Abstract
The hygrolides, a family of 16-member-ring-containing plecomacrolides produced by Actinobacteria, exhibit numerous reported bioactivities. Using HR-MS/MS, nucleophilic 1,4-addition-based labeling, NMR, and bioinformatic analysis, we identified Streptomyces varsoviensis as a novel producer of JBIR-100, a fumarate-containing hygrolide, and elucidated the previously unknown stereochemistry of the natural product. We investigated the antimicrobial activity of JBIR-100, with preliminary insight into mode of action indicating that it perturbs the membrane of Bacillus subtilis. S. varsoviensis is known to produce compounds from multiple hygrolide sub-families, namely hygrobafilomycins (JBIR-100 and hygrobafilomycin) and bafilomycins (bafilomycin C1 and D). In light of this, we identified the biosynthetic gene cluster for JBIR-100, which, to our knowledge, represents the first reported for a hygrobafilomycin. Finally, we performed a bioinformatic analysis of the hygrolide family, describing clusters from known and predicted producers. Our results indicate that potential remains for the Actinobacteria to yield novel hygrolide congeners, perhaps with differing biological activities.
Keywords: Streptomyces varsoviensis; polyketide; plecomacrolide; hygrolide; hygrobafilomycin; JBIR-100; TS155-2; nucleophilic 1,4-addition; antimicrobial; antifungal; bioinformatics; whole-genome sequencing; resistance mapping
Graphical abstract
1. Introduction
The plecomacrolide natural products feature 16- or 18-membered macrolactone rings (hygrolides and concanamycins, respectively) containing two conjugated diene units and a tetrahydropyran ring [1]. The hygrolide subfamily (Fig. 1a) contains nearly 50 known members, including the bafilomycins [2–8], hygrolidin [9, 10], and hygrobafilomycin [11], among others, all of which are produced by Actinobacteria from the Streptomyces, Kitasatospora, Micromonospora, and Nocardia genera. Macrolide natural products such as the hygrolides arise from the linkage of acyl thioester monomers through decarboxylative Claisen condensations mediated by type I polyketide synthases (PKSs). Prototypical PKSs are large, multimodular enzymes consisting of several catalytic domains with specific functions that act in an assembly line-like fashion to select and successively reduce polyketide monomer units [12]. The polyketide monomer sequence typically follows the domain sequence of the associated PKS, which enables homology-based in silico prediction of the polyketide structure as well as functional prediction based on biosynthetic precedence [13, 14]. Conversely, the structure of an isolated compound, coupled with polyketide biosynthetic logic, can be leveraged to identify the corresponding BGC [14, 15].
Figure 1.
Structural variety of the hygrolide family. (A) Sites of known structural variability in hygrolide antibiotics are indicated. Variations include stereochemistry, glycosylation, acylation, methylation, oxidation state, and ring-opening, among others. (B) Six main scaffolds of the hygrolide antibiotics. Names under the drawings indicate scaffold name rather than the exact name of the natural product(s). (C) Structure of JBIR-100 with the fumarate moiety containing an electrophilic alkene suitable for nucleophilic addition highlighted with a yellow circle.
A multitude of biological activities have been ascribed to hygrolide subfamily members, including antibacterial and antifungal [2], antiplasmodial [16], antiparasitic [17], insecticidal [3], herbicidal [18, 19], antiviral [20–23], anti-atherosclerotic [24–26], and neuroprotective effects [27, 28]. A number of hygrolides inhibit the transmembrane vacuolar-type H+-ATPases (V-ATPases), a family of ATP-driven proton pumps that regulate the pH of intracellular compartments, including lysosomes, phagosomes, and secretory vesicles [29]. The most well-studied hygrolide, bafilomycin A1, potently and selectively inhibits V-ATPases, but not P-ATPases or F-ATPases [30]. Thus, it has found use as a tool for both distinguishing V-ATPases and determining their cellular functions [31]. As well as being critical for various physiological processes, V-ATPases have been implicated in a number of diseases [32]. As a result, hygrolides have attracted considerable research attention as potential therapeutics, e.g. to block the excessive bone resorption associated with osteoporosis [33] and to slow tumor cell growth and metastasis [34–36]. Bafilomycin A1 has been extensively used in the study of autophagy due to its ability to impede fusion between autophagosomes and lysosomes by inhibition of the sarco/endoplasmic reticulum Ca2+-ATPase (SERCA) pump rather than V-ATPase [37]. Additionally, bafilomycin A1 has been shown to disrupt mitochondrial function by acting as carrier type K+ ionophore [38].
Although etiologically linked to several diseases, the essential role of V-ATPases in various physiological processes means that therapeutic application of the plecomacrolides, including the hygrolides, may be limited [39]. However, there is hope that toxic effects can be overcome by identifying or designing inhibitors to target specific V-ATPase subunit isoforms. To this end, chemical modification of the hygrolides has been undertaken to provide insights into structure-activity relationships (SAR) and perhaps even improve pharmacokinetic properties and efficacy [40, 41]. In addition to total- and semi-synthetic approaches, accessing natural chemical diversity helps to shape our understanding of promising pharmacophores. Indeed, screening of actinobacterial extracts has been successful in yielding hygrolide congeners with differing functional groups. After more than 30 years of investigation, the production of a hygrolide was definitively linked to a biosynthetic gene cluster (BGC) in 2012 [42–44]. Armed with this knowledge, genome mining can allow prioritization of strains for investigation. Furthermore, knowledge of these BGCs raises the possibility that rational bioengineering can be applied to produce new hygrolide analogues [45, 46].
In the present study, we used HR-MS/MS, reactivity-based metabolite labeling, NMR, and bioinformatics to identify Streptomyces varsoviensis as a novel producer of the hygrolide JBIR-100. Comparative analysis of 16S rRNA sequences indicated that S. varsoviensis is distinct from the previously known JBIR-100 producer, Streptomyces sp. IR027-SDHV6 [47]. Nucleophilic 1,4-addition-based labeling proved useful in connecting the JBIR-100 BGC to its corresponding metabolite, vastly simplifying the structure elucidation process. We confirmed the structure of JBIR-100, and for the first time, propose its complete stereochemistry using a combination of NMR and bioinformatic analysis. In terms of its core scaffold, JBIR-100 most closely resembles hygrobafilomycin, also produced by S. varsoviensis [11]. Unfortunately, hygrolide nomenclature is not unified in the literature, and similar names do not always denote a common core scaffold. For clarity, we refer to hygrolides with a hygrobafilomycin-type scaffold (defined by 2-C methyl group, a 23-C isopropyl group, and 16-membered macrolactone ring), including JBIR-100, as hygrobafilomycins (Fig. 1b). We used purified JBIR-100 to gain preliminary insights into its antimicrobial activity, particularly against Bacillus subtilis. Finally, we sought to reconcile the available bioinformatic data on the hygrolide family of natural products. Thus, we surveyed the hygrolide genomic landscape and identified and contextualized BGCs for numerous known and predicted hygrolide producers, with several of the newly described BGCs potentially encoding novel compounds.
2. Results and Discussion
2.1. Identification of a novel producer of JBIR-100 by nucleophilic 1,4-addition
In addition to powerful genetic approaches, a number of chemical strategies can aid in connecting a BGC to its product(s). For example, in the so-called genomisotopic approach, target molecules can be labeled by biosynthetic incorporation of 13C- or 15N-enriched precursors followed by NMR detection [48]. A similar methodology involves growing the microorganism of interest in fully labeled 13C or 15N medium supplemented with a non-labeled precursor, with precursor incorporation into specific molecules being detected by mass spectrometry [49]. Another method targets genetically encoded functional groups with a suitable probe, then uses comparative MS to indicate the masses in a microbial extract that may represent the molecule(s) of interest. For example, dehydrated amino acids, which are common to several classes of ribosomally synthesized and post-translationally modified peptide (RiPP) natural products, can be covalently labeled at activated alkenes (i.e., α,β-unsaturated carbonyl/imine moieties) with a thiol probe [50]. Coupled with bioinformatic analysis of putative BGCs, this strategy allowed targeted screening of strains and resulted in the identification of a novel RiPP of the thiopeptide class, the antibiotic cyclothiazomycin C [50].
As part of a screen based on thiol-mediated nucleophilic labeling [50], we treated a cell surface extract of S. varsoviensis NRRL B-3589 with dithiothreitol (DTT) in the presence of a mild base. MALDI-TOF MS analysis revealed the production of an exported compound that was covalently modified by DTT (Fig. 2a). We surveyed the S. varsoviensis genome using the web-based tool antiSMASH 3.0 [51] and the MiBIG database [52] for BGCs corresponding to potential electrophile-containing natural products. Indeed, we noticed that one of the BGCs showed high similarity to that of bafilomycin C1, a hygrolide bearing a fumarate moiety [53]. Using HR-MS/MS and NMR, we confirmed that the natural product labeled with DTT was identical to the fumarate-containing hygrolide JBIR-100 (also known as TS155-2) (Fig. 1c) [47, 54, 55]; the structure was corroborated by comparison of its spectroscopic data with those in the literature and by bioinformatic analysis (see Section 2.2) [47].
Figure 2.
Mass spectrometry characterization of JBIR-100 and DTT-labeled JBIR-100. (A) MALDI-TOF MS of JBIR-100 nucleophilic labeling performed in the context of an organic, cell surface extract of Streptomyces varsoviensis NRRL B-3589. The black spectrum (top) is an unreacted control, while the red spectrum (bottom) resulted from DTT-labeling. JBIR-100 was visibly labeled by one DTT molecule. (B) High resolution LTQ-FT-MS mass spectrum of the [M+Na]+ species of JBIR-100, which was used to calculate exact mass. (C) CID spectrum of the [M+Na]+ species of JBIR-100. Labeled peaks correspond to identified fragments of JBIR-100, shown on the structure in panel (D). (E) High resolution LTQ-FT-MS mass spectrum of the [M+Na]+ species of DTT-labeled JBIR-100, which was used to calculate exact mass. (F) CID spectrum of the [M+Na]+ species of DTT-labeled JBIR-100. Labeled peaks correspond to identified fragments of DTT-labeled JBIR-100, shown on the structure in panel (G).
A sample of the unlabeled isolated material was subjected to HR-ESI-MS. The observed mass (m/z 727.4055 Da, [M+Na]+) was consistent with a molecular formula of C39H60O11 (theoretical m/z 727.4033 Da, [M+Na]+, error 3.0 ppm) possessing 10 degrees of unsaturation (Fig. 2b – d). Nucleophilic labeling of JBIR-100 with DTT suggested at least one degree of unsaturation to be due to an electrophilic alkene (Fig. 2a); accordingly, collision-induced dissociation (CID)-based fragmentation yielded a prominent fragment consistent with the loss of a fumaric acid motif (observed m/z 611.3911 Da, [M+Na]+; theoretical m/z 611.3924 Da, [M+Na]+; error 2.1 ppm) relative to the parent ion (Fig. 2b – d). As predicted, LTQ-FT-MS/MS analysis of DTT-labeled JBIR-100 indicated that DTT was incorporated in the fragment containing the fumarate moiety (Fig. 2e – g). This result extends reactivity-based labeling [50] beyond RiPPs to members of the polyketide natural product class and demonstrates its utility to quickly narrow down a list of candidate BGCs. Additionally, the data suggest that the fumarate moiety, or other similar electrophiles, are viable chemical handles for future discovery efforts.
2.2 Structure determination of the hygrobafilomycin JBIR-100
Full structural characterization by NMR was performed using one- and two-dimensional experiments (1H, 1H-1H DQF-COSY, 1H-1H TOCSY, 1H-13C HSQC, and 1H-13C HMBC) with CD3OD as solvent (Table S1 & Figs. S1–6, Supplementary data). Spin systems were assigned by 1H, DQF-COSY, and TOCSY. Carbons directly attached to protons were assigned by multiplicity-edited HSQC, and unattached carbons were assigned by HMBC. Exchangeable peaks from three hydroxyl protons and one carboxyl proton were not detected/assigned. Ten methyl groups and one methoxy group were detected; each was localized using HMBC, DQF-COSY, and TOCSY. Attachment of the fumarate at 21-C was verified by HMBC peaks between 21-H and 1′-C. The site of macrocyclization was confirmed by correlations between 15-H and 1-C. Unlike prototypical bafilomycins (e.g. B1 and C1, Fig. S8b, Supplementary data), the molecule has a methyl rather than methoxy group attached to 2-C. Only weak 1H-1H correlations were observed between 6-H/7-H, 15-H/16-H, 17-H/18-H, and 23-H/24-H, although coupling was confirmed by close inspection of the DQF-COSY, as well as analysis of J-values; 1H-13C HMBC correlations were also observed for these positions. Overall, the NMR data were consistent with a structure with atomic connectivity identical to JBIR-100, a known metabolite from Streptomyces sp. IR027-SDHV6 [47].
It was noted by Ueda et al. that the configurations at the 12 stereocenters of JBIR-100 were not known [47]. We thus analyzed the vicinal coupling constants for the protons at these positions using 1H and DQF-COSY NMR (Fig. S1d, Supplementary data). The coupling between protons 22-H/23-H (J22-H/23-H = 10.4 Hz) and 21-H/22-H (J21-H/22-H = 10.9 Hz) in the six-membered ring indicated a trans-diaxial relative configuration, which we propose as 21R,22S,23R. Due to ambiguity in J-values, we have assigned the two diastereomeric protons of 20-C primarily on chemical shift basis, given the propensity for equatorial protons in six-membered rings to usually display downfield shifts (equatorial: 2.30 ppm, J20-Hax/21H = 4.8 Hz; axial: 1.31 ppm, no detected coupling to 21-H). For analysis of the stereocenters at 6-C, 7-C, 8-C, 14-C, 15-C, 16-C, 17-C, and 18-C, we compared vicinal coupling constants of these sites in JBIR-100 to dihedral angles obtained from the crystal structure of bafilomycin A1 [Cambridge Crystallographic Data Centre (CCDC) 1159422] and 21-O-methyl-24-demethyl-bafilomycin A1 (CCDC 707561) using the Karplus equation [7, 56, 57]. The high consistency between these values enabled us to assign the structure as 6R,7S,8S,14S,15R,16S,17R,18S. Coupling constants and chemical shifts of stereogenic centers were also consistent with those previously observed for bafilomycin-type compounds with the above stereochemistry [2, 9, 58]. J-values for the opposite configuration were predicted to be substantially different from those observed at each stereocenter, with the exception of 8-C, for which the difference between predicted and observed J7-H/8-H was small (ΔJ < 2 Hz). However, Hertweck and coworkers assigned 8S configuration to the co-metabolites bafilomycin C1, C2, and D, which are also produced by S. varsoviensis; the co-metabolite hygrobafilomycin showed NOESY correlations also suggestive of this stereochemistry [11]. Thus, due to their common biosynthetic origin and closely aligned chemical shifts, we also assigned this center as 8S.
On the basis of coupling constants, the fumaryl alkene (J2′-H/3′-H = 15.8 Hz) and the disubstituted alkene at 12-C/13-C (J12-H/13-H = 15.0 Hz) were assigned as E-configuration. Assignment of the trisubstituted alkenes at 2-C/3-C, 4-C/5-C, and 10-C/11-C as E was made in analogy to the chemical shifts at this position for hygrobafilomycin, which is consistent with other members of the family [2, 11].
We assigned the configuration of the acetal at 19-C as R based on similarity of chemical shifts in the immediate vicinity to those of previously-characterized bafilomycins [11, 56]; in the crystal structure of bafilomycin A1 (CCDC 1159422), the hydroxyl moiety (R-configuration) at this site is involved in intramolecular hydrogen bonding, which, combined with the anomeric effect as well as the equatorial placement of the remaining substituents, is likely to favor this stereoconfiguration [56]. This is also consistent with the biosynthetic origin of this stereocenter, which arises from nucleophilic attack of the 23-C hydroxyl moiety into a ketone at 19-C. We also bioinformatically substantiated this assignment (see Section 2.4).
Interestingly, we noted by NMR a tendency of JBIR-100 to gradually isomerize or degrade over several days at ~22 °C in CD3OD. By ESI-MS we observed that this corresponded to an incorporation of CD3OD into the molecule (data not shown). Under mildly acidic conditions in chloroform, we also observed rapid elimination of the fumarate moiety and gradual decomposition of the molecule as a pink solution that eventually became pale yellow with time. Intermediates observed by NMR during decomposition were similar to several reported isolated bafilomycin-like compounds, including bafilomycins L and I, suggesting that these might be degradation artifacts rather than bona fide biosynthetic products [4–7]. For instance, it has been noted that hygrolidin K2 forms a methyl acetal in methanol at reflux [17], the methyl acetal of the hygrolide leucanicidin results during isolation with methanol and is not detectable in culture [59], and the original disclosure of bafilomycins A, B, and C ascribed the methyl acetals to isolation artifacts [2].
2.3. Insights into the antimicrobial activity of JBIR-100
Many questions remain regarding the features that dictate the diverse bioactivities of hygrolides. To date, the main focus of investigations into hygrolide bioactivity has been on eukaryotic targets, with the result that little is known about antibacterial SAR. The first report on the bioactivity of bafilomycin A1 (which does not contain a fumarate moiety), B1 and C1 (fumarate-containing) showed that all three compounds have antifungal activity, indicating that the fumarate might be dispensable for this bioactivity. However, while bafilomycin B1 and bafilomycin C1 have antibacterial activity, bafilomycin A1 displays no detectable antibacterial activity [2]. Similarly, a recent report found that bafilomycin L, which does not contain a fumarate moiety, has no antibiotic activity [6]. These observations lead us to speculate that the fumarate might contribute to antibacterial activity and we thus determined the minimum inhibitory concentration (MIC) of JBIR-100 against a number of bacteria. We determined that Mycobacterium smegmatis and the Proteobacteria tested were insensitive to JBIR-100 up to the maximum concentration used (64 µM) (Table 1). The lack of activity towards Gram-negative bacteria is in agreement with previous observations for hygrolides. In contrast, JBIR-100 showed antimicrobial activity against all Firmicutes examined, with MICs as low as 4 µM observed against Bacillus sp. Al Hakam and Staphylococcus aureus USA300 (Table 1). In addition, we tested five fungi for sensitivity to JBIR-100 and only observed significant activity towards Debaryomyces hansenii (Table 1). It is notable that hygrobafilomycin was far more pronounced in its antifungal than antibacterial effects [11], while in the case of JBIR-100, the opposite was true. Although not the only structural difference between hygrobafilomycin and JBIR-100, the former lacks the fumarate moiety.
Table 1.
Spectrum of antimicrobial activity for JBIR-100. MICs are reported as the modal value obtained from three experiments with three independent cultures each.
| Microorganism | MIC (µM) |
|---|---|
| Firmicutes (Gram-positive) | |
| Bacillus anthracis Sterne ΔLF | 4 |
| Bacillus cereus ATCC 4342 | 4 |
| Bacillus subtilis subsp. subtilis str. 168 | 8 |
| Bacillus subtilis ATCC 47096 | 8 |
| Bacillus sp. Al Hakam | 4 |
| Enterococcus faecium U503 (VRE1) | 16 |
| Staphylococcus aureus USA300 (MRSA2) | 4 |
| Actinobacterium | |
| Mycobacterium smegmatis B-14616 | > 64 |
| Proteobacteria (Gram-negative) | |
| Escherichia coli DH5α | > 64 |
| Escherichia coli MC1061 | > 64 |
| Pseudomonas putida KT2440 | > 64 |
| Ascomycota (Fungi) | |
| Candida albicans | > 128 |
| Debaryomyces hansenii | 8 |
| Kluyveromyces lactis | > 64 |
| Saccharomyces cerevisiae | > 64 |
| Zygosaccharomyces rouxii | 32 |
Vancomycin-resistant Enterococcus.
Methicillin-resistant Staphylococcus aureus
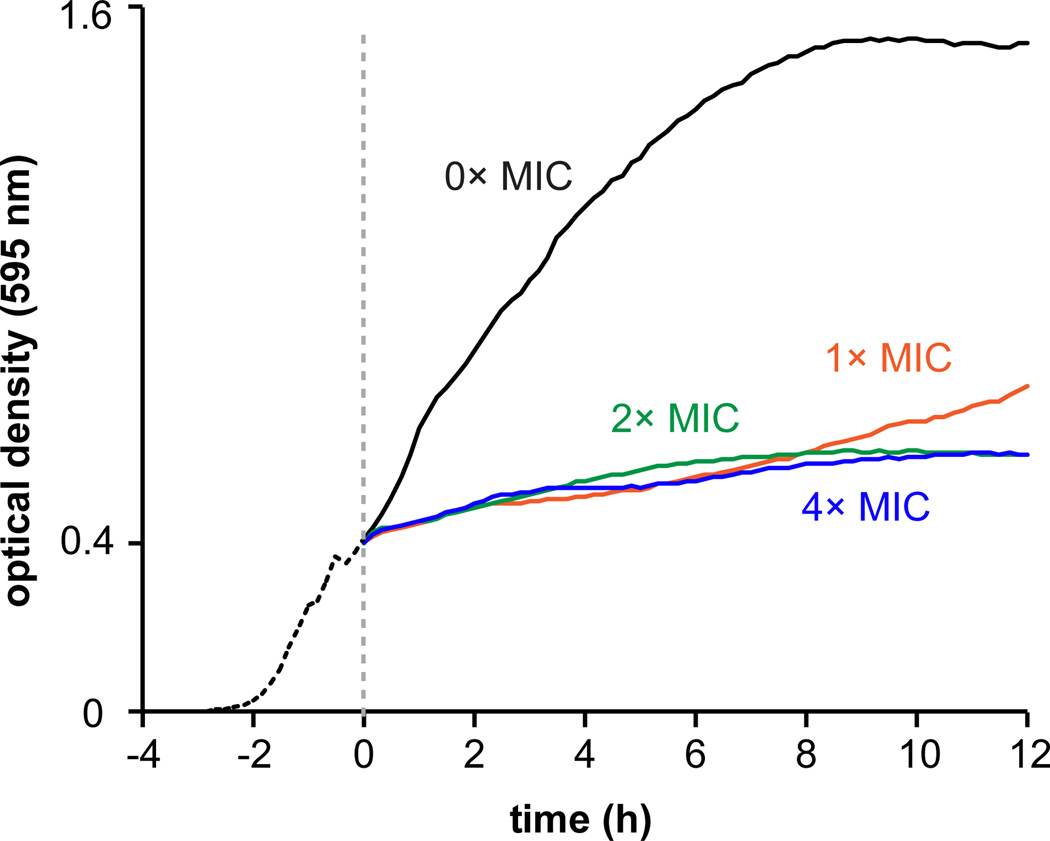
To further evaluate antibacterial activity, a growth curve of B. subtilis (MIC = 8 µM) challenged at mid-exponential phase with various concentrations of JBIR-100 showed a sharp reduction in growth immediately upon addition of the compound (Fig. 3). The minimum bactericidal concentration (MBC = 16 µM) was also determined against B. subtilis, classifying JBIR-100 as bactericidal towards this organism [60].
Figure 3.
Growth curve (optical density at 595 nm) depicting the effect of addition of JBIR-100 to B. subtilis ATCC 47096. The dashed line denotes the point at which JBIR-100 was added at concentrations of 1× (8 µM), 2× (16 µM), and 4× (32 µM) MIC as indicated; 0× MIC curve represents the vehicle (DMSO)-treated control. JBIR-100 appeared to be growth-suppressive at all concentrations tested. Data depicted is the average of three independent replicates.
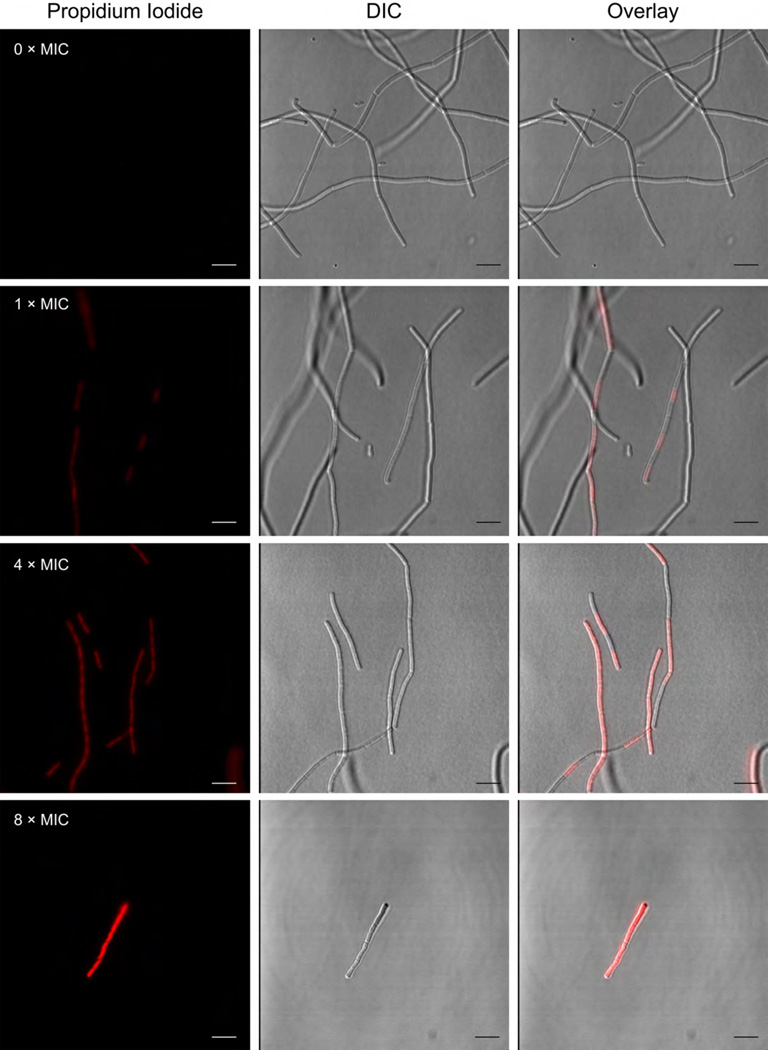
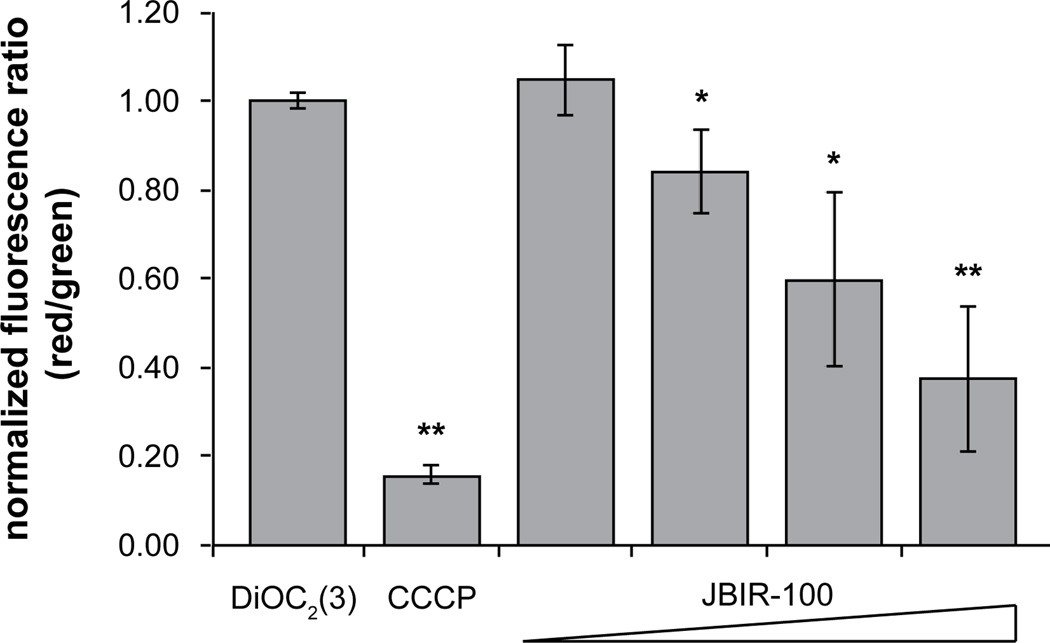
Despite several reports on the antibacterial activity of the hygrolides, to our knowledge there are no published data on the bacterial molecular targets or mode of action (MOA). In light of this, we chose B. subtilis as an investigative model for further study. The cationic, cell-impermeable, fluorescent dye propidium iodide (PI) was used as an indicator of membrane integrity. PI is well known to enter cells that have suffered membrane damage, causing the membrane-compromised cells to become fluorescent. After treatment with JBIR-100, uptake of PI by B. subtilis was visualized by confocal fluorescence microscopy. While untreated cells did not display any PI fluorescence, we observed a dose-dependent increase in the number and brightness of B. subtilis cells stained with PI, indicating membrane permeabilization by JBIR-100 (Fig. 4). Flow cytometry was then used to examine possible changes in the polarization of the B. subtilis membrane upon exposure to sub-MIC levels of JBIR-100 (up to 1 µM or 0.125 × MIC) using the membrane potential-sensitive dye 3,3′-diethyloxacarbocyanine iodide [DiOC2(3)] [61]. Treatment of B. subtilis with concentrations of JBIR-100 greater than 0.2 µM (0.025 × MIC) resulted in a significant decrease in fluorescence intensity, with the data showing a dose-dependent depolarization of the B. subtilis membrane (Fig. 5). Taken together, these experiments indicate that JBIR-100 perturbs the B. subtilis cell membrane, although it remains to be determined if this is a direct or indirect consequence of its MOA.
Figure 4.
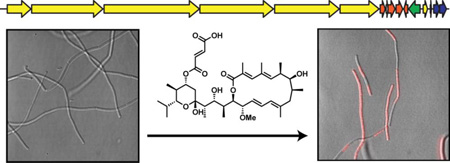
Fluorescence microscopy reveals the uptake of propidium iodide (PI) by B. subtilis ATCC 47096 after treatment with JBIR-100. Cells were exposed at mid-log phase to vehicle alone (DMSO), 1× (8 µM), 4× (32 µM), or 8× (64 µM) MIC of JBIR-100, which was followed by washing and treatment with PI. Left panels, fluorescence of PI; middle panels, differential interference contrast (DIC); right panel, overlay of PI and DIC channels.
Figure 5.
JBIR-100 depolarizes the B. subtilis ATCC 47096 cellular membrane. Red/green ratios were calculated using mean fluorescence intensities of cells treated for 30 min at ~22 °C with 0.1 µM DiOC2(3) and vehicle (DMSO) (negative control), 5.0 µM CCCP (positive control), 0.1 µM JBIR-100, 0.2 µM JBIR-100, 0.5 µM JBIR-100, 1.0 µM JBIR-100. Data were normalized to the negative control sample and are depicted from three independent replicates. Error is given as standard deviation with n = 3. P-values are given relative to the negative control with * indicating < 0.05 and ** indicating < 0.005. Abbreviations: DiOC2(3), 3,3′-diethyloxacarbocyanine iodide; CCCP, carbonyl cyanide m-chlorophenyl hydrazone.
In order to gain further insight into the antibacterial action of JBIR-100, we selected and mapped resistance-conferring polymorphisms in B. subtilis [62]. Our initial attempts to obtain spontaneous mutants of B. subtilis resistant to JBIR-100 were unsuccessful, perhaps indicating that there is no straightforward path to resistance and that JBIR-100 might interact with multiple bacterial targets. We therefore employed a dose-escalation approach to gradually select for JBIR-100 resistance. We began by culturing B. subtilis in the presence of a sublethal concentration of JBIR-100 in triplicate, then gradually increased the concentration with the number of passages [63]. After ten serial passages, the resistance of the resulting B. subtilis strains (P10A, P10B, P10C) to JBIR-100 was only modestly increased (MICs = 16 µM) compared to the parent B. subtilis (MIC = 8 µM). There were no major differences in morphology of strains P10A, P10B, and P10C compared to the parent by differential interference contrast (DIC) microscopy (data not shown). As is often the case for antibiotic resistant bacteria [64], all three strains incurred fitness costs as a result of the polymorphisms accrued. Not only was a significant delay observed for the onset of exponential phase for strains P10A–C, we also noted that strains P10B and P10C failed to reach a stationary phase density comparable to the parent (Fig. S7, Supplementary data).
We next isolated genomic DNA from the parent and JBIR-100-resistant strains for whole genome sequencing (WGS). Polymorphisms detected in P10A, P10B, and P10C relative to the parent strain were mapped and confirmed by Sanger sequencing (Table 2). A number of polymorphisms were identified in each resistant strain, as expected in mutants derived from extensive passaging in the presence of antibiotic. Interestingly, all three mutants contained a frameshift mutation in yusO, a putative transcriptional regulator of the MarR (multiple antibiotic resistance repressor) family. MarR functions as a repressor in E. coli until bound by structurally diverse compounds including antibiotics, organic solvents, and other toxic chemicals [65]. Subsequent expression of the transcriptional activator MarA then leads to upregulation of a multidrug efflux system. Moreover, loss of function mutations in MarR or MarR homologues are known to cause clinically relevant drug-resistant phenotypes, in particular towards fluoroquinolones [66–69]. While these results potentially implicate the mar locus in JBIR-100 resistance, the general nature of this resistance mechanism means mutation of yusO is unlikely to give meaningful insight into biological target(s) of JBIR-100. It also complicates our interpretation of the potential contribution of the other polymorphisms to the resistance phenotypes of these strains.
Table 2.
Description of polymorphisms identified in JBIR-100-resistant mutants of B. subtilis ATCC 47096. Position of polymorphism corresponds to DNA location in reference genome (AL009126.3).
| Strain | Polymorphism Position and Type |
Locus Tag / Gene | Annotation | Consequence |
|---|---|---|---|---|
| P10C | 1,234,377_1,234,378insT | BSU11550 / yjbH | putative thiol management oxidoreductase component |
p.I46fsX67 (nonsense at aa 67 of 299 aa protein) |
| P10A | 1,741,429C>T | BSU16690 / pnpA | polynucleotide phosphorylase | p.Q683X (nonsense at aa 683 of 705 aa protein) |
| P10B | 2,451,421_2,451,422insA | Between BSU23540 / yqkK & BSU23550 / mleA |
hypothetical protein & NAD- dependent malic enzyme |
N/A – intergenic region |
| P10A | 3,118,362C>T | BSU30450 / ytrB | ABC transporter | p.V165M (missense, conserved) |
| P10A, P10B & P10C |
3,374,721delA | BSU32870 / yusO | putative transcriptional regulator (MarR family) |
p.Q77fsX92 (nonsense at aa 92 of 155 aa protein) |
| P10B | 3,947,564C>A | BSU38460 / tyrZ | tyrosyl-tRNA synthetase | p.P136H (missense, non- conserved) |
Abbreviations: >, substitution; del, deletion; ins, insertion, r., RNA sequence; p., protein sequence; fs, frameshift; X, stop codon; aa, amino acid.
To determine whether resistance to JBIR-100 was concurrent with resistance to other antibiotics of known mechanism, and to provide insight into the fitness of the resistant mutants, JBIR-100-resistant strains P10A, P10B, and P10C were challenged with a panel of structurally and mechanistically diverse antibiotics (Table 3). Susceptibility was unchanged for ampicillin, triclosan, and ciprofloxacin. Some mutants displayed modest increases in susceptibility (2-fold) to chloramphenicol and vancomycin, and all three mutants were more susceptible (2- to 4-fold) than the parent strain to kanamycin. Interestingly, the mutations present in P10A, P10B, and P10C conferred cross-resistance only against rifampicin, with a 2- to 4-fold increase in MIC.
Table 3.
MICs of B. subtilis JBIR-100-resistant mutants for various antibiotics compared to the parental B. subtilis ATCC 47096. MICs are reported in µM as the modal value obtained from three experiments with three independent cultures each.
| Antibiotic | ATCC 470961 |
P10A1 | P10B1 | P10C1 |
|---|---|---|---|---|
| Ampicillin | 0.125 | 0.125 | 0.125 | 0.125 |
| Chloramphenicol | 8 | 4 | 8 | 8 |
| Ciprofloxacin | 0.125 | 0.125 | 0.125 | 0.125 |
| Kanamycin | 256 | 128 | 64 | 64 |
| Rifampicin | 0.0625 | 0.25 | 0.125 | 0.125 |
| Triclosan | 8 | 8 | 8 | 8 |
| Vancomycin | 0.125 | 0.125 | 0.0625 | 0.0625 |
MIC values are given in µM
2.4. Biosynthetic origin of hygrobafilomycins
To our knowledge, no hygrobafilomycin BGC has been reported. Additionally, we believe S. varsoviensis marks the first sequenced microorganism found to produce multiple hygrolide sub-families—in this case, hygrobafilomycins (JBIR-100 and hygrobafilomycin) and bafilomycins (bafilomycins C1 and D) [11]. We wondered whether this phenomenon was due to the presence of multiple BGCs or the promiscuity within a single BGC. We thus sought to identify the biosynthetic origin of JBIR-100/hygrobafilomycin. We used antiSMASH 3.0 [51] to survey two S. varsoviensis genomes, NRRL ISP-5346 and NRRL B-3589. Both contained a BGC that was logically consistent with the structure of JBIR-100. Although the BGC was fragmented between small contigs in the NRRL B-3589 genome (GenBank accessions NZ_JOFN01000036, NZ_JOFN01000065, and NZ_JOFN01000038), the ISP-5346 genome harbored the entire BGC within a single contig (GenBank accession NZ_JOBF01000033). All coding sequences in the BGC were identical at the protein level between NRRL ISP-5346 and NRRL B-3589, with the exception of HbaAII (99% protein sequence identity). PRISM [70], BLAST [71], and antiSMASH 3.0 [51] were used to annotate the functions for each gene in the ISP-5346 BGC (Table 4).
Table 4.
Genes in the S. varsoviensis NRRL ISP-5346 biosynthetic gene cluster (hba) for the hygrobafilomycin JBIR-100. Genes are given with their homologs from the baf cluster from S. lohii. For flanking genes, where no baf homolog was found, the nearest BLAST hit is provided.
| ORF | Product | Accession (or locus) |
Length (aa) | Assignment |
S. lohii Bafhomolog |
Accession | Length (aa) | Identity (%) | Top BLAST hit in GenBank, non-redundant database | Accession | Length (aa) | Identity (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | WP_030890302 | 183 | - | - | - | - | hypothetical protein QR77_18815 [Streptomyces sp. 150FB] | KIF75396 | 187 | 63 | ||
| 2 | WP_048832934 | 266 | - | - | - | - | phytanoyl-CoA dioxygenase [Streptomyces sp. 150FB] | WP_052488889 | 265 | 73 | ||
| 3 | WP_030890308 | 452 | - | - | - | - | hypothetical protein [Streptomyces aureocirculatus] | WP_030560004 | 446 | 69 | ||
| 4 | WP_030890312 | 169 | - | - | - | - | hypothetical protein [Allokutzneria albata] | WP_030432205 | 164 | 42 | ||
| 5 | HbaAIa | IF55_RS32365 (locus) | 1325 | polyketide synthase, loading module | BafAI | ADC79616 | 4884 | 68 | ||||
| HbaAIb | 3866 | polyketide synthase, module 1+2+3 | ||||||||||
| 6 | HbaAII | WP_048832936 | 5105 | polyketide synthase, module 4+5+6 | BafAII | ADC79617 | 5145 | 69 | ||||
| 7 | HbaAIII | IF55_RS32375 (locus) | 3935 | polyketide synthase, module 7+8 | BafAIII | ADC79618 | 3968 | 67 | ||||
| 8 | HbaAIV | WP_037966908 | 3473 | polyketide synthase, module 9+10 | BafAIV | ADC79619 | 3511 | 68 | ||||
| 9 | HbaAV | IF55_RS32385 (locus) | 2098 | polyketide synthase, module 11 | BafAV | ADC79620 | 2103 | 62 | ||||
| 10 | HbaB | WP_048832937 | 294 | glyceryl-ACP oxidase | BafB | ADC79621 | 296 | 78 | ||||
| 11 | HbaC | WP_048832938 | 100 | acyl carrier protein (ACP) | BafC | ADC79622 | 93 | 73 | ||||
| 12 | HbaD | IF55_RS32400 (locus) | 385 | acyl-ACP dehydrogenase | BafD | ADC79623 | 363 | 73 | ||||
| 13 | HbaE | WP_048832952 | 374 | glycerate ACP ligase | BafE | ADC79624 | 365 | 80 | ||||
| 14 | HbaF | WP_030890472 | 220 | O-methyl transferase | BafF | ADC79625 | 220 | 77 | ||||
| 15 | HbaG | WP_030890475 | 637 | AfsR family regulator | BafG | ADC79626 | 609 | 67 | ||||
| 16 | HbaH | WP_030890478 | 249 | thioesterase | BafH | ADC79627 | 253 | 67 | ||||
| 17 | HbaI | WP_037966912 | 72 | LuxR family regulator | orf1 | ADC79628 | 117 | 72 | ||||
| 18 | HbaJ | WP_030890486 | 319 | putative malonyl transferase | orf2 | ADC79629 | 320 | 79 | ||||
| 19 | HbaK | WP_048832939 | 366 | putative CoA-ligase | orf3 | ADC79630 | 332 | 82 | ||||
| 20 | WP_030890492 | 162 | - | - | - | - | hypothetical protein [Saccharothrix syringae] | WP_033433816 | 185 | 33 | ||
| 21 | WP_030890495 | 270 | - | - | - | - | alpha/beta hydrolase [Streptomyces violaceorubidus] | WP_030191810 | 269 | 68 | ||
| 22 | WP_048832953 | 268 | - | - | - | - | ABC transporter permease [Streptomyces violaceorubidus] | WP_051777733 | 255 | 53 | ||
| 23 | WP_030890502 | 315 | - | - | - | - | daunorubicin resistance protein DrrA family ABC transporter ATP-binding protein [Saccharothrix sp. NRRL B-16314] | WP_033438336 | 309 | 65 | ||
| 24 | WP_030890505 | 138 | - | - | - | - | aspartate decarboxylase [Streptomyces decoyicus] | WP_030076920 | 138 | 90 | ||
| 25 | WP_048832940 | 270 | - | - | - | - | hypothetical protein [Streptomyces globisporus] | WP_037675397 | 270 | 77 |
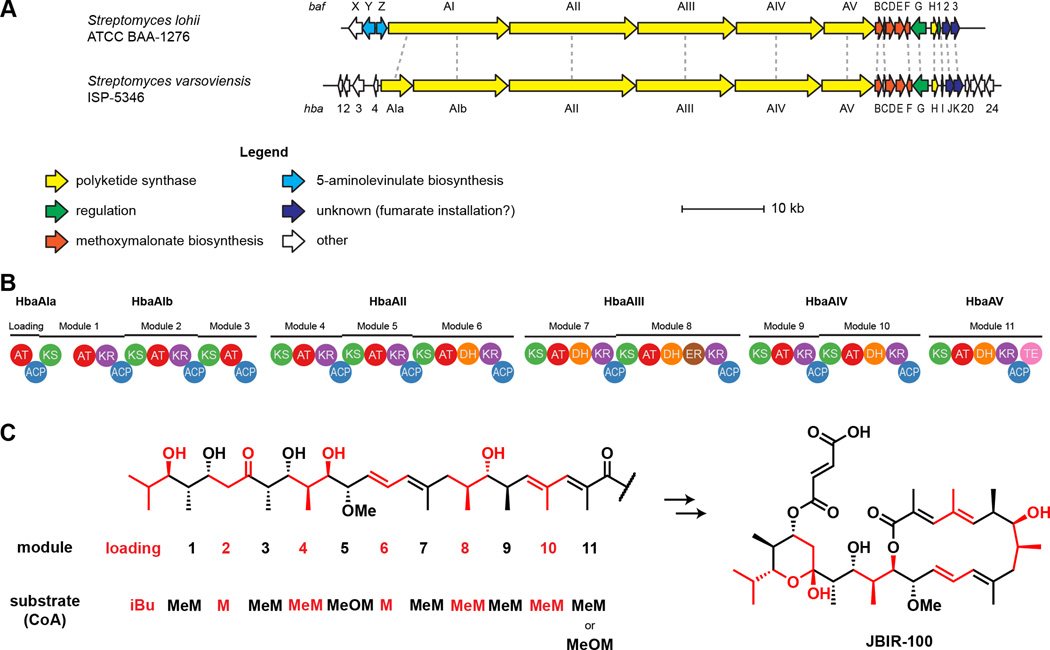
The JBIR-100/hygrobafilomycin BGC hbaA–hbaK consists of 15 open reading frames (ORFs), which we have named analogously to the bafilomycin BGC from Streptomyces lohii (Table 4, Fig. 6a) [42]. The cluster contains several PKS genes, hbaAI (68% protein sequence identity, 74% similarity), hbaII (69% id, 76% sim), hbaAIII (67% id, 74% sim), hbaAIV (68% id, 76% sim), and hbaAV (62% id, 70% sim), homologous to the five PKS genes from S. lohii. Although hbaAI is annotated in GenBank as a single pseudogene of comparable length to bafAI, analysis with PRISM and antiSMASH indicated this region was split into two smaller ORFs, hbaAIa (consisting of the PKS loading module) and hbaAIb (consisting of PKS modules 1–3). The modifications encoded by individual modules in the PKS domains in hbaAIa–hbaAV exactly coincide with the linear scaffold of JBIR-100 and hygrobafilomycin (Fig. 6b,c). The PRISM-predicted acyltransferase (AT) substrate specificity for each extender module (malonyl-, methylmalonyl-, or methoxymalonyl-CoA) matched that of the linear scaffold. Interestingly, PRISM predicts not only AT5 but also AT11 to be specific for methoxymalonyl-CoA, despite AT11’s utilization of methylmalonyl-CoA in hygrobafilomycin biosynthesis; however, S. varsoviensis also produces compounds with a methoxy substitution at the 2-C position, indicating AT11 substrate promiscuity [11].
Figure 6.
Biosynthetic gene cluster for hygrobafilomycin/JBIR-100. (A) The newly-identified cluster for JBIR-100 and related bafilomycin/hygrobafilomycin compounds, identified in Streptomyces varsoviensis NRRL ISP-5346, is shown aligned to the reported bafilomycin C1 and B1 gene cluster from Streptomyces lohii. Notably, the S. varsoviensis cluster is missing the bafXYZ trio thought to be involved in installation of the 5-aminolevinulate-derived 2-amino-3-hydroxy-cyclopent-2-enone moiety found in bafilomycin B1 and similar compounds; this substructure has not been found in any polyketides from S. varsoviensis. (B) The baf cluster and hba cluster share the same PKS domain organization. Both clusters harbor 11 type I PKS modules and one loading module within 6 distinct ORFs. AT, acyltransferase. ACP, acyl carrier protein. KS, ketosynthase. KR, ketoreductase. DH, dehydratase. ER, enoylreductase. TE, thioesterase. (C) The domain architecture shown in (B) is consistent with the linear structure (left) and mature structure (right) of JBIR-100. Modules responsible for installation of each section of the scaffold are indicated along with the extender substrate utilized by that module. iBu, isobutryl-CoA. MeM, methylmalonyl-CoA. M, malonyl-CoA. MeOM, methoxymalonyl-CoA. Based on the isolation of compounds containing either Me or MeO at the module-11 site, this module can accept MeM (giving hygrobafilomycin compounds like hygrobafilomycin or JBIR-100) or MeOM (giving bafilomycin compounds like bafilomycin C1).
Immediately downstream of hbaAV are five genes, hbaB–hbaF, which are homologous to bafB–bafF. Homologues of bafB–bafF have been implicated in methoxymalonyl PKS extender unit biosynthesis from 1,3-bisphosphoglycerate [72, 73], and their function was previously assigned as methoxymalonyl-CoA generation in the biosynthesis of bafilomycin A–C [42, 43]. Schuhmann and Grond used feeding studies to determine that this unit originates as glycerate in bafilomycin A1 [74]. In this case, HbaE (80% protein sequence identity to BafE, 86% similarity) would be responsible for ligating glycerate to the acyl carrier protein (ACP) HbaC (73% identity to BafC, 80% similarity). HbaB (78% identity to BafB, 83% similarity) and HbaD (73% identity to BafD, 79% similarity) could then oxidize the glycerate, and HbaF (77% identity to BafF, 86% similarity) would O-methylate the resulting hydroxymalonyl-ACP to yield the methoxymalonyl unit.
Two regulatory proteins are encoded in the hba cluster: the AfsR-family regulator HbaG (67% protein sequence identity to BafG, 76% similarity) and the LuxR-family regulator HbaI (72% identity to the protein encoded by orf1 in S. lohii, 85% similarity).
HbaH encodes a type II thioesterase (TEII) [75] with similarity to BafH (67% protein sequence identity, 76% similarity). HbaJ encodes an AT homologue of the S. lohii gene product of orf2 (79% identity, 86% similarity). The hbaK gene encodes a putative acyl-CoA ligase that is a homologue of orf3 (82% identity, 87% similarity). It has been speculated that homologues of HbaJ and HbaK could be involved in fumaryl attachment, although this has not been investigated experimentally [43]. Interestingly, Hertweck et al. reported that hygrobafilomycin bears an unusual fumaryl-like monoalkylmaleic anhydride moiety [11], which may be installed by the same machinery as the fumarate.
In contrast to any other known hygrolide BGC, homologues of bafX, bafY, and bafZ are absent from the JBIR-100 cluster. Although tailoring genes can sometimes be located distal to the core polyketide BGC (e.g. the methoxymalonate subcluster in galbonolide biosynthesis) [76], there are no obvious homologues of this trio in any S. varsoviensis genome. Homologues of the bafXYZ subcluster have been linked to installation of a 5-aminolevulinate-derived 2-amino-3-hydroxy-cyclopent-2-enone ring [42–44, 77], which is consistent with the absence of this moiety from any hygrolide metabolites isolated from S. varsoviensis [11]. Finally, the genes flanking hbaAI–hbaK share no similarity to genes adjacent to any of several other identified bafilomycin, hygrolidin, or concanamycin gene clusters (Table 4), suggesting the cluster boundaries are the same as for the baf BGC.
Another important aspect of JBIR-100 biosynthesis is the resulting configuration of its 12 stereocenters, which we elucidated using NMR (see Section 2.2). To corroborate this assignment and to furnish the absolute configuration, we bioinformatically analyzed several of the ketoreductase (KR) domains. McDaniel and Caffrey previously determined key sequence signatures that separated KR domains into A- and B-type [78, 79], and this approach has been further expanded by more sophisticated methods including hidden Markov model (HMM)-based sequence classification [80]. This sequence-based approach has been used successfully to assign the absolute configuration of several polyketides and has been corroborated by analysis of the crystal structures of several representative KRs [14, 81–83]. Using PRISM [70], we extracted the KR sequences from modules 1, 2, 4, 5, and 9 (KR1-KR9) of the S. varsoviensis BGC, which are responsible for setting the stereocenters at 7-C, 15-C, 17-C, 21-C, and 23-C, respectively. KR1 and KR5 are B-type (d alcohol configuration), whereas KR2, KR4, and KR9 are A-type (l alcohol configuration), as determined by the presence of signature sequence motifs in each (Fig. S9, Supplementary data). Kitsche and Kalesse demonstrated that KR sequence could also occasionally distinguish methyl stereochemistry [80]; indeed, using their HMM-based classification program ScoreDiff, KR9 is confidently predicted to give rise to an l α-methyl branch, although KR2, KR4, and KR5 methyl stereochemistry could not be reliably predicted using this method. The KR9 stereospecificity classification is especially important in the context of the spectroscopic ambiguity of the 8-C stereocenter. These sequence-based classifications provide a strong prediction of the absolute stereochemistry and corroborated our NMR-derived assignments, as well as those of hygrobafilomycin [11] (Fig. S9b,c, Supplementary data).
2.5. Comparison of known hygrolide biosynthetic gene clusters and structures
Reports have shown that a hygrolide scaffold from a single producer can be decorated with a variety of structural patterns, including ring-opening, methyl acetal formation, aminolevinulate attachment, fumaryl attachment, etc., possibly resulting from premature termination of biosynthesis, combinatorial biosynthesis, or degradation [2, 3]. However, other structural aspects, such as variation within the macrocyclic ring scaffold or at the terminus of the molecule, are likely to be genetic in origin. In light of the structural diversity present among the more than 48 members of the hygrolide family, we sought to compare the known and predicted BGCs of bafilomycin, hygrolidin, hyrobafilomycin, and concanamycin (although the biosynthesis of concanamycin is similar, it is not a technically a hygrolide as it contains an 18-membered rather than 16-membered macrolactone). We posited that this would provide insight into what structural variation could be genetic in nature, with the intent of guiding novel discovery efforts and providing a phylogenetic context for (hygro)bafilomycin biosynthesis.
We thus assembled the sequences from reported clusters as well as some clusters newly identified herein (Fig. S8 & Table S2, Supplementary data). We analyzed 48 hygrolide structures from 30 reported producers to gain insight into the molecular phylogeny. The hygrolides group structurally into what we classify as six main families, based on substitutions at 2-C and 23-C: bafilomycins (2-OMe/23-iPr), demethylbafilomycins (2-OMe/23-Et), hygrobafilomycins (2-Me/23-iPr), hygrolidins (2-Me/23-Et), micromonospolides (2-OMe/23-pentadiene), and didemethylbafilomycins (2-OMe/23-Me) (Fig. 1b). Within each family, numerous substitutions are found at various sites, including 7-O, 17-O, 19-O, 21-O, as well as a number of apparent elimination or ring-opening products, sometimes giving a diversity of compounds from the same organism (bafilomycin A, B, and C, for instance, are typically seen together) (Fig. 1a). However, reports of multiple families being isolated from the same organism are restricted to S. varsoviensis, justifying the above family grouping. Compared to the BGCs from other known hygrolide-producing Actinomycetes, which share high amino acid identity to each other and form a distinct clade in a phylogenetic tree, the S. varsoviensis BGC shows considerably more sequence divergence (Fig. S10 & Table S2, Supplementary data).
Obtaining direct relationships between structure and phylogeny in the hygrolidin family is difficult because (i) typically only new compounds are reported from a given organism, and thus knowledge of the full biosynthetic complement of each is presumed incomplete; and (ii) genomic information, or even 16S rRNA sequence, for many reported producers is not available. In spite of these knowledge gaps, general conclusions can be drawn from our bioinformatic analysis.
The hygrolidin BGC from Streptomyces halstedii was fragmented between three small contigs; of note, S. halstedii produces leucanicidin, a bafilomycin-scaffold compound bearing a glycan instead of a fumaryl appendage, and its cluster was not previously known [84]. In the course of writing this manuscript, it was reported that Kitasatospora sp. MBT66 also produces leucanicidin. The BGC for leucanicidin was revealed, including the glycosyltransferase responsible for rhamnose installation [59]. The three known leucanicidin producers (including also Streptomyces olivaceus) do not tightly cluster by 16S rRNA sequence nor by BafD/ORF3 similiarity (Figs. S10 and S11, Supplementary data); they appear to be more closely related to other hygrolide producers than to each other. In light of the leucanicidin cluster disclosure, we briefly surveyed the genetic potential for each BGC to append sugars to its cognate natural product(s) by analyzing the local genomic regions for the presence of glycosyltransferases using PF00201 or PF00534 profile HMMs (pHMMs) in HMMER3, revealing six total BGCs harboring these enzymes [59, 85] (Fig. S10, Supplementary data); this suggests that hygrolide glycosylation may be relatively infrequent. However, the BGC from S. halstedii, a known leucanicidin producer, does not encode a predicted glycosyltransferase, which shows that our prediction of glycosylation frequency is probably underestimated. Encoded elsewhere in the S. halstedii genome is a glycosyltransferase that might be responsible for hygrolide glycosylation (GenBank accession WP_051422100), given its 34% protein sequence identity and 49% similarity to the K. sp. MBT66 leucanicidin glycosyltransferase. This homologue is also found in the genome of S. sp. DpondAA-B6, an organism closely related to S. halstedii (GenBank accession WP_051422100).
BafBCDEF are responsible for the biosynthesis of the methoxymalonate extender unit that is incorporated once in hygrobafilomycins and hygrolidins, and twice in bafilomycins. The absence of the methyoxymalonyl extender unit biosynthetic subcluster in the hygrolidin BGC from S. hygroscopicus is thus puzzling [44]. However, there is precedence for this subcluster to be found elsewhere in the genome, as is the case for galbonolide produced by S. galbus [76]. Moreover, some S. hygroscopicus strains produce FK520, whose BGC harbors this subcluster [86]. Markedly, the S. hygroscopicus hygrolidin cluster also lacks a bafX homologue. Unfortunately, the genome sequence of the S. hygroscopicus strain that produces hygrolidin is not available, so it cannot be determined if bafBCDEF and bafX lie elsewhere in the genome.
While some of the structural diversity found within the hygrolide family can be explained bioinformatically, there is no clear genetic basis for some other structural aspects, including acetal methylation, ring-opening, and elimination. Instead, these variants may represent biosynthetic intermediates, off-pathway compounds, combinatorial products due to enzyme promiscuity, or even degradation products. For example, isobutyrylleucanicidin bears a 7-O-acylation, for which no corresponding genes have been identified [87]. There is currently also no confirmed BGC for the micromonospolides, which are so far exclusively derived from Nocardia and Micromonospora spp., and further genome sequencing-driven investigation of these clusters is warranted [88, 89].
Unlike RiPPs, whose modifications are easy to predict based on the BGC [90], facilitating rational labeling of functional groups, the link between genes and structure is less direct for other types of natural products, such as the polyketides. For example, the biosynthetic origin of the fumaryl appendage that is present in many hygrolide natural products is unclear. By identifying or postulating signature genes that would reliably result in the installation of this functional group, strains encoding fumarate-containing hygrolides could be prioritized for thiol labeling with the aim of identifying new, biologically active metabolites. Indeed, HbaJ/HbaK (ORF2/ORF3 in S. lohii) may be the mostly likely candidates for fumaryl activation into CoA (HbaK) and transfer (HbaJ), as proposed by Hwang et al. [43]. This is supported by the presence of orf2/orf3 homologues in all known fumarate-bearing hygrolide BGCs. One known hygrolide cluster—the recently disclosed leucanicidin BGC from Kitasatospora sp. MBT66—lacks orf2/orf3 homologues, and this is consistent with the fact that the known hygrolides from this organism are fumarate-free, instead functionalized with a sugar at the same site [59]. The presence of orf2/orf3 homologues in the concanamycin BGC is not consistent with the lack of a fumaryl appendage in concanamycin A, but is worth noting that virustomycin and viranamycin, members of the concanamycin family also isolated from Streptomyces spp., indeed bear a fumarate, and these proteins may be responsible for their biosynthesis [91, 92]. The reveromycins are antifungal spiroacetals which are, like some hygrolides, ester-substituted at an alcohol. Although the originally-reported compound reveromycin A contains a succinyl group [93], it was found recently that the parent organism produces an array of compounds, among them reveromycins H–K, which instead bear a fumaryl moiety [94]. Additionally, the reveromycin BGC contains a pair of proteins including the transferase RevK (36% protein sequence identity, 55% similarity to S. lohii ORF2) and acyl-CoA ligase RevL (61% identity, 73% similarity to S. lohii ORF3) [95]. Although the functions of these two proteins were not interrogated in the original report [95], it is most probable that they install succinyl and fumaryl moieties into the corresponding reveromycin congeners.
2.6. Bioinformatic survey of known and unknown hygrolide biosynthetic gene clusters
We next sought to determine the phylogenetic context of the JBIR-100 (hygrobafilomycin-type scaffold) BGC and survey the hygrolide biosynthetic sequence space. Using representative genes from the bafilomycin, hygrolidin, hygrobafilomycin, and concanamycin BGCs (Table S2, Supplementary data), we created pHMMs of the conserved biosynthetic machinery from each using HMMER3 [85]. Using BafD and ORF3 sequences from S. lohii, S. varsoviensis, and S. neyagawaensis as BLAST queries, we identified the most closely-related homologues of these genes and inspected the local genomic region using HMMER3 and our pHMMs. On the basis of HMM hits and cluster organization, we thus assembled a list of 34 predicted and known hygrolide BGCs. In cases where only a partial BGC sequence was available due to the presence of small contigs, clustering of BafD homologues with confirmed BGCs on a BafD homologue sequence similarity network was used for classification (data not shown) [96]. Each BGC was assigned to a structural family by pairwise comparison of the BafD and ORF3 homologues to those from known producers (Table S3, Supplementary data). In most cases, this allowed clear assignment, although some organisms harbored genes not close to any of the standards (<75% identity at the protein level). Interestingly, no clusters with sequence identities >90% to the S. varsoviensis cluster were found, despite many other hygrolide clusters having >90% identity to S. lohii and to each other.
Using phylogenetic trees built on 16S rRNA, BafD, and ORF3 homologue alignments, we compared phylogeny and molecular scaffold (Figs. S10 & S11, Supplementary data). In general, the hygrolide producers are highly related to each other and group closely, but this is unsurprising given that most are part of the Streptomyces or Kitasatospora genera. Interestingly, 16S rRNA sequence comparison showed that the known JBIR-100 producers S. varsoviensis and S. sp. IR027-SDHV6 are not only distinct organisms, but are also more closely related to other hygrolide producers of different structural subfamilies than to each other. The concanamycin producer Streptomyces neyagawaensis and the micromonospolide producers do not group closely by 16S rRNA sequence (Fig. S11, Supplementary data), which was somewhat unexpected given the structural similarity of the alkene terminus of the acetal ring in the two scaffolds. We found that the phylogenic relationship of BafD and ORF3 homologues seemed to parallel the overall hygrolide scaffold from each producing organism (Fig. S10, Supplementary data). In particular, the BGCs for eight known classical bafilomycins formed a clade that includes 12 other putative bafilomycin producers. The BafD and ORF3 homologues from the hygrobafilomycin cluster from S. varsoviensis are near, but outside of, this highly-related bafilomycin clade; also outside are one known (S. neyagawaensis) and two predicted (S. scabiei and S. stelliscabiei) concanamycin clusters, one known (S. hygroscopicus subsp. hygroscopicus) and three predicted (S. sp. 150FB, S. ghanaensis, and S. viridosporus) hygrolidin producers, as well as several organisms for which a confident scaffold prediction could not be made.
We identified a number of organisms not previously known to harbor hygrolide BGCs (Table S3, Supplementary data). To our knowledge, the biosynthetic capabilities of these organisms, with respect to hygrolide biosynthesis, have not been investigated. The BGCs from a number of organisms (Table S3, Supplementary data) were not highly related to any known BGC but still appear hygrolide-like in gene architecture, suggesting that the products of these BGCs may represent novel hygrolide scaffolds. Given the elusiveness of micromonospolide, demethylbafilomycin, and didemethylbafilomycin BGCs, it is possible that future investigation of these organisms will shed light into the genomic origin of these compounds or could yield new scaffolds entirely. Overall, we identified 22 BGCs to which natural products have not yet been linked.
3. Conclusions
There is a pressing need to discover new compounds to combat the increasing number of infections caused by drug resistant bacteria. To this end, natural products and derivatives thereof have served as a primary source of leads for the development of antibacterial agents [97]. In the present study, we identified Streptomyces varsoviensis as a novel producer of the hygrobafilomycin JBIR-100, described its BGC, and provided spectral and bioinformatic evidence for the previously unknown stereochemistry of the natural product. Currently, the antibacterial MOA(s) of the hygolides is not known. This study shows that JBIR-100 has promising antibacterial activity and that treatment of B. subtilis with JBIR-100 leads to cell membrane permeabilization and depolarization, although other bactericidal mechanisms have yet to be investigated. The modestly JBIR-100 resistant mutants contained a number of polymorphisms but the genomic locations of the polymorphisms and the phenotypes of the mutants did not permit a precise biological target for JBIR-100 to be proposed in the present work. Future work can focus on further elucidation of the MOA; however, our data suggest that hygrolide scaffolds could be investigated as a potential avenue for antibiotic development.
The successful identification of JBIR-100 by nucleophilic 1,4-addition in a S. varsoviensis extract indicates that this simple reaction will be useful in discovering other fumarate-containing plecomacrolides and other natural products containing electron-deficient alkenes. Importantly, fumaryl substitution is not limited to macrolides, but is present in other polyketides, including reveromycin H–J [94], the antibiotic fumimycin [98], and some nonribosomal peptide natural products such as the thrombin inhibitor Ro 09–1679 [99]. Our attempt to reconcile the molecular and genetic phylogenies of the hygrolide family resulted in the identification of 22 orphan hygrolide gene clusters, indicating that potential remains to find novel hygrolide variants among the Actinomycetes.
4. Material and methods
4.1. Bacterial strains and culture
S. varsoviensis NRRL B-3589 was grown in 5 mL of MMS (mannitol-malt-soy) broth (1 L contains 10 g mannitol [BDH Chemicals], 10 g malt extract [BD, Franklin Lakes, NJ], 10 g roasted soybean flour [Wel-Pac]) at 30 °C for 3 days with aeration, before subculturing to MMS agar followed by incubation at 30 °C for 7 days. Firmicutes and Mycobacterium smegmatis were grown in LB (Luria-Bertani) broth (BD) and incubated at 37 °C with aeration. In the case of M. smegmatis, broth contained 0.05% Tween 80. Media was solidified by adding 1.5% agar (BD) where relevant. Ascomycota were grown in YPD (yeast-peptone-dextrose) broth (1 L contains 10 g yeast extract, 20 g peptone and 20 g D-glucose) at 30 °C with aeration.
4.2. Isolation of JBIR-100
For production of JBIR-100, S. varsoviensis NRRL B-3589 was grown on 1 L MMS agar for 7 days. The cell mass was scraped from the solid medium and exported metabolites were extracted by gentle agitation in n-BuOH for 4 h at ~22 °C. The solid medium was frozen, thawed, and the resulting liquid was removed by filtration through cheesecloth with pressure and extracted similarly. After centrifugation (4000 × g, 5 min) of the crude extract, the organic layer was removed from the intact, harvested cells. Solvent was removed and the residue was suspended in MeCN (100 mL) and EtOAc (20 mL) before being concentrated onto 10 g of Celite 545 (Sigma-Aldrich) by rotary evaporation (25 °C). The dried suspension was purified by MPLC (TeleDyne Isco 130 g C18 cartridge; 15–100% gradient of MeCN in aq. 10 mM NH4HCO3, pH 8). Fractions containing JBIR-100, as determined by MALDI-TOF MS, were combined and concentrated. The combined fractions were redissolved in 50% aq. MeCN (4 mL), centrifuged (17,000 × g, 5 min), and purified by HPLC (Teledyne Isco C18Aq column; 5 µm particle size; 20 × 150 mm; 19 mL/min; 20–100% gradient of MeCN in aq. 10 mM NH4HCO3, pH 8). The residual undissolved material was resuspended, sonicated, centrifuged, and purified analogously until no more JBIR-100 was detected by MALDI in the eluent. Combined fractions from all 4 HPLC batches, when combined and evaporated, yielded 150 mg of light yellow solid. This was then dissolved in 75% aq. MeCN (3.5 mL) and purified by HPLC (Teledyne Isco C18Aq column; 5 µm particle size; 20 × 150 mm; 19 mL/min; 35–55% gradient of MeCN in aq. 10 mM NH4HCO3, pH 8). Fractions containing JBIR-100 were combined and evaporated overnight. The isolated material was analyzed by HPLC (Thermo Betasil C18; 5 µm particle size; 4.6 × 150 mm; 1 mL/min; gradient of MeCN (B) in aq. 10 mM NH4HCO3, pH 8 (A); 5% B for 3 min, 15 min ramp to 95% B, 10 min hold at 95% B). Suspension in H2O, flash-freezing, and lyophilization yielded JBIR-100 as a colorless powder (30 mg).
4.3. Thiol labeling
JBIR-100 was subjected to labeling as previously described [50]. An aliquot (14 µL) of a crude n-BuOH extract of S. varsoviensis NRRL B-3589 or of isolated material in n-BuOH was mixed with DTT (in MeOH) and DIPEA (N,N-diisopropylethylamine; in MeOH) to a final volume of 20 µL. The final concentration of DTT was 500 mM and that of DIPEA was 10 mM in 7:3 n-BuOH/MeOH. The mixture was allowed to react for 16 h at ~22 °C. The sample was analyzed for DTT incorporation by MALDI-TOF MS.
4.4. Mass Spectrometry
4.4.1. MALDI-TOF MS analysis
An aliquot (1 µL) of extract was mixed with 9 µL of saturated α-cyano-4-hydroxycinnamic acid matrix solution in 1:1 MeCN/H2O containing 0.1% trifluoroacetic acid. Samples (1.5 µL) were spotted on a steel plate and air-dried at ~22 °C. Mass spectra were obtained using a Bruker Daltonics UltrafleXtreme MALDI-TOF/TOF mass spectrometer in positive reflector mode. The instrument was calibrated using a peptide calibration kit (AnaSpec Peptide Mass Standard Kit). Data were analyzed using flexAnalysis 3.3 (Bruker).
4.4.2. FT-MS/MS analysis
Samples were dried by centrifugal vacuum concentration and dissolved in 50% aq. MeCN containing 1% (v/v) acetic acid. The diluted samples were directly infused using an Advion Nanomate 100 to an LTQ-FT hybrid linear ion trap-FTMS system (ThermoFisher) operating at 11 T. The MS was calibrated weekly using calibration mixture following the manufacturer’s instructions, and tuned daily with Pierce LTQ Velos ESI Positive Ion Calibration Solution (ThermoFisher). Spectra were collected in profile mode with a resolution of 100,000. The singly charged ions were targeted for CID using an isolation width of 2 m/z, a normalized collision energy of 35, an activation q value of 0.4, and an activation time of 30 ms. Data analysis was performed using Thermo Xcalibur software.
4.5. NMR spectroscopy
HPLC-purified and lyophilized JBIR-100 (ca. 5 mg, 12 mM) was dissolved in CD3OD (99.96% D, Sigma-Aldrich; 600 µL). NMR spectra were obtained using an Agilent VNMRS 750 MHz narrow bore magnet spectrometer equipped with a 5 mm triple resonance (1H-13C-15N) triaxial gradient probe and pulse-shaping capabilities. Samples were held at 296 K during acquisition. Standard Varian pulse sequences were used for each of the following experiments: 1H (s2pul; 32768 points; 64 scans), 1H-1H DQF-COSY (gDQCOSY; 1234 × 256 points; 4 scans), 1H-1H TOCSY (zTOCSY; 80 ms mixing time; 1234 × 256 points; 8 scans), multiplicity-edited 1H-13C HSQC (HSQCAD; 1234 × 256 points; 32 scans), and 1H-13C HMBC (gHMBCAD; optimized for 8 Hz heteronuclear couplings; 1234 × 256 points; 64 scans). Spectra were recorded with VNMRJ 3.2A software, and data was processed using MestReNova 8.1.1. Chemical shifts (δ, ppm) were referenced internally to the solvent peak (MeOH, 3.30 ppm).
4.6. Biological activity of JBIR-100
4.6.1. Minimum inhibitory concentration determination
MIC determinations were performed by microbroth dilution assay as described by Clinical and Laboratory Standards Institute [100]. Briefly, JBIR-100 was added at a starting concentration of 64 µM (except in the case of C. albicans at a starting concentration of 128 µM) and two-fold serial dilutions added to wells containing ~5 × 104 colony forming units of test strain in the relevant medium (see Section 4.1). Wells were inspected for turbidity after 16–18 h incubation (40–42 h in the case of Z. rouxii) under the relevant conditions (see Section 4.1) and the MIC was recorded as the lowest compound concentration causing inhibition of visible growth. The procedure was identical for kanamycin (starting concentration 256 µM), chloramphenicol and triclosan (starting concentration 64 µM), and ampicillin, ciprofloxacin, rifampicin, and vancomycin (starting concentration 8 µM).
4.6.2. Minimum bactericidal concentration determination
The MBC is the lowest concentration of antibacterial agent that reduces the viability of an initial bacterial inoculum by ≥99.9% [60]. After performance of the B. subtilis 47096 MIC, the MBC of JBIR-100 was determined by subculturing those wells with no visible growth to LB agar. After 16 h incubation at 37 °C, the plates were inspected for the presence of colonies; the MBC was recorded as the lowest compound concentration required to kill B. subtilis (i.e. no colonies recovered) in three independent replicates.
4.6.3. Growth curve
Three independent stationary cultures of B. subtilis ATCC 47096 were diluted 1000-fold in fresh LB broth and grown to mid-exponential phase (OD600 of 0.4). The cultures were then distributed into a microtiter plate and treated with DMSO (vehicle), 1× (8 µM), 2× (16 µM), or 4× (32 µM) MIC JBIR-100. The plate was covered with a clear plastic lid and placed in a pre-warmed (37 °C) plate reader (Molecular Devices FilterMax F5). After shaking for 5 s in linear mode at medium intensity, the initial density (t = 0) was read. The plate was then monitored at 595 nm every 10 min for 16 h with 565 s of linear mode, medium intensity shaking between reads. OD values were averaged and normalized to an initial read value of 0.4 at t = 0.
4.6.4. Fluorescence microscopy
Aliquots (200 µL) of stationary phase cultures of B. subtilis ATCC 47096 were used to inoculate fresh LB (5 mL). After growing cells to OD600 of 0.5 at 37 °C with shaking, aliquots (1 mL) were centrifuged and resuspended in sterile PBS (phosphate-buffered saline; pH 7.4; 1 L contains 8 g NaCl, 0.2 g KCl, 1.44 g NaH2PO4, 0.24 g KH2PO4) containing vehicle alone (DMSO), 1× (8 µM), 4× (32 µM), or 8× (64 µM) MIC of JBIR-100. Cells were exposed for 30 min at ~22 °C before being washed and subsequently treated with 10 µM PI in PBS for 15 min at ~22 °C. After washing in PBS, cells were resuspended in PBS and immobilized on glass slides by mixing 1:1 (v/v) cell suspensions with liquefied low gelling temperature agarose (Sigma-Aldrich, 2% w/v in sterile H2O). Images were obtained on the same day using a Zeiss LSM 700 confocal microscope with a 63×/1.4 Oil DIC objective. A 405 nm laser was used for DIC and a 555 nm laser was used to excite PI (emission detected at 585 nm). Laser intensity and gain were kept to a minimum and held constant for all experiments. Images were processed using Zen 2012 software. Linear contrast was equally applied during image processing.
4.6.5. Membrane depolarization assay
Three independent stationary phase cultures of B. subtilis ATCC 47096 were used to inoculate fresh LB and grown at 37 °C with shaking to OD600 = 0.5. Aliquots (10 µL) were diluted to 1 mL in PBS containing 0.1 mM 3,3′-diethyloxacarbocyanine iodide and compounds (DMSO – vehicle, 5 µM carbonyl cyanide m-chlorophenyl hydrazone – positive control, 0.1 µM JBIR-100, 0.2 µM JBIR-100, 0.5 µM JBIR-100, 1.0 µM JBIR-100). Cells were mixed at ~22 °C for 30 min prior to analysis by flow cytometry (BD LSR II Flow Cytometry Analyzer). Voltages for fluorescein isothiocyanate and PI fluorescence were set so that average counts per cell were between 103 and 104 for the vehicle control samples and kept constant for all subsequent samples. Cells were gated with forward scatter of 1100 and side scatter of 1800. Geometric means for fluorescence ratios were normalized to the vehicle control samples.
4.6.6. Isolation of JBIR-100-resistant mutants
For each attempt to obtain spontaneous JBIR-100-resistant mutants of B. subtilis ATCC 47096, three independent cultures were grown overnight in LB medium at 37 °C. From each culture, a portion containing ~ 106 cells was spread on LB agar containing 4× MIC (32 µM) JBIR-100, resulting in no growth after 48 h at 37 °C. In a second and third attempt, ~ 107 and 108 cells were spread from each culture on LB containing 2× MIC (16 µM) JBIR-100, again resulting in no growth after 48 h at 37 °C.
Dose-escalation mutants of B. subtilis ATCC 47096 were isolated by serial passaging in the presence of increasing concentrations of JBIR-100. Three independent cultures (A, B & C) were started at an OD600 of 0.1 in LB containing 0.5× MIC JBIR-100. After incubation for 24 h at 37 °C, the cultures were diluted to an OD600 of 0.1 in LB containing the same concentration of JBIR-100 as the originating culture, as well as in LB containing a 0.5× MIC increase in JBIR-100. For subsequent passages, the inoculum was taken from the culture from the previous passage containing the higher concentration of JBIR-100, if growth was obtained. Again, the cultures were diluted to an OD600 of 0.1 in LB containing the same concentration of JBIR-100 as the originating culture, as well as in LB containing a 0.5× MIC increase in JBIR-100. In this way, the B. subtilis ATCC 47096 cultures were subjected to increasing concentrations of JBIR-100 over 10 passages. These cultures were then streaked on LB agar lacking JBIR-100 and incubated for 24 h at 37 °C. For each replicate, an isolated colony was picked and restreaked onto fresh LB agar plates lacking JBIR-100. This process was repeated a further two times to confirm mutant stability. The three mutant strains (named P10A, P10B and P10C) were tested for JBIR-100 resistance via MIC as described above (see Section 4.6.1).
4.7. Whole-genome sequencing
Genomic DNA from the parent B. subtilis ATCC 47096 and three JBIR-100-resistant mutants was isolated using an UltraClean Microbial DNA Isolation Kit (MO BIO). Shotgun genomic libraries were constructed and sequenced by the Roy J. Carver Biotechnology Center (University of Illinois, Urbana, IL, USA). The genomic libraries were prepared using a KAPA Hyper Prep Kit (Kapa Biosystems), with average gDNA fragment of 400 bp (from 80 bp to 700 bp). The libraries were quantified by qPCR, and sequenced on one lane on an Illumina HiSeq2500 with single-reads 100 nt in length using a HiSeq SBS sequencing kit version 4. Fastq files were generated and demultiplexed with the bcl2fastq v1.8.4 Conversion Software (Illumina). The parent genome was assembled relative to a reference genome (AL009126.3) using both CLC Genomics Workbench (CLC bio) and Geneious (Biomatters). Polymorphisms in the genomes of the JBIR-100-resistant mutants (P10A, P10B and P10C) were mapped relative to the parent genome and verified by PCR amplification followed by Sanger sequencing.
4.8. Bioinformatic analysis
4.8.1. Ketoreductase domain classification
KR sequences were extracted from the JBIR-100 BGC of S. varsoviensis NRRL ISP-5346 using PRISM (http://magarveylab.ca/prism/) [70]. KR sequences were aligned using MUSCLE (v. 3.8.31, http://www.ebi.ac.uk/Tools/msa/muscle/) on default settings [101, 102]. Classification was performed by the absence or presence of diagnostic motifs identified by previous studies as well as by use of the ScoreDiff web tool (https://akitsche.shinyapps.io/profileHMM_App) [78–80]. Alignments with indicated motifs are given in Fig. S9, Supplementary data.
4.8.2. Analysis of gene clusters
Clusters were analyzed using PRISM [70], antiSMASH [51], HMMER3 [85], and BLAST [71]. The function of genes was inferred by homology as identified by BLAST against the nr database from GenBank [71, 103]. PKS domains were identified with antiSMASH [51] and PRISM [70]. To generate bafilomycin biosynthetic protein pHMMs, the sequences of homologues of BafB–H, BafX–Z, and ORF1–3 from S. lohii, K. setae, S. varsoviensis, S. hygroscopicus subsp. hygroscopicus, and S. neyagawaensis were aligned using MUSCLE [101] on default settings; pHMMs were generated from the alignments using HMMER3. For analysis of predicted hygrolide producers, the local genomic region (+/− 16 genes) of S. lohii BafD or ORF3 homologues, as identified by BLAST, were analyzed using HMMER3 against a set of custom pHMMs. Clusters containing several pHMM hits were then analyzed by BLAST to determine similarity of BafD and ORF3 homologues to those from known BGCs. Clusters were classified on the basis of overall architecture and amino acid percent identity of BafD and ORF3 homologues to the corresponding proteins from S. lohii (bafilomycin), K. setae (bafilomycin), S. varsoviensis (hygrobafilomycin), S. hygroscopicus subsp. hygroscopicus (hygrolidin), and S. neyagawaensis (concanamycin). 16S rRNA sequences were not available for some organisms, nor were genomes and/or BafD/ORF3 sequences available for all organisms for which 16S rRNA sequences were obtained. As no micromonospolide BGC has been reported, producers were grouped together by 16S rRNA sequence alone.
4.8.3. Generation of phylogenetic trees
16S rRNA sequences were downloaded from the SILVA database [104]. Alignment of sequences was done using MUSCLE with default parameters within MEGA6 [101, 105]. Phylogenetic trees were created in MEGA6 using the maximum-likelihood method with default parameters [105]. Trees were annotated using FigTree (v.1.4.2; http://tree.bio.ed.ac.uk/software/figtree/) and Adobe Illustrator. The actinomycete Frankia alni was used as an outgroup to root the tree in all cases.
Supplementary Material
Acknowledgments
This work was supported in part by a NIH Director’s New Innovator Award Program (DP2 OD008463 to D.A.M.), the David and Lucile Packard Fellowship for Science and Engineering (to D.A.M.), the Robert C. and Carolyn J. Springborn Endowment (to J.I.T.). P.M.B. and J.I.T. were supported in part supported by ACS Division of Medicinal Chemistry predoctoral fellowships. The Bruker UltrafleXtreme MALDI-TOF/TOF mass spectrometer was purchased in part with a grant from the National Center for Research Resources, National Institutes of Health (S10 RR027109 A). We thank Parth Patel for contribution of bioinformatics code used for gene cluster analysis. We also thank Dr. Courtney L. Cox for helpful bioinformatics advice and Dr. Lingyang Zhu (Univ. Illinois NMR Laboratory) for assistance with collection of NMR spectra.
Abbreviations
- DiOC2(3)
3,3′-diethyloxacarbocyanine iodide
- ACP
acyl carrier protein
- AT
acyltransferase
- BGC
biosynthetic gene cluster
- CCDC
Cambridge Crystallographic Data Centre
- CID
collision-induced dissociation
- DIC
differential interference contrast
- DTT
dithiothreitol
- HMM
hidden Markov model
- KR
ketoreductase
- MBC
minimum bactericidal concentration
- MIC
minimum inhibitory concentration
- MOA
mode of action
- ORF
open reading frame
- PKS
Polyketide synthase
- pHMM
profile hidden Markov model
- PI
propidium iodide
- RiPP
ribosomally synthesized and post-translationally modified peptide
- SERCA
sarco/endoplasmic reticulum Ca2+-ATPase
- SAR
structure-activity relationships
- TEII
type II thioesterase
- V-ATPase
vacuolar-type H+-ATPase
- WGS
whole-genome sequencing
Footnotes
Supplementary Data
Supplementary data associated with this article can be found, in the online version, at: http doi link
Contributor Information
Evelyn M. Molloy, Email: emolloy@illinois.edu.
Jonathan I. Tietz, Email: tietz2@illinois.edu.
Patricia M. Blair, Email: pmblair2@illinois.edu.
Douglas A. Mitchell, Email: douglasm@illinois.edu.
References
- 1.Bindseil KU, Zeeck A. Liebigs Ann. Chem. 1994:305–312. [Google Scholar]
- 2.Werner G, Hagenmaier H, Drautz H, Baumgartner A, Zahner H. J. Antibiot. 1984;37:110–117. doi: 10.7164/antibiotics.37.110. [DOI] [PubMed] [Google Scholar]
- 3.Kretschmer A, Dorgerloh M, Deeg M, Hagenmaier H. Agric. Biol. Chem. 1985;49:2509–2511. [Google Scholar]
- 4.Carr G, Williams DE, Diaz-Marrero AR, Patrick BO, Bottriell H, Balgi AD, Donohue E, Roberge M, Andersen RJ. J. Nat. Prod. 2010;73:422–427. doi: 10.1021/np900632r. [DOI] [PubMed] [Google Scholar]
- 5.Zhang DJ, Wei G, Wang Y, Si CC, Tian L, Tao LM, Li YG. J. Antibiot. 2011;64:391–393. doi: 10.1038/ja.2011.12. [DOI] [PubMed] [Google Scholar]
- 6.Kobayashi K, Fukuda T, Usui T, Kurihara Y, Kanamoto A, Tomoda H. J. Antibiot. 2015;68:126–132. doi: 10.1038/ja.2014.100. [DOI] [PubMed] [Google Scholar]
- 7.Li J, Lu C, Shen Y. J. Antibiot. 2010;63:595–599. doi: 10.1038/ja.2010.95. [DOI] [PubMed] [Google Scholar]
- 8.Moon SS, Hwang WH, Chung YR, Shin J. J. Antibiot. 2003;56:856–861. doi: 10.7164/antibiotics.56.856. [DOI] [PubMed] [Google Scholar]
- 9.Seto H, Akao H, Furihata K, Otake N. Tetrahedron Lett. 1982;23:2667–2670. [Google Scholar]
- 10.Seto H, Tajima I, Akao H, Furihata K, Otake N. J. Antibiot. 1984;37:610–613. doi: 10.7164/antibiotics.37.610. [DOI] [PubMed] [Google Scholar]
- 11.Tchize N, dejouong Ble S, Sattler I, Maier A, Kelter G, Menzel KD, Fiebig HH, Hertweck C. J. Antibiot. 2010;63:359–363. doi: 10.1038/ja.2010.52. [DOI] [PubMed] [Google Scholar]
- 12.Hertweck C. Angew. Chem. Int. Ed. Engl. 2009;48:4688–4716. doi: 10.1002/anie.200806121. [DOI] [PubMed] [Google Scholar]
- 13.Johnston C, Ibrahim A, Magarvey N. Medchemcomm. 2012;3:932–937. [Google Scholar]
- 14.Tietz JI, Mitchell DA. Curr. Top. Med. Chem. 2015 doi: 10.2174/1568026616666151012111439. [DOI] [PubMed] [Google Scholar]
- 15.Nett M. In: Progress in the Chemistry of Organic Natural Products. Kinghorn AD, Falk H, Kobayashi J, editors. Vol. 99. Switzerland: Springer; 2014. pp. 199–245. [DOI] [PubMed] [Google Scholar]
- 16.van Schalkwyk DA, Chan XW, Misiano P, Gagliardi S, Farina C, Saliba K. J. Biochem. Pharmacol. 2010;79:1291–1299. doi: 10.1016/j.bcp.2009.12.025. [DOI] [PubMed] [Google Scholar]
- 17.Goetz MA, Mccormick PA, Monaghan RL, Ostlind DA, Hensens OD, Liesch JM, Albersschonberg G. J. Antibiot. 1985;38:161–168. doi: 10.7164/antibiotics.38.161. [DOI] [PubMed] [Google Scholar]
- 18.Robinson DG, Albrecht S, Moriysu Y. Protoplasma. 2004;224:255–260. doi: 10.1007/s00709-004-0070-6. [DOI] [PubMed] [Google Scholar]
- 19.Crevelin EJ, Canova SP, Melo IS, Zucchi TD, da Silva RE, Moraes LA. Appl. Biochem. Biotechnol. 2013;171:1602–1616. doi: 10.1007/s12010-013-0418-5. [DOI] [PubMed] [Google Scholar]
- 20.Bayer N, Schober D, Prchla E, Murphy RF, Blaas D, Fuchs R. J. Virol. 1998;72:9645–9655. doi: 10.1128/jvi.72.12.9645-9655.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yeganeh B, Ghavami S, Kroeker AL, Mahood TH, Stelmack GL, Klonisch T, Coombs KM, Halayko A. J. Am. J. Physiol. Lung Cell. Mol. Physiol. 2015;308:L270–L286. doi: 10.1152/ajplung.00011.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ochiai H, Sakai S, Hirabayashi T, Shimizu Y, Terasawa K. Antiviral Res. 1995;27:425–430. doi: 10.1016/0166-3542(95)00040-s. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki T, Yamaya M, Sekizawa K, Hosoda M, Yamada N, Ishizuka S, Nakayama K, Yanai M, Numazaki Y, Sasaki H. Am. J. Physiol. Lung Cell. Mol. Physiol. 2001;280:L1115–L1127. doi: 10.1152/ajprenal.2001.280.6.F1115. [DOI] [PubMed] [Google Scholar]
- 24.Furuchi T, Aikawa K, Arai H, Inoue K. J. Biol. Chem. 1993;268:27345–27348. [PubMed] [Google Scholar]
- 25.Naganuma S, Kuzuya N, Sakai K, Hasumi K, Endo A. Biochim. Biophys. Acta. 1992;1126:41–48. doi: 10.1016/0005-2760(92)90214-g. [DOI] [PubMed] [Google Scholar]
- 26.Woo JT, Shinohara C, Sakai K, Hasumi K, Endo A. Eur. J. Biochem. 1992;207:383–389. doi: 10.1111/j.1432-1033.1992.tb17061.x. [DOI] [PubMed] [Google Scholar]
- 27.Pivtoraiko VN, Harrington AJ, Mader BJ, Luker AM, Caldwell GA, Caldwell KA, Roth KA, Shacka JJ. J. Neurochem. 2010;114:1193–1204. doi: 10.1111/j.1471-4159.2010.06838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shacka JJ, Klocke BJ, Roth KA. Autophagy. 2006;2:228–230. doi: 10.4161/auto.2703. [DOI] [PubMed] [Google Scholar]
- 29.Ma B, Xiang Y, An L. Cell Signal. 2011;23:1244–1256. doi: 10.1016/j.cellsig.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 30.Bowman EJ, Siebers A, Altendorf K. Proc. Natl. Acad. Sci. U.S.A. 1988;85:7972–7976. doi: 10.1073/pnas.85.21.7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drose S, Altendorf K. J. Exp. Biol. 1997;200:1–8. doi: 10.1242/jeb.200.1.1. [DOI] [PubMed] [Google Scholar]
- 32.Forgac M. Nat. Rev. Mol. Cell Biol. 2007;8:917–929. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- 33.Qin A, Cheng TS, Pavlos NJ, Lin Z, Dai KR, Zheng MH. Int. J. Biochem. Cell Biol. 2012;44:1422–1435. doi: 10.1016/j.biocel.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 34.Lu X, Chen L, Chen Y, Shao Q, Qin W. Exp. Ther. Med. 2015;10:1829–1834. doi: 10.3892/etm.2015.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan N, Song L, Zhang S, Lin W, Cao Y, Xu F, Fang Y, Wang Z, Zhang H, Li X, Wang Z, Cai J, Wang J, Zhang Y, Mao X, Zhao W, Hu S, Chen S, Wang J. Haematologica. 2015;100:345–356. doi: 10.3324/haematol.2014.113324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hernandez A, Serrano-Bueno G, Perez-Castineira JR, Serrano A. Curr Pharm Des. 2012;18:1383–1394. doi: 10.2174/138161212799504821. [DOI] [PubMed] [Google Scholar]
- 37.Mauvezin C, Nagy P, Juhasz G, Neufeld TP. Nat Commun. 2015;6:7007. doi: 10.1038/ncomms8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teplova VV, Tonshin AA, Grigoriev PA, Saris NE, Salkinoja-Salonen MS. J Bioenerg Biomembr. 2007;39:321–329. doi: 10.1007/s10863-007-9095-9. [DOI] [PubMed] [Google Scholar]
- 39.Niikura K. Drug News Perspect. 2006;19:139–144. doi: 10.1358/dnp.2006.19.3.977442. [DOI] [PubMed] [Google Scholar]
- 40.Gagliardi S, Rees M, Farina C. Curr Med Chem. 1999;6:1197–1212. [PubMed] [Google Scholar]
- 41.Drose S, Bindseil KU, Bowman EJ, Siebers A, Zeeck A, Altendorf K. Biochemistry. 1993;32:3902–3906. doi: 10.1021/bi00066a008. [DOI] [PubMed] [Google Scholar]
- 42.Zhang W, Fortman JL, Carlson JC, Yan J, Liu Y, Bai F, Guan W, Jia J, Matainaho T, Sherman DH, Li S. ChemBioChem. 2013;14:301–306. doi: 10.1002/cbic.201200743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hwang JY, Kim HS, Kim SH, Oh HR, Nam DH. AMB Express. 2013;3:24. doi: 10.1186/2191-0855-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim EY, Han JW, Lee JY, Kim BS. Folia Microbiol. 2012;57:543–550. doi: 10.1007/s12223-012-0173-y. [DOI] [PubMed] [Google Scholar]
- 45.Hertweck C. Trends Biochem Sci. 2015;40:189–199. doi: 10.1016/j.tibs.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 46.Weissman K. J. Nat Prod Rep. 2016;33:203–230. doi: 10.1039/c5np00109a. [DOI] [PubMed] [Google Scholar]
- 47.Ueda JY, Hashimoto J, Yamamura H, Hayakawa M, Takagi M, Shin-ya K. J. Antibiot. 2010;63:627–629. doi: 10.1038/ja.2010.104. [DOI] [PubMed] [Google Scholar]
- 48.Gross H, Stockwell VO, Henkels MD, Nowak-Thompson B, Loper JE, Gerwick WH. Chem. Biol. 2007;14:53–63. doi: 10.1016/j.chembiol.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 49.Bode HB, Reimer D, Fuchs SW, Kirchner F, Dauth C, Kegler C, Lorenzen W, Brachmann AO, Grun P. Chem. Eur. J. 2012;18:2342–2348. doi: 10.1002/chem.201103479. [DOI] [PubMed] [Google Scholar]
- 50.Cox CL, Tietz JI, Sokolowski K, Melby JO, Doroghazi JR, Mitchell DA. ACS Chem. Biol. 2014;9:2014–2022. doi: 10.1021/cb500324n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, Lee SY, Fischbach MA, Muller R, Wohlleben W, Breitling R, Takano E, Medema MH. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Medema MH, et al. Nat. Chem. Biol. 2015;11:625–631. doi: 10.1038/nchembio.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Werner G, Hagenmaier H, Albert K, Kohlshorn H, Drautz H. Tetrahedron Letters. 1983;24:5193–5196. [Google Scholar]
- 54.Chikanishi T, Koike Y, Washimi Y, Takeuchi T. JP2000302782A. 2000:7. [Google Scholar]
- 55.Mahmoud KAS. Univ. Göttingen. 2008 [Google Scholar]
- 56.Baker GH, Brown PJ, Dorgan RJJ, Everett JR, Ley SV, Slawin AMZ, Williams DJ. Tetrahedron Lett. 1987;28:5565–5568. [Google Scholar]
- 57.Karplus MJ. Am. Chem. Soc. 1963;85:2870. [Google Scholar]
- 58.Corey EJ, Ponder JW. Tetrahedron Lett. 1984;25:4325–4328. [Google Scholar]
- 59.Wu C, Medema MH, Lakamp RM, Zhang L, Dorrestein PC, Choi YH, van Wezel GP. ACS Chem Biol. 2016;11:478–490. doi: 10.1021/acschembio.5b00801. [DOI] [PubMed] [Google Scholar]
- 60.French GL. J Antimicrob Chemother. 2006;58:1107–1117. doi: 10.1093/jac/dkl393. [DOI] [PubMed] [Google Scholar]
- 61.Novo D, Perlmutter NG, Hunt RH, Shapiro HM. Cytometry. 1999;35:55–63. doi: 10.1002/(sici)1097-0320(19990101)35:1<55::aid-cyto8>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 62.Silver LL. Clin Microbiol Rev. 2011;24:71–109. doi: 10.1128/CMR.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Friedman L, Alder JD, Silverman JA. Antimicrob Agents Chemother. 2006;50:2137–2145. doi: 10.1128/AAC.00039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Melnyk AH, Wong A, Kassen R. Evol Appl. 2015;8:273–283. doi: 10.1111/eva.12196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alekshun MN, Levy SB. Asm News. 2004;70:451–456. [Google Scholar]
- 66.Warner DM, Yang QW, Duval V, Chen MJ, Xu YC, Levy SB. Antimicrob Agents Chemother. 2013;57:1935–1937. doi: 10.1128/AAC.02445-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sun J, Deng Z, Yan A. Biochem Biophys Res Commun. 2014;453:254–267. doi: 10.1016/j.bbrc.2014.05.090. [DOI] [PubMed] [Google Scholar]
- 68.Sato T, Yokota S, Uchida I, Okubo T, Usui M, Kusumoto M, Akiba M, Fujii N, Tamura Y. Front Microbiol. 2013;4:125. doi: 10.3389/fmicb.2013.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tavio MM, Aquili VD, Vila J, Poveda JB. J Med Microbiol. 2014;63:56–65. doi: 10.1099/jmm.0.063727-0. [DOI] [PubMed] [Google Scholar]
- 70.Skinnider MA, Dejong CA, Rees PN, Johnston CW, Li H, Webster AL, Wyatt MA, Magarvey NA. Nucleic Acids Res. 2015;43:9645–9662. doi: 10.1093/nar/gkv1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 72.Wenzel SC, Williamson RM, Grunanger C, Xu J, Gerth K, Martinez RA, Moss SJ, Carroll BJ, Grond S, Unkefer CJ, Muller R, Floss HG. J. Am. Chem. Soc. 2006;128:14325–14336. doi: 10.1021/ja064408t. [DOI] [PubMed] [Google Scholar]
- 73.Sun Y, Hong H, Gillies F, Spencer JB, Leadlay PF. ChemBioChem. 2008;9:150–156. doi: 10.1002/cbic.200700492. [DOI] [PubMed] [Google Scholar]
- 74.Schuhmann T, Grond S. J. Antibiot. 2004;57:655–661. doi: 10.7164/antibiotics.57.655. [DOI] [PubMed] [Google Scholar]
- 75.Kotowska M, Pawlik K. Appl. Microbiol. Biotechnol. 2014;98:7735–7746. doi: 10.1007/s00253-014-5952-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Karki S, Kwon SY, Yoo HG, Suh JW, Park SH, Kwon HJ. FEMS Microbiol. Lett. 2010;310:69–75. doi: 10.1111/j.1574-6968.2010.02048.x. [DOI] [PubMed] [Google Scholar]
- 77.Zhang WJ, Bolla ML, Kahne D, Walsh CT. J. Am. Chem. Soc. 2010;132:6402–6411. doi: 10.1021/ja1002845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Caffrey P. ChemBioChem. 2003;4:654–657. doi: 10.1002/cbic.200300581. [DOI] [PubMed] [Google Scholar]
- 79.Reid R, Piagentini M, Rodriguez E, Ashley G, Viswanathan N, Carney J, Santi DV, Hutchinson CR, McDaniel R. Biochemistry. 2003;42:72–79. doi: 10.1021/bi0268706. [DOI] [PubMed] [Google Scholar]
- 80.Kitsche A, Kalesse M. ChemBioChem. 2013;14:851–861. doi: 10.1002/cbic.201300063. [DOI] [PubMed] [Google Scholar]
- 81.Zheng J, Taylor CA, Piasecki SK, Keatinge-Clay AT. Structure. 2010;18:913–922. doi: 10.1016/j.str.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 82.Keatinge-Clay AT, Stroud RM. Structure. 2006;14:737–748. doi: 10.1016/j.str.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 83.Keatinge-Clay AT. Chem. Biol. 2007;14:898–908. doi: 10.1016/j.chembiol.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 84.Isogai A, Sakuda S, Matsumoto S, Ogura M, Furihata K, Seto H, Suzuki A. Agric. Biol. Chem. 1984;48:1379–1381. [Google Scholar]
- 85.Finn RD, Clements J, Arndt W, Miller BL, Wheeler TJ, Schreiber F, Bateman A, Eddy SR. Nucleic Acids Res. 2015;43:W30–W38. doi: 10.1093/nar/gkv397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu K, Chung L, Revill WP, Katz L, Reeves CD. Gene. 2000;251:81–90. doi: 10.1016/s0378-1119(00)00171-2. [DOI] [PubMed] [Google Scholar]
- 87.Sakuda S, Isogai A, Matsumoto S, Ogura M, Furihata K, Seto H, Suzuki A. Agric. Biol. Chem. 1987;51:2841–2842. [Google Scholar]
- 88.Ohta E, Ohta S, Kubota NK, Suzuki M, Ogawa T, Yamasaki A, Ikegami S. Tetrahedron Lett. 2001;42:4179–4181. [Google Scholar]
- 89.Kumagai K, Fukui A, Tanaka S, Ikemoto M, Moriguchi K, Nabeshima S. J. Antibiot. 1993;46:1139–1144. doi: 10.7164/antibiotics.46.1139. [DOI] [PubMed] [Google Scholar]
- 90.Arnison PG, et al. Nat. Prod. Rep. 2013;30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Omura S, Imamura N, Hinotozawa K, Otoguro K, Lukacs G, Faghih R, Tolmann R, Arison BH, Smith JL. J. Antibiot. 1983;36:1783–1786. doi: 10.7164/antibiotics.36.1783. [DOI] [PubMed] [Google Scholar]
- 92.Hayakawa Y, Takaku K, Furihata K, Nagai K, Seto H. J. Antibiot. 1991;44:1294–1299. doi: 10.7164/antibiotics.44.1294. [DOI] [PubMed] [Google Scholar]
- 93.Osada H, Koshino H, Isono K, Takahashi H, Kawanishi G. J. Antibiot. 1991;44:259–261. doi: 10.7164/antibiotics.44.259. [DOI] [PubMed] [Google Scholar]
- 94.Fremlin L, Farrugia M, Piggott AM, Khalil Z, Lacey E, Capon R. J. Org. Biomol. Chem. 2011;9:1201–1211. doi: 10.1039/c0ob00654h. [DOI] [PubMed] [Google Scholar]
- 95.Takahashi S, Toyoda A, Sekiyama Y, Takagi H, Nogawa T, Uramoto M, Suzuki R, Koshino H, Kumano T, Panthee S, Dairi T, Ishikawa J, Ikeda H, Sakaki Y, Osada H. Nat. Chem. Biol. 2011;7:461–468. doi: 10.1038/nchembio.583. [DOI] [PubMed] [Google Scholar]
- 96.Gerlt JA, Bouvier JT, Davidson DB, Imker HJ, Sadkhin B, Slater DR, Whalen KL. Biochim. Biophys. Acta. 2015;1854:1019–1037. doi: 10.1016/j.bbapap.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Newman DJ, Cragg GM. Journal of Natural Products. 2012;75:311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kwon YJ, Sohn MJ, Zheng CJ, Kim WG. Org. Lett. 2007;9:2449–2451. doi: 10.1021/ol0703231. [DOI] [PubMed] [Google Scholar]
- 99.Kamiyama T, Umino T, Nakayama N, Itezono Y, Satoh T, Yamashita Y, Yamaguchi A, Yokose K. J. Antibiot. 1992;45:424–427. doi: 10.7164/antibiotics.45.424. [DOI] [PubMed] [Google Scholar]
- 100.Clinical and Laboratory Standards Institute. CLSI document M07-A9. Wayne, PA, USA: CLSI; 2012. [Google Scholar]
- 101.Edgar RC. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li W, Cowley A, Uludag M, Gur T, McWilliam H, Squizzato S, Park YM, Buso N, Lopez R. Nucleic Acids Res. 2015;43:W580–W584. doi: 10.1093/nar/gkv279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Benson DA, Karsch-Mizrachi I, Clark K, Lipman DJ, Ostell J, Sayers EW. Nucleic Acids Res. 2012;40:D48–D53. doi: 10.1093/nar/gkr1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.