Abstract
Five cultivars of tomato having different levels of salt stress tolerance were exposed to different treatments of NaCl (0, 3 and 6 g L−1) and ZnO-NPs (0, 15 and 30 mg L−1). Treatments with NaCl at both 3 and 6 g L−1 suppressed the mRNA levels of superoxide dismutase (SOD) and glutathione peroxidase (GPX) genes in all cultivars while plants treated with ZnO-NPs in the presence of NaCl, showed increments in the mRNA expression levels. This indicated that ZnO-NPs had a positive response on plant metabolism under salt stress. Superior expression levels of mRNA were observed in the salt tolerant cultivars, Sandpoint and Edkawy while the lowest level was detected in the salt sensitive cultivar, Anna Aasa. SDS–PAGE showed clear differences in patterns of protein expression among the cultivars. A negative protein marker for salt sensitivity and ZnO-NPs was detected in cv. Anna Aasa at a molecular weight of 19.162 kDa, while the tolerant cultivar Edkawy had two positive markers at molecular weights of 74.991 and 79.735 kDa.
Abbreviations: cDNA, complementary DNA; GPX, glutathione peroxidase; MW, molecular weight; NPs-ZnO, nanoparticles of zinc oxide; RF, relative factor; RT-PCR, real time polymerase chain reaction; SDS–PAGE, sodium dodecyl sulfate–poly acrylamide gel electrophoresis; SOD, superoxide dismutase
Keywords: Tomato, Salt stress, Nanoparticles, Gene expression, Real-time PCR, Polymorphism
1. Introduction
In the face of a rapidly growing world population and against a background of decreasing arable area and increasing global environmental changes an increased production of high-quality foods with reduced inputs is urgently needed and technological solutions are required. Many cellular functions of plants are severely affected by environment stress such as drought, salinity, frost and heat which ultimately exert a negative impact on plant growth and reproduction (Noaman et al., 2004). With regard to tomato (Solanum lycopersicum L. formerly Lycopersicon esculentum Mill.), germplasm improvement through either classical breeding or by modern biotechnologies is becoming more important for world tomato production since several important production regions, such as in Mediterranean countries like Italy, Spain, Egypt, and Turkey are increasingly suffering from periods of drought and increased salinity in irrigation water (Rinaldi et al., 2011). One of the technologies which has emerged recently is nanotechnology and the development of nano-devices and nanomaterials is beginning to open up novel applications in agriculture and plant biotechnology (Nair et al., 2010). Applications of nanomaterials can help faster seed germination, improved plant tolerance to abiotic and biotic stress, efficient nutrient utilization and enhanced plant growth with reduced environmental impact compared to traditional approaches of fertilizers and pesticides (Reynolds, 2002, Sheikh et al., 2009). ZnO-NPs appear to play a strong role in arranging several mechanisms included in recognition and response to abiotic stresses in plants (Prasad et al., 2012). There are an increasing number of reports regarding the interaction between salinity and ZnO in higher plants but there is currently no information available about the possible beneficial effects of ZnO-NPs application to reduce damage from salt stress.
Under salt stress, increases in intracellular levels of Reactive Oxygen Species (ROS) were found to cause significant damage to cell structures (Bhattachrjee, 2005) and influence the expression of a number of genes such as SOD and GPX (Gill and Tuteja, 2010). With respect to nanoparticles, few experiments have been performed to show the effects of nanoparticles that may affect the growth, development, and gene expression in plants (Burklew et al., 2012). Methods that detect and quantify gene expression such as real-time PCR (RT-PCR), have been developed and have become more rapid, sensitive, showing minimal changes due to changes in environmental conditions of plants (Sturzenbaum and Kille, 2001). RT-PCR provides advantages, such as precise quantification of mRNA levels of genes of interest when expression levels are compared under different conditions or treatments. RT-PCR, does not however provide information about the transcriptional activity of genes, but measures quantification of RNA levels of gens in the cells under different condition of treatments (Soydam et al., 2013). Nevertheless, with good experimental design including sequential time course sampling, RT-PCR is becoming the technique of choice for gene expression quantification.
While several physiological and growth parameters may be directly used to evaluate the stress-tolerance of cultivars, several workers have more recently undertaken to study the proteome of the plant cells. By such means, a number of proteins have been shown to be induced by abiotic stresses which reflect the complexity of the biochemical and physiological responses of plants to stress (Chen and Tabaeizadeh, 1992, Arefian et al., 2014). These changes in protein expression are directly associated with biological change, being responsible for the increased performance of stress-tolerant cultivars. A previous study (Azeez and Morakinyo, 2004), using leaf samples of different tomato cultivars succeeded in the detection of inter-cultivar qualitative as well as quantitative protein band patterns that depict some degree of genetic relationship among the tomato cultivars studied. Even early research (Cooke, 1984) indicated that electrophoresis marker could be useful in providing an indirect method for genome probing by exposing structural variation in protein banding patterns. However, unambiguous identification of potential protein markers indicating the salt tolerance of a tomato cultivar is still pending.
The objective of the current research was to examine the effectiveness of the application of ZnO-NPs in the evaluation of mRNA expression of SOD and GPX genes and proteins in tomato germplasm under different treatments of NaCl.
2. Material and methods
2.1. Germplasm used
Experiments were conducted in a glasshouse at the Faculty of Science, King Abdulaziz University, Jeddah, Saudi Arabia in cooperation with the Biotechnology Laboratory, Faculty of Science, Jeddah University, Jeddah, Saudi Arabia during the period February 2014 to May 2015. Seeds of four tomato (L. esculentum Mill) cultivars were kindly provided by the Leibniz Institute of Plant Genetics and Crop Plant Research (IPK), Gatersleben, Germany, while seeds of the Egyptian tomato cultivar Edkawy were obtained from the Agriculture Research Center (ARC), Giza, Egypt (Table 1).
Table 3.
Ideogram of protein banding pattern in cv. Sandpoint under different concentrations of NaCl and ZnO-NPs.
| RF | MW | Lane a | Lane b | Lane c | Lane d | Lane e | Lane f | Frequency | Polymorphism |
|---|---|---|---|---|---|---|---|---|---|
| 0.310 | 122.575 | + | + | − | − | + | + | 0.667 | Polymorphic |
| 0.055 | 113.711 | + | + | − | − | + | − | 0.500 | Polymorphic |
| 0.084 | 81.640 | − | + | + | − | + | + | 0.667 | Polymorphic |
| 0.212 | 76.327 | − | + | + | − | + | + | 0.667 | Polymorphic |
| 0.544 | 72.288 | − | + | + | + | − | + | 0.667 | Polymorphic |
| 0.502 | 63.348 | + | + | + | + | + | + | 1.000 | Monomorphic |
| 0.416 | 51.900 | − | + | + | − | + | + | 0.667 | Polymorphic |
| 0.387 | 48.147 | − | + | + | − | + | + | 0.667 | Polymorphic |
| 0.259 | 38.538 | − | + | + | − | − | + | 0.500 | Polymorphic |
| 0.238 | 34.568 | − | + | + | − | + | + | 0.667 | Polymorphic |
| 0.591 | 30.608 | + | + | + | + | + | + | 1.000 | Monomorphic |
| 0.626 | 27.957 | + | + | + | + | + | + | 1.000 | Monomorphic |
| 0.657 | 25.801 | − | + | + | + | + | + | 0.833 | Polymorphic |
| 0.679 | 24.373 | − | + | − | + | + | + | 0.667 | Polymorphic |
| 0.738 | 22.669 | + | + | + | + | + | + | 1.000 | Monomorphic |
| 0.707 | 20.921 | − | − | + | + | + | + | 0.667 | Polymorphic |
| 0.774 | 19.059 | − | + | + | + | + | + | 0.833 | Polymorphic |
| 0.814 | 17.185 | − | − | + | − | + | + | 0.500 | Polymorphic |
| 0.884 | 15.778 | − | − | + | − | + | + | 0.500 | Polymorphic |
| 0.847 | 14.337 | + | + | − | + | + | + | 0.833 | Polymorphic |
+, present band; −, absent band; lane a, T1 = control; lane b, T3 = 6 g L−1 NaCl; lane c, T4 = 15 mg L−1 ZnO-NPs; lane d, T 5 = 30 mg L−1 ZnO-NPs; lane f, T8 = 6 g L−1 NaCl + 15 mg L−1 ZnO-NPs; lane b, T9 = 6 g L−1 NaCl + 30 mg L−1 ZnO-NPs.
2.2. Preparation of ZnO-NPs suspension
Nanoparticles of ZnO with an average primary particle size of 30 nm were purchased from Sigma–Aldrich Company, California, USA. In order to prepare different concentrations of ZnO-NPs at 15 and 30 mg L−1, a bulk solution was first prepared where 1.5 g of solid ZnO-NPs was dissolved in 1000 mL distilled water and a sonicator was used to homogenize the solution and then diluted to the desired strengths. The nanoparticle suspensions were then centrifuged (3000g for 1 h) and filtered (0.7 μm glass filter) prior to being added to culture media (Helaly et al., 2014).
2.3. Plant growing and experimental treatments
Plants were prepared by sowing the seeds in a nursery (beginning of September 2014) in module trays (10 ⩽ cm depth) filled with peat moss and irrigated with half strength Hoagland solution (Hoagland and Arnon, 1983). After 45 days (middle of March 2015), tomato plants were transplanted to the glasshouse into 1.1 L (30 cm diameter) pots filled with a mixture of peat moss and quartz sand (1:3 ratio by volume). Pots were set up in rows and laid out in split plot combinations of treatments with three replicates. Different levels of NaCl and NPs-ZnO treatments were applied as the main plots and tomato cultivars were assigned as the subplots. Each treatment was represented by three pots each with three plants, giving a total of 27 plants per cultivar per treatment. Plants irrigated with 600 mL of tap water three times a week were considered as the control treatment and a 3 (NaCl at 0, 3 and 6 g L−1) × 3 (ZnO-NPs at 0, 15 and 30 mg L−1) treatment factorial combination was established (T1 = control; T2 = 3 g L−1 NaCl; T3 = 6 g L−1 NaCl; T4 = 15 mg L−1 ZnO-NPs; T5 = 30 mg L−1 ZnO-NPs; T6 = 3 g L−1 NaCl + 15 mg L−1 ZnO-NPs; T7 = 3 g L−1 NaCl + 30 mg L−1 ZnO-NPs; T8 = 6 g L−1 NaCl + 15 mg L−1 ZnO-NPs; T9 = 6 g L−1 NaCl + 30 mg L−1 ZnO-NPs). The plants were maintained at 22/16 °C (day/night) under a relative humidity of 60% for the entire growth period. All pots were fertilized twice; the first dose was at the end of October and the second in mid-December, using liquid fertilizer (NPK15-10-5%).
2.4. Gene expression assays
After 70 days from transplanting, leaves were harvested at random from each treatment and frozen in liquid nitrogen and stored at −80 °C for future RNA extraction. Frozen samples were ground in a mortar and pestle under liquid nitrogen and RNA extracted using RNeasy Plant Mini Kits (50) from Qiagen, USA Catalog No. 74904. Chosen primer sequences for plant GPX, SOD and β-actin (constitutively expressed and used as a housekeeping gene for normalization) were chosen in common with previous assays (Miao and Gaynor, 1993, Medeiros et al., 2009) and are given in Table 2. PCR reactions were carried out in a Rotor gene (Biometra, Germany) thermocycler. The quantitative fold changes in mRNA expression were determined relative to β-actin mRNA levels in each corresponding group and calculated using the 2−DDCT method (Livak and Scmittgen, 2001).
Table 4.
Monomorphic bands, polymorphic bands, total polypeptides bands and polymorphism percentage for the 5 different tomato (Lycopersicon esculentum Mill) cultivars growing under different concentrations of NaCl and ZnO-NPs.
| Cultivars | Sandpoint | Anna Aasa | Australische Rosen | Sankt Ignatius | Edkawy |
|---|---|---|---|---|---|
| Monomorphic bands | 18 | 21 | 26 | 17 | 4 |
| Polymorphic bands | 8 | 1 | 1 | 1 | 16 |
| Unique bands | 0 | 1 | 0 | 0 | 0 |
| Poly. + Uniq. bands | 8 | 1 | 1 | 1 | 16 |
| Total number of bands | 26 | 2 | 27 | 18 | 20 |
| Polymorphism% | 30.76 | 8.696 | 3.704 | 5.556 | 80 |
| Mean of band frequency | 0.936 | 0.935 | 0.988 | 0.963 | 0.725 |
Polymorphism % = polymorphic bands/total number of bands.
2.5. Protein assay
Leaves of all cultivars were collected from 8-week old plants grown under control (T1), one level of salt stress (6 g L−1 NaCl) (T3), two levels of ZnO-NPs (15 (T4) and 30 (T5) mg L−1) and the combinations between NaCl and ZnO-NPs (6 g L−1 NaCl + 15 mg L−1 ZnO-NPs (T8); 6 g L−1 NaCl + 30 mg L−1 ZnO-NPs (T9)) and stored at −80 °C until protein analysis. Briefly, 0.5 g of frozen leaf tissue was used to extract soluble protein according to Bradford (1976). SDS–PAGE of leaf protein extracts were carried out in a vertical slab gel using 12% acrylamide according to Laemmli (1970) and a volume of 15–20 μL was applied to each well. In a separate lane of the gel, a protein ladder ranging from 10 to 250 kDa (Thermo Fisher Scientific, Waltham, MA, USA) was loaded in order to allow the estimation of the molecular masses of the separated proteins. Electrophoresis was run in a protein II electrophoresis system (Bio-Rad, California, USA) for about one hour in running buffer at 150 V/100 mA. The gels obtained were photographed with a gel documentation system (Syngene, Cambridge, UK). The molecular weights of the dissociated or unknown protein bands were determined using the standard curve obtained from the Rf-values and molecular weights of the protein ladder (10–250 kDa) and calculated using the gel analyzer version 3 software program.
2.6. Statistical analysis
The alterations in expression patterns of mRNA of SOD and GPX genes by RT-PCR were checked for statistical significance and represented as a mean ± SD, n = 10 according to ANOVA using SPSS version 20, statistical packages (IBM, New York, NY, USA). The results were considered statistically significant if the p value was ⩽ 0.05 according to Duncan (1955). The percentage of polymorphism was calculated according to the formula: polymorphism % = no. polymorphic bands/total no. of bands.
3. Results
3.1. Expression levels of mRNA of SOD and GPX genes
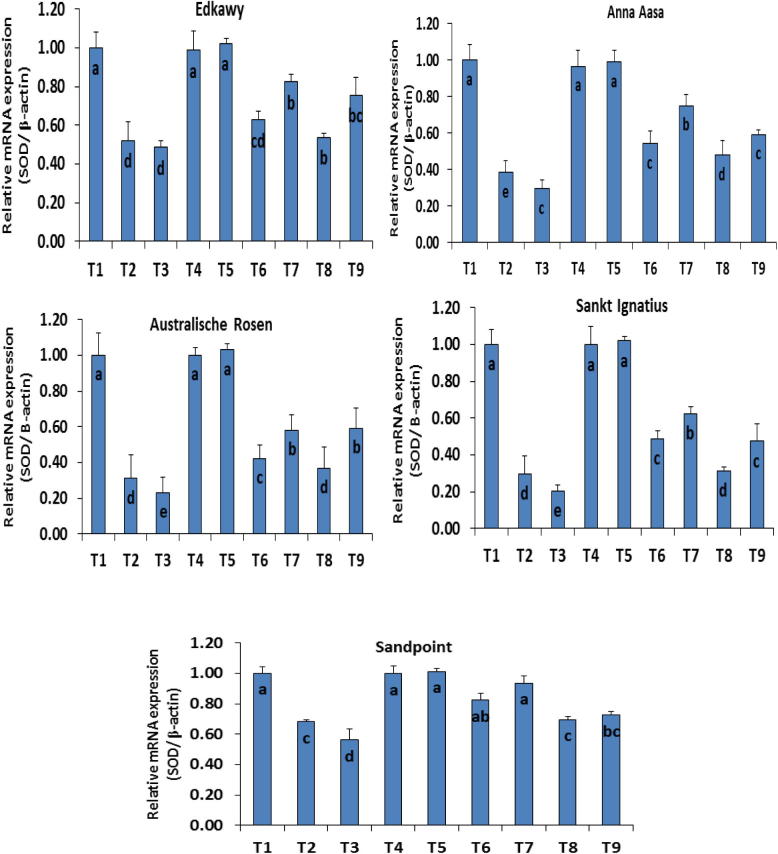
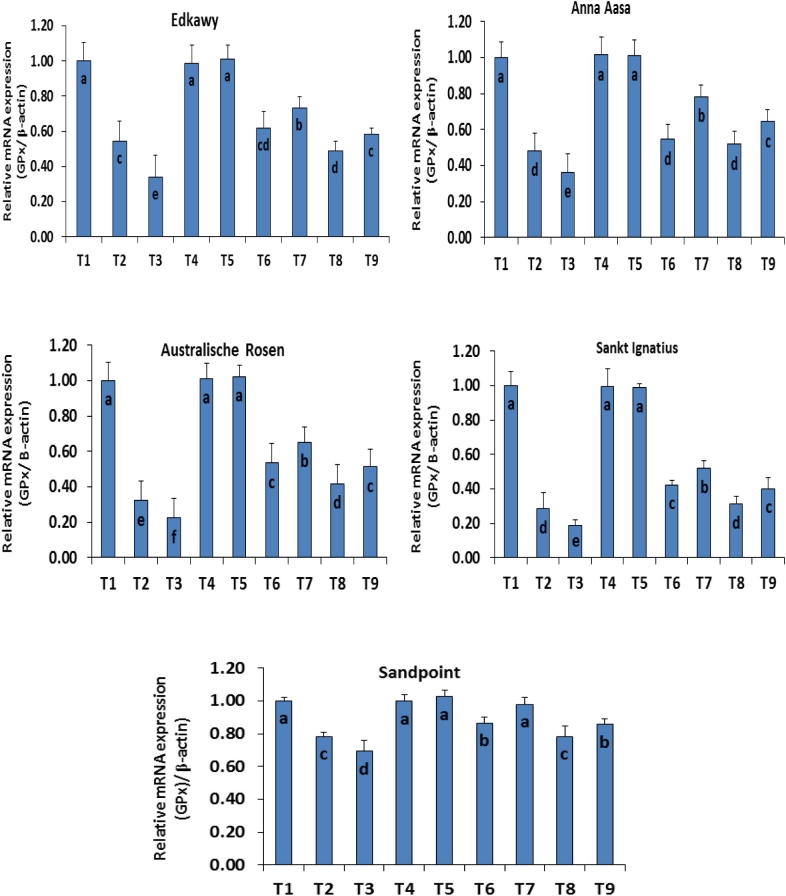
With regard to the control and to each other, the results recorded different expression levels of mRNA of both SOD and GPX genes. The highest expression levels of mRNA were observed in control (T1) and ZnO-NPs treatments (T4 and T5) with values ranging between 0.97 and 1.3 SOD/β-actin; 0.99 and 1.2 GPX/β-actin and showed non-significant differences (Figure 1, Figure 2). Treatment with NaCl either at 3.0 g L−1 (T2) or 6.0 g L−1 (T3) suppressed the mRNA levels of SOD and GPX genes in all the cultivars. Cultivar Sandpoint had the highest values (0.78, 0.69 and 0.68, 0.56), while cv. Anna Aasa recorded the lowest values (0.30, 0.20 and 0.28, 0.18), under T2 and T3 NaCl levels respectively. ZnO-NPs at both levels (15 mg L−1 (T4) and 30 mg L−1 (T5)) showed amelioration of the mRNA expression in all cultivars especially at the higher dose. Values of treatments T6 (3 g L−1 + 15 mg L−1 ZnO-NPs), T7 (3 g L−1 + 30 mg L−1 ZnO-NPs), T8 (6 g L−1 + 15 mg L−1 ZnO-NPs) and T9 (6 g L−1 + 15 mg L−1 ZnO-NPs) showed significant differences when compared to other treatments especially T4 (15 mg L−1 ZnO-NPs) and T5 (30 mg L−1 ZnO-NPs). Cultivars showed different responses to treatments T6–T9 where superior expression levels of mRNA were observed in cv. Sandpoint (0.94 ± 0.05; 0.98 ± 0.04) followed by cv. Edkawy (0.82 ± 0.04; 0.78 ± 0.06) under treatment T7, while the worst was cv. Sankt Ignatius (0.31 ± 0.02; 0.31 ± 0.05) under treatment T8 for SOD and GPX genes respectively.
Figure 1.
Expression level of mRNA for SOD gene using (RT-PCR) different cultivars of tomato (Lycopersicon esculentum Mill) exposed to different NaCl concentrations (0.0, 3.0 and 6.0 g L−1) and ZnO-NPs (0.0, 15 and 30 mg L−1) individually or in different combinations. T1 = control; T2 = 3 g L−1 NaCl; T3 = 6 g L−1 NaCl; T4 = 15 mg L−1 ZnO-NPs; T5 = 30 mg L−1 ZnO-NPs; T6 = 3 g L−1 NaCl + 15 mg L−1 ZnO-NPs; T7 = 3 g L−1 NaCl + 30 mg L−1 ZnO-NPs; T8 = 6 g L−1 NaCl + 15 mg L−1 ZnO-NPs; T9 = 6 g L−1 NaCl + 30 mg L−1 ZnO-NPs.
Figure 2.
Expression level of mRNA for GPX gene using (RT-PCR) different cultivars of tomato (Lycopersicon esculentum Mill) exposed to different NaCl concentrations (0.0, 3.0 and 6.0 g L−1) and ZnO-NPs (0.0 15 and 30 mg L−1) individually or in different combinations. T1 = control; T2 = 3 g L−1 NaCl; T3 = 6 g L−1 NaCl; T4 = 15 mg L−1 ZnO-NPs; T5 = 30 mg L−1 ZnO-NPs; T6 = 3 g L−1 NaCl + 15 mg L−1 ZnO-NPs; T7 = 3 g L−1 NaCl + 30 mg L−1 ZnO-NPs; T8 = 6 g L−1 NaCl + 15 mg L−1 ZnO-NPs; T9 = 6 g L−1 NaCl + 30 mg L−1 ZnO-NP.
3.2. Protein analysis
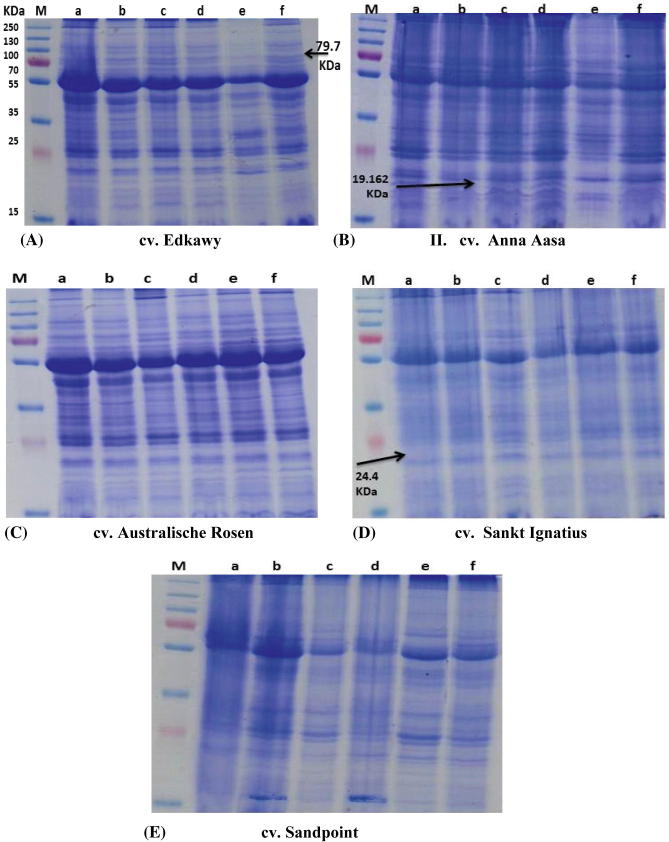
SDS–PAGE gave protein bands with molecular weights ranging from 14.337 kDa to 134.220 kDa (data not show). The total number of bands ranged from 7 to 26 for the control and 10 to 27 for other treatments. Newly synthesized protein bands of NaCl salt and ZnO-NPs treated cultivars were observed (Fig. 3). Two bands at molecular weights of 74.991 and 79.735 kDa were present only in the sensitive cv. Edkawy while the tolerant cultivar Sandpoint exhibited new bands at molecular weights 19.059, 24.373, 25.801, 34.568, 38.538, 48.147, 51.900, 72.288, 76.327, and 81.640 kDa (Table 3). Our results indicated that the protein at 19.162 kDa was NaCl salt enhanced in cv. Anna Aasa. The other cultivars, Australische Rosen and Sankt Ignitus, showed no synthesis of new bands under the different treatments compared to the control (Fig. 3). The number of polymorphic protein bands varied between the cultivars. The highest value (15 polymorphic bands) was recorded for salt tolerant cultivar cv. Sandpoint followed by cv. Edkawy (8 polymorphic bands) (Table 4). The highest level of polymorphism (80% and 30.8%) was recorded for resistant cultivars Sandpoint and Edkawy respectively, followed by Anna Aasa (8.70%), Sankt Ignatius (5.60%) and Australische Rosen (3.70%).
Figure 3.
SDS–PAGE protein patterns of 5 tomato (Lycopersicon esculentum Mill) cultivars in response to NaCl and ZnO-NPs. (M) Protein marker; lane (A) control; lane (B) 6 g L−1 NaCl; (C) 15 mg L−1; (D) 30 mg L−1; (E) 6 g L−1 + 15 mg L−1 and lane (F) 6 g L−1 NaCl + 30 mg L−1.
Table 1.
Accession code, commercial name, botanical name and origin of 5 tomato cultivars.
| IPK Accession code⁎ | Commercial name | Botanical name# | Origin |
|---|---|---|---|
| LYC3028 | Edkawy | Lycopersicon esculentum Mill | Egypt |
| LYC4112 | Anna Aasa | Lycopersicon esculentum Mill. Convar. infiniens Lehm. Var. flammatum | Russia |
| LYC3152 | Australische Rosen | Lycopersicon esculentum Mill | Australia |
| LYC4079 | Sankt Ignatius | Lycopersicon esculentum Mill. Convar. infiniens Lehm. Var. commune | Italy |
| LYC2493 | Sandpoint | Lycopersicon esculentum Mill. Convar. fruticosum Lehm. Var. pygmaeum Lehm. | USA |
Accession code of the Leibniz Institute of Plant Genetics and Crop Plant Research (IPK).
The botanical name Lycopersicon esculentum is used in the database of IPK and, thus, being used here.
Table 2.
Primer oligonucleotide sequences of GPX, SOD and β-actin.
| Gene | Oligonucleotide sequences 5′–3′ | Gen ID | References | |
|---|---|---|---|---|
| GPX | F | ACGGAGCAAGCGACAATTGACAAC | SGN-U213351 | Medeiros et al. (2009) |
| R | CGATTGATTCACCGCAAAGCTCGT | |||
| SOD | F | CACGTCTTCAAAGCAAGTGG | Miao and Gaynor (1993) | |
| R | CTAAGAAGAAGGGCATTCTTTGGCAT | |||
| β-actin | F | TTGACTGAGGCACCACTTAACCCT | SGN-U226051 | Medeiros et al. (2009) |
| R | GCTTTCAGGTGGTGCAACGACTTT | |||
F, forward primer; R, reserve primer.
4. Discussion
Reactive oxygen species (ROS) are known to influence the expression of a number of genes and contribute to many processes in abiotic stress responses induced by salinity. Several other mRNA studies have shown mediation of abiotic stress responses to drought and salinity in plants by altered gene expression (Burklew et al., 2012). It has been observed by (Ursini et al., 1995, Aydın et al., 2014) that SOD and GPX playing an important role in keeping the cells healthy under stress environmental by scavenging super-oxidase radicals, catalyzing their conversion to O2, reduces H2O2 and organic hydro-peroxides to water and alcohols using reduced glutathione (GSH). The current research is the first study which has reported and investigated the effect of NPs-ZnO on the expression mRNA levels of SOD and GPX genes under salinity stress and confirmed that a decrease in mRNA expression of SOD and GPX genes occurred during exposure to NaCl (Hernandez et al., 2000, Wang et al., 2007). A decrease in SOD and GPX activities under NaCl stress has been previously postulated by Khodary (2004) as an inhibition of nitrogen uptake which then affects peptide synthesis and causes enzyme limitation leading to inevitable decrease in amounts of enzyme. Nevertheless, several other studies have shown the opposite of our results and point to an increased expression of mRNA levels of the SOD and GPX genes in tomato under salinity (Srinineng et al., 2015, Mohamed et al., 2015).
Tomato plants treated with ZnO-NPs at both levels (15 and 30 mg L−1) under NaCl stress showed increases in the mRNA expression level of SOD and GPX genes (Figure 1, Figure 2). It seems that the presence of ZnO-NPs can alter the activity of mRNA in tomato plants and this could ameliorate the effect of salinity. A possible explanation for this has been previously put forward (Laware and Raskar, 2014) indicating that the low and/or appropriate dose of ZnO-NPs has a positive response on plant metabolism, enhancing absorption of essential nutrients such as nitrogen which then affects ion homeostasis, osmolytic biosynthesis, protein content and toxic radical scavenging. The increases in the mRNA levels of SOD and GPX genes however could also be as a result of increased stability of transcribed mRNAs (Soydam et al., 2013).
SDS–PAGE of Seed or leaf protein is a practical biochemical technique and has been used as a reliable method to detect the biochemical markers for the differentiation of tomato cultivars (Furdi, 2012). Our SDS–PAGE results demonstrated differences in patterns of protein changes between tolerant cultivars and moderate or sensitive cultivars and represented protein banding patterns with different molecular weights as appositive markers and demonstrated more changes in protein profile and a higher percentage of polymorphism in tolerant cultivars Sandpoint and Edkawy compared to Anna Aasa, Sankt Ignatius and Australische Rosen (Fig. 3E and Table 3). Our explanation of these results is that the tolerant cultivars are able to successfully adapt to saline environments by adjusting their biochemical processes and consequently the accumulation or depletion of certain metabolite activities which led to a repression of pre-existing protein synthesis and an enhanced or de novo synthesis of proteins which facilitate resistance mechanisms. This theory or explanation is also supported by previous results (Abu Hena et al., 2010, Ullah et al., 2014) which indicated that salt adaptive changes rely largely on alteration in gene expression and transcriptional activators and transcription factor function in the expression of stress inducible gene. Various investigators have indicated that the decline in the number of bands in sensitive genotypes compared with tolerant genotypes is associated with the denaturing of enzymes involved in amino acid and protein synthesis under abiotic stress (Dubey and Ranu, 1989). Both sensitive and moderately resistant cultivars in this study showed irregular changes in protein profile and an inability to rapidly accumulate antioxidant proteins as an indicator of sensitivity. A negative molecular marker associated with salt tolerance and NPs-ZnO was detected in cv. Anna Aasa at a molecular weight of 19.162 kDa (Fig. 3B) and this protein could correspond to stress damaging mechanisms. This result agrees with that of Bayoumi et al. (2008) who indicated that sensitive plants exposed to abiotic stress conditions frequently exhibit a characteristic set of cellular and metabolic responses including a decrease or an increase in the synthesis of protein. The protein bands at molecular weight 74.991 kDa in cv. Edkawy and 25.801 and 19.059 kDa in cv. Sandpoint can be considered as positive markers for stress, and it was noted that these bands exist under salinity treatment and ZnO-NPs treatment as well as salinity together with ZnO-NPs treatments and were not apparent under the control treatment. Ali et al. (2007) indicated that salt tolerance genotypes under salt treatment were characterized by a specific band (No. 10) with an approximate molecular weight of 17.54 kDa and suggested that such specific bands may use as markers for the identification of resistant genotypes under salt stress. Similarly, the band at 19.162 kDa in the salt sensitive cv. Anna Aasa can be considered as a negative molecular marker for the identification of sensitive genotypes. Other workers have indicated that a 32 kDa protein band was salt enhanced in sensitive barley genotypes (Bendary, 2000). The similarity between the protein bands under salt treatment and ZnO-NPs was not discussed in previous studies but can be explained as a similarity in biological action caused by both NaCl and ZnO-NPs within the cell and the ability of plants to successfully adapt to saline or ZnO-NPs treatments by adjusting biochemical processes which lead to enhanced protein synthesis. This observation requires further work which needs to focus on the toxicity of salinity, action of ZnO-NPs and the interaction between salinity and ZnO-NPs inside the cell.
Acknowledgements
This project was funded by the Deanship of Scientific Research (DSR) at King Abdulaziz University, Jeddah, under Grant No. (12/130/36/RG). The authors, therefore, acknowledge with thanks DSR technical and financial support. The Leibniz Institute of Plant Genetics and Crop Plant Research (IPK, Gatersleben, Germany) is acknowledged for generously providing the seeds of the tomato cultivars.
Footnotes
Peer review under responsibility of King Saud University.
References
- Abu Hena M., Ki-Hyun K., Kwang-Hyun S., Jong-Soon C., Byung-Kee B., Hisashi T., Hwa Young H., Chul-Soo P., Sun-Hee W. Abiotic stress responsive proteins of wheat grain determined using proteomics technique. Aust. J. Crop Sci. 2010;4:196–208. [Google Scholar]
- Ali A., Mageed M., Ahmed I., Mariey S. Genetic and molecular studies on barley salt tolerance. Afr. Crop Sci. Conf. Proc. 2007;8:669–682. [Google Scholar]
- Arefian M., Saeedreza V., Abdolreza B. Biochemical changes and SDS–PAGE analyses of chickpea (Cicer arietinum L.) genotypes in response to salinity during the early stages of seedling growth. J. Biol. Environ. Sci. 2014;8:99–109. [Google Scholar]
- Aydın S., Büyük İ., Aras E.S. Expression of SOD gene and evaluating its role in stress tolerance in NaCl and PEG stressed Lycopersicum esculentum. Turk. J. Bot. 2014;38:89–98. [Google Scholar]
- Azeez M.A., Morakinyo J.A. Electrophoretic characterization of crude leaf proteins in Lycopersicon and Trichosanthes cultivars. Afr. J. Biotecnol. 2004;3:585–587. [Google Scholar]
- Bayoumi T., Manal E., Metwali E. Application of physiological and biochemical indices as a screening technique for drought tolerance in wheat genotypes. Afr. J. Biotechnol. 2008;7:2341–2352. [Google Scholar]
- Bendary M.H.A. Faculty of Agriculture, Ain Shams University; Egypt: 2000. Genetic Studies on the Molecular Biology Bases of Salinity Stress on Barley (Hordeum vulgare L.) [Google Scholar]
- Bhattachrjee S. Reactive oxygen species and oxidative burst: roles in stress, senescence and signal transduction in plant. Curr. Sci. 2005;89:1113–1121. [Google Scholar]
- Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye) binding. Ann. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Burklew C.E., Ashlock J., Winfrey W.B., Zhang B. Effects of aluminum oxide nanoparticles on the growth, development, and microRNA expression of tobacco (Nicotiana tabacum) PLoS One. 2012;7:1–9. doi: 10.1371/journal.pone.0034783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.D., Tabaeizadeh Z. Alteration of gene expression in tomato plants (Lycopersicon esculentum) by drought and salt stress. Genome. 1992;35:385–391. [Google Scholar]
- Cooke R. The characterization and identification of crops by electrophoresis. Electrophoresis. 1984;5:59–72. [Google Scholar]
- Dubey R., Ranu M. Influence of NaCl salinity on growth and metabolic status of proteins and amino acids in Rice seedlings. J. Agron. Crop Sci. 1989;162:97–106. [Google Scholar]
- Duncan D.B. Multiple range and multiple F tests. Biometrics. 1955;11:1–42. [Google Scholar]
- Furdi F. Assessment of genetic diversity in a collection of local tomatoes by SDS-PAGE method. J. Hortic. Forestry Biotechnol. 2012;16:133–136. [Google Scholar]
- Gill S.S., Tuteja N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010;48:909–930. doi: 10.1016/j.plaphy.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Helaly M.N., El-Metwally M.A., El-Hoseiny H., Omar S.A., El-Sheery N.I. Effect of nanoparticles on biological contamination of in vitro cultures and organogenic regeneration of banana. Aust. J. Crop Sci. 2014;8:612–624. [Google Scholar]
- Hernandez J.A., Jimerez A., Mullineaux P., Sevilla F. Tolerance of pea (Pisum sativum) to long term salt stress is associated with induction of antioxidant defenses. Plant Cell Biol. 2000;23:853–862. [Google Scholar]
- Hoagland D.R., Arnon D.I. The water-culture method for growing plants without soil. Calif. Agric. Exp. Station Circular. 1983;347:39. [Google Scholar]
- Khodary S.E.A. Effect of NaCl salinity on improvement of nitrogen metabolism and some ions uptake in lupine plants subjected to gamma irradiation. Int. J. Agric. Biol. 2004;6:1–4. [Google Scholar]
- Laemmli U.K. Cleavage of structural proteins during assembly of head bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laware S., Raskar S. Influence of zinc oxide nanoparticles on growth, flowering and seed productivity in onion. Int. J. Curr. Microbiol. Appl. Sci. 2014;3:874–881. [Google Scholar]
- Livak K.J., Scmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Medeiros F.C.L., Resende M.L.V., Medeiros F.H.V., Zhang H.M., Pare P.W. Defense gene expression induced by a coffee-leaf extracts formulation in tomato. Physiol. Mol. Plant Pathol. 2009;74:175–183. [Google Scholar]
- Miao Z.H., Gaynor J.J. Molecular cloning, characterization and expression of Mn-superoxide dismutase from the rubber tree (Hevea brasiliensis) Plant Mol. Biol. 1993;23:267–277. doi: 10.1007/BF00029003. [DOI] [PubMed] [Google Scholar]
- Mohamed H.E., Hemeida A.E., Mohamed A.G. Role of hydrogen peroxide pretreatment on developing antioxidant capacity in the levels of tomato plant (Lycopersicon esculentum) grown under saline stress. Int. J. Adv. Res. 2015;3:878–879. [Google Scholar]
- Nair R., Varghese S.H., Nair B.G., Maekawa T., Yoshida Y., Kumar D.S. Nanoparticulate material delivery to plants. Plant Sci. 2010;179:154–163. [Google Scholar]
- Noaman S.H., Lamis D.S., El-Sayed A.H., Eman E.S. In vitro selection for water stress tolerant callus line of Helianthus annus L. Cv. Myak. Int. J. Agric. Biol. 2004;6:13–18. [Google Scholar]
- Prasad T., Sudhakar P., Sreenivasulu Y., Latha P., Munaswamy V., Raja K., Sreeprasad T.S., Sajanlal P.R., Pradeep T. Effect of nanoscale zinc oxide particles on the germination, growth and yield of peanut. J. Plant Nutr. 2012;35:905–927. [Google Scholar]
- Reynolds G.H. Forward to the future nanotechnology and regulatory policy. Pacific Res. Inst. 2002;24:1–23. [Google Scholar]
- Rinaldi M., Garofalo P., Rubino P., Steduto P. Processing tomatoes under different irrigation regimes in Southern Italy: agronomic and economic assessments in a simulation case study. Ital. J. Agrometeorol. 2011;3:39–56. [Google Scholar]
- Sheikh N., Hassanzadeh G., Baghestani M., Zabd B. Study the effect of zinc foliar application on the quantitative yield of grain maize under water stress. Electron. J. Crop Prod. 2009;2:59–74. [Google Scholar]
- Soydam S., Büyük İ., Aras S. Relationships among lipid peroxidation, SOD enzyme activity, and SOD gene expression profile in Lycopersicum esculentum L. exposed to cold stress. Genet. Mol. Res. 2013;12:3220–3229. doi: 10.4238/2013.August.29.6. [DOI] [PubMed] [Google Scholar]
- Srinineng K., Salsavoey T., Karnchanatat A. Effect of salinity stress on antoxidantive enzyme activities in tomato in vitro. Pak. J. Bot. 2015;47:1–10. [Google Scholar]
- Sturzenbaum S.R., Kille P. Control genes in quantitative molecular biological techniques: the variability of invariance. Comp. Biochem. Physiol. 2001;13:281–289. doi: 10.1016/s1096-4959(01)00440-7. [DOI] [PubMed] [Google Scholar]
- Ullah M., Akhtar N., Mehmood N., Shah I. Effect of mannitol induced drought stress on seedling traits and protein profile of two wheat cultivars. J. Animal Plant Sci. 2014;24:1246–1251. [Google Scholar]
- Ursini F., Maiorino M., Brigelius-Flohé R., Aumann K.D., Roveri A., Schomburg D., Flohé L. Diversity of glutathione peroxidases. Methods Enzymol. 1995;252:38–53. doi: 10.1016/0076-6879(95)52007-4. [DOI] [PubMed] [Google Scholar]
- Wang Y., Meilan R., Wisniewski M., Uratsu S.L., Cui M., Dandekar A., Fuchigami L. Ectopic expression of Mn-SOD in Lycopersicon esculentum leads to enhanced tolerance to salt and oxidative stress. J. Appl. Hortic. 2007;9:3–8. [Google Scholar]