Abstract
Peroxisomal biogenesis disorders (PBD) are caused by mutations in PEX genes, and are typically diagnosed with biochemical testing in plasma followed by confirmatory testing. Here we report the unusual diagnostic path of a child homozygous for PEX1 p.G843D. The patient presented with sensorineural hearing loss, pigmentary retinopathy, and normal intellect. After testing for Usher syndrome was negative, he was found to have PBD through a research sequencing panel. When evaluating a patient with hearing loss and pigmentary retinopathy, mild PBD should be on the differential regardless of cognitive function.
Keywords: PEX1 p.G843D, Zellweger syndrome spectrum, Peroxisomal biogenesis disorders, Usher syndrome
1. Introduction
Peroxisomal biogenesis disorders (PBD) are caused by mutations in the PEX genes and are divided into two distinct syndromes, Zellweger syndrome spectrum (PBD-ZSS) and rhizomelic chondodysplasia puctata spectrum (PBD-RCDP). With the exception of PEX7, mutations in PEX genes result in generalized peroxisomal dysfunction and the PBD-ZSS phenotype [1]. Peroxisomes play a fundamental role in many biochemical pathways including very long chain fatty acid (VLCFA) metabolism and plasmalogen synthesis. Patients with PBD-ZSS exhibit altered plasma levels of these molecules [2].
Peroxisomal dysfunction leads to multisystem disease, including neurological symptoms. Severe cases can have refractory epilepsy, neuronal migration disorders, and leukodystrophy [1]. Mild cases can include sensorineural hearing loss, retinal abnormalities, leukodystrophy, and developmental and cognitive delay [1]. While severe cases present in the neonatal period and result in death before one year of age, patients with mild disease present after newborn period and can live into adulthood [1].
Pathogenic variants in PEX1 are the most common cause of PBD-ZSS and are found in almost 60% of affected individuals [3], [4]. The most common European disease-causing allele of PEX1 is p.G843D [5], [6], [7], [8], [9], [10], and the high frequency of this variant is likely due to a founder effect [11]. Generally, patients homozygous for p.G843D have milder phenotypes compared to patients with PEX1 truncation mutations, insertions, deletions, and other PEX1 pathogenic variants [9], [12]. Additionally, patients homozygous for p.G843D had residual PEX1 protein levels on fibroblast culture. In contrast, patients with other pathologic variants presented with more severe phenotypes and lacked any PEX1 protein on fibroblast culture [8]. However, disease severity varies between patients homozygous for p.G843D [13], including neurologic findings [14], making it difficult to predict outcomes solely from genotype.
PBD-ZSS are typically diagnosed through detection of elevated VLCFA in plasma and confirmed by genetic analysis or biochemical analysis of fibroblast culture [1]. Diagnosis of patients with mild or atypical PBD-ZSS can be challenging [15], [16], [17]. Recently neonatal screening for X-linked adrenoleukodystrophy has become available. This screening also identifies PBD-ZSS and may be useful in early diagnosis of patients with mild PBD-ZSS with elevated lyso-PCs who are asymptomatic at birth [18], [19].
Here we report an atypical path to diagnosis of a child homozygous for PEX1 p.G843D in a particularly mild patient. After genetic testing for Usher syndrome was negative, he was found to have PBD-ZSS through a next generation sequencing (NGS) research protocol for retinal dystrophy. With NGS, the spectrum of PBD-ZSS phenotypes is expanding as are the diagnostic paths [15], [16], [17]. This case draws attention to the unusual diagnostic journey of a patient with a very mild PBD-ZSS.
2. Materials and methods
2.1. Ethics statement
Informed consent for research and publication was obtained prior to participation for the subject who was recruited under the Institutional Review Board approved Biochemical Correlates of Peroxisomal Biogenesis Defects Study (H-32837) at Baylor College of Medicine.
2.2. Electroretinograms and optical coherence tomography
Full-field electroretinograms were obtained at 19 months from dilated eyes according to International Society for Clinical Electrophysiology of Vision standards [20] (LKC Technologies, Inc.). Frequency-domain optical coherence tomography (Spectralis; Heidelberg Engineering, Heidelberg, Germany) retinal scans were obtained at 6 years of age from the horizontal midline of dilated eyes.
2.3. DNA library preparation and next generation sequencing
Approximately 1 μg of genomic DNA was sheared into 200-500 bp fragments which were blunt-end repaired. A single adenine base was added using Klenow exonuclease. Illumina adapters were ligated to the repaired ends. DNA fragments were polymerase chain reaction (PCR) amplified for 8 cycles after ligation. The targeted DNA was captured with a customized gene panel [21] for sequencing to screen for variants in known retinal disease causing genes.
2.4. Bioinformatics analysis
Paired-end sequencing reads were obtained for each sample and mapped to human reference genome hg19 using Burrows–Wheeler aligner [22]. Base-quality recalibration, local realignment, and variant calling were performed as previously described [23]. In screening for Mendelian disorders, variants with a frequency > 1/200 in a series of public and internal databases [24] were filtered out. After frequency-based filtering, synonymous variants were filtered out, and a known retinal disease-causing variant was identified.
2.5. Sanger sequencing
A repeat patient DNA sample and additional samples from both unaffected parents were obtained. A 500 bp flanking sequence covering the candidate variant was obtained from the UCSC genome browser (hg19 assembly). Primer 3 [25] was used to design primers for PCR. The PCR amplicons were sequenced on an Applied Biosystems 3500xL Genetic Analyzer.
2.6. Peroxisomal biochemical studies
Plasma VLCFA at 6 years of age were measured by the Peroxisomal Disease Lab at the Kennedy Krieger Institute using capillary gas chromatography/mass spectrometry of pentafluorobenzyl bromide fatty acid esters as described [26], [27].
C26:0 lysoPC content was measured in a whole venous blood spot obtained at age eight by LC-MS/MS as described [18].
3. Results
3.1. Case report
The patient (Fig. 1A) was born at 39 weeks gestation to unrelated parents with no prior pregnancies. He attended physical and occupational therapy from 7 to 15 months for low muscle tone but adequately met both language and motor developmental milestones. He was noted to have dysphagia and used a milk thickener until age two. Growth percentile at 0, 2, 9, 12, and 15 months were as follows: length (25%, 85%, 55%, 75%, 75%), weight (63%, 25%, 20%, 37%, 45%), head circumference (10%, 10%, 8%, 7%, 10%). At 19 months, he presented for evaluation of progressive sensorineural hearing loss. An eye exam revealed pigmentary retinopathy and optic nerve hypoplasia. Visual acuity at the age of 18.8 months was 20/90 (OD) and 20/105 (OS). MRI brain revealed no abnormalities. Subsequent to receiving the molecular diagnosis, he was started on Cholic Acid for a mild transaminitis.
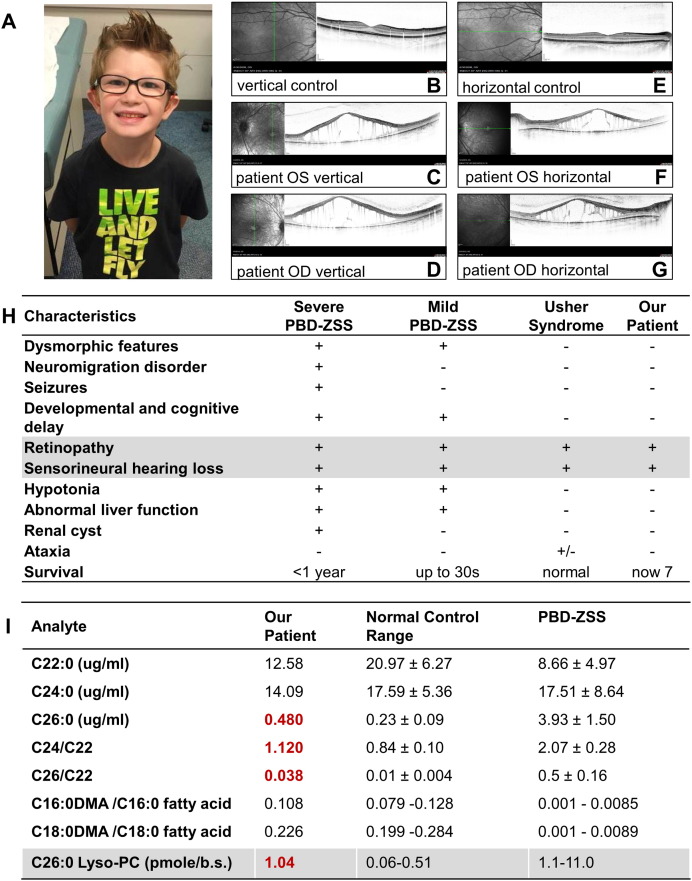
Fig. 1.
A. The patient at 8 years of age. B–G. Optical coherence tomography shows cystoid macular edema and absence of photoreceptor layer compared to age matched controls. H. The patient's clinical characteristics compared to mild and severe peroxisome biogenesis disorder Zellweger spectrum (PBD-ZSS) phenotypes and Usher syndrome. I. Red bold values indicate results outside of normal control range. For C22:0, C24:0, C26:0, C24/C22 ratio, and C26/C22 ratio, the patient's result is shown, and the mean ± standard deviation is shown for normal controls and PBD-ZSS. For C16:0DMA/C16:0 fatty acid and C18:0DMA/C18:0 fatty acid ratios, patient's mean is shown, and a range is shown for normal controls and PBS-ZSS. For C26:0 Lyso-PC, the patient's result is shown, and a range is shown for normal controls and PBD-ZSS. C26:0 Lyso-PC values are reported in pmole per 1/8th inch dried whole venous blood spot.
3.2. Ophthalmologic evaluation
The patient was evaluated through full-field electroretinogram (ERG) and optical coherence tomography (OCT). ERG responses showed non-detectable rod components and no reliable cone responses. OCT showed cystoid macular edema and absence of the photoreceptor layer in the macula in both eyes (Fig. 1B–G).
3.3. Genetic findings
Based on clinical features (Fig. 1H), Usher syndrome was suspected. However DNA testing for Usher syndrome and hearing loss (OtoGenome™) was negative. Surprisingly, through a research panel of known retinal disease causing genes [21], he was found to have a homozygous known pathogenic variant p.G843D in PEX1 (chr7:92130876C > T [hg19]: NM_000466.2:c. G2528A:p.G843D).
3.4. Biochemical analysis
Subsequent to the molecular diagnosis, biochemical testing for VLCFA and plasmalogens was performed. Biochemical testing showed C22:0 12.58 μg/ml (upper limit of normal range (ULN) 33.51 μg/ml), C24:0 14.09 μg/ml (ULN 28.31 μg/ml), C26:0 0.480 μg/ml (ULN 0.41 μg/ml), C24/22 1.120 (ULN 1.04), C26/22 ratio 0.038 (ULN 0.018), C16:DMA/C16:0 fatty acid 0.108 (normal range 0.079–0.128), and C18:DMA/C18:0 fatty acid 0.226 (normal range 0.199–0.284). C22:0 is reported for the purpose of C24/22 and C26/22 ratio calculation. In summary, the VLCFA show slight abnormalities in C26:0, C24/C22, and C26/C22 (Fig. 1I, red bold values) but milder than those in typically seen in PBD-ZSS.
The C26:0-lyso-PC level measured using methodology currently employed for X-ALD neonatal screens was just below the PBD-ZSS range at 1.04 pmole per 1/8th inch spot (normal range 0.06–0.51; PBD-ZSS range 1.1–11.0).
Liver function tests were performed after the molecular diagnosis. Aspartate transaminase (AST) was elevated at 55, 55, and 62 U/L at 6, 7, and 8 years of age respectively (normal 12–32 U/L). Similarly alanine transaminase (ALT) was elevated at 42, 32, and 47 at 6, 7, and 8 years of age respectively (normal 12–32 U/L). Total bilirubin, direct bilirubin, alkaline phosphatase, albumin, and total protein were normal at 7 and 8 years of age. Total bile acids were normal at the age at 7 years of age.
4. Discussion
In this report, a patient with apparently isolated hearing and visual loss was found to be homozygous for the PEX1 p.G843D allele. The patient's clinical phenotype was sensorineural hearing loss with pigmentary retinopathy, mildly decreased muscle tone, and small head circumference in an otherwise healthy and cognitively normal child. Based on the sensorineural hearing loss, retinal changes, and normal cognition, Usher syndrome was suspected, and both diagnostic and subsequent research genetic testing for retinal dystrophy genes were undertaken. It was therefore surprising to identify PBD as the cause of this patient's phenotype. While there is significant overlap between Usher and PBD-ZSS [28], the patient's diagnostic path was unusual.
Typically, the primary step in making PBD-ZSS diagnosis is measuring serum VLCFA [29]. However, the first diagnostic step of this case was sequencing. Although the diagnostic path was atypical, our patient's retinal phenotype is consistent with a mild PBD-ZSS. The decreased visual acuity, absent ERG responses, and pigmentary retinopathy we observed are consistent with PBD-ZSS [30], [31], including the PEX1 p.G843D allele. However our patient had cystoid edema in contrast to foveal thinning previously observed in the patient homozygous for the PEX1 p.G843D allele [16], although cystoid edema has previously been reported in PBD-ZSS patients [32].
The biochemical analysis showed the patient's C26:0, C24/22 ratio, and C26/22 ratio were elevated compared to normal controls but not within the range of control PBD-ZSS patients. These mild biochemical findings are consistent with a mild PBD-ZSS, as VLCFA levels correlate with severity of phenotype [8], [26]. C26:0 Lyso-PC almost fell into the range for PBD-ZSS samples but was outside of normal control range.
Although PBD-ZSS are typically diagnosed first by detecting biochemical abnormalities, NGS is useful in identifying specific PBD-ZSS patients with very mild [17] and atypical phenotypes with mildly elevated or normal VLCFAs [33], [34], [35]. Additionally, NGS has confirmed the PBD-ZSS diagnosis in patients initially diagnosed with other retinal disorders such as Leber congenital amaurosis [16]. Therefore, NGS has allowed a wider range of PBD-ZSS phenotypes to be diagnosed.
While NGS and biochemical testing are prompted by clinical findings, neonatal screening could provide early diagnosis prior symptom onset in mild PBD-ZSS's [18]. Early diagnosis would allow for close surveillance of systems affected by PBD-ZSS [15], [29]. Our patient's C26:0-lyso-PC level on blood spot at eight years of age was outside of normal range and may have prompted further testing if this result was obtained on a neonatal screen. Although his value as a newborn is not known, it seems possible that neonatal screening could have led to an early diagnosis for this mild PBD-ZSS patient.
This case is an example of the importance of genetic testing in the diagnosis of very mild PBD-ZSS and further emphasizes the need to consider mild PBD-ZSS when evaluating a patient with retinopathy and hearing loss regardless of cognitive function.
Conflict of interest
Michael Wangler and Ann Moser have served as advisors on PBD treatment for Retrophin Pharmaceuticals.
Acknowledgements
The authors thank the family for their participation. This study was funded by NIH K08NS076547 to MFW, the Simmons Family Foundation Collaborative Research Grant, the Clayton Murphy Peroxisomal Disorders Research Grant, NIH EY007142, the Hermann Eye Fund, and the William Stamps Farish Fund.
References
- 1.Braverman N.E., D’Agostino M.D., Maclean G.E. Peroxisome biogenesis disorders: biological, clinical and pathophysiological perspectives. Dev. Disabil. Res. Rev. 2013;17:187–196. doi: 10.1002/ddrr.1113. [DOI] [PubMed] [Google Scholar]
- 2.Wanders R.J.A. Metabolic and molecular basis of peroxisomal disorders: a review. Am. J. Med. Genet. A. 2004;126A:355–375. doi: 10.1002/ajmg.a.20661. [DOI] [PubMed] [Google Scholar]
- 3.Ebberink M.S., Mooijer P.A.W., Gootjes J., Koster J., Wanders R.J.A., Waterham H.R. Genetic classification and mutational spectrum of more than 600 patients with a Zellweger syndrome spectrum disorder. Hum. Mutat. 2011;32:59–69. doi: 10.1002/humu.21388. [DOI] [PubMed] [Google Scholar]
- 4.Yik W.Y., Steinberg S.J., Moser A.B., Moser H.W., Hacia J.G. Identification of novel mutations and sequence variation in the Zellweger syndrome spectrum of peroxisome biogenesis disorders. Hum. Mutat. 2009;30:E467–E480. doi: 10.1002/humu.20932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reuber B.E., Germain-Lee E., Collins C.S., Morrell J.C., Ameritunga R., Moser H.W., Valle D., Gould S.J. Mutations in PEX1 are the most common cause of peroxisome biogenesis disorders. Nat. Genet. 1997;17:445–448. doi: 10.1038/ng1297-445. [DOI] [PubMed] [Google Scholar]
- 6.Imamura A., Tamura S., Shimozawa N., Suzuki Y., Zhang Z., Tsukamoto T., Orii T., Kondo N., Osumi T., Fujiki Y. Temperature-sensitive mutation in PEX1 moderates the phenotypes of peroxisome deficiency disorders. Hum. Mol. Genet. 1998;7:2089–2094. doi: 10.1093/hmg/7.13.2089. [DOI] [PubMed] [Google Scholar]
- 7.Maxwell M.A., Allen T., Solly P.B., Svingen T., Paton B.C., Crane D.I. Novel PEX1 mutations and genotype-phenotype correlations in Australasian peroxisome biogenesis disorder patients. Hum. Mutat. 2002;20:342–351. doi: 10.1002/humu.10128. [DOI] [PubMed] [Google Scholar]
- 8.Walter C., Gootjes J., Mooijer P.A., Portsteffen H., Klein C., Waterham H.R., Barth P.G., Epplen J.T., Kunau W.H., Wanders R.J. Disorders of peroxisome biogenesis due to mutations in PEX1: phenotypes and PEX1 protein levels. Am. J. Hum. Genet. 2001;69:35–48. doi: 10.1086/321265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Preuss N., Brosius U., Biermanns M., Muntau A.C., Conzelmann E., Gartner J. PEX1 mutations in complementation group 1 of Zellweger spectrum patients correlate with severity of disease. Pediatr. Res. 2002;51:706–714. doi: 10.1203/00006450-200206000-00008. [DOI] [PubMed] [Google Scholar]
- 10.Steinberg S., Chen L., Wei L., Moser A., Moser H., Cutting G., Braverman N. The PEX Gene Screen: molecular diagnosis of peroxisome biogenesis disorders in the Zellweger syndrome spectrum. Mol. Genet. Metab. 2004;83:252–263. doi: 10.1016/j.ymgme.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 11.Collins C.S., Gould S.J. Identification of a common PEX1 mutation in Zellweger syndrome. Hum. Mutat. 1999;14:45–53. doi: 10.1002/(SICI)1098-1004(1999)14:1<45::AID-HUMU6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 12.Crane D.I., Maxwell M.A., Paton B.C. PEX1 mutations in the Zellweger spectrum of the peroxisome biogenesis disorders. Hum. Mutat. 2005;26:167–175. doi: 10.1002/humu.20211. [DOI] [PubMed] [Google Scholar]
- 13.Poll-The B.T., Gootjes J., Duran M., De Klerk J.B.C., de Wenniger-Prick L.J.M.B., Admiraal R.J.C., Waterham H.R., Wanders R.J.A., Barth P.G. Peroxisome biogenesis disorders with prolonged survival: phenotypic expression in a cohort of 31 patients. Am. J. Med. Genet. A. 2004;126A:333–338. doi: 10.1002/ajmg.a.20664. [DOI] [PubMed] [Google Scholar]
- 14.Barth P.G., Majoie C.B.L.M., Gootjes J., Wanders R.J.A., Waterham H.R., van der Knaap M.S., de Klerk J.B.C., Smeitink J., Poll-The B.T. Neuroimaging of peroxisome biogenesis disorders (Zellweger spectrum) with prolonged survival. Neurology. 2004;62:439–444. doi: 10.1212/01.wnl.0000106943.40848.03. [DOI] [PubMed] [Google Scholar]
- 15.Klouwer F.C.C., Huffnagel I.C., Ferdinandusse S., Waterham H.R., Wanders R.J.A., Engelen M., Poll-The B.T. Clinical and biochemical pitfalls in the diagnosis of peroxisomal disorders. Neuropediatrics. 2016;47:205–220. doi: 10.1055/s-0036-1582140. [DOI] [PubMed] [Google Scholar]
- 16.Majewski J., Wang Z., Lopez I., Humaid S.A., Ren H., Racine J., Bazinet A., Mitchel G., Braverman N., Koenekoop R.K. A new ocular phenotype associated with an unexpected but known systemic disorder and mutation: novel use of genomic diagnostics and exome sequencing. J. Med. Genet. 2011;48:593–596. doi: 10.1136/jmedgenet-2011-100288. [DOI] [PubMed] [Google Scholar]
- 17.Ratbi I., Falkenberg K.D., Sommen M., Al-Sheqaih N., Guaoua S., Vandeweyer G., Urquhart J.E., Chandler K.E., Williams S.G., Roberts N.A. Heimler syndrome is caused by hypomorphic mutations in the peroxisome-biogenesis genes PEX1 and PEX6. Am. J. Hum. Genet. 2015;97:535–545. doi: 10.1016/j.ajhg.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hubbard W.C., Moser A.B., Liu A.C., Jones R.O., Steinberg S.J., Lorey F., Panny S.R., Vogt R.F., Macaya D., Turgeon C.T. Newborn screening for X-linked adrenoleukodystrophy (X-ALD): validation of a combined liquid chromatography-tandem mass spectrometric (LC-MS/MS) method. Mol. Genet. Metab. 2009;97:212–220. doi: 10.1016/j.ymgme.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 19.Theda C., Gibbons K., Defor T.E., Donohue P.K., Golden W.C., Kline A.D., Gulamali-Majid F., Panny S.R., Hubbard W.C., Jones R.O. Newborn screening for X-linked adrenoleukodystrophy: further evidence high throughput screening is feasible. Mol. Genet. Metab. 2014;111:55–57. doi: 10.1016/j.ymgme.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marmor M.F., Fulton A.B., Holder G.E., Miyake Y., Brigell M., Bach M., International Society for Clinical Electrophysiology of Vision ISCEV standard for full-field clinical electroretinography (2008 update) Doc. Ophthalmol. 2009;118:69–77. doi: 10.1007/s10633-008-9155-4. [DOI] [PubMed] [Google Scholar]
- 21.Xu M., Eblimit A., Wang J., Li J., Wang F., Zhao L., Wang X., Xiao N., Li Y., Wong L.-J.C. ADIPOR1 Is mutated in syndromic retinitis pigmentosa. Hum. Mutat. 2016;37:246–249. doi: 10.1002/humu.22940. (Li and Durbin, 2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang F., Wang H., Tuan H.-F., Nguyen D.H., Sun V., Keser V., Bowne S.J., Sullivan L.S., Luo H., Zhao L. Next generation sequencing-based molecular diagnosis of retinitis pigmentosa: identification of a novel genotype-phenotype correlation and clinical refinements. Hum. Genet. 2014;133:331–345. doi: 10.1007/s00439-013-1381-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tajiguli A., Xu M., Fu Q., Yiming R., Wang K., Li Y., Eblimit A., Sui R., Chen R., Aisa H.A. Next-generation sequencing-based molecular diagnosis of 12 inherited retinal disease probands of Uyghur ethnicity. Sci. Report. 2016;6:21384. doi: 10.1038/srep21384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B.C., Remm M., Rozen S.G. Primer3 - new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moser A.B., Kreiter N., Bezman L., Lu S., Raymond G.V., Naidu S., Moser H.W. Plasma very long chain fatty acids in 3,000 peroxisome disease patients and 29,000 controls. Ann. Neurol. 1999;45:100–110. doi: 10.1002/1531-8249(199901)45:1<100::aid-art16>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 27.Lagerstedt S.A., Hinrichs D.R., Batt S.M., Magera M.J., Rinaldo P., McConnell J.P. Quantitative determination of plasma c8-c26 total fatty acids for the biochemical diagnosis of nutritional and metabolic disorders. Mol. Genet. Metab. 2001;73:38–45. doi: 10.1006/mgme.2001.3170. [DOI] [PubMed] [Google Scholar]
- 28.Raas-Rothschild A., Wanders R.J.A., Mooijer P.A.W., Gootjes J., Waterham H.R., Gutman A., Suzuki Y., Shimozawa N., Kondo N., Eshel G. A PEX6-defective peroxisomal biogenesis disorder with severe phenotype in an infant, versus mild phenotype resembling Usher syndrome in the affected parents. Am. J. Hum. Genet. 2002;70:1062–1068. doi: 10.1086/339766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Braverman N.E., Raymond G.V., Rizzo W.B., Moser A.B., Wilkinson M.E., Stone E.M., Steinberg S.J., Wangler M.F., Rush E.T., Hacia J.G. Peroxisome biogenesis disorders in the Zellweger spectrum: an overview of current diagnosis, clinical manifestations, and treatment guidelines. Mol. Genet. Metab. 2016;117:313–321. doi: 10.1016/j.ymgme.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weleber R.G., Tongue A.C., Kennaway N.G., Budden S.S., Buist N.R. Ophthalmic manifestations of infantile phytanic acid storage disease. Arch. Ophthalmol. 1984;102:1317–1321. doi: 10.1001/archopht.1984.01040031067026. [DOI] [PubMed] [Google Scholar]
- 31.Folz S.J., Trobe J.D. The peroxisome and the eye. Surv. Ophthalmol. 1991;35:353–368. doi: 10.1016/0039-6257(91)90185-i. [DOI] [PubMed] [Google Scholar]
- 32.Pakzad-Vaezi K.L., Maberley D.A.L. Infantile Refsum disease in a young adult: case presentation and brief review. Retin Cases Brief Rep. 2014;8:56–59. doi: 10.1097/ICB.0000000000000004. [DOI] [PubMed] [Google Scholar]
- 33.Régal L., Ebberink M.S., Goemans N., Wanders R.J.A., De Meirleir L., Jaeken J., Schrooten M., Van Coster R., Waterham H.R. Mutations in PEX10 are a cause of autosomal recessive ataxia. Ann. Neurol. 2010;68:259–263. doi: 10.1002/ana.22035. [DOI] [PubMed] [Google Scholar]
- 34.Sevin C., Ferdinandusse S., Waterham H.R., Wanders R.J., Aubourg P. Autosomal recessive cerebellar ataxia caused by mutations in the PEX2 gene. Orphanet J. Rare Dis. 2011;6:8. doi: 10.1186/1750-1172-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mignarri A., Vinciguerra C., Giorgio A., Ferdinandusse S., Waterham H., Wanders R., Bertini E., Dotti M.T., Federico A. Zellweger spectrum disorder with mild phenotype caused by PEX2 gene mutations. JIMD Rep. 2012;6:43–46. doi: 10.1007/8904_2011_102. [DOI] [PMC free article] [PubMed] [Google Scholar]