Abstract
The JNK and P38α pathways play an important role in the sensitivity and outcomes of chemotherapy. We hypothesize that functional single nucleotide polymorphisms (SNPs) of genes of these pathways modulate outcomes of patients with advanced non–small cell lung cancer (NSCLC) treated with first-line platinum-based chemotherapy (PBC). We selectively genotyped 11 independent, potentially functional SNPs of 9 genes in the JNK and P38α pathways first in a discovery group of 355 patients with advanced NSCLC treated with PBC, and we evaluated their associations with progression-free survival (PFS) and overall survival (OS) by Cox proportional hazards regression analysis. Then, resultant significant SNPs were further validated in a replication group of 355 patients. In both discovery and validation groups as well as their combined analysis, the MAPK14 rs3804451GA/AA genotypes showed a strong association with a reduced PFS (adjusted hazards ratio [HR] = 1.39; 95% confidence interval [CI] = 1.16–1.66; P = .0003) and OS (adjusted HR = 1.41; 95% CI = 1.11-1.80; P = .005) compared with the wild-type GG genotype. In contrast, patients with or without the MAPK14 rs3804451A allele had no significant difference in OS in response to tyrosine-kinase inhibitor treatment (adjusted HR = 0.86; 95% CI = 0.56-1.33; P = .505). The present study provides evidence that the MAPK14 rs3804451 G>A variant may modulate survival outcomes in patients with advanced NSCLC treated with PBC. Larger studies of additional patient populations are needed to validate our findings.
Introduction
Non–small cell lung cancer (NSCLC), accounting for more than 80% of all lung cancer cases, is one of the most fatal human malignancies [1], [2]. In particular, a large number of patients are diagnosed at an advanced stage (i.e., stage III or IV) [3] because there were no obvious symptoms that could draw medical attention at their early stages [4]. Platinum-based (cisplatin or carboplatin) double-agent chemotherapies are widely used as the standard first-line treatment for patients with advanced NSCLC without sensitive EGFR mutations (NCCN guidelines, http://www.nccn.org/professionals/), which have led to improved survival outcomes. However, the efficacy of different combinations is not satisfying because the response rate of most clinical trials has been no more than 40% [5].
The main action site of platinum compounds is thought to be DNA, causing inter- and intrastrand cross-links that lead to damages of DNA, activation of apoptosis signaling pathways, and death of cancer cells [6]. Nonetheless, a significant fraction of lung cancer patients bears neoplastic lesions that are intrinsically resistant to genocytotoxic effects of platinum compounds [7]. This suggests that intrinsic resistance to platinum-based chemotherapy (PBC) is an important limiting factor in the treatment of NSCLC. As a matter of fact, numerous clinical studies have elucidated that genetic factors may be associated with the response and outcomes of PBC [8], [9], [10]. Thus, interindividual genetic variation may play an important role in determining outcomes of NSCLC patients treated with PBC [11].
Stress-activated MAPKs, such as P38 MAPK and JNK, are well known for their effects on controlling apoptosis and autophagic death after genotoxic stresses [12], and thus, they seem to play an important role in the sensitivity and outcomes of PBC [13]. Several studies have shown that the posttarget cisplatin resistance was associated with defects in several apoptotic signal transducers, including JNK and P38 pathways [7]. In cisplatin-resistant NSCLC cell lines, for example, functional inhibition of the JNK/ATF2 pathway could restore the sensitivity to cisplatin [14], but the upregulating of P38α MAPK might promote chemoresistance in lung adenocarcinoma cells [15]. These results indicate that the activation of the JNK and P38α pathways probably affect the outcomes of NSCLC patients treated with PBC. Previous studies also reported that genetic variants in genes involved in DNA repair and apoptosis were associated with survival of NSCLC patients treated by PBC [11], [16], [17], [18]. Currently, there is still a lack of studies on associations between genetic variants in genes of the JNK and P38α pathways and chemotherapy outcome. Therefore, we performed a retrospective two-stage analysis in a cohort of patients with advanced NSCLC treated with PBC to assess the effects of genetic variants of genes in JNK and P38α pathways on the response to PBC and survival outcomes in these patients.
Materials and Methods
Study Subjects and Follow-Up
In the present study, we recruited patients with histologically confirmed advanced NSCLC from our institution. The discovery group consisted of 355 NSCLC patients recruited between February 2009 and February 2012, whereas the replication group included 355 NSCLC patients recruited between March 2012 and November 2013. All participants were genetically unrelated ethnic Han Chinese, and none had blood transfusions in the last 6 months. Recruitment criteria were as follows: inoperable TNM stages III to IV tumors; no prior history of cancer except for an in situ carcinoma; receiving PBC as the first-line treatment; Eastern Cooperative Oncology Group (ECOG) performance status 0 to 2; required laboratory values for blood tests and uronoscopy in normal range and no active infection or serious medical (e.g., recent myocardial infarction, active congestive heart failure, cardiac arrhythmia, or cerebral apoplexy, etc.) or psychological (e.g., crankiness or depression etc.) factors that might prevent patients from adherence to treatment schedule.
Demographic and other clinical data (i.e., age at treatment, sex, smoking history, ECOG performance status, TNM stage, histologic grade, histological type, chemotherapy regimens, radiotherapy, tyrosine-kinase inhibitor [TKI] treatment and grade 3/4 overall chemotherapy toxicity [including hematological toxicity and gastrointestinal toxicity]) were collected from patients' medical records. The initial chemotherapy response was evaluated after the second cycle of chemotherapy by computed tomography and magnetic resonance imaging scans using the Response Evaluation Criteria in Solid Tumor guidelines [19]. In the present study, progressive disease (PD) was defined as drug resistance, whereas complete response (CR), partial response (PR), and stable disease (SD) were identified as non–drug resistance.
Survival data were collected from patients' next of kin through a telephone follow-up and inpatient and outpatient clinical medical records. Overall survival (OS) time was calculated from the date when patients first received chemotherapy until the date of the last follow-up or death. Progression-free survival (PFS) time was calculated from the date when patients first received chemotherapy until the date of progression or the last follow-up or death. The median follow-up period was 32.1 months. The present study was approved by our Institutional Review Board, and all participants provided a written informed consent.
Chemotherapy Regimens
All patients enrolled in the study were given the first-line platinum-based chemotherapy, that is, cisplatin (75 mg/m2) or carboplatin (area under the curve [AUC] 6 mg/ml·min), administered on day 1 every 3 weeks, in combination with paclitaxel (175 mg/m2) on day 1 every 3 weeks, docetaxel (75 mg/m2) on day 1 every 3 weeks, gemcitabine (1250 mg/m2) on days 1 and 8 every 3 weeks, pemetrexed (500 mg/m2) on day 1 every 3 weeks or vinorelbine (25 mg/m2) on days 1 and 8 every 3 weeks, and cisplatin (100 mg/m2) or carboplatin (AUC 6 mg/ml·min) administered on day 1 every 4 weeks, in combination with etoposide (100 mg/m2) on days 1 to 3 every 4 weeks. All chemotherapeutic drugs were administered intravenously.
Single Nucleotide Polymorphism (SNP) Selection and Genotyping
All blood samples were collected from the tissue bank of our institution. Genomic DNA was extracted from blood samples of all the study subjects by using the DNA Blood Mini Kit (Qiagen, Valencia, CA). Preselected 11 independent, potentially functional SNPs of 9 key genes (GADD45A, GADD45B, GADD45G, MAP2K7, MAP2K4, MAP3K4, MAPK8, MAPK9, and MAPK14) [20], [21] involved in the JNK and P38α pathways were genotyped by using the TaqMan allelic discrimination assay on the ABI7900 HT sequence detector system (Applied Biosystems, Foster City, CA) [22]. More detailed information is provided in Supplemental Table S1. We further randomly selected 10% samples for each of the selected SNPs for regenotyping, and the results were 100% concordant.
Tissue Preparation, and RNA and DNA Extraction
Non–small cell lung cancer tumor tissues were dissected and evaluated by two pathologists after surgical removal, and the leftover tumors tissues were transferred into liquid nitrogen immediately after resection and stored at −80°C until use at the Department of Pathology and Tissue Bank. For the present study, 170 tumor tissue samples were randomly selected for mRNA expression analysis. Both RNA and DNA were extracted from tumor tissue samples with the TRIzol reagent (Invitrogen, Carlsbad, CA) and the DNA kit (Tiangen, Beijing, China). RNA samples were used to synthesize complementary DNA with the PrimeScript RT Master Mix system (Takara, Otsu, Japan), as previously described [23]. The DNA samples were used for genotyping as mentioned above.
Real-Time Polymerase Chain Reaction (PCR)
The samples of total RNA from the 170 target tissues were used for the real-time reverse transcriptase PCR to detect mRNA expression, which was performed using the ABI-Prism 7900 sequence detection system (Applied Biosystems, Foster City, CA) as previously described [23], [24]. The primers used for the real-time reverse transcriptase PCR were 5′-AACCTGTCTCCAGTGGGCTCT-3′ and 5′-CGTAACCCCGTTTTTGTGTCA-3′ for MPAK14[25] and 5′-GAACGGGAAGCTCACTGG-3′ and 5′-GCCTGCTTCACCACCTTCT-3′ for GAPDH[26].
Statistical Analysis
To minimize false-positive report probability [27] and to show the internal consistency, a two-stage analysis was performed to investigate associations between the selected SNPs and PFS/OS. The association between each genetic variant and PFS/OS was estimated by using the Cox proportional hazards regression models with adjustment for confounders and depicted by the Kaplan-Meier method. All 11 SNPs were first evaluated in the discovery group, and those SNPs that had a P value <.05 by either the trend or genotype tests for associations with PFS/OS in Cox regression models were further validated in the replication group. Finally, SNPs that met the same criteria in the replication group were further subjected to the combined analyses for all subjects. The observed associations were also evaluated in subgroup analyses, stratified by selected demographic and clinical variables. The heterogeneity between subgroups was assessed with the χ2-based Q test. Multiplicative interactions were assessed by Cox regression models. Associations between SNPs and chemotherapy response were also estimated by comparing individuals with drug resistance to those without in unconditional logistic regression models. Predictive values of both the selected variables only and their combination with SNPs were evaluated by incident/dynamic (I/D) AUC of the receiver-operator characteristic (ROC) curves for censored data and concordance index (C index) for comparison of survival models [28]. Variables in the predictive model were selected using a backward-stepwise Cox regression process with a maximum iteration of 20 and an entry possibility of .05 as well as an excluded possibility of .10 at each step. The I/D ROC and I/D AUC were calculated and plotted by using risksetROC package [29] of R software (version 3.2.3;The R Foundation for Statistical Computing), whereas 95% confidence intervals (CIs) of C index and AUCs of time-dependent ROC curves were calculated by using the bootstrap method with 1000 replications. Linear regression analysis and Student's t tests were applied to the comparison of mRNA levels between samples by genotypes. To control for the multiple testing, the conservative Bonferroni correction was used. Unless otherwise specified, all tests were two-sided by using the SAS software (version 9.3; SAS Institute, Cary, NC) and GraphPad Prism 5 (GraphPad Software, Inc., San Diego, CA). P < .05 was considered statistically significant.
Results
Characteristics of the Study Population
The distributions of demographic variables and clinical information are described in Supplemental Table S2. Overall, 710 patients were included in the analysis, among which 508 were males and 202 were females. The median age was 58 (23-83) years, 334 (47.0%) were nonsmokers, 41 (5.8%) were former smokers, and 335 (47.2%) were current smokers. The chemotherapy regimen consisted of docetaxel/paclitaxel, etoposide, gemcitabine, vinorelbine, pemetrexed, and cisplatin or carboplatin. A total of 30.8% of patients received palliative radiotherapy, and 36.2% received TKI treatment.
PFS and OS by Clinical Characteristics of Patients with NSCLC
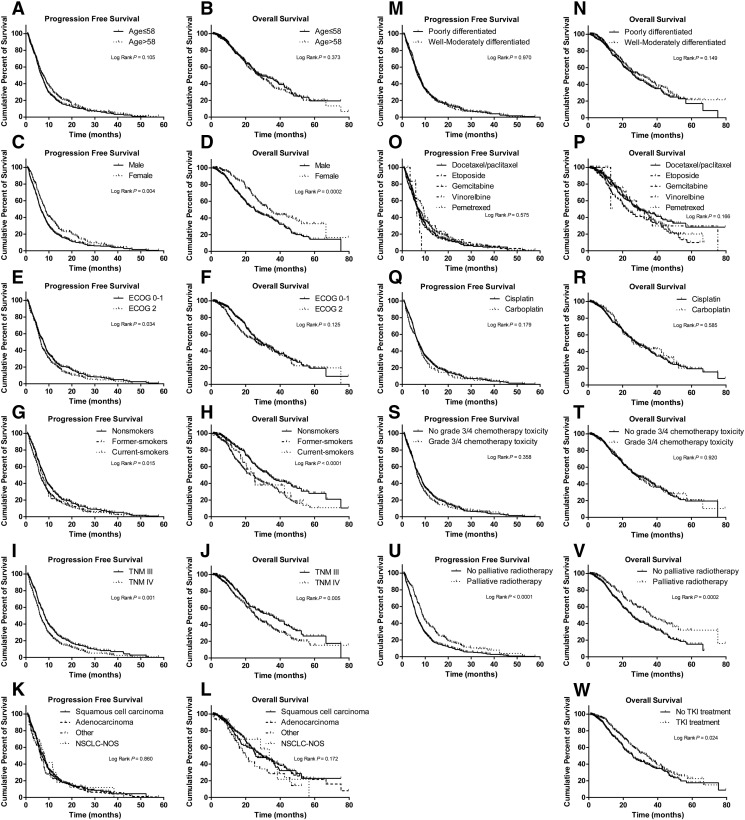
Supplemental Table S3 and Supplemental Figure S1 show the associations between clinical characteristics and patients' survival. The TKI treatment was used only after the patients presented disease progression; therefore, it was excluded from the confounding factors of PFS. In the univariate Cox regression model, sex (Supplemental Figure S1, C and D), smoking status (Supplemental Figure S1, G and H), TNM stages (Supplemental Figure S1, I and J), and palliative radiotherapy (Supplemental Figure S1, U and V) were significant for PFS and OS. The ECOG performance status (0-1 vs 2; HR = 1.19; 95% CI = 1.01-1.40; P = .035) (Supplemental Figure S1E) showed a significant association with PFS, and it also showed a similar tendency toward significance for OS (0-1 vs 2; HR = 1.19; 95% CI = 0.95-1.48; P = .035) (Supplemental Figure S1F). Other possible confounding factors such as different chemotherapy regimens (docetaxel/paclitaxel, etoposide, gemcitabine, vinorelbine, and pemetrexed) (Supplemental Figure S1O) did not show significance associations with PFS (log-rank P = .575), but patients treated with gemcitabine seemed to have a shorter OS (HR = 1.46; 95% CI = 1.08-1.96; P = .014) when compared with those treated with docetaxel/paclitaxel (Supplemental Figure S1P). In addition, TKI treatment was associated with significantly increased OS of patients (25.5 vs 33.7 months; HR = 0.77; 95% CI = 0.62-0.97; P = .024) (Supplemental Figure S1W). However, to control for any possible confounding on the main effects of the studied SNPs on OS, age at treatment, sex, TNM stage, smoking status, histological type, histologic grade, ECOG performance status, chemotherapy regimens, grade 3/4 chemotherapy toxicity, palliative radiotherapy, and TKI treatment were all further adjusted in the subsequent multivariate Cox regression analysis.
Supplementary Figure S1.
Selected SNPs and NSCLC Survival
The distributions of genotypes of all the investigated 11 SNPs in the discovery group were in accordance with Hardy-Weinberg equilibrium (P > .05), and the observed minor allele frequency of each SNP was consistent with that previously reported in HapMap database. In the discovery group, among the SNPs evaluated, four SNPs (GADD45A rs581000, MAP2K7 rs3679, and MAPK14 rs3804451) and four SNPs (GADD45A rs581000, MAP3K4 rs1488, MAP3K4 rs678290 and MAPK14 rs3804451) were associated with PFS and OS, respectively (P < .05 by the trend test or genotype test in Cox proportional hazards regression models) (Supplemental Tables S4 and 5); however, only MAPK14 rs3804451 was found to be associated with a shorter PFS and OS by using the criteria previously described in the replication group and subjected to the combined analysis (Table 1).
Table 1.
Associations of MAPK14 rs3804451G>A with PFS and OS in Patients with Advanced NSCLC in a Chinese Population
| PFS |
OS |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients | MAPK14 SNP rs3804451 | PHWE⁎ | Event/No. | MST (mo) | P†, ⁎ | Adjusted HR‡, † (95% CI) | P‡, † | Event/No. | MST (mo) | P†, ⁎ | Adjusted HR§, ‡ (95% CI) | P§, ‡ |
| Discovery group | .594 | .014¶ | .0003¶, # | |||||||||
| GG | 212/254 | 7.2 | .067 | 1.00 (ref.) | 119/254 | 35.2 | .053 | 1.00 (ref.) | ||||
| GA | 85/91 | 6.2 | 1.26 (0.97-1.65) | .080 | 53/91 | 25.3 | 1.71 (1.19-2.43) | .003#, ¶ | ||||
| AA | 8/10 | 4.1 | 2.29 (1.10-4.76) | .027 | 5/10 | 10.8 | 3.72 (1.41-9.78) | .008 | ||||
| GA/AA | 93/101 | 6.1 | .032 | 1.32 (1.02-1.70) | .034 | 58/101 | 25.3 | .023 | 1.79 (1.27-2.53) | .001#, ¶ | ||
| Replication group | .422 | .001¶, # | .049¶ | |||||||||
| GG | 210/258 | 7 | .022 | 1.00 (ref.) | 111/258 | 26.8 | .434 | 1.00 (ref.) | ||||
| GA | 79/87 | 7.7 | 1.45 (1.10-1.91) | .008 | 38/87 | 23.5 | 1.28 (0.86-1.9) | .232 | ||||
| AA | 10/10 | 3.8 | 2.30 (1.16-4.55) | .017 | 6/10 | 24.9 | 2.55 (0.99-6.6) | .053 | ||||
| GA/AA | 89/97 | 6.6 | .008 | 1.51 (1.16-1.97) | .002#, ¶ | 44/97 | 23.5 | .483 | 1.37 (0.94-2.00) | .104 | ||
| All combined | .346 | <.0001¶, # | .001¶, # | |||||||||
| GG | 422/512 | 7.2 | .002 | 1.00 (ref.) | 230/512 | 32.1 | .034 | 1.00 (ref.) | ||||
| GA | 164/178 | 6.6 | 1.34 (1.12-1.62) | .002#, ¶ | 91/178 | 24.4 | 1.34 (1.04-1.72) | .024 | ||||
| AA | 18/20 | 3.4 | 2.07 (1.27-3.35) | .003#, ¶ | 11/20 | 19.8 | 2.67 (1.42-5.00) | .002#, ¶ | ||||
| GA/AA | 182/198 | 6.5 | .001 | 1.39 (1.16-1.66) | .0003#, ¶ | 102/198 | 24.4 | .033 | 1.41 (1.11-1.80) | .005 | ||
The results were in bold if P < .05.
PHWE: P value for Hardy-Weinberg equilibrium test.
P: value for log-rank tests.
Data were calculated using Cox regression, adjusted by age at treatment, sex, smoking status, TNM stage, histological type, histologic grade, ECOG performance status, chemotherapy regimens, grade 3/4 chemotherapy toxicity, and palliative radiotherapy.
Data were calculated using Cox regression, adjusted by age at treatment, sex, smoking status, TNM stage, histological type, histologic grade, ECOG performance status, chemotherapy regimens, grade 3/4 chemotherapy toxicity, palliative radiotherapy, and TKI treatment.
Ptrend: P value for trend.
Significance remained after Bonferroni correction.
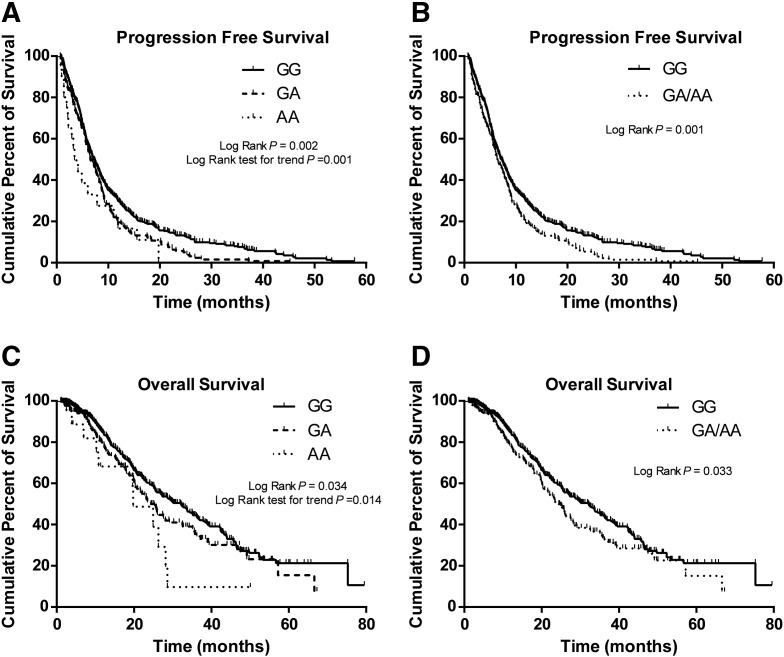
In the combined analysis of all subjects, the MAPK14 rs3804451 A allele was found to be significantly associated with poor PFS and OS in NSCLC patients. Patients with rs3804451GA/AA genotypes showed a reduced PFS compared with the wild-type GG homozygotes (6.5 vs 7.2 months; log-rank P = .001; HR = 1.39; 95% CI = 1.16-1.66; adjusted P = .0003) (Figure 1B). The median survival time (MST) for patients with rs3804451GA/AA genotypes (MST =24.4 months) was shorter than that for those with the GG genotype (32.1 months; log-rank P = .033; HR = 1.41; 95% CI = 1.11-1.80; adjusted P = .005) (Figure 1D), and the significance remained after Bonferroni correction (Table 1).
Figure 1.
Kaplan-Meier curves of PFS and OS by the MAPK14 rs3804451 genotype in all patients combined. PFS in an additive genetic model (A) and a dominant genetic model (B) and OS in an additive genetic model (C) and a dominant genetic model (D).
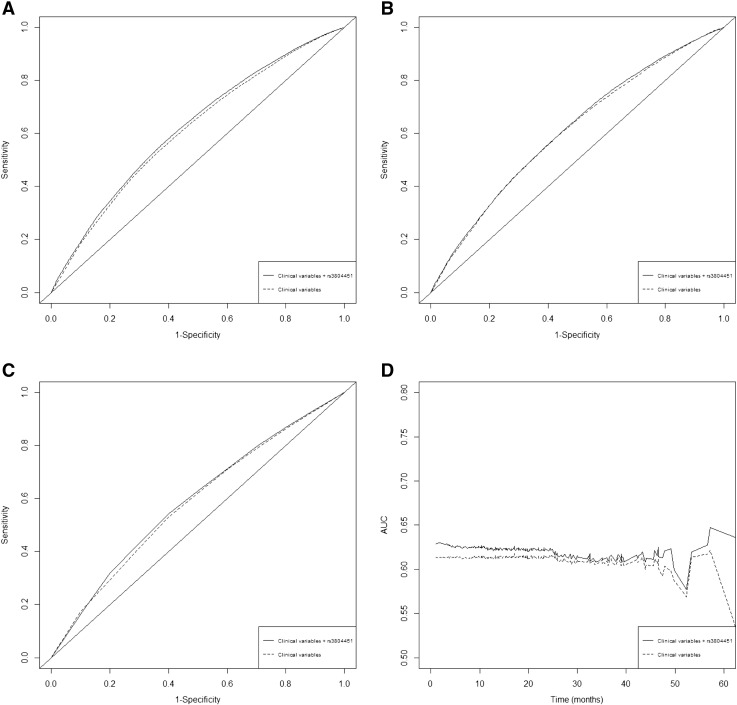
We compared I/D AUC of the time-dependent ROC curves and estimated C index to assess the discriminative accuracy of the OS prediction model by removing the rs3804451 SNP from the model as selected by a backward-stepwise Cox regression process. The rank of the I/D AUC prediction model including all selected variables (i.e., rs3804451, sex, smoking status, TNM stage, palliative radiotherapy, and TKI treatment) was higher than the model that only included clinical variables at 1 (Figure 2A), 3 (Figure 2B), and 5 (Figure 2C) years for OS (Supplemental Table S6), whereas the C index also decreased from 0.621 (95% CI = 0.586-0.646) to 0.612 (95% CI = 0.580-0.635) (Figure 2D), although the difference was not statistically significant (P = .321) (Supplemental Table S6).
Figure 2.
Time-dependent ROC curves for the prediction of 1-year (A), 3-year (B), and 5-year OS rate (C) based on only selected variables (sex, smoking status, TNM stages, palliative radiotherapy, and TKI treatment) and the combined rs3804451 genotypes along with clinical variables; incident/dynamic AUC plots for the present study data (D) based on only selected variables and combined variables.
The rs3804451 SNP and Chemotherapy Response
The association between the MAPK14 rs3804451 SNP and chemotherapy responses is summarized in Supplemental Table S7. Compared with the rs3804451GG genotype, variant GA/AA genotypes were associated with a higher risk of PBC resistance (GA/AA versus GG; adjusted OR =1.91; 95% CI = 1.24-2.93; P = .003), assuming a dominant effect of the A allele (Supplemental Table S7).
Stratified Analysis of the Associations of MAPK14 rs3804451 with PFS and OS
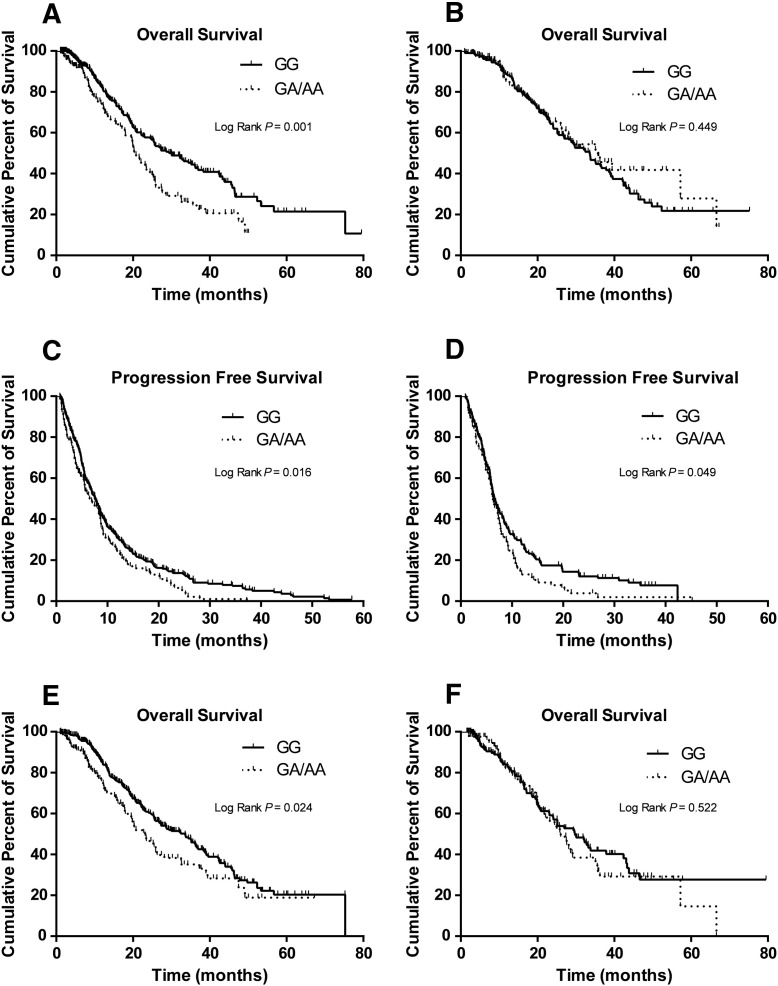
In stratification analyses, by assuming a dominant genetic model, the effect of the rs3804451A allele was still statistically significant in most of the subgroups, except for those subgroups with a small number of subjects; however, further homogeneity tests did not support any evidence for differences between the strata (Supplemental Table S8). Similarly, rs3804451GA/AA carriers also had a shorter OS in most of the subgroups (Supplemental Table S8). There was a difference in HRs between subgroups with or without the TKI treatment for MAPK14 rs3804451 (P = .001) (Table 2) by the homogeneity test, as shown in Kaplan-Meier survival curves (Figure 3, A and B), which were consistent with the results from the Cox regression analysis of interaction (P = .014) between rs3804451 and TKI treatment with adjustment for age at treatment, sex, smoking status, TNM stage, histological type, histologic grade, ECOG performance status, chemotherapy regimens, grade 3/4 chemotherapy toxicity, and palliative radiotherapy.
Table 2.
Associations of MAPK14 rs3804451G>A with OS in Advanced NSCLS Patients with or without TKI Treatment
| rs3804451 (Event/No.) |
OS |
||||
|---|---|---|---|---|---|
| Variables | GG | GA/AA | Adjusted HR⁎(95% CI) | P⁎ | Phom |
| TKI treatment | |||||
| No | 136/323 | 72/130 | 1.84 (1.36-2.48) | <.0001 | |
| Yes | 94/189 | 30/68 | 0.86 (0.56-1.33) | .505 | .001 |
Abbreviation: hom, heterogeneity test. The results were in bold if P < .05.
Adjusted by age at treatment, sex, smoking status, TNM stage, histological type, histologic grade, ECOG performance status, chemotherapy regimens, grade 3/4 chemotherapy toxicity, and palliative radiotherapy.
Figure 3.
Kaplan-Meier curves of OS by the dichotomized rs3804451GA/AA genotypes for patients without (A) and with (B) TKI; Kaplan-Meier curves of PFS by the dichotomized rs3804451GA/AA genotypes for patients without (C) and with (D) grade 3/4 chemotherapy toxicity; Kaplan-Meier curves of OS by the dichotomized rs3804451GA/AA genotypes for patients without (E) and with (F) grade 3/4 chemotherapy toxicity.
Our previous study demonstrated that carriers of the MAPK14 rs3804451A variant allele had a significantly higher risk for grade 3/4 overall chemotherapy toxicity [22]; nevertheless, the effects of rs3804451GA/AA genotypes on PFS (Figure 3, C and D) and OS (Figure 3, E and F) between different toxicity subgroups showed no difference in the present study (P = .676 and .090, respectively) (Supplemental Table S8), suggesting that there was a direct effect of the A allele on PFS and OS, which was not mediated by the toxicity.
Correlation between MAPK14 Genotypes and mRNA Expression Levels
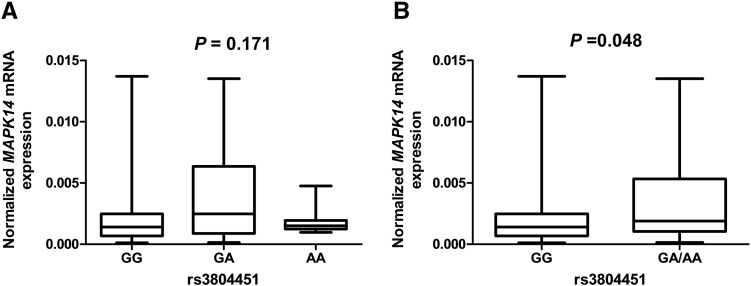
Finally, we used experimental data of MAPK14 mRNA expression levels and SNP genotypes of the target tumor tissues from 170 unrelated Chinese NSCLC patients to evaluate the effect of the rs3804451A allele on the mRNA expression. There were 108 GG carriers, 55 GA carriers, and 7 AA carriers (Figure 4A). MAPK14 mRNA expression levels of tumor tissues with rs3804451 GA/AA genotypes were higher than those with the GG genotype (P = .048) (Figure 4B).
Figure 4.
mRNA expression levels by MAPK14 rs3804451 genotypes in tumor tissues of 170 patients. (A) An additive genetic model for rs3804451 (108, 55, and 7 for GG, GA, and AA, respectively; P = .171); (B) a dominant genetic model for rs3804451 (108 and 62 for GG and GA/AA, respectively; P = .048).
Discussion
In the present study, we investigated, in a two-step study design, the associations of potentially functional 11 SNPs of 9 genes in the JNK and P38α pathways with PBC response and survival outcomes in patients with advanced NSCLC treated by PBC in a Chinese population. The MAPK14 rs3804451A allele was found to be significantly associated with PBC resistance and a shorter PFS and OS of the patients. We also observed that MAPK14 mRNA expression levels of tumor tissues with rs3804451 GA/AA genotypes were higher than those with the GG genotype, which provides additional biological support for the observed associations with survival in response to PBC in the patients.
PBC is still the first-line treatment for most patients with advanced NSCLC, although target therapy and immunotherapy are recently developed and used to treat NSCLC patients. However, platinum-based double-agent chemotherapy has a low response rate but a high toxicity occurrence rate. Its main effect is thought to enhance cell apoptosis and autophagy [6], [30]. Many studies have focused on the roles of JNK and P38α pathways in the response of tumors to chemotherapeutic agents, such as platinum compounds. It has been shown that cisplatin sensitivity depends on a long-term cisplatin-induced activation of JNK and P38α pathways [31], [32]. If the P38a pathway of tumor cells is constitutively activated under normal conditions, tumor cells will attenuate to further activate P38 MAPK, which is essential to induce cell apoptosis in response to cisplatin [33]. Indeed, it has been shown that cancer cell lines with high baseline levels of the P38 MAPK activity tend to be more resistant to cisplatin [34].
The rs3804451 SNP is located on the 3′ untranslated region of MAPK14 (P38a) and likely to be functional as predicted by SNPInfo [22]. Indeed, in the present study, we showed that the high expression levels of P38a in tumor tissues were associated with the A allele. In fact, studies have shown that baseline hyperactivation of the P38a pathway exists in lung cancer samples [35] and is correlated with chemoresistance [33]. The fact that patients with the rs3804451A variant allele have a significantly increased P38a mRNA expression levels provides a biological plausibility that rs3804451A variant carriers may tend to be more resistant to PBC than patients without the A allele. However, this finding needs to be tested through additional in vivo and in vitro functional studies.
In the stratification analysis, the effect of the rs3804451A allele was still statistically significant in most of the subgroups. Because our previous study suggested that carriers of MAPK14 rs3804451GA/AA genotypes had a significantly higher risk for grade 3/4 overall chemotherapy toxicity [22], it is likely that the rs3804451A allele may reduce survival time by increasing chemotherapy toxicity. However, further homogeneity tests between subgroups with or without grade 3/4 overall chemotherapy toxicity did not appear to support any difference between these two subgroups. This may suggest that the rs3804451 SNP is associated with toxicity of normal tissue and sensitivity of tumor tissue in response to PBC. Interestingly, we found that there was a significant difference in HRs between subgroups with or without the TKI treatment, likely because the rs3804451 may only modulate the chemotherapy response of tumor tissues; as the TKI treatment significantly prolongs survival time of patients with sensitive EGFR mutations, the effect of the rs3804451A allele in these patients may be greatly masked by the TKI treatment. This may also explain why there was no significant increase in the efficacy of the OS prediction model when adding the rs3804451 SNP to the same model including other selected variables. Nevertheless, these results need to be validated in future studies with large sample sizes.
In the context of cancer, mechanisms of the resistance to platinum compounds are complex [7], [36] because tumors may have disease-specific mutations and carry germline variants that affect drugs' pharmacokinetics and pharmacodynamics, which will collectively affect drug response [37]. Therefore, it would lead to bias if we performed the chemotherapy resistance risk assessment or prognosis prediction with either genetic or clinical factors, let alone a single SNP from one gene. However, a tentative predictive model with an increased prediction accuracy by adding both genetic and clinical factors with known significant associations with chemotherapy sensitivity has been established [38]. There is room for additional improvement by identifying other important genomic information (including somatic mutations, germline variants, epigenetic factors, etc.) of tumor tissues in response to PBC, which will lead to a better predictive model to distinguish patients who tend to benefit from PBC from those who do not.
However, the present study has some limitations. First, all patients were treated at the same hospital and may have selection bias and information bias. Studies from other patient populations are needed to validate our findings. Also, because of the retrospective nature of the original study design, EGFR mutation information of the patients only treated by chemotherapy was largely missing, which may influence patients' survival. Finally, this study was only performed in a Chinese patient population, and the results may not be generalizable to the general populations or other ethnic groups. Therefore, well-designed, large studies in other ethnic populations with advanced-NSCLC patients are necessary to validate our findings.
Conclusions
The present study provides evidence that the MAPK14 rs3804451 G>A SNP may modulate PBC-related survival outcomes in Chinese patients with advanced NSCLC. Such information about genetic factors could be used to guide the selection of proper treatment in the future, if validated in large-scale, perspective studies.
The following are the supplementary data related to this article.
Supplementary Tables S1–8
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by funding from China Recruitment Program of Global Experts at Fudan University, Shanghai, China.
Acknowledgements
We thank Dr. Haiquan Chen of the Department of Thoracic Surgery for contributing to the tissue collection, the staff members of the Tissue Bank for their continued support in providing the stored blood samples and tissue samples, and the hospital informatics office for collecting patients' clinical information.
Footnotes
Funding: This work was supported by funding from China Recruitment Program of Global Experts at Fudan University, Shanghai, China.
Contributor Information
Ming Jia, Email: 14111230007@fudan.edu.cn.
Yuan Xu, Email: 14111230023@fudan.edu.cn.
Meiling Zhu, Email: anniezhu_79@163.com.
Mengyun Wang, Email: wangmengyun1120@163.com.
Menghong Sun, Email: menghongs@163.com.
Ji Qian, Email: qianji2012@126.com.
Jianhua Chang, Email: changjianhua@163.com.
Qingyi Wei, Email: weiqingyi@yahoo.com, qingyi.wei@duke.edu.
References
- 1.Bender E. Epidemiology: the dominant malignancy. Nature. 2014;513:S2–S3. doi: 10.1038/513S2a. [DOI] [PubMed] [Google Scholar]
- 2.Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F., Jemal A., Yu X.Q., He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016 doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Zell J.A., Ignatius Ou S.H., Ziogas A., Anton-Culver H. Validation of the proposed International Association for the Study of Lung Cancer non–small cell lung cancer staging system revisions for advanced bronchioloalveolar carcinoma using data from the California Cancer Registry. J Thorac Oncol. 2007;2:1078–1085. doi: 10.1097/JTO.0b013e31815ba260. [DOI] [PubMed] [Google Scholar]
- 4.Wistar-led research team discovers genetic pattern that indicates early stage lung cancerCancer Biol Ther. 2009;8:iii–iiv. doi: 10.4161/cbt.8.24.11004. [DOI] [PubMed] [Google Scholar]
- 5.Yang Z.Y., Liu L., Mao C., XY W., Huang Y.F., XF H., Tang J.L. Chemotherapy with cetuximab versus chemotherapy alone for chemotherapy-naive advanced non–small cell lung cancer. Cochrane Database Syst Rev. 2014;11:CD009948. doi: 10.1002/14651858.CD009948.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Danesi R., de Braud F., Fogli S., de Pas T.M., Di Paolo A., Curigliano G., Del Tacca M. Pharmacogenetics of anticancer drug sensitivity in non–small cell lung cancer. Pharmacol Rev. 2003;55:57–103. doi: 10.1124/pr.55.1.4. [DOI] [PubMed] [Google Scholar]
- 7.Galluzzi L., Vitale I., Michels J., Brenner C., Szabadkai G., Harel-Bellan A., Castedo M., Kroemer G. Systems biology of cisplatin resistance: past, present and future. Cell Death Dis. 2014;5:e1257. doi: 10.1038/cddis.2013.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu L., Wu C., Zhao X., Heist R., Su L., Zhao Y., Han B., Cao S., Chu M., Dai J. Genome-wide association study of prognosis in advanced non–small cell lung cancer patients receiving platinum-based chemotherapy. Clin Cancer Res. 2012;18:5507–5514. doi: 10.1158/1078-0432.CCR-12-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan X.L., Moyer A.M., Fridley B.L., Schaid D.J., Niu N., Batzler A.J., Jenkins G.D., Abo R.P., Li L., Cunningham J.M. Genetic variation predicting cisplatin cytotoxicity associated with overall survival in lung cancer patients receiving platinum-based chemotherapy. Clin Cancer Res. 2011;17:5801–5811. doi: 10.1158/1078-0432.CCR-11-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu C., Xu B., Yuan P., Ott J., Guan Y., Liu Y., Liu Z., Shen Y., Yu D., Lin D. Genome-wide examination of genetic variants associated with response to platinum-based chemotherapy in patients with small-cell lung cancer. Pharmacogenet Genomics. 2010;20:389–395. doi: 10.1097/FPC.0b013e32833a6890. [DOI] [PubMed] [Google Scholar]
- 11.Han B., Gao G., Wu W., Gao Z., Zhao X., Li L., Qiao R., Chen H., Wei Q., Wu J. Association of ABCC2 polymorphisms with platinum-based chemotherapy response and severe toxicity in non–small cell lung cancer patients. Lung Cancer. 2011;72:238–243. doi: 10.1016/j.lungcan.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 12.Chen Y.R., Wang X., Templeton D., Davis R.J., Tan T.H. The role of c-Jun N-terminal kinase (JNK) in apoptosis induced by ultraviolet C and gamma radiation. Duration of JNK activation may determine cell death and proliferation. J Biol Chem. 1996;271:31929–31936. doi: 10.1074/jbc.271.50.31929. [DOI] [PubMed] [Google Scholar]
- 13.Sui X., Kong N., Ye L., Han W., Zhou J., Zhang Q., He C., Pan H. p38 and JNK MAPK pathways control the balance of apoptosis and autophagy in response to chemotherapeutic agents. Cancer Lett. 2014;344:174–179. doi: 10.1016/j.canlet.2013.11.019. [DOI] [PubMed] [Google Scholar]
- 14.Lo Iacono M., Monica V., Vavala T., Gisabella M., Saviozzi S., Bracco E., Novello S., Papotti M., Scagliotti G.V. ATF2 contributes to cisplatin resistance in non–small cell lung cancer and celastrol induces cisplatin resensitization through inhibition of JNK/ATF2 pathway. Int J Cancer. 2015;136:2598–2609. doi: 10.1002/ijc.29302. [DOI] [PubMed] [Google Scholar]
- 15.Chung L.Y., Tang S.J., Sun G.H., Chou T.Y., Yeh T.S., SL Y., Sun K.H. Galectin-1 promotes lung cancer progression and chemoresistance by upregulating p38 MAPK, ERK, and cyclooxygenase-2. Clin Cancer Res. 2012;18:4037–4047. doi: 10.1158/1078-0432.CCR-11-3348. [DOI] [PubMed] [Google Scholar]
- 16.Qian J., Qu H.Q., Yang L., Yin M., Wang Q., Gu S., Wu Q., Zhao X., Wu W., Wu J. Association between CASP8 and CASP10 polymorphisms and toxicity outcomes with platinum-based chemotherapy in Chinese patients with non–small cell lung cancer. Oncologist. 2012;17:1551–1561. doi: 10.1634/theoncologist.2011-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rabik C.A., Dolan M.E. Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat Rev. 2007;33:9–23. doi: 10.1016/j.ctrv.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qian J., Liu H., Gu S., Wu Q., Zhao X., Wu W., Wang H., Wang J., Chen H., Zhang W. Genetic variants of the MDM2 gene are predictive of treatment-related toxicities and overall survival in patients with advanced NSCLC. Clin Lung Cancer. 2015;16:e37–e53. doi: 10.1016/j.cllc.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 19.Therasse P., Arbuck S.G., Eisenhauer E.A., Wanders J., Kaplan R.S., Rubinstein L., Verweij J., Van Glabbeke M., van Oosterom A.T., Christian M.C. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 20.Gupta M., Gupta S.K., Hoffman B., Liebermann D.A. Gadd45a and Gadd45b protect hematopoietic cells from UV-induced apoptosis via distinct signaling pathways, including p38 activation and JNK inhibition. J Biol Chem. 2006;281:17552–17558. doi: 10.1074/jbc.M600950200. [DOI] [PubMed] [Google Scholar]
- 21.Abell A.N., Granger D.A., Johnson G.L. MEKK4 stimulation of p38 and JNK activity is negatively regulated by GSK3beta. J Biol Chem. 2007;282:30476–30484. doi: 10.1074/jbc.M705783200. [DOI] [PubMed] [Google Scholar]
- 22.Jia M., Zhu M., Wang M., Sun M., Qian J., Ding F., Chang J., Wei Q. Genetic variants of GADD45A, GADD45B and MAPK14 predict platinum-based chemotherapy-induced toxicities in Chinese patients with non–small cell lung cancer. Oncotarget. 2016;7:25291–25303. doi: 10.18632/oncotarget.8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang M., Zhang R., He J., Qiu L., Li J., Wang Y., Sun M., Yang Y., Wang J., Yang J. Potentially functional variants of PLCE1 identified by GWASs contribute to gastric adenocarcinoma susceptibility in an Eastern Chinese population. PLoS One. 2012;7:e31932. doi: 10.1371/journal.pone.0031932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo H., Liu H., Wei J., Li Y., Yu H., Guan X., Li E.W., Li G., Sturgis E.M., Wei Q. Functional single nucleotide polymorphisms of the RASSF3 gene and susceptibility to squamous cell carcinoma of the head and neck. Eur J Cancer. 2014;50:582–592. doi: 10.1016/j.ejca.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hale K.K., Trollinger D., Rihanek M., Manthey C.L. Differential expression and activation of p38 mitogen-activated protein kinase alpha, beta, gamma, and delta in inflammatory cell lineages. J Immunol. 1999;162:4246–4252. [PubMed] [Google Scholar]
- 26.Tang B., Du J., Wang J., Tan G., Gao Z., Wang Z., Wang L. Alpinetin suppresses proliferation of human hepatoma cells by the activation of MKK7 and elevates sensitization to cis-diammined dichloridoplatium. Oncol Rep. 2012;27:1090–1096. doi: 10.3892/or.2011.1580. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Wacholder S., Chanock S., Garcia-Closas M., El Ghormli L., Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96:434–442. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanvetyanon T., Padhya T., McCaffrey J., Zhu W., Boulware D., Deconti R., Trotti A. Prognostic factors for survival after salvage reirradiation of head and neck cancer. J Clin Oncol. 2009;27:1983–1991. doi: 10.1200/JCO.2008.20.0691. [DOI] [PubMed] [Google Scholar]
- 29.Heagerty P.J., Zheng Y. Survival model predictive accuracy and ROC curves. Biometrics. 2005;61:92–105. doi: 10.1111/j.0006-341X.2005.030814.x. [DOI] [PubMed] [Google Scholar]
- 30.Siddik Z.H. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22:7265–7279. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- 31.Brozovic A., Fritz G., Christmann M., Zisowsky J., Jaehde U., Osmak M., Kaina B. Long-term activation of SAPK/JNK, p38 kinase and fas-L expression by cisplatin is attenuated in human carcinoma cells that acquired drug resistance. Int J Cancer. 2004;112:974–985. doi: 10.1002/ijc.20522. [DOI] [PubMed] [Google Scholar]
- 32.Mansouri A., Ridgway L.D., Korapati A.L., Zhang Q., Tian L., Wang Y., Siddik Z.H., Mills G.B., Claret F.X. Sustained activation of JNK/p38 MAPK pathways in response to cisplatin leads to Fas ligand induction and cell death in ovarian carcinoma cells. J Biol Chem. 2003;278:19245–19256. doi: 10.1074/jbc.M208134200. [DOI] [PubMed] [Google Scholar]
- 33.Galan-Moya E.M., de la Cruz-Morcillo M.A., LlanosValero M., Callejas-Valera J.L., Melgar-Rojas P., Hernadez Losa J., Salcedo M., Fernandez-Aramburo A., Ramon y Cajal S., Sanchez-Prieto R. Balance between MKK6 and MKK3 mediates p38 MAPK associated resistance to cisplatin in NSCLC. PLoS One. 2011;6:e28406. doi: 10.1371/journal.pone.0028406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pereira L., Igea A., Canovas B., Dolado I., Nebreda A.R. Inhibition of p38 MAPK sensitizes tumour cells to cisplatin-induced apoptosis mediated by reactive oxygen species and JNK. EMBO Mol Med. 2013;5:1759–1774. doi: 10.1002/emmm.201302732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greenberg A.K., Basu S., Hu J., Yie T.A., Tchou-Wong K.M., Rom W.N., Lee T.C. Selective p38 activation in human non–small cell lung cancer. Am J Respir Cell Mol Biol. 2002;26:558–564. doi: 10.1165/ajrcmb.26.5.4689. [DOI] [PubMed] [Google Scholar]
- 36.Galluzzi L., Senovilla L., Vitale I., Michels J., Martins I., Kepp O., Castedo M., Kroemer G. Molecular mechanisms of cisplatin resistance. Oncogene. 2012;31:1869–1883. doi: 10.1038/onc.2011.384. [DOI] [PubMed] [Google Scholar]
- 37.Wheeler H.E., Maitland M.L., Dolan M.E., Cox N.J., Ratain M.J. Cancer pharmacogenomics: strategies and challenges Nature reviews. Genetics. 2013;14:23–34. doi: 10.1038/nrg3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yin J.Y., Li X., Li X.P., Xiao L., Zheng W., Chen J., Mao C.X., Fang C., Cui J.J., Guo C.X. Prediction models for platinum-based chemotherapy response and toxicity in advanced NSCLC patients. Cancer Lett. 2016;377:65–73. doi: 10.1016/j.canlet.2016.04.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables S1–8