Abstract
Much remains unknown regarding the regulatory networks formed by transcription factors in mature, differentiated mammalian cells in vivo, despite many studies of individual DNA-binding transcription factors. We report a constellation of feed-forward loops formed by the pancreatic transcription factors MIST1 and PTF1 that govern the differentiated phenotype of the adult pancreatic acinar cell. PTF1 is an atypical basic helix-loop-helix transcription factor complex of pancreatic acinar cells and is critical to acinar cell fate specification and differentiation. MIST1, also a basic helix-loop-helix transcription factor, enhances the formation and maintenance of the specialized phenotype of professional secretory cells. The MIST1 and PTF1 collaboration controls a wide range of specialized cellular processes, including secretory protein synthesis and processing, exocytosis, and homeostasis of the endoplasmic reticulum. PTF1 drives Mist1 transcription, and MIST1 and PTF1 bind and drive the transcription of over 100 downstream acinar genes. PTF1 binds two canonical bipartite sites within a 0.7-kb transcriptional enhancer upstream of Mist1 that are essential for the activity of the enhancer in vivo. MIST1 and PTF1 coregulate target genes synergistically or additively, depending on the target transcriptional enhancer. The frequent close binding proximity of PTF1 and MIST1 in pancreatic acinar cell chromatin implies extensive collaboration although the collaboration is not dependent on a stable physical interaction.
INTRODUCTION
The mammalian pancreas is a mixed exocrine and endocrine gland consisting of acini, ducts, and islets. Approximately 90% of the mass of the adult pancreas is exocrine acinar tissue. Pancreatic acinar cells are highly specialized for the synthesis and secretion of digestive enzymes that are flushed via ducts to the intestine for digestion of complex nutrients (1). Abundant rough endoplasmic reticulum (ER) supports an extraordinary level of secretory protein synthesis (2). Maintenance of ER homeostasis without the accumulation of misfolded and unfolded proteins is especially important for acinar function and viability (3). Acinar cells are polarized, with basal rough ER and an extensive supranuclear Golgi apparatus for sorting and condensing newly synthesized secretory proteins into secretory vesicles (zymogen granules) that fill the apical cellular domain nearest the luminal plasma membrane. Secretion is regulated to ensure an appropriate surge of digestive enzyme release in response to feeding (1).
Several transcription factors, including PTF1 and MIST1, are known to play crucial roles in the specification, differentiation, and maturation of pancreatic acinar cells (4–6). PTF1 is a complex of three tightly associated DNA-binding subunits: the cell-type-restricted basic helix-loop-helix (bHLH) protein PTF1A, one of the common bHLH E proteins (e.g., E47) (7), and RBPJ or its paralog RBPJL (8, 9). All three subunits contribute to the recognition of an extended bipartite binding sequence consisting of an E box bound by the PTF1A-E protein heterodimer and a TC box bound by the RBP subunit (8, 10, 11). Early pancreatic development requires the RBPJ form of the PTF1 complex (12), designated PTF1-J. With the onset of acinar development, PTF1-J activates Rbpjl, and RBPJL gradually replaces RBPJ in the complex (12). The RBPJL form (PTF1-L) is restricted to pancreatic acinar cells and controls the final stage of acinar differentiation. The expression of genes encoding digestive enzymes and proteins involved in regulated exocytosis is reduced in Rbpjl-null pancreatic acinar cells (13).
MIST1 (encoded by Bhlha15) is a bHLH transcription factor present selectively in serous secretory cells, including pancreatic acinar cells (14, 15). In the pancreas, Ptf1a and Mist1 are expressed selectively in acinar cells, and inactivation of either gene affects the differentiated acinar phenotype (16, 17). Mist1-null pancreatic acinar cells have aberrant calcium signaling, reduced content of digestive enzymes, and decreased expression of critical genes of the secretory pathway, such as Rab genes (18–20). These deficiencies cause the disruption of apical-basal polarity, the failure of intercellular communication due to defective gap junctions, and a severe impairment of secretagogue-stimulated secretion. Furthermore, the absence of functional MIST1 causes progressive acinar lesions and the acquisition of some ductal properties (15). Mice lacking MIST1 have a greatly reduced ability to recover from caerulein-induced acute pancreatitis (21) and increased sensitivity to KRAS-driven adenocarcinoma (22).
Little is known of the regulatory networks formed by transcription factors in pancreatic acinar cells (23, 24). Large-scale studies of transcription factors in unicellular organisms, including Escherichia coli and Saccharomyces cerevisiae, identified regulatory motifs that recur in transcriptional networks, especially autoregulatory and feed-forward loops (25, 26). For autoregulation, a regulator binds and affects the transcriptional regulatory region of its own gene. Positive autoregulatory loops can create a stable, complex biological system to maintain a cellular identity. Dual, positive autoregulation of Ptf1a and Rbpjl by the PTF1-L complex in pancreatic acinar cells sustains the level of PTF1 (12, 27). A feed-forward loop consists of a pair of regulators in which one controls the other and the two jointly control a third (target) gene. Feed-forward loops are highly represented in transcriptional networks and control 10% of yeast genes (25) and more than 30% of E. coli genes (28). Feed-forward loops respond to sustained rather than transient stimuli, provide precise temporal control for developmental decisions (29), and can be particularly effective in support of the stability of a differentiated state.
Here, we report a multioutput, feed-forward regulatory system consisting of PTF1, MIST1, and a set of shared target genes in pancreatic acinar cells. PTF1 binds and directly regulates Mist1 transcription, and together MIST1 and PTF1 bind and coregulate genes for a wide range of specialized pancreatic acinar cellular processes. We show that MIST1 and PTF1 coactivate additively or synergistically, depending on the nature of the regulatory sequences of the target gene.
MATERIALS AND METHODS
Generation of transient transgenic embryos and detection of transgene activities.
Transient transgenic embryos bearing an ∼0.7-kb genomic region 6 kb upstream of Mist1 (Mus musculus build 10 [mm10], chromosome 5 [chr5] positions 144183992 to 144184700) were generated, and transgene activities were detected as previously described (27). Briefly, the enhancer of interest was placed 5′ of the lacZ reporter gene and sent to the Transgenic Core Facility of the University of Texas Southwestern Medical Center to generate transient transgenic embryos. Embryos were sacrificed at embryonic day 12.5 (E12.5), E14.5, and E15.5 and stained for β-galactosidase activity. There are two PTF1-binding sites in the Mist1 enhancer. To examine the ability of PTF1 to regulate the enhancer activity in vivo, the TC boxes were modified from TTCCCA to TTCAAT, and the E boxes were modified from CANNTG to TCNNTA (mutations are underlined). Transient transgenic embryos carrying the enhancer with either the two TC boxes or the two E boxes mutated were generated, collected at E15.5, and stained for β-galactosidase activity. The University of Texas Southwestern Institutional Animal Care and Use Committee approved all of the animal experiments.
Immunohistochemistry.
Whole-mount β-galactosidase-stained E15.5 transient transgenic embryos were embedded in paraffin and sectioned. Sections were deparaffinized and blocked with 0.3% H2O2 for 30 min and 10% normal donkey serum buffer (Jackson ImmunoResearch, West Grove, PA) for 1 h. Primary antibodies included goat anti-CPA1 (used at 1:200; R&D Systems, Minneapolis, MN), rat anti-cytokeratin 19 (CK19) (1:10; Developmental Studies Hybridoma Bank, Iowa City, IA), guinea pig anti-insulin (1:1,000; Millipore, Billerica, MA), and guinea pig anti-glucagon (1:1,000; Millipore). Primary antibodies, except anti-CK19, were applied overnight at 4°C, followed by the application of secondary antibodies (Jackson ImmunoResearch) for 30 min at room temperature. Anti-CK19 was applied for 1 h at room temperature. Immunolocalization was visualized by diaminobenzidine (DAB) staining (Roche, Indianapolis, IN) and then eosin Y counterstaining (Sigma-Aldrich, St. Louis, MO). The percentage of immunostaining-positive cells in the β-galactosidase-positive (blue) cell population was counted for each section. The mean and standard deviation of the percentage were calculated for each marker.
Coimmunoprecipitation.
The nuclear extract was derived from a whole pancreas (300 mg) of an ICR mouse by an NE-PER nuclear and cytoplasmic protein extraction kit (Thermo Fisher Scientific, Waltham, MA), except that the nuclear extraction reagent (NER) buffer was replaced with a lysis buffer (20 mM HEPES, 1% Triton X-100, 150 mM NaCl, 5 mM EDTA, 5% glycerol, pH 7.4). The lysate was precleared and then immunoprecipitated with 3 μg of affinity-purified rabbit anti-mouse PTF1A (8). The immunoprecipitate was resolved by SDS-PAGE and blotted with goat anti-mouse PTF1A, rabbit anti-mouse RBPJL (8), or rabbit anti-human MIST1 (30). Goat anti-mouse PTF1A was a gift from Christopher Wright (Vanderbilt University Medical Center, Nashville, TN).
ChIP-Seq.
Chromatin immunoprecipitation with high-throughput sequencing (ChIP-Seq) was performed with chromatin isolated from adult mouse pancreas, as previously described (13), using rabbit antibodies against either rat or human MIST1 (30, 31). Two independent ChIP-Seq assays (one pancreas per ChIP-Seq experiment) were performed with each MIST1 antibody. The sequence reads were aligned with Bowtie (32). The ChIP-Seq assays using anti-human MIST1 had 13.5 million and 54.9 million aligned reads, and the ones using anti-rat MIST1 had 11.5 million and 18.6 million aligned reads. MIST1-binding peaks were called using HOMER (33) with a false discovery rate (FDR) of <0.01 for the combined anti-rat MIST1 data sets and confirmed if overlapping peaks were present for the combined anti-human MIST1 data sets with an FDR of <0.05. The de novo discovery of MIST1 DNA-binding motifs was performed using MEME (34). MIST1-bound regions were assigned to nearby genes using GREAT (35) with its default settings.
Four ChIP-Seq assays were also performed with rabbit anti-mouse PTF1A (8) and with rabbit anti-mouse RBPJL (8). The raw data are available through the NCBI Gene Expression Omnibus (GEO) database under accession number GSE86262 (17). Identification of regions bound by either PTF1A or RBPJL followed the pipeline of MIST1 ChIP-Seq analysis described above. PTF1-bound regions were identified by the overlap of PTF1A and RBPJL binding (peak width, 250 bp; overlap, ≥1 bp). The probability of overlap between PTF1- and MIST1-bound regions was calculated with the phyper function of the R project (www.r-project.org) and based on 20.3 million base pairs of open chromatin, which is the median value for DNase I-hypersensitive sites of a variety of mouse tissues (36).
RNA sequencing.
Total RNA samples from two control (Mist1+/−) and three Mist1-KO (Mist1−/−) adult mouse pancreases were subjected to RNA sequencing (RNA-Seq) as previously described (13). The Mist1 knockout (Mist1-KO) mouse (Bhlha15TM1Skz) has been described previously (15). The sequence reads were aligned with TopHat (37). The control libraries have 35 million and 24.4 million aligned reads, and the Mist1-KO libraries have 33.1 million, 30.6 million, and 35.3 million aligned reads. HTSeq (38) was used to count the number of reads mapped to each gene. Differential gene expression analysis was then performed with edgeR (39) (FDR of <0.05). Cufflinks (37) calculated the number of reads per kilobase per million mapped reads (RPKM), a measure of the relative level of a transcript normalized to 1 million reads. RNA-Seq results for three control and four Ptf1a conditional knockout (Ptf1a-cKO) mouse pancreases were deposited in the NCBI GEO database under accession number GSE86261 (17). Differential gene expression analysis and RPKM calculation followed the pipeline described above.
In silico functional analysis of genes controlled by both PTF1 and MIST1.
Among potential PTF1 and MIST1 target genes assigned by GREAT (26), our functional analysis focused on those with meaningful expression in the normal pancreas (RPKM of ≥1). A value of 1 RPKM is equivalent to ∼1 mRNA molecule per cell, based on an RNA/DNA ratio of 9 for normal pancreas and 6 pg of DNA per cell.
Three important gene sets were collected, including pancreas-restricted, acinus-related, and exocytosis genes. (i) The results of gene expression profiling of 91 adult mouse cell types and tissues (NCBI accession number GSE10246) (40) were analyzed using the affy package of R (41). Pancreas-restricted genes with the top two probe intensities in pancreas and with values at least 10-fold higher than those in the other tissues were chosen. (ii) A PubMed query for acinus-related genes was performed with an NCBI eUtils pipeline using a homemade PERL script. The keywords were the following: (gene symbol OR gene synonyms) AND (acin*[TIAB] OR “gastric chief cell”[TIAB] OR “zymogenic cell”[TIAB] OR “plasma cell”[TIAB]). Gastric chief cells (also referred to as zymogenic cells) and plasma cells are acinar cell-like. Our PubMed search added these cell types to the keywords because genes functioning in these cells might also play a role in pancreatic acinar cells. (iii) Exocytosis-related genes were collected from the Gene Ontology Database (http://geneontology.org), from proteomic studies of mouse and rat pancreatic zymogen granules (42–44), and from reviews on exocytosis (45, 46). Experimental evidence for involvement in exocytosis was collected for each gene with an NCBI eUtils pipeline using a homemade PERL script. Genes without the support of experimental evidence or gene ontology were removed from the exocytosis gene list. Hypergeometric tests were performed with the phyper function of R to examine the statistical significance of enrichment of the potential MIST1 and PTF1 targets in the three gene sets. Qiagen's Ingenuity Pathway Analysis (IPA; Qiagen, Redwood City, CA) and WebGestalt analysis (47) were performed to discover pathways enriched in the MIST1 and PTF1 target gene sets.
Cell transfection assays of enhancer function.
The enhancers of Rab26 (+739 to +250), Cabp2 (−1172 to −502 relative to the transcription start site [TSS]), Nfe2l2 (−740 to +1198 and +67896 to +67648), and Tead2 (+825 to +1893) were amplified from the C57BL/6 mouse genomic DNA and cloned into pGL3 (Promega, San Luis Obispo, CA). Mutagenesis of the transcription factor binding sites was performed with overlap extension PCR. For PTF1-binding sites, the TC boxes were modified from TCCCA to TCAAT, and the E boxes were modified from CANNTG to TCNNTA (mutations are underlined). MIST1-preferred E boxes CA(G/T)(C/A)TG were modified to TC(G/T)(C/A)TA. The cDNA expression plasmids of mouse Ptf1a and Rbpjl have been described previously (8). The full-length protein coding region with part of the 3′ untranslated region of Mist1 (bp 125 to 818) was amplified from mouse genomic DNA, linked downstream of the 5′ untranslated region of Xenopus laevis β-globin mRNA, and inserted into the pcDNA3.1(+) vector between HindIII and XhoI restriction sites (Invitrogen). Transfection of 293 human embryonic kidney cells (HEK293; American Type Culture Collection CRL-1573) was performed using FuGene 6 (Promega) as previously described (48). Each transfection was repeated three times. The enhancer-driven firefly luciferase activity was normalized to the Renilla luciferase activity of cotransfected pRL-CMV (Promega).
Accession numbers.
Raw data have been deposited in the NCBI GEO database under accession numbers GSE86288 and GSE86289.
RESULTS
The PTF1 complex directs pancreas-specific transcription of Mist1 via an upstream enhancer.
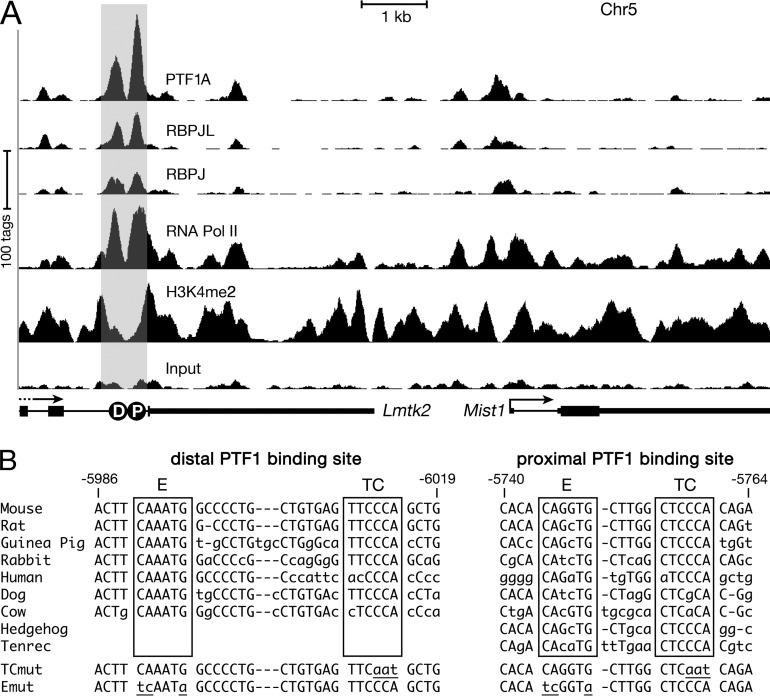
PTF1 is crucial for pancreatic organogenesis, including the differentiation of acinar cells (4, 13, 49). However, knowledge of the downstream genes controlled by PTF1 in mature acinar cells is largely limited to the genes of the secretory digestive enzymes (8, 13). Inactivation of Ptf1a in the adult pancreas (17) reduced Mist1 mRNA to 24% of the normal level after 6 days (0.24 ± 0.13; P = 3.4E−4). To determine whether Mist1 is a direct target of PTF1, we performed ChIP-Seq experiments with chromatin from adult pancreas using antibodies against the PTF1 subunit PTF1A, RBPJ, or RBPJL. Two sites approximately 300 bp apart with strong binding for all three proteins were identified 6 kb upstream of Mist1 (Fig. 1A), indicating a mix of PTF1-J and PTF1-L complexes at both sites in the acinar cell population. PTF1A, RBPJ, and RBPJL were also present at these sites in pancreatic chromatin during fetal development (see Fig. S1A in the supplemental material). Consistent with the previous report that early pancreatic development requires PTF1-J and that RBPJL replaces RBPJ in the complex during late development (12), preferential occupancy by RBPJ at E15.5 (RBPJ/RBPJL, 11:1) changed to a preference for RBPJL at E17.5 (1:6) and in adult mice (1:3). The presence of RNA polymerase II and histone 3 dimethylated at lysine 4 (H3K4me2) at and surrounding these two sites in embryonic and adult pancreatic chromatin indicates that this region is likely a transcriptional enhancer (50, 51).
FIG 1.
PTF1 binds to an enhancer present 6 kb upstream of Mist1 in the adult mouse pancreas. (A) Representative ChIP-Seq for binding of PTF1A, RBPJ, RBPJL, RNA polymerase II (Pol II), and H3K4me2 at the Mist1 locus in chromatin from adult mouse pancreas. Distal (D) and proximal (P) PTF1-binding sites are present in the last intron of Lmtk2, 6 kb upstream of Mist1. The gray shading indicates the ∼0.7-kb enhancer. (B) The nucleotide sequences of the two PTF1-binding sites are conserved in several mammals. Lowercase letters and dashes represent nucleotide differences and additions or deletions, respectively, compared to the mouse sequence. Mutations (lowercase and underlined) in the TC and E boxes (TCmut and Emut, respectively) of the PTF1 binding sites analyzed in this study are shown at the bottom.
We note that this potential enhancer is located in the last intron of Lmtk2. The expression of Lmtk2 during embryonic pancreatic development is constant, unlike the increasing expression of known PTF1 targets, including Mist1, as Ptf1a expression increases (unpublished data). In addition, the continuous distributions of RNA polymerase II and H3K4me2 from the enhancer to the Mist1 promoter indicate that the enhancer and Mist1 promoter are in the same regulatory block. Thus, this enhancer controls Mist1; however, we cannot rule out some effect on Lmtk2.
The RBPJ and RBPJL forms of PTF1 recognize the same DNA-binding sequence, which consists of an E box (CANNTG) and a TC box (TTCCCA) spaced one, two, or (infrequently) three helical DNA turns apart (8, 10, 52). The PTF1-binding sequences present near the center of each of the two PTF1-bound regions in the Mist1 enhancer are well conserved (including the spacing between the E and TC boxes) in mouse, rat, human, and four other mammals. The proximal binding sequence is further conserved, except for a single nucleotide variation in the TC box, across eight additional mammalian species (see Fig. S1B in the supplemental material). Conservation extends throughout the proposed enhancer and the gene arrangement of this region of mouse chr5 (see Fig. S1C).
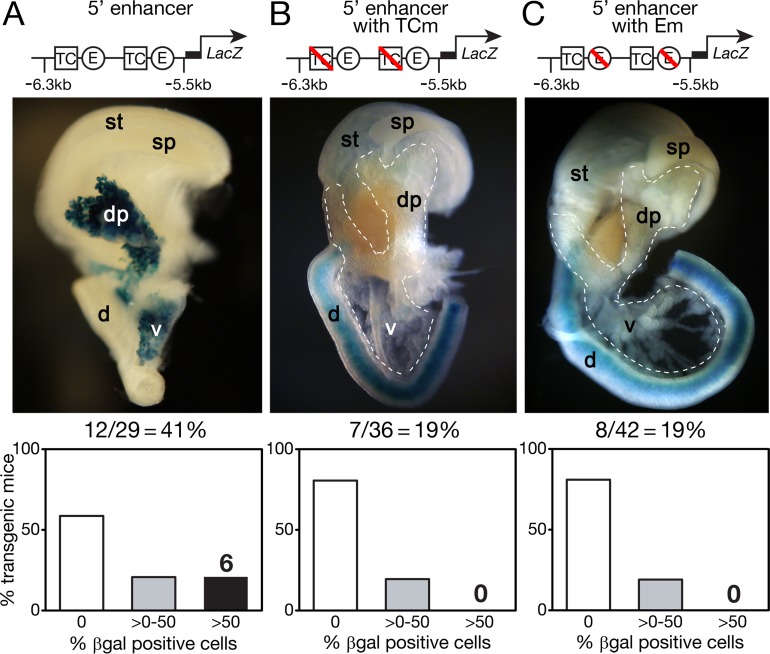
To test whether the conserved PTF1-bound sites are part of an acinar transcriptional enhancer, the 0.7-kb region containing the two sites (Fig. 1A, gray shading) was linked to a lacZ reporter gene and injected into fertilized mouse eggs to examine activity in fetal mouse tissues (Fig. 2). Enhancer-driven β-galactosidase activity was detected selectively in the pancreas for 12 of 29 (41%) E15.5 founder transgenic embryos (Fig. 2A) and in 5 of 14 (36%) E14.5 transgenic embryos (see Fig. S2 in the supplemental material) but only very faintly in 1 of 10 E12.5 transgenic embryos (data not shown). Thus, the enhancer was activated specifically and within the same developmental window as endogenous Mist1 expression between E12.5 and E13.5, shortly after the onset of acinar differentiation (15, 53). Transgenic β-galactosidase activity was detected in most acinar cells but not in ductal or endocrine cells (Fig. 3). The activity of the enhancer in the acinar cells of the pancreas and not elsewhere in the gut is the same as that of the endogenous Mist1 gene (15). Thus, the proper temporal and cell-type-restricted expression is further evidence that the enhancer drives Mist1 transcription in pancreatic acinar cells.
FIG 2.
In vivo activity of Mist1 enhancer at E15.5. The enhancer without alterations (A) or with mutations (B and C) in either the TC boxes (TCm) or E boxes (Em) was ligated upstream of a lacZ reporter and injected into fertilized mouse eggs. These eggs were then implanted in pseudopregnant female mice. Embryos at E15.5 were stained for β-galactosidase (β-Gal) activity. A representative staining photograph and the percentage of transgenic embryos showing pancreatic β-galactosidase activity are displayed for each transgene. A bar graph at the bottom summarizes the pancreas staining results for transgenic embryos. dp, dorsal pancreas; vp, ventral pancreas; d, duodenum; sp, spleen; st, stomach.
FIG 3.
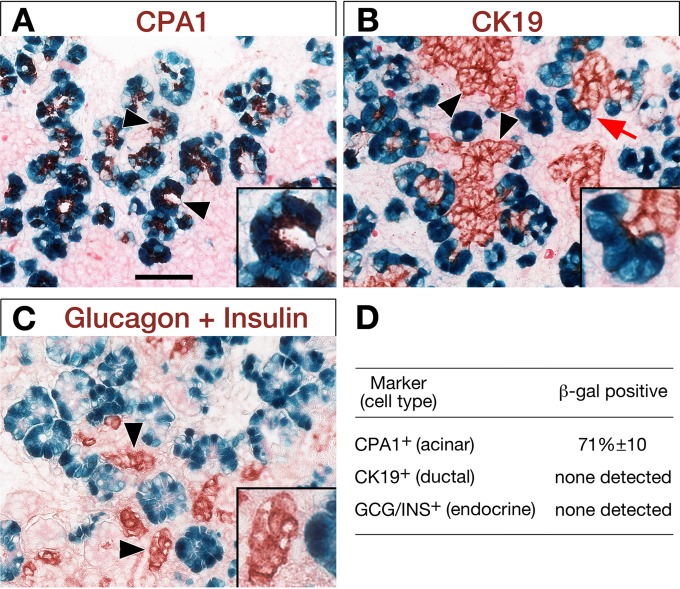
The enhancer activity is restricted to the acinar cells. To study cell specificity of the Mist1 enhancer, immunohistochemistry was performed on sections from whole-mount β-galactosidase staining (blue) of two E15.5 transient transgenic embryos bearing the wild-type enhancer. (A) CPA1 marks acinar cells. Arrowheads, clear examples of forming acini. (B) CK19 (cytokeratin 19) marks ductal cells. Arrowheads, ductal complexes; arrow and inset, ductal/acinar junction. (C) Glucagon plus insulin identifies differentiated endocrine cells. Arrowheads and inset, pre-islet endocrine clusters. Insets are higher-magnification images. Scale bar, 50 μm. (D) Summary of the selective presence or absence of β-galactosidase in acinar cells (9,505/13,392, or 71%), ductal cells, and endocrine cells. GCG/INS, glucagon plus insulin.
To test whether the activity of the Mist1 enhancer requires PTF1 binding, we examined the effect of specific binding-site mutations known to eliminate PTF1 binding of other pancreatic gene control regions (8, 27, 54). Minimal 3-bp mutations in both E boxes or both TC boxes largely eliminated the activity of the enhancer in vivo (Fig. 2B and C). No mice with transgenic mutant enhancers had extensive β-galactosidase staining of the pancreas; any residual activity was limited to much less than 1% of the pancreas. These results show that the enhancer requires binding of the trimeric PTF1 complex; that is, binding of the PTF1A-E protein heterodimer or an RBP alone is not sufficient.
MIST1 prefers binding to GC, TA, and GA/TC E boxes.
MIST1 plays a critical role in regulating the secretory function of the exocrine pancreas (18, 19); however, there is limited knowledge of MIST1 target genes (19). We performed four ChIP-Seq experiments using two independently derived antibodies against MIST1 to identify MIST1-bound sites in pancreatic chromatin. The reproducibility among the four experiments across biological and experimental replicates and two independent MIST1 antibodies (see Materials and Methods) was high (Pearson correlation coefficients between 0.64 and 0.99) (see Table S1 in the supplemental material). Both MIST1 antibodies detected 11,384 MIST1-bound regions. Thirteen of 19 MIST1-bound regions previously identified by ChIP-PCR (19) were also detected in our ChIP-Seq analysis.
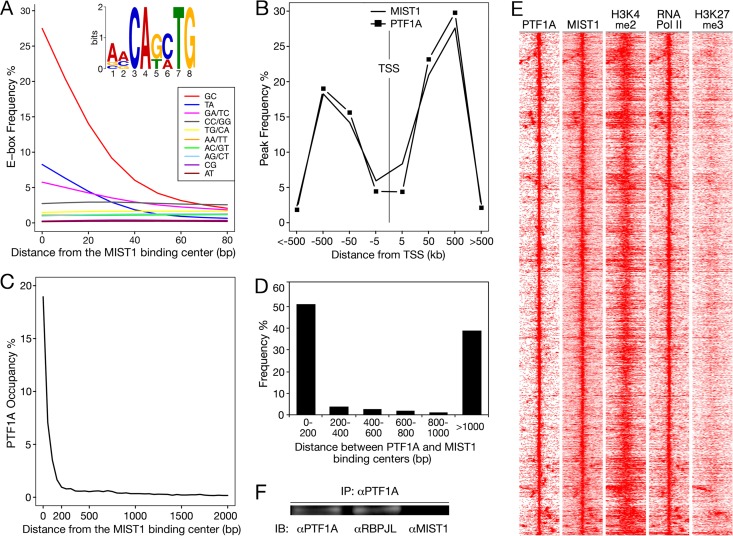
The predominant sequence motif present at MIST1-bound sites (E value, <E−3,000) (Fig. 4A) includes three possible E boxes, CAGCTG (termed GC E box), CATATG (TA E box), and CAGATG (GA/TC E box). Greater than 95% of MIST1-bound regions contain one of these three versions. MIST1 protein binds GC and TA E boxes in electrophoretic mobility shift assays (31, 55). The GC E box is the prevailing MIST1 DNA binding motif in vivo despite its lower affinity for MIST1 protein than the TA E box in vitro (55). The three E boxes are enriched at the center of MIST1-bound regions (Fig. 4A), the preferred position of binding sequences for regions predicted from ChIP-Seq data. The seven other E-box versions were distributed uniformly across the length of MIST1-bound sites, indicating that their occurrence is unspecific. We note that the sequence CAGCAG(G/C) was present in 523 MIST1-bound regions without a MIST1-preferred E box (E value, <E−140). However, this motif often occurs far away from the center of MIST1-bound regions and therefore is unlikely to be a novel MIST1-binding sequence. It may be the preferred binding motif of an unknown factor that collaborates with MIST1. Furthermore, the preferred RBP-binding TC boxes (TTYCY) were also enriched in the MIST1-binding regions (E value, 2E−120) and consistent with nearby binding of PTF1.
FIG 4.
Close localization between PTF1- and MIST1-bound regions. (A) MIST1 DNA-binding motifs. Distribution of the 10 possible E-box sequences relative to the centers of MIST1-bound regions. The occurrence was calculated for 10-bp windows. E boxes are named by their central two nucleotides. The inset shows the MIST1-binding motif identified with MEME (E value, 7E−3,310). (B) Distributions of PTF1- and MIST1-bound sites relative to transcription start sites (TSSs). (C) Distribution of PTF1-bound sites relative to the centers of MIST1-bound sites. (D) Distance between PTF1- and MIST1-bound sites associated with the MIST1- and PTF1-coregulated genes. (E) Heat map signatures (±2 kb) of PTF1A peak centers with MIST1 binding within 1 kb. Frequent colocalization of PTF1A and MIST1 with H3K4me2 and RNA polymerase II and the lack of H3K27me3 are consistent with preferential binding at transcriptional enhancers and promoters. (F) Coimmunoprecipitation of nuclear extract from adult mouse pancreas did not detect a stable physical association of MIST1 and PTF1. IP, immunoprecipitation; IB, immunoblotting.
MIST1 and PTF1 appear to coregulate gene expression independent of a direct, stable physical interaction.
The colocalization of MIST1 and PTF1 selectively in pancreatic acinar cells (8) and the similar effects of PTF1 and MIST1 deficiency on the acinar cell phenotype (13, 15) suggest that these two factors collaborate to establish and maintain cell-type-specific acinar cell functions. To investigate the potential for direct collaboration, we compared the genome-wide binding of MIST1 and PTF1A. The results of ChIP-Seq analyses for PTF1A and RBPJL identified a total of 6,750 cobound sites representing the presence of the trimeric PTF1-L complex. Most of the PTF1-bound sites are between 50 kb and 500 kb upstream or downstream of transcriptional start sites (TSSs) (Fig. 4B). The distribution of MIST1-bound regions is similar. MIST1 and PTF1 had a high incidence of juxtaposed binding: 23% of PTF1-binding centers had MIST1 bound within 200 bp (Fig. 4C) (hypergeometric test P value, <E−300).
The enrichment of PTF1- and MIST1-bound regions with active chromatin marks (H3K4me2, acetylated histone 3 [H3ac], and RNA polymerase II [50, 51, 56]) as well as the lack of the repressive mark H3K27me3 (histone 3 trimethylated at lysine 27) (51, 56) (Fig. 4E; see also Fig. S3 in the supplemental material) indicates that these two transcription factors mainly activate gene transcription. Totals of 9,082 and 5,365 genes are associated with MIST1- and PTF1-bound sites, respectively. Approximately half of PTF1-associated (51%) and MIST1-associated (47%) genes are expressed in the normal mouse pancreas (RNA-Seq RPKM of ≥1).
To identify genes cobound by MIST1 and PTF1 with expression dependent on both factors, we compared the effects of the inactivation of Ptf1a and Mist1 on the pancreatic mRNA population. RNA-Seq analyses revealed that the mRNAs of 33% of the cobound genes decreased in adult pancreas after Ptf1a inactivation, and the mRNAs of 8.9% of the cobound genes decreased in Mist1-KO pancreas. A total of 157 genes were cobound by MIST1 and PTF1 and also codependent on the presence of both factors (see Table S2 in the supplemental material). Cobound and codependent target genes are designated coregulated. The binding of PTF1 and MIST1 was within 200 bp at more than half of the coregulated genes (Fig. 4D).
Frequent close proximity of PTF1 and MIST1 binding suggested a physical interaction between the transcription factors. A coimmunoprecipitation experiment with pancreatic nuclear extracts readily detected the association between PTF1A and RBPJL but not of PTF1A with MIST1 (Fig. 4F). Thus, PTF1 and MIST1 coregulation does not require the formation of a stable complex containing the two factors.
The collaboration of MIST1 and PTF1 is tied to the specialized characteristics of the acinar pancreas.
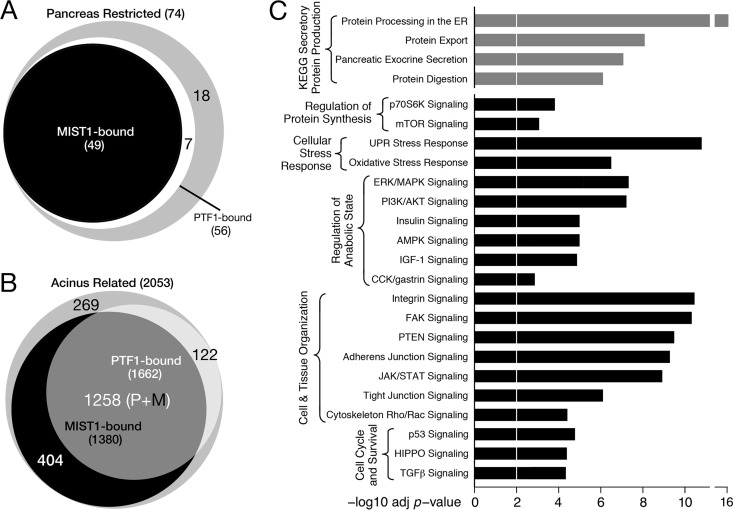
To inquire about the relevance of PTF1-MIST1 coregulation, we identified pancreatic acinus-specific genes preferentially controlled by MIST1 and PTF1. In addition to RNA-Seq results (this report), data from other sources were gleaned to provide a wide view of gene expression. From an analysis of the microarray hybridization data for 91 mouse tissues and cell types (31), we identified 74 genes with expression highly restricted to the pancreas (see Table S3 in the supplemental material). Of these, six are restricted to islet tissue and therefore excluded from the PTF1 and MIST1 regulatory domain. For the remaining 68, 70% (hypergeometric test P value, 2E−20) had both PTF1 and MIST1 bound nearby (Fig. 5A), and 40% were also codependent on the two factors. None of the endocrine genes were coregulated. Among the pancreas-restricted genes are those of the secretory digestive enzymes and their secreted accessory proteins. More than half of these (17 of 30) are both cobound and codependent (see Fig. S4 in the supplemental material), indicating the importance of the collaboration between the two transcription factors in support of the quintessential function of pancreatic acinar cells.
FIG 5.
Collaboration between MIST1 and PTF1 in the maintenance of the pancreatic acinar phenotype. (A and B) Pancreas-restricted genes (A) and acinus-related genes (B) were enriched for associated MIST1 and PTF1 binding. P+M, PTF1 and MIST1 bound. (C) Cellular pathways overrepresented in the set of MIST1- and PTF1-cobound genes, identified by IPA or KEGG pathway (via WebGestalt) analyses (Benjamini-Hochberg-adjusted Fisher's exact test P value, <0.01). Gray bars, KEGG pathways (73); black bars, IPA pathways (Qiagen, Redwood City, CA). ERK, extracellular signal-regulated kinase; MAPK, mitogen-activated protein kinase; PI3K, phosphatidylinositol 3-kinase; AMPK, AMP-activated protein kinase; FAK, focal adhesion kinase; TGF-β, transforming growth factor β.
Casting a wider net, PubMed database searches identified 4,895 genes active in secretory cells. Of these, 2,053 were expressed in the normal pancreas (RPKM of ≥1), and 69% of these genes were associated with either PTF1 or MIST1 binding in pancreatic chromatin (Fig. 5B). Moreover, a significant fraction had both transcription factors bound in close proximity (38%; hypergeometric test P value, 2E−98). These results reveal the great extent of MIST1 and PTF1 collaboration in regulating acinar cell processes.
We used Ingenuity Pathway Analysis and WebGestalt analysis of the 2,285 genes with occupancy of both MIST1 and PTF1 and mRNA levels greater than 1 RPKM in normal pancreas to identify cellular pathways potentially coregulated by the two factors. This analysis revealed that collaboration between PTF1 and MIST1 might control a wider range of pancreatic acinar functions, such as components of the protein synthetic machinery, regulation of translational efficiency, signal recognition particle (SRP)-dependent transfer of secretory proteins into the ER, ER stress, intracellular transport, redox homeostasis, protein degradation, and cell/tissue organization (Fig. 5C). Particularly enriched were genes for processes that compose the synthesis, processing and packaging of the secretory enzymes (see Fig. S4 in the supplemental material), the oxidative stress response (see Fig. S5), and exocytosis (see Table S4).
MIST1 and PTF1 coactivate gene transcription additively or synergistically.
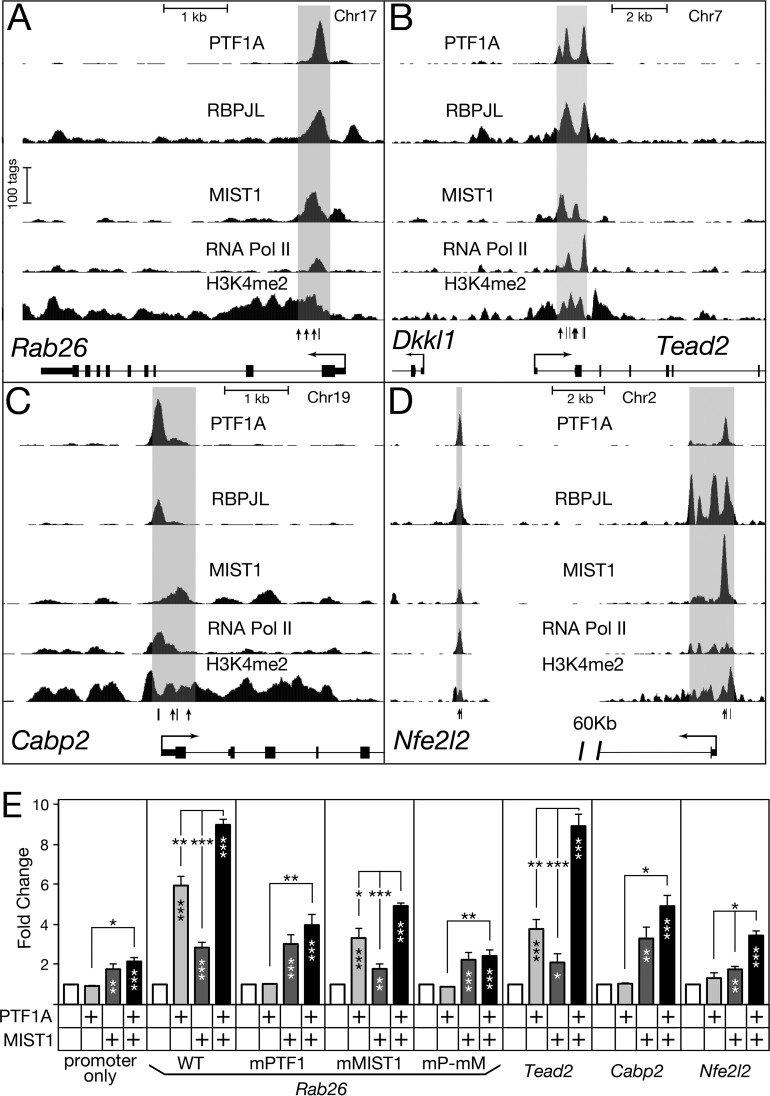
To explore the collaborative activities of MIST1 and PTF1, we examined their actions at the control regions of four representative target genes by cell transfection analyses. The enhancer-like control regions of Rab26, Tead2, Cabp2, and Nfe2l2 have characteristic in vivo binding of PTF1A, RBPJL, and RNA polymerase II, as well as the presence of H3K4me2 (Fig. 6A to D). Each of these control regions could be activated by PTF1A or MIST1 expression in transfected HEK293 cells (Fig. 6E). HEK293 cells are devoid of PTF1A, MIST1, and RBPJL but have RBPJ and common E proteins, which can form active PTF1 complexes with added PTF1A (5).
FIG 6.
MIST1 and PTF1 coregulate gene expression either additively or synergistically. (A to D) Representative ChIP-Seq for PTF1A, RBPJL, MIST1, and RNA polymerase II binding and H3K4me2 in adult pancreatic chromatin at the Rab26, Tead2, Cabp2, and Nfe2l2 loci. Potential binding sites of MIST1 and PTF1 are indicated with vertical lines and arrows, respectively. A schematic of MIST1- and PTF1-binding sites is shown in Fig. S6D in the supplemental material. The gray shading indicates genomic regions cloned and used in transfections. (E) Luciferase reporter constructs were transfected into HEK293 cells together with the expression vector for PTF1A and/or MIST1. The normalized luciferase activities were compared with those of the corresponding controls without addition of PTF1A and MIST1 to measure effects on the regulatory regions. WT, unmodified Rab26 enhancer; mMIST1, mutation of the three MIST1 sites; mPTF1, mutation of the E and TC boxes of the PTF1 site; mP-mM, combined mutations of MIST1 and PTF1 sites (see Materials and Methods). Error bars indicate standard errors of the means. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Asterisks within the bars, added factors versus the cognate control without the factors (open bars); asterisks above the bars, combined factors versus individual factors.
PTF1A and MIST1 activated the Rab26 control region in an additive manner (Fig. 6E). Although the promoter-only control was induced 2-fold by the combination of both factors, the induction with the enhancer present was greater than 9-fold (t test P value, <0.01) and independent of the effects on the promoter (see Fig. S6B in the supplemental material). Mutation of the PTF1 or MIST1 binding sites in the Rab26 control region abolished the induction by the corresponding factor as well as the additive effect (Fig. 6E). Varying the level of cotransfected expression vectors over a 9-fold range altered the magnitude of the induction proportionally without changing the characteristic response patterns of the mutants (see Fig. S6A), demonstrating that the levels of PTF1A and MIST1 tested were below saturation and within an appropriate response range. These results showed that MIST1 and PTF1 coregulation of the Rab26 enhancer is dependent on their binding to defined sites.
PTF1A and MIST1 together induced the activity of Tead2, Cabp2, and Nfe2l2 control regions to an extent greater than the sum of their separate effects (Fig. 6E). PTF1A and MIST1 activated the Tead2 enhancer 3.8- and 2.1-fold, respectively; together, the induction was 8.9-fold. Total induction greater than the simple sum of the two individual activation values reflects MIST1-PTF1A synergy at this enhancer. For the Cabp2 enhancer, PTF1A did not increase reporter activity, yet the addition of PTF1A enhanced the activation by MIST1; this increase depended on the presence of intact PTF1-binding sites (see Fig. S6C in the supplemental material). In cotransfection of RBPJL and PTF1A expression plasmids, PTF1-L was no more effective than PTF1-J on the Cabp2 enhancer, and both were equally effective in combination with MIST1 (see Fig. S6C). Consistent with the augmented expression of Nfe2l2 in acinar cells and the presence of MIST1 and PTF1 binding at the Nfe2l2 locus, both factors activated the enhancer modestly, and together their effect was synergistic (Fig. 6E).
DISCUSSION
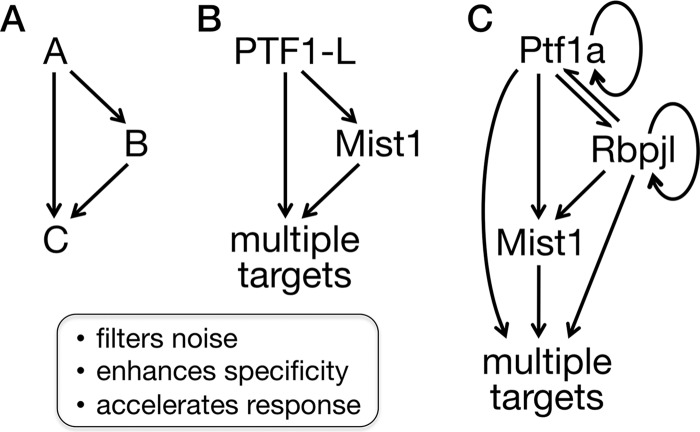
The long-term stability of a cellular phenotype is maintained through the transcriptional control of cell-type-specific gene expression programs. In this report, we describe an extensive feed-forward loop system that helps maintain cellular physiology and differentiation in a mature, terminally differentiated cell type in vivo. Feed-forward loop regulation is an effective and widespread strategy of transcriptional control (57). A feed-forward loop in a gene regulatory network comprises two transcriptional regulators, one of which binds and activates the gene of the second, and a target gene bound and regulated by both transcriptional regulators (Fig. 7). Positive coherent feed-forward loops in which both transcriptional regulators are activators are the most prevalent (58). The utility of feed-forward loops has been described for prokaryotic gene transcription networks (26), in which they rapidly provide maximal levels of induced gene expression in response to carbon and energy sources and other environmental stimuli and resist responses to biologic noise (59). Feed-forward loops are also prominent in developmental programs of metazoans (60), where they provide regulatory resistance to transient inductive signals and thereby reduce spurious cellular fate changes, help stabilize fate decisions once made, and enforce differentiation programs (61). In this study, we showed that feed-forward regulation also occurs in mature, differentiated mammalian cells. PTF1 and MIST1 form reiterative feed-forward loops with a large battery of target genes that function prominently in the maintenance of the acinar cell phenotype and homeostasis.
FIG 7.
PTF1A, RBPJL, and MIST1 collaborate via a two-stage, multioutput feed-forward loop system. (A) A simple positive feed-forward loop with two transcription factors. (B) Feed-forward loop considering the PTF1-L complex as a single factor. (C) Underlying regulatory relationships illustrate two feed-forward stages (PTF1A collaborating with RBJL and MIST1) as well as autoregulatory components (PTF1 regulates both Ptf1a and Rbpjl).
Whereas PTF1A is the master transcriptional regulator of pancreatic acinar cell identity and differentiation (62, 63), MIST1 has a more restricted role in the completion and optimization of acinar differentiation. MIST1 provides this specialization function for serous exocrine cell types of multiple organs (64). Within this context, PTF1 and MIST1 control two broad classes of genes: those with expression highly restricted to acinar cells of the pancreas and others more widely expressed and highly induced in other professional secretory cells. The highly restricted genes include the hallmark secretory enzymes and cofactors. The more broadly expressed genes encode proteins involved in secretory protein production, processing, protein folding, and cellular stress responses. The PTF1-MIST1 partnership extends to both gene classes. The PTF1-MIST1 loop is integrated in a more extended regulatory motif with PTF1A and RBPJL, providing additional properties that are expected to stabilize the acinar gene expression pattern (Fig. 7C).
The collaboration of MIST1 and the PTF1 complex satisfies the criteria for a reiterative system of feed-forward control (Fig. 7) as follows. (i) PTF1 controls Mist1 directly. PTF1 binds and activates Mist1 transcription through two sites in the Mist1 pancreatic enhancer; those sites are required for the activity of the enhancer in pancreatic acinar cells in vivo. (ii) MIST1 and PTF1 coregulate target genes. We identified genes with PTF1 and MIST1 bound less than 200 bp apart within regions with chromatin properties of transcriptional enhancers and tested four of the regions in cell transfection assays. Tests in vitro showed that all possessed enhancer activity, that they were activated by PTF1A and MIST1 in either an additive or synergistic manner, and that the activation depended on the retention of PTF1 and MIST1 binding site sequences. (iii) The PTF1-MIST1 loop is reiterated. MIST1 and PTF1 cobind more than 2,000 target genes. Ptf1a inactivation affects (increases or decreases) nearly half of these genes, and the absence of MIST1 affects one-seventh; 157 genes are affected in common.
The numerous MIST1-PTF1 target genes broadly define the pancreatic acinar phenotype. The cobound genes compose large fractions of acinus-enhanced differentiated processes and pathways (Fig. 5C), including 17 of the 30 genes for acinar secretory enzymes and their accessory factors as well as 20 to 50% of the genes for KEGG and IPA pathways of secretory protein production, translational regulation, the unfolded protein response (UPR) and oxidative stress response, control of the anabolic state, cell and tissue organization, and cell cycle and survival. Other acinus-like secretory cells that depend on MIST1 (i.e., gastric chief cells, plasma cells, and salivary gland acinar cells) (65–67) do not express Ptf1a (or Rbpjl) (our unpublished results). We propose that additional, organ-specific transcriptional regulators with equivalent responsibilities to PTF1 form similar reiterated feed-forward regulatory systems with MIST1 to provide the homologous secretory mechanisms for these acinus-like cells as well. It seems likely that this general regulatory strategy extends to other mature cell types using additional scaling factors analogous to MIST1 for the coregulation of other differentiation states.
The mechanisms that maintain the phenotype of adult cells are of potential biomedical relevance since the disruption of cellular phenotype and identity is a common basis of organ failure and cancer. Indeed, the induction of chronic or acute pancreatitis causes the loss of PTF1A and MIST1 and concomitant decrease of acinar differentiation (68–70). Moreover, PTF1A and MIST1 disappear in mouse and human pancreatic preneoplastic lesions (PanINs) of cancer patients, and inactivation of Ptf1a or Mist1 greatly enhances activated KRAS-driven transformation of pancreatic acinar cells in mouse models of pancreatic adenocarcinoma (22, 71, 72). An understanding of the mechanisms for the maintenance of the acinar phenotype may inform interventions to prevent further degradation of acinar cells during pancreatitis or to resist the neoplastic transformation of pancreatic adenocarcinoma.
Supplementary Material
ACKNOWLEDGMENTS
We thank Robert Hammer and the University of Texas Southwestern Transgenic Core for transgenic embryos, Jessica Williams and the Molecular Pathology Core for histological services, and Quan-Zhen Li and Ward Wakeland of the Genomics and Microarray Core for RNA and ChIP sequencing. We are grateful to Christopher Wright for goat anti-mouse PTF1A and Jason Mills for rabbit anti-human MIST1 antibodies. We also thank fellow lab members Mike Hale and Jumin Xue for their generous advice and expert assistance.
This study was supported by National Institutes of Health grants R01 DK55489 (S.F.K.), R01 CA124586 (S.F.K.), and R01 DK61220 (R.J.M.).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00370-16.
For a companion article on this topic, see doi:10.1128/MCB.00366-16.
REFERENCES
- 1.Gorelick FS, Jamieson JD. 2012. Structure-function relationships in the pancreatic acinar cell, p 1341–1360. In Johnson LR, Ghishan FK, Kaunitz JD, Merchant JL, Said HM, Wood JD (ed), Physiology of the gastrointestinal tract, 5th ed, vol 2 Academic Press, London, United Kingdom. [Google Scholar]
- 2.Case M. 1978. Synthesis, intracellular transport and discharge of exportable proteins in the pancreatic acinar cell and other cells. Biol Rev Camb Philos Soc 53:211–354. doi: 10.1111/j.1469-185X.1978.tb01437.x. [DOI] [PubMed] [Google Scholar]
- 3.Lee AH, Chu GC, Iwakoshi NN, Glimcher LH. 2005. XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. EMBO J 24:4368–4380. doi: 10.1038/sj.emboj.7600903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.MacDonald RJ, Swift GH, Real FX. 2010. Transcriptional control of acinar development and homeostasis. Prog Mol Biol Transl Sci 97:1–40. doi: 10.1016/B978-0-12-385233-5.00001-5. [DOI] [PubMed] [Google Scholar]
- 5.Magri E, Baldoni G, Grazi E. 1975. On the biosynthesis of creatine. Intramitochondrial localization of transamidinase from rat kidney. FEBS Lett 55:91–93. [DOI] [PubMed] [Google Scholar]
- 6.Cleveland MH, Sawyer JM, Afelik S, Jensen J, Leach SD. 2012. Exocrine ontogenies: on the development of pancreatic acinar, ductal and centroacinar cells. Semin Cell Dev Biol 23:711–719. doi: 10.1016/j.semcdb.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rose SD, Swift GH, Peyton MJ, Hammer RE, MacDonald RJ. 2001. The role of PTF1-P48 in pancreatic acinar gene expression. J Biol Chem 276:44018–44026. doi: 10.1074/jbc.M106264200. [DOI] [PubMed] [Google Scholar]
- 8.Beres TM, Masui T, Swift GH, Shi L, Henke RM, MacDonald RJ. 2006. PTF1 is an organ-specific and Notch-independent basic helix-loop-helix complex containing the mammalian Suppressor of Hairless (RBP-J) or its paralogue, RBP-L. Mol Cell Biol 26:117–130. doi: 10.1128/MCB.26.1.117-130.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Obata J, Yano M, Mimura H, Goto T, Nakayama R, Mibu Y, Oka C, Kawaichi M. 2001. p48 subunit of mouse PTF1 binds to RBP-Jk/CBF1, the intracellular mediator of Notch signalling, and is expressed in the neural tube of early stage embryos. Genes Cells 6:345–360. doi: 10.1046/j.1365-2443.2001.00422.x. [DOI] [PubMed] [Google Scholar]
- 10.Cockell M, Stevenson BJ, Strubin M, Hagenbuchle O, Wellauer PK. 1989. Identification of a cell-specific DNA-binding activity that interacts with a transcriptional activator of genes expressed in the acinar pancreas. Mol Cell Biol 9:2464–2476. doi: 10.1128/MCB.9.6.2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roux E, Strubin M, Hagenbuchle O, Wellauer PK. 1989. The cell-specific transcription factor PTF1 contains two different subunits that interact with the DNA. Genes Dev 3:1613–1624. doi: 10.1101/gad.3.10.1613. [DOI] [PubMed] [Google Scholar]
- 12.Masui T, Long Q, Beres TM, Magnuson MA, MacDonald RJ. 2007. Early pancreatic development requires the vertebrate Suppressor of Hairless (RBPJ) in the PTF1 bHLH complex. Genes Dev 21:2629–2643. doi: 10.1101/gad.1575207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masui T, Swift GH, Deering T, Shen C, Coats WS, Long Q, Elsasser HP, Magnuson MA, MacDonald RJ. 2010. Replacement of Rbpj with Rbpjl in the PTF1 complex controls the final maturation of pancreatic acinar cells. Gastroenterology 139:270–280. doi: 10.1053/j.gastro.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pin CL, Bonvissuto AC, Konieczny SF. 2000. Mist1 expression is a common link among serous exocrine cells exhibiting regulated exocytosis. Anat Rec 259:157–167. doi:. [DOI] [PubMed] [Google Scholar]
- 15.Pin CL, Rukstalis JM, Johnson C, Konieczny SF. 2001. The bHLH transcription factor Mist1 is required to maintain exocrine pancreas cell organization and acinar cell identity. J Cell Biol 155:519–530. doi: 10.1083/jcb.200105060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hess DA, Strelau KM, Karki A, Jiang M, Azevedo-Pouly AC, Lee A-H, Deering TG, Hoang CQ, MacDonald RJ, Konieczny SF. 2016. MIST1 links secretion and stress as both target and regulator of the unfolded protein response. Mol Cell Biol 36:2931–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoang CQ, Hale MA, Azevedo-Pouly A, Elsasser HP, Deering TG, Willet SG, Pan FC, Magnuson MA, Wright CVE, Swift GH, MacDonald RJ. 3 October 2016. Transcriptional maintenance of pancreatic acinar identity, differentiation, and homeostasis by PTF1A. Mol Cell Biol doi: 10.1128/MCB.00358-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo X, Shin DM, Wang X, Konieczny SF, Muallem S. 2005. Aberrant localization of intracellular organelles, Ca2+ signaling, and exocytosis in Mist1 null mice. J Biol Chem 280:12668–12675. doi: 10.1074/jbc.M411973200. [DOI] [PubMed] [Google Scholar]
- 19.DiRenzo D, Hess DA, Damsz B, Hallett JE, Marshall B, Goswami C, Liu Y, Deering T, MacDonald RJ, Konieczny SF. 2012. Induced Mist1 expression promotes remodeling of mouse pancreatic acinar cells. Gastroenterology 143:469–480. doi: 10.1053/j.gastro.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian X, Jin RU, Bredemeyer AJ, Oates EJ, Blazewska KM, McKenna CE, Mills JC. 2010. RAB26 and RAB3D are direct transcriptional targets of MIST1 that regulate exocrine granule maturation. Mol Cell Biol 30:1269–1284. doi: 10.1128/MCB.01328-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kowalik AS, Johnson CL, Chadi SA, Weston JY, Fazio EN, Pin CL. 2007. Mice lacking the transcription factor Mist1 exhibit an altered stress response and increased sensitivity to caerulein-induced pancreatitis. Am J Physiol Gastrointest Liver Physiol 292:G1123–G1132. [DOI] [PubMed] [Google Scholar]
- 22.Shi G, Zhu L, Sun Y, Bettencourt R, Damsz B, Hruban RH, Konieczny SF. 2009. Loss of the acinar-restricted transcription factor Mist1 accelerates Kras-induced pancreatic intraepithelial neoplasia. Gastroenterology 136:1368–1378. doi: 10.1053/j.gastro.2008.12.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arda HE, Benitez CM, Kim SK. 2013. Gene regulatory networks governing pancreas development. Dev Cell 25:5–13. doi: 10.1016/j.devcel.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacDonald RJ, Swift GH, Real FX. 2010. Transcriptional control of exocrine pancreas development and homeostasis, p 1–40. In Kaestner KH. (ed), Development, differentiation and disease of the para-alimentary tract. Progress in molecular biology and translations science, vol 97 Academic Press, London, United Kingdom. [Google Scholar]
- 25.Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, Zeitlinger J, Jennings EG, Murray HL, Gordon DB, Ren B, Wyrick JJ, Tagne JB, Volkert TL, Fraenkel E, Gifford DK, Young RA. 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 26.Shen-Orr SS, Milo R, Mangan S, Alon U. 2002. Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet 31:64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- 27.Masui T, Swift GH, Hale MA, Meredith DM, Johnson JE, Macdonald RJ. 2008. Transcriptional autoregulation controls pancreatic Ptf1a expression during development and adulthood. Mol Cell Biol 28:5458–5468. doi: 10.1128/MCB.00549-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma HW, Kumar B, Ditges U, Gunzer F, Buer J, Zeng AP. 2004. An extended transcriptional regulatory network of Escherichia coli and analysis of its hierarchical structure and network motifs. Nucleic Acids Res 32:6643–6649. doi: 10.1093/nar/gkh1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alon U. 2007. Network motifs: theory and experimental approaches. Nat Rev Genet 8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 30.Lennerz JK, Kim SH, Oates EL, Huh WJ, Doherty JM, Tian X, Bredemeyer AJ, Goldenring JR, Lauwers GY, Shin YK, Mills JC. 2010. The transcription factor MIST1 is a novel human gastric chief cell marker whose expression is lost in metaplasia, dysplasia, and carcinoma. Am J Pathol 177:1514–1533. doi: 10.2353/ajpath.2010.100328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemercier C, To RQ, Swanson BJ, Lyons GE, Konieczny SF. 1997. Mist1: a novel basic helix-loop-helix transcription factor exhibits a developmentally regulated expression pattern. Dev Biol 182:101–113. doi: 10.1006/dbio.1996.8454. [DOI] [PubMed] [Google Scholar]
- 32.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bailey TL, Williams N, Misleh C, Li WW. 2006. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vierstra J, Rynes E, Sandstrom R, Zhang M, Canfield T, Hansen RS, Stehling-Sun S, Sabo PJ, Byron R, Humbert R, Thurman RE, Johnson AK, Vong S, Lee K, Bates D, Neri F, Diegel M, Giste E, Haugen E, Dunn D, Wilken MS, Josefowicz S, Samstein R, Chang KH, Eichler EE, De Bruijn M, Reh TA, Skoultchi A, Rudensky A, Orkin SH, Papayannopoulou T, Treuting PM, Selleri L, Kaul R, Groudine M, Bender MA, Stamatoyannopoulos JA. 2014. Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. Science 346:1007–1012. doi: 10.1126/science.1246426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lattin JE, Schroder K, Su AI, Walker JR, Zhang J, Wiltshire T, Saijo K, Glass CK, Hume DA, Kellie S, Sweet MJ. 2008. Expression analysis of G protein-coupled receptors in mouse macrophages. Immunome Res 4:5. doi: 10.1186/1745-7580-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gautier L, Cope L, Bolstad BM, Irizarry RA. 2004. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 42.Chen X, Walker AK, Strahler JR, Simon ES, Tomanicek-Volk SL, Nelson BB, Hurley MC, Ernst SA, Williams JA, Andrews PC. 2006. Organellar proteomics: analysis of pancreatic zymogen granule membranes. Mol Cell Proteomics 5:306–312. [DOI] [PubMed] [Google Scholar]
- 43.Rindler MJ, Xu CF, Gumper I, Smith NN, Neubert TA. 2007. Proteomic analysis of pancreatic zymogen granules: identification of new granule proteins. J Proteome Res 6:2978–2992. doi: 10.1021/pr0607029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun X, Jiang X. 2013. Combination of FASP and fully automated 2D-LC-MS/MS allows in-depth proteomic characterization of mouse zymogen granules. Biomed Chromatogr 27:407–408. doi: 10.1002/bmc.2805. [DOI] [PubMed] [Google Scholar]
- 45.Williams JA, Chen X, Sabbatini ME. 2009. Small G proteins as key regulators of pancreatic digestive enzyme secretion. Am J Physiol Endocrinol Metab 296:E405–E414. doi: 10.1152/ajpendo.90874.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Porat-Shliom N, Milberg O, Masedunskas A, Weigert R. 2013. Multiple roles for the actin cytoskeleton during regulated exocytosis. Cell Mol Life Sci 70:2099–2121. doi: 10.1007/s00018-012-1156-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang B, Kirov S, Snoddy J. 2005. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res 33:W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y, MacDonald RJ, Swift GH. 2001. DNA binding and transcriptional activation by a PDX1.PBX1b.MEIS2b trimer and cooperation with a pancreas-specific basic helix-loop-helix complex. J Biol Chem 276:17985–17993. doi: 10.1074/jbc.M100678200. [DOI] [PubMed] [Google Scholar]
- 49.Kawaguchi Y, Cooper B, Gannon M, Ray M, MacDonald RJ, Wright CVE. 2002. The role of the transcriptional regulator PTF1a in converting intestinal to pancreatic progenitors. Nat Genet 32:128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- 50.Natoli G, Andrau JC. 2012. Noncoding transcription at enhancers: general principles and functional models. Annu Rev Genet 46:1–19. doi: 10.1146/annurev-genet-110711-155459. [DOI] [PubMed] [Google Scholar]
- 51.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 52.Meredith DM, Borromeo MD, Deering TG, Casey B, Savage TK, Mayer PR, Hoang C, Tung KC, Kumar M, Shen C, Swift GH, Macdonald RJ, Johnson JE. 2013. Program specificity for Ptf1a in pancreas versus neural tube development correlates with distinct collaborating cofactors and chromatin accessibility. Mol Cell Biol 33:3166–3179. doi: 10.1128/MCB.00364-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hale MA, Swift GH, Hoang CQ, Deering TG, Masui T, Lee YK, Xue J, MacDonald RJ. 2014. The nuclear hormone receptor family member NR5A2 controls aspects of multipotent progenitor cell formation and acinar differentiation during pancreatic organogenesis. Development 141:3123–3133. doi: 10.1242/dev.109405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rose SD, MacDonald RJ. 1997. Evolutionary silencing of the human elastase I gene (ELA1). Hum Mol Genet 6:897–903. doi: 10.1093/hmg/6.6.897. [DOI] [PubMed] [Google Scholar]
- 55.Tran T, Jia D, Sun Y, Konieczny SF. 2007. The bHLH domain of Mistl is sufficient to activate gene transcription. Gene Expr 13:241–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tessarz P, Kouzarides T. 2014. Histone core modifications regulating nucleosome structure and dynamics. Nat Rev Mol Cell Biol 15:703–708. doi: 10.1038/nrm3890. [DOI] [PubMed] [Google Scholar]
- 57.Alon U. 2007. Introduction to systems biology: design principles of biological circuits. CRC, Boca Raton, FL. [Google Scholar]
- 58.Mangan S, Alon U. 2003. Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci U S A 100:11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kittisopikul M, Suel GM. 2010. Biological role of noise encoded in a genetic network motif. Proc Natl Acad Sci U S A 107:13300–13305. doi: 10.1073/pnas.1003975107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davidson EH. 2010. Emerging properties of animal gene regulatory networks. Nature 468:911–920. doi: 10.1038/nature09645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mullen AC, Orlando DA, Newman JJ, Loven J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, DeKoter RP, Young RA. 2011. Master transcription factors determine cell-type-specific responses to TGF-β signaling. Cell 147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hesselson D, Anderson RM, Stainier DY. 2011. Suppression of Ptf1a activity induces acinar-to-endocrine conversion. Curr Biol 21:712–717. doi: 10.1016/j.cub.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pan FC, Bankaitis ED, Boyer D, Xu X, Van de Casteele M, Magnuson MA, Heimberg H, Wright CV. 2013. Spatiotemporal patterns of multipotentiality in Ptf1a-expressing cells during pancreas organogenesis and injury-induced facultative restoration. Development 140:751–764. doi: 10.1242/dev.090159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mills JC, Taghert PH. 2012. Scaling factors: transcription factors regulating subcellular domains. Bioessays 34:10–16. doi: 10.1002/bies.201100089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Capoccia BJ, Lennerz JK, Bredemeyer AJ, Klco JM, Frater JL, Mills JC. 2011. Transcription factor MIST1 in terminal differentiation of mouse and human plasma cells. Physiol Genomics 43:174–186. doi: 10.1152/physiolgenomics.00084.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ramsey VG, Doherty JM, Chen CC, Stappenbeck TS, Konieczny SF, Mills JC. 2007. The maturation of mucus-secreting gastric epithelial progenitors into digestive-enzyme secreting zymogenic cells requires Mist1. Development 134:211–222. doi: 10.1242/dev.02700. [DOI] [PubMed] [Google Scholar]
- 67.Johnson CL, Kowalik AS, Rajakumar N, Pin CL. 2004. Mist1 is necessary for the establishment of granule organization in serous exocrine cells of the gastrointestinal tract. Mech Dev 121:261–272. doi: 10.1016/j.mod.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 68.Molero X, Adell T, Skoudy A, Padilla MA, Gomez JA, Chalaux E, Malagelada JR, Real FX. 2007. Pancreas transcription factor 1α expression is regulated in pancreatitis. Eur J Clin Invest 37:791–801. doi: 10.1111/j.1365-2362.2007.01856.x. [DOI] [PubMed] [Google Scholar]
- 69.Pinho AV, Rooman I, Reichert M, De Medts N, Bouwens L, Rustgi AK, Real FX. 2011. Adult pancreatic acinar cells dedifferentiate to an embryonic progenitor phenotype with concomitant activation of a senescence programme that is present in chronic pancreatitis. Gut 60:958–966. doi: 10.1136/gut.2010.225920. [DOI] [PubMed] [Google Scholar]
- 70.Karki A, Humphrey SE, Steele RE, Hess DA, Taparowsky EJ, Konieczny SF. 2015. Silencing Mist1 gene expression is essential for recovery from acute pancreatitis. PLoS One 10:e0145724. doi: 10.1371/journal.pone.0145724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Krah NM, De La OJ, Swift GH, Hoang CQ, Willet SG, Chen Pan F, Cash GM, Bronner MP, Wright CV, MacDonald RJ, Murtaugh LC. 2015. The acinar differentiation determinant PTF1A inhibits initiation of pancreatic ductal adenocarcinoma. eLife 4:e07125. doi: 10.7554/eLife.07125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shi G, DiRenzo D, Qu C, Barney D, Miley D, Konieczny SF. 2013. Maintenance of acinar cell organization is critical to preventing Kras-induced acinar-ductal metaplasia. Oncogene 32:1950–1958. doi: 10.1038/onc.2012.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.