Abstract
Unexpected abscission of flowers or fruits is a major limiting factor for crop productivity. Key genes controlling abscission in plants, especially in popular fruit trees, are largely unknown. Here we identified a litchi (Litchi chinensis Sonn.) IDA-like (INFLORESCENCE DEFICIENT IN ABSCISSION-like) gene LcIDL1 as a potential key regulator of abscission. LcIDL1 encodes a peptide that shows the closest homology to Arabidopsis IDA, and is localized in cell membrane and cytoplasm. Real-time PCR analysis showed that the expression level of LcIDL1 accumulated gradually following flower abscission, and it was obviously induced by fruit abscission-promoting treatments. Transgenic plants expressing LcIDL1 in Arabidopsis revealed a role of LcIDL1 similar to IDA in promoting floral organ abscission. Moreover, ectopic expression of LcIDL1 in Arabidopsis activated the expression of abscission-related genes. Taken together, our findings provide evidence that LcIDL1 may act as a key regulator in control of abscission.
Abscission is characterized by a coordination of biochemical events that take place in abscission zones (AZ) to shed vegetative or reproductive organs1. Currently it is well known that abscission involves multiple changes in cell structure, metabolism and gene expression, and the process is divided into four major steps1,2: (i) the differentiation of AZ at a specific position, (ii) the acquisition by the AZ of competence, (iii) the onset of the cell separation within the AZ, (iv) the differentiation of a protective layer at the plant’s side. Over the past decades, a few genes have been found to play important roles in these four steps, which are mainly reported in model plants tomato and Arabidopsis3,4,5,6,7,8,9,10,11,12,13,14,15,16. In tomato, the MADS box transcriptional factors JOINTLESS and MACROCALYX form a complex to regulate the formation of the pedicel AZ together with the VHIID protein LATERAL SUPPRESSOR11,17. In Arabidopsis, two genes, BLADEON-PETIOLE 1 and 2 (BOP1, BOP2) which encode proteins belonging to a family containing BTB/POZ domains and ankyrin repeats, are necessary for AZ development. bop1bop2 double mutants retain all floral organs indefinitely and do not form cytologically distinct and active floral AZ cells7. NtBOP2, a homologue of the Arabidopsis BOP2 protein, also showed a specific role in the differentiation of the corolla AZ16. When AZ differentiation is completed, phytohormones are thought to be important abscission signals. In general, ethylene and jasmonic acid accelerate abscission, while auxin, gibberellins, and brassinosteroids inhibit abscission1. In addition, many studies have shown that the rate and degree of abscission are largely dependent on the balance between the levels of auxin and ethylene in AZ, especially on changes in auxin gradients10,18,19,20. Once the abscission process is activated, many components start to function in the AZ. For the third phase, an IDA-HAESA (HAE)/HAESA-LIKE2 (HSL2) signaling system has been characterized during floral organ abscission in Arabidopsis4,8,9,15,21. Briefly, IDA encodes a peptide ligand that forms a complex with redundant receptor-like kinases HAE and HSL2, which presumably activates a MITOGEN-ACTIVATED PROTEIN KINASE (MAPK) cascade that acts to regulate downstream KNOX-like transcription factor BP/KNAT1. Recently, it has been proposed that the involvement of this signaling module in abscission processes is conserved in other plant species22,23. However, so far, the characterization of key regulators in control of abscission has been reported mainly in the model plants tomato and Arabidopsis; these factors are largely unknown in perennial woody plants, especially in important horticultural cultivated fruit crops, such as litchi.
Litchi (Litchi chinensis Sonn.), a famous fruit tree originating from South China, has been widely cultivated in Southeast Asia due to its delicious and nutritional fruits. The litchi tree is easily subject to massive fruit drop, leading to low yield and heavy economic loss24,25. For example, a medium-sized tree may produce close to 60,000 female flowers but less than 5% of flowers can develop into mature fruits26,27. There are three to four waves of physiological fruit drop throughout fruit development depending on cultivar. Wave I, wave II, and wave III of abscission occur around 1 week, 3 weeks, and 6–7 weeks after full bloom, respectively, but wave IV is cultivar-specific and occurs at 2–3 weeks before harvest26,27. Thus, to reduce unexpected fruit abscission in litchi, a number of studies have been conducted on endogenous hormones27,28,29 and carbohydrates30,31,32,33,34,35, which are proposed to play vital roles in the regulation of fruit abscission. Recently, genome-wide digital transcript analysis further revealed that a range of fruitlet abscission-related genes are regulated by ethylene and carbohydrate stress36,37. In fact, massive transcriptomic and metabonomic data about abscission were also obtained in other fruit trees, such as citrus leaf and shoot tips38,39, apple young fruits40,41 and olive mature fruits42. A deeper understanding of the mechanisms underlying abscission would be helpful not only for the regulation of fruit crop production and the improvement of current cultural and management practices, but also for the elucidation of new molecular markers to improve genetic breeding43.
Here we identified three IDA-like genes in litchi, of which LcIDL1 shows the closest homology to Arabidopsis IDA and is found to be involved in litchi male flower and fruitlet abscission. Ectopic expression of LcIDL1 in Arabidopsis revealed that LcIDL1 is able to function to induce floral organ abscission. Our data suggest that LcIDL1 may act as a key regulator in control of abscission.
Results
Identification of key candidate genes regulating fruit abscission in litchi
Previously, in an effort to identify the components of fruit abscission in litchi, we screened for genes that were differentially expressed during abscission from a RNA-seq database. Through GO (Gene Ontology) and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis, a number of genes were identified as the candidate components involved in fruit abscission process. These genes were clustered into diverse metabolic processes and pathways, including carbohydrate metabolism, plant hormone synthesis and signaling, transcription factor activity and cell wall modification36,37. But which is a master one? We found Litchi_GLEAN_10054315 encoding an IDA (INFLORESCENCE DEFICIENT IN ABSCISSION)-like gene and being highly expressed before fruit abscission. Here we designated it as LcIDL1. It appears that key regulators, such as IDA/IDL, may have conserved function in control of cell separation in different plant species4, so LcIDL1 attracted much more attention in our lab.
LcIDL1 is an IDA-like gene and shows closest homology to Arabidopsis IDA
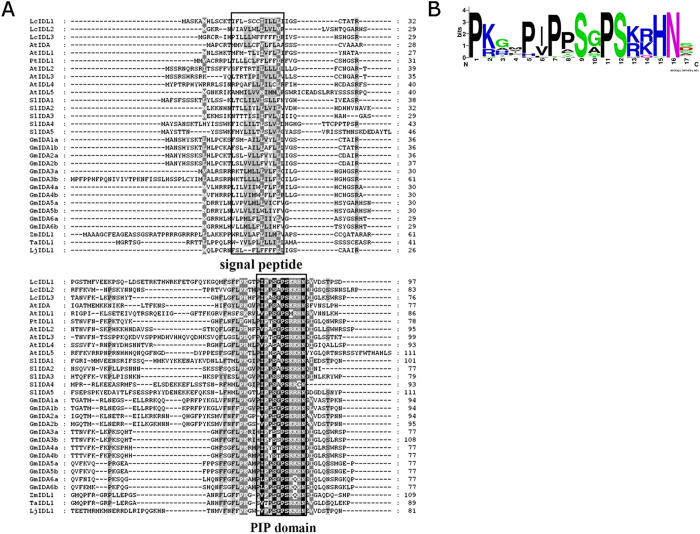
To acquire all the IDA-like gene members in litchi, we used the conserved domain of IDA to blast against the litchi genome database. We obtained two more IDA-like genes, Litchi_GLEAN_10027620 and Litchi_GLEAN_10009234, and we named them as LcIDL2 and LcIDL3. IDA protein alignment between litchi and other plant species was conducted based on their full-length protein sequences. As shown in Fig. 1A, all members of the IDL gene family are characterized by an N-terminal secretory signal peptide (SP), a variable region and a conserved C-terminal region called the PIP domain4. The N-terminal of SP contains a stretch of aliphatic residues, a typical motif of secreted proteins. A sequence consensus logo of the C-terminal conserved PIP domain showed that this domain consists of a PSGPS motif and four highly conserved residues [(R/K)(R/K)HN] (Fig. 1B).
Figure 1. Alignment of LcIDLs with other plant IDA/IDL proteins.
(A) Alignment was made using the ClustalW2 program (http://www.ebi.ac.uk/Tools/clustalw2/) with manual adjustment using GeneDoc (http://www.psc.edu/biomed/genedoc/). The amino acid sequences of LcIDL1 (Litchi_GLEAN_10054315), LcIDL2 (Litchi_GLEAN_10027620), LcIDL3 (Litchi_GLEAN_10009234), AtIDA (At1g68765), AtIDL1 (AT3G25655), AtIDL2 (AT5G64667), AtIDL3 (AT5G09805), AtIDL4 (AT3G18715), AtIDL5 (AT1G76952), PtIDL1 (BU889756), ljIDL1(AW719486), ZmIDL1(BI430572), TaIDL1 (BM135459), SlIDAs and GmIDAs (see reference Tucker and Yang, 2012) were aligned. The conserved domains of signal peptide and PIP are marked with rectangles. (B) A sequence logo representation of the conserved C-terminal PIP domain of IDA/IDL peptides.
In order to examine the relationship of litchi IDL genes with those in other species, a phylogenetic analysis using the full-length IDA/IDL protein sequences from Arabidopsis (At), tomato (Sl), lotus (Lj), soybean (Gm), maize (Zm), poplar (Pt) and wheat (Ta) was performed. As shown in Fig. 2, the IDL family is divided into two subgroups, one containing LcIDL1 and the other containing LcIDL2 and LcIDL3. In contrast to LcIDL2 and LcIDL3, LcIDL1 shows the closest relationship to Arabidopsis IDA.
Figure 2. Phylogenetic tree of IDA/IDL proteins.
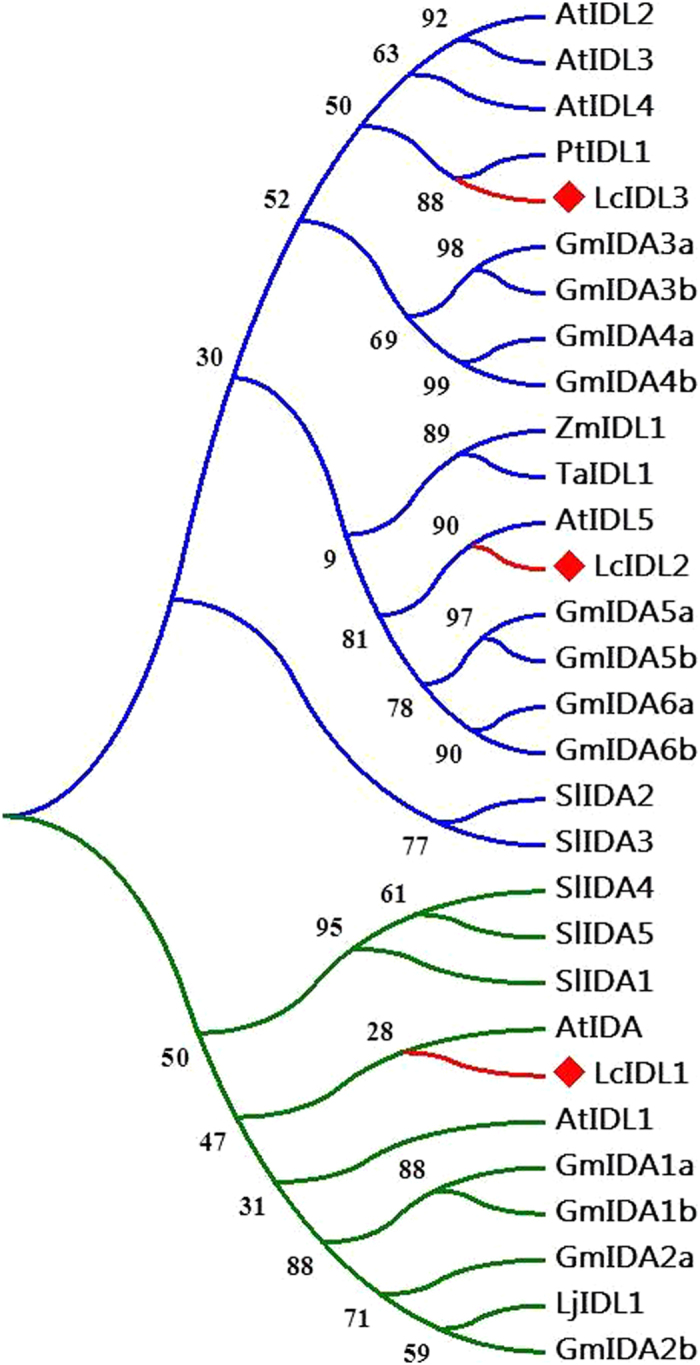
A maximum likelihood phylogenetic tree was constructed based on the protein alignment of the IDA/IDL from Arabidopsis (At), tomato (Sl), lotus (Lj), soybean (Gm), maize (Zm), poplar (Pt) and wheat (Ta). The bootstrap consensus tree was inferred from 1000 replicates. The two top-level sub-branches are represented by blue and green color, respectively. The red lines and diamonds indicate the LcIDLs.
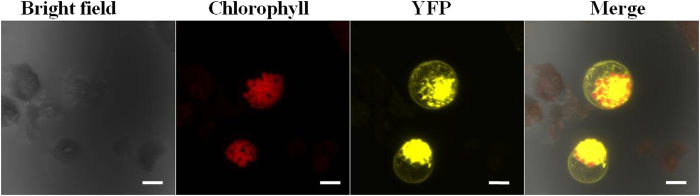
To investigate the subcellular localization of the LcIDL1 protein, the full-length coding sequence of LcIDL1 fused with yellow fluorescent protein (YFP) was delivered into the Arabidopsis protoplasts. LcIDL1-YFP protein was observed to be localized to the cell membrane and cytoplasm (Fig. 3).
Figure 3. Subcellular localization analysis of LcIDL1.
Constructs of LcIDL1-YFP were transfected into Arabidopsis protoplasts; and the fluorescence signal was detected with the laser scanning confocal microscope. The yellow color indicates fluorescence of YFP, while the red color indicates the auto-fluorescence of chlorophyll; the bar is 25 μm.
LcIDL1 is an abscission-associated gene during the male flower and fruitlet drop in litchi
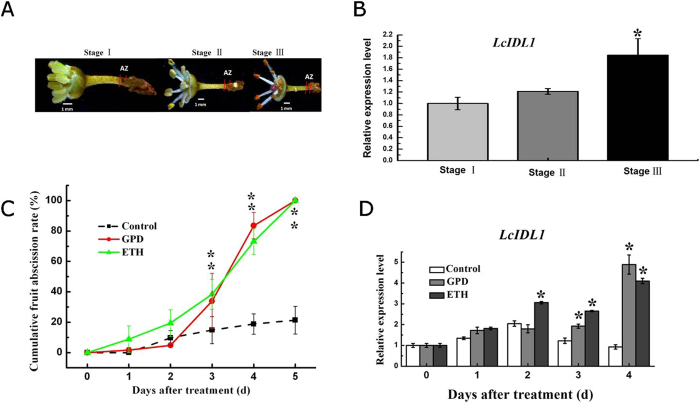
The expression level of LcIDL1 in pedicel AZ tissues of male flowers was determined using a real-time PCR assay. Generally, the male flowers will drop at the pedicel AZ within one week after the stamens protruding out. AZ tissues of 1st day (stage I), 3rd day (stage II) and 7th day (stage III) after stamens protruding out were harvested for analysis (Fig. 4A). The transcription level of LcIDL1 at stage III was increased significantly compared to the other two stages (Fig. 4B), which was consistent with the degree of flower abscission. Next, we examined the effect of two abscission-inducing treatments, girdling plus defoliation (GPD) and ethephon (ETH)37 on the transcription of LcIDL1 during litchi fruitlet abscission. Both GPD and ETH treatments significantly promoted the fruitlet drop starting from the third day, the cumulative fruit abscission rate was up to 100% on the fifth day (Fig. 4C). The expression level of LcIDL1 in pedicel AZ tissues was significantly induced after GPD and ETH treatments, with about 5-fold and 4-fold higher in GPD and ETH-treated AZ tissues than that in control on the fourth day (Fig. 4D). Collectively, these data suggested that LcIDL1 is an abscission-associated gene in litchi.
Figure 4. LcIDL1 is an abscission-associated gene in litchi.
(A) Magnified images of stage I, stage II and stage III of litchi male flower; the region between two short red lines indicates the AZ. (B) qRT-PCR analysis of LcIDL1 expression in pedicel AZ cells of litchi male flower during abscission. The Y-axis is fold-change, the expression levels are relative to stage I. (C) Girdling plus defoliation (GPD) and ethephon (ETH) treatments significantly induced fruitlet abscission. (D) qRT-PCR analysis of LcIDL1 expression in peduncle AZ cells after GPD and ETH treatments. LcEF-1a was used as an internal control. The Y-axis is fold-change, the expression levels are relative to control 0 d. Data shown are means ± SD. One-way ANOVA (Tukey-Kramer test) analysis was performed, and statistically significant differences (P < 0.05) were indicated by asterisks.
Ectopic expression of LcIDL1 driven by the 35S promoter strongly activated the floral organ abscission in Arabidopsis
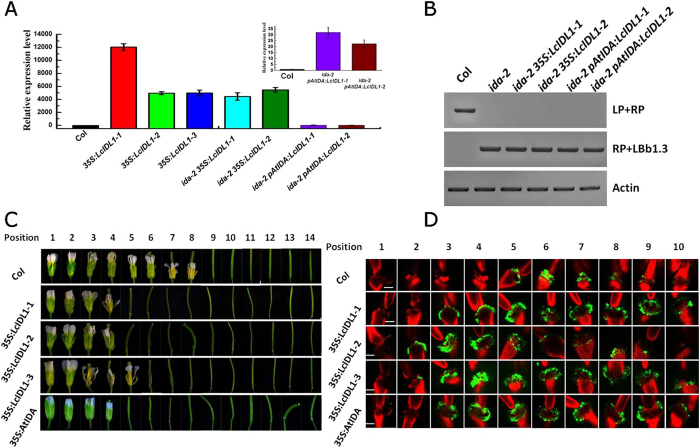
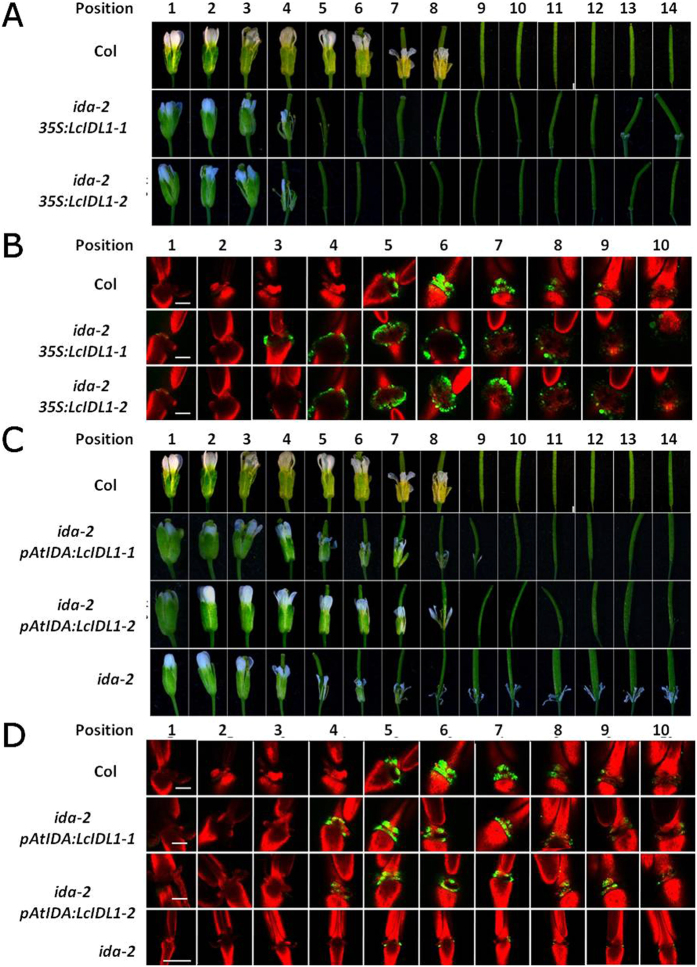
To examine the possible role of LcIDL1 in abscission, LcIDL1 was ectopically expressed in the model plant Arabidopsis. We constructed transgenic plants expressing LcIDL1 under the control of the 35S promoter by Agobacterium-mediated plant transformation44. Meanwhile, transgenic Arabidopsis plants over-expressing AtIDA were also generated and used as a control. Homozygous 35S:LcIDL1 and 35S:IDA transgenic lines were obtained by antiabiotic selection. Further, real-time PCR analysis showed that the transcript of LcIDL1 was highly expressed in these transgenic plants (Fig. 5A). The phenotypes of the flowers or siliques at specific positions along the inflorescence of wild-type Col, 35S:LcIDL1, and 35S:IDA transgenic plants were examined. Similar to what has been reported previously6, the 35S:AtIDA plants exhibited earlier abscission of floral organs (Fig. 5C), suggesting our construct is valid. Interestingly, we showed that the 35S:LcIDL1 plants abscised their flowers first at position 5, whereas the wild-type dropped their flowers first at position 9 (Fig. 5C), which suggested that LcIDL1 is able to induce floral organ abscission in transgenic Arabidopsis.
Figure 5. Overexpression of LcIDL1 caused earlier floral organ abscission in Arabidopsis.
(A) Expression level of LcIDL1 in different transgenic Arabidopsis lines. 35S:LcIDL1-1, 35S:LcIDL1-2, and 35S:LcIDL1-3 lines were ectopic expression of LcIDL1 under the control of the 35S promoter in wild type Col; ida-2 35S:LcIDL1-1 and ida-2 35S:LcIDL1-2 lines were ectopic expression of LcIDL1 under the control of the 35S promoter in ida-2; ida-2 pAtIDA:LcIDL1-1 and ida-2 pAtIDA:LcIDL1-2 lines were ectopic expression of LcIDL1 under the control of the Arabidopsis IDA promoter in ida-2. AtUBQ was used as an internal control for qRT-PCR analysis. The Y-axis is fold-change, the expression levels are relative to wild type Col. Data shown are means ± SD. (B) Genotyping analysis of IDA in Col, ida-2, ida-2 35S:LcIDL1-1, ida-2 35S:LcIDL1-1, ida-2 pAtIDA:LcIDL1-1 and ida-2 pAtIDA:LcIDL1-1. The T-DNA insertion mutant ida-2 are analyzed with LP (Left border primer of the T-DNA insertion), RP (Right border primer of the T-DNA insertion), and LBb1.3 (used for Salk genotyping project). (C) Phenotype of floral organ abscission in transgenic lines. Position numbers were counted from the first flower with visible white petals on the top of the inflorescence. (D) BCECF fluorescence micrographs of floral organ AZ of Arabidopsis Col, 35S:LcIDL1-1, 35S:LcIDL1-2, and 35S:LcIDL1-3. Inflorescences were sampled separately, incubated in BCECF solution, and examined by a confocal laser scanning microscope. The microscopic fluorescence images represent merged images of BCECF fluorescence with chlorophyll autofluorescence images. The increase in pH is shown by green fluorescence, which is distinguished from the red chlorophyll autofluorescence. Scale bars are 100 μm in length. The images presented for each plant and positions are representative images out of 3–4 replicates.
Previous work revealed that the cytosolic pH increase in AZ cells occurs concomitantly with the execution of organ abscission45. We further observed the pH-value in the abscission zone of the floral organs using a pH-sensitive indicator, BCECF45. The BCECF-treated floral organs were imaged using a confocal microscope. As shown in Fig. 5D, obvious green fluorescence was observed in the abscission zone of the position 5 flowers in the wild-type. In contrast, the 35S:LcIDL1 and 35S:AtIDA overexpression lines not only displayed much earlier BCECF green fluorescence which can be detected at P3, but also showed much stronger BCECF green fluorescence and maintained a relatively higher signal density prolonged to position 10 (Fig. 5D). These data confirmed that LcIDL1 is able to function to promote floral organ abscission in Arabidopsis.
LcIDL1 has a similar role to AtIDA in control of floral organ abscission in Arabidopsis
Phylogenetic analysis revealed that LcIDL1 is a close homologue of AtIDA. To further compare the role of LcIDL1 and AtIDA in floral organ abscission, we generated transgenic plants expressing LcIDL1 driven by 35S or AtIDA promoters under an ida-2 mutant background9. Homozygous ida-2 35S:LcIDL1 and ida-2 pAtIDA:LcIDL1 transgenic lines were selected and validated by real-time PCR analysis (Fig. 5A,B). As described previously9, ida-2, the T-DNA insertion allele of ida, is deficient in floral organ abscission (Fig. 6C). We showed that the floral organs of ida-2 35S:LcIDL1–1 and ida-2 35S:LcIDL1-2 dropped at position 5, which is earlier than the wild-type Col (position 9) and ida-2 mutant (Fig. 6A). Consistently, the BCECF green fluorescence in AZ cells of ida-2 35S:LcIDL1 plants was observed earlier, stronger and longer than that in Col plants (Fig. 6B).
Figure 6. LcIDL1 has a similar role as AtIDA in control of floral organ abscission.
(A) ida-2 35S:LcIDL1-1 and ida-2 35S:LcIDL1-2 displayed earlier floral organ abscission. Position numbers were counted from the first flower with visible white petals on the top of the inflorescence. (B) BCECF fluorescence micrographs of floral organ AZ of Arabidopsis Col, ida-2 35S:LcIDL1-1 and ida-2 35S:LcIDL1-2. Scale bars are 100 μm. The images presented for each plant and positions are representative images out of 3–4 replicates. (C) ida-2 pAtIDA:LcIDL1-1 and ida-2 pAtIDA:LcIDL1-2 displayed normal floral organ abscission. Position numbers were counted from the first flower with visible white petals on the top of the inflorescence. (D) BCECF fluorescence micrographs of floral organ AZ of Arabidopsis Col, ida-2 pAtIDA:LcIDL1-1 and ida-2 pAtIDA:LcIDL1-2. Scale bars are 100 μm. The images presented for each plant and position are representative images out of 3–4 replicates.
Furthermore, we showed that ida-2 pAtIDA:LcIDL1 transgenic lines ida-2 pAtIDA:LcIDL1-1 and ida-2 pAtIDA:LcIDL1-2 displayed the same floral organ abscission process as the wild-type, with the floral organs dropped at position 9 (Fig. 6C). In addition, the BCECF green fluorescence in AZ cells of ida-2 pAtIDA:LcIDL1-1 and ida-2 pAtIDA:LcIDL1-2 transgenic lines was also recovered (Fig. 6D), which suggested that ectopic expression of LcIDL1 driven by the Arabidopsis IDA promoter completely rescued the ida-2 floral organ abscission phenotype. Collectively, these data confirmed that LcIDL1 is capable of functioning in place of AtIDA to induce floral organ abscission in Arabidopsis.
Ectopic expression of LcIDL1 in Arabidopsis activated abscission-related gene expression
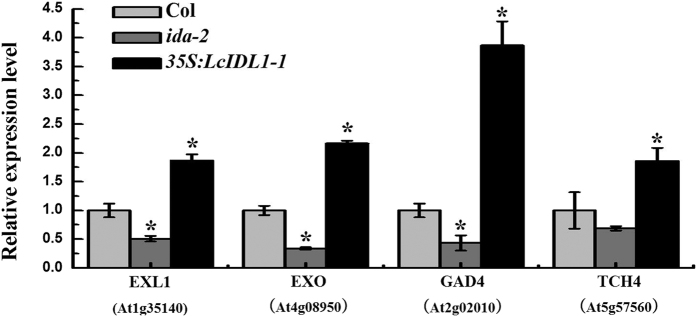
A previous study reported a positive role of IDA in regulating abscission-related gene expression14. In the present work, the expression levels of a subset of abscission-related genes, such as cell-wall remodelling (TCH4/XTH22)3, cell expansion (EXO and EXL1)46, Ca2+ binding (GAD4)47 were tested with qRT-PCR in Col, 35S:LcIDL1-1 and ida-2. AZ regions undergoing cell-wall loosening and organ separation (positions 3–8) were harvested for analysis. As shown in Fig. 7, the expression level of TCH4, EXO, EXL1 and GAD4 was decreased in ida-2 compared with wild-type. In contrast, the transcription of these genes was significantly up-regulated in 35S:LcIDL1-1 compared with the wild-type. These data suggested that LcLDL1 may induce floral organ abscission by activating abscission-related gene expression.
Figure 7. Expression levels of abscission-related cell wall remodeling genes in Col, ida-2 and 35S:LcIDL1-1.
AZ tissues of position 3–8 were collected for testing. AtUBQ was used as an internal control for qRT-PCR analysis. The Y-axis is fold-change, the expression levels are relative to wild type Col. Data shown are means ± SD. One-way ANOVA (Tukey-Kramer test) analysis was performed, and statistically significant differences (P < 0.05) were indicated by asterisks.
Discussion
To date, IDA-like genes have been found in Arabidopsis15, soybean (Glycine max), tomato (Solanum lycopersicum)12 and citrus (sweet orange and clementine)22. However, only Arabidopsis IDA has been tested for subcellular localization and found localized in the cell membrane, supporting that IDA, as a signal peptide, can be secreted4. In our report, LcIDL1 can be detected in the cytoplasm (Fig. 3). AtIDA is, in contrast to LcIDL1, not found in the cytoplasm. One possible reason is that AtIDA and LcIDL1 do not function completely similarly, the other one is that different systems were used for signal detection, since AtIDA was bombarded into the onion cells and Arabidopsis protoplasts were transfected in the present assay. Thus further study on IDA delivered to Arabidopsis protoplasts can tell more.
That ethylene acts an important factor inducing organ shedding has been established for many years48. Exposure to exogenous ethylene has no impact on the inhibition of floral organ abscission and ethylene sensitivity in ida mature plants, suggesting that IDA acts downstream of ethylene in the pathway by which ethylene controls abscission4. Consistent with this, the promoter fusion construct IDA:GUS, which is primarily expressed in the floral AZ and floral organs, is restricted only to the nectaries in ethylene insensitive mutant etr1-1, indicating that ethylene acts at least partly through ETR1 to transcriptionally induce IDA expression which then instigates abscission. Here our report further supports that IDA/IDL genes seem to be influenced by an ethylene-response pathway. When treated with ethylene, the expression level of LcIDL1 significantly increased in AZ cells. Additionally, GPD treatment induced much higher expression of LcIDL1 (Fig. 4), we inferred that this is likely also associated with ethylene emission induced by GPD treatment36. Similar results were also obtained in Populus and oil palm, which showed that PtIDA and PtIDL1 were expressed more highly in leaves after shade treatment (a similar treatment to GPD resulting in carbohydrate stress), and EgIDA2 and EgIDA5 could be induced by ethylene23, further supporting that IDA functions in the ethylene-response pathway.
Genetic studies have already revealed a framework of cell signalling, membrane trafficking, and transcriptional networks in the later stages of floral organ abscission in Arabidopsis, of which IDA has been found as a critical regulator as ida mutant plants are deficient in floral organ abscission in Arabidopsis4, while opposite to what is observed in the ida mutant, Arabidopsis plants overexpressing IDA cause precocious floral organ abscission6. Interestingly, transgenic plants expressing LcIDL1 in Arabidopsis also caused earlier floral organ abscission, suggesting that LcIDL1 is able to function to induce floral abscission in Arabidopsis (Fig. 5). In addition, beyond our expectation, transgenic plants expressing LcIDL1 under the control of the 35S promoter in ida-2 also displayed earlier floral organ abscission (Fig. 6), suggesting a similar role of LcIDL1 to AtIDA. More importantly, transgenic plants expressing LcIDL1 under the control of the Arabidopsis IDA native promoter in ida-2 can completely restore the deficiency in floral organ abscission, further supporting the notion that LcIDL1 shares the same function with AtIDA of promoting floral organ abscission in Arabidospsis (Fig. 6). Similarly, that citrus CitIDA3 is able to function to promote floral organ abscission in transgenic Arabidopsis has also been reported, and ectopic expression of CitIDA3 could also complement the abscission deficiency of the ida mutant22. Taken together, these results indicate that IDA, as a regulator of abscission, is functionally conserved in different species.
It has been suggested that pH changes in AZ cells are associated with abscission progress. Before onset of organ abscission, the alkalization of the cytosol in AZ cells will occur and stronger BCECF fluorescence will be detected in AZ cells of mutants that promote abscission. Mutants that inhibit abscission will maintain a low pH value and weak BCECF fluorescence in AZ cells45. In the present study, we showed that an earlier, stronger and longer BCECF fluorescence can be detected in transgenic plants overexpressing LcIDL1 which displayed significantly earlier floral organ abscission (Figs 5 and 6), indicating that there is a strong correlation between pH changes in AZ cells and the abscission process. Further, genes involved in pH changes in AZ cells need to be identified.
Once the abscission process is activated, the pectin in the cell walls between the AZ cell layers starts to degrade followed by an expansion in the size of the AZ cells. A concerted effort of research on many plant species has identified cell wall remodeling enzymes acting on structural polysaccharides leading to the hydrolysis of the middle lamella and cell walls of the AZ cells1. It has also revealed that IDA not only regulates floral abscission but also regulates lateral root emergence through changing the CWR enzyme activity13. In this study, higher expression levels of four abscission-related cell wall remodeling genes, GAD4, TCH4, EXO and EXL1, were detected in LcIDL1 overexpression lines (Fig. 7). In addition, our previous report showed that a cluster of cell wall degradation and loosening genes including β-1,3-glucanase, β-D-xylosidase, endoglucanase, xyloglucan endotransglucosylase/hydrolase, pectinesterase, polygalacturonase, and β-D-xylosidase were induced in AZ cells during litchi fruit abscission. Collectively, it can be proposed that IDA/IDL regulates initial cell wall loosening and separation of AZ cells to induce abscission, probably by controlling the expression of cell wall remodelling genes.
In conclusion, our findings revealed that LcIDL1 is likely involved in the regulation of male flower and fruitlet abscission in litchi. Furthermore, LcIDL1 functions similarly as AtIDA to promote floral organ abscission in Arabidopsis, which may operate through regulating pH changes and cell wall remodelling genes in AZ cells.
Materials and Methods
Plant materials and treatments
For litchi, three 9-year-old litchi trees (Litchi chinensis Sonn. cv. Feizixiao) grown in an orchard in South China Agricultural University (Guangzhou, China) were randomly selected. Thirty fruit bearing shoots with similar diameter (about 5–8 mm) growing in different directions from each tree were tagged. Ten of them were treated with girdling (a ring of bark about 0.5 cm in width and cambium was removed from the branch base) followed by defoliation (removing all leaves above the girdle) at 35 days after anthesis (GPD treatment); Ten of them were dipped in 250 mg L−1 ethephon solution (containing 0.05% Tween-80 surfactant) for 1 min (ETH treatment), while the remaining untreated shoots were used as controls. Three out of each set of ten treated shoots were used to monitor fruit abscission dynamics for 5 days and the others were used for sampling. Cumulative fruit abscission rate (CFAR) was calculated according to our previous method49. Samples were collected at 0, 1, 2, 3 and 4 days after treatment. AZ was excised by cutting around 2 mm at each side of the abscission fracture plane. After separation, all tissues were quickly frozen in liquid nitrogen and stored at −80 °C for future analysis. Each tree was treated as a biological replicate. For the flower assay, male flowers were selected and flowers with stamens just spreading were designed as stage I, the 3rd day after stamens spreading was designed as stage II, and the 7th day after stamens spreading was designed as stage III. Male flower AZ tissues were sampled as mentioned above.
For Arabidopsis materials, Arabidopsis ecotype Col-0 was used in all experiments and the ida-2 mutant (SALK_133209)9, which is in the Col background, was obtained from the Nottingham Arabidopsis Stock Centre (NASC). For generation of LcIDL1 and AtIDA overexpression lines in Col or in ida-2, the full-length open reading frame (ORF) of LcIDL1 and AtIDA were subcloned into vector pCAMBIA1302 under the control of the 35S promoter with Clontech’s In-Fusion system primers (Supplemental Table), then these constructs were transformed into Col or ida-2 plants following the floral dip method44. For generation of ida-2 pAtIDA:LcIDL1 transgenic plants, the full-length ORF of LcIDL1 was subcloned into vector pCAMBIA1302 with the 35S promoter replaced with the Arabidopsis IDA promoter (−1 to −1490 bp), then this vector was transformed into ida-2 plants following the floral dip method. The T3 homozygous transgenic plants were used for phenotypic analysis. All the Arabidopsis plants were grown at 22 °C under long-day (16 h light/8 h dark) conditions. To reduce variation, all genotypes tested in each experiment were grown together.
Subcellular Localization Analysis
The coding sequence of LcIDL1 without the stop codon was amplified by PCR primers (listed in Supplemental Table) and then subcloned into the pSAT6-EYFP-N1 vector and fused in-frame with the Yellow Fluorescent Protein (YFP) sequence under the control of the 35S promoter. The fusion constructs were introduced into Arabidopsis protoplasts by using 40% polyethylene glycol (PEG) as described previously50. YFP fluorescence was observed with a confocal laser scanning microscope (LSM 7 DUO, ZEISS, Germany). The transient expression assay was repeated three times.
Quantitative RT-PCR Analysis
Total RNA was isolated from litchi AZ tissues or Arabidopsis leaves (20-day-old) using 1 mL Trizol reagent (Invitrogen). The first strand cDNA synthesis was generated using 2 μg total RNA according to the manufacturer’s instructions of the TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix Kit (TransGen, Beijing). 100 ng synthesized cDNA was used as a template to perform quantitative RT-PCR analysis. PCR reactions were performed in a total volume of 20 μL, with 0.5 μL for each primer (10 mM, final concentration 100 nM) and 10 μL for SYBR Green PCR Supermix (Bio-Rad) on an ABI7500 Real-Time PCR System (Applied Biosystems). The PCR program included an initial denaturation step at 94 °C for 3 min, followed by 40 cycles of 5 s at 94 °C and 1 min at 60 °C. Each sample was quantified at least in triplicate and normalized using Ubiquitin 10 (UBQ) or EF-1a as an internal control for Arabidopsis and litchi51, respectively. The gene-specific primer pairs for quantitative Real-Time PCR are listed in Supplemental Table. All PCR reactions were normalized using a Ct value corresponding to the reference gene. The relative expression levels of target gene were calculated with the formula 2−ddCt 52. Values represented the average of three biological replicates.
Phenotypic Analysis
Litchi and Arabidopsis flowers were carefully removed from the plant body and then were imaged using a stereoscope (ZEISS, SV11). The whole Arabidopsis plant and a single leaf placed on a black cloth were photographed by digital camera (D3200, Nikon).
BCECF fluorescence analyses
BCECF fluorescence analysis was conducted according to a previous detailed method with some modification45. Inflorescences with flowers located at various positions along the inflorescence were carefully removed from the plant body and immersed in 10 μM BCECF-AM (B1150, Thermo Scientific™) solution under darkness for 20 min, then the inflorescences were rinsed four times with phosphate-buffered saline (PBS, pH 7.4) to remove excess BCECE-AM. Before imaging, flowers at different positions were excised separately from the inflorescences, and each flower’s sepals, petals, and stamens were removed using forceps without damaging the carpels, receptacle, and peduncle. The images were snapped with a confocal laser scanning microscope (LSM 7 DUO, ZEISS, Germany). Samples were excited by both 488 nm and 633 nm light, then BCECF fluorescence and chlorophyll auto-fluorescence were detected through 494–598 and 647–721 filters, respectively. All BCECF experiments were repeated three times with different biological samples from different inflorescences, and representative images are presented.
Additional Information
How to cite this article: Ying, P. et al. Identification and molecular characterization of an IDA-like gene from litchi, LcIDL1, whose ectopic expression promotes floral organ abscission in Arabidopsis. Sci. Rep. 6, 37135; doi: 10.1038/srep37135 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by the Natural Science Foundation of Guangdong Province, China (grant no. 2016A030310451) and the Outstanding Talent Program of the Ministry of Agriculture.
Footnotes
Author Contributions Minglei Zhao, Jianguo Li and Peiyuan Ying contributed to designing the experiments. Minglei Zhao, Peiyuan Ying and Caiqin Li performed the experiments, and collected and analyzed the data. Minglei Zhao, Jianguo Li, Xuncheng Liu and Rui Xia contributed to data interpretation and preparation of the manuscript. All authors reviewed the manuscript.
References
- Estornell L. H., Agusti J., Merelo P., Talon M. & Tadeo F. R. Elucidating mechanisms underlying organ abscission. Plant Sci. 199, 48–60 (2013). [DOI] [PubMed] [Google Scholar]
- Patterson S. E. Cutting loose. Abscission and dehiscence in Arabidopsis. Plant Physiol. 126, 494–500 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W. et al. Arabidopsis TCH4, regulated by hormones and the environment, encodes a xyloglucan endotransglycosylase. Plant Cell 7, 1555–1567 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butenko M. A. et al. Inflorescence deficient in abscission controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. Plant Cell 15, 2296–2307 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C. M. et al. AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 132, 4563–4574 (2005). [DOI] [PubMed] [Google Scholar]
- Stenvik G. E., Butenko M. A., Urbanowicz B. R., Rose J. K. & Aalen R. B. Overexpression of INFLORESCENCE DEFICIENT IN ABSCISSION activates cell separation in vestigial abscission zones in Arabidopsis. Plant Cell 18, 1467–1476 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim S. M. et al. The BLADE-ON-PETIOLE genes are essential for abscission zone formation in Arabidopsis. Development 135, 1537–1546 (2008). [DOI] [PubMed] [Google Scholar]
- Stenvik G. E. et al. The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell 20, 1805–1817 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. K. et al. Regulation of floral organ abscission in Arabidopsis thaliana. Proc. Natl. Acad. Sci. 105, 15629–15634 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meir S. et al. Microarray analysis of the abscission-related transcriptome in the tomato flower abscission zone in response to auxin depletion. Plant Physiol. 154, 1929–1956 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T. et al. MACROCALYX and JOINTLESS interact in the transcriptional regulation of tomato fruit abscission zone development. Plant Physiol. 158, 439–450 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M. L. & Yang R. IDA-like gene expression in soybean and tomato leaf abscission and requirement for a diffusible stelar abscission signal. Aob Plants 2012, pls035–pls035 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumpf R. P. et al. Floral organ abscission peptide IDA and its HAE/HSL2 receptors control cell separation during lateral root emergence. Proc. Natl. Acad. Sci. 60, 2289–2296 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B. et al. NEVERSHED and INFLORESCENCE DEFICIENT IN ABSCISSION are differentially required for cell expansion and cell separation during floral organ abscission in Arabidopsis thaliana. J. Exp. Bot. 64, 5345–5357 (2013). [DOI] [PubMed] [Google Scholar]
- Vie A. K. et al. The IDA/IDA-LIKE and PIP/PIP-LIKE gene families in Arabidopsis: phylogenetic relationship, expression patterns, and transcriptional effect of the PIPL3 peptide. J. Exp. Bot. 66, 5351–5365 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X. M. et al. The tobacco BLADE-ON-PETIOLE2 gene mediates differentiation of the corolla abscission zone by controlling longitudinal cell expansion. Plant Physiol. 159, 835–850 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher K., Schmitt T., Rossberg M., Schmitz G. & Theres K. The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc. Natl. Acad. Sci. 96, 290–295 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson S. E. Cutting loose. Abscission and dehiscence in Arabidopsis. Plant Physiol. 126, 494–500 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J. A., Elliott K. A. & Gonzalez-Carranza Z. H. Abscission, dehiscence, and other cell separation processes. Annu. Rev. Plant Biol. 53, 131–158 (2002). [DOI] [PubMed] [Google Scholar]
- Meir S., Hunter D. A., Chen J. C., Halaly V. & Reid M. S. Molecular changes occurring during acquisition of abscission competence following auxin depletion in Mirabilis jalapa. Plant Physiol. 141, 1604–1616 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C. L. et al. Arabidopsis class I KNOTTED-like homeobox proteins act downstream in the IDA-HAE/HSL2 floral abscission signaling pathway. Plant Cell 23, 2553–2567 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estornell L. H. et al. The IDA Peptide Controls Abscission in Arabidopsis and Citrus. Front. Plant Sci. 6, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sto I. M. et al. Conservation of the abscission signaling peptide IDA during Angiosperm evolution: withstanding genome duplications and gain and loss of the receptors HAE/HSL2. Front. Plant Sci. 6, 1152–1158 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S. K., Pereira L. S., Pathak P. K. & Majumdar D. Fruit abscission pattern of lychee cultivars. Acta Hortic. 665, 215–218 (2005). [Google Scholar]
- Roe D. J., Menzel C. M., Oosthuizen J. H. & Doogan V. J. Effects of Current CO2 Assimilation and Stored Reserves On Lychee Fruit Growth. J. Hortic. Sci. 72, 397–405 (1997). [Google Scholar]
- Stern R. A., Kigel J., Tomer E. & Gazit S. Mauritius lychee fruit-development and reduced abscission after treatment with the auxin 2,4,5-TP. J. Am. Soc. Hortic. 120, 65 (1995). [Google Scholar]
- Yuan R. & Huang H. Litchi fruit abscission: its patterns, effect of shading and relation to endogenous abscisic acid. Sci. Hortic. 36, 281–292 (1988). [Google Scholar]
- Xiang X., Qiu Y. & Zhang Z. Endogenous Hormones in the Fruit of Litichi chinensis cv. Nuomici Relating to Furit Abscission. J. Fruit Sci (1995). [Google Scholar]
- Jian-Guo L. I. & Liu S. Z. Changes in Endogenous Polyamine Contents During Fruit Development of Litchi (Litchi chinensis). Plant Physiol. Commun. 40, 153–156 (2004). [Google Scholar]
- Yuan R. Improvement of fruit-set in litchi chinensis sonn. through regulation of source-sink relationships. J. South China Agric. Univ (1992). [Google Scholar]
- Yuan R. & Huang H. Regulation of Root and Shoot Growth and Fruit-dorp of Young Litchi Trees by Trunk Girdling in View of Source-Sink Relationships. J. Fruit Sci (1993). [Google Scholar]
- Zhou X., Huang D., Huang H. & Dingyao A. W. Carbohydrate and Endohormone Status in Relation to Fruit Set as Influenced by Trunk Spiral Girdling of Young Litchi Trees. Acta Hortic. Sinica (1999). [Google Scholar]
- Hieke S., Menzel C. M., Doogan V. J. & Lüdders P. The relationship between yield and assimilate supply in lychee (Litchi chinensis Sonn.). J. Hortic. Sci. Biotech. 77, 326–332 (2002). [Google Scholar]
- Chang J. C. & Lin T. S. Gas exchange in litchi under controlled and field conditions. Sci. Hortic. 114, 268–274 (2007). [Google Scholar]
- Yuan W. Q. et al. Seasonal Changes in Carbon Nutrition Reserve in Nuomici Litchi Trees and Its Relation to Fruit Load. Acta Hortic. Sinica (2009). [Google Scholar]
- Li C. et al. An improved fruit transcriptome and the identification of the candidate genes involved in fruit abscission induced by carbohydrate stress in litchi. Front. Plant Sci. 6, 439 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Wang Y., Ying P., Ma W. & Li J. Genome-wide digital transcript analysis of putative fruitlet abscission related genes regulated by ethephon in litchi. Front. Plant Sci. 6, 1937–1942 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agusti J., Merelo P., Cercos M., Tadeo F. R. & Talon M. Comparative transcriptional survey between laser-microdissected cells from laminar abscission zone and petiolar cortical tissue during ethylene-promoted abscission in citrus leaves. BMC Plant Biol. 9, 1–20 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Z., Zhao K., Ai X. Y. & Hu C. G. Involvements of PCD and changes in gene expression profile during self-pruning of spring shoots in sweet orange (Citrus sinensis). BMC Genomics 15, 1–16 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botton A. et al. Signaling pathways mediating the induction of apple fruitlet abscission. Plant Physiol. 155, 185–208 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H. et al. Transcriptomics of shading-induced and NAA-induced abscission in apple (Malus domestica) reveals a shared pathway involving reduced photosynthesis, alterations in carbohydrate transport and signaling and hormone crosstalk. BMC Plant Biol. 11, 1–20 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilamado J. A. & Gomezjimenez M. C. Transcriptome analysis of mature fruit abscission control in olive. Plant & Cell Physiol. 54, 244–269 (2013). [DOI] [PubMed] [Google Scholar]
- Tadeo F. R. et al. “To fall or not to fall, that’s the question!” molecular mechanisms underlying organ abscission in citrus. Acta Hortic. 1189–1195 (2015). [Google Scholar]
- Clough S. J. & Bent A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998). [DOI] [PubMed] [Google Scholar]
- Sundaresan S. et al. Abscission of flowers and floral organs is closely associated with alkalization of the cytosol in abscission zone cells. J. Exp. Bot. 66, 913–919 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder F., Lisso J., Lange P. & Mussig C. The extracellular EXO protein mediates cell expansion in Arabidopsis leaves. BMC Plant Biol. 9, 1–12 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delk N. A., Johnson K. A., Chowdhury N. I. & Braam J. CML24, regulated in expression by diverse stimuli, encodes a potential Ca2+ sensor that functions in responses to abscisic acid, daylength, and ion stress. Plant Physiol. 139, 240–253 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K. M. Ethylene and abscission. Physiol. Plantarum 100, 567–576 (1997). [Google Scholar]
- Kuang J. F. et al. Carbohydrate stress affecting fruitlet abscission and expression of genes related to auxin signal transduction pathway in litchi. Int. J. Mol. Sci. 13, 16084–16103 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S. D., Cho Y. H. & Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572 (2007). [DOI] [PubMed] [Google Scholar]
- Zhong H. Y. et al. Selection of reliable reference genes for expression studies by reverse transcription quantitative real-time PCR in litchi under different experimental conditions. Plant Cell Rep. 30, 641–653 (2011). [DOI] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.