Abstract
Transcriptional networks that govern secretory cell specialization, including instructing cells to develop a unique cytoarchitecture, amass extensive protein synthesis machinery, and be embodied to respond to endoplasmic reticulum (ER) stress, remain largely uncharacterized. In this study, we discovered that the secretory cell transcription factor MIST1 (Bhlha15), previously shown to be essential for cytoskeletal organization and secretory activity, also functions as a potent ER stress-inducible transcriptional regulator. Genome-wide DNA binding studies, coupled with genetic mouse models, revealed MIST1 gene targets that function along the entire breadth of the protein synthesis, processing, transport, and exocytosis networks. Additionally, key MIST1 targets are essential for alleviating ER stress in these highly specialized cells. Indeed, MIST1 functions as a coregulator of the unfolded protein response (UPR) master transcription factor XBP1 for a portion of target genes that contain adjacent MIST1 and XBP1 binding sites. Interestingly, Mist1 gene expression is induced during ER stress by XBP1, but as ER stress subsides, MIST1 serves as a feedback inhibitor, directly binding the Xbp1 promoter and repressing Xbp1 transcript production. Together, our findings provide a new paradigm for XBP1-dependent UPR regulation and position MIST1 as a potential biotherapeutic for numerous human diseases.
INTRODUCTION
Professional secretory cells are functionally defined by their ability to synthesize, modify, concentrate, and release vast quantities of protein products, with some cells (e.g., activated B cells and pancreatic acinar cells) capable of releasing as much as 70% of their total protein content (1). Despite extensive specialization and adaptation for high protein throughput, these cells often exceed their synthesis machinery's capacity, resulting in disruptions of protein production and accumulation of misfolded proteins within the endoplasmic reticulum (ER), collectively termed ER stress (2). In order to avoid a potentially fatal disruption in protein processing, secretory cells have developed a coordinated response that involves three distinct ER membrane-embedded sensor proteins (ATF6, PERK, and IRE1) that activate three unique pathways via separate master regulator transcription factors (nuclear ATF6 [nATF6], ATF4, and XBP1) (3). This physiological reaction to excess protein production, termed the unfolded protein response (UPR), can reduce/resolve ER stress via expression of downstream gene targets that expand ER capacity, increase misfolded protein degradation, enhance chaperone expression, and generally increase protein throughput (2). In the case of unresolvable stress, the UPR can activate a proapoptotic program and trigger cell death, thus requiring cells to maintain tight control over the master regulators and their associated target genes. Indeed, disruption of the UPR and its corresponding response pathways has been linked to cancer progression as well as to other human diseases (4–7), highlighting the need to discover new regulatory and effector UPR mechanisms that can be exploited in designing strategic biotherapeutics.
Regulation of both secretion and the UPR involves coordination between many distinct cellular compartments. Secretory cells utilize transcription factor networks to alter their cytoskeletal arrangement, polarity, membrane protein composition, and organelle makeup to support the proper generation, storage, and release of protein products (8). Recently, the basic helix-loop-helix (bHLH) transcription factor MIST1 (encoded by Bhlha15) has emerged as a potent regulator of a wide variety of secretory cell functions and responses to stress (9–19). Secretory cells lacking MIST1 exhibit structural and functional defects, including altered cytoskeletal organization, a change in cell polarity, and improper cell-to-cell communication (12, 17, 19–21). Interestingly, the most severe phenotype observed in Mist1 knockout (Mist1KO) cells is a nearly complete loss of secretory capacity (21, 22), a tantalizing finding that hints at a much more extensive role for MIST1 and its target genes beyond mere regulation of cargo-cytoskeletal interactions. While several studies have investigated small numbers of MIST1 gene targets (12–14, 18, 19, 23), no comprehensive analysis of MIST1 genes has been performed that fully explains the severe secretory dysfunction observed in the absence of MIST1.
Previous studies from our group showed that pancreatic acinar cells undergoing ER stress dramatically increase expression of MIST1 but only in cells that retain XBP1 activity (24), suggesting that MIST1 and XBP1 may function coordinately to regulate cell secretion and to handle instances of UPR activity. To examine this further, we investigated the role of MIST1 as a facilitator of the UPR in pancreatic cell lines and in primary acinar cells from wild-type (WT) mice and from mice conditionally null for Mist1 (Mist1cKO) to address three crucial questions. (i) Is MIST1 a key regulator of target genes involved throughout the protein synthesis-exocytosis network? (ii) Does MIST1 transcriptionally regulate specific ER stress gene targets? (iii) Do XBP1 and MIST1 function within a shared UPR regulatory pathway? Our results demonstrate that MIST1 induction during ER stress is a common feature in pancreatic acinar cells and that its elevated expression is governed specifically by XBP1. Whole-genome chromatin immunoprecipitation with high-throughput sequencing (ChIP-Seq) coupled with transcriptome sequencing (RNA-Seq) studies identified novel MIST1 gene targets that function throughout the secretory and UPR networks, establishing MIST1 as a central regulator in these pathways. Finally, utilizing both cell lines and genetically engineered mouse models, we show for the first time that MIST1 functions as a feedback regulator of the Xbp1 gene, silencing Xbp1 expression once ER stress has resolved. Together, these results establish a unique pathway by which XBP1 and MIST1 coregulate a complex network of genes that are involved in normal secretory function and are required for correcting ER stress conditions.
MATERIALS AND METHODS
Cell culture and transfection.
Mouse embryonic fibroblasts (MEFs) were generated by the Purdue University Transgenic Mouse Core Facility. MEFs were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum, 1% l-glutamine, 1% nonessential amino acids, 0.5% penicillin-streptomycin, and 0.001% β-mercaptoethanol on standard tissue culture treated plates. Mouse acinar 266-6 cells (CRL-2151; ATCC) were maintained in high-glucose DMEM (hgDMEM) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin on 0.1% gelatin-coated tissue culture plates.
ER stress was induced in cells at 70 to 80% confluence by addition of thapsigargin (catalog number T9003; Sigma-Aldrich, St. Louis, MO) to growth medium at a final concentration of 250 nM. Cells were harvested at designated times for protein or RNA via mechanical disruption with cell scrapers. RNA was harvested in TRK lysis buffer (Omega Bio-Tek, Norcross, GA), followed by passage through homogenizer minicolumns (HCR003; Omega Bio-Tek, Norcross, GA) and subsequent isolation using E.Z.N.A. Total RNA Kit 1 (R6834; Omega Bio-Tek, Norcross, GA). Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assays were performed using an In Situ Cell Death Detection kit (11684910; Roche, Indianapolis, IN) on MEFs affixed to glass coverslips. Chromatin immunoprecipitation experiments were performed as previously described (12) utilizing the following reagents or antibodies: rabbit Ig (2 μg) (sc-2027; Santa Cruz Biotechnology, Santa Cruz, CA), MIST1 (2 μg; 5859) (25), and XBP1S (2 μg) (sc-7160; Santa Cruz Biotechnology, Santa Cruz, CA).
Transfection of MEFs was performed using XtremeGene 9 DNA transfection reagent (06365787001; Roche, Indianapolis, IN) at the recommended conditions. Transfection of 266-6 cells was accomplished using a Lonza Nucleofector 2b device (AAB-1001; Lonza, Allendale, NJ) following optimization with a Cell Line Optimization kit (VCO-1001N; Lonza, Allendale, NJ) or using Xfect transfection reagent (631317; Clontech, Mountain View, CA).
Primary acinar cell isolation and ER stress induction.
Adult (6- to 8-week-old) C57BL/6 wild-type and Mist1cKO (26) mice were given tamoxifen (Tam; 200 μl of 20 mg/ml) via oral gavage for two consecutive days, and then primary mouse acinar cells were harvested 24 h after the last Tam addition. Briefly, whole pancreatic tissue was excised and digested in 10 mg/ml collagenase P for 1 h. Acinar cells were isolated from cell homogenates via size-based filtration through a 100-μm-pore-size sterile mesh filter followed by separation based on density through a saturated bovine serum albumin (BSA) solution. Acinar cells were cultured in hgDMEM supplemented with 0.1% BSA to which either dimethyl sulfoxide (DMSO) vehicle or 500 nM thapsigargin was added. Cells were harvested by centrifugation and homogenized in TRK lysis buffer before isolation of RNA.
Protein immunoblot assays.
Whole-cell protein extracts were quantified, and 20 μg of extracts was run on 12% polyacrylamide gels, followed by transfer to polyvinylidene difluoride (PVDF) membranes and incubation with antigen-specific primary antibodies as follows: ATF6α/nATF6 (1:1,000) (sc-22799; Santa Cruz Biotechnology, Santa Cruz, CA), CHOP (1:500) (2895S; Cell Signaling, Boston, MA), HSP90 (1:4,000) (sc-7947; Santa Cruz Biotechnology, Santa Cruz, CA), LC3B (1:1,000) (3868S; Cell Signaling, Boston, MA), MIST1 (1:1,000; 5859) (25), S6 (1:1,000) (sc-74459; Santa Cruz Biotechnology, Santa Cruz, CA), XBP1S (1:1,000) (sc-7160; Santa Cruz Biotechnology, Santa Cruz, CA). Immunoblots were visualized using an enhanced chemiluminescence kit (34076; Thermo Scientific, Rockford, IL).
Reverse transcription-quantitative PCR (RT-qPCR) gene expression analysis.
Whole-cell RNA isolates were reverse transcribed using a cDNA synthesis kit (170-8891; Bio-Rad, Hercules, CA). Relative gene expression was assayed using gene-specific primer sets (sequences available upon request), FastStart Universal SYBR green (04913914001; Roche, Indianapolis, IN), and a LightCycler 96 real-time PCR system (Roche, Indianapolis, IN).
Luc reporter assays.
Cells were grown on gelatin-coated six-well plates and cotransfected with 50 ng of Renilla luciferase (Luc) expression vector and a total of 3 μg of activator/reporter plasmid DNA. Cells were harvested at 48 h posttransfection in passive lysis buffer (E153A; Promega, Madison, WI). Luciferase expression was analyzed using a Renilla luciferase assay system (E2820; Promega, Madison, WI) and a firefly luciferase assay system (E1501; Promega, Madison, WI). Relative luciferase activity was determined following normalization of firefly luciferase output to Renilla luciferase output.
MIST1 enrichment analysis.
A complete description of the ChIP-Seq and analyses procedures can be found in Jiang et al. (27). Briefly, ChIP-Seq was conducted on whole murine pancreatic lysates (total, n = 4) using two separate anti-MIST1 antibodies to minimize the effects of spurious, nonspecific antibody binding. In order to avoid bias, peak calling using the HOMER software package was performed on raw reads from each individual antibody data set, and the peak positions from the two sets were intersected to identify common peaks. Gene proximity annotation was accomplished using the MochiView software suite (28) to identify the three gene transcription start sites closest to each called peak (hard limit, 50 kbp). Enriched motifs were identified via MEME-CHIP analysis of peak data using the MEME suite of tools (29). KEGG and Swiss-Prot/UniProt keyword annotations were performed using the WebGestalt (30, 31) and DAVID (32) bioinformatics analysis platforms.
Animal studies.
Acute pancreatitis (AP) was triggered at 7 days post-Tam administration in adult (6- to 8-week-old) C57BL/6 wild-type, Mist1cKO (26), and constitutively overexpressing-Mist1 (Mist1OE) (12) mice via intraperitoneal injection of caerulein (Sigma-Aldrich, St. Louis, MO) at 50 μg/kg body weight in eight hourly injections for two consecutive days. Control mice received phosphate-buffered saline (PBS). Pancreata were harvested for RNA at 0, 2, and 4 days after the final caerulein injection. All data represent 3 to 7 mice per time point/genotype/condition. All animal studies were conducted in strict compliance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and the Purdue University IACUC guidelines under approved protocol number 1110000037.
Statistical analyses.
Statistical analyses of RT-qPCR was accomplished using multiple t test analyses and, where appropriate, corrected for multiple comparisons using the Holm-Sidak method. Graphed outputs represent mean values ± standard errors of the means unless otherwise indicated.
Accession numbers.
Raw data have been deposited in the NCBI GEO database under accession numbers GSE86288 and GSE86289.
RESULTS
ER stress induces the three UPR branches and the secretory transcription factor MIST1.
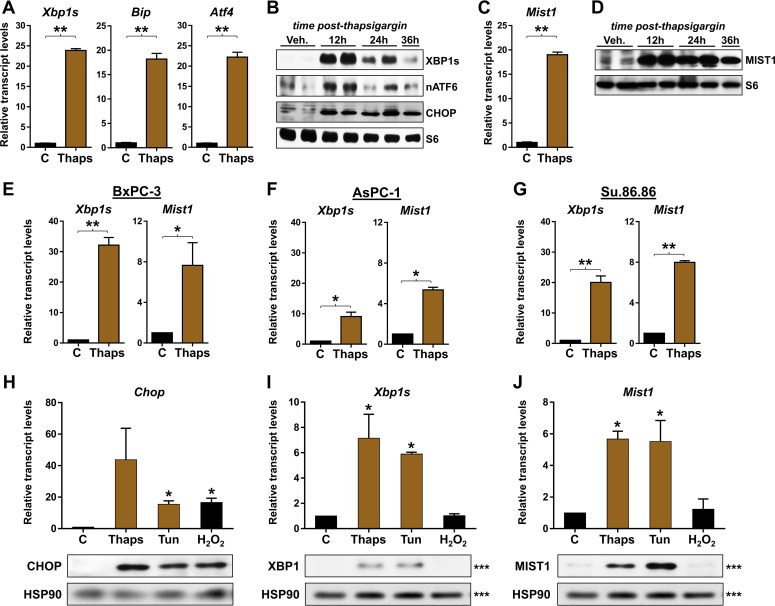
To begin investigating if the MIST transcription network directly influences both secretory and stress functions, we examined Mist1 gene expression in cells that were induced to activate the unfolded protein response (UPR) pathway. The UPR is divided into three branches (IRE1, ATF6, and PERK) that activate both unique and shared gene targets to alleviate ER stress (33). ER stress was induced by exposing 266-6 pancreatic acinar cells (34–37) to thapsigargin, a sesquiterpene that blocks calcium flux into the ER (38). As expected, thapsigargin treatment led to activation of the IRE1 branch of the UPR with elevated expression of Xbp1s, the ER stress-induced form of the Xbp1 primary transcript (Fig. 1A). Dramatic increases in Bip and Atf4, direct targets of the ATF6 and PERK UPR pathways, respectively, were also induced by thapsigargin (Fig. 1A). Consistent with published reports (39), the proteins XBP1, nuclear ATF6, and CHOP (a downstream target of the PERK pathway) were induced by thapsigargin, followed by gradual decreases in the IRE/XBP1 and ATF6 UPR branches (Fig. 1B), confirming the expected activation and execution of the entire UPR under these conditions. Interestingly, acinar cells undergoing ER stress also increased Mist1 transcripts (∼17-fold) and MIST1 protein in thapsigargin-treated cells (Fig. 1C and D). Indeed, whereas both the IRE and ATF6 branches began to resolve by 24 h postthapsigargin, MIST1 levels remained high (Fig. 1B to D), suggesting that elevated MIST1 may be critical for acinar cells to overcome ER stress conditions.
FIG 1.
MIST1 is expressed during ER stress in pancreatic cells. (A) RT-qPCR on RNA isolated from 266-6 acinar cells treated with thapsigargin (Thaps). Thapsigargin treatment leads to increased expression of the ER stress markers Xbp1s, Bip, and Atf4. (B) Immunoblotting (IB) of ER stressed cells reveals elevated levels of XBP1s, nuclear ATF6 (nATF6), and CHOP protein, demonstrating activation of all three branches of the UPR. (C and D) RT-qPCR and immunoblot analysis show the induction of Mist1 expression. (E to G) Expression analysis of the BxPC-3, AsPC-1, and Su.86.86 pancreatic ductal cell lines, as indicated, following induced ER stress. All exhibit significant induction of Xbp1s and Mist1. (H to J) RT-qPCR and immunoblot analysis of acinar cells treated with various stress agents. (H) CHOP is induced by both ER stress (thapsigargin and tunicamycin [Tun]) and oxidative stress (H2O2) agents. (I) XBP1 expression is triggered by both ER stress agents but not by oxidative stress. (J) MIST1 is induced by ER stress but not oxidative stress. *, P ≤ 0.05; **, P ≤ 0.01. Note that the XBP1, MIST1, and HSP90 immunoblots shown in panels I and J were run on a single gel (denoted by ***). The HSP90 blot is duplicated for clarity. C, control.
Because MIST1 is restricted to professional secretory cells (e.g., acinar cells), we asked if nonsecretory cells could similarly induce Mist1 expression upon ER stress. For these studies, we utilized a number of human pancreatic ductal cell lines that normally do not express Mist1 (14, 20, 25, 40, 41). In all cases, treatment of cells with thapsigargin led to induction of Xbp1s and Mist1 transcripts (Fig. 1E to G). Similar responses were observed in a number of additional nonsecretory cells, including HeLa and HEK293 cells (data not shown), implying that MIST1 induction during ER stress is a common phenomenon that extends to cells outside the secretory cell lineage.
Several studies have shown that MIST1 is essential for appropriate responses to diverse cell stimuli, including alcohol and drug-induced inflammation, development, and response to secretagogues (9, 10, 12, 26). Thus, we evaluated if induced Mist1 expression was restricted to ER stress conditions or whether other physiological stresses could influence expression of the Mist1 gene. For these studies, acinar cells were treated with three different stress-inducing agents: thapsigargin (ER stress), tunicamycin (ER stress), and hydrogen peroxide (oxidative stress). As shown in Fig. 1H, CHOP expression was induced by all treatments, consistent with activation of the PERK pathway in response to both ER stress and oxidative stress (42). In contrast, Xbp1s transcript and protein levels were induced only in cells treated with the two ER stress agents (Fig. 1I). Importantly, Mist1 expression was similarly induced by the ER stressors but not in response to oxidative stress (Fig. 1J), demonstrating that MIST1 induction occurs in an ER stress-specific manner.
XBP1 activates Mist1 gene expression.
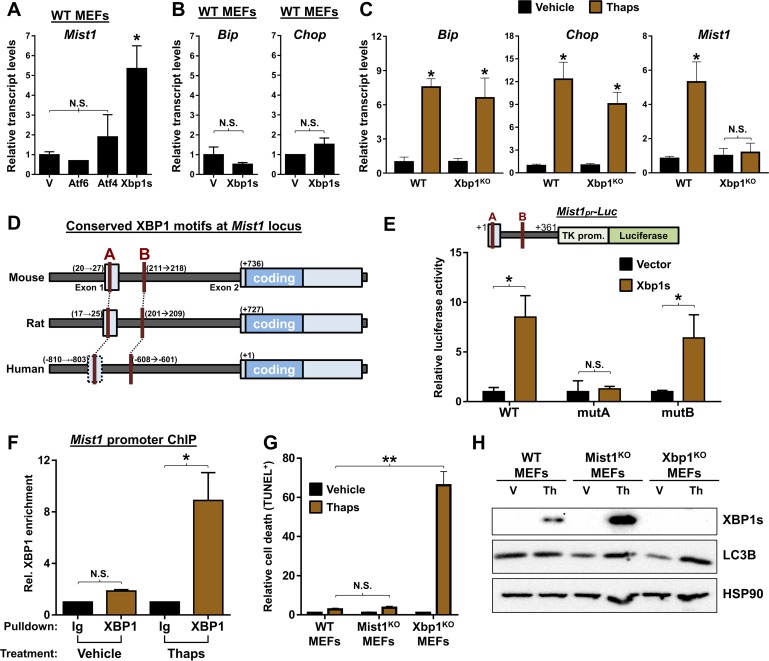
The three branches of the UPR exhibit extensive overlap in both their downstream targets and in the transcriptional networks that become activated in response to ER stress (43–46). Although previous studies have linked the UPR to MIST1 activity (19, 24, 47), the extensive cross talk and overlap between distinct UPR branches has complicated identifying which UPR branch specifically leads to induction of MIST1. To address this shortcoming, we utilized mouse embryonic fibroblasts (MEFs) to take advantage of genetic models and to examine UPR responses independently of the complex protein processing machinery found in secretory cells. As a first step, WT MEFs were transfected with expression plasmids encoding each of the three master regulator transcription factors (nATF6, ATF4, and XBP1s). As shown in Fig. 2A, only XBP1 generated an endogenous Mist1 gene response even though XBP1 alone failed to elicit a UPR signature for other downstream effectors (Bip and Chop) (Fig. 2B). Mist1 gene expression was then examined in Xbp1KO MEFs following thapsigargin treatment. Consistent with pancreatic acinar cells, Xbp1KO MEFs exhibited full UPR induction with no difference in resting or induced Bip or Chop levels compared to those in WT MEFs (Fig. 2C). However, despite induced ER stress and activation of the ATF6 and PERK pathways, Mist1 gene expression failed to be induced in the Xbp1KO MEFs (Fig. 2C).
FIG 2.
Mist1 is a direct transcriptional target of XBP1 during ER stress. (A) RT-qPCR analysis of WT mouse embryo fibroblasts (MEFs) transfected with expression plasmids for Atf6, Atf4, or Xbp1s. Mist1 levels are significantly increased only in response to Xbp1s. V, empty vector. (B) RT-qPCR analysis of Xbp1s transfected WT MEFs. ER stress indicators Bip and Chop are not induced, indicating lack of UPR activation/ER stress. (C) Expression analysis of WT and Xbp1KO MEFs treated with thapsigargin. Cells lacking Xbp1 induce Bip and Chop but cannot trigger increased expression of Mist1. (D) Schematic of the Mist1 promoter in mice, rats, and humans reveals two conserved unfolded protein response elements (red A and B), a known DNA binding site for XBP1. (E) Cotransfection of an Xbp1 expression vector with luciferase reporter constructs containing the Mist1 promoter (Mist1pr-Luc) in 266-6 cells. The WT Mist1 promoter is sufficient to drive luciferase expression in the presence of XBP1. In contrast, mutation of the site A UPRE (mutA) prevents XBP1-mediated luciferase expression. TK prom, thymidine kinase promoter. (F) Anti-XBP1 chromatin immunoprecipitation (ChIP) and quantitative PCR reveal significant enrichment of XBP1 at the Mist1 promoter in ER stressed cells. Ig, control anti-IgG. (G) Quantification of TUNEL staining in MEFs derived from WT, Mist1KO, and Xbp1KO mice. Xbp1KO MEFs exhibit significantly more cell death upon induction of ER stress via thapsigargin. (H) Immunoblots of XBP1 and LC3B in WT, Mist1KO, and Xbp1KO MEFs. Mist1KO MEFs induce high levels of XBP1 upon ER stress induction. Mist1KO and Xbp1KO MEFs also show elevated LC3B in ER stressed cells compared to levels in WT MEFs. *, P ≤ 0.05; **, P ≤ 0.01; N.S., not significant.
Given the genetic evidence that Xbp1 is required for ER stress-mediated Mist1 induction, we searched for XBP1 binding sites, termed unfolded protein response elements (UPREs), within the Mist1 promoter. Sequence analysis of the mouse Mist1 gene revealed two UPREs at positions +20 (site A) and +211 (site B) that are conserved in sequence, location, and spacing in most mammalian Mist1 genes (Fig. 2D). To determine if these sites are required for XBP1-mediated induction of Mist1, individual Mist1 gene regions were tested by cell transfection assays. As shown in Fig. 2E, cotransfection of 266-6 cells with an Xbp1s expression plasmid with a WT Mist1-Luc reporter containing both UPREs led to high luciferase expression. Similar results were obtained when the site B UPRE was mutated (mutB), leaving only site A intact. However, XBP1 failed to induce Mist1-Luc expression when site A was mutated (mutA) (Fig. 2E), demonstrating that the UPRE at +20 is required for XBP1-driven expression of Mist1 in vitro. Chromatin immunoprecipitation (ChIP) was then employed with primers flanking the site A UPRE. As predicted, no appreciable binding of XBP1 in cells under basal conditions occurred (Fig. 2F). However, when ER stress was induced by thapsigargin, robust XBP1 binding to the Mist1 site A UPRE was readily detected (Fig. 2F).
Finally, to assess whether MIST1 could be operating as a component of the IRE1/XBP1 branch of the UPR, we examined MEFs derived from WT, Mist1KO, and Xbp1KO mice. Thapsigargin was used to induce ER stress, and then cells were monitored for cell survival and levels of autophagy, a cellular process known to be increased in the absence of XBP1 (24, 48). Quantification of TUNEL staining in stressed MEFs (Fig. 2G) revealed no significant difference between stressed WT and Mist1KO cells, while ablation of Xbp1 resulted in a large increase in apoptotic events, consistent with previous work demonstrating the necessity of XBP1 for cell survival (24). Interestingly, analysis of protein extracts from these cells (Fig. 2H) revealed substantially increased expression of XBP1 in Mist1KO MEFs as well as a marked increase in induction of LC3B, a marker of cellular autophagy. Collectively, these studies demonstrate that XBP1 triggers increased Mist1 expression during ER stress and that loss of MIST1 (or its targets) causes aberrant XBP1 expression and increased autophagy.
MIST1 associates with genes that function within the entire protein synthesis, processing, and secretory network.
Pancreatic acinar cells lacking MIST1 lose nearly all ability to secrete digestive enzymes and have significantly increased basal cellular autophagy, characteristics that are found under ER stress conditions as well as in cells lacking Xbp1 (12, 21, 24, 49, 50). The observed XBP1-mediated induction of Mist1 during ER stress, coupled with the marked reduction in secretion associated with Mist1KO acinar cells (12, 21), prompted us to investigate if MIST1 might regulate a previously uncharacterized set of gene targets needed for both basal and ER stress-induced maintenance of the secretory machinery. For these studies, chromatin from WT mouse pancreata was isolated using two independently derived MIST1 antibodies and subsequently analyzed via ChIP-Seq strategies. Peak calling on each data set was cross-referenced to identify only conserved binding regions using both MIST1 antibodies (see Materials and Methods). Consistent with previous reports on MIST1 DNA binding (18, 51), CAGCTG (GC) and CATATG (TA) E-box sites were the most enriched motifs within all MIST1 peaks, with an expected enhanced distribution centered over the transcription start site (TSS), mirroring the genome-wide MIST1 binding distribution. As predicted, several previously confirmed MIST1 targets were well represented in this analysis, including members of the RAB family of cytoskeletal adapters (18) and the proapoptotic gene Htra2 (12).
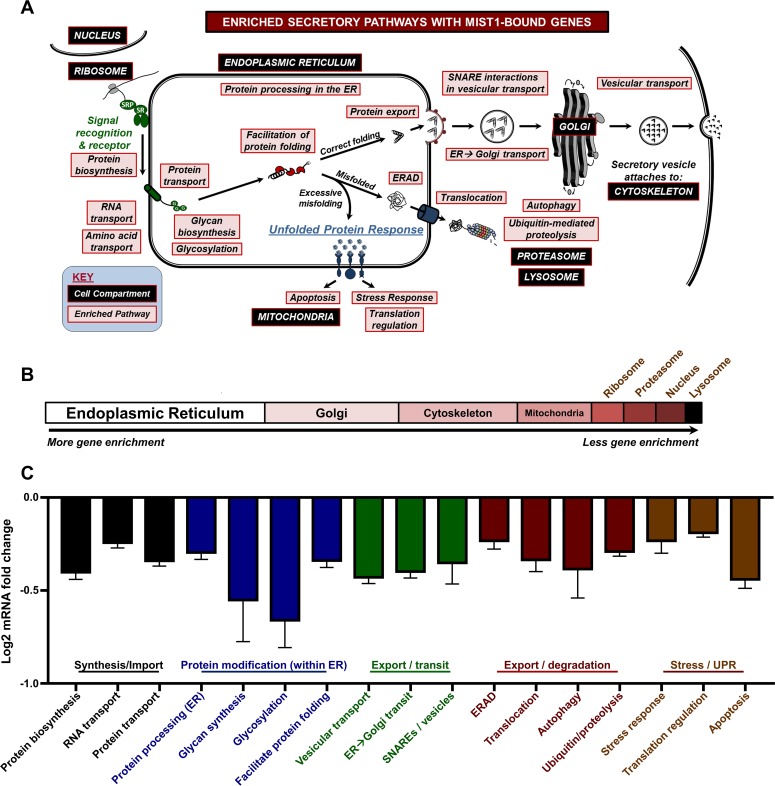
The list of proximally bound MIST1 genes was annotated using two separate databases (KEGG and Swiss-Prot Protein Information Resource [SP-PIR]) to determine whether the putative targets were statistically overrepresented in specific compartments or pathways associated with secretion. Remarkably, MIST1-bound genes encode proteins that function within critical pathways across the entire spectrum of the secretory pathway, having essential roles in protein biosynthesis, RNA, protein and amino acid transport, protein modification, protein translocation, vesicular transport and packaging, autophagy, ubiquitin-mediated proteolysis, and secretory vesicle-regulated exocytosis (Fig. 3A). A subset of these genes also function within the ER to facilitate protein folding and ER protein export, to remove misfolded proteins through the ER-associated protein degradation (ERAD) pathway, and to function within the UPR to relieve stress or induce apoptosis. Analysis of the secretory-related gene enrichment terms revealed that the highest represented single cellular compartment was the ER, followed by the Golgi compartment, cytoskeleton, and mitochondria (Fig. 3B). To further investigate these pathways, we utilized RNA-Seq to evaluate global gene expression differences in adult pancreata isolated from WT and Mist1KO mice (27). As shown in Fig. 3C, putative target genes throughout the secretory pathway (identified via ChIP-Seq) were substantially reduced in Mist1KO samples. Indeed, a high percentage of these genes (∼40%) had proximal MIST1 binding peaks as determined by ChIP-Seq. Together, these data reveal that MIST1 binds and regulates a large set of genes whose products have a significant function in many aspects of the protein synthesis, processing, and secretory pathways.
FIG 3.
Genome-wide identification of MIST1 targets reveals significant association with components of the entire cell secretory machinery and the ER. (A) Cellular processes and compartments involved in protein throughput and maintenance of the secretory machinery. MIST1 targets are associated with each indicated pathway (red boxes) across the entire spectrum of the secretory network. (B) The distribution of MIST1-enriched pathways. The ER is shown as the top gene enrichment category. (C) Relative expression of MIST1-bound targets in Mist1KO acinar cells. Analysis of Mist1KO RNA-Seq data revealed similarly enriched pathways throughout the secretory pathway. For each category, a majority of MIST1 targets are downregulated in Mist1KO samples. Only categories and expression results with a P value of ≤0.01 are included.
MIST1 is bound to secretory genes known to be effectors of XBP1.
Analysis of MIST1-associated genes within the full spectrum of the secretory network identified the ER as the top enriched pathway in acinar cells (Fig. 3B). This was particularly interesting because our studies also showed that XBP1, a master regulator of ER stress through the UPR, directly elevates Mist1 gene expression under ER stress conditions and is itself elevated in the absence of MIST1, suggesting that both transcription factors function within overlapping regulatory networks. Indeed, MIST1 and XBP1 have a role in a number of shared pathways outside the UPR, including controlling the maturation of secretory cell lineages for immunoglobulin-producing B cells (52), gastric zymogenic cells (19), and pancreatic acinar cells (53), and in regulating numerous secretory pathway-related genes (12, 54). As such, Mist1- and Xbp1-deficient exocrine cells share similar phenotypes, including altered expression of terminal differentiation markers, erroneous secretory granule transport, increased autophagy, and decreased rough endoplasmic reticulum (12, 19, 24, 47). This phenotypic similarity, coupled with the identification of MIST1 gene targets that are enriched in ER biology, prompted us to investigate if XBP1 and MIST1 coregulate a shared set of genes. As a first step, we performed a comprehensive analysis of microarray data from Mist1KO pancreata (12), known UPR-induced secretory genes (54, 55), and MIST1 called peaks from our ChIP-Seq studies. Interestingly, 44% of all UPR-induced secretory genes exhibited altered expression in Mist1KO pancreata (Fig. 4A), and within this gene set 61% contained MIST1 peaks in their regulatory regions (Fig. 4B). To narrow the broad UPR pathway, we next screened XBP1-dependent secretory effector genes that previously were identified in XBP1 transduced fibroblasts (54). A full 70% of the identified XBP1 effector genes (80 out of 115) contained MIST1 peaks within their respective control regions or within the control regions of closely related proteins of the same family (Fig. 4C). Classification of the XBP1 effector genes revealed substantial MIST1 binding in every secretory pathway category (Fig. 4D), again demonstrating that MIST1-bound genes operate throughout the secretory spectrum, including participating within a large swath of the XBP1 target network. Indeed, promoter analysis of known XBP1 targets for XBP1 and MIST1 DNA-binding motifs showed a strong correlation across a large number of MIST1-bound targets where 33% of XBP1 effectors contain both UPRE sites and MIST1 motifs within 500 bp of each other and within 1 kb of the TSS (Fig. 4E). These data strongly support a model of direct regulation of shared gene targets by XBP1 and MIST1.
FIG 4.
MIST1-bound genes include XBP1 secretory effectors. (A) Venn diagram of previously identified UPR-induced secretory genes that exhibit altered expression in Mist1KO pancreata. (B) MIST1 ChIP-Seq peaks are associated with 61% of known UPR-induced secretory genes. (C) Analysis of known XBP1 secretory effector genes reveals that 70% exhibit MIST1 binding. (D) Representation of XBP1 effectors versus MIST1-bound genes categorized by function within the protein secretion pathway. MIST1 binding is common in all classes. (E) The presence of MIST1 and/or XBP1 DNA-binding motifs and bound MIST1 in the promoters of known XBP1 secretory targets. Approximately one-third of all promoters contain either MIST1 only, XBP1 only, or MIST1 and XBP1 motifs, with nearly half of the promoters with MIST1 motifs containing bound MIST1.
MIST1 contributes to the regulation of ER stress response genes.
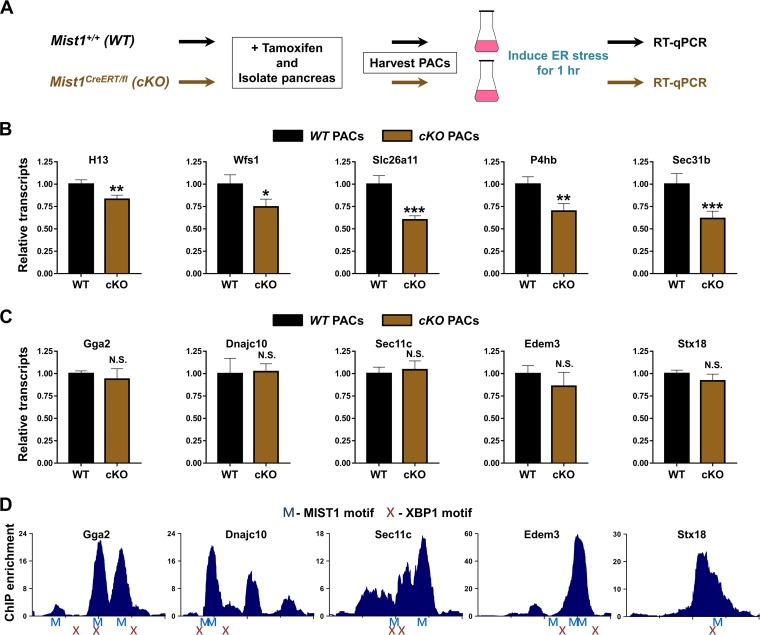
Our studies have shown that MIST1 controls target genes that are involved throughout the protein synthesis/processing spectrum under normal secretory basal conditions. Bioinformatics analyses also suggested that MIST1 regulates key genes during ER stress in response to activation of the IRE pathway. In order to examine this in greater detail, we took advantage of a newly developed Mist1 conditional mouse model where one Mist1 allele is flanked by loxP sites while the other Mist1 allele encodes CreERT2 (26). Tamoxifen-treated Mist1CreERT/fl mice rapidly delete the remaining Mist1 coding allele, generating an acinar-specific Mist1 null pancreas. These mice, termed Mist1cKO, permitted the study of immediate effects of Mist1 ablation exclusively in mature acinar cells. To minimize islet, duct, and stromal cell contaminants, pancreatic acinar cells were first purified (56) from tamoxifen-treated WT and Mist1cKO mice. Isolated acinar cells were then subjected to ER stress for 1 h, and RNA was isolated for expression profiling to evaluate how loss of MIST1 affects immediate secretory gene expression upon ER stress induction (Fig. 5A).
FIG 5.
MIST1 regulates a subset of secretory genes in response to ER stress. (A) Schematic of the experimental design. Mist1 WT and Mist1cKO (cKO) acinar cells were isolated from mouse pancreata and subjected to ER stress by thapsigargin treatment for 1 h prior to RNA isolation. (B and C) MIST1 target gene expression in acinar samples during ER stress as determined by RT-qPCR analysis. For all genes shown in panel B, Mist1cKO samples exhibited reduced responses to ER stress. In contrast, none of the genes shown in panel C exhibited significant differences in expression levels between WT and Mist1cKO samples. (D) MIST1 ChIP-Seq peaks (−1000 to +1000 centered at the TSS) for the genes shown in panel C. Identified MIST1 (M) and XBP1 (X) motifs are indicated below each trace. All XBP1 sites are located within 500 bp of a MIST1 site. *, P ≤ 0.1; **, P ≤ 0.05; ***, P ≤ 0.01; N.S., not significant.
As predicted, a number of MIST1-bound secretory target genes across a range of functional classes exhibited significantly reduced expression in purified Mist1cKO acinar cells subjected to ER stress (Fig. 5B). These MIST1-regulated genes included the signal peptidase and putative Xbp1-regulatory molecule H13 (57), the stress-induced transcription factor Wfs1 (58), and the plasma membrane-bound transporter Slc26a11 (59). The loss of MIST1 also resulted in reduced expression of canonical UPR-induced target classes, including chaperones (P4hb) and protein exporters (Sec31b) (54, 60). These results support the previous genome-wide observations (Fig. 3 and 4) regarding the breadth of MIST1's transcriptional activity.
Despite the clear requirement for MIST1 in maintaining proper gene regulation during times of ER stress, we were surprised to identify a number of MIST1-bound targets that were equivalently expressed in both WT and Mist1cKO cells under stress conditions (Fig. 5C). In an attempt to explain this observation, we screened the promoters of genes with normal expression (TSS ±1,000 bp) to verify the presence of MIST1 GC/TA E-box motifs. As shown in Fig. 5D, MIST1 motifs (blue Ms) were always found in close proximity to both the TSS and the local MIST1 ChIP-Seq maxima (blue histogram) within these genes. Interestingly, sequence analyses also revealed coenriched XBP1 binding motifs (Fig. 5D, red X) in close proximity (<200 bp) to both the MIST1 motif sites and MIST1 ChIP-Seq peaks (Fig. 5D). Thus, the lack of MIST1-dependent regulation during ER stress in this class of genes is most likely due to induced XBP1, which can itself regulate genes with UPRE sites. Importantly, MIST1 genes that exhibit significantly reduced expression in Mist1cKO samples under ER stress (Fig. 5B) are devoid of proximal XBP1 motifs. Combined with our findings regarding MIST1 activity in basal pancreatic acinar cells, these data support a model of MIST1 as a regulator of multiple genes in the basal secretory pathway but with additional transcriptional targets and activities that are controlled only during ER stress.
MIST1 acts within a feedback regulatory pathway that suppresses Xbp1.
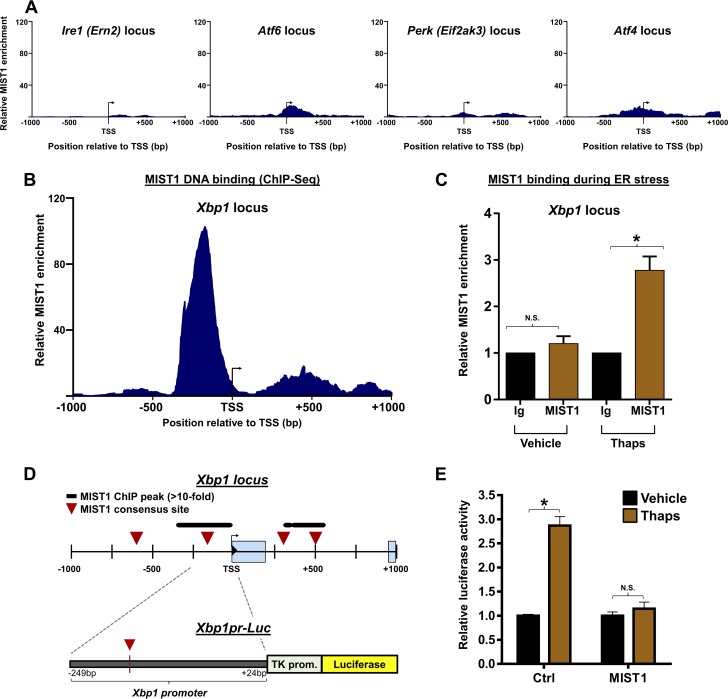
The discovery that MIST1 functions as an active transcriptional regulator under ER stress conditions prompted us to investigate if MIST1 itself could be responsible for altering expression of any of the three master regulator transcription factors (ATF6, ATF4, and XBP1) or their effector proteins within the UPR. Analysis of MIST1 ChIP-Seq data indicated that MIST1 was not significantly enriched at TSS regions of any of the primary sensors (Ire1, Atf6, and Perk) or Atf4 (Fig. 6A). However, the Xbp1 promoter (Xbp1pr) contained two prominent MIST1 peaks centered over E-box sites proximal to the Xbp1 TSS (Fig. 6B). MIST1 binding to Xbp1 was subsequently confirmed by small-scale ChIP using chromatin isolates from cells treated with thapsigargin (Fig. 6C). Thus, ER stress results in increased MIST1 expression followed by direct MIST1 binding to the promoter of its activator, Xbp1.
FIG 6.
MIST1 binds to the Xbp1 promoter. (A) ChIP-Seq plots showing the absence of MIST1 enrichment in the TSS regions of the major UPR activator proteins. (B) ChIP-Seq plot of MIST1 binding at the Xbp1 locus. MIST1 binding is centered on an E-box site immediately upstream of the Xbp1 TSS. (C) Anti-MIST1 ChIP confirms elevated MIST1 binding on the Xbp1 promoter in cells undergoing ER stress. Anti-IgG (Ig) served as a control. (D) Schematic of the Xbp1 locus with MIST1-bound peaks and consensus sites shown (top) and the Xbp1pr-Luc gene construct (bottom). (E) Xbp1pr-Luc activity in cells without or with exogenous MIST1 and treated with vehicle or thapsigargin to induce ER stress. MIST1 suppresses the ER stress-induced expression of the Xbp1pr-Luc reporter gene. *, P ≤ 0.05; N.S., not significant.
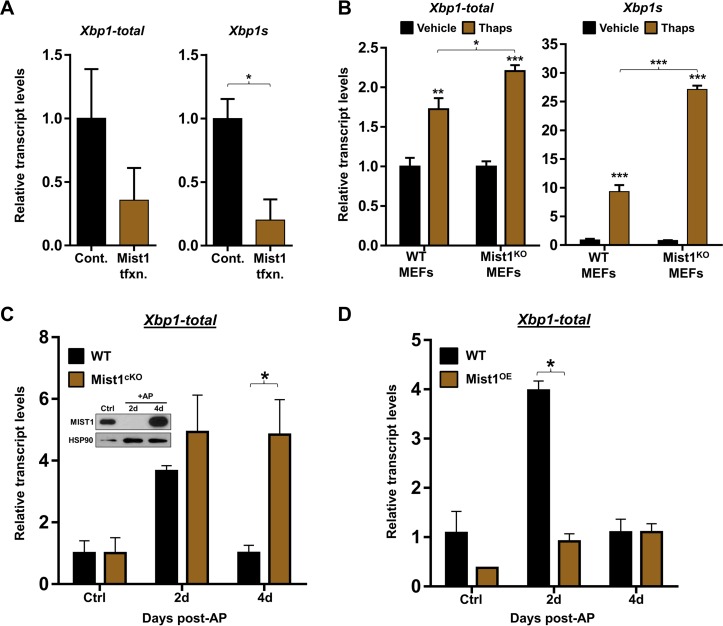
To elucidate how MIST1 affects Xbp1 gene expression, we generated an Xbp1pr-Luc plasmid that contained the upstream MIST1 consensus site at position −163 (Fig. 6D). Cells were then transfected with Xbp1pr-Luc and a control or Mist1 expression plasmid and subjected to ER stress by thapsigargin treatment. As predicted, Xbp1pr-Luc expression was induced in cells undergoing ER stress (Fig. 6E). However, cells cotransfected with Mist1 failed to activate Xbp1pr-Luc expression upon ER stress induction (Fig. 6E). In complementary experiments, we monitored endogenous Xbp1 transcript levels in ER stress-induced cells with and without transfected Mist1. As shown in Fig. 7A, MIST1 similarly inhibited endogenous Xbp1 gene expression, suggesting that MIST1 functions as a transcriptional repressor when bound to the Xbp1 locus. As a second confirmation, WT and Mist1KO MEFs were subjected to ER stress induction and monitored for induced Xbp1 expression. As predicted, Mist1KO MEFs undergoing ER stress produced significantly higher levels of Xbp1 transcripts (both total Xbp1 and spliced Xbp [Xbp1s]) than WT MEFs (Fig. 7B), again supporting the idea that MIST1 negatively regulates Xbp1 expression.
FIG 7.
MIST1 represses Xbp1 gene expression. (A) RT-qPCR analysis of acinar cells transfected with a Mist1 expression construct. MIST1 inhibits expression of total Xbp1 and Xbp1s transcript levels. Cont, control; tfxn, transfection. (B) RT-qPCR analysis of RNA harvested from WT and Mist1KO MEFs treated with thapsigargin for 12 h. Mist1KO MEFs express significantly higher total Xbp1 and Xbp1s transcript levels than WT MEFs under ER stress conditions. (C) RT-qPCR analysis of total Xbp1 expression in WT and Mist1cKO pancreatic isolates following caerulein-induced acute pancreatitis (AP). The inset shows loss of MIST1 protein at 2 days post-AP in WT pancreas (26). Loss of Mist1 is correlated with increased expression of total Xbp1 at 2 days post-AP, the time of maximum tissue damage. During AP recovery (4 days post-AP), total Xbp1 levels return to control basal levels in WT mice when the MIST1 protein is reexpressed (inset). However, total Xbp1 expression remains high in Mist1cKO cells. d, day. (D) RT-qPCR analysis of total Xbp1 expression in WT and Mist1-overexpressing (Mist1OE) pancreata during AP. In contrast to the Mist1cKO results, Mist1OE pancreata fail to induce total Xbp1 expression at 2 days post-AP. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Finally, we examined how the presence or absence of MIST1 influenced Xbp1 gene expression during recovery from pancreatitis, a condition that can lead to persistently high ER stress levels (61, 62). Studies by Lin et al. (39) have shown that sustained ER stress results in the eventual downregulation of Xbp1, a necessary step as cells transition from a prosurvival UPR (governed primarily by XBP1 and ATF6 targets) to a proapoptotic UPR (governed primarily by PERK targets). To determine if MIST1 alters Xbp1 gene expression during pancreatitis, two transgenic mouse models were used in which MIST1 was either conditionally deleted (Mist1cKO) or constitutively overexpressed (Mist1OE) via acinar-specific Cre-mediated recombination (12, 26). Acute pancreatitis (AP) was induced in WT, Mist1cKO, and Mist1OE mice by caerulein injection, and pancreatic RNAs were isolated 2 and 4 days post-AP, corresponding to the maximum cell damage and initial recovery phases, respectively (26). Importantly, Mist1 is transiently downregulated at 2 days post-AP in this model (Fig. 7C, inset) (26). As expected, WT mice produced a marked increase in Xbp1 levels by 2 days post-AP, which fully returned to control levels by 4 days post-AP when Mist1 expression resumed and the pancreas recovered (Fig. 7C). In contrast, Mist1cKO pancreata demonstrated a significantly reduced ability to repress Xbp1 expression during AP recovery. Indeed, Mist1cKO pancreata had nearly five times the levels of Xbp1 transcripts at 4 days post-AP compared to levels in WT pancreata under the same conditions (Fig. 7C). As predicted, Mist1OE mice exhibited the opposite result. In this instance, sustained Mist1 expression inhibited induced Xbp1 transcripts at 2 days post-AP (Fig. 7D). Importantly, there was no significant difference in the expression level of the ER stress marker Bip or Chop in WT, Mist1cKO, or Mist1OE mice induced to undergo AP (data not shown). Together, these paired data sets support a model where XBP1-dependent induction of Mist1 expression during ER stress triggers a subsequent negative feedback pathway in which MIST1 silences Xbp1 gene expression.
DISCUSSION
Higher eukaryotes utilize complex systems that allow individual cells to coordinate and respond to the demands of an ever-changing environment. Secretory cells and the products they manufacture are often at the center of these systems, facilitating the generation and release of response molecules ranging from digestive enzymes to antibodies and hormones. Appropriate safeguards to monitor synthesis, processing, and exocytosis in these cells are critical to human health as they prevent and mitigate multiple disease states resulting from ineffective quality control (e.g., Alzheimer's, Huntington's, and irritable bowel syndrome) or failure to execute apoptotic programs in response to stress (e.g., malignancy) (6, 63–65). These safety mechanisms are driven by response systems coupling protein stress sensors with downstream transcriptional networks. Multiple transcription factors then must coordinately regulate the expression of effector molecules that are essential for ensuring normal protein production/transport as well as regulating complex modifications to the secretory apparatus to relieve stress. In this study, we investigated the importance of the secretory cell transcription factor MIST1 as a regulator of normal protein-secreting function and of the unfolded protein response, the fundamental pathway by which secretory cells adapt and survive in the face of extreme protein demand. Our results uncovered a unique role for MIST1 as both a target and enhancer of the UPR, serving as a transcriptional regulator of genes utilized throughout the secretory and ER stress response pathways and functioning as a previously uncharacterized feedback inhibitor of XBP1, a master regulator of the unfolded protein response.
Pancreatic acinar cells under ER stress rapidly increase Mist1 gene expression in an XBP1-dependent fashion, in which XBP1 binds to an evolutionarily conserved UPRE motif adjacent to the Mist1 TSS. This observation is consistent with previous studies showing that gastric chief cells also induce Mist1 during ER stress (19), and it supports the hypothesis that transient ER stress and Xbp1s expression may be the developmental trigger that activates Mist1 within regenerating stomach crypts. This is in contrast to pancreatic acinar cells, in which developmental expression of Mist1 is achieved by the acinar transcription factor complex PTF1 (27) while the ER stress-induced enhancement of MIST1 reported here is fully dependent on XBP1. Interestingly, a modest 2-fold increase in Ptf1a expression is also observed during thapsigargin-induced ER stress (unpublished results), suggesting that the PTF1 complex might play a minor role in augmenting Mist1 gene expression in ER-stressed pancreatic acinar cells. The absence of Ptf1a expression in gastric chief cells could also account for why disruption of the XBP1-MIST1 network results in dissimilar outcomes in survival and secretion in gastric chief cells and pancreatic acinar cells (19, 24), implying that unique transcriptional targets and mechanisms exist in these two cell populations. The concept that development and ER stress utilize different approaches to control Mist1 expression supports a model in which MIST1 regulates a large but variable set of downstream target genes that depend on diverse cellular contexts. Importantly, this finding positions MIST1 as a potential target molecule for pancreas-specific and other secretory cell malignancies and diseases.
MIST1 can function as a transcriptional activator or repressor for individual targets (12, 23, 66), giving it enormous capability to modulate and fine-tune cellular responses. Indeed, the breadth of MIST1's influence throughout the cell has hindered previous attempts to determine why cells lacking MIST1 exhibit a crippled capacity to synthesize and secrete protein products (12, 21). Our recent ChIP-Seq studies, coupled with genome-wide screening of MIST1-bound gene promoters, revealed that MIST1 is uniformly associated with nearly every step of protein production, from initial protein biosynthesis to RNA, protein and amino acid transport, protein modification, ER export, vesicular packaging, and secretory vesicle exocytosis. Indeed, Mist1-deficient acinar cells have significantly reduced expression of a large number of genes that are responsible for basal maintenance of the secretory machinery as well as genes that are important in resolving ER stress. MIST1's widespread transcriptional regulatory role throughout the secretory network establishes it as a key component in ensuring the proper function of these pathways. This broad role also explains, for the first time, the complex defective exocytosis phenotype associated with Mist1KO mice (12, 18, 19, 21, 67).
The transcriptional role of MIST1 during ER stress is primarily to augment the secretory pathway rather than to initiate new expression of target genes, consistent with previous classifications of MIST1 as a “scaling factor,” a unique class of transcriptional regulators that serve to enhance the effects of other transcription complexes (67). Despite multiple examples of MIST1's scaling effects, the specific means by which MIST1 achieves this end have not been established. Here, we identified a subset of MIST1-bound secretory target genes that contain adjacent XBP1 and MIST1 DNA-binding consensus motifs within their control regions, suggesting that XBP1 and MIST1 jointly regulate these genes upon activation of the UPR pathway. Importantly, MIST1's transcriptional activity alongside XBP1 is stress dependent, again suggesting that cell state plays a critical role in how MIST1 functions. Thus, in the context of ER stress, the scaling effect of MIST1 is achieved by amplifying an initial transcriptional activation achieved by XBP1. This model is supported by additional findings showing that MIST1 similarly collaborates with PTF1 to jointly regulate developmental gene targets via proximal DNA binding (27). The many observations that MIST1's transcriptional activity typically produces moderate changes in gene expression levels rather than functioning as a more customary binary on/off switch support this supposition.
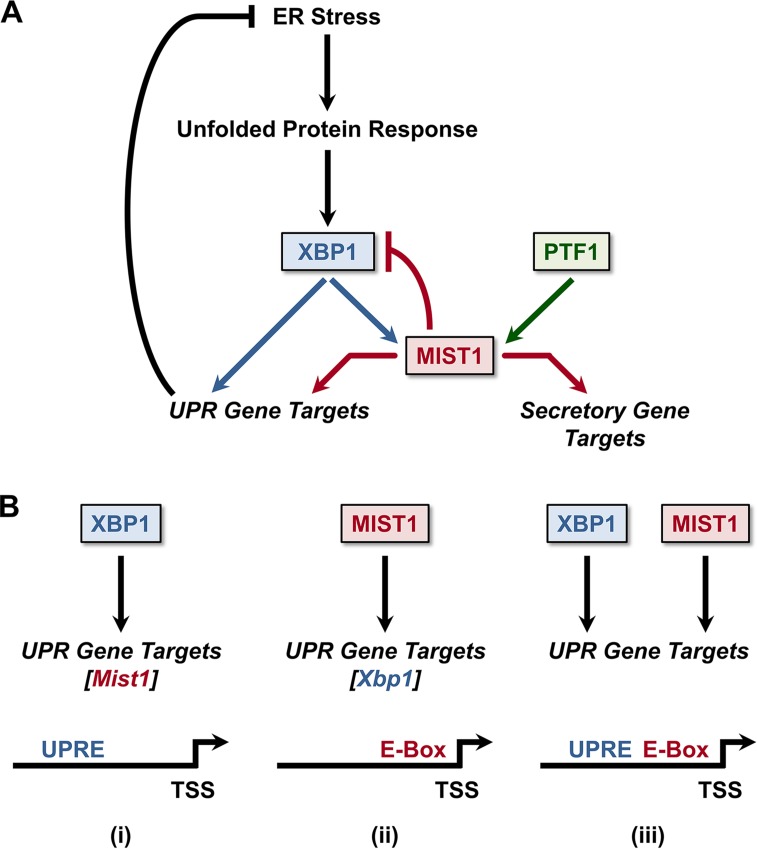
Collectively, we propose that MIST1 facilitates protein production and viability of secretory cells during both basal and stressed states (Fig. 8A). In this model, Mist1 gene expression is induced by PTF1 as part of an acinar cell program that regulates shared and unique MIST1 targets throughout the broad secretory pathway. However, upon induction by XBP1 during ER stress, MIST1 enhances the UPR response by coregulating a subset of secretory genes and UPRE-containing XBP1 target genes. Our studies have also revealed that MIST1 functions within a negative feedback loop by directly binding the Xbp1 promoter and repressing its expression. Importantly, XBP1 levels can also influence other UPR branches and regulate the switch between prosurvival and proapoptotic components of the UPR (33, 39, 68). Thus, MIST1 may have an additional role in ultimately regulating the PERK and ATF6 UPR pathways, consistent with published reports of aberrant ER stress responses outside the XBP1 branch in Mist1KO animals (16). Additional studies will be needed to determine if added layers of regulation of XBP1, MIST1, and/or downstream MIST1 targets operate under different physiological states.
FIG 8.
The XBP1/MIST1 gene regulatory network. (A) MIST1's basal expression in the pancreas is maintained by PTF1, the master regulator of acinar cell identity, to orchestrate full secretory activity in pancreatic acinar cells. During ER stress, XBP1 expression leads to a further increase in MIST1 levels as well as a switch in target gene preferences, allowing MIST1 to directly regulate expression of additional genes whose products facilitate recovery from ER stress. MIST1 subsequently binds to the Xbp1 promoter to negatively limit Xbp1 expression as ER stress is resolved. (B) ER stress response genes can be regulated by three distinct mechanisms: (i) XBP1 binding to UPRE-containing genes, (ii) MIST1 binding to E-box-containing genes, or (iii) XBP1 and MIST1 interacting with genes that contain both UPRE and E-box motifs.
The proposed model also predicts three distinct classes of UPR effector genes regulated by XBP1 or MIST1. One target class (including Mist1) contains only UPRE sites within its control regions and is induced by direct XBP1 binding (Fig. 8B). A second class (including Xbp1) contains promoter-localized GC/TA E-box MIST1 binding sites and is induced by direct MIST1 binding while a third class contains promoter regulatory regions that harbor both UPRE and E-box sites in close proximity (<200 bp) to each other. This subset likely is activated by XBP1, with MIST1 providing a scaling enhancement during ER stress. In support of these predictions, analysis of known XBP1 target genes (54) confirmed that roughly 30% fall within each of the three categories (XBP1 alone, MIST1 alone, and XBP1 and MIST1), suggesting that the varied regulatory mechanisms associated with these pathways are indeed part of the broad UPR network. Future studies will examine if XBP1 and MIST1 function as a single transcription complex in UPRE/E-box-containing genes or whether they operate independently but in an additive fashion to provide the appropriate high levels of gene transcription needed during ER stress induction.
These studies have uncovered a novel XBP1/MIST1 cross talk network that functions within normal secretory pathways as well as within the UPR. A greater understanding of the regulation of the unfolded protein response has already proved critical in designing enhanced therapies for the prevention of tumor development and for generating new chemotherapeutics (69–71). Indeed, multiple approaches have investigated the XBP1 branch and its targets as a means to control human diseases (7, 64, 72, 73). The inclusion of MIST1 as both an XBP1 target with wide-ranging effects and as a potential negative regulator of Xbp1 expression greatly augments the number of possibilities for developing novel therapeutics for treating protein-processing diseases and disorders.
ACKNOWLEDGMENTS
We thank the Purdue University Center for Cancer Research Transgenic Mouse Core Facility (supported by P30 CA023168) for producing the genetically engineered mouse strains and the mouse embryo fibroblasts used in this study.
This work was supported by grants to S.F.K. (NIH DK55489 and NIH CA124586), a grant from the Indiana Clinical and Translational Sciences Institute (ICTSI 22-808-25), a Purdue Research Foundation Fellowship to D.A.H., and a grant to R.J.M. (NIH DK61220).
We declare that we have no conflicts of interest.
D.A.H., K.M.S., and A.K. designed, performed, and analyzed experiments; T.G.D., C.Q.H., and R.J.M. designed and performed ChIP-Seq experiments; M.J. and A.C.A.-P. analyzed data; A.-H.L. provided cellular reagents; D.A.H. and S.F.K. wrote the manuscript.
Footnotes
For a companion article on this topic, see doi:10.1128/MCB.00370-16.
REFERENCES
- 1.Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell J. 2000. Overview of the secretory pathway. W. H. Freeman, New York, NY. [Google Scholar]
- 2.Hetz C, Martinon F, Rodriguez D, Glimcher LH. 2011. The unfolded protein response: integrating stress signals through the stress sensor IRE1α. Physiol Rev 91:1219–1243. doi: 10.1152/physrev.00001.2011. [DOI] [PubMed] [Google Scholar]
- 3.Lee A-H, Iwakoshi NN, Glimcher LH. 2003. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol 23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kubisch CH, Sans MD, Arumugam T, Ernst SA, Williams JA, Logsdon CD. 2006. Early activation of endoplasmic reticulum stress is associated with arginine-induced acute pancreatitis. Am J Physiol Gastrointest Liver Physiol 291:G238–G245. doi: 10.1152/ajpgi.00471.2005. [DOI] [PubMed] [Google Scholar]
- 5.Suh DH, Kim M-K, Kim HS, Chung HH, Song YS. 2012. Unfolded protein response to autophagy as a promising druggable target for anticancer therapy. Ann N Y Acad Sci 1271:20–32. doi: 10.1111/j.1749-6632.2012.06739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vidal RL, Figueroa A, Court FA, Thielen P, Molina C, Wirth C, Caballero B, Kiffin R, Segura-Aguilar J, Cuervo AM, Glimcher LH, Hetz C. 2012. Targeting the UPR transcription factor XBP1 protects against Huntington's disease through the regulation of FoxO1 and autophagy. Hum Mol Genet 21:2245–2262. doi: 10.1093/hmg/dds040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang S, Kaufman RJ. 2012. The impact of the unfolded protein response on human disease. J Cell Biol 197:857–867. doi: 10.1083/jcb.201110131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burgess TL, Kelly RB. 1987. Constitutive and regulated secretion of proteins. Annu Rev Cell Biol 3:243–293. doi: 10.1146/annurev.cb.03.110187.001331. [DOI] [PubMed] [Google Scholar]
- 9.Alahari S, Mehmood R, Johnson CL, Pin CL. 2011. The absence of MIST1 leads to increased ethanol sensitivity and decreased activity of the unfolded protein response in mouse pancreatic acinar cells. PLoS One 6:e28863. doi: 10.1371/journal.pone.0028863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Capoccia BJ, Lennerz JKM, Bredemeyer AJ, Klco JM, Frater JL, Mills JC. 2011. Transcription factor MIST1 in terminal differentiation of mouse and human plasma cells. Physiol Genomics 43:174–186. doi: 10.1152/physiolgenomics.00084.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chikada H, Ito K, Yanagida A, Nakauchi H, Kamiya A. 2015. The basic helix-loop-helix transcription factor, Mist1, induces maturation of mouse fetal hepatoblasts. Sci Rep 5:14989. doi: 10.1038/srep14989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DiRenzo D, Hess DA, Damsz B, Hallett JE, Marshall B, Goswami C, Liu Y, Deering T, Macdonald RJ, Konieczny SF. 2012. Induced Mist1 expression promotes remodeling of mouse pancreatic acinar cells. Gastroenterology 143:469–480. doi: 10.1053/j.gastro.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garside VC, Kowalik AS, Johnson CL, DiRenzo D, Konieczny SF, Pin CL. 2010. MIST1 regulates the pancreatic acinar cell expression of Atp2c2, the gene encoding secretory pathway calcium ATPase 2. Exp Cell Res 316:2859–2870. doi: 10.1016/j.yexcr.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jia D, Sun Y, Konieczny SF. 2008. Mist1 regulates pancreatic acinar cell proliferation through p21 CIP1/WAF1. Gastroenterology 135:1687–1697. doi: 10.1053/j.gastro.2008.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson CL, Kowalik AS, Rajakumar N, Pin CL. 2004. Mist1 is necessary for the establishment of granule organization in serous exocrine cells of the gastrointestinal tract. Mech Dev 121:261–272. doi: 10.1016/j.mod.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 16.Kowalik AS, Johnson CL, Chadi SA, Weston JY, Fazio EN, Pin CL. 2007. Mice lacking the transcription factor Mist1 exhibit an altered stress response and increased sensitivity to caerulein-induced pancreatitis. Am J Physiol Gastrointest Liver Physiol 292:G1123–G1132. [DOI] [PubMed] [Google Scholar]
- 17.Shi G, Zhu L, Sun Y, Bettencourt R, Damsz B, Hruban RH, Konieczny SF. 2009. Loss of the acinar-restricted transcription factor Mist1 accelerates Kras-induced pancreatic intraepithelial neoplasia. Gastroenterology 136:1368–1378. doi: 10.1053/j.gastro.2008.12.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tian X, Jin RU, Bredemeyer AJ, Oates EJ, Błazewska KM, McKenna CE, Mills JC. 2010. RAB26 and RAB3D are direct transcriptional targets of MIST1 that regulate exocrine granule maturation. Mol Cell Biol 30:1269–1284. doi: 10.1128/MCB.01328-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huh WJ, Esen E, Geahlen JH, Bredemeyer AJ, Lee AH, Shi G, Konieczny SF, Glimcher LH, Mills JC. 2010. XBP1 controls maturation of gastric zymogenic cells by induction of MIST1 and expansion of the rough endoplasmic reticulum. Gastroenterology 139:2038–2049. doi: 10.1053/j.gastro.2010.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pin CL, Rukstalis JM, Johnson C, Konieczny SF. 2001. The bHLH transcription factor Mist1 is required to maintain exocrine pancreas cell organization and acinar cell identity. J Cell Biol 155:519–530. doi: 10.1083/jcb.200105060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo X, Shin DM, Wang X, Konieczny SF, Muallem S. 2005. Aberrant localization of intracellular organelles, Ca2+ signaling, and exocytosis in Mist1 null mice. J Biol Chem 280:12668–12675. doi: 10.1074/jbc.M411973200. [DOI] [PubMed] [Google Scholar]
- 22.DiRenzo DM. 2012. Identifying Mist1 regulated genes in pancreatic acinar cell homeostasis and carcinogenesis. PhD dissertation. Purdue University, West Lafayette, IN. [Google Scholar]
- 23.Rukstalis JM, Kowalik A, Zhu L, Lidington D, Pin CL, Konieczny SF. 2003. Exocrine specific expression of Connexin32 is dependent on the basic helix-loop-helix transcription factor Mist1. J Cell Sci 116:3315–3325. doi: 10.1242/jcs.00631. [DOI] [PubMed] [Google Scholar]
- 24.Hess DA, Humphrey SE, Ishibashi J, Damsz B, Lee AH, Glimcher LH, Konieczny SF. 2011. Extensive pancreas regeneration following acinar-specific disruption of Xbp1 in mice. Gastroenterology 141:1463–1472. doi: 10.1053/j.gastro.2011.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pin CL, Bonvissuto AC, Konieczny SF. 2000. Mist1 expression is a common link among serous exocrine cells exhibiting regulated exocytosis. Anat Rec 259:157–167. doi:. [DOI] [PubMed] [Google Scholar]
- 26.Karki A, Humphrey SE, Steele RE, Hess DA, Taparowsky EJ, Konieczny SF. 2015. Silencing Mist1 gene expression is essential for recovery from acute pancreatitis. PLoS One 10:e0145724. doi: 10.1371/journal.pone.0145724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang M, Azevedo-Pouly A, Deering TG, Hoang CQ, DiRenzo D, Hess DA, Konieczny SF, Swift GH, MacDonald R. 2016. MIST1 and PTF1 collaborate in feed-forward regulatory loops that maintain the pancreatic acinar phenotype in adult mice. Mol Cell Biol 36:2945–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Homann OR, Johnson AD. 2010. MochiView: versatile software for genome browsing and DNA motif analysis. BMC Biol 8:49. doi: 10.1186/1741-7007-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J, Duncan D, Shi Z, Zhang B. 2013. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res 41:W77–W83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang B, Kirov S, Snoddy J. 2005. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res 33:W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 33.Chakrabarti A, Chen AW, Varner JD. 2011. A review of the mammalian unfolded protein response. Biotechnol Bioeng 108:2777–2793. doi: 10.1002/bit.23282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ornitz DM, Palmiter RD, Messing A, Hammer RE, Pinkert CA, Brinster RL. 1985. Elastase I promoter directs expression of human growth hormone and SV40 T antigen genes to pancreatic acinar cells in transgenic mice. Cold Spring Harb Symp Quant Biol 50:399–409. doi: 10.1101/SQB.1985.050.01.050. [DOI] [PubMed] [Google Scholar]
- 35.Rose SD, Swift GH, Peyton MJ, Hammer RE, MacDonald RJ. 2001. The role of PTF1-P48 in pancreatic acinar gene expression. J Biol Chem 276:44018–44026. doi: 10.1074/jbc.M106264200. [DOI] [PubMed] [Google Scholar]
- 36.Swift GH, Kruse F, MacDonald RJ, Hammer RE. 1989. Differential requirements for cell-specific elastase I enhancer domains in transfected cells and transgenic mice. Genes Dev 3:687–696. doi: 10.1101/gad.3.5.687. [DOI] [PubMed] [Google Scholar]
- 37.Kruse F, Komro CT, Michnoff CH, MacDonald RJ. 1988. The cell-specific elastase I enhancer comprises two domains. Mol Cell Biol 8:893–902. doi: 10.1128/MCB.8.2.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foldi I, Toth AM, Szabo Z, Mozes E, Berkecz R, Datki ZL, Penke B, Janaky T. 2013. Proteome-wide study of endoplasmic reticulum stress induced by thapsigargin in N2a neuroblastoma cells. Neurochem Int 62:58–69. doi: 10.1016/j.neuint.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 39.Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, Lavail MM, Walter P. 2007. IRE1 signaling affects cell fate during the unfolded protein response. Science 318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu L, Tran T, Rukstalis JM, Sun P, Damsz B, Konieczny SF. 2004. Inhibition of Mist1 homodimer formation induces pancreatic acinar-to-ductal metaplasia. Mol Cell Biol 24:2673–2681. doi: 10.1128/MCB.24.7.2673-2681.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deer EL, Gonzalez-Hernandez J, Coursen JD, Shea JE, Ngatia J, Scaife CL, Firpo MA, Mulvihill SJ. 2010. Phenotype and genotype of pancreatic cancer cell lines. Pancreas 39:425–435. doi: 10.1097/MPA.0b013e3181c15963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Avivar-Valderas A, Salas E, Bobrovnikova-Marjon E, Diehl JA, Nagi C, Debnath J, Aguirre-Ghiso JA. 2011. PERK integrates autophagy and oxidative stress responses to promote survival during extracellular matrix detachment. Mol Cell Biol 31:3616–3629. doi: 10.1128/MCB.05164-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bommiasamy H, Back SH, Fagone P, Lee K, Meshinchi S, Vink E, Sriburi R, Frank M, Jackowski S, Kaufman RJ, Brewer JW. 2009. ATF6α induces XBP1-independent expansion of the endoplasmic reticulum. J Cell Sci 122:1626–1636. doi: 10.1242/jcs.045625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J, Song B, Yau GDY, Kaufman RJ. 2007. ATF6α optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell 13:351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 45.Yamamoto K, Yoshida H, Kokame K, Kaufman RJ, Mori K. 2004. Differential contributions of ATF6 and XBP1 to the activation of endoplasmic reticulum stress-responsive cis-acting elements ERSE, UPRE and ERSE-II. J Biochem 136:343–350. doi: 10.1093/jb/mvh122. [DOI] [PubMed] [Google Scholar]
- 46.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107:881–891. doi: 10.1016/S0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 47.Acosta-Alvear D, Zhou Y, Blais A, Tsikitis M, Lents NH, Arias C, Lennon CJ, Kluger Y, Dynlacht BD. 2007. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol Cell 27:53–66. doi: 10.1016/j.molcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 48.Hetz C, Thielen P, Matus S, Nassif M, Court F, Kiffin R, Martinez G, Cuervo AM, Brown RH, Glimcher LH. 2009. XBP-1 deficiency in the nervous system protects against amyotrophic lateral sclerosis by increasing autophagy. Genes Dev 23:2294–2306. doi: 10.1101/gad.1830709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schroder M. 2008. Endoplasmic reticulum stress responses. Cell Mol Life Sci 65:862–894. doi: 10.1007/s00018-007-7383-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao Y, Li X, Cai MY, Ma K, Yang J, Zhou J, Fu W, Wei FZ, Wang L, Xie D, Zhu WG. 2013. XBP-1u suppresses autophagy by promoting the degradation of FoxO1 in cancer cells. Cell Res 23:491–507. doi: 10.1038/cr.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tran T, Jia D, Sun Y, Konieczny SF. 2007. The bHLH domain of Mistl is sufficient to activate gene transcription. Gene Expr 13:241–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, Friend D, Grusby MJ, Alt F, Glimcher LH. 2001. Plasma cell differentiation requires the transcription factor XBP-1. Nature 412:300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- 53.Lee AH, Chu GC, Iwakoshi NN, Glimcher LH. 2005. XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. EMBO J 24:4368–4380. doi: 10.1038/sj.emboj.7600903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sriburi R, Bommiasamy H, Buldak GL, Robbins GR, Frank M, Jackowski S, Brewer JW. 2007. Coordinate regulation of phospholipid biosynthesis and secretory pathway gene expression in XBP-1(S)-induced endoplasmic reticulum biogenesis. J Biol Chem 282:7024–7034. [DOI] [PubMed] [Google Scholar]
- 55.Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101:249–258. doi: 10.1016/S0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 56.Williams JA. 2010. Isolation of rodent pancreatic acinar cells and acini by collagenase digestion. Pancreapedia: exocrine pancreas knowledge base. doi: 10.3998/panc.2010.18. [DOI]
- 57.Chen CY, Malchus NS, Hehn B, Stelzer W, Avci D, Langosch D, Lemberg MK. 2014. Signal peptide peptidase functions in ERAD to cleave the unfolded protein response regulator XBP1u. EMBO J 33:2492–2506. doi: 10.15252/embj.201488208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fonseca SG, Fukuma M, Lipson KL, Nguyen LX, Allen JR, Oka Y, Urano F. 2005. WFS1 is a novel component of the unfolded protein response and maintains homeostasis of the endoplasmic reticulum in pancreatic beta-cells. J Biol Chem 280:39609–39615. doi: 10.1074/jbc.M507426200. [DOI] [PubMed] [Google Scholar]
- 59.Xu J, Barone S, Li H, Holiday S, Zahedi K, Soleimani M. 2011. Slc26a11, a chloride transporter, localizes with the vacuolar H+-ATPase of A-intercalated cells of the kidney. Kidney Int 80:926–937. doi: 10.1038/ki.2011.196. [DOI] [PubMed] [Google Scholar]
- 60.Takayanagi S, Fukuda R, Takeuchi Y, Tsukada S, Yoshida K. 2013. Gene regulatory network of unfolded protein response genes in endoplasmic reticulum stress. Cell Stress Chaperones 18:11–23. doi: 10.1007/s12192-012-0351-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sah RP, Garg SK, Dixit AK, Dudeja V, Dawra RK, Saluja AK. 2014. Endoplasmic reticulum stress is chronically activated in chronic pancreatitis. J Biol Chem 289:27551–27561. doi: 10.1074/jbc.M113.528174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kubisch CH, Logsdon CD. 2008. Endoplasmic reticulum stress and the pancreatic acinar cell. Expert Rev Gastroenterol Hepatol 2:249–260. doi: 10.1586/17474124.2.2.249. [DOI] [PubMed] [Google Scholar]
- 63.Kaser A, Lee A-H, Franke A, Glickman JN, Zeissig S, Tilg H, Nieuwenhuis EES, Higgins DE, Schreiber S, Glimcher LH, Blumberg RS. 2008. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell 134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin JH, Walter P, Yen TSB. 2008. Endoplasmic reticulum stress in disease pathogenesis. Annu Rev Pathol 3:399–425. doi: 10.1146/annurev.pathmechdis.3.121806.151434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brown MK, Naidoo N. 2012. The endoplasmic reticulum stress response in aging and age-related diseases. Front Physiol 3:263. doi: 10.3389/fphys.2012.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lemercier C, To RQ, Carrasco RA, Konieczny SF. 1998. The basic helix-loop-helix transcription factor Mist1 functions as a transcriptional repressor of myoD. EMBO J 17:1412–1422. doi: 10.1093/emboj/17.5.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mills JC, Taghert PH. 2012. Scaling factors: transcription factors regulating subcellular domains. Bioessays 34:10–16. doi: 10.1002/bies.201100089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tabas I, Ron D. 2011. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol 13:184–190. doi: 10.1038/ncb0311-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nawrocki ST, Carew JS, Pino MS, Highshaw RA, Dunner K Jr, Huang P, Abbruzzese JL, McConkey DJ. 2005. Bortezomib sensitizes pancreatic cancer cells to endoplasmic reticulum stress-mediated apoptosis. Cancer Res 65:11658–11666. doi: 10.1158/0008-5472.CAN-05-2370. [DOI] [PubMed] [Google Scholar]
- 70.Nawrocki ST, Carew JS, Dunner K Jr, Boise LH, Chiao PJ, Huang P, Abbruzzese JL, McConkey DJ. 2005. Bortezomib inhibits PKR-like endoplasmic reticulum (ER) kinase and induces apoptosis via ER stress in human pancreatic cancer cells. Cancer Res 65:11510–11519. doi: 10.1158/0008-5472.CAN-05-2394. [DOI] [PubMed] [Google Scholar]
- 71.Romero-Ramirez L, Cao H, Nelson D, Hammond E, Lee AH, Yoshida H, Mori K, Glimcher LH, Denko NC, Giaccia AJ, Le QT, Koong AC. 2004. XBP1 is essential for survival under hypoxic conditions and is required for tumor growth. Cancer Res 64:5943–5947. doi: 10.1158/0008-5472.CAN-04-1606. [DOI] [PubMed] [Google Scholar]
- 72.Romero-Ramirez L, Cao H, Regalado MP, Kambham N, Siemann D, Kim JJ, Le QT, Koong AC. 2009. X box-binding protein 1 regulates angiogenesis in human pancreatic adenocarcinomas. Transl Oncol 2:31–38. doi: 10.1593/tlo.08211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Florez JC, Jablonski KA, McAteer J, Sandhu MS, Wareham NJ, Barroso I, Franks PW, Altshuler D, Knowler WC, Diabetes Prevention Program Research G. 2008. Testing of diabetes-associated WFS1 polymorphisms in the Diabetes Prevention Program. Diabetologia 51:451–457. doi: 10.1007/s00125-007-0891-x. [DOI] [PMC free article] [PubMed] [Google Scholar]