Abstract
ZAP70 [zeta-chain (TCR)-associated protein kinase, 70-kDa], is required for T cell activation. ZAP70 deficiencies in humans and null mutations in mice lead to severe combined immune deficiency. Here, we describe a zap70 loss-of-function mutation in zebrafish (zap70y442) that was created using transcription activator-like effector nucleases (TALENs). In contrast to what has been reported for morphant zebrafish, zap70y442 homozygous mutant zebrafish displayed normal development of blood and lymphatic vasculature. Hematopoietic cell development was also largely unaffected in mutant larvae. However, mutant fish had reduced lck:GFP+ thymic T cells by 5 days postfertilization that persisted into adult stages. Morphological analysis, RNA sequencing, and single-cell gene expression profiling of whole kidney marrow cells of adult fish revealed complete loss of mature T cells in zap70y442 mutant animals. T cell immune deficiency was confirmed through transplantation of unmatched normal and malignant donor cells into zap70y442 mutant zebrafish, with T cell loss being sufficient for robust allogeneic cell engraftment. zap70 mutant zebrafish show remarkable conservation of immune cell dysfunction as found in mice and humans and will serve as a valuable model to study zap70 immune deficiency.
INTRODUCTION
ZAP70 [zeta-chain (TCR) associated protein kinase, 70 kDa], is an important mediator of T cell differentiation and function. ZAP70 is a cytoplasmic protein kinase consisting of two amino-terminal Src homology 2 (SH2) domains and a carboxy-terminal kinase domain that plays a key role in regulating the T cell receptor (TCR) phosphorylation cascade. TCRs are activated by interacting with peptide antigens bound to major histocompatibility complexes (MHCs), causing the recruitment of CD4 or CD8 costimulatory receptors. The CD4 and CD8 receptors are bound by intracellular lymphocyte-specific protein kinase (LCK), which, in turn, phosphorylates the TCR immunoreceptor tyrosine-based activation motifs (ITAMs) of CD3ζ. Phosphorylated ITAMs recruit ZAP70 by binding its SH2 domains with high affinity. LCK phosphorylates ITAM-bound ZAP70, releasing it from an autoinhibited conformation and promoting its catalytic activity. Further phosphorylation of the TCR-associated scaffolding proteins by ZAP70 allows for the recruitment of additional signaling molecules and ultimately results in T cell activation, proliferation, and differentiation (1).
Human ZAP70 deficiencies result in severe combined immune deficiency (SCID) with specific defects in T cell maturation (2–4). Patients with inactivating mutations in ZAP70 lack mature CD8+ cytotoxic T cells and produce nonfunctional CD4+ helper T cells. ZAP70 null CD4+ T cells exit the thymus, yet they have dysfunctional T cell signaling and cannot mount effective T cell responses. Zap70 mutant mice also have T cell deficiencies, but they exhibit key differences compared with humans (5, 6). Zap70 mutant mice have a more severe block in thymocyte maturation, with T cells arresting at the CD4+/CD8+ cortical stage of development. Because of this, Zap70-deficient mice lack all mature T cell subsets, including mature CD4 and CD8 single positive effector cells. The differential effects of ZAP70 deficiency in humans and mice have been largely attributed to gene compensation by spleen tyrosine kinase (SYK) in humans. Elder et al. showed that SYK could partially rescue the developmental requirements of ZAP70 in CD4 single positive cells, though it could not phosphorylate the downstream ZAP70 targets necessary for TCR signaling and activation (7). In mice, Syk is not expressed in late-stage thymocytes, likely accounting for the full ablation of CD4+ T cells in knockout animals. Taken together, these results suggest a divergent requirement for ZAP70 in thymocyte development in mice and humans and underscore the strikingly conserved functional requirement for ZAP70 in TCR signaling and effector cell function in mature T cells. Roles for zap70 in regulating T cell development in zebrafish have not yet been described.
Morpholino-based studies with zebrafish have shown that sprouting and development of the early vasculature are regulated by zap70 and syk (8). In addition to its roles in regulating B and T cell development, SYK has been shown to have an important role in lymphatic vascular development (9–14). While at least one report has implicated SYK in endothelial-cell proliferation and migration in vitro (15), its primary role in regulating vascular development in vivo is to maintain blood-lymphatic vascular separation by functioning in a nonautonomous manner within platelets (16). Defects in lymphatic or blood endothelial specification have not been reported for ZAP70-deficient mice or humans. Given the discrepancies between studies involving morphant zebrafish (8) and knockout mice and humans with ZAP70 deficiencies, a role for ZAP70 in vessel and lymphatic system development remains controversial.
Here, we describe the generation and characterization of novel zap70 mutant zebrafish. Characterization of larval-stage zebrafish revealed no defects in vascular and lymphatic development. Further characterization of zap70 mutant zebrafish revealed reductions in thymic T cells and a lack of mature T cells in the whole kidney marrow. Zebrafish zap70 mutants robustly engrafted nonmatched, allogeneic tissues, validating functional defects in T cell responses and inability to mount effective immune rejection. Our analysis of mutant zap70-deficient zebrafish highlights the conservation of Zap70 function in T cell signaling throughout vertebrate evolution and underscores the utility of zebrafish to study immunodeficiencies.
MATERIALS AND METHODS
Generation of zap70y442 TALEN-induced mutants.
Transcription activator-like effector nucleases (TALENs) were constructed to target the second exon of zap70 and recognize the following sequences: 5′ arm target, GTTCCTCCTGCGACAGTGC, and 3′ arm target, CCAGATCATAGACAGCACATA. One hundred picograms of each in vitro-transcribed zap70 TALEN arm was injected into one-cell-stage embryos in the Tg(fli1a:eGFP)y1, Tg(gata1:dsRED)sd2 zebrafish background. F0 injected embryos were raised to adulthood and incrossed. The F1 generation was fin clipped to identify germ line mutations. Induced mutations were identified by visualization of PCR products amplified using the forward primer 5′ GTATGGGAGACGGCCTGTTC 3′ and reverse primer 5′ TCCAGGTTCCAGATCATAGACA 3′ on a 3% agarose gel by electrophoresis. The molecular lesion was confirmed by sequencing PCR-amplified genomic DNA fragments.
Imaging zap70y442 embryonic vascular morphology.
Zebrafish larvae were anesthetized at 30 hours postfertilization (hpf) or 5 days postfertilization (dpf) with 0.168 mg/ml of Tricaine, mounted in 0.8% agarose, and imaged with an Olympus FV 1000 or a Leica upright TCS-sp5 II two-photon confocal microscope and a ProgRes C14 camera mounted on a Leica MZ12 stereomicroscope. Images in Fig. 1 show only Tg(fl1a:eGFP) expression to facilitate ease of viewing vessel development, while Fig. S1 in the supplemental material shows both vessels and erythroid cells present in Tg(fli1a:eGFP)y1, Tg(gata1:dsRED)sd2, zap70y442 homozygous mutant zebrafish at 30 hpf.
FIG 1.
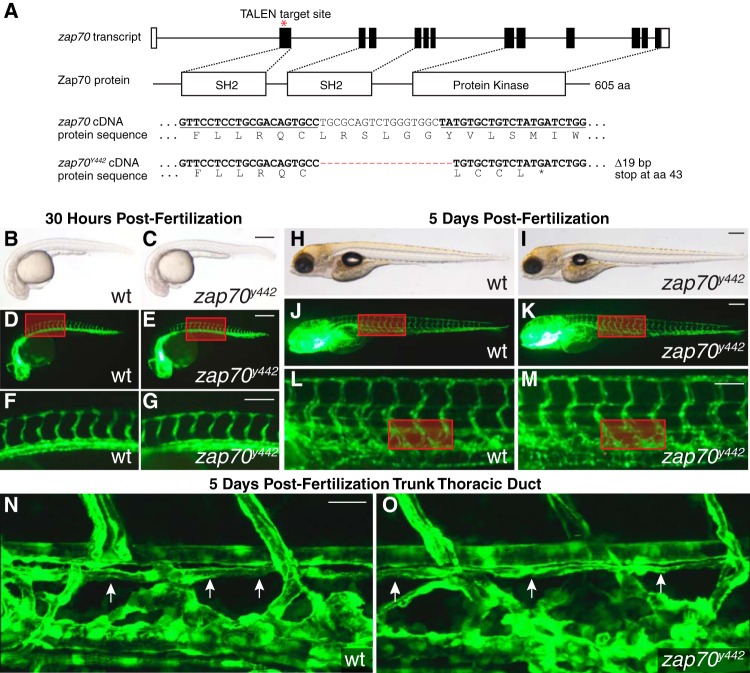
zap70y442 mutant zebrafish have normal vascular and lymphatic development. (A) Zebrafish zap70 genomic locus with exons indicated by boxes and the TALEN binding site marked by an asterisk. Zap70 protein domains corresponding to exons are labeled by white boxes. Zap70 cDNA and amino acid (aa) sequences are shown with the TALEN binding sites underlined and 19-bp deletion corresponding to the zap70y442 mutation indicated by red dashes. (B to O) Analysis of vascular patterning and thoracic duct formation in Tg(fli1a:eGFP)y1 embryos and larvae. (B to G) Vascular development in sibling wild-type (B, D, and F) and zap70y442 mutant (C, E, and G) zebrafish at 30 hpf. (F and G) Magnified views of the regions boxed in panels D and E (n = 60 per genotype). (H to O) Vascular development in wild-type sibling (H, J, L, and N) and zap70y442 mutant (I, K, M, and O) zebrafish at 5 dpf. (L and M) Magnified views of the regions boxed in panels J and K. (N and O) Higher-magnification composites of fluorescent confocal z-stacks of regions boxed in panels L and M (n = 60 per genotype). Arrows indicate thoracic duct (N and O). Scale bars represent 250 μm in panels B, D, H, and J, 100 μm in panels F and I, and 25 μm in panel N.
WISH.
Whole-mount in situ hybridization (WISH) was performed as described previously, with the modification of using BM purple (Roche; 11442074001) as the substrate for alkaline phosphatase (41). Digoxigenin-labeled probes were transcribed in vitro using the Roche SP6 in vitro transcription kits on PCR amplicons produced using the primers in Table S4 in the supplemental material. The rag1 (17) and mpx (18) probes were transcribed in vitro from plasmid vectors as previously described.
Transgenic zebrafish.
In vitro-transcribed zap70 TALENs were injected into and maintained on the Tg(fli1a:eGFPy1); Tg(gata1:dsRED)sd2 zebrafish background. Isolated mutants were also outcrossed and maintained in the Tg(lck:eGFP)cz1 (19) or Tg(cd41:eGFP) (20) transgenic background.
Quantification of lck:eGFP thymocytes.
The volume of green fluorescent protein (GFP)-positive thymocytes was quantified in 5-day-old wild-type and zap70y442 mutant larvae using confocal z-stacks of the region and volumetric analysis using Fiji (ImageJ) (n = 12 larvae per genotype). Significance was calculated by Student's t test.
Histology and thymus size quantification.
Thymus histology was performed as previously described (21). Briefly, 3-month-old fish were sacrificed and fixed in 4% paraformaldehyde, embedded in paraffin, and step sectioned. Infection and inflammation analysis was performed by preparing 5- to 7-month-old healthy and moribund fish as described above. Slides were stained with hematoxylin and eosin by Specialized Histopathology Services at Massachusetts General Hospital. Thymus sections were imaged at a magnification of ×200 using an Olympus BX41 compound microscope. Images were imported into Fiji (ImageJ), the thymus was traced using the “Freehand Selection” tool, and the area was quantified. The average area of thymi (n ≥ 10) was graphed, and standard errors of the means (SEM) were determined. Significance was calculated by Student's t test.
Cytospins.
Three-month-old wild-type, heterozygous, and mutant sibling fish were euthanized by Tricaine overdose. The whole kidney marrow (WKM) was isolated via surgical extraction and placed in 250 μl of 0.9× phosphate-buffered saline (PBS) plus 5% fetal bovine serum (FBS). WKM was triturated 8 to 10 times using a 200-μl pipette, passed through a 40-μm cell strainer (Corning; 352340), and washed. A total of 100 to 250 μl of cell suspension was added per Cytofunnel-equipped Cytoslide (Thermo Scientific; A78710003 and 5991056) and spun at 500 rpm for 5 min. The slides were dried briefly and stained with May-Grünwald solution (Sigma-Aldrich; 63590-500ML) for 5 min and then with diluted Giemsa stain (1:20 dilution; Fluka Analytical; 48900-500ML-F) for 10 min. After staining, the slides were rinsed, dried, and sealed with a coverslip. A total of ≥5 fields at a magnification of ×1,000 were analyzed per slide, and ≥200 cells were counted per animal. A clinical hematopathologist and hematologist (J. N. Berman and R. S. Liwski) performed the cell counts in a blinded manner on deidentified slides. Changes in relative numbers of each blood cell lineage between the genotypes were assed via two-tailed Student's t test; a P value of ≤0.05 was considered significant.
tcrb and igm gene expression.
A standard RNA extraction protocol RNAeasy minikit (Qiagen; 7416) was used to isolate RNA from the WKM of 3-month-old wild-type, heterozygous, and mutant siblings (1 kidney/350 μl of RLT buffer). Upon extraction, RNA was made into cDNA using SuperScript III reverse transcriptase (Invitrogen; 18080-051). tcrb and igm receptor rearrangements were PCR amplified using a nested protocol, as previously described (21). The first PCR utilized 2 ng of cDNA and was performed using an optimized 23.75-μl reaction mixture with Choice Taq Blue master mix (Denville Scientific; CB4065-8). Briefly, 12.5 μl of Choice Blue master mix was mixed with 1.25 μl of forward and reverse primers (PCR 1; see Table S4 in the supplemental material; 10 μM each), 9 μl of water, and 1 μl of cDNA (2 ng/μl).
PCR cycle parameters were (i) denaturation at 94°C for 45 s, (ii) annealing at 56°C for 30 s, and (iii) elongation at 72°C for 1 min, repeated for 35 cycles. One microliter of the original PCR mixture was used in a second PCR along with nested primers specific to each transcript (PCR 2; see Table S4 in the supplemental material). The cycling parameters were identical to those of the first reaction, except that the steps were repeated for 25 cycles. The final products of this nested PCR are visualized on a 2% Tris-acetate-EDTA (TAE)–agarose gel containing ethidium bromide.
WKM gene expression as assessed by RNA sequencing.
The WKM of 3-month-old wild-type, heterozygous, and zap70y442 mutant siblings was dissected and placed in 350 μl of Qiagen RLT buffer (supplemented with 1% 2-mercaptoethanol; Bio-Rad; 161-0710). The RNeasy minikit (Qiagen; 7416) was used to isolate RNA from these samples. During isolation, an on-column DNase (Qiagen; 79254) digestion was performed, in accordance with the manufacturer's recommendations. The total RNA samples corresponding to three animals for each genotype underwent rRNA depletion using the RiboZero kit (Illumina; MRZH116) followed by next-generation sequence library construction using the NEBNext Ultra directional RNA library prep kit for Illumina (New England BioLabs; E7420S) using 15 cycles of PCR amplification. Sequencing was performed on an Illumina HiSeq 2500 instrument, and reads were mapped to the Danio rerio reference genome (Zv9 build) using STAR software (22), resulting in an average of 18 million aligned pairs of 50-bp reads per sample. Read counts over transcripts were calculated using HTSeq v.0.6.0 (23) based on the most current Ensembl annotation file for Zv9. The EdgeR package (version 3.8.6) (24) was used to analyze differential gene expression between the samples based on the criteria of a >1.75-log2 fold change (FC) in expression value and false-discovery rates (FDR) (Benjamini-Hochberg test) of <0.05.
WKM single-cell isolation and quantitative PCR using the Fluidigm BioMark HD platform.
Cell extraction, sorting, gene expression reactions, quantification, and analysis were performed as described previously (25). Briefly, WKM from 2.5- to 3-month-old wild-type and zap70y442 homozygous mutant fish was isolated, individually placed in 5% FBS–1× PBS, manually dissociated by trituration, and filtered through a 40-μm cell strainer. Single cells were sorted into 96-well plates, where each well contained 5 μl of lysis buffer, using a BD FACSAria Fusion cell sorter. Plates were then heated at 65°C for 90 s to lyse cell membranes. Fluidigm reverse transcription mix (Fluidigm; 100-6299) was added to each well in order to make cDNA. Next, samples were preamplified using gene target-specific nested outer primers (166 nM) and 10× PreAmp master mix (Fluidigm; 100-5581) (a list of gene-specific primers used is provided in reference 25). Unincorporated primers were removed by exonuclease treatment (New England BioLabs; M0293L). cDNA samples were then diluted with 36 μl of 1× TE, resulting in a total 50 μl of preamplified, PCR-ready cDNA. Three microliters of preamplified cDNA was mixed with 3.5 μl of 2× SSo Fast EvaGreen Supermix with low ROX (Bio-Rad; 172-5212), 0.35 μl of 20× DNA binding dye sample loading reagent (Fluidigm; 100-7609), and 0.15 μl of water, resulting in a 5-μl reaction mixture which was subsequently loaded into a primed 96.96 dynamic array chip for gene expression (Fluidigm; BMK-M-96.96). Five microliters of 96 separate inner primer pairs (5 μM) were also loaded onto the 96.96 dynamic array chip and mixed in the Fluidigm IFC controller HX. The dynamic array chip was then loaded into the Fluidigm BioMark HD by following the manufacturer's protocol. Thermal cycling was completed using protocol GE 96 × 96 PCR melt v2.pcl.
Statistical analysis and display of single-cell data.
Threshold cycle (CT) values were recovered from the BioMark HD. The quality threshold was set to 0.65, and a linear derivative was used as a baseline correction. The limit of detection was set to a CT value of 28, and expression was calculated using default settings in SINGuLAR. Wells that failed to express either reference genes (eef1a1l1 and actb1) or other lineage-specific markers were eliminated from further analysis. Unsupervised hierarchical clustering was performed using SINGuLAR software. Lineage determinations were made after hierarchical clustering. The principal-component analysis (PCA), hierarchical clustering, and 3-dimensional PCA were also generated using the SINGuLAR R package.
Muscle and tumor cell transplantation in zebrafish.
Muscle and tumor cell isolation and transplantation were performed as previously described (21, 26). In short, donor fish were euthanized using Tricaine (Western Chemical; MS-222), and donor tissues were placed in a 10-cm petri dish with 500 μl of 0.9× PBS plus 5% FBS solution. Tissues were dissociated mechanically with a razor blade, as well as by trituration using a 200-μl pipette. Cells were then filtered through a 40-μm strainer (Falcon; 352340), centrifuged at 1,000 × g for 10 min, and resuspended in the desired volume. For intramuscular transplantation of allogeneic muscle tissue, 1.2 × 106 cells were resuspended in a volume of 2 μl and injected into the dorsal musculature of 2- to 4-month-old sibling recipients. For zebrafish tumor cell transplantation, 7 × 105 to 1 × 106 tumor cells were injected into the peritoneal cavity of sibling recipients in a total volume of 5 μl. A 26-gauge Hamilton (80366) syringe was used to perform the transplants. Engraftment of transplanted tissues was assessed by epifluorescence microscopy at 10, 20, 30, and 45 days posttransplantation (dpt). Recipient fish were euthanized when moribund or at 45 dpt if animals showed no signs of engraftment. These fish were either fixed in 4% paraformaldehyde and sent out for sectioning or used to prepare cytospins of peripheral blood for histological analysis.
RESULTS AND DISCUSSION
Generation of a zebrafish zap70y442 mutant.
In order to generate zebrafish harboring mutations in the zap70 gene, we constructed TALENs targeting the second exon of this gene (Fig. 1A). F0 zebrafish embryos were injected with in vitro-transcribed mRNA encoding these TALENs, raised to adulthood, and incrossed. We identified F1 founder fish that contained a mutant zap70 allele harboring a 19-bp deletion in exon 2 of the coding sequence (referred to as zap70y442). This mutation caused a frameshift, resulting in the presence of a premature stop codon at the 43rd amino acid (Fig. 1A). This allele is predicted to be a null mutation, as the truncated protein would lack the SH2 binding domains and the catalytic protein kinase domain.
zap70 mutants exhibit normal angiogenesis and lymphangiogenesis.
Since a previous study using a morpholino knockdown of zebrafish zap70 had reported an early defect in developmental angiogenesis (8), we examined formation of the trunk vasculature in Tg(fli1a:eGFP)y1, Tg(gata1:dsRED)sd2 transgenic, zap70y442 homozygous mutant zebrafish larvae (see Fig. S1 in the supplemental material). Tg(fli1a:eGFP)y1, Tg(gata1:dsRED)sd2 animals express enhanced green fluorescent protein (EGFP) in the blood and lymphatic vasculature (27) and the Discosoma sp. red fluorescent protein (dsRED) in the circulating blood (28). At 30 h postfertilization (hpf), when zap70 morphants were reported to exhibit defects in intersegmental vessel (ISV) growth, homozygous zap70y442 mutants displayed no such defects. The ISVs of homozygous zap70y442 mutants reached the dorsal regions of the fish and began forming the dorsal longitudinal anastomotic vessel (DLAV) with timing and morphology indistinguishable from those of their wild-type siblings (Fig. 1B to G) (n = 60 fish analyzed per genotype).
Homozygous zap70y442 mutants also displayed no visible defects in lymphangiogenesis during early development (n = 60 fish analyzed per genotype). At 5 days postfertilization (dpf), development of trunk lymphatic vessels in zap70y442 mutants looked identical to that in their wild-type siblings, including the intersegmental lymphatic vessels (ISLV), which extend dorsally and ventrally from the parachordal lines, and the thoracic duct (TD), which extends anteriorly and posteriorly along the length of the trunk just ventral to the dorsal aorta (Fig. 1H to O; see also Fig. S1 in the supplemental material). Together, these results indicate that Zap70 function is not required for vascular development in zebrafish, which is consistent with reports of Zap70 knockouts in mice (5) and human ZAP70-deficient patients (4). The lack of vascular phenotypes in zap70y442 mutants contrasts with previous observations in zap70 morphant zebrafish (8), suggesting that the antisense morpholino injections resulted in off-target effects on vessel growth, such as those described in previous reports (29, 30).
zap70 y442 larvae have decreased thymocytes.
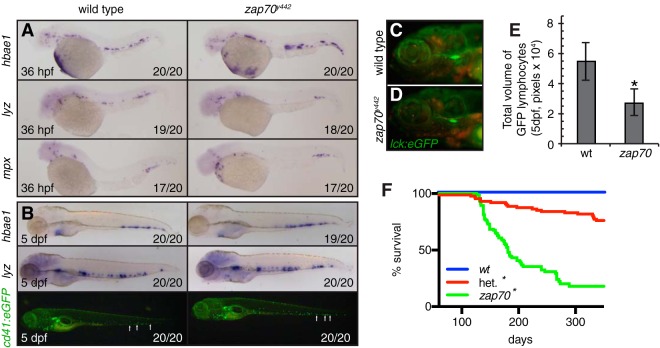
Human patients and mice with ZAP70 loss of function have defects in T lymphocyte differentiation and TCR signaling (2–6). Thus, we investigated whether zap70y442 mutants have early hematopoietic defects in erythroid, myeloid, and lymphoid cell types. Whole-mount in situ hybridization (WISH) was performed at 36 hpf and 5 dpf to assess definitive hematopoiesis using probes for alpha embryonic hemoglobin (hbae1), lysozyme C (lyz), and myeloperoxidase (mpx), which are expressed in erythrocytes, macrophages, and neutrophils, respectively. WISH for these factors showed no differences between zap70 mutants and their wild-type siblings, suggesting that initial specification of these cell types was normal in zap70y442 mutants (n > 17) (Fig. 2A and B). Normal numbers of erythrocytes labeled by the gata1:dsRED transgene were also observed circulating in both larval and adult zap70y442 mutant fish, suggesting that erythrocyte development and maintenance were not affected in mutants at later stages. zap70y442 mutants were also crossed into the Tg(cd41:eGFP) background to determine if there were any defects in the formation of hematopoietic stem cells (HSC) or thrombocytes in mutant zebrafish (20, 28, 31). During the first 5 days of development, there were no obvious defects seen in HSC or thrombocytes in zap70y442 fish (n = 20) (Fig. 2B, bottom images). These data are consistent with mouse models in which ZAP70 deficiency has no effect on the development of these lineages (5, 6).
FIG 2.
zap70y442 mutant zebrafish have largely normal larval hematopoiesis but have reduced survival as adults. (A) Images of whole-mount in situ hybridization comparing hematopoietic marker gene expression between wild-type and zap70y442 mutant zebrafish at 30 hpf. (B) Analysis of larvae at 5 dpf. Whole-mount in situ hybridization (top two rows) and epifluorescence images of Tg(cd41:eGFP) expression in wild-type and zap70y442 mutant zebrafish at 5 dpf (bottom row). The number of animals with depicted phenotype/total number of animals analyzed is indicated. (C to E) Total volumes of GFP-positive thymocytes in Tg(lck:eGFP) transgenic wild-type (wt) (C) and zap70y442 mutant (D) fish, quantified in panel E (n = 12 larvae analyzed per genotype; *, P < 0.00001, Student's t test). Error bars indicate standard deviations. (F) Kaplan-Meier survival analysis for wild-type, heterozygous, and zap70y442 homozygous mutant fish (n > 57 animals/arm; *, P < 0.0001, log rank test, comparing wt to heterozygous [het.], het. to zap70y442, and wt to zap70y442). “zap70” in panels E and F refers to the homozygous mutant.
Because Zap70-deficient mice and humans have prominent defects in T cell specification and development, we examined T lymphoid cells in zap70y442 mutants. Expression of rag1 and rag2 was detected in thymocytes of zap70y442 mutant zebrafish by WISH, indicating that early lymphocyte progenitors were properly specified in these mutants (see Fig. S2 in the supplemental material; n = 20 fish per genotype analyzed). To examine T cell development more closely, zap70y442 mutants were crossed into the Tg(lck:eGFP)cz1 transgenic line to label developing thymocytes and mature T cells (19). Following imaging at 5 dpf, the volume of GFP-positive thymocytes was quantified using ImageJ, which revealed a significant reduction in thymocytes in zap70y442 mutants relative to that in matched wild-type siblings (P < 0.0001; Student's t test [Fig. 2C to E]). These data are consistent with a role for ZAP70 in mammalian T cell development, as ZAP70-deficient mice have a severe reduction in thymocytes and an absence of mature T cells (5).
Adult zap70y442 mutant zebrafish have T cell deficiencies and increased mortality.
Mouse and human patients with loss of ZAP70 function have defects in T cell differentiation and activation (2–6). ZAP70-deficient patients are also susceptible to opportunistic infections and have a shortened life span if they do not receive an allogeneic bone marrow transplant (7). Although zap70y442 mutants did not display defects in angiogenesis or definitive hematopoiesis, adult homozygous zap70 y442 fish had elevated mortality beginning after 4 months of age (Fig. 2F). Only 9% of homozygous zap70y442 mutants remained alive by 10 months postfertilization, compared to 82% of heterozygous mutants and 100% of wild-type siblings (P < 0.0001, log rank statistic [Fig. 2F]). Histological analysis revealed that 9 out of 11 moribund adult zap70y442 homozygous mutants had prominent infections with Pseudoloma neurophilia (see Fig. S3 in the supplemental material). This common zebrafish pathogen is present in 74% of zebrafish aquatic facilities and was likely unmasked by the zap70y442 homozygous T cell deficiency phenotype (32). Disease manifested in 5- to 7-month-old homozygous mutant animals as xenomas found throughout the musculature. In contrast, 7-month-old clinically “healthy”-looking zap70y442 mutant fish had no signs of infection (n = 11) and were similar to both wild-type (n = 3) and zap70y442 heterozygous (n = 3) sibling control fish. Analysis of the intestine and gills of homozygous zap70y442 mutants revealed no overt signs of chronic inflammation prior to onset of externally visible signs of infection (see Fig. S3).
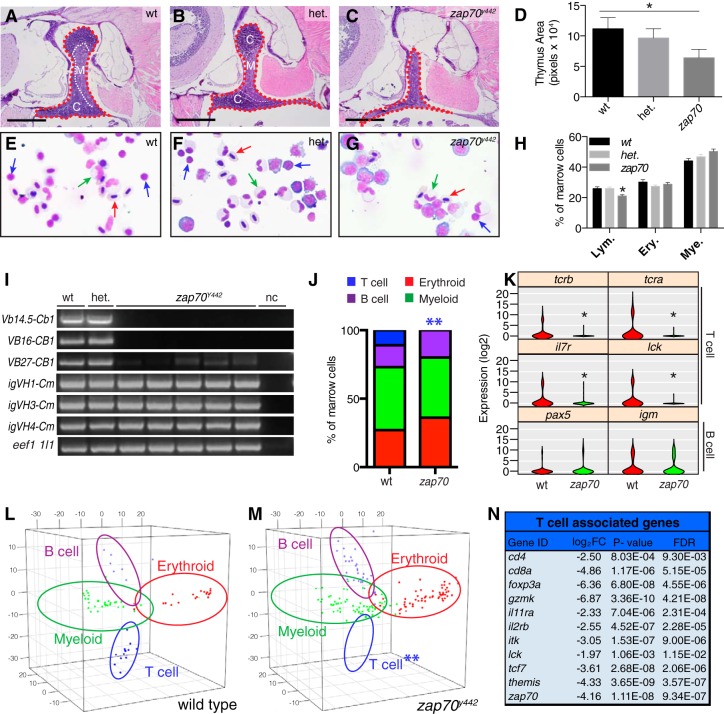
To investigate whether the early-mortality phenotype was attributable to immune deficiency, we examined whether zap70y442 zebrafish mutants had thymus or blood cell defects in adulthood. Thymus morphology was assessed on histological sections of 90-day-old zap70y442 mutant zebrafish, revealing a slight but significant reduction in thymus size between wild-type and zap70y442 mutant zebrafish (P = 0.047, Student's t test; n ≥ 10) (Fig. 3A to D). Moreover, the thymi of zap70y442 mutants displayed atypical loss of defined medullary and cortex regions and a reduction in thymocyte number (Fig. 3C), a thymic phenotype shared with Zap70 mutant mice (5).
FIG 3.
Adult zap70y442 mutant zebrafish have reduced thymocytes and loss of mature T cells in the marrow. (A to C) Images of hematoxylin-and-eosin-stained thymus sections from 3-month-old wild-type (A), heterozygous (B), and zap70y442 (C) mutant zebrafish. Thymi are outlined by red dashed line, with the medulla (M) and cortex (C) boundaries indicated by a white dashed line. (D) Quantification of thymus size (*, P < 0.047, Student's t test, n ≥ 10 animals per group analyzed, wt versus zap70y442; error bars show SEM). (E to G) Whole kidney marrow cytospins of wild-type (E), heterozygous (F), and zap70y442 mutant (G) zebrafish. Lymphocytes are indicated by blue arrows, erythrocytes by red arrows, and granulocytes by green arrows. (H) Quantitation of blood cell types following manual cell counts on cytospins (*, P < 0.01, Student's t test, comparing zap70y442 to wt and het. fish; error bars show SEM; n ≥ 200 cell analyzed per individual marrow and ≥4 animals per genotype). Lym., lymphocytes; Ery., erythroid cells; Mye., myeloid cells. (I) tcrb and igm transcripts detected following quantitative real-time PCR of whole kidney marrow from wt, het., and zap70y442 zebrafish. nc, negative control. Specific variable (V) and constant (C) regions and igm transcripts spanning the variable H (VH) and constant mu (Cm) regions are noted. The positive control is eef1a1l1 (elongation factor 1 alpha, like 1) (bottom row). (J) Quantification of blood cells contained in the marrow as assessed by Fluidigm single-cell quantitative PCR (*, P < 0.0001, Fisher's exact test). (K) Violin plots showing distribution of T cell-specific gene expression (top two rows) and B cell-specific gene expression (bottom row) within individual marrow cells. (L and M) Principal-component analysis showing hematopoietic lineages assigned to wild-type (L) and zap70y442 mutant (M) marrow. Significant reductions in T cells are indicated by asterisks (P = 0.0001, Fisher's exact test). Blood cell lineages are denoted by ovals. (N) T cell-associated genes identified by transcriptome sequencing analysis that are significantly downregulated in the marrow of zap70y442 mutants compared to that of wild-type fish. ID, identifier; FDR, false-discovery rate. Scale bars equal 100 μm (A to C) and 20 μm (E to G). “zap70” in panels D, H, J, and K refers to the homozygous mutant.
We next assessed blood cell populations in the whole kidney marrow (WKM), the site of hematopoiesis in the adult zebrafish. WKM cells were first assessed morphologically by cytospin quantification, revealing relatively equal proportions of erythroid and myeloid cells in all genotypes surveyed (Fig. 3E to H). zap70y442 mutant zebrafish had a slight but significant reduction in the total numbers of marrow lymphocytes. The marrow of wild-type fish was comprised of 25.9% ± 2.4% lymphocytes, compared to 21.0% ± 2.2% lymphocytes in zap70y442 mutant fish (P = 0.015, Student's t test; n ≥ 4 fish assessed per genotype [Fig. 3H]). Cytospin and morphological analysis are unable to delineate T versus B cells; thus, we analyzed T cell receptor β (tcrb) and immunoglobulin heavy chain (igm) gene expression in WKM cells. tcrb and igm transcripts were robustly amplified in both wild-type and zap70y442 heterozygous zebrafish, while zap70y442 homozygous mutant fish had nearly undetectable tcrb expression but normal igh transcript expression in the kidney (Fig. 3I). These data suggested that zap70y442 mutant fish had specific defects in mature T cells but not B cells. In order to differentiate lymphocyte populations with single-cell resolution, we next utilized a recently developed Fluidigm microfluidics platform that accurately identifies blood cell lineages based on single-cell transcriptional analysis by PCR with 96 gene-specific primer pairs (25). Using gene expression, unsupervised hierarchical clustering, and principal-component analysis, we quantified the percentage of myeloid, erythroid, T, and B cell populations that comprise the kidney marrow. Wild-type fish marrow consisted of 11% T lymphocytes, whereas zap70y442 homozygous mutants were devoid of mature T cells (P < 0.0001, Fisher's exact test [Fig. 3J, L, and M]). We observed no significant difference in B cell numbers (P = 0.61, Fisher's exact test [Fig. 3J to M]). As expected, T cell-specific gene expression (tcra, tcrb, lck, and il7r) was greatly reduced in zap70y442 mutants, while B cell markers (pax5 and igm) were expressed normally in mutant fish (Fig. 3K). Gene expression for additional erythroid, myeloid, and B cell markers was also normal compared to that in wild-type fish (see Fig. S4 in the supplemental material).
As an independent confirmation of T cell defects in zap70y442 mutants, we performed RNA sequencing on 3-month-old wild-type and mutant whole kidney marrow. Transcriptome sequencing analysis identified 444 significantly downregulated genes in zap70y442 mutants (log2 FC > 1.75; FDR < 0.05 [see Table S1 in the supplemental material]). Downregulated genes were compared for overlap with gene sets found in the Molecular Signatures Database (MsigDB) (33), providing independent and unbiased assessment of losses of specific blood cell types. This analysis confirmed that a significant number of genes downregulated in zap70y442 mutants are associated with T cell identity and maturation (P ≤ 1.68 × 10−7; FDR ≤ 4.88 × 10−6 [Fig. 3N; see also Fig. S5 and Table S2]). In addition, genes downregulated in zap70y442 are highly associated with gene ontology (GO) terms that define functions of lymphocytes and mediators of TCR signaling (see Fig. S5 and Table S3). For example, gene classes like “enzyme linked receptor protein signaling pathway,” “tyrosine kinase signaling pathway,” and “lymphocyte activation,” which are associated with proper function of TCR signaling in lymphocytes, were all downregulated in zap70y442 mutants. A smaller thymus, reduced lymphocytes in the marrow, and lack of T cell-specific gene expression in the marrow cells all demonstrate a functional defect in T cell differentiation and confirm the loss of mature T cells in zap70y442 zebrafish mutants.
zap70y442 mutants are immunocompromised and accept allografts.
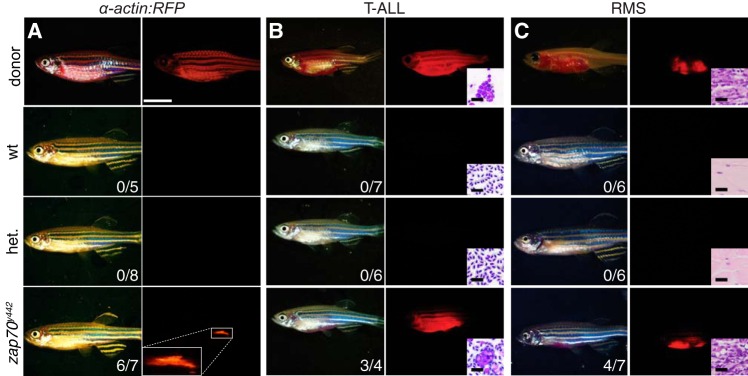
One of the principal phenotypic characteristics of immunocompromised mice and zebrafish is the loss of adaptive immune responses, resulting in an enhanced ability to engraft non-immune-matched tissue without the need for preconditioning (21, 34, 35). To assess the ability of zap70y442 mutant fish to accept allografts, we orthotopically transplanted muscle from Tg(alpha:actin-RFP) fish by intramuscular injection into 2- to 4-month-old zap70y442 mutant fish (26, 36). As expected, neither wild-type nor heterozygous animals with an intact immune system were able to accept allografts. In contrast, transgenic muscle durably engrafted into homozygous zap70y442 mutants, with donor muscle fibers remaining visible past 45 days posttransplantation (dpt) (n = 6 out of 7 [Fig. 4A]). Similar results were obtained following engraftment of malignant Myc-induced T cell acute lymphoblastic leukemia (T-ALL) (37, 38) and kRASG12D-induced embryonal rhabdomyosarcoma (ERMS) (39, 40). Tumors successfully engrafted into zap70y442 homozygous mutant fish but not wild-type or heterozygous mutant fish. Tumor engraftment persisted past 30 dpt with tumors having histology similar to that of donor tumors (T-ALL, n = 3 out of 4; ERMS, n = 4 out of 7 [Fig. 4B and C]). The ability to engraft allogeneic, unmatched cells into homozygous zap70y442 zebrafish validates T cell immune deficiency in this model.
FIG 4.
Adult zap70y442 mutant fish are immune deficient and engraft allogeneic fluorescence-labeled normal and malignant tissues. (A) Engraftment of normal muscle cells from donor alpha-actin-RFP transgenic fish (upper images) into recipient zebrafish (lower images). (B) Engraftment of mCherry-labeled Myc-induced T cell acute lymphoblastic leukemia (T-ALL) from CG1 donor fish (upper images) into recipient zebrafish (lower three images). Insets show cytospins of peripheral blood isolated from representative fish. (C) Engraftment of mCherry-labeled embryonal rhabdomyosarcoma (ERMS) from donor fish (upper images) to recipient zebrafish (lower three images). Insets show histology of muscle and tumors invading muscle. The numbers of animals with successful engraftment are indicated within the images. Scale bars equal 1 cm in panels A to C and 20 μm in insets.
Our work describes the generation and characterization of a novel zap70y442 mutant zebrafish model with a remarkably conserved T cell deficiency. Thymic histology, loss of mature T cell gene expression in the marrow, and allogeneic cellular transplantation confirm a T cell-specific immunodeficiency akin to that described for mice and humans. As with mouse models, zap70 y442 mutant zebrafish have a complete lack of mature T cells (5). This contrasts with ZAP70 deficiency in humans, where CD4+ cells are specified due to the partially redundant role of SYK in regulating T cell maturation, but these cells are nonfunctional due to an inability to sufficiently activate downstream TCR activation signaling (4, 7). Our data also indicate that similar to the case with mice, Syk does not compensate for the loss of Zap70 function in zebrafish, which results in the loss of mature T cells. zap70y442 mutant zebrafish will be a valuable tool to better understand the role of Zap70 in the immune system and T cell differentiation in vivo. Moreover, these and other immune deficiency models will be useful in chemical and genetic screens to identify drugs and pathways that restore immune cell function. Finally, our work underscores the remarkable conservation of immune cell signaling and developmental pathways regulating immune cell function, credentialing the zebrafish as a new and powerful model for studying immune deficiencies.
Supplementary Material
ACKNOWLEDGMENTS
We thank Andre Bernard for help with gene annotation, the Massachusetts General Hospital (MGH) Next Generation Sequencing Core (P30 DK040561), MGH Specialized Histopathology Services, the Dana-Farber/Harvard Cancer Center (P30 CA06516), MGH Cancer Center/Molecular Pathology Confocal Core, the MGH Pathology Flow and Image Cytometry Research Core, which obtained support from the NIH Shared Instrumentation program with 1S10OD012027-01A1, 1S10OD016372-01, S10RR020936-01, and 1S10RR023440-01A1.
J.C.M., N.T.Y., Q.T., and R.L. were supported by NIH grants R24OD016761, R01CA154923, and U54CA168512. Q.T. is funded by the China Scholarship Council. T.S.M., D.C., V.N.P., and B.M.W. were supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (ZIA-HD008808).
J.C.M., T.S.M., N.T.Y., Q.T., B.M.W., and D.M.L. designed and performed experiments. J.C.M., T.S.M., B.M.W., and D.M.L. wrote the manuscript. J.C.M., T.S.M., N.T.Y., Q.T., R.L., A.A., R.S.L., V.N.P., D.C., J.N.B., R.I.S., B.M.W., and D.M.L. performed data analysis and interpretation.
We declare no conflict of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00281-16.
REFERENCES
- 1.Wang H, Kadlecek TA, Au-Yeung BB, Goodfellow HE, Hsu LY, Freedman TS, Weiss A. 2010. ZAP-70: an essential kinase in T-cell signaling. Cold Spring Harb Perspect Biol 2:a002279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arpaia E, Shahar M, Dadi H, Cohen A, Roifman CM. 1994. Defective T cell receptor signaling and CD8+ thymic selection in humans lacking zap-70 kinase. Cell 76:947–958. doi: 10.1016/0092-8674(94)90368-9. [DOI] [PubMed] [Google Scholar]
- 3.Chan AC, Kadlecek TA, Elder ME, Filipovich AH, Kuo WL, Iwashima M, Parslow TG, Weiss A. 1994. ZAP-70 deficiency in an autosomal recessive form of severe combined immunodeficiency. Science 264:1599–1601. doi: 10.1126/science.8202713. [DOI] [PubMed] [Google Scholar]
- 4.Elder ME, Lin D, Clever J, Chan AC, Hope TJ, Weiss A, Parslow TG. 1994. Human severe combined immunodeficiency due to a defect in ZAP-70, a T cell tyrosine kinase. Science 264:1596–1599. doi: 10.1126/science.8202712. [DOI] [PubMed] [Google Scholar]
- 5.Negishi I, Motoyama N, Nakayama K, Nakayama K, Senju S, Hatakeyama S, Zhang Q, Chan AC, Loh DY. 1995. Essential role for ZAP-70 in both positive and negative selection of thymocytes. Nature 376:435–438. doi: 10.1038/376435a0. [DOI] [PubMed] [Google Scholar]
- 6.Kadlecek TA, van Oers NS, Lefrancois L, Olson S, Finlay D, Chu DH, Connolly K, Killeen N, Weiss A. 1998. Differential requirements for ZAP-70 in TCR signaling and T cell development. J Immunol 161:4688–4694. [PubMed] [Google Scholar]
- 7.Elder ME, Skoda-Smith S, Kadlecek TA, Wang F, Wu J, Weiss A. 2001. Distinct T cell developmental consequences in humans and mice expressing identical mutations in the DLAARN motif of ZAP-70. J Immunol 166:656–661. doi: 10.4049/jimmunol.166.1.656. [DOI] [PubMed] [Google Scholar]
- 8.Christie TL, Carter A, Rollins EL, Childs SJ. 2010. Syk and Zap-70 function redundantly to promote angioblast migration. Dev Biol 340:22–29. doi: 10.1016/j.ydbio.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 9.Abtahian F, Guerriero A, Sebzda E, Lu MM, Zhou R, Mocsai A, Myers EE, Huang B, Jackson DG, Ferrari VA, Tybulewicz V, Lowell CA, Lepore JJ, Koretzky GA, Kahn ML. 2003. Regulation of blood and lymphatic vascular separation by signaling proteins SLP-76 and Syk. Science 299:247–251. doi: 10.1126/science.1079477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng AM, Rowley B, Pao W, Hayday A, Bolen JB, Pawson T. 1995. Syk tyrosine kinase required for mouse viability and B-cell development. Nature 378:303–306. doi: 10.1038/378303a0. [DOI] [PubMed] [Google Scholar]
- 11.Cornall RJ, Cheng AM, Pawson T, Goodnow CC. 2000. Role of Syk in B-cell development and antigen-receptor signaling. Proc Natl Acad Sci U S A 97:1713–1718. doi: 10.1073/pnas.97.4.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sebzda E, Hibbard C, Sweeney S, Abtahian F, Bezman N, Clemens G, Maltzman JS, Cheng L, Liu F, Turner M, Tybulewicz V, Koretzky GA, Kahn ML. 2006. Syk and Slp-76 mutant mice reveal a cell-autonomous hematopoietic cell contribution to vascular development. Dev Cell 11:349–361. doi: 10.1016/j.devcel.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 13.Turner M, Mee PJ, Costello PS, Williams O, Price AA, Duddy LP, Furlong MT, Geahlen RL, Tybulewicz VL. 1995. Perinatal lethality and blocked B-cell development in mice lacking the tyrosine kinase Syk. Nature 378:298–302. doi: 10.1038/378298a0. [DOI] [PubMed] [Google Scholar]
- 14.Yanagi S, Inatome R, Ding J, Kitaguchi H, Tybulewicz VL, Yamamura H. 2001. Syk expression in endothelial cells and their morphologic defects in embryonic Syk-deficient mice. Blood 98:2869–2871. doi: 10.1182/blood.V98.9.2869. [DOI] [PubMed] [Google Scholar]
- 15.Inatome R, Yanagi S, Takano T, Yamamura H. 2001. A critical role for Syk in endothelial cell proliferation and migration. Biochem Biophys Res Commun 286:195–199. doi: 10.1006/bbrc.2001.5355. [DOI] [PubMed] [Google Scholar]
- 16.Bertozzi CC, Schmaier AA, Mericko P, Hess PR, Zou Z, Chen M, Chen CY, Xu B, Lu MM, Zhou D, Sebzda E, Santore MT, Merianos DJ, Stadtfeld M, Flake AW, Graf T, Skoda R, Maltzman JS, Koretzky GA, Kahn ML. 2010. Platelets regulate lymphatic vascular development through CLEC-2-SLP-76 signaling. Blood 116:661–670. doi: 10.1182/blood-2010-02-270876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willett CE, Zapata AG, Hopkins N, Steiner LA. 1997. Expression of zebrafish rag genes during early development identifies the thymus. Dev Biol 182:331–341. doi: 10.1006/dbio.1996.8446. [DOI] [PubMed] [Google Scholar]
- 18.Lieschke GJ, Oates AC, Crowhurst MO, Ward AC, Layton JE. 2001. Morphologic and functional characterization of granulocytes and macrophages in embryonic and adult zebrafish. Blood 98:3087–3096. doi: 10.1182/blood.V98.10.3087. [DOI] [PubMed] [Google Scholar]
- 19.Langenau DM, Ferrando AA, Traver D, Kutok JL, Hezel JP, Kanki JP, Zon LI, Look AT, Trede NS. 2004. In vivo tracking of T cell development, ablation, and engraftment in transgenic zebrafish. Proc Natl Acad Sci U S A 101:7369–7374. doi: 10.1073/pnas.0402248101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin HF, Traver D, Zhu H, Dooley K, Paw BH, Zon LI, Handin RI. 2005. Analysis of thrombocyte development in CD41-GFP transgenic zebrafish. Blood 106:3803–3810. doi: 10.1182/blood-2005-01-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang Q, Abdelfattah NS, Blackburn JS, Moore JC, Martinez SA, Moore FE, Lobbardi R, Tenente IM, Ignatius MS, Berman JN, Liwski RS, Houvras Y, Langenau DM. 2014. Optimized cell transplantation using adult rag2 mutant zebrafish. Nat Methods 11:821–824. doi: 10.1038/nmeth.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore FE, Garcia EG, Lobbardi R, Jain E, Tang Q, Moore JC, Cortes M, Molodtsov A, Kasheta M, Luo CC, Garcia AJ, Mylvaganam R, Yoder JA, Blackburn JS, Sadreyev RI, Ceol CJ, North TE, Langenau DM. 2 May 2016. Single-cell transcriptional analysis of normal, aberrant, and malignant hematopoiesis in zebrafish. J Exp Med doi: 10.1084/jem.20152013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tenente IM, Tang Q, Moore JC, Langenau DM. 2014. Normal and malignant muscle cell transplantation into immune compromised adult zebrafish. J Vis Exp 2014(94):52597. doi: 10.3791/52597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lawson ND, Weinstein BM. 2002. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol 248:307–318. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- 28.Traver D, Paw BH, Poss KD, Penberthy WT, Lin S, Zon LI. 2003. Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat Immunol 4:1238–1246. doi: 10.1038/ni1007. [DOI] [PubMed] [Google Scholar]
- 29.Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC. 2007. p53 activation by knockdown technologies. PLoS Genet 3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kok FO, Shin M, Ni CW, Gupta A, Grosse AS, van Impel A, Kirchmaier BC, Peterson-Maduro J, Kourkoulis G, Male I, DeSantis DF, Sheppard-Tindell S, Ebarasi L, Betsholtz C, Schulte-Merker S, Wolfe SA, Lawson ND. 2015. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev Cell 32:97–108. doi: 10.1016/j.devcel.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bertrand JY, Kim AD, Violette EP, Stachura DL, Cisson JL, Traver D. 2007. Definitive hematopoiesis initiates through a committed erythromyeloid progenitor in the zebrafish embryo. Development 134:4147–4156. doi: 10.1242/dev.012385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murray KN, Dreska M, Nasiadka A, Rinne M, Matthews JL, Carmichael C, Bauer J, Varga ZM, Westerfield M. 2011. Transmission, diagnosis, and recommendations for control of Pseudoloma neurophilia infections in laboratory zebrafish (Danio rerio) facilities. Comp Med 61:322–329. [PMC free article] [PubMed] [Google Scholar]
- 33.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang Q, Moore JC, Ignatius MS, Tenente IM, Hayes MN, Garcia EG, Torres Yordan N, Bourque C, He S, Blackburn JS, Look AT, Houvras Y, Langenau DM. 2016. Imaging tumour cell heterogeneity following cell transplantation into optically clear immune-deficient zebrafish. Nat Commun 7:10358. doi: 10.1038/ncomms10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hess I, Iwanami N, Schorpp M, Boehm T. 2013. Zebrafish model for allogeneic hematopoietic cell transplantation not requiring preconditioning. Proc Natl Acad Sci U S A 110:4327–4332. doi: 10.1073/pnas.1219847110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Higashijima S, Okamoto H, Ueno N, Hotta Y, Eguchi G. 1997. High-frequency generation of transgenic zebrafish which reliably express GFP in whole muscles or the whole body by using promoters of zebrafish origin. Dev Biol 192:289–299. doi: 10.1006/dbio.1997.8779. [DOI] [PubMed] [Google Scholar]
- 37.Blackburn JS, Liu S, Wilder JL, Dobrinski KP, Lobbardi R, Moore FE, Martinez SA, Chen EY, Lee C, Langenau DM. 2014. Clonal evolution enhances leukemia-propagating cell frequency in T cell acute lymphoblastic leukemia through Akt/mTORC1 pathway activation. Cancer Cell 25:366–378. doi: 10.1016/j.ccr.2014.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langenau DM, Traver D, Ferrando AA, Kutok JL, Aster JC, Kanki JP, Lin S, Prochownik E, Trede NS, Zon LI, Look AT. 2003. Myc-induced T cell leukemia in transgenic zebrafish. Science 299:887–890. doi: 10.1126/science.1080280. [DOI] [PubMed] [Google Scholar]
- 39.Langenau DM, Keefe MD, Storer NY, Guyon JR, Kutok JL, Le X, Goessling W, Neuberg DS, Kunkel LM, Zon LI. 2007. Effects of RAS on the genesis of embryonal rhabdomyosarcoma. Genes Dev 21:1382–1395. doi: 10.1101/gad.1545007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ignatius MS, Chen E, Elpek NM, Fuller AZ, Tenente IM, Clagg R, Liu S, Blackburn JS, Linardic CM, Rosenberg AE, Nielsen PG, Mempel TR, Langenau DM. 2012. In vivo imaging of tumor-propagating cells, regional tumor heterogeneity, and dynamic cell movements in embryonal rhabdomyosarcoma. Cancer Cell 21:680–693. doi: 10.1016/j.ccr.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thisse C, Thisse B. 2008. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc 3:59–69. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.