Abstract
Background
Epigenetic factors, including DNA methylation, play an important role in the etiology of alcohol use disorders. Non-candidate based methylome-wide studies leveraging multiple tissue types are needed to identify a set of CpG targets that reliably differentiate alcohol use disorder (AUD) patients from controls and strongly correlate across brain tissue and more commonly collected tissue types (e.g., buccal cells).
Methods
Post-mortem precuneus brain tissue samples were collected from 49 alcohol dependent (AD) cases and 47 controls (sample I), and DNA was extracted from precuneus and putamen brain tissue and buccal cells in 24 post-mortem subjects (sample II). Methylation levels were analyzed at over 450,000 CpG sites in both samples. CpGs that demonstrated significant methylation differences between cases and controls were advanced for further analysis with the goal of identifying CpGs that also demonstrated consistent correlations across tissue type.
Results
In the primary analysis, 244 hypomethylated and 188 hypermethylated CpGs, met a priori criteria for both significant differences between cases and controls as well as significant correlation across brain and buccal cell tissue types, employing stringent Bonferroni p-value correction. Many of these CpGs were involved in gene networks related to lipid metabolism, immune response, inflammatory response/disease, and gastro intestinal disease.
Conclusions
More than four hundred CpGs demonstrated differences in methylation between AD cases and controls and showed significant correlation across tissue types. Several genes and pathways (e.g., inflammation and immune functioning) that have been previously associated with AUD were identified in the current analyses.
Keywords: Methylation, Alcohol Use Disorder, Epigenetics, Addiction, inflammation
Introduction
The pathogenesis of alcohol use disorder (AUD) is highly complex, including genetic, developmental, and cultural factors. In addition, mounting evidence also implicates epigenetic factors in the neurobiology of addiction (Nestler, 2014) and theoretical models have been revised to include epigenetic processes that may account for an important portion of the variance in the etiology of alcohol use disorders (Karoly et al., 2013). Epigenetics refer to molecular processes that alter gene expression without altering the DNA sequence itself (Nestler, 2014). Through these processes, the protein complex that organizes DNA (e.g. chromatin) is modified, which alters the accessibility of genes to transcription factors, the proteins that bind to specific DNA sequences and regulate gene transcription. Specifically, methylation is a particular epigenetic modification involving the addition of a methyl group to the fifth carbon of cytosine residues. Methylation is associated with downstream gene expression changes and has been shown to elicit long-lasting alterations to phenotypes, including those related to AUDs (Ponomarev et al., 2012).
Emerging evidence suggests that aberrant methylation patterns might be associated with heavy alcohol consumption and alcohol use disorders. For example, recent studies discussed below, have found methylation differences between AUD patients and controls. Broadly, AUD is associated with differential methylation patterns (e.g. Weng et al., 2014). Studies have demonstrated that CpG hypermethylation is associated with AUDs. Specifically, DNA methylation levels in promoter regions of several theoretically important genes have been associated with AUD, including the dopamine transporter gene (SLC6A3) (Hillemacher et al., 2009a), atrial natriuretic peptide (ANP) and vasopressin precursor genes (Hillemacher et al., 2009b), and the alpha synuclein gene (Bönsch et al., 2005). An analysis of 384 CpGs in the promoter regions of 82 candidate genes revealed that a number of CpGs, demonstrated significant hypermethylation between AUD cases and controls (Zhang et al., 2013a). Likewise a methylome-wide analysis demonstrated that 865 hypomethylated and 716 hypermethylated CG sites differentiated AUD patients from controls (Zhao et al., 2013). However, a number of studies have reported no methylation differences between cases versus controls at specific gene sites (Muschler et al., 2010; Park et al., 2011; Philibert et al., 2008).
Importantly, methylation is often cell type specific (Khavari, Sen & Rinn, 2010). Therefore, methylation patterns should ideally be measured in the tissue type that is most proximal to the phenotype of interest (Harlaar et al., 2013). Clearly, human brain tissue would be the most ideal tissue type by which to study methylation patterns in AUD. Unfortunately, practical constraints do not allow for brain tissue sampling from live human subjects. Thus, methodological approaches for examining methylation patterns are limited to peripheral tissue samples, such as blood and saliva and post mortem brain tissue samples. One reason for inconsistent results across studies to date may be that methylation at particular loci is consistent across tissue types while methylation at other loci is not.
The aims of the study were two-fold. The first aim was to conduct an exploratory methylome-wide investigation of DNA from post-mortem brain tissue, comparing 49 alcohol dependent subjects to 47 age and gender matched controls in order to identify CpG sites that differentiated cases and controls. The second aim was to examine the consistency of methylation across those CpGs in DNA extracted from post-mortem brain tissue and buccal cells in a separate sample. Therefore, this study is uniquely poised to identify methylated areas in the genome that are both implicated in AUD and consistent across brain and peripheral tissue types.
Materials and methods
Study Samples
The first sample set (sample set I) consisted of frozen precuneus brain tissue samples from the 49 cases that met a DSM-IV diagnosis of alcohol dependence and 47 controls. Precuneus brain tissue was chosen based on previous analyses done in our lab showing associations between precuneus activation and AUD phenotypes (Liu, Calhoun, Chen, Claus & Hutchison, 2013). The sample was predominantly Caucasian (97.9 % in AUD group; 87.2% in control group). The sample groups were matched on age and gender (see table 1). Samples were obtained from the New South Wales and Victorian Tissue Resource Centre. The clinical diagnosis of lifetime alcohol dependence was confirmed by physician interviews, review of hospital medical records, questionnaires to next-of-kin, and from pathology, radiology and neuropsychology reports. Cause of death was determined by medical records and given to us along with the samples by the New South Wales and Victorian Tissue Resource Centre. Information regarding cause of death by diagnosis category can be found in table 2.
Table 1.
Matched Characteristics of Sample Set I
| AD (n=49) | Control (n=47) | Test | P | |
|---|---|---|---|---|
| Age | M= 47.76 SD=7.78 |
M= 48.43 SD= 8.38 |
t(92.8)=.41 | .69 |
| Sex | n female = 11 n male = 38 |
n female = 12 n male = 35 |
χ2(1, N=96) = .013 |
.91 |
| PMI | M= 34.58 SD= 11.05 |
M= 27.98 SD=13.98 |
t(90.7)= −2.57 | .01 |
Sample Set I consists of frozen precuneus brain tissue samples from 49 alcohol dependent cases and 47 neurologically-normal controls obtained from the New South Wales and Victorian Tissue Resource Centre; AD, alcohol dependent cases; PMI, post mortem interval in hours; M, mean; SD, standard deviation; p, p-values obtained from Welch’s t-test/Pearson’s χ2.
Table 2.
Cause of Death Frequency Counts Within the Australian Samples.
| Cardiac | Toxicity | Respiratory | Trauma | Hepatic | Blood Loss |
Vascular | Obesity | Neurological | Other/ Unknown |
|
|---|---|---|---|---|---|---|---|---|---|---|
| AD Cases |
14 | 12 | 9 | 1 | 5 | 3 | 1 | 1 | 3 | |
| Controls | 37 | 1 | 4 | 1 | 1 | 3 |
Cause of death frequency counts from 49 alcohol dependent cases and 47 neurologically-normal controls obtained from the New South Wales and Victorian Tissue Resource Centre; AD Cases, alcohol dependent cases; Controls, non alcohol dependent control samples.
To examine consistency across tissue type, postmortem brain samples were obtained through collaboration with The Office of the Medical Investigator (OMI) in Albuquerque, NM. Criteria for selection of descendants were as follows: age range 25 to 50 years old, post mortem interval less than 72 hours and no obvious trauma to the head or brain. As part of the normal autopsy process, bilateral precuneus and putamen tissues were dissected from brain slices and cheek cells were obtained using cytology brushes also at time of autopsy. The sample included 12 males and 12 females, and an ethnicity breakdown as follows: 18 Caucasian, 5 Hispanic, 1 American Indian. Age ranged from 37 to 50 years (M= 41.38, SD=7.48). The average reported post mortem interval for this sample was 25.89 hours (SD=21.05).
DNA preparation and methylation assay
Tissue samples were stored frozen at −80°C until they were pulled for nucleic acid preparation. Genomic DNA was isolated from the brain tissue and cheek cell samples using Qiagen®’s Puregene DNA prep kit (Qiagen®, Venlo, Limburg). DNA yields were determined via PicoGreen® and fluorimetry (Qubit®, Life Technologies). Whole genome methylation interrogation was performed using The Illumina® Infinium® Assay Platform in conjunction with the Infinium HumanMethylation450 BeadChip. The array interrogates approximately 480,000 CpG sites and covers 99% of RefSeq genes. The array covers an average of 17 CpG sites per gene region distributed across the promoter, 5'UTR, first exon, gene body, and 3'UTR. Given the exploratory nature of our study, we included all interrogated sites in our analyses. 600 ng of genomic DNA was treated with sodium bisulfite using the Zymo EZ DNA Methylation Kit to convert unmethylated cytosines to uracil, while methylated cytosines remain unchanged. The DNA was then purified and quantified by measuring absorbance at 260 nm in preparation for whole genome amplification, followed by fragmentation and ethanol precipitation. The DNA was then resuspended in hybridization buffer and applied to the bead chip array for an overnight incubation. Following hybridization, the arrays were washed to eliminate un-hybridized and non-specifically hybridized DNA. The samples then underwent single base extension and staining followed by more washing. The arrays were allowed to dry and then scanned using the Illumina iScan system.
Methylation interrogation was accomplished via two probe types. Type I probes incorporated florescent labels for detection at allele-specific single base extensions. Type II probes employed green and red dye colors in order to detect M and U signals, respectively, using one probe per CpG locus. Illumina’s GenomeStudio software was used to quantify results and annotate each site (e.g. transcription site proximity, gene name, and presence of a single nucleotide polymorphism (SNP)). Summaries of the probe interrogations yield average signals for methylated (M) and unmethylated (U) alleles at each CpG site, which are used to compute a β-value, such that:
For example, a β-value of 0 would indicate an unmethylated CpG site, while a β-value of 1 indicates a fully methylated site. These methylation β values were based on precuneus tissue samples for sample set I. In sample set II, methylation β values were calculated from the brain tissue from sample set II, and a separate estimate was given for buccal cells.
Using sample set I, differential methylation between the case and control groups was calculated for each CpG (DiffScore). In order to quantify significant methylation differences between cases and controls across CpGs, the DiffScores were converted to p values (Illumina Genome Studio Support, 2015). For a more detailed explanation of the equations used to make these calculations, see the GenomeStudio Methylation Module v1.8 User Guide by Illumina.
Analysis Plan
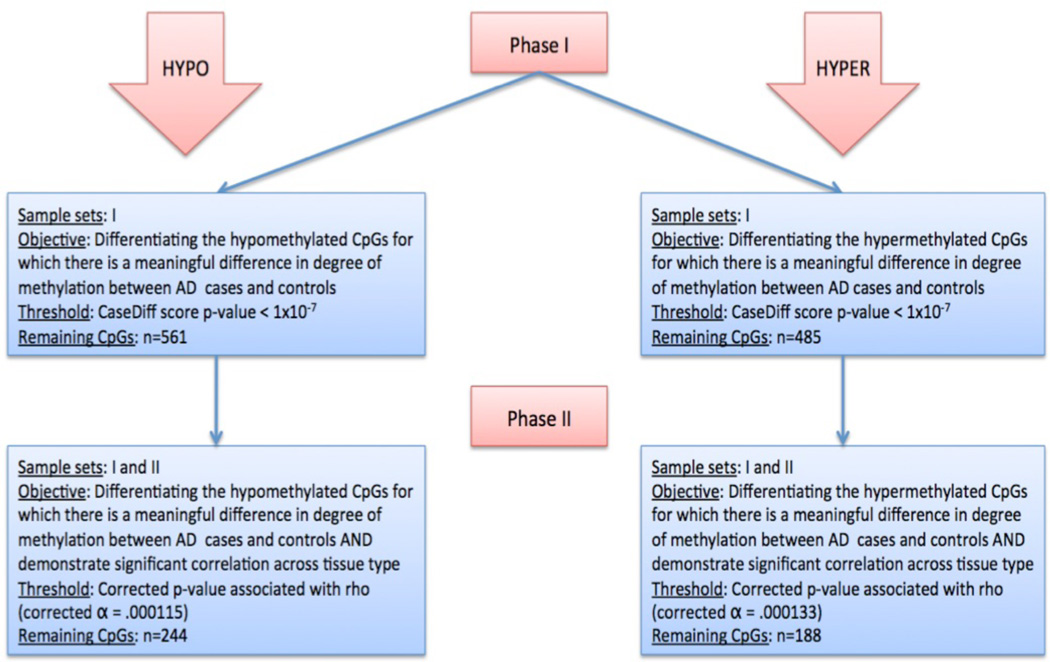
The aims of the present study were twofold: 1) to identify CpGs methylome-wide for which methylation at the CpG level is significantly different between cases versus controls, 2) to use a separate sample to determine whether these phenotypically relevant CpGs demonstrate consistent levels of methylation across tissue types (e.g. brain tissue and buccal cells). The analyses followed the outline provided below (also see figure 1 for a flow chart of the primary analysis).
Figure 1.
Flow chart outlining the objectives, samples, statistical thresholds, and number of CpGs tested at each phase of the primary analysis.
Phase I
The objective of phase I was to detect CpGs for which their case difference score demonstrated a meaningful difference of degree of methylation between cases and controls. The a priori p-value threshold was set at 1×10−7, corresponding to a case difference score of |70|.
Phase II
For the significantly hypomethylated and hypermethylated CpG sites in cases vs. controls, phase II examined the correlation of methylation β values between buccal cells and the methylation β value from the brain tissue at each CpG site in sample set II. The objective of this phase was to identify the remaining hypomethylated and hypermethylated CpGs that demonstrated significant correlation across tissue type. That is, hypomethylated and hypermethylated CpGs that advanced through phase I were tested in phase II against an a priori cross tissue type correlation threshold, stringently corrected for multiple tests. The p-value threshold for the hypomethylated and hypermethylated CpG sets was set at a Bonferroni corrected alpha based on the number of significantly hypomethylated and hypermethylated CpGs identified in phase 1.
The Australian samples are meant to provide insight into methylation differences relative to the AUD phenotype, and the NM samples are meant to provide insight into within-subject methylation consistency across tissue types and are not related to the AUD phenotype (e.g. neither classified as cases nor controls). Therefore, we would not necessarily expect consistent methylation measurements between the Australian samples and the New Mexico samples. For this reason, testing the methylation consistency across the two sample sets is not relevant to our study aims.
In order to understand how the genes identified by inferential statistics in phases I and II are functionally relevant and interrelated, functional network analyses were executed using Ingenuity Pathways Analysis tools (Ingenuity Systems, Mountain View, CA), a web-delivered application that enables the discovery, visualization, and exploration of interactional networks in genetic data. The CpG sites identified using phases I and II of the analysis were uploaded into the Ingenuity pathway analysis. Each CpG site was mapped to its corresponding gene symbol location and derivative molecule in the Ingenuity pathway knowledge base. These genes were then used as a starting point for generating biological networks, where genes from our CpG lists were combined based on their interconnectedness with each other relative to all genes and derivative molecules within the Ingenuity Knowledge base. A score was then computed for each network according to the degree of fit or connectedness of the identified gene set. This score reflects the negative logarithm of the P that indicates the likelihood of the focus genes in a network being found together due to random chance and was calculated with the right-tailed Fisher's Exact Test. Significance was then assigned to each network by determining a P for the enrichment of the genes in the network for such functions compared with the whole Ingenuity pathway knowledge base as a reference set. As an example, suppose that a network of 35 genes has a Fisher Exact Test result of 1×10−6. The network’s Score = −log(Fisher's Exact test result) = 6. In other words, there is a 1 in a million chance of getting a network containing at least the same number of connected genes by chance when randomly picking 35 genes that can be in networks from the Ingenuity Knowledge Base (Ingenuity Systems, Mountain View, CA). Using a 99% confidence level, scores of >3 were considered significant.
Results
Phase I
The goal of phase 1 was to identify hypermethylated and hypomethylated sites that were significantly different between cases and controls. Using the a priori threshold of p < 1×10−7 to differentiate between cases and controls, 561 hypomethylated CpGs and 485 hypermethylated CpGs were identified that demonstrated significant methylation differences between alcohol dependent cases and controls.
Phase II
Of the 561 hypomethylated CpGs, 436 were also available for analysis in sample set II, and 377 of the 485 hypermethylated CpGs were available for analysis. These hypomethylated and hypermethylated CpG sites identified in phase I were then separately examined for significant correlations among the average methylation β from the brain tissue and methylation β from buccal cells at the corresponding CpG site. The Rho value is a rank order correlation measurement. That is, the Rho represents the degree to which the methylation beta value of a CpG site in the brain tissue is correlated with the methylation beta value in the buccal cells. We used the p-value associated with the Rho in order to determine for which CpG sites methylation beta values were significantly correlated. The Bonferroni corrected alpha threshold for cross-tissue type correlation in the hypomethylated CpG set was .000115 (.05/436), and the corrected alpha threshold cross-tissue type correlation in the hypermethylated CpG set was .000133 (.05/377). Based on these respective thresholds, a total of 244 hypomethylated CpGs (see supplementary table 1) and 188 hypermethylated CpGs (see supplementary table 2) that had been identified in phase 1, also met the criteria for significant correlation across both peripheral and neural tissues in phase II. That is to say, data converged from two independent samples (e.g. sample set I and II) to suggest that 432 total CpG sites (244 hypomethylated and 188 hypermethylated) demonstrated significant methylation differences between cases and controls as well as significant cross tissue type correlation between brain tissue and buccal cells. Tables containing the resulting hypomethylated and hypermethylated CpGs, can be found in the supplementary materials. Within these supplementary tables, readers will find a comprehensive information on each CpG marker, including: the location of the SNP (if applicable), gene name corresponding to the CpG marker, the distance to the transcription site (measured in base pairs), and the chromosome and coordinate information. Additionally, these supplementary tables contain the average methylation β values for AD cases and controls, the methylation difference score, the corresponding difference score p-value, the correlation of methylation across tissue types, and the corresponding p-value.
Functional network analysis
To understand how the CpG sites identified through phases I and II may be functionally relevant and interrelated, the hyper- and hypo- methylated CpG sets from the primary analysis were then placed in the context of present knowledge about gene networks and molecular interactions, using the Ingenuity knowledge base. Based on the computed network scores from the primary analysis, several functional networks were found to be significant in the hyper- and hypo- methylated gene sets at significance level ranging from P = 17–51 (see table 3). The ability to rank the networks based on their functional connectedness over and above random chance allowed for rapid prioritization of networks with highest empirical relevance. Across the hyper- and hypo- methylated sets that emerged from the primary analysis, the top-ranking functional networks were most commonly related to neurodevelopment and neurobehavior, lipid metabolism, and inflammatory immune responses.
Table 3.
Functional network analyses of hypermethylated and hypomethlyated gene sets derived from the primary analysis
| ID | Genes Related to the Identified Network | Score | Number of Genes | Top Diseases and Functions |
|---|---|---|---|---|
| Hypermethylated-related networks | ||||
| 1 | AHSP, BOC, CBX7, CCND1, DBH, DHX9, MBD5, MMP17, NCR2, PCGF3, PNLIPRP2, PPP1R16B, PSMD13, SMAD6, SMYD2, TMEM165, TP53INP2 |
34 | 18 | Cellular Function and Maintenance, Neurodevelopmental Disorder, Hereditary Disorder |
| 2 | DAP, DOCK5, ELOVL5, INPP5A, IRS1, KLF4, LRAT, MYO15A, NDFIP1, PAH, PDGFA, RRAGC, SCD, SLC6A3, THBS2 |
30 | 16 | Lipid Metabolism, Molecular Transport, Small Molecule Biochemistry |
| 3 | ACOT1, CPAMD8, FAM101B, FARS2, FCHO1, KCNH5, MICAL3, MYEOV, NTPCR, PLEKHG4B, REEP3, SEC24D, TM9SF1, TSPAN10, TUBAL3, XXYLT1 |
30 | 16 | Lipid Metabolism, Nucleic Acid Metabolism, Small Molecule Biochemistry |
| 4 | AHRR, ARID3A, BATF, CCR6, CYB561, CYP2E1, HS6ST1, IRF8, MAP2K3, NOX4, TLR4, TLR6, TRIM29, ZBP1 |
27 | 15 | Humoral Immune Response, Protein Synthesis, Cell Morphology |
| 5 | B3GALT1, BAHD1, CYBRD1, DPF3, DTX2, FTCD, HCG4, HHLA2, RGPD4 (includes others), SLC26A1, SNORA70, UBL4B, WRB |
23 | 13 | Lipid Metabolism, Small Molecule Biochemistry, Cell Morphology |
| 6 | ABR, CDK18, CHST8, DRD4, GABRB3, GALNT9, GPR123, KCNN1, MSRA, NUDT1, PFDN4, RHPN1, SSTR5, STK25 |
22 | 14 | Behavior, Developmental Disorder |
| 7 | ATCAY, CLEC4F, FADS3, FAM120B, FAM173B, GRAMD4, HLA-DPA1, LMF1, NEGR1, OR2L13, UMODL1 |
19 | 12 | Cell-To-Cell Signaling and Interaction, Inflammatory Response |
| 8 | EPHA10, FEM1B, HOOK2, LRRK1, MICALL2, MRGPRX2, MYT1L, RP1L1, SDK1 |
17 | 11 | Cell-To-Cell Signaling and Interaction, Nervous System Development and Function |
| Hypomethylated-related networks | ||||
| 1 | AKR7A2, AP2A2, B4GALT6, calpain, CAPN8, DGKZ, DUSP5, GYPA, HMGCS1, HSPG2, HTATIP2, MAP2, Mek, MUC2, NCOA5, OCA2, OVGP1, PCSK9, PDGFRA, RFTN1, SCN1A, SCN4B, SORL1, SPRY1, SPTBN4, SULF2, TPO, |
51 | 25 | Cancer, Gastrointestinal Disease, Hepatic System Disease |
| 2 | AGPAT1, ATXN7L1, B3GNT7, C1orf109, CLSTN2, FMN2, GBX2, HAAO, ITIH3, KCNK17, LHX8, LRRN4, PDE11A, TAS1R2, ZNF607 |
30 | 17 | Cellular Development, Embryonic Development, Organismal Development |
| 3 | BAK1, BRDT, COX4I1, FANCA, FHIT, GALNT1, GLRX3, HIPK2, HISTONE, HLA-DQA1, HLA-DQB1, HLA-DRA, HLA-DRB5, HLA-L, NFATC1, REL, SDHB, SKP2 |
29 | 18 | Inflammatory Response, Endocrine System Disorders |
| 4 | ACSF3, ATP5G1, DYNC2H1, HERC6, KIF26B, MRTO4, MYOM2, NUPL1, PGRMC2, PHC1, SDK1, TRIM31, UBTD1, WDR41 |
24 | 14 | Energy Production, Nucleic Acid Metabolism, Small Molecule Biochemistry |
| 5 | EAPP, FGF22, KIAA1804, KRT82, MXRA7, SLC35E2, SLC39A8, SLC6A13, SOX11, SPATA18, TRIML2, VTRNA2-1, WDR60 |
22 | 14 | Organ Morphology, Cancer, Cell Cycle |
| 6 | CCDC57, DFNA5, FAM213B, FEM1B, GDAP2, KIAA0319, LRRC20, PYROXD1, RAB20, SLC12A8, SLC25A46, THUMPD1, TMEM9, TMEM100 |
21 | 13 | Neurodevelopmental Disorder, Neurological Disease |
Network connections – Functional network analysis results based on hypo- and hyper-methylated gene sets remaining after phases I and II of the primary analysis; ID, identification number of significant gene networks; Score, computed network score based on the Ingenuity knowledge base, used to rank the networks based on their functional connectedness over and above random chance; Number of Genes, number of genes in each network; Top Diseases and Functions, the top diseases and functions associated with each network based on the Ingenuity knowledge base.
Discussion
Due to the exploratory nature of the present study, it is important to first acknowledge the strengths and limitations of our study design and samples. One limitation of the current study is that the investigation is limited to the sites that are interrogated on the 450K array. It is therefore important to acknowledge that a truly methylome-wide approach would require approaches such as whole genome bisulfite sequencing (WGBS). Additionally, the current study was not able to look at cell-specific methylation in brain tissue. Importantly, some studies have shown that brain cellular heterogeneity may bias DNA methylation patterns (e.g. Guintivano, Aryee & Kaminsky, 2013). It is also worth noting that bisulfite conversion precludes testing multiple forms of DNA methylation (e.g., hydroxymethylation). Finally, only limited clinical information was available on the subjects who provided the tissue samples. Additional information regarding the severity/duration of alcohol use and additional information about substance use, disease states, and psychopathology would provide a more comprehensive picture of these samples and lend additional justification for case vs. control classification. However, information regarding the Australian brain samples is limited to that collected and compiled by the New South Wales Tissue Resource Centre (NSW TRC).
An additional limitation of our study was the fact that two separate samples were used to accomplish both aims of the study. First, the Australian brain samples were used in order to determine which CpG sites differentiated AD cases from controls (i.e. which CpG sites were phenotypically relevant). A separate sample was used to determine which CpG sites demonstrated consistent methylation beta values across brain and peripheral tissue types. We did not have both brain and peripheral tissue samples available to us through the NSW TRC, and thus, were limited by practical constraints of the samples that were available. DNA methylation differs across populations (Fraser, Lam, Neumann, Kobor, 2012) and our two sample sets are racially and ethnically divergent. However, there is no evidence to date to suggest that the consistency of DNA methylation across tissue types differs dramatically across populations. In any case, our approach should be seen as a preliminary proof of concept. Future studies may collect DNA from multiple tissues in the same sample to evaluate both the effect of the methylation on the phenotype as well as the consistency across tissue types.
One strength of the present study is that type I error was controlled with conservative p values. Although there have been very few studies of this nature published to date, two studies published using similar methodologies and designs incorporated less conservative thresholds than those employed here. For example, a recent methylome-wide study analyzed differences between cases and controls at the CpG evaluated 27,578 CpGs using a p value threshold of < .005 (Zhang et al., 2013b). Similarly, another methylome-wide study interrogated over 450,000 CpGs using an absolute value DiffScore of less than 20 as the threshold to differentiate between cases and controls, which correspondes to a p-value threshold of < .01 (Zhao et al. 2013). In contrast to these studies, the present study employed a more conservative approach of a threshold of abs(DiffScore) greater than 70 and a p-value of 1×10−7. Given the limited sample size and statistical power combined with conservative p values, the primary limitation of the current study is a high probability of a Type II error where the analyses failed to identify a number of CpGs that may differentiate alcohol cases from controls. Keeping in mind our exploratory study objectives and the current lack of statistical threshold consensus within this emerging literature, we acknowledge this limitation to our approach and look to future studies to replicate and probe further into our findings.
Findings and Impressions
Using two independent samples, the present study was designed to identify CpGs that 1) demonstrate methylation differences between alcohol dependent cases and controls and 2) to determine which of these phenotypically relevant CpGs demonstrate significant correlation across tissue type. The results of the primary analysis identified a total of 432 CpGs that passed rigorous statistical thresholds. Of these CpGs, 244 were classified as hypomethylated and 188 were classified as hypermethylated in cases as compared to controls and were significantly correlated across brain and buccal cell tissue types. Gene network analyses were used to identify the functional relevance of our identified gene sets from the primary analysis. These results suggested that the top-ranking functional networks were most commonly related to inflammation, immune system regulation, lipid metabolism, and gastrointestinal disease. These pathways and diseases have been previously associated with AUDs in the extant literature (Goral, Karavitiss & Kovacs, 2008; Cook, 1998; Baraona & Lieber, 1979; Schuckit, 2009), which adds credibility to the results of these exploratory analyses.
A number of the individual genes that emerged in the analyses have also been previously linked with AUDs in previous genetic and epigenetic studies, including dopamine receptor D4 (DRD4), dopamine beta hydroxylase (DBH), dopamine transporter (SLC6A3), cytochrome P450 2E1(CYP2E1) and toll-like receptor 4 (TLR4). DRD4, DBH, and SLC6A3 represent three genes from this list that fell within the top hypermethylated results. Previous studies suggest that these genes are all involved in the neurobiology of reward response. SLC6A3 functions as the dopamine transporter gene and is responsible for the reuptake of extracellular synaptic dopamine into presynaptic neurons, and therefore governs termination of dopaminergic transmission (Giros et al., 1992). Thus, it follows that different levels of SLC6A3 expression may have a significant impact on dopaminergic reward circuits (Drgon et al., 2006). Additionally, Hillemacher et al. (2009a) demonstrated that compared to healthy controls, the dopamine transporter (SLC6A3) promoter was significantly hypermethylated in alcohol dependent patients, and hypermethylation of SLC6A3 was associated with obsessive alcohol craving. Thus, the fact that a CpG within the SLC6A3 gene was identified as a top hypermethylated site with a high level of consistency across neural and peripheral tissues in our samples, lends evidence regarding the potential importance of epigenetic regulation of this gene.
Additionally, several studies suggest that the DBH gene is associated with AUD (Kato et al., 1979; Schuckit et al., 1981; Bagdy & Arató, 1987; Köhnke et al., 2006), and ethanol induced regulation of DBH has been observed (Hassan et al., 2003). In contrast to our hypermethylated finding, hypomethylation of the DBH gene was found to be higher in discordant twins who exhibited alcohol-dependent tendencies (Zhao et al., 2013). DBH is responsible for catalyzing the conversion of dopamine into to noradrenaline or norepinephrine (Joh & Hwang, 1987); thus, hypomethylation of DBH resulting from chronic alcohol exposure is suggested to contribute to alcohol tolerance by way of dampening the dopaminergic reward response (Zhao et al., 2013). Interestingly, a CpG within the DBH gene emerged in the hypermethylated final list of our primary results. These conflicting results suggest the potential importance of the DBH gene and highlight the complexity of methylation in the context of AUD, and implore the need for future replication studies. Finally, analysis of DNA from peripheral blood revealed that a CpG site in the DRD4 gene was significantly hypermethylated in cases compared with controls (Zhang et al., 2013a). This finding is consistent with the present results, and future studies might investigate how methylation of the DRD4 gene contributes to networks and disease pathways associated with AUD.
Consistent with our findings implicating inflammatory and immune networks, both animal and human research in recent years have empirically demonstrated the association between heavy alcohol exposure and the activation of key neuroinflammatory mediators in the brain. In our primary analysis, TLR4 emerged as a hypermethylated site. Previous studies indicate that alcohol exposure can trigger neuroinflammatory signaling cascades through the activation of the toll-like receptor-4 (TLR4) pathway (Guerri & Pascual, 2013; Crews et al., 2015). Evidence suggests that activation of this pathway results in prolonged microglial activation and the downstream production of additional proinflammatory cytokine mediators, which impact neuroimmue functioning. These neuroinflammatory processes lead to deleterious neurotoxic effects, including neurodegeneration (Fernandez-Lizarbe et al., 2009). Consistent with theoretical models of alcohol dependence, inflammation-induced neurotoxicity is a potential mechanism by which the control network becomes weaker throughout the course of disease progression, resulting in increased consumption and reduced inhibitions (Karoly, Harlaar & Hutchison, 2013). Given the prominent role of TLR4 in the neuroinflammatory response to alcohol, our results suggesting hypermethylation within TLR4 may be a promising target in terms of elucidating the epigenetic mechanisms underlying neuroadaptations in AUD.
The CYP2E1 gene, a significantly hypermethylated CpG site in the network associated with humoral immune response, protein synthesis, and cell morphology, demonstrates high cross tissue correlation in the current study, and has also garnered interest in the alcohol literature. CYP2E1 appears to be involved in the metabolism of exogenous compounds, including alcohol and is regulated by various transcriptional and post-transcriptional factors (Jones et al., 1992). Evidence suggests that ethanol induces CYP2E1, which in turn catalyzes ethanol oxidation and results in the formation of acetaldehyde. Increased CYP2E1 leads to accelerated metabolism, which might be partially responsible for developing tolerance to alcohol and other drugs (Lieber, 1997). In support of this theory, one study demonstrated that human alcoholics had elevated levels of CYP2E1 in particular brain regions, which may be partially responsible for the neurotoxic effects and metabolic tolerance that are characteristic of chronic alcohol exposure (Howard et al., 2003). Although it does not appear that CYP2E1 has emerged in the results of other methylome studies on alcohol dependence, our results suggest that future methylation studies should interrogate CpGs within this gene. Future research might explore how involvement in the network that emerged from our results might be associated with more downstream mechanisms associated with AUD.
Additionally, we examined whether or not the CpGs sites that emerged from our analyses reflected patterns that were present across multiple CpGs within that genetic region, further supporting their importance as candidates for targeted replication in future studies. To this end, we examined whether there were additional CpGs within 2000 base pairs of transcription start sites of the genes that emerged from our analyses (i.e. supplementary tables 1 and 2) and used a relaxed DiffScore threshold (DiffScore > |40|) in order to detect regions were there were more than one CpG site differentially methylated between AD cases and controls. Within our hypomethylated results (see notion in supplementary table 1), several sites fell within these differentially methylated regions of interest, including VTRNA2-1, LHX8, and KLHL35. Several sites within our hypermethylated results (see notation supplementary table 2) fell within differentially methylated regions of interest, including, FAM59B, OR2L13, PM20D1, SEPT7L, and WRB. Future studies may be useful for follow-up on these specific genes.
Given the budding evidence base for epigenetic mechanisms in the genesis and progression of alcohol use disorders, our results can be used by scientists to inform future study designs and hypotheses. Future studies could use the derived lists of CpGs in order to test and replicate these findings in other independent samples of AUD cases with greater power. In addition, future studies should seek to understand whether methylation of DNA isolated from buccal cells performs as well as DNA from blood. These efforts would provide more evidence surrounding the open question of tissue type specificity in the context of methylation and AUD. Alternatively, the biological pathways that emerged from the current study might similarly inform the continued refinement of theoretical models and understanding of the underlying mechanisms of AUD.
Supplementary Material
Acknowledgments
Sources of Support: This material is based upon work supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE 1144083 awarded to Sarah Hagerty; NIH K23DA033302 to LCB; NIH R01AA012238 to KEH; NIH K99AA020536 to NH; New South Wales and Victorian Tissue Resource Centre.
References
- Bagdy G, Arató M. Serum dopamine-beta-hydroxylase activity and alcohol withdrawal symptoms. Drug Alcohol Depend. 1987;19:45–50. doi: 10.1016/0376-8716(87)90086-x. [DOI] [PubMed] [Google Scholar]
- Baraona E, Lieber CS. Effects of ethanol on lipid metabolism. J Lipid Res. 1979;20:289–315. [PubMed] [Google Scholar]
- Bönsch D, Lenz B, Kornhuber J, Bleich S. DNA hypermethylation of the alpha synuclein promoter in patients with alcoholism. Neuroreport. 2005;16:167–170. doi: 10.1097/00001756-200502080-00020. [DOI] [PubMed] [Google Scholar]
- Cook RT. Alcohol abuse, alcoholism, and damage to the immune system—a review. Alcohol Clin Exp Res. 1998;22:1927–1942. [PubMed] [Google Scholar]
- Crews FT, Sarkar DK, Qin L, Zou J, Boyadjieva N, Vetreno RP. Neuroimmune Function and the Consequences of Alcohol Exposure. Alcohol Res. 2015;37:e1–e20. doi: 10.35946/arcr.v37.2.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drgon T, Lin Z, Wang GJ, Fowler J, Pablo J, Mash DC, Volkow N, Uhl GR. Common human 5' dopamine transporter (SLC6A3) haplotypes yield varying expression levels in vivo. Cell Mol Neurobiol. 2006;26:875–889. doi: 10.1007/s10571-006-9014-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Lizarbe S, Pascual M, Guerri C. Critical role of TLR4 response in the activation of microglia induced by ethanol. J Immunol. 2009;183:4733–4744. doi: 10.4049/jimmunol.0803590. [DOI] [PubMed] [Google Scholar]
- Fraser H, Lam L, Neumann S, Kobor M. Population-specificity of human DNA methylation. Genome Biol. 2012;13:R8. doi: 10.1186/gb-2012-13-2-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giros B, El Mestikawy S, Godinot N, Zheng K, Han H, Yang-Feng T, Caron MG. Cloning, pharmacological characterization, and chromosome assignment of the human dopamine transporter. Mol Pharmacol. 1992;42:383–390. [PubMed] [Google Scholar]
- Goral J, Karavitis J, Kovacs EJ. Exposure-dependent effects of ethanol on the innate immune system. Alcohol. 2008;42:237–247. doi: 10.1016/j.alcohol.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerri C, Pascual M. Role of Toll-Like Receptor 4 in Alcohol Induced Neuroinflammation and Behavioral Dysfunctions. In: Cui, Changhai, Grandison, Lindsey, Noronha, Antonio, editors. Neural-Immune Interactions in Brain Function and Alcohol Related Disorders. Springer, US: 2013. pp. 279–306. [Google Scholar]
- Guintivano J, Aryee MJ, Kaminsky ZA. A cell epigenotype specific model for the correction of brain cellular heterogeneity bias and its application to age, brain region and major depression. Epigenetics. 2013;8:290–302. doi: 10.4161/epi.23924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlaar N, Hutchison KE. Alcohol and the methylome: design and analysis considerations for research using human samples. Drug Alcohol Depend. 2013;133:305–316. doi: 10.1016/j.drugalcdep.2013.07.026. [DOI] [PubMed] [Google Scholar]
- Hassan S, Duong B, Kim KS, Miles MF. Pharmacogenomic analysis of mechanisms mediating ethanol regulation of dopamine beta-hydroxylase. J Biol Chem. 2003;278:38860–38869. doi: 10.1074/jbc.M305040200. [DOI] [PubMed] [Google Scholar]
- Hillemacher T, Frieling H, Hartl T, Wilhelm J, Kornhuber J, Bleich S. Promoter specific methylation of the dopamine transporter gene is altered in alcohol dependence and associated with craving. J Psychiatr Res. 2009a;43:388–392. doi: 10.1016/j.jpsychires.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Hillemacher T, Frieling H, Luber K, Yazici A, Muschler MA, Lenz B, Wilhelm J, Kornhuber J, Bleich S. Epigenetic regulation and gene expression of vasopressin and atrial natriuretic peptide in alcohol withdrawal. Psychoneuroendocrinology. 2009b;34:555–560. doi: 10.1016/j.psyneuen.2008.10.019. [DOI] [PubMed] [Google Scholar]
- Howard LA, Miksys S, Hoffmann E, Mash D, Tyndale RF. Brain CYP2E1 is induced by nicotine and ethanol in rat and is higher in smokers and alcoholics. Br J Pharmacol. 2003;138:1376–1386. doi: 10.1038/sj.bjp.0705146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2 February 2015];Illumina, GenomeStudio Support, Questions & Answers. 2015 Available from: < https://support.illumina.com/array/array_software/genomestudio/questions.html>. [Google Scholar]
- Joh TH, Hwang O. Dopamine beta-hydroxylase: biochemistry and molecular biology. Ann N Y Acad Sci. 1987;493:342–350. doi: 10.1111/j.1749-6632.1987.tb27217.x. [DOI] [PubMed] [Google Scholar]
- Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- Jones SM, Boobis AR, Moore GE, Stanier PM. Expression of CYP2E1 during human fetal development: methylation of the CYP2E1 gene in human fetal and adult liver samples. Biochem Pharmacol. 1992;43:1876–1879. doi: 10.1016/0006-2952(92)90726-y. [DOI] [PubMed] [Google Scholar]
- Karoly HC, Harlaar N, Hutchison KE. Substance use disorders: a theory-driven approach to the integration of genetics and neuroimaging. Ann N Y Acad Sci. 2013;1282:71–91. doi: 10.1111/nyas.12074. [DOI] [PubMed] [Google Scholar]
- Kato N, Takahashi S, Tani N, Iwase N, Odani K. Changes in the metabolism of biogenic amines in alcoholism—especially regarding CSF monoamine metabolites and serum DBH activity. Alcohol Clin Exp Res. 1979;3:24–27. doi: 10.1111/j.1530-0277.1979.tb04761.x. [DOI] [PubMed] [Google Scholar]
- Kerkel K, Spadola A, Yuan E, Kosek J, Jiang L, Hod E, Li K, Murty VV, Schupf N, Vilain E, Morris M, Haghighi F, Tycko B. Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat Genet. 2008;40:904–908. doi: 10.1038/ng.174. [DOI] [PubMed] [Google Scholar]
- Khavari DA, Sen GL, Rinn JL. DNA methylation and epigenetic control of cellular differentiation. Cell Cycle. 2010;9:3880–3883. doi: 10.4161/cc.9.19.13385. [DOI] [PubMed] [Google Scholar]
- Köhnke MD, Kolb W, Köhnke AM, Lutz U, Schick S, Batra A. DBH*444G/A polymorphism of the dopamine-beta-hydroxylase gene is associated with alcoholism but not with severe alcohol withdrawal symptoms. J Neural Transm. 2006;113:869–876. doi: 10.1007/s00702-005-0365-6. [DOI] [PubMed] [Google Scholar]
- Laine TP, Ahonen A, Torniainen P, Heikkilä J, Pyhtinen J, Räsänen P, Niemelä O, Hillbom M. Dopamine transporters increase in human brain after alcohol withdrawal. Mol Psychiatry. 1999;4:189–191. 104–185. doi: 10.1038/sj.mp.4000514. [DOI] [PubMed] [Google Scholar]
- Lieber CS. Cytochrome P-4502E1: its physiological and pathological role. Physiol Rev. 1997;77:517–544. doi: 10.1152/physrev.1997.77.2.517. [DOI] [PubMed] [Google Scholar]
- Liu J, Calhoun VD, Chen J, Claus ED, Hutchison KE. Effect of homozygous deletions at 22q13. 1 on alcohol dependence severity and cue elicited BOLD response in the precuneus. Addict Biol. 2013;18:584–558. doi: 10.1111/j.1369-1600.2011.00393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschler MA, Hillemacher T, Kraus C, Kornhuber J, Bleich S, Frieling H. DNA methylation of the POMC gene promoter is associated with craving in alcohol dependence. J Neural Transm. 2010;117:513–519. doi: 10.1007/s00702-010-0378-7. [DOI] [PubMed] [Google Scholar]
- Nestler EJ. Epigenetic mechanisms of drug addiction. Neuropharmacology. 2014;76(Pt B):259–268. doi: 10.1016/j.neuropharm.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BY, Lee BC, Jung KH, Jung MH, Park BL, Chai YG, Choi IG. Epigenetic changes of serotonin transporter in the patients with alcohol dependence: methylation of an serotonin transporter promoter CpG island. Psychiatry Investig. 2011;8:130–133. doi: 10.4306/pi.2011.8.2.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philibert RA, Sandhu H, Hollenbeck N, Gunter T, Adams W, Madan A. The relationship of 5HTT (SLC6A4) methylation and genotype on mRNA expression and liability to major depression and alcohol dependence in subjects from the Iowa Adoption Studies. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:543–549. doi: 10.1002/ajmg.b.30657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J Neurosci. 2012;32:1884–1897. doi: 10.1523/JNEUROSCI.3136-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA. Alcohol-use disorders. Lancet. 2009;373:492–501. doi: 10.1016/S0140-6736(09)60009-X. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, O'Connor DT, Duby J, Vega R, Moss M. Dopamine-beta-hydroxylase activity levels in men at high risk for alcoholism and controls. Biol Psychiatry. 1981;16:1067–1075. [PubMed] [Google Scholar]
- Weng JT, Wu LS, Lee CS, Hsu PW, Cheng AT. Integrative epigenetic profiling analysis identifies DNA methylation changes associated with chronic alcohol consumption. Comput Biol Med. 2014 doi: 10.1016/j.compbiomed.2014.12.003. [DOI] [PubMed] [Google Scholar]
- Wilhelm-Benartzi CS, Koestler DC, Karagas MR, Flanagan JM, Christensen BC, Kelsey KT, Marsit J, Houseman EA, Brown R. Review of processing and analysis methods for DNA methylation array data. Br J Cancer. 2013;109:1394–1402. doi: 10.1038/bjc.2013.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Herman AI, Kranzler HR, Anton RF, Zhao H, Zheng W, Gelernter J. Array-based profiling of DNA methylation changes associated with alcohol dependence. Alcohol Clin Exp Res. 2013a;37(Suppl 1):E108–E115. doi: 10.1111/j.1530-0277.2012.01928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Miao Q, Wang C, Zhao R, Li W, Haile CN, Hao W, Zhang XY. Genome-wide DNA methylation analysis in alcohol dependence. Addict Biol. 2013b;18:392–403. doi: 10.1111/adb.12037. [DOI] [PubMed] [Google Scholar]
- Zhao R, Zhang R, Li W, Liao Y, Tang J, Miao Q, Hao W. Genome-wide DNA methylation patterns in discordant sib pairs with alcohol dependence. Asia Pac Psychiatry. 2013;5:39–50. doi: 10.1111/appy.12010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.