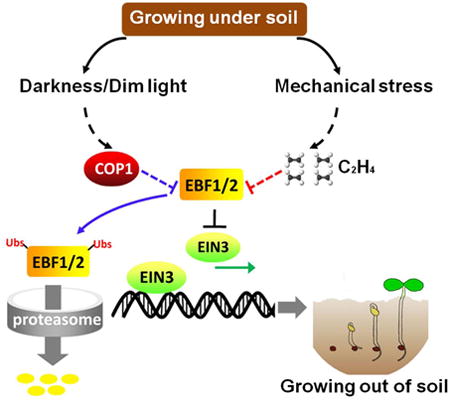
Summary
The survival of seed plants in natural environments requires the successful emergence from the soil. In this process, the ethylene signaling pathway is utilized by plants to sense and respond to the mechanical resistance of the soil. Here, we report that CONSTITUTIVE PHOTOMORPHOGENESIS 1 (COP1), a central repressor of light signaling, is a key component required for seedlings to sense the depth of soil overlay. Mutation in COP1 causes severe defects in penetrating soil, due to decreased level of EIN3, a master transcription factor in ethylene pathway that mediates seedling emergence. We show that COP1 directly targets the F-box proteins EBF1 and EBF2 for ubiquitination and degradation, thus stabilizing EIN3. As seedlings grow towards the surface, the depth of soil overlay decreases, resulting in a gradual increase of light fluences. COP1 channels the light signals while ethylene transduces the information on soil mechanical conditions, which cooperatively control EIN3 protein levels to promote seedling emergence from the soil. The COP1-EBF1/2-EIN3 module reveals a mechanism by which plants sense the depth to surface and uncovers a novel regulatory paradigm of an ubiquitin E3 ligase cascade.
Keywords: Seedling emergence, COP1, ethylene signaling, EBF1 and EBF2, EIN3
Graphical abstract
Introduction
To colonize the land, terrestrial flowering plants (angiosperms) have evolved tremendous developmental plasticity to adapt to the subterranean environments. The seeds buried under soil or litter can survive through hostile conditions and germinate when the environments become favorable. After germination, seedlings need to adjust their growth in accordance with the subterranean environment to reach the soil surface [1, 2]. Soil overlay brings about at least two consequences to the germinating seedlings: a dark environment and mechanical pressure. To grow in the dark, seedlings adopt a developmental strategy known as skotomorphogenesis, which is characterized by long hypocotyls, small and closed cotyledons, and curved apical hooks, an optimal shape to vigorously grow towards the surface [1, 3-5]. In addition, the etioplast development in cotyledons allows the seedlings to make a rapid transition to autotrophic growth upon light radiation at the soil surface [2, 5]. Meanwhile, mechanical impedance boosts seedlings ethylene production, which suppresses hypocotyl elongation and increases radical expansion, a stature fit for enhancing the lifting capacity of etiolated seedlings and protecting against mechanical injuries [6-9]. The crucial roles of ethylene in this process have been demonstrated by the observations that mutants lacking ethylene responses (ethylene-insensitive) showed defects in soil emergence [10, 11]. These studies show that both light and mechanical pressure (ethylene) of the soil provide important cues that instruct plants how to grow morphologically and physiologically in the soil. However, whether and how the signals on lights and mechanical pressure (ethylene) are integrated to modulate plant growth in complex soil conditions is largely unknown.
Ethylene is a gaseous plant hormone that performs wide-ranging and dramatic effects on plant growth, development, and stress responses [9, 12, 13]. Ethylene is perceived by a family of five endoplasmic-reticulum (ER)-localized receptors. In the absence or low ethylene, the receptors are in an uncharged active state associated with CTR1, which represses the downstream signaling pathway. Upon binding to ethylene, the receptors disassociate from CTR1, causing de-repression of CTR1 on EIN2, resulting ultimately in EIN3 and EILs accumulation. EIN3 and EIL1 are master transcription factors in conducting a myriad of ethylene responses [14-17], and EIN3 level is tightly regulated by the 26S proteasome-mediated degradation pathway through two F-box proteins, EBF1 and EBF2 (EBF1/2) [18-20]. Ethylene triggers EIN2 cleavage and translocation, inducing EIN2-dependent proteasomal degradation of EBF1/2 to stabilize EIN3 proteins [21-24]. Our previous studies have further shown that EIN3 protein levels are quantitatively increased in response to soil overlay. EIN3 specifically activates two downstream pathways in the hypocotyl and cotyledons, respectively mediated by ERF1 and PIF3, to coordinately regulate hypocotyl elongation and chlorophyll biosynthesis in response to soil conditions during seedling emergence [11].
CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1) was originally identified as a central repressor of light-induced plant morphogenetic changes, evidenced by cop1 mutant seedlings exhibiting constitutive photomorphogenesis in the dark [25-27]. COP1 is a RING E3 ubiquitin ligase and comprises three protein-protein interaction domains: a N-terminal RING-finger region followed by a Coiled-coil domain, and seven WD40 repeats at its C-terminus [28-30]. The COP1 protein is evolutionally conserved in both plants and animals, and it acts as a vital regulatory point of diverse developmental processes. In plants, COP1 functions as a master switch of light signaling pathway, by maintaining skotomorphogenesis and repressing photomorphogenesis [27, 31]. Downstream of COP1, there are at least two groups of transcription factors conducting light-induced developmental transition: phytochrome-interacting factors (PIFs) in promoting skotomorphogenesis, and regulators including HY5, LAF1 and HFR1 in promoting photomorphogenesis [5, 31]. Extensive studies have consolidated that in the dark, COP1 targets HY5, LAF1 and HFR1 for ubiquitin-proteasome mediated degradations and stabilized PIFs through unknown mechanisms [5, 29, 32-36]. In mammalian cells, COP1 E3 ubiquitin ligase targets c-Jun and ETS transcription factors for degradation, thereby serving as a tumor suppressor [37, 38]. Based on current evidence, the major biochemical scheme that COP1 employs to regulate biological processes is direct protein interaction and ubiquitination of the key factors.
Although seedling etiolated growth and ethylene signaling transduction have been extensively studied in the past decades, most genetic works were conducted under soil-free growth conditions. Only in recent years, we utilized Arabidopsis genetic tools to study soil responses of plants. Our previous works have uncovered the prominent role of the ethylene pathway in responding to mechanical pressure of the soil, and have identified critical regulatory factors in this response such as EIN3, ERF1, and PIF3 [11]. In this study, we identify COP1 as a crucial regulator for seedling soil emergence that specifically transduces the information, not on mechanical pressure of the soil, but on the light fluences in the soil. Mechanistically, COP1 directly targets EBF1/2 upstream of EIN3 to mediate soil responses.
Results
COP1 functions upstream of EIN3 to promote seedling emergence
Skotomorphogenesis is a developmental strategy adopted by plant seedlings in terrestrial environments [1, 2]. To determine whether etiolated growth is functionally involved in seedling emergence from soil overlay, we examined the role of COP1 in regulating seedlings emergence phenotypes. In the assay, we plated seeds on 1/2 MS agar medium and evenly covered them with a layer of soil. After 7 days of growth in white light, the emergence frequencies were recorded. Our results showed that without soil covering, the cop1 mutants grew similarly to wild-type (WT, Col-0) (Figure 1A and 1B). However, when covered under a layer of soil (SiO2 powder), most of cop1 mutant seedlings failed to emerge from the soil (Figure 1A and 1B). We further examined the seeding emergence with different particle size sands. Similar to the SiO2 powder, the cop1 mutants exhibited severe defects in emergence from either 50-70 or 120-200 mesh sands covering, while WT grew well in the same soil conditions (Figure 1C and 1D). Moreover, for a specific sand type, the survival rate of cop1 mutants dramatically decreased with increasing soil depths (Figure 1A to 1D). To verify the requirement for the COP1 gene in seedling emergence from the soil, we used the 50-70 mesh sands as the standard soil conditions and investigated the emergence phenotypes of another cop1 mutant allele cop1-6. Like cop1-4, cop1-6 grew similarly to WT without soil covering, but showed severe defects in seedling emergence when covered by a layer of soil (Figure 1E and 1F). In addition, expressing COP1 in cop1 mutant background (35S:COP1/cop1-6) rescued the emergence defect and restored the emergence frequency of cop1 to wild-type levels (Figure 1E and 1F). We further examined the soil penetration ability of seedlings under complete darkness. The results showed that almost all the cop1 mutants failed to emerge from the soil, and this severely impaired ability could be fully restored by expressing COP1 in cop1 mutants (Figure 1G). These observations demonstrate that COP1 is essential for seedling emergence from the soil.
Figure 1. COP1 acts upstream of EIN3 to promote seedling emergence from the soil.
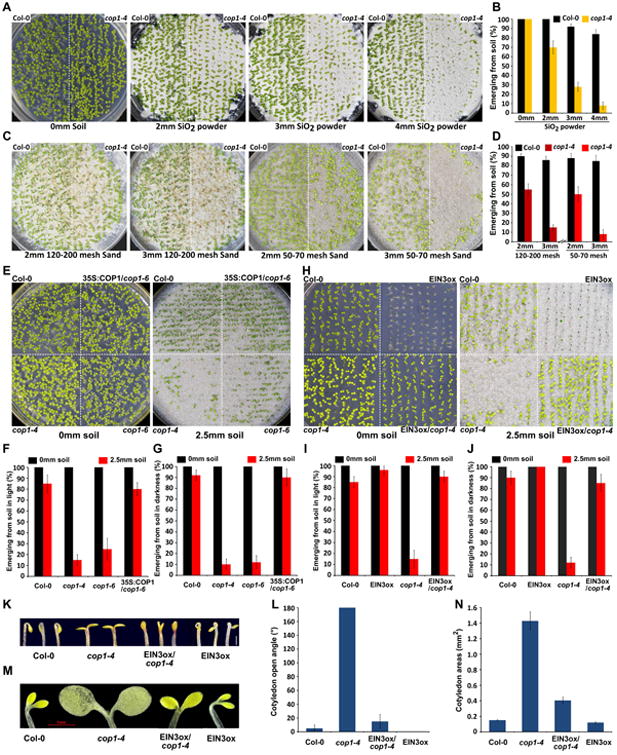
(A-D) Soil emergence phenotypes (A and C) and quantitative analysis (B and D) of Col-0 (wild type) and cop1-4 mutant. Seedlings were grown under continuous white light for 7 days without (0 mm soil) or with increasing depths overlay of SiO2 powder (A and B) and various particle size sands (C and D). Mean ±s.d., n=3.
(E-F) Soil emergence phenotypes (E) and quantitative analysis (F) of Col-0, cop1-4, cop1-6 and 35S:COP1/cop1-6. Seedlings were grown under continuous white light for 7 days without (0 mm soil) or with 2.5mm soil (50-70 mesh sands) overlay. Mean ±s.d., n=3.
(G) Quantitative analysis of 4-day old etiolated Col-0, cop1-4, cop1-6 and 35S:COP1/cop1-6 seedlings without (0 mm soil) or with 2.5mm soil (50-70 mesh sands) overlay. Mean ±s.d., n=3.
(H-I) Soil emergence phenotypes (H) and quantitative analysis (I) of Col-0, cop1-4, EIN3ox and EIN3ox/cop1-4. Seedlings were grown under continuous white light for 7 days without (0 mm soil) or with 2.5mm soil (50-70 mesh sands) overlay. Mean ±s.d., n=3.
(J) Quantitative analysis of 4-day old etiolated Col-0, cop1-4, EIN3ox and EIN3ox/cop1-4 seedlings without (0 mm soil) or with 2.5mm soil overlay. Mean ±s.d., n=3.
(K-L) Images (K) and angles (L) of cotyledon opening of 3-day old etiolated Col-0, cop1-4, EIN3ox and EIN3ox/cop1-4 seedlings. Mean ±s.d., n>20.
(M-N) Images (M) of cotyledons and areas (N) of single cotyledons of 7-day old etiolated Col-0, cop1-4, EIN3ox and EIN3ox/cop1-4 seedlings. Mean ±s.d., n>20.
It has been recently shown that EIN3, the master transcription factor of the ethylene signaling pathway, plays a crucial role in promoting seedling emergence [11]. Thus we analyzed the genetic relationship of COP1 and EIN3. We crossed the EIN3 over-expressing transgene EIN3ox into cop1 (EIN3ox/cop1), and examined the emergence frequencies of EIN3ox, cop1 and EIN3ox/cop1. The results showed that all the lines grew well without soil covering (Figure 1H and 1I). Once buried under soil, while WT and EIN3ox were able to penetrate the soil, most of the cop1 mutants failed to emerge as in previous experiments (Figure 1H and 1I). However, almost all of the EIN3ox/cop1 seedlings successfully grew out of soil, indicating that over-expression of EIN3 can rescue the soil emergence defect of cop1 (Figure 1H and 1I). These data suggest that COP1 functions upstream of EIN3 to promote seedlings emergence.
Similarly, when grown under complete darkness, overexpression of EIN3 can also rescue the cop1's phenotype in emerging from the soil (Figure 1J). By closely examining the phenotypes of seedlings grown in the dark, we found that consistent with previous studies [25], the cotyledons of cop1 were fully opened (Figure 1K and 1L), and dramatically expanded (Figure 1M and 1N). However, over-expressing EIN3 in cop1 (EIN3ox/cop1) largely restored the cotyledon phenotypes of cop1, resulting in closed and much less expanded cotyledons, similar to WT (Figure 1K to 1N). Therefore, the impaired seedling emergence of cop1 associated with an abnormal developmental pattern in the dark underneath the soil surface, which can be partially corrected by over-production of EIN3.
EIN3 activity is eliminated in cop1 mutant
Since cop1 displayed severe defects in seedling emergence similar to the ein3eil1 mutant [11] and can be rescued by overexpressing EIN3 (Figure 1H to 1J), the function of EIN3 is possibly impaired in cop1 mutants. To investigate the activity of EIN3 in cop1, we introduced a 5 × EBS:GUS reporter line into different backgrounds by genetic crossing. This transgene harbors five tandem repeats of the EIN3 Binding Sequence (EBS) in the promoter fused with a GUS reporter gene and can be used to monitor the transcription activity of EIN3 in plants [39]. We found that EIN3ox notably increased the 5 × EBS:GUS expression, whereas the elevation in EIN3ox was dramatically decreased by the mutation of COP1 (Figure 2A). It was previously reported that several genes, including ERF1, EBF2 and PIF3, are directly activated by EIN3 [40-42]. Our results showed that the expression patterns of all of these genes were down-regulated in cop1 to the extend similar to ein3eil1 and cop1ein3eil1 triple mutant (Figure 2B to 2D). Moreover, overexpressing EIN3 in the cop1 mutant largely restored these genes' expression to wild-type level (Figure 2B to 2D). We further examined the expression of some EIN3-repressed genes. Both cop1 and cop1ein3eil1 evidently upregulated these genes similar to ein3eil1, while EIN3ox/cop1 largely suppressed the expression of these genes to a level similar to wild-type (Figure 2E and 2F). Therefore, EIN3 activity is dramatically suppressed in cop1 mutant, indicating that COP1 is required to maintain EIN3 function in etiolated seedlings.
Figure 2. EIN3 transcriptional actions are largely abolished by COP1 mutation.
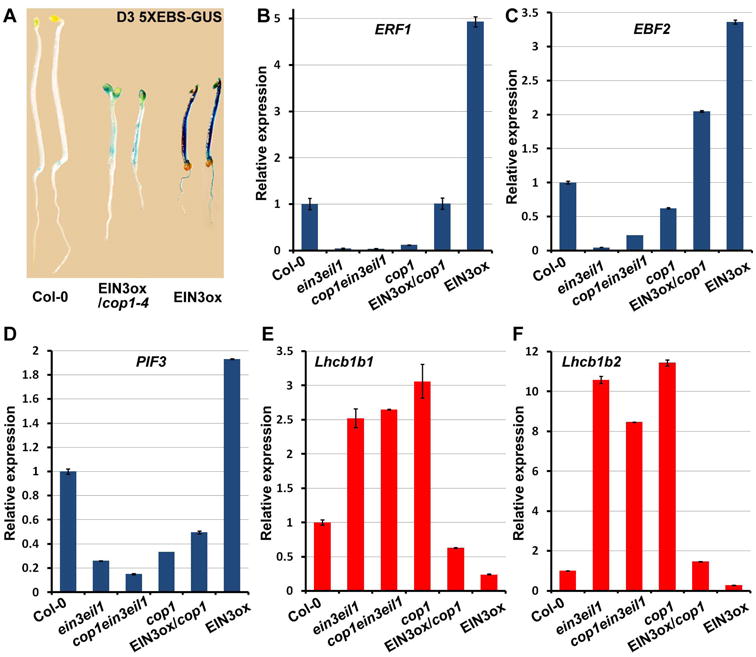
(A) GUS staining images of 3-day old etiolated seedlings of 5XEBS-GUS in Col-0, EIN3ox and EIN3ox/cop1-4 backgrounds.
(B-F) qRT-PCR results showing the gene expressions of ERF1 (B), EBF2 (C), PIF3 (D), Lhcb1b1 (E) and Lhcb1b2 (F) in 4-day old dark-grown seedlings. The expressions were normalized to PP2A. Mean ±s.d., n=3.
COP1 is required to prevent 26S proteasome-mediated degradation of EIN3 protein in seedlings
To investigate the regulation of COP1 on EIN3, we used a transgenic Arabidopsis line that expresses EIN3-Luciferase fused proteins driven by the native EIN3 promoter. We found that in cop1 mutants, EIN3 protein levels were notably decreased in etiolated seedlings (Figure 3A), while the transcript level of EIN3 gene did not significantly change (Figure S1). To study posttranscriptional regulation of EIN3, we generated a transgenic line that constitutively expressed EIN3-Myc fusion proteins in the eil3eil1 mutant (35S:EIN3-Myc/ein3eil1), and crossed it into cop1 mutant background. Immunoblot results showed that the EIN3-Myc protein levels were markedly decreased in cop1ein3eil1 compared to ein3eil1 (Figure 3B). Treatment with MG132, a 26S proteasome-specific inhibitor, significantly restored EIN3 protein abundance in cop1ein3eil1 seedlings (Figure 3B), suggesting that EIN3 is post-translationally protected by COP1 from 26S proteasome-mediated degradation in plants. Moreover, the repression of EIN3 protein degradation by COP1 can be reconstituted in a cell-free assay by using purified full-length EIN3-His recombinant proteins. When added into the cell extracts of WT seedlings, EIN3 proteins were gradually degraded (Figure 3C). However, when added into cop1 cell extracts, the EIN3 proteins were extremely unstable and were rapidly degraded to an undetectable level within half an hour (Figure 3C). In both cases, MG132 prevented EIN3 degradation, confirming EIN3 degradation was mediated by the 26S proteasome in the cell extracts (Figure 3C). Additionally, fluorescence microscopy showed that the constitutively expressed EIN3-GFP proteins were dramatically decreased in the nucleus of cop1 (Figure 3D). Together, these results indicate that COP1 is essential for EIN3 protein accumulation by repressing the 26S proteasome-mediated EIN3 degradation.
Figure 3. COP1 stabilizes EIN3 protein.
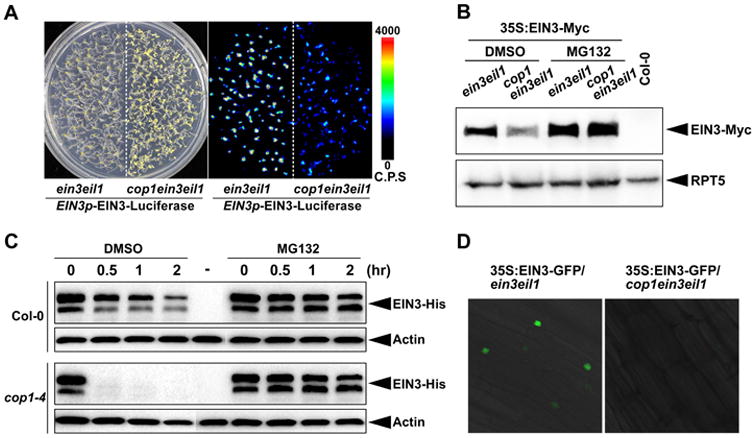
(A) White light (Left) and bioluminescence (Right) images of 4-day old etiolated seedlings of EIN3p-EIN3-Luciferase in ein3eil1 and cop1ein3eil1 backgrounds. The color-coded bar indicates the intensity of lucifersase activity. C.P.S stands for Counts Per Second.
(B) Western blot analysis of EIN3 protein levels. Seedlings over-expressing EIN3-Myc in ein3eil1 and cop1ein3eil1 backgrounds were grown on 1/2MS medium in the dark for 4 days without (DMSO) or with MG132 pre-treatment for 12 h before harvesting. Col-0 was used as a negative control. RPT5 was used as a loading control.
(C) Cell-free degradation of recombinant EIN3-His proteins in 4-day old etiolated Col-0 (top) and cop1-4 (bottom) seedlings. Equal amount of EIN3-His proteins were added into the cell extracts and incubated for the indicated periods of time, and then analyzed by immunoblots. “-” stands for the no EIN3-His protein control. Actin was used as a loading control.
(D) Fluorescence microscopic analysis of the EIN3-GFP protein levels. Seedlings over-expressing EIN3-GFP in ein3eil1 and cop1ein3eil1 backgrounds were grown on 1/2MS medium for 4 days in the dark.
COP1 promotes proteasomal degradation of both EBF1 and EBF2
Previous studies have shown that two F-box proteins EBF1 and EBF2 target EIN3 for 26S proteasome-mediated degradation by acting as substrate receptors for the SCF E3 ligase complex [18-20]. We wanted to know whether COP1 affects the accumulation of EBF1 and EBF2, given the E3 ligase activity of COP1 and our finding that COP1 stabilizes EIN3. We crossed the constitutively expressed EBF1-TAP or EBF2-TAP transgenic lines into the cop1 mutant background. Immunodetection of EBF1-TAP and EBF2-TAP showed that levels of both proteins were highly elevated in the cop1 mutants (Figure 4A and 4B), suggesting that COP1 functions to destabilize EBF1 and EBF2. Moreover, MG132 treatment visibly elevated the protein levels of both EBF1 and EBF2 in WT, but no obvious changes were observed in the cop1 mutant (Figure 4A and 4B). Quantitatively analyzing the protein levels of EBF1 and EBF2 indicated that they were five times and three times higher in cop1 than in WT, respectively, while MG132 treatment increased their protein abundances in WT to comparable levels as in cop1 (Figure 4A and 4B). Therefore, COP1 acts to decrease EBF1 and EBF2 proteins through the 26S proteasome pathway.
Figure 4. COP1 is responsible for the 26S proteasome-mediated EBF1 and EBF2 protein degradation.
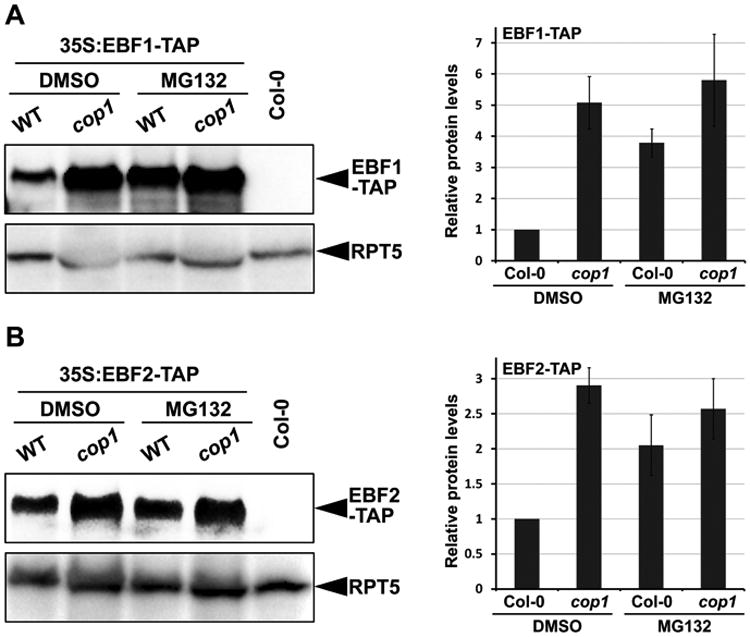
Western blots showing the EBF1(A, left panel) or EBF2 (B, left panel) protein levels. Seedlings over-expressing EBF1-TAP of EBF2-TAP in Col-0 (WT) and cop1-4 mutant backgrounds were grown on 1/2MS medium in the dark for 4 days without (DMSO) or with MG132 pre-treatment for 12 h before harvesting. Col-0 was used as a negative control. RPT5 was used as a loading control. Right panel shows the quantification analysis of the three biological replicates of EBF1 (A, right panel) or EBF2 (B, right panel) protein levels after normalizing to RPT5. The protein levels EBF1-TAP/EBF2-TAP in Col-0 backgrounds without MG132 treatment (DMSO) was set as 1. Mean ±s.d., n=3.
COP1 directly interacts with EBF1 and EBF2
To assess whether COP1 is an E3 ligase for EBF1 and EBF2, we first examined if COP1 directly interacts with EBF1 and EBF2 proteins. COP1 protein contains three recognizable domains: a RING-finger domain (Ring), a Coiled-coil domain (Coil) and seven WD40 repeats (WD40) (Figure 5A). In our yeast two-hybrid assays, both EBF1 and EBF2 were shown to directly interact with the N-terminal 282-amino acid fragment of COP1, which contains the Ring and Coil domains (Figure 5A). To further identify the domains that interact with EBF1 and EBF2, we used a series of deletion constructs of the N-terminal region of COP1 in the assays. Both EBF1 and EBF2 interacted with several mutant derivatives of COP1, including the Coil domain or the Ring domain alone, and deletion mutants lacking either the Coil or the Ring domain (Figure 5A). No interaction was detected when both the Coil and Ring domains were deleted (Figure 5A), indicating that these two domains are responsible for the interactions with EBF1 and EBF2. Interestingly, the C-terminal WD40 domain of COP1, which has been shown to target several known transcription factors [30], did not interact with either EBF1 or EBF2 in the assays (Figure 5A). These data suggest that the RING-finger and the coiled-coil domains within the N-terminal region of COP1 are involved in the interactions with EBF1 and EBF2.
Figure 5. EBF1 and EBF2 physically interact with COP1 in vitro and in vivo.
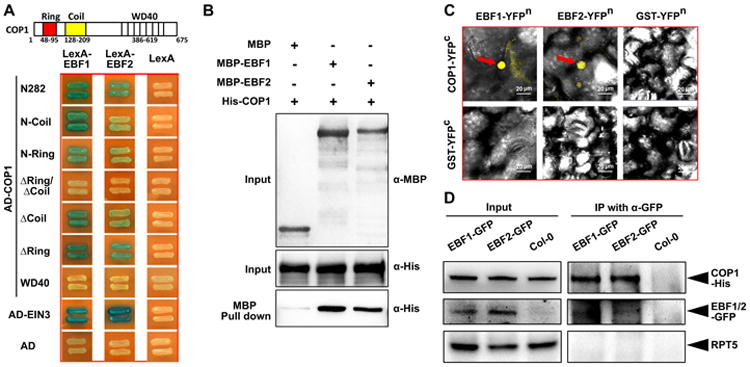
(A) N-terminal fragments of COP1 directly interact with EBF1 and EBF2 in yeast two-hybrid assays. Left diagrams indicate the various fragments of COP1 fused with the activation domain. Full length EBF1 and EBF2 fused with LexA DNA binding domain were the prey constructs in the assay.
(B) COP1 and EBF1/2 directly interact in pull-down assays. Purified COP1-His was used as prey and was pulled down by the baits EBF1-MBP, EBF2-MBP and MBP, respectively. Anti-MBP and anti-His were used for the immunoblot analysis.
(C) BiFC assay reveals that COP1 interacts with EBF1 and EBF2 in the nucleus of Nicotiana benthamiana leaf cells. Full length COP1 or EBF1 and EBF2 were fused to the split N-terminal or C-terminal (YFPn or YFPc) fragments of YFP. GST fused to YFPn or YFPc fragments were used as negative controls. Red arrow indicates the position of YFP speckles. Bar = 20 μm.
(D) Semi-in vivo co-immunoprecipitation assay of COP1 with EBF1 and EBF2 proteins. EBF1-GFP and EBF2-GFP overexpression transgenic plants and Col-0 control were grown in the dark for 4 days. Equal amount of COP1-His proteins were added into the cell extracts and immunoprecipitated using anti-GFP antibody and immunoblotted using indicated antibodies.
Furthermore, an in vitro pull-down assay using recombinant fusion proteins further showed that COP1-His directly interacted with both MBP-EBF1 and MBP-EBF2, but not MBP alone (Figure 5B). To verify their interactions in vivo, we performed bimolecular fluorescence complementation (BiFC) assays in tobacco leaves. The results demonstrated that coexpression of COP1-YFPc with either EBF1-YFPn or EBF2-YFPn reconstituted functional YFP proteins in the nucleus, but GST protein controls did not (Figure 5C), indicating that COP1 directly interacts with EBF1 and EBF2 in nucleus of plant cells. Consistently, firefly luciferase complementation imaging (LCI) assay further supported the observation that EBF1 and EBF2 primarily interacted with the N-terminal region of COP1 in vivo by reconstituting the activity of luciferase in plants (Figure S2). Moreover, co-immunoprecipitation assays using transgenic plants expressing EBF1-GFP or EBF2-GFP and purified COP1-His recombinant proteins confirmed their interaction in Arabidopsis seedlings (Figure 5D). These in vitro and in vivo results demonstrate that COP1 directly interacts with both EBF1 and EBF2 proteins.
EBF1 and EBF2 proteins are directly ubiquitinated by COP1
The regulation of EBF1 and EBF2 by COP1 and their direct interaction in vitro and in vivo suggest that COP1 may act as an E3 ligase for EBF1 and EBF2. We performed an in vitro ubiquitination assay in which we used the EBF1-MBP and EBF2-MBP recombinant proteins purified from E. coli as substrates, and a purified COP1-His recombinant protein as the E3 ligase. In the presence of the universal E1, E2 and ubiquitin, both EBF1 and EBF2 were polyubiquitinated by COP1 (Figure 6A and 6B). Moreover, with an increase of COP1 in the reaction, the polyubiquitinated levels of both EBF1 and EBF2 proteins were accordingly elevated (Figure 6A and 6B). Taken together, our results indicate that both EBF1 and EBF2 can be directly ubiquitinated by COP1 E3 ubiquitin ligase.
Figure 6. COP1 directly ubiquitinates both EBF1 and EBF2 proteins.
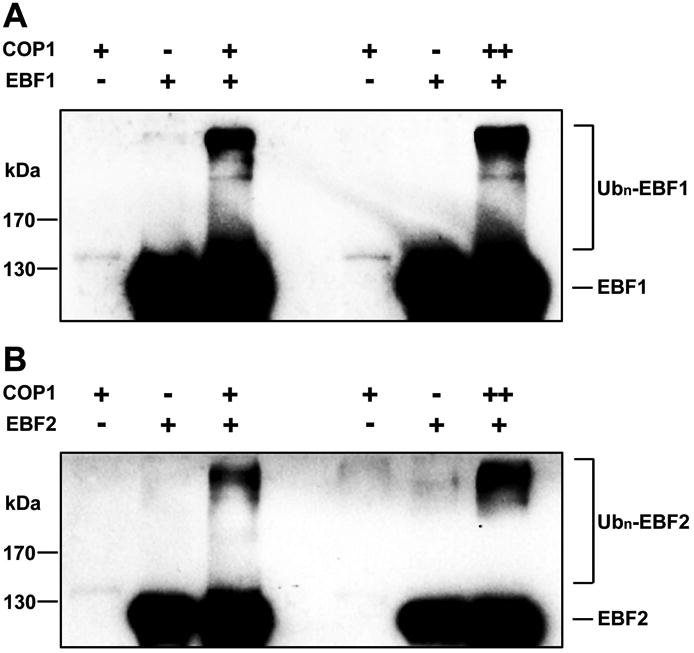
(A-B) In vitro ubiquitination assays of EBF1 (A) or EBF2 (B) by COP1. Purified recombinant COP1-His and EBF1-MBP or EBF2-MBP were used in the assays. The ubiquitination of EBF1 or EBF2 was analyzed by western blot using an anti-MBP antibody.
COP1 is not required for ethylene and mechanical pressure induced stabilization of EIN3
Since both COP1 and ethylene stabilize EIN3 by negatively regulating EBF1 and EBF2, we investigated whether COP1 is involved in ethylene-regulated EIN3 protein accumulation. We examined the EIN3 levels in etiolated seedlings under various ethylene-related chemical treatments, including 1-aminocyclopropane-1-carboxylic acid (ACC), a precursor of ethylene biosynthesis, as well as Ag+ and aminoethoxyvinylglycine (AVG), the inhibitors of ethylene action and biosynthesis, respectively. The results showed that the EIN3 protein levels were notably elevated with ACC treatment, and decreased in Ag+ or AVG supplemented medium, and this occurred even in the cop1 mutants (Figure S3A and S3B). We further examined the stability of EIN3 in response to increasing concentrations of ACC in the medium. Immunoblot analysis showed that EIN3-Myc accumulation was induced by increasing ACC concentrations in both ein3eil1 and cop1ein3eil1 mutant seedlings (Figure S3C and S3D). Although the quantitative levels of EIN3 in these mutants were different, cop1 mutant nonetheless displayed normal patterns of ethylene responsive regulation on EIN3 protein, suggesting that the regulation of EIN3 by ethylene does not require COP1.
In order to specify the role of COP1-EBF1/2-EIN3 pathway in seedling emergence, we examined the light effect on soil-induced EIN3 accumulation. Consistent with our previous report [11], soil covering of dark-grown seedlings enhanced the constitutively expressed EIN3-Myc proteins levels about two to three fold in both ein3eil1 and cop1ein3eil1 (Figure 7A). Notably, when the seedlings were grown in light, soil overlay caused a greater impact as it dramatically elevated the EIN3-Myc level in ein3eil1 for over ten fold (Figure 7B). However, in light-grown cop1ein3eil1, the soil covering-induced increase of EIN3-Myc was substantially weaker, with about four fold, even though cop1ein3eil1 and ein3eil1 had similar EIN3-Myc protein levels without soil covering (Figure 7B). These results suggest that light is a critical factor in soil-induced EIN3 protein accumulation, and that the light factor of the soil response is largely dependent on the action of COP1.
Figure 7. COP1 modulates the level of EIN3 proteins in response to the increased light fluences as seedlings grow towards the surface.
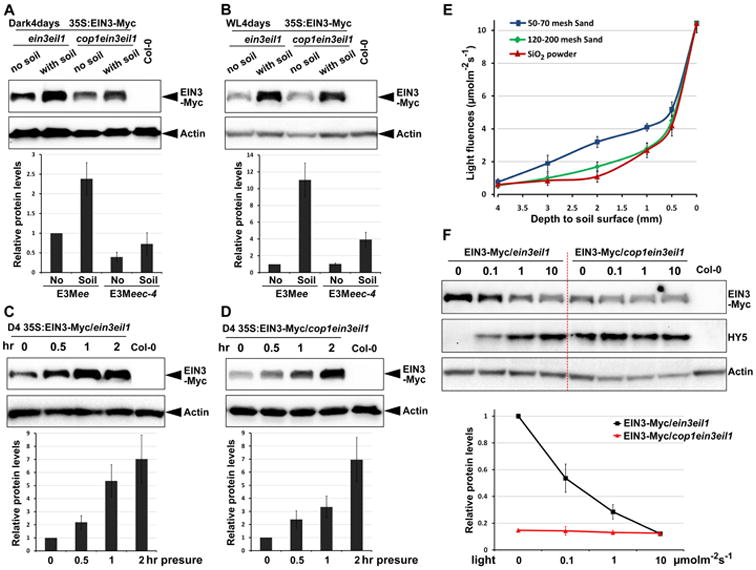
(A-B) Western blots showing EIN3 protein levels in the 4-day old seedlings grown in the dark (A) or white light (B). Seedlings over-expressing EIN3-Myc in ein3eil1 (E3Mee) and cop1ein3eil1 (E3Meec-4) backgrounds were grown on 1/2MS medium with or without 2.5mm soil (50-70 mesh Sand) covering. Bottom panels show the quantification analysis of the three biological replicates of EIN3-Myc protein levels after normalizing to Actin. The protein levels of EIN3-Myc/ein3eil1 without soil covering was set as 1. Mean ±s.d., n=3.
(C-D) Western blots showing the EIN3-Myc protein levels in ein3eil1 (C) or cop1ein3eil1 (D) background seedlings upon mechanical stresses. Seedlings were grown on 1/2MS medium for 4 days in the dark, then were pressed by a glass plate with the pressure of about 150Pa for the indicated periods of time. Bottom panels show the quantification analysis of the three biological replicates of EIN3-Myc protein levels after normalizing to Actin. The protein level of EIN3-Myc/ein3eil1 without mechanical stress was set as 1. Mean ±s.d., n=3.
(E) Light fluence in the soil increases as seedlings grow towards the soil surface. The light fluence was measured under the water-saturated soils with indicated particle sizes and depths.
(F) Western blots showing the EIN3 and HY5 protein levels in ein3eil1 and cop1ein3eil1 seedlings grown under indicated light fluences (μmolm2s-1) for 4 days. Dark-grown 4 day-old Col-0 was used as a negative control. Bottom panel shows the quantification analysis of the three biological replicates of EIN3-Myc protein levels after normalizing to Actin. The protein level of EIN3-Myc/ein3eil1 in the dark was set as 1. Mean ±s.d., n=3.
Differ from soil overlay in the dark, soil overlay in the light can reduce the light irradiation in addition to imposing mechanical pressure. To delineate the role of COP1 in soil-induced EIN3 stabilization under light conditions, we designed experiments to investigate the responses to light and mechanical pressure separately. We first pressed the etiolated seedlings with glass plates in the dark to achieve mechanical pressure independently of light, and found that the level of EIN3 was remarkably up-regulated and progressively increased upon longer periods of pressing (Figure 7C). Moreover, the accumulation of EIN3-Myc caused by mechanical pressure was similar in cop1ein3eil1 to ein3eil1 (Figure 7D), suggesting that COP1 is not involved in mechanical pressure alone. Thus, it is likely that the mechanical pressure response is mediated via the ethylene pathway independently of COP1.
COP1 is primarily responsible to modulate EIN3 levels according to the changing light fluences during soil emergence
It has been shown that light transmission is affected by both the texture and depth of soil [43-45]. In our assay, the seeds were covered by water-saturated soil of different particle sizes. By measuring the light fluences under water-saturated soil overlay, we found that regardless of the particle sizes, the light intensity dramatically decreased when the soil depth increased (Figure 7E). Interestingly, light transmission in dry sands was much lower than that in water-saturated status, and the light intensity decreased even more rapidly with increasing depths of soil (Figure S4A), consistent with the previous reports [43-45].
Next, we examined the effect of light intensity changes on EIN3 stability. During seedling emergence, the distance to soil surface decreases, resulting in gradually increased light fluences (Figure 7E). Therefore, the series of increasing light intensities from darkness to strong light simulate the light fluence changes during growth towards the soil surface. We found that with the increase of light intensity, the levels of EIN3-Myc in ein3eil1 gradually decreased (Figure 7F). Quantitative analysis showed that the EIN3-Myc protein level in the dark was eight fold greater compared with the strong light condition (Figure 7F). However, EIN3-Myc accumulation in cop1ein3eil1 mutant failed to change with light fluences, but rather displaying a constitutively low level similar to ein3eil1 grown under strong light (Figure 7F). This result suggests that COP1 is required for EIN3 stability in the dark and for quantitative modulation of EIN3 levels in response to light intensity changes. Moreover, the levels of HY5, a well-characterized substrate of COP1 [29], were progressively elevated in 35:EIN3-Myc/ein3eil1 but not in 35:EIN3-Myc/cop1ein3eil1 when light intensity was increased (Figure 7F), indicating that the activity of COP1 gradually decreases with increasing light fluences. Therefore, light signals quantitatively act to reduce EIN3 levels through negatively regulating COP1 activity. This may be a mechanism to turn down EIN3 activity according to the changes of soil depth when emerging seedlings grow towards the soil surface and perceive increased amounts of light.
Discussion
Successfully emerging from soil is crucial for the survival of seed plants in natural environments and farm systems. However, the underlying molecular mechanism by which plants overcome soil barrier to grow out is poorly understood. By using a set of soil assays, our previous study has identified the ethylene signaling pathway as a key soil pressure sensory and response system that modulates morphogenesis of germinating seedlings under the soil environments [11]. In this process, the master transcription factor EIN3 plays an essential role by activating two separate pathways through PIF3 and ERF1 in the cotyledons and the hypocotyl, respectively, that coordinate the organ-specific soil responses in germinated seedlings [11].
In this study, we report that COP1, a central regulator of etiolated plant growth, is required for seedlings to emerge from the soil. In this pathway, COP1 primarily mediates light rather than mechanical stress signals to modulate EIN3 levels in the soil. The mechanism by which COP1 stabilizes EIN3 is by direct interaction and ubiquitination of EBF1 and EBF2 (EBF1/2), the two F-box proteins that target EIN3 for degradation. Taken together, we propose a model in which COP1 and ethylene are recruited to respond to soil-imposed darkness and mechanical stress effects, respectively (Figure S4B). These two pathways converge on EBF1/2 to cooperatively adjust EIN3 levels, allowing the seedling to rapidly and precisely respond to the changing soil and light environments as it grows towards the surface (Figure S4B). Interestingly, although cop1 mutants respond to both the precursor and inhibitor of ethylene in a similar pattern as WT, the fold-changes of EIN3 protein level in cop1 mutants are greater than in WT. We think this may be caused by the higher levels of EBF1/2 protein in cop1 mutants. As ethylene inhibits EBF1/2's ability to down-regulate EIN3 at both the protein stability and protein activity of EBF1/2 [22-24, 46], the greater range of changes in EBF1/2 by ethylene in cop1 mutants would presumably bring about a stronger effect compared to WT that has lower levels of EBF1/2. Moreover, considering the complex regulation of EBF1/2 at multiple-levels by ethylene [41, 46-48], there might be other points of signaling interaction between ethylene and COP1 upstream EIN3. Nevertheless, our results not only support the long-assumed hypothesis that skotomorphogenesis is an adaptive developmental program evolved to cope with terrestrial survival, but also reveal a genetic pathway, defined by the COP1-EBF1/2-EIN3 module, which enables the plants to coordinate growth and physiological activities according to light fluence changes in the soil during emergence. We have learned from this study that plants can sense the depth of the soil covering and timing of their emergence through combined information on mechanical pressure and on dim light (light fluences) of the soil.
Studying the mechanism of seedling emergence is critical in order to develop method to increase plant survival and crop yield. The key question in seedling emergence is how plants sense and integrate these environmental factors to precisely adjust their developmental patterns. During soil emergence, one of the first responses is the production of the gaseous phytohormone ethylene, which is stimulated upon mechanical impedance [6, 7, 11]. Light is another major factor in regulating seedling penetration of the soil. Germinated seedlings grown in the dark adopt a distinct morphogenetic program termed skotomorphogenesis, which provides an adaptive advantage to grow under the soil where light is low or absent [1, 3, 5]. Recently, mechanical stress was also found to activate the biosynthesis of other plant hormones, such as jasmonate and gibberallin acids [49-51]. It will be of interest to explore whether these hormones can also modulate seedling emergence. In addition, growth in waterlogged soil could cause hypoxia, which was recently reported to regulate skotomorphogenesis [52]. Considering our and other recent studies, seedling emergence can serve an effective model to study the basic question in biology of how plants integrate various signals to adapt to changing environments.
Our findings also offer novel insights into the mechanism of how key transcription factors are stabilized. In light-regulated seedling morphogenesis, COP1 functions as a central repressor by degrading positive transcription factors, such as HY5, as well as stabilizing negative transcription factors, like PIF3 [5, 31]. Although extensive studies have revealed that COP1 directly degrades HY5 and other positive regulators by the ubiquitin-proteasome system, it remains poorly understood how COP1 positively regulates the accumulation of key negative transcription factors. EIN3 was previously shown to promote hypocotyl elongation and repress cotyledon development in the light, which negatively regulates seedling photomorphogenesis [21, 42, 53]. In addition, EIN3 cooperates with PIF1 in regulating seedling greening [54, 55]. The scheme that COP1 promotes accumulation of EIN3 by targeting its negative regulators EBF1/2 could be applicable to other COP1-promoting factors. Besides ethylene, SCF F-box proteins-mediated protein degradation has emerged as a universal mechanism in response to various plant hormones, including auxin, abscisic acid, gibberellins and jasmonate [56, 57]. Because seedling morphogenesis is coordinately regulated by environmental and various hormonal signals, it is appealing to hypothesize that the COP1 E3 ligase-mediated degradation of other E3 ligases might be a common mechanism by which light signaling is integrated with other hormonal pathways.
Experimental Procedures
Plant material and growth condition
The wild type (WT) Arabidopsis seedlings used in this study is Columbia-0 ecotype. EIN3ox, ein3eil1, cop1-4, cop1-6, 35S:COP1/cop1-6, 5XEBS-GUS/Col-0, EIN3p-EIN3-Luciferase/ein3eil1, EIN3-GFP/ein3eil1, EBF1-TAP/Col-0, EBF2-TAP/Col-0 were previously reported [11, 39, 46, 54, 58]. Multiple mutants were generated by crossing and homozygous lines were genotyped. Plants were grown under long-day photoperiod white light at 22 °C.
Soil experiment
The soil experiment was performed as previously described [11]. Surface-sterilized seeds were plated on 1/2 MS medium (2.2 g/L MS salts, 0.5% sucrose, pH 5.7, and 8g/L agar). The silicon dioxide (SiO2) powder (Sigma), soil (50-70 mesh particle size sand, Sigma), or 120-200 mesh particle size sand was autoclaved and then evenly spread onto the medium. After stratification in 4 °C for 4 days, the plates were transferred to white light for 6h to induce germination. The plates were then incubated under white light or in darkness for the indicated time. At least 100 seeds were used for each experimental treatment and three biological replicates were used for statistical analysis.
Supplementary Material
Acknowledgments
We greatly appreciate Dr. Cynthia D. Nezames from Yale University for critically reading and commenting on the manuscript; Dr. Hongwei Guo from Peking University for generously providing us the EBF1/2-TAP and EBF1/2-GFP transgenic plant seeds, and valuable suggestions on the study. We also want to thank Lei Wang and Dr. Yanli Li from Peking University for experimental assistance; Xing Wen and Xing Zhang from Peking University for advices on the protein purification experiments. This work was supported by grants from National Natural Science Foundation (31330048), National Basic Research Program of China (973 Program) (2012CB910900), and US NIH grant (GM047850) to X.W.D.; National Science Foundation of China (31570188), 985 project and the Recruitment Program of Global Youth Experts of China to S.Z.; H.S. was supported by China Postdoctoral Science Foundation Grants (2015T80014).
Footnotes
Supplemental Information: Supplemental Information includes Supplemental Experimental Procedures and four figures and can be found with this article online.
Author contributions: S.Z., X.W.D. and H.S. designed research; H.S., R.L., C.X. and X.S. performed research; S.Z., X.W.D., H.S. and N.W. analyzed data; and S.Z., H.S., N.W. and X.W.D. wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.McNellis TW, Deng XW. Light control of seedling morphogenetic pattern. Plant Cell. 1995;7:1749–1761. doi: 10.1105/tpc.7.11.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huq E, Al-Sady B, Hudson M, Kim C, Apel K, Quail PH. Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science. 2004;305:1937–1941. doi: 10.1126/science.1099728. [DOI] [PubMed] [Google Scholar]
- 3.Von Arnim A, Deng XW. Light Control of Seedling Development. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:215–243. doi: 10.1146/annurev.arplant.47.1.215. [DOI] [PubMed] [Google Scholar]
- 4.Chen M, Chory J, Fankhauser C. Light signal transduction in higher plants. Annual Review of Genetics. 2004;38:87–117. doi: 10.1146/annurev.genet.38.072902.092259. [DOI] [PubMed] [Google Scholar]
- 5.Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH. Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol. 2008;18:1815–1823. doi: 10.1016/j.cub.2008.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goeschl JD, Rappaport L, Pratt HK. Ethylene as a factor regulating the growth of pea epicotyls subjected to physical stress. Plant Physiol. 1966;41:877–884. doi: 10.1104/pp.41.5.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kays SJ, Nicklow CW, Simons DH. Ethylene in Relation to Response of Roots to Physical Impedance. Plant and Soil. 1974;40:565–571. [Google Scholar]
- 8.Morgan PW, Drew MC. Ethylene and plant responses to stress. Physiologia Plantarum. 1997;100:620–630. [Google Scholar]
- 9.Ecker JR. The ethylene signal transduction pathway in plants. Science. 1995;268:667–675. doi: 10.1126/science.7732375. [DOI] [PubMed] [Google Scholar]
- 10.Harpham NVJ, Berry AW, Knee EM, Rovedahoyos G, Raskin I, Sanders IO, Smith AR, Wood CK, Hall MA. The Effect of Ethylene on the Growth and Development of Wild-Type and Mutant Arabidopsis-Thaliana (L) Heynh. Annals of Botany. 1991;68:55–61. [Google Scholar]
- 11.Zhong S, Shi H, Xue C, Wei N, Guo H, Deng XW. Ethylene-orchestrated circuitry coordinates a seedling's response to soil cover and etiolated growth. Proc Natl Acad Sci USA. 2014;111:3913–3920. doi: 10.1073/pnas.1402491111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo H, Ecker JR. The ethylene signaling pathway: new insights. Curr Opin Plant Biol. 2004;7:40–49. doi: 10.1016/j.pbi.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 13.Yoo SD, Cho YH, Sheen J. Emerging connections in the ethylene signaling network. Trends in Plant Science. 2009;14:270–279. doi: 10.1016/j.tplants.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kieber JJ. The ethylene response pathway in arabidopsis. Annual Review of Plant Physiology and Plant Molecular Biology. 1997;48:277–296. doi: 10.1146/annurev.arplant.48.1.277. [DOI] [PubMed] [Google Scholar]
- 15.Solano R, Ecker JR. Ethylene gas: perception, signaling and response. Curr Opin Plant Biol. 1998;1:393–398. doi: 10.1016/s1369-5266(98)80262-8. [DOI] [PubMed] [Google Scholar]
- 16.Chang C, Shockey JA. The ethylene-response pathway: signal perception to gene regulation. Curr Opin Plant Biol. 1999;2:352–358. doi: 10.1016/s1369-5266(99)00004-7. [DOI] [PubMed] [Google Scholar]
- 17.Chen YF, Etheridge N, Schaller GE. Ethylene signal transduction. Annals of Botany. 2005;95:901–915. doi: 10.1093/aob/mci100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo H, Ecker JR. Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell. 2003;115:667–677. doi: 10.1016/s0092-8674(03)00969-3. [DOI] [PubMed] [Google Scholar]
- 19.Potuschak T, Lechner E, Parmentier Y, Yanagisawa S, Grava S, Koncz C, Genschik P. EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell. 2003;115:679–689. doi: 10.1016/s0092-8674(03)00968-1. [DOI] [PubMed] [Google Scholar]
- 20.Gagne JM, Smalle J, Gingerich DJ, Walker JM, Yoo SD, Yanagisawa S, Vierstra RD. Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc Natl Acad Sci USA. 2004;101:6803–6808. doi: 10.1073/pnas.0401698101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science. 1999;284:2148–2152. doi: 10.1126/science.284.5423.2148. [DOI] [PubMed] [Google Scholar]
- 22.Qiao H, Shen Z, Huang SS, Schmitz RJ, Urich MA, Briggs SP, Ecker JR. Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science. 2012;338:390–393. doi: 10.1126/science.1225974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ju C, Yoon GM, Shemansky JM, Lin DY, Ying ZI, Chang J, Garrett WM, Kessenbrock M, Groth G, Tucker ML, et al. CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc Natl Acad Sci USA. 2012;109:19486–19491. doi: 10.1073/pnas.1214848109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wen X, Zhang C, Ji Y, Zhao Q, He W, An F, Jiang L, Guo H. Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus. Cell Res. 2012;22:1613–1616. doi: 10.1038/cr.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng XW, Caspar T, Quail PH. cop1: a regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev. 1991;5:1172–1182. doi: 10.1101/gad.5.7.1172. [DOI] [PubMed] [Google Scholar]
- 26.Deng XW, Matsui M, Wei N, Wagner D, Chu AM, Feldmann KA, Quail PH. COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell. 1992;71:791–801. doi: 10.1016/0092-8674(92)90555-q. [DOI] [PubMed] [Google Scholar]
- 27.Osterlund MT, Ang LH, Deng XW. The role of COP1 in repression of Arabidopsis photomorphogenic development. Trends Cell Biol. 1999;9:113–118. doi: 10.1016/s0962-8924(99)01499-3. [DOI] [PubMed] [Google Scholar]
- 28.Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell. 1998;1:213–222. doi: 10.1016/s1097-2765(00)80022-2. [DOI] [PubMed] [Google Scholar]
- 29.Osterlund MT, Hardtke CS, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405:462–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- 30.Yi C, Deng XW. COP1 - from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol. 2005;15:618–625. doi: 10.1016/j.tcb.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 31.Lau OS, Deng XW. The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 2012;17:584–593. doi: 10.1016/j.tplants.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 32.Seo HS, Yang JY, Ishikawa M, Bolle C, Ballesteros ML, Chua NH. LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature. 2003;423:995–999. doi: 10.1038/nature01696. [DOI] [PubMed] [Google Scholar]
- 33.Duek PD, Elmer MV, van Oosten VR, Fankhauser C. The degradation of HFR1, a putative bHLH class transcription factor involved in light signaling, is regulated by phosphorylation and requires COP1. Curr Biol. 2004;14:2296–2301. doi: 10.1016/j.cub.2004.12.026. [DOI] [PubMed] [Google Scholar]
- 34.Yang J, Lin R, Sullivan J, Hoecker U, Liu B, Xu L, Deng XW, Wang H. Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell. 2005;17:804–821. doi: 10.1105/tpc.104.030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jang IC, Yang JY, Seo HS, Chua NH. HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev. 2005;19:593–602. doi: 10.1101/gad.1247205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bauer D, Viczian A, Kircher S, Nobis T, Nitschke R, Kunkel T, Panigrahi KCS, Adam E, Fejes E, Schafer E, et al. Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell. 2004;16:1433–1445. doi: 10.1105/tpc.021568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Migliorini D, Bogaerts S, Defever D, Vyas R, Denecker G, Radaelli E, Zwolinska A, Depaepe V, Hochepied T, Skarnes WC, et al. Cop1 constitutively regulates c-Jun protein stability and functions as a tumor suppressor in mice. J Clin Invest. 2011;121:1329–1343. doi: 10.1172/JCI45784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vitari AC, Leong KG, Newton K, Yee C, O'Rourke K, Liu J, Phu L, Vij R, Ferrando R, Couto SS, et al. COP1 is a tumour suppressor that causes degradation of ETS transcription factors. Nature. 2011;474:403–406. doi: 10.1038/nature10005. [DOI] [PubMed] [Google Scholar]
- 39.Stepanova AN, Hoyt JM, Hamilton AA, Alonso JM. A Link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell. 2005;17:2230–2242. doi: 10.1105/tpc.105.033365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Solano R, Stepanova A, Chao QM, Ecker JR. Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 1998;12:3703–3714. doi: 10.1101/gad.12.23.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Konishi M, Yanagisawa S. Ethylene signaling in Arabidopsis involves feedback regulation via the elaborate control of EBF2 expression by EIN3. Plant J. 2008;55:821–831. doi: 10.1111/j.1365-313X.2008.03551.x. [DOI] [PubMed] [Google Scholar]
- 42.Zhong S, Shi H, Xue C, Wang L, Xi Y, Li J, Quail PH, Deng XW, Guo H. A molecular framework of light-controlled phytohormone action in Arabidopsis. Curr Biol. 2012;22:1530–1535. doi: 10.1016/j.cub.2012.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woolley JT, Stoller EW. Light Penetration and Light-induced Seed Germination in Soil. Plant Physiol. 1978;61:597–600. doi: 10.1104/pp.61.4.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tester M, Morris C. The Penetration of Light through Soil. Plant Cell and Environment. 1987;10:281–286. [Google Scholar]
- 45.Mandoli DF, Ford GA, Waldron LJ, Nemson JA, Briggs WR. Some Spectral Properties of Several Soil Types - Implications for Photomorphogenesis. Plant Cell and Environment. 1990;13:287–294. [Google Scholar]
- 46.An F, Zhao Q, Ji Y, Li W, Jiang Z, Yu X, Zhang C, Han Y, He W, Liu Y, et al. Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell. 2010;22:2384–2401. doi: 10.1105/tpc.110.076588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Merchante C, Brumos J, Yun J, Hu Q, Spencer KR, Enriquez P, Binder BM, Heber S, Stepanova AN, Alonso JM. Gene-Specific Translation Regulation Mediated by the Hormone-Signaling Molecule EIN2. Cell. 2015;163:684–697. doi: 10.1016/j.cell.2015.09.036. [DOI] [PubMed] [Google Scholar]
- 48.Li W, Ma M, Feng Y, Li H, Wang Y, Ma Y, Li M, An F, Guo H. EIN2-Directed Translational Regulation of Ethylene Signaling in Arabidopsis. Cell. 2015;163:670–683. doi: 10.1016/j.cell.2015.09.037. [DOI] [PubMed] [Google Scholar]
- 49.Braam J. In touch: plant responses to mechanical stimuli. New Phytol. 2005;165:373–389. doi: 10.1111/j.1469-8137.2004.01263.x. [DOI] [PubMed] [Google Scholar]
- 50.Chehab EW, Yao C, Henderson Z, Kim S, Braam J. Arabidopsis touch-induced morphogenesis is jasmonate mediated and protects against pests. Curr Biol. 2012;22:701–706. doi: 10.1016/j.cub.2012.02.061. [DOI] [PubMed] [Google Scholar]
- 51.Lange MJP, Lange T. Touch-induced changes in Arabidopsis morphology dependent on gibberellin breakdown. Nature Plants. 2015;1:14025. doi: 10.1038/nplants.2014.25. [DOI] [PubMed] [Google Scholar]
- 52.Abbas M, Berckhan S, Rooney DJ, Gibbs DJ, Vicente Conde J, Sousa Correia C, Bassel GW, Marin-de la Rosa N, Leon J, Alabadi D, et al. Oxygen sensing coordinates photomorphogenesis to facilitate seedling survival. Curr Biol. 2015;25:1483–1488. doi: 10.1016/j.cub.2015.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smalle J, Haegman M, Kurepa J, VanMontagu M, VanderStraeten D. Ethylene can stimulate Arabidopsis hypocotyl elongation in the light. Proc Natl Acad Sci USA. 1997;94:2756–2761. doi: 10.1073/pnas.94.6.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhong S, Zhao M, Shi T, Shi H, An F, Zhao Q, Guo H. EIN3/EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. Proc Natl Acad Sci USA. 2009;106:21431–21436. doi: 10.1073/pnas.0907670106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhong S, Shi H, Xi Y, Guo H. Ethylene is crucial for cotyledon greening and seedling survival during de-etiolation. Plant Signal Behav. 2010;5:739–42. doi: 10.4161/psb.5.6.11698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dreher K, Callis J. Ubiquitin, hormones and biotic stress in plants. Annals of Botany. 2007;99:787–822. doi: 10.1093/aob/mcl255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smalle J, Vierstra RD. The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol. 2004;55:555–590. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- 58.Rubio V, Shen Y, Saijo Y, Liu Y, Gusmaroli G, Dinesh-Kumar SP, Deng XW. An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J. 2005;41:767–778. doi: 10.1111/j.1365-313X.2004.02328.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


