Abstract
The loss-of-function mutations of serine protease inhibitor, Kazal type 1 (SPINK1) gene are associated with human chronic pancreatitis, but the underlying mechanisms remain unknown. We previously reported that mice lacking Spink3, the murine homologue of human SPINK1, die perinatally due to massive pancreatic acinar cell death, precluding investigation of the effects of SPINK1 deficiency. To circumvent perinatal lethality, we have developed a novel method to integrate human SPINK1 gene on the X chromosome using Cre-loxP technology and thus generated transgenic mice termed “X-SPINK1“. Consistent with the fact that one of the two X chromosomes is randomly inactivated, X-SPINK1 mice exhibit mosaic pattern of SPINK1 expression. Crossing of X-SPINK1 mice with Spink3+/− mice rescued perinatal lethality, but the resulting Spink3−/−;XXSPINK1 mice developed spontaneous pancreatitis characterized by chronic inflammation and fibrosis. The results show that mice lacking a gene essential for cell survival can be rescued by expressing this gene on the X chromosome. The Spink3−/−;XXSPINK1 mice, in which this method has been applied to partially restore SPINK1 function, present a novel genetic model of chronic pancreatitis.
Trypsin is a major serine protease produced in pancreatic acinar cells as inactive zymogen (trypsinogen). In physiological conditions, trypsinogen is secreted by the acinar cells and is cleaved/activated in the duodenum by enterokinase, resulting in generation of trypsin1,2. Human serine protease inhibitor, Kazal type 1 (SPINK1) and its murine homologue Spink3 play a critical role in suppression of aberrant, intra-acinar/intrapancreatic activation of trypsinogen, which is considered a key mechanism preventing the development of pancreatitis3,4. Consistent with this concept, loss-of-function mutations of SPINK1 gene are associated with various forms of human chronic pancreatitis; however, the mechanisms through which SPINK1 mutations predispose to pancreatitis remain elusive5,6. We have previously reported3 that Spink3−/− mice spontaneously develop severe pancreatic damage and die within two weeks after birth. The histopathological changes start gradually at embryonic day (E) 16.5 and are restricted to pancreatic acinar, but not ductal or islet, cells. The cytoplasm of acinar cells of Spink3−/− mice is filled with numerous autophagic vacuoles3, suggesting that Spink3 deletion interferes with autophagy, a key cellular, lysosome-driven process that degrades and recycles damaged or unneeded organelles, long-lived proteins, and lipids7. The aberrant autophagy could trigger acinar cell death in Spink3−/− mice (it is, however, worth noting that these cells do not display chromatin condensation, a hallmark of apoptosis)3. The early death of Spink3−/− mice precludes investigation into the mechanisms of long-term effects of SPINK1 deficiency.
X-chromosome inactivation is a process by which one of the two X chromosomes in female mammals is randomly inactivated by packaging into transcriptionally inactive heterochromatin8. Once the X chromosome is inactivated, it will remain inactive throughout the lifetime of the cell. During screening of a gene trap library, we found that one ES cell line (B210) possessed a trap vector on the X chromosome. By utilizing X-chromosome inactivation and the B210 ES cells, we here present a novel method to efficiently integrate a target gene on the X chromosome, resulting in a mosaic pattern of the target protein expression. This strategy enabled us to rescue perinatal lethality of Spink3−/− mice. The resulting Spink3−/−;XXSPINK1 mice developed spontaneous pancreatitis, thus representing a novel genetic model for this disease.
Results
Integration of SPINK1 gene at a locus on X chromosome by using Cre-loxP technology results in a mosaic pattern of its expression
Gene-trap mutagenesis is a technique that randomly generates loss-of-function mutations in many genes9. During gene-trap mutagenesis screening, we found that in one ES cell line (designated B210 ES cells) the integrated trap vector inactivated Diaphanous homolog 2 (Diap2) gene on the X chromosome10 (Fig. S1a). Diap2, also known as murine Dia3, is one of three members of the Diap family and considered to play a role in de novo actin filament formation11. Subsequent plasmid rescue and PCR analysis revealed that the trap vector was integrated into intron 23 of the Diap2 gene at 50 kbs downstream of exon 23 and 130 kbs upstream of exon 27, resulting in deletion of exons 24 to 26 (Fig. S1a,b). We next investigated whether the trap vector integration affected the expression of Diap2 protein in B210 ES cells. While Diap2 protein was detected in wild-type ES cells, Diap2 protein completely disappeared in B210 cells (Fig. S1c). This suggests that truncated mRNA of Diap2 is unstable, causing complete depletion of Diap2 protein. Taken into account that Diap2 (that is, Dia3)-deficient mice develop without abnormalities and are fertile11, we reckoned that integration and expression of a target gene in the Diap2 locus on X chromosome might be feasible by using Cre-loxP technology; and further, that this approach would allow us to express the target gene in a mosaic pattern due to random inactivation of one of the two X chromosomes in females.
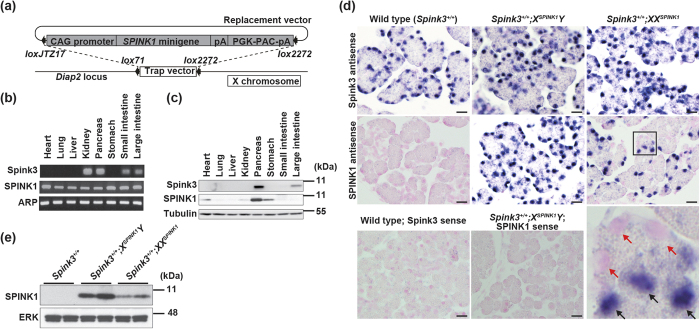
To this end, we generated a replacement vector containing CAG promoter-human SPINK1 minigene12 flanked by two mutated loxP sites, and co-transfected this vector along with a Cre recombinase expression vector13 into B210 ES cells (Fig. 1a). We obtained 11 ES cell lines harboring SPINK1 minigene on X chromosome with high frequency, and used these ES cells to generate mice termed “X-SPINK1”. Male and female X-SPINK1 mice were healthy, fertile, and did not show any abnormalities. These knock-in mice are henceforth referred to as, respectively, XSPINK1Y (male), XSPINK1XSPINK1 (female), and XXSPINK1 (female) mice. We first verified the expression of SPINK1 mRNA and protein in XSPINK1Y mice by RT-PCR and immunoblot (IB) analysis. Because SPINK1 expression in these mice is driven by the ubiquitous CAG promoter, SPINK1 mRNA was ubiquitously expressed in various tissues we examined (Fig. 1b). Interestingly, SPINK1 protein expression was restricted to pancreas and, to a much lesser extent, stomach and heart (Fig. 1c). The endogenous Spink3 mRNA expression is restricted to kidney, pancreas, small and large intestines (Fig. 1b), whereas Spink3 protein is predominantly expressed in the pancreas (and to a much lesser extent, large intestine; Fig. 1c).
Figure 1. Generation of human SPINK1 X-chromosome knock-in (“X-SPINK1”) mice.
(a) A replacement vector containing SPINK1 minigene under the CAG promoter was introduced into Diap2 locus on the X chromosome by Cre-loxP technology. (b) RT-PCR analysis of Spink3 and SPINK1 mRNAs in various tissues of X-SPINK1 mice at 8 weeks. Acidic ribosomal phosphoprotein P0 (ARP), a “housekeeping” gene control. (c) The levels of Spink3 and SPINK1 proteins in various tissues of X-SPINK1 mice at 8 weeks (immunoblot). Representative of two independent experiments. (d) ISH analysis of SPINK1 mRNA expression in pancreas of mice of the indicated genotype at P0.5. Pancreatic tissue sections were hybridized with Spink3 (upper panels) or SPINK1 (middle panels) antisense riboprobes (blue stain). Nuclei stained with Nuclear Fast Red have a pink appearance. The bottom right panel is an enlarged image of the boxed area in the panel above. Black and red arrows indicate acinar cells with and without SPINK1 expression, respectively. The bottom left and center panels show ISH background control using SPINK1 and Spink3 sense riboprobes. Scale bars, 20 μm. Representative of two independent experiments. (e) Pancreas homogenates from mice of the indicated genotype at 8 weeks were analyzed by immunoblot analysis. In this and other figures, ERK or tubulin serve as loading control; each lane represents an individual animal; and the numbers to the right are protein molecular mass markers in kDa.
We next analyzed the expression of SPINK1 mRNA in pancreas of X-SPINK1 mice at 0.5 days after birth (P0.5) by in situ hybridization (ISH; Fig. 1d). As expected, all pancreatic acinar cells expressed SPINK1 mRNA in XSPINK1Y mice (the wild type Spink3+/+ mice served as negative control). Notably, in XXSPINK1 mice approximately half of acinar cells expressed SPINK1 mRNA (Fig. 1d), consistent with the mosaic pattern of SPINK1 mRNA expression. The endogenous Spink3 was expressed in all pancreatic acinar cells in XSPINK1Y and XXSPINK1 mice, same as in Spink3+/+ mice (Fig. 1d). The sense Spink3 or SPINK1 riboprobes were used as a background ISH control and did not show significant staining in any of the mouse strains examined. Consistent with mRNA expression, the level of SPINK1 protein in pancreas of XXSPINK1 mice was one-third to one-half of those in XSPINK1Y mice (Fig. 1e).
The data in Fig. 1 indicate that integration of SPINK1 gene on one of the two X chromosomes results in a mosaic pattern of SPINK1 expression through X-chromosome inactivation.
Crossing Spink3 deficient mice with X-SPINK1 rescues the resultant Spink3 −/− ;XX SPINK1 mice from perinatal lethality
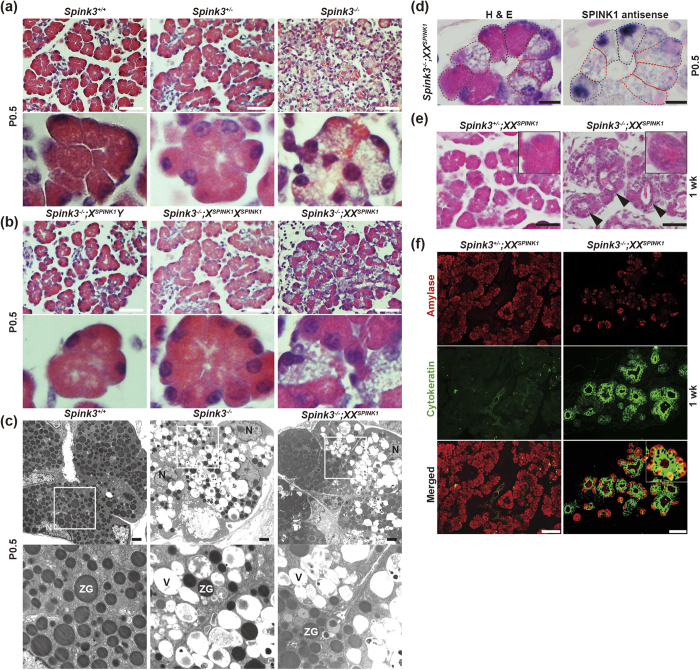
Spink3−/− mice die perinatally3, making it impossible to investigate long-term effects of Spink3 deficiency. To circumvent this problem, we crossed Spink3+/− mice with X-SPINK1 mice. As we previously reported3, pancreatic acinar cells of Spink3−/− mice exhibited prominent vacuolization at P0.5, which is not seen in pancreas of Spink3+/− mice (Fig. 2a). The acinar cell vacuolization was prevented in Spink3−/−;XSPINK1Y and Spink3−/−;XSPINK1XSPINK1 mice (Fig. 2b), suggesting that ectopic expression of SPINK1 gene completely compensated for a defect caused by Spink3 deficiency. Notably, in Spink3−/−;XXSPINK1 mice at P0.5 approximately half of all acinar cells exhibited vacuolization, whereas the other half appeared normal (Fig. 2b). We further used transmission electron microscopy to examine acinar cell morphology in Spink3−/− and Spink3−/−;XXSPINK1 mice at P0.5 in greater detail. The cytoplasmic vacuolization was prominent in all acinar cells in Spink3−/− pancreas, whereas in the pancreas of Spink3−/−;XXSPINK1 mice approximately one-half of acinar cells exhibited vacuolization and the rest appeared normal (Fig. 2c). As we previously reported3,14,15, the fact that large numbers of these vacuoles contain cytoplasmic constituents, in particular organelle remnants, indicates their autophagic nature. Further, many vacuoles contain poorly/incompletely degraded cargo, suggesting a defect in later stages of the autophagy process.
Figure 2. Crossing Spink3 deficient mice with X-SPINK1 rescues the resultant Spink3−/−;XXSPINK1 mice from perinatal lethality.
(a,b) Hematoxylin and eosin (H&E)-stained pancreatic tissue sections from mice of the indicated genotype at P0.5. The lower rows in (a,b) show enlarged images of the upper panels. Scale bars, 50 μm. (c) Electron micrographs of pancreata of mice of the indicated genotype at P0.5. The lower row shows enlarged images of the areas designated by white boxes in the upper panels. ZG, zymogen granule; V, vacuole; N, nucleus. Scale bars, 2 μm. (d) Adjacent pancreatic tissue sections from Spink3−/−;XXSPINK1 at P0.5 were analyzed by H&E staining and ISH. Acinar cells expressing SPINK1 are delineated by black dashed lines, and those with no SPINK1 mRNA expression, by red dashed lines. Scale bars, 10 μm. (e) H&E staining of pancreatic tissue sections from mice of the indicated genotype at 1 week. Arrowheads indicate tubular structures. Insets show 2.5x-enlarged areas. Scale bars, 50 μm. (f) Cells positive for both amylase and pan-cytokeratin in pancreas of Spink3−/−;XXSPINK1 mice at 1 week. Pancreatic tissue sections from mice of the indicated genotype were immunostained using anti-amylase (red) or anti-pan-cytokeratin (green) antibodies. Scale bars, 50 μm.
Of note, the histologically normal (i.e., without vacuoles) acinar cells in P0.5 Spink3−/−;XXSPINK1 pancreas expressed SPINK1 mRNA as shown by ISH (Fig. 2d; delineated by black dashed line); in contrast, acinar cells that exhibited massive vacuolization did not express SPINK1 mRNA (Fig. 2d; delineated by red dashed line). Together with our previous findings3, these results indicate that the one-half of acinar cells in Spink3−/−;XXSPINK1 mice that do not express SPINK1 (due to X-chromosome inactivation) develop prominent vacuolization whereas the other half, which do express SPINK1, are devoid of these vacuoles. Surprisingly, acinar cells filled with numerous vacuoles in pancreas of Spink3−/−;XXSPINK1 mice at P0.5 disappeared at one week after birth (Fig. 2e), suggesting that such cells might have been eliminated during this period. How exactly these cells die out remains to be determined; with TUNEL, we did not detect apoptotic acinar cells in either Spink3−/− or Spink3−/−;XXSPINK1 mice (Fig. S2).
Of note, the number of ductal-like cells forming so-called “tubular structures” dramatically increased in pancreas of 1-week-old Spink3−/−;XXSPINK1 mice (Fig. 2e). Moreover, these cells were positive for both amylase and cytokeratins (Fig. 2f), suggesting they derived from SPINK1-positive acinar cells through acinar-ductal metaplasia16.
Spink3 −/− ;XX SPINK1 mice spontaneously develop pancreatitis characterized by chronic inflammation and fibrosis
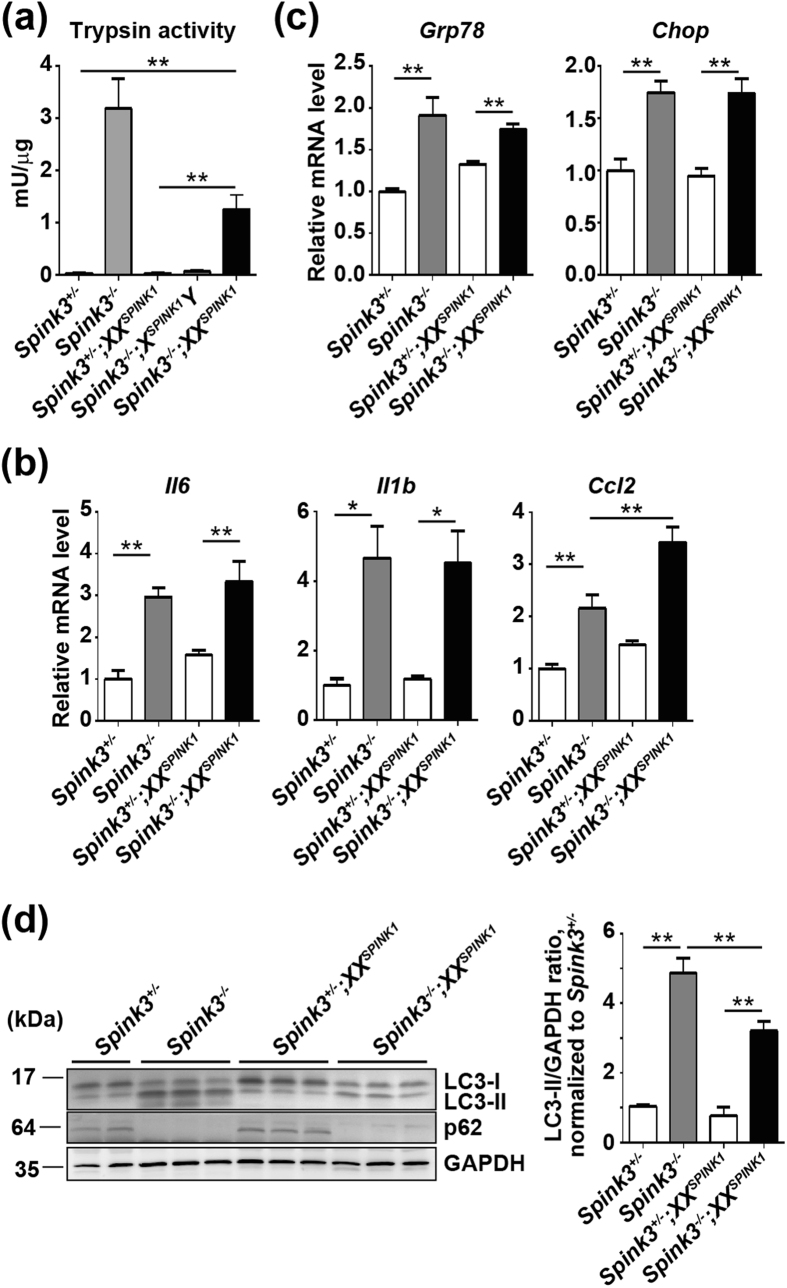
We investigated the development of pancreatic damage, and the underlying mechanisms, in Spink3−/− and Spink3−/−;XXSPINK1 mice. The inappropriate increase in pancreatic trypsin activity is a hallmark of both human and experimental pancreatitis17,18. We find that pancreatic trypsin activity was dramatically upregulated in Spink3−/− mice at P0.5 (compared to wild type or Spink3+/− mice) and that this increase was ~3 times less in Spink3−/−;XXSPINK1 mice (Fig. 3a). Moreover, there was essentially no increase in pancreatic trypsin activity in Spink3−/−;XSPINK1Y mice (Fig. 3a), indicating that human SPINK1 can compensate for the loss of trypsin inhibition in Spink3−/− mice.
Figure 3. Pancreas damage in Spink3−/−;XXSPINK1 mice at birth.
(a) Trypsin activity in pancreas of mice of the indicated genotype at P0.5 was measured by a fluorogenic enzymatic assay (see Methods). Values are means ± SEM (n = 4–6 mice). **P < 0.01. (b,c) mRNA expression of proinflammatory cytokines and chemokines, and ER stress markers in pancreas of mice of the indicated genotype was analyzed by qPCR. Values are means ± SEM (n = 4 mice). *P < 0.05; **P < 0.01. (d) Autophagy markers LC3 and p62/SQSTM1 were analyzed by immunoblotting in pancreas of mice of the indicated genotype. The LC3-II/LC3-I band intensity ratio was measured by densitometry and normalized to that in Spink3+/− pancreas. Values are means ± SEM (n = 4–6 mice). **P < 0.01.
Proinflammatory cytokines, such as Il6 and Il1b, and the chemokine Ccl2/Mcp-1 (monocyte chemotactic protein-1) were upregulated in pancreas of Spink3−/− and Spink3−/−;XXSPINK1 mice at P0.5 (Fig. 3b). In addition, the expression of genes associated with ER stress, such as Bip/Grp78 and Chop, was upregulated in pancreas of Spink3−/− and Spink3−/−;XXSPINK1 mice compared to Spink3+/− or Spink3+/−;XXSPINK1 mice (Fig. 3c). In these (and other) measurements we used Spink3+/−;XXSPINK1and Spink3+/− mice as controls for, respectively, Spink3−/−;XXSPINK1 and Spink3−/− to analyze the effects of mosaic expression of SPINK1 in rescuing Spink3−/− mice from perinatal lethality and the development of pancreatic injury in Spink3−/−;XXSPINK1 mice; and because Spink3+/− mice have normal phenotype. Other mouse strains which can also serve as controls, such as Spink3−/−;XSPINK1Y or Spink3−/−; XSPINK1XSPINK1, express SPINK1 in all cells; as noted, these mice do not show any histological or other abnormalities (Figs 2b and 3a; and data below).
We next measured the changes in autophagy markers in pancreas of Spink3−/−;XXSPINK1 mice. The conversion of microtubule-associated protein 1 light chain 3 (LC3) from the cytosolic LC3-I to the membrane LC3-II form that uniquely localizes to autophagic vacuoles7 was increased in pancreas of Spink3−/− and (to a lesser extent) Spink3−/−;XXSPINK1 mice at P0.5, compared to that in Spink3+/− and Spink3+/−;XXSPINK1 mice (Fig. 3d). These data are consistent with acinar cell vacuolization we find in Spink3−/− and Spink3−/−;XXSPINK1 mice (Fig. 2b–d). At the same time, p62/SQSTM1, a protein which both mediates autophagy and is degraded by autophagy19, disappeared in pancreas of Spink3−/− and Spink3−/−;XXSPINK1 mice (Fig. 3d). Of note, a recent study20 has shown that p62 accumulates in pancreas of mice with pancreas-specific knockout of the kinase IKKα (IkkaΔpan mice), mediating the development of spontaneous pancreatitis in this genetic model. Our data in Fig. 3d indicate that, in contrast, p62 does not mediate the development of pancreatitis in Spink3−/−;XXSPINK1 mice.
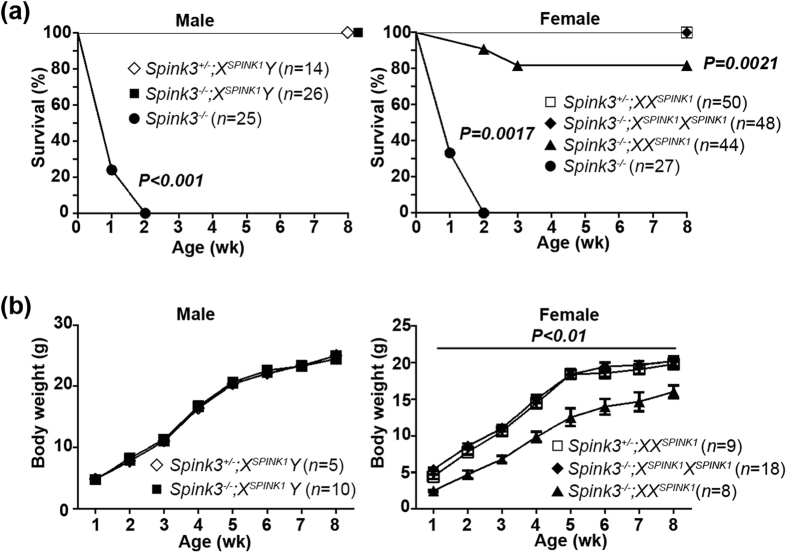
Although Spink3−/− mice die within 2 weeks after birth3, all Spink3−/−;XSPINK1Y (male) and Spink3−/−;XSPINK1XSPINK1 (female) mice grew normally and were healthy at 8 weeks (Fig. 4a,b). Approximately 80% of Spink3−/−;XXSPINK1 (female) mice survived and exhibited mildly, but significantly, retarded body weight gain compared to Spink3+/− and Spink3−/−;XSPINK1XSPINK1 mice (Fig. 4a,b). Macroscopically, the pancreas of Spink3−/−;XXSPINK1 mice at 8 weeks was atrophic compared to that of Spink3+/−;XXSPINK1 mice (Fig. 5a). Spink3−/−;XXSPINK1 mice gradually developed chronic pancreatitis, manifested by loss of acinar cells, intralobular fibrosis, and dilatation of interlobular ducts with protein plugs (Figs 5b and S3, and Supplementary Table 1). In contrast to Spink3−/−;XXSPINK1 mice, there were no histopathological alterations in pancreas of Spink3−/−;XSPINK1Y and Spink3−/−;XSPINK1X mice even at later age (13 weeks; Fig. S4).
Figure 4. Spink3−/−;XXSPINK1 mice exhibit mild growth retardation.
(a) Survival curves of male and female mice of the indicated genotype. P value, as compared to the wild type (Spink3+/+), was calculated by the log rank test. (b) Body weight gain of mice of the indicated genotype. Values are means ± SEM. *P < 0.05.
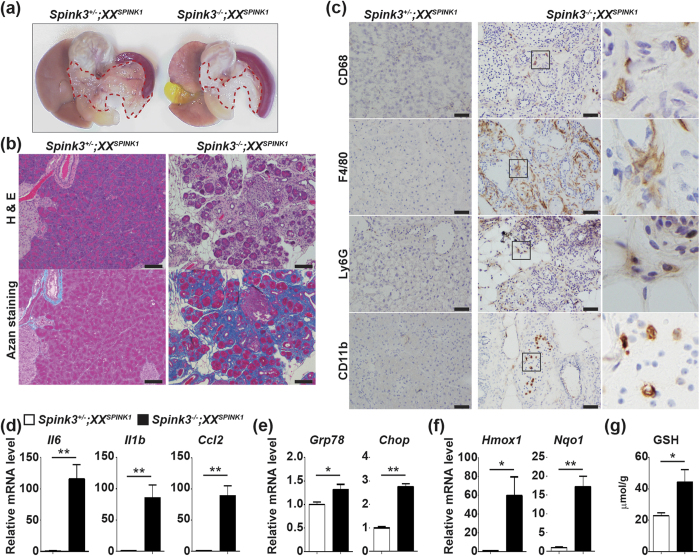
Figure 5. Spink3−/−;XXSPINK1 mice spontaneously develop chronic pancreatitis.
(a) Macroscopic analysis of Spink3+/−;XXSPINK1 and Spink3−/−;XXSPINK1 mice at 8 weeks. Dashed lines delineate the pancreas. (b) Pancreatic tissue sections from mice of the indicated genotype at 8 weeks were stained with H&E (upper panels) or Azan [for collagen (dark blue); lower panels]. Scale bars, 100 μm. (c) Pancreatic cryosections from mice of the indicated genotype were stained for immune cell markers CD68, F4/80, Ly-6G, or CD11b (brown). Right-column panels show enlarged images of the boxed areas in the corresponding middle-column panels. Scale bars, 50 μm. (d–f) qPCR analysis of gene expression for (d) proinflammatory cytokines and chemokines, (e) ER and (f) oxidative stress markers. (g) Reduced glutathione (GSH) levels in pancreas of mice of the indicated genotype at 8 weeks. Values are means ± SEM (n = 4–6 mice). *P < 0.05; **P < 0.01.
Large number of inflammatory cells including macrophages, neutrophils, and monocytes infiltrated the pancreas, consistent with dramatic upregulation of proinflammatory cytokines and chemokines (Fig. 5c,d). Moreover, the expression of genes induced by ER stress (Bip/Grp78 and Chop), and those induced by oxidative stress (such as Hmox1 and Nqo1) increased in pancreas of Spink3−/−;XXSPINK1 mice compared to Spink3+/−;XXSPINK1mice (Fig. 5e,f). Of note, the upregulation of all these genes was much greater at 8 weeks (Fig. 5d–f) than at P0.5 (Fig. 3b,c), illustrating progressive development of pancreatitis in Spink3−/−;XXSPINK1 mice. The reduced form of glutathione (GSH) protects cellular components from reactive oxygen species such as free radicals and peroxides. The pancreatic GSH level increased in Spink3−/−;XXSPINK1 mice (Fig. 5g), indicating a protective response.
As stated above (Fig. 4b), body weight of Spink3−/−;XXSPINK1 mice was somewhat lower than that of Spink3+/−;XXSPINK1and Spink3−/−;XSPINK1XSPINK1 mice, suggesting a mild exocrine pancreas dysfunction. We did not observe dramatic changes in blood biochemical parameters of Spink3−/−;XXSPINK1 mice at 8 weeks (Fig. S5), except for 50% decrease in blood triglycerides. Glucose level in blood was not altered; a modest decrease in serum amylase (Fig. S5) may reflect loss of acinar tissue.
Precancerous changes in the pancreas of Spink3 −/−;XX SPINK1 mice
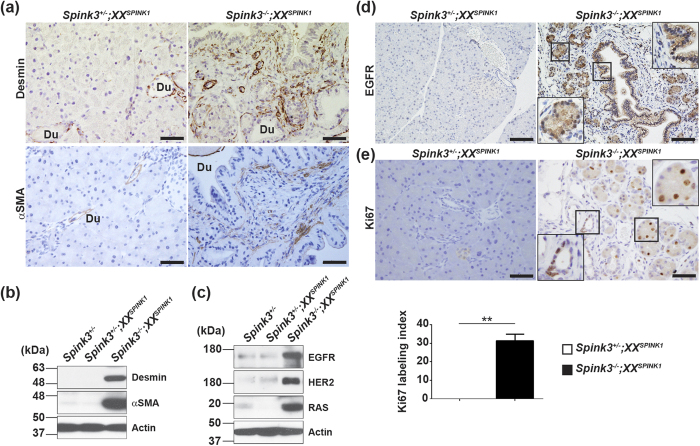
Chronic pancreatitis is associated with a stromal reaction including proliferation and activation of pancreatic stellate cells (PSCs), a type of mesenchymal cells activation of which mediates the development of pancreatic fibrosis21. PSCs are desmin-positive; the number of desmin-positive cells greatly increased in pancreas of Spink3−/−;XXSPINK1 mice compared to Spink3+/−;XXSPINK1 mice (Fig. 6a), and they accumulated around ducts. We further found increased staining for α-smooth muscle actin (αSMA), a marker of activated PSCs (Fig. 6a). IB analysis (Fig. 6b) confirmed the increases in desmin and αSMA. Moreover, the levels of products of proto-oncogenes implicated in the development of pancreatic cancer, such as EGFR, HER2, and RAS, dramatically increased in pancreas of Spink3−/−;XXSPINK1 mice at 8 weeks (Fig. 6c). The expression of EGFR was not only detected in acinar-like cells but also in the epithelial cells of hyperplastic ducts (Fig. 6d). Staining for Ki67, a proliferation marker, also dramatically increased in 8-week-old Spink3−/−;XXSPINK1 mice (Fig. 6e). Together, these data indicate that chronic pancreatitis that develops in Spink3−/−;XXSPINK1 mice induces pre-cancerous changes, suggestive of PanIN lesions’ formation. However, we did not detect the development of pancreatic adenocarcinoma even in 12-month-old Spink3−/−;XXSPINK1 mice (data not shown).
Figure 6. Pre-cancerous changes in pancreas of Spink3−/−;XXSPINK1 mice.
(a) Pancreatic tissue sections from mice of the indicated genotype at 8 weeks were immunostained for desmin or αSMA (brown). Scale bars, 50 μm. Du, duct. (b,c) Up-regulation of (b) stellate cell markers and (c) proto-oncogene products in pancreas of Spink3−/−;XXSPINK1 mice at 8 weeks (immunoblot). (d) Pancreatic tissue sections from mice of the indicated genotype at 8 weeks were immunostained for EGFR (brown). Scale bars, 100 μm. (e) Ki67 staining of pancreatic tissue sections and Ki67 labeling index at 8 weeks. Scale bars, 50 μm. Values are means ± SEM (n = 5 mice). **P < 0.01.
Discussion
In the present study, we have developed a novel method to express a target gene in a mosaic pattern through its’ specific integration onto the X-chromosome by using Cre-loxP technology. Consistent with our hypothesis, female X-SPINK1 mice, in which one of the two copies of X-chromosome is inactivated, express human SPINK1 mRNA in a mosaic pattern. Crossing Spink3 deficient mice with X-SPINK1 rescues the resultant Spink3−/−;XXSPINK1 mice from perinatal lethality, but they develop chronic pancreatitis. Our results indicate that the one- half of acinar cells in pancreas of Spink3−/−;XXSPINK1 mice that do not express SPINK1 (due to X chromosome inactivation) display prominent vacuolization, whereas the other half harboring SPINK1 on the active X chromosome are histologically normal. In general, the method we have developed should be useful to rescue mice lacking an essential survival gene, and to monitor long-term effects of insufficient gene function.
A number of recent studies have indicated the involvement of lysosomal and autophagic dysfunction in the pathogenesis of pancreatitis, and in particular, the development of chronic inflammation15,20,22,23,24,25,26. For example, a recent study20 has shown that pancreas specific deletion of Ikka causes defective autophagy completion, resulting in accumulation of p62 which mediates the ER and oxidative stresses, leading to chronic inflammation and other pancreatitis responses in the IkkaΔpan genetic model. In contrast, we here find that p62 protein is dramatically down-regulated in pancreas of Spink3−/− and Spink3−/−;XXSPINK1 mice (at P0.5) and, therefore, is unlikely to mediate the development of chronic pancreatitis in the “X-SPINK1” genetic model. Interestingly, recent studies (e.g., ref. 26 and our unpublished data) show that p62 level in pancreas can be regulated not only through its autophagic degradation but also through changes in mRNA expression. Taken together, the accumulation of abnormally large autophagic vacuoles (many of which contain poorly degraded cargo), decrease in p62, and increased LC3-II level suggest that autophagy in pancreas of Spink3−/−;XXSPINK1 mice might be upregulated but also impaired. Of note, these two effects on autophagy are not mutually exclusive; in particular, both occur in acute cerulein pancreatitis15,24,27. The characterization of pancreatic autophagy in Spink3−/−;XXSPINK1 mice requires detailed investigation.
Accumulating studies have shown that acute acinar cell injury caused by aberrant activation of trypsin is not sufficient to develop chronic pancreatitis in mice28. Also, we did not find evidence for apoptotic death of acinar cells in pancreas of Spink3−/−;XXSPINK1 mice; it remains to be determined how exactly the cells that do not express SPINK1 are eliminated in these mice. Our findings presented in another study (manuscript in preparation) indicate contribution of RIPK3-dependent necroptosis29 to this process. In addition to loss of acinar tissue, chronic pancreatitis is characterized by persistent inflammation and fibrosis, along with tissue remodeling5. Activated PSCs are considered critical for the development of fibrosis in chronic pancreatitis, by producing extracellular matrix proteins as well as proinflammatory cytokines and chemokines5. Importantly, we found positive staining for αSMA, a PSCs activation marker, indicating that PSCs mediate fibrosis in Spink3−/−;XXSPINK1 mice. Thus, Spink3−/−;XXSPINK1 mice reproduce key responses of human chronic pancreatitis. Further, chronic pancreatitis that develops in Spink3−/−;XXSPINK1 mice is associated with increases in proliferation and pancreatic levels of proteins implicated in the development of pancreatic cancer, such as EGFR, HER2, and RAS.
In conclusion, we have developed a novel method thereby mice lacking a gene essential for cell survival can be rescued by expressing this gene on the X chromosome. We applied this method, utilizing X-chromosome inactivation, to generate Spink3−/−;XXSPINK1 mice in which perinatal lethality is rescued and SPINK1 function is partially restored. These mice develop spontaneous pancreatitis, revealing a critical role of SPINK1 in regulating normal autophagy in the exocrine pancreas. Our results, together with recent findings from our groups as well as others3,14,15,20,24,26,27, indicate that defects in autophagy function lead to pancreatitis.
Methods
Antibodies
The antibodies used in this study were against insulin (Santa Cruz; SC; sc-9168), amylase (SC; sc-12821), Dia3 (SC; sc-10892), glucagon (Dako, A0565), desmin (Dako, M0760), αSMA (Dako, A0851), cytokeratin (Nichirei, 422061), LC3 (Cell Signaling Technology; CST, #2775), p62 (CST, #5114), ERK (CST, #9102), Spink1 (CST, #2744), CD68 (Serotec, MCA1957), CD11b (eBioscience, 11-0112-81), F4/80 (eBioscience, 14-4801-81), Ly-6G (TONBO biosciences, 70-5931), Ki-67 (Novocastra, NCL-Ki67p), EGFR (Proteintech, 18986-1-AP), HER2 (Proteintech, 18299-1-AP), SPINK1 (Abnova, H00006690-M01), Ras (Millipore, #05-516), glyceraldehyde-3-phosphate dehydrogenase (Abcam, ab8245), actin (Sigma, 20-33), and tubulin (Sigma, T5168).
Animal use
Mice were kept under specific-pathogen-free (SPF) conditions with free access to food and water in a 12 hours light/dark cycle. Spink3−/− mice were described previously3. C57BL/6 N mice were purchased from the Clea Japan. All animal experiments were performed with the approval of the Kumamoto University Institutional Animal Care and Use Committee (A27-092). All methods were carried out in accordance with the relevant guidelines of the Kumamoto University, including any relevant details.
Generation of SPINK1 minigene X-chromosome knock-in mice
SPINK1 minigene was described elsewhere12. For the present study, we have generated a replacement vector30 containing loxJTZ17-CAG-promoter (pCAGGS)- SPINK1 minigene-polyA (pA)- phosphoglycerate kinase-1 promoter (PGK)-puromycin resistance gene (PAC)-pA (PGK-PAC-pA)-lox2272. The TT2 ES cell line contains a knocked-out Diap2 allele located on X-chromosome, in which Diap2 was targeted by the PGK-neomycin resistance gene (neo) fragment flanked by the mutant lox sites, lox71 and lox2272 (gene trap vector) (Fig. 1a). For co-electroporation with pCAGGS-Cre (a Cre recombinase gene expression vector13) and replacement vector plasmids, we used 20 μg of each plasmid and 1 × 107 Diap2+/− ES cells. The cells were co-electroporated using a Bio-Rad Gene Pulser, and then cultured for 48 hours in a standard medium supplemented with 2 μg/ml puromycin (Sigma). Selection was maintained for 5 days, and then colonies were picked into 48-well plates and expanded for storage. The puromycin-resistant colonies were analyzed by Southern blotting and by PCR to select ES cell lines showing successful integration of pCAGGS-SPINK1 minigene-pA sequence. Positive clones were aggregated with ICR morula according to the previously described protocol30. Germline transmission was obtained in three mouse lines, resulting in X-SPINK1 mice, which were back-crossed onto C57BL/6 N mice for at least 5 generations.
Reverse transcriptase (RT)-PCR analysis
Total RNA was isolated using the RNeasy Mini Kit (Qiagen). cDNA was synthesized using qPCR RT Kit (Toyobo). For the detection of Ccl2 (Mm00441242), Il6 (Mm00446190), and Il1b (Mm00434228) mRNA, TaqMan Gene Expression Assays on the AB 7500 Real Time PCR System (Applied Biosystems) was used. Other PCR primers are described in the Supplementary Table 2. Densitometric quantification was done using Image J software (http://rsb.info.nih.gov/ij/).
In situ hybridization (ISH)
Pancreases were fixed in 4% paraformaldehyde phosphate buffer solution for 48 hours at room temperature and cut into 4-μm sections. The sections were mounted on glass slides and processed sequentially according to a standard protocol. To prepare the digoxigenin (DIG)-labeled RNA sense (control) and antisense riboprobes (Roche), total RNA was extracted from human or mouse pancreas and then each complementary DNA (cDNA) template was amplified using reverse transcriptase (RT)-PCR. Primers applied in the RT-PCR reactions were as follows: primer hPSTI1 (CGTGGTAAGTGCGGTGCAGT) located in the first exon of SPINK1 gene; primer hPSTI2 (CGCGGTGACCTGATGGGATT) located in the fourth exon of SPINK1 gene, and primer mPsti1 (AGTTCTTCTGGCTTTTGCACCC) located in the first exon of Spink3 gene; primer mPsti25 (TTCAACGAACCCACGTTGCCTT) located in the fourth exon of Spink3 gene. ISH analysis was performed with a Ventana XT System Discovery (Roche). Nuclear Fast Red staining was performed after ISH.
Histological and Immunohistochemical analysis
For histological analysis, tissues were fixed overnight in 15% formalin (Wako), embedded in paraffin, sectioned, and stained using standard procedure for H&E, Azan, or Sirius red. Immunohistochemistry was performed by using the antibodies listed above.
Transmission electron microscopy
Anesthetized mice were fixed by intracardial perfusion with 2% glutaraldehyde and 2% paraformaldehyde in 0.1 M phosphate buffer, pH 7.4. Slices of thus fixed tissues were postfixed with 2% OsO4, dehydrated in ethanol and embedded in Epok 812 (Okenshoji Co.). Ultrathin sections were cut with a ultramicrotome (ultracut N or UC6: Leica), stained with uranyl acetate and lead citrate, and were examined on a Hitachi HT7700 or JEOL JEM-1230 electron microscope.
Immunoblot analysis
Pancreas or other tissues were disrupted in the Multi-Beads Shocker system (Yasui-Kikai), and homogenized in a RIPA buffer [150 mM M NaCl, 50 mM Tris-HCl (pH 7.2), 1% deoxycholic acid, 1% Triton X-100, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail (1:100 dilution; Nacalai tesque)]. The homogenates were subjected to SDS-PAGE and transferred onto Immobilon polyvinylidene difluoride membranes (Millipore). The membranes were blotted using the indicated antibodies, developed with ECL Western Blotting Detection Reagents, and analyzed in a LAS4000 (GE Healthcare Life Sciences). Quantification of the LC3-II to LC3-I ratio was performed with ImageJ software.
Trypsin activity
To measure trypsin activity, mouse pancreas was disrupted using the Multi-Beads Shocker system (Yasui Kikai), and in buffer containing 5 mM 2-Morpholinoethanesulfonic acid (pH 6.5), 1 mM MgSO4, and 250 mM sucrose. Trypsin activity in homogenates was measured fluorimetrically using Boc-Gln-Ala-Arg-MCA (Peptide Institute. Inc.) as a substrate, in the Infinite 200 PRO (Tecan), according to the method of Kawabata et al.31. Trypsin activity in each sample was determined using a standard curve for purified trypsin (T1426, Sigma).
Terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick-end labeling (TUNEL) assay
This was performed by using in situ apoptosis detection kit (Wako).
GSH content measurement
Pancreas were homogenized in 5% methacholic acid buffer. After homogenization, the lysates were centrifuged at 2800 g and the supernatants were collected and used for GSH assays. Determination of GSH was performed with the GSH assay kit (Oxis International) according to the manufacturer’s instructions. To normalize GSH contents per mg protein of liver extracts, we solubilized the pellets in a RIPA buffer and measured protein content with the Bradford assay (Pierce).
Statistical analysis
Data in graphs are expressed as means ± standard error of mean (SEM) from 4 or more experiments per group, and each experiment was performed at least twice. Statistical analysis was performed by using unpaired Student’s t test or one-way analysis of variance (ANOVA) test, as appropriate, with GraphPad Prism 6 (GraphPad Software, Inc.). P < 0.05 was considered to be statistically significant.
Additional Information
How to cite this article: Sakata, K. et al. Novel method to rescue a lethal phenotype through integration of target gene onto the X-chromosome. Sci. Rep. 6, 37200; doi: 10.1038/srep37200 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank Y. Fukuchi and M. Nakata for technical assistance. This work was supported by MEXT (Ministry of Education, Culture, Sports, Science and Technology) grants (#26111516 and 26110003), KAKENHI Scientific Research (B and C) and Challenging Exploratory Research from Japan Society for the Promotion of Science (JSPS), and the Takeda Science Foundation, the Naito Science Foundation, the Uehara Science Foundation, and Novartis Science Foundation. M.O. is especially grateful to the American Pancreatic Association for a mini-sabbatical award that made possible for him to spend several months at the Gukovskaya laboratory in Los Angeles.
Footnotes
Author Contributions Conception and design: H. Nakano, H. Baba, M. Ohmuraya. Development of methodology: K. Sakata, K. Araki, D. Hashimoto, M. Ohmuraya. Acquisition of data (performed experiments, provided animals, provided facilities, etc.): K. Sakata, K. Araki, T. Nishina, S. Komazawa-Sakon, S. Murai, G.E. Lee, D. Hashimoto, C. Suzuki, K. Notohara. Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): Y. Uchiyama, K.-I. Yamamura, A.S. Gukovskaya, I. Gukovsky, M. Ohmuraya Writing, review, and/or revision of the manuscript: H. Nakano, H. Baba, A.S. Gukovskaya, I. Gukovsky, M. Ohmuraya. Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): M. Ohmuraya. Study supervision: H. Nakano, Y. Uchiyama, K-I.Yamamura, M. Ohmuraya.
References
- Whitcomb D. C. Value of genetic testing in the management of pancreatitis. Gut 53, 1710–1717 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitcomb D. C. Clinical practice. Acute pancreatitis. N Engl J Med 354, 2142–2150 (2006). [DOI] [PubMed] [Google Scholar]
- Ohmuraya M. et al. Autophagic cell death of pancreatic acinar cells in serine protease inhibitor Kazal type 3-deficient mice. Gastroenterology 129, 696–705 (2005). [DOI] [PubMed] [Google Scholar]
- Ohmuraya M., Hirota M., Araki K., Baba H. & Yamamura K. Enhanced trypsin activity in pancreatic acinar cells deficient for serine protease inhibitor kazal type 3. Pancreas 33, 104–106 (2006). [DOI] [PubMed] [Google Scholar]
- Witt H., Apte M. V., Keim V. & Wilson J. S. Chronic pancreatitis: challenges and advances in pathogenesis, genetics, diagnosis, and therapy. Gastroenterology 132, 1557–1573 (2007). [DOI] [PubMed] [Google Scholar]
- Chen J. M. & Ferec C. Genetics and pathogenesis of chronic pancreatitis: the 2012 update. Clin Res Hepatol Gastroenterol 36, 334–340 (2012). [DOI] [PubMed] [Google Scholar]
- Mizushima N. & Komatsu M. Autophagy: renovation of cells and tissues. Cell 147, 728–741 (2011). [DOI] [PubMed] [Google Scholar]
- Lyon M. F. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190, 372–373 (1961). [DOI] [PubMed] [Google Scholar]
- Yamamura K. & Araki K. Gene trap mutagenesis in mice: new perspectives and tools in cancer research. Cancer science 99, 1–6 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniwaki T. et al. Characterization of an exchangeable gene trap using pU-17 carrying a stop codon-beta geo cassette. Dev Growth Differ 47, 163–172 (2005). [DOI] [PubMed] [Google Scholar]
- Thumkeo D. et al. Deficiency of mDia, an actin nucleator, disrupts integrity of neuroepithelium and causes periventricular dysplasia. PLoS One 6, e25465 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ida S. et al. SPINK1 Status in Colorectal Cancer, Impact on Proliferation, and Role in Colitis-Associated Cancer. Mol Cancer Res. 13, 1130–1138 (2015). [DOI] [PubMed] [Google Scholar]
- Niwa H., Yamamura K. & Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108, 193–199 (1991). [DOI] [PubMed] [Google Scholar]
- Hashimoto D. et al. Involvement of autophagy in trypsinogen activation within the pancreatic acinar cells. J Cell Biol 181, 1065–1072 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mareninova O. A. et al. Impaired autophagic flux mediates acinar cell vacuole formation and trypsinogen activation in rodent models of acute pancreatitis. J Clin Invest 119, 3340–3355 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert M. & Rustgi A. K. Pancreatic ductal cells in development, regeneration, and neoplasia. J Clin Invest 121, 4572–4578 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sah R. P., Dawra R. K. & Saluja A. K. New insights into the pathogenesis of pancreatitis. Current opinion in gastroenterology 29, 523–530 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmuraya M., Sugano A., Hirota M., Takaoka Y. & Yamamura K. Role of Intrapancreatic SPINK1/Spink3 Expression in the Development of Pancreatitis. Frontiers in physiology 3, 126 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M. et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131, 1149–1163 (2007). [DOI] [PubMed] [Google Scholar]
- Li N. et al. Loss of acinar cell IKKalpha triggers spontaneous pancreatitis in mice. J Clin Invest 123, 2231–2243 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omary M. B., Lugea A., Lowe A. W. & Pandol S. J. The pancreatic stellate cell: a star on the rise in pancreatic diseases. J Clin Invest 117, 50–59 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halangk W. et al. Role of cathepsin B in intracellular trypsinogen activation and the onset of acute pancreatitis. J Clin Invest 106, 773–781 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wartmann T. et al. Cathepsin L inactivates human trypsinogen, whereas cathepsin L-deletion reduces the severity of pancreatitis in mice. Gastroenterology 138, 726–737 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gukovsky I., Li N., Todoric J., Gukovskaya A. & Karin M. Inflammation, autophagy, and obesity: common features in the pathogenesis of pancreatitis and pancreatic cancer. Gastroenterology 144, 1199–1209 e1194 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gukovsky I. & Gukovskaya A. S. Impaired autophagy triggers chronic pancreatitis: lessons from pancreas-specific atg5 knockout mice. Gastroenterology 148, 501–505 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diakopoulos K. N. et al. Impaired autophagy induces chronic atrophic pancreatitis in mice via sex- and nutrition-dependent processes. Gastroenterology 148, 626–638 e617 (2015). [DOI] [PubMed] [Google Scholar]
- Gukovskaya A. S. & Gukovsky I. Autophagy and pancreatitis. Am J Physiol Gastrointest Liver Physiol 303, G993–G1003 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiser S. et al. Intracellular activation of trypsinogen in transgenic mice induces acute but not chronic pancreatitis. Gut 60, 1379–1388 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S. et al. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell 137, 1100–1111 (2009). [DOI] [PubMed] [Google Scholar]
- Araki K., Araki M. & Yamamura K. Site-directed integration of the cre gene mediated by Cre recombinase using a combination of mutant lox sites. Nucleic Acids Res 30, e103 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata S. et al. Highly sensitive peptide-4-methylcoumaryl-7-amide substrates for blood-clotting proteases and trypsin. Eur J Biochem 172, 17–25 (1988). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.