAX (PAX-2, PAX-3, PAX-5, PAX-6 and PAX-7 and PAX-8) and NKX6 (NKX6.2) homeodomain factors are known to play a prominent role in the development and function of the CNS (66-70). This finding supports our results showing tissue-specific activity of ECR47.
The risk allele substitution of rs2271338 disrupts YY1 binding to ECR47. YY1 is a controversial and versatile transcriptional regulator (71); it can either activate or repress gene expression depending on the cofactors it recruits (72). The potential function of YY1 in the developing nervous system was first suggested by the phenotypic analysis of Yy1+/− mice. Null Yy1 mice show early embryonic lethality, but a subset of Yy1+/− mice (~20%) display growth retardation and neural tube defects. The brains of Yy1+/− mouse embryos show exencephaly, asymmetric structure and the presence of pseudo-ventricles (73). Morpholino-knockdown studies in Xenopus reveal similar neurulation defects (74). While a significant reduction of XYY1 protein levels results in early embryonic lethality, a partial depletion results in antero-posterior patterning defects and reduction of head structures (74, 75). At the molecular level, Xyy1 ablation in Xenopus embryos reveals downregulation of transcription factors involved in neural patterning (i.e. homeobox genes, engrailed2, otx2, and krox20) and neural crest cell specification and migration (i.e. slug, snail) (76). Studies using other systems have also shown dysregulation of neurotransmitter signaling and metabolism genes, such as dynamin-1 and dopamine β-hydroxylase in neurons, and Glast glutamate/aspartate transporter in astrocytes (76).
Analysis of the effect of rs2271338 risk allele on ADGRL3 expression in cultured human cells and post-mortem brain tissue showed apparently contradictory results. While YY1 downregulation did not affect ADGRL3 expression in neuroblastoma cells, the rs2271338 risk allele was associated with reduced expression in the adult thalamus. The thalamus was one of the earliest brain areas considered in the pathophysiology of ADHD (77) due to its key role in filtering information and stimulus processing (78). Morphological abnormalities of the thalamus (79) and thalamic volume reduction (80) have been demonstrated previously in children with ADHD. Various methods, including functional connectivity analysis, have uncovered thalamic abnormalities in ADHD (81, 82). Morphological abnormalities have also been found across a number of cortical regions in children and adolescents with ADHD (83) supporting the view that cortico-striato-thalamo-cortical loops (84) play a key role in the pathogenesis of ADHD.
Although additional studies are required to establish the precise role of ECR47, the results presented here suggest an important neurological function. While brain expression data indicate an ADGRL3 expression maximum across fetal and infant stages (Figure S6 in Supplement 1) relatively high expression levels are maintained throughout life, suggesting that this gene is necessary for proper brain function from conception to demise. The lack of effect of YY1 knockdown on endogenous ADGRL3 expression in differentiated cells suggests that ECR47 may only be active during developmental stages; however, the association between rs2271338 and reduced ADGRL3 expression levels in the adult thalamus hampers our understanding of ADGRL3 spatiotemporal regulation by ECR47-YY1. Elucidation of this genetic interaction will help to decipher the molecular mechanisms underlying ADHD pathogenesis.
The experimental methodology used in this research could be a paradigm for the evaluation of non-coding risk variants associated with complex traits.
LIMITATIONS
In this study we tested risk variants only for their effects on transcriptional enhancer activity. While other transcriptional regulatory functions (e.g. those mediated by silencer and insulator elements) were not investigated, functional testing of these types of sequence have proven difficult to design and there are currently no robust in vivo assays in widespread use (85, 86). We must also consider the possible effect of risk variants on ADGRL3 mRNA splicing—especially synonymous changes and non-coding variants within 50 bp of exon-intron junctions—and on the expression and function of overlapping non-coding RNAs.
Supplementary Material
ACKNOWLEDGEMENTS
This research was conducted using intramural resources from the National Human Genome Research Institute (NHGRI) of the U.S. National Institutes of Health (NIH). We thank Dr. Paul Kruszka for the detailed revision of the manuscript and their helpful comments. This study used the computational capabilities of a demo license to Genomatix Software Suite (Genomatix Software GmbH, Munich, Germany).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting galley proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
FINANCIAL DISCLOSURES
The authors declare no biomedical financial interests or potential conflicts of interest.
REFERENCES
- 1.Faraone SV, Asherson P, Banaschewski T, Biederman J, Buitelaar JK, Ramos-Quiroga JA, et al. Attention-deficit/hyperactivity disorder. Nat Rev Dis Primers. 2015;1:15020. doi: 10.1038/nrdp.2015.20. [DOI] [PubMed] [Google Scholar]
- 2.Visser SN, Danielson ML, Bitsko RH, Holbrook JR, Kogan MD, Ghandour RM, et al. Trends in the parent-report of health care provider-diagnosed and medicated attention-deficit/hyperactivity disorder: United states, 2003-2011. J Am Acad Child Adolesc Psychiatry. 2014;53:34–46. e32. doi: 10.1016/j.jaac.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Visser SN, Bitsko RH, Danielson ML, Perou R. Increasing prevalence of parent-reported attention-deficit/hyperactivity disorder among children --- united states, 2003 and 2007. MMWR Morb Mortal Wkly Rep. 2010;59:1439–1443. [PubMed] [Google Scholar]
- 4.American Psychiatric Association . Diagnostic and statistical manual of mental disorders. American Psychiatric Association; Washington, D.C.: 1994. [Google Scholar]
- 5.Biederman J, Faraone SV. Attention deficit hyperactivity disorder: A worldwide concern. J Nerv Ment Dis. 2004;192:453–454. doi: 10.1097/01.nmd.0000131803.68229.96. [DOI] [PubMed] [Google Scholar]
- 6.Jain M, Palacio LG, Castellanos FX, Palacio JD, Pineda D, Restrepo MI, et al. Attention-deficit/hyperactivity disorder and comorbid disruptive behavior disorders: Evidence of pleiotropy and new susceptibility loci. Biol Psychiatry. 2007;61:1329–1339. doi: 10.1016/j.biopsych.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 7.Faraone SV, Perlis RH, Doyle AE, Smoller JW, Goralnick JJ, Holmgren MA, et al. Molecular genetics of attention-deficit/hyperactivity disorder. Biol Psychiatry. 2005;57:1313–1323. doi: 10.1016/j.biopsych.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 8.Akutagava-Martins GC, Salatino-Oliveira A, Kieling CC, Rohde LA, Hutz MH. Genetics of attention-deficit/hyperactivity disorder: Current findings and future directions. Expert Rev Neurother. 2013;13:435–445. doi: 10.1586/ern.13.30. [DOI] [PubMed] [Google Scholar]
- 9.Arcos-Burgos M, Velez JI, Solomon BD, Muenke M. A common genetic network underlies substance use disorders and disruptive or externalizing disorders. Hum Genet. 2012;131:917–929. doi: 10.1007/s00439-012-1164-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ribases M, Ramos-Quiroga JA, Sanchez-Mora C, Bosch R, Richarte V, Palomar G, et al. Contribution of lphn3 to the genetic susceptibility to adhd in adulthood: A replication study. Genes Brain Behav. 2011;10:149–157. doi: 10.1111/j.1601-183X.2010.00649.x. [DOI] [PubMed] [Google Scholar]
- 11.Arcos-Burgos M, Jain M, Acosta MT, Shively S, Stanescu H, Wallis D, et al. A common variant of the latrophilin 3 gene, lphn3, confers susceptibility to adhd and predicts effectiveness of stimulant medication. Mol Psychiatry. 2010;15:1053–1066. doi: 10.1038/mp.2010.6. [DOI] [PubMed] [Google Scholar]
- 12.Fallgatter AJ, Ehlis AC, Dresler T, Reif A, Jacob CP, Arcos-Burgos M, et al. Influence of a latrophilin 3 (lphn3) risk haplotype on event-related potential measures of cognitive response control in attention-deficit hyperactivity disorder (adhd). Eur Neuropsychopharmacol. 2013;23:458–468. doi: 10.1016/j.euroneuro.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Acosta MT, Velez JI, Bustamante ML, Balog JZ, Arcos-Burgos M, Muenke M. A two-locus genetic interaction between lphn3 and 11q predicts adhd severity and long-term outcome. Transl Psychiatry. 2011;1:e17. doi: 10.1038/tp.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jain M, Velez JI, Acosta MT, Palacio LG, Balog J, Roessler E, et al. A cooperative interaction between lphn3 and 11q doubles the risk for adhd. Mol Psychiatry. 2012;17:741–747. doi: 10.1038/mp.2011.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choudhry Z, Sengupta SM, Grizenko N, Fortier ME, Thakur GA, Bellingham J, et al. Lphn3 and attention-deficit/hyperactivity disorder: Interaction with maternal stress during pregnancy. J Child Psychol Psychiatry. 2012;53:892–902. doi: 10.1111/j.1469-7610.2012.02551.x. [DOI] [PubMed] [Google Scholar]
- 16.Labbe A, Liu A, Atherton J, Gizenko N, Fortier ME, Sengupta SM, et al. Refining psychiatric phenotypes for response to treatment: Contribution of lphn3 in adhd. Am J Med Genet B Neuropsychiatr Genet. 2012;159B:776–785. doi: 10.1002/ajmg.b.32083. [DOI] [PubMed] [Google Scholar]
- 17.Hwang IW, Lim MH, Kwon HJ, Jin HJ. Association of lphn3 rs6551665 a/g polymorphism with attention deficit and hyperactivity disorder in korean children. Gene. 2015;566:68–73. doi: 10.1016/j.gene.2015.04.033. [DOI] [PubMed] [Google Scholar]
- 18.Bruxel EM, Salatino-Oliveira A, Akutagava-Martins GC, Tovo-Rodrigues L, Genro JP, Zeni CP, et al. Lphn3 and attention-deficit/hyperactivity disorder: A susceptibility and pharmacogenetic study. Genes Brain Behav. 2015;14:419–427. doi: 10.1111/gbb.12224. [DOI] [PubMed] [Google Scholar]
- 19.Song J, Kim SW, Hong HJ, Lee MG, Lee BW, Choi TK, et al. Association of snap-25, slc6a2, and lphn3 with oros methylphenidate treatment response in attention-deficit/hyperactivity disorder. Clin Neuropharmacol. 2014;37:136–141. doi: 10.1097/WNF.0000000000000045. [DOI] [PubMed] [Google Scholar]
- 20.Domene S, Stanescu H, Wallis D, Tinloy B, Pineda DE, Kleta R, et al. Screening of human lphn3 for variants with a potential impact on adhd susceptibility. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:11–18. doi: 10.1002/ajmg.b.31141. [DOI] [PubMed] [Google Scholar]
- 21.Bravo ML, Valenzuela CY, Arcos-Burgos OM. Polymorphisms and phyletic relationships of the paisa community from antioquia (colombia). Gene Geogr. 1996;10:11–17. [PubMed] [Google Scholar]
- 22.Arcos-Burgos M, Castellanos FX, Pineda D, Lopera F, Palacio JD, Palacio LG, et al. Attention-deficit/hyperactivity disorder in a population isolate: Linkage to loci at 4q13.2, 5q33.3, 11q22, and 17p11. Am J Hum Genet. 2004;75:998–1014. doi: 10.1086/426154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palacio JD, Castellanos FX, Pineda DA, Lopera F, Arcos-Burgos M, Quiroz YT, et al. Attention-deficit/hyperactivity disorder and comorbidities in 18 paisa colombian multigenerational families. J Am Acad Child Adolesc Psychiatry. 2004;43:1506–1515. doi: 10.1097/01.chi.0000142279.79805.dc. [DOI] [PubMed] [Google Scholar]
- 24.Pineda DA, Lopera F, Puerta IC, Trujillo-Orrego N, Aguirre-Acevedo DC, Hincapie-Henao L, et al. Potential cognitive endophenotypes in multigenerational families: Segregating adhd from a genetic isolate. Atten Defic Hyperact Disord. 2011;3:291–299. doi: 10.1007/s12402-011-0061-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ovcharenko I, Nobrega MA, Loots GG, Stubbs L. Ecr browser: A tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 2004;32:W280–286. doi: 10.1093/nar/gkh355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol. 2010;28:817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, Zhuang J, Iyer S, Lin XY, Greven MC, Kim BH, et al. Factorbook.Org: A wiki-based database for transcription factor-binding data generated by the encode consortium. Nucleic Acids Res. 2013;41:D171–176. doi: 10.1093/nar/gks1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- 31.Ovcharenko I, Stubbs L, Loots GG. Interpreting mammalian evolution using fugu genome comparisons. Genomics. 2004;84:890–895. doi: 10.1016/j.ygeno.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 32.Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: From properties to genome-wide predictions. Nat Rev Genet. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- 34.Birnbaum RY, Clowney EJ, Agamy O, Kim MJ, Zhao J, Yamanaka T, et al. Coding exons function as tissue-specific enhancers of nearby genes. Genome Res. 2012;22:1059–1068. doi: 10.1101/gr.133546.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Birnbaum RY, Patwardhan RP, Kim MJ, Findlay GM, Martin B, Zhao J, et al. Systematic dissection of coding exons at single nucleotide resolution supports an additional role in cell-specific transcriptional regulation. PLoS Genet. 2014;10:e1004592. doi: 10.1371/journal.pgen.1004592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Visel A, Minovitsky S, Dubchak I, Pennacchio LA. Vista enhancer browser--a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mazei-Robison MS, Bowton E, Holy M, Schmudermaier M, Freissmuth M, Sitte HH, et al. Anomalous dopamine release associated with a human dopamine transporter coding variant. J Neurosci. 2008;28:7040–7046. doi: 10.1523/JNEUROSCI.0473-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakrikar D, Mazei-Robison MS, Mergy MA, Richtand NW, Han Q, Hamilton PJ, et al. Attention deficit/hyperactivity disorder-derived coding variation in the dopamine transporter disrupts microdomain targeting and trafficking regulation. J Neurosci. 2012;32:5385–5397. doi: 10.1523/JNEUROSCI.6033-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kovtun O, Sakrikar D, Tomlinson ID, Chang JC, Arzeta-Ferrer X, Blakely RD, et al. Single-quantum-dot tracking reveals altered membrane dynamics of an attention-deficit/hyperactivity-disorder-derived dopamine transporter coding variant. ACS Chem Neurosci. 2015;6:526–534. doi: 10.1021/cn500202c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mergy MA, Gowrishankar R, Gresch PJ, Gantz SC, Williams J, Davis GL, et al. The rare dat coding variant val559 perturbs da neuron function, changes behavior, and alters in vivo responses to psychostimulants. Proc Natl Acad Sci U S A. 2014;111:E4779–4788. doi: 10.1073/pnas.1417294111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKinney J, Johansson S, Halmoy A, Dramsdahl M, Winge I, Knappskog PM, et al. A loss-of-function mutation in tryptophan hydroxylase 2 segregating with attention-deficit/hyperactivity disorder. Mol Psychiatry. 2008;13:365–367. doi: 10.1038/sj.mp.4002152. [DOI] [PubMed] [Google Scholar]
- 42.McKinney JA, Turel B, Winge I, Knappskog PM, Haavik J. Functional properties of missense variants of human tryptophan hydroxylase 2. Hum Mutat. 2009;30:787–794. doi: 10.1002/humu.20956. [DOI] [PubMed] [Google Scholar]
- 43.Mavroconstanti T, Johansson S, Winge I, Knappskog PM, Haavik J. Functional properties of rare missense variants of human cdh13 found in adult attention deficit/hyperactivity disorder (adhd) patients. PLoS ONE. 2013;8:e71445. doi: 10.1371/journal.pone.0071445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mastronardi CA, Pillai E, Pineda DA, Martinez AF, Lopera F, Velez JI, et al. Linkage and association analysis of adhd endophenotypes in extended and multigenerational pedigrees from a genetic isolate. Mol Psychiatry. 2015 doi: 10.1038/mp.2015.172. doi: 10.1038/mp.2015.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krain AL, Castellanos FX. Brain development and adhd. Clin Psychol Rev. 2006 doi: 10.1016/j.cpr.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 46.Yamagishi S, Hampel F, Hata K, Del Toro D, Schwark M, Kvachnina E, et al. Flrt2 and flrt3 act as repulsive guidance cues for unc5-positive neurons. EMBO J. 2011;30:2920–2933. doi: 10.1038/emboj.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O'Sullivan ML, de Wit J, Savas JN, Comoletti D, Otto-Hitt S, Yates JR, 3rd, et al. Flrt proteins are endogenous latrophilin ligands and regulate excitatory synapse development. Neuron. 2012;73:903–910. doi: 10.1016/j.neuron.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wallis D, Hill DS, Mendez IA, Abbott LC, Finnell RH, Wellman PJ, et al. Initial characterization of mice null for lphn3, a gene implicated in adhd and addiction. Brain Res. 2012;1463:85–92. doi: 10.1016/j.brainres.2012.04.053. [DOI] [PubMed] [Google Scholar]
- 49.Lange M, Norton W, Coolen M, Chaminade M, Merker S, Proft F, et al. The adhd-susceptibility gene lphn3.1 modulates dopaminergic neuron formation and locomotor activity during zebrafish development. Mol Psychiatry. 2012;17:946–954. doi: 10.1038/mp.2012.29. [DOI] [PubMed] [Google Scholar]
- 50.Molina BS, Pelham WE., Jr. Childhood predictors of adolescent substance use in a longitudinal study of children with adhd. J Abnorm Psychol. 2003;112:497–507. doi: 10.1037/0021-843x.112.3.497. [DOI] [PubMed] [Google Scholar]
- 51.Molina BS, Hinshaw SP, Eugene Arnold L, Swanson JM, Pelham WE, Hechtman L, et al. Adolescent substance use in the multimodal treatment study of attention-deficit/hyperactivity disorder (ADHD) (MTA) as a function of childhood adhd, random assignment to childhood treatments, and subsequent medication. J Am Acad Child Adolesc Psychiatry. 2013;52:250–263. doi: 10.1016/j.jaac.2012.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Biederman J, Wilens T, Mick E, Milberger S, Spencer TJ, Faraone SV. Psychoactive substance use disorders in adults with attention deficit hyperactivity disorder (adhd): Effects of adhd and psychiatric comorbidity. Am J Psychiatry. 1995;152:1652–1658. doi: 10.1176/ajp.152.11.1652. [DOI] [PubMed] [Google Scholar]
- 53.Nogueira M, Bosch R, Valero S, Gomez-Barros N, Palomar G, Richarte V, et al. Early-age clinical and developmental features associated to substance use disorders in attention-deficit/hyperactivity disorder in adults. Compr Psychiatry. 2014;55:639–649. doi: 10.1016/j.comppsych.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 54.Barkley RA. Behavioral inhibition, sustained attention, and executive functions: Constructing a unifying theory of adhd. Psychol Bull. 1997;121:65–94. doi: 10.1037/0033-2909.121.1.65. [DOI] [PubMed] [Google Scholar]
- 55.Castellanos FX, Tannock R. Neuroscience of attention-deficit/hyperactivity disorder: The search for endophenotypes. Nat Rev Neurosci. 2002;3:617–628. doi: 10.1038/nrn896. [DOI] [PubMed] [Google Scholar]
- 56.Liu L, Cheng J, Su Y, Ji N, Gao Q, Li H, et al. Deficiency of sustained attention in adhd and its potential genetic contributor maoa. J Atten Disord. 2015 doi: 10.1177/1087054715574832. pii: 1087054715574832. [DOI] [PubMed] [Google Scholar]
- 57.Pineda DA, Puerta IC, Aguirre DC, Garcia-Barrera MA, Kamphaus RW. The role of neuropsychologic tests in the diagnosis of attention deficit hyperactivity disorder. Pediatr Neurol. 2007;36:373–381. doi: 10.1016/j.pediatrneurol.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 58.Shin MS, Park SY, Park SR, Seol SH, Kwon JS. Clinical and empirical applications of the rey-osterrieth complex figure test. Nat Protoc. 2006;1:892–899. doi: 10.1038/nprot.2006.115. [DOI] [PubMed] [Google Scholar]
- 59.Bayer SA, Altman J. Neocortical development. Illustrated ed. Raven Press; New York: 1991. [Google Scholar]
- 60.Shen Q, Wang Y, Dimos JT, Fasano CA, Phoenix TN, Lemischka IR, et al. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nat Neurosci. 2006;9:743–751. doi: 10.1038/nn1694. [DOI] [PubMed] [Google Scholar]
- 61.Barres BA. The mystery and magic of glia: A perspective on their roles in health and disease. Neuron. 2008;60:430–440. doi: 10.1016/j.neuron.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 62.Yamamuro K, Kimoto S, Rosen KM, Kishimoto T, Makinodan M. Potential primary roles of glial cells in the mechanisms of psychiatric disorders. Front Cell Neurosci. 2015;9:154. doi: 10.3389/fncel.2015.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sandau US, Alderman Z, Corfas G, Ojeda SR, Raber J. Astrocyte-specific disruption of syncam1 signaling results in adhd-like behavioral manifestations. PLoS ONE. 2012;7:e36424. doi: 10.1371/journal.pone.0036424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Erokhin M, Vassetzky Y, Georgiev P, Chetverina D. Eukaryotic enhancers: Common features, regulation, and participation in diseases. Cell Mol Life Sci. 2015;72:2361–2375. doi: 10.1007/s00018-015-1871-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, et al. Chip-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Andersen B, Schonemann MD, Pearse RV, 2nd, Jenne K, Sugarman J, Rosenfeld MG. Brn-5 is a divergent pou domain factor highly expressed in layer iv of the neocortex. J Biol Chem. 1993;268:23390–23398. [PubMed] [Google Scholar]
- 67.Cui H, Bulleit RF. Expression of the pou transcription factor brn-5 is an early event in the terminal differentiation of cns neurons. J Neurosci Res. 1998;52:625–632. doi: 10.1002/(SICI)1097-4547(19980615)52:6<625::AID-JNR1>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 68.Eisenstat DD, Liu JK, Mione M, Zhong W, Yu G, Anderson SA, et al. Dlx-1, dlx-2, and dlx-5 expression define distinct stages of basal forebrain differentiation. J Comp Neurol. 1999;414:217–237. doi: 10.1002/(sici)1096-9861(19991115)414:2<217::aid-cne6>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 69.Thompson JA, Ziman M. Pax genes during neural development and their potential role in neuroregeneration. Prog Neurobiol. 2011;95:334–351. doi: 10.1016/j.pneurobio.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 70.Burbach JP, Smidt MP. Molecular programming of stem cells into mesodiencephalic dopaminergic neurons. Trends Neurosci. 2006;29:601–603. doi: 10.1016/j.tins.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 71.Deng Z, Cao P, Wan MM, Sui G. Yin yang 1: A multifaceted protein beyond a transcription factor. Transcription. 2010;1:81–84. doi: 10.4161/trns.1.2.12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yant SR, Zhu W, Millinoff D, Slightom JL, Goodman M, Gumucio DL. High affinity yy1 binding motifs: Identification of two core types (acat and ccat) and distribution of potential binding sites within the human beta globin cluster. Nucleic Acids Res. 1995;23:4353–4362. doi: 10.1093/nar/23.21.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Donohoe ME, Zhang X, McGinnis L, Biggers J, Li E, Shi Y. Targeted disruption of mouse yin yang 1 transcription factor results in peri-implantation lethality. Mol Cell Biol. 1999;19:7237–7244. doi: 10.1128/mcb.19.10.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morgan MJ, Woltering JM, In der Rieden PM, Durston AJ, Thiery JP. Yy1 regulates the neural crest-associated slug gene in xenopus laevis. J Biol Chem. 2004;279:46826–46834. doi: 10.1074/jbc.M406140200. [DOI] [PubMed] [Google Scholar]
- 75.Kwon HJ, Chung HM. Yin yang 1, a vertebrate polycomb group gene, regulates antero-posterior neural patterning. Biochem Biophys Res Commun. 2003;306:1008–1013. doi: 10.1016/s0006-291x(03)01071-4. [DOI] [PubMed] [Google Scholar]
- 76.He Y, Casaccia-Bonnefil P. The yin and yang of yy1 in the nervous system. J Neurochem. 2008;106:1493–1502. doi: 10.1111/j.1471-4159.2008.05486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Denhoff E, Laufer MW, Solomons G. Hyperkinetic impulse disorder in children's behavior problems. Psychosom Med. 1957;19:38–49. doi: 10.1097/00006842-195701000-00005. [DOI] [PubMed] [Google Scholar]
- 78.Gaudreau JD, Gagnon P. Psychotogenic drugs and delirium pathogenesis: The central role of the thalamus. Med Hypotheses. 2005;64:471–475. doi: 10.1016/j.mehy.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 79.Ivanov I, Bansal R, Hao X, Zhu H, Kellendonk C, Miller L, et al. Morphological abnormalities of the thalamus in youths with attention deficit hyperactivity disorder. Am J Psychiatry. 2010;167:397–408. doi: 10.1176/appi.ajp.2009.09030398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xia S, Li X, Kimball AE, Kelly MS, Lesser I, Branch C. Thalamic shape and connectivity abnormalities in children with attention-deficit/hyperactivity disorder. Psychiatry Res. 2012;204:161–167. doi: 10.1016/j.pscychresns.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cao X, Cao Q, Long X, Sun L, Sui M, Zhu C, et al. Abnormal resting-state functional connectivity patterns of the putamen in medication-naive children with attention deficit hyperactivity disorder. Brain Res. 2009;1303:195–206. doi: 10.1016/j.brainres.2009.08.029. [DOI] [PubMed] [Google Scholar]
- 82.Qiu MG, Ye Z, Li QY, Liu GJ, Xie B, Wang J. Changes of brain structure and function in adhd children. Brain Topogr. 2011;24:243–252. doi: 10.1007/s10548-010-0168-4. [DOI] [PubMed] [Google Scholar]
- 83.Seidman LJ, Valera EM, Makris N. Structural brain imaging of attention-deficit/hyperactivity disorder. Biol Psychiatry. 2005;57:1263–1272. doi: 10.1016/j.biopsych.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 84.Castellanos FX, Sonuga-Barke EJ, Milham MP, Tannock R. Characterizing cognition in adhd: Beyond executive dysfunction. Trends Cogn Sci. 2006;10:117–123. doi: 10.1016/j.tics.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 85.Petrykowska HM, Vockley CM, Elnitski L. Detection and characterization of silencers and enhancer-blockers in the greater cftr locus. Genome Res. 2008;18:1238–1246. doi: 10.1101/gr.073817.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bessa J, Tena JJ, de la Calle-Mustienes E, Fernandez-Minan A, Naranjo S, Fernandez A, et al. Zebrafish enhancer detection (zed) vector: A new tool to facilitate transgenesis and the functional analysis of cis-regulatory regions in zebrafish. Dev Dyn. 2009;238:2409–2417. doi: 10.1002/dvdy.22051. [DOI] [PubMed] [Google Scholar]
- 87.Kim JD, Hinz AK, Bergmann A, Huang JM, Ovcharenko I, Stubbs L, et al. Identification of clustered yy1 binding sites in imprinting control regions. Genome Res. 2006;16:901–911. doi: 10.1101/gr.5091406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wilanowski T, Caddy J, Ting SB, Hislop NR, Cerruti L, Auden A, et al. Perturbed desmosomal cadherin expression in grainy head-like 1-null mice. EMBO J. 2008;27:886–897. doi: 10.1038/emboj.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Uv AE, Thompson CR, Bray SJ. The drosophila tissue-specific factor grainyhead contains novel DNA-binding and dimerization domains which are conserved in the human protein cp2. Mol Cell Biol. 1994;14:4020–4031. doi: 10.1128/mcb.14.6.4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cross SH, McKie L, West K, Coghill EL, Favor J, Bhattacharya S, et al. The opdc missense mutation of pax2 has a milder than loss-of-function phenotype. Hum Mol Genet. 2011;20:223–234. doi: 10.1093/hmg/ddq457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Daily K, Patel VR, Rigor P, Xie X, Baldi P. Motifmap: Integrative genome-wide maps of regulatory motif sites for model species. BMC Bioinformatics. 2011;12:495. doi: 10.1186/1471-2105-12-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.