Abstract
The present report is about Streptomyces sp. isolate ERI-26 isolated from the soil sample of Nilgiri forest, Western Ghats. The methanol extract of ERI-26 showed good antimicrobial activity against tested microbes. The antimicrobial novel anthraquinones were purified by bioactivity-guided fractionation using a silica gel column and preparative HPLC. The compound was characterized and identified by UV, IR, NMR and MASS spectral data. The compound named as 6,61-bis (1,5,7-trihydroxy-3-hydroxymethylanthraquinone), showed significant antimicrobial activities against tested microbes. The isolated compound inhibited the tested bacterial growth, Staphylococcus aureus at 62.5 μg/ml, Staphylococcus epidermidis at 15.62 μg/m, Bacillus subtilis at 62.5 μg/ml, fungi; Trichophyton mentagrophytes at 15.62 μg/m Trichophyton simii at 15.62 μg/ml, Aspergillus niger at. 7.81 μg/ml, Aspergiller flavus at 3.90 μg/ml, Trichophyton rubrum 296 at 62.5 μg/ml, T. rubrum 57/01 at 7.81 μg/ml, Magnaporthe grisea at 15.62 μg/ml. and Botrytis cinerea at 3.90 μg/ml. Isolated anthraquinone compound and its antimicrobial activity were newly reported.
Keywords: Streptomyces sp., Antimicrobial, 16s rRNA, Anthraquinones, NMR, HPLC
1. Introduction
The emerging microbial infection and drug resistant pathogens are creating a major health problem throughout the world. In order to rectify these problems, searching for an effective, new antibiotic compound with significant drug mode of action is very important. The secondary metabolites from actinomycetes, particularly the members those belonging to the phylum Actinobacteria are having a wide variety of chemical metabolites possessing strong biological activities (Spizek et al., 2010). Streptomyces is GC rich, Grampositive, filamentous member of the phylum, Actinobacteria and it produces many pharmaceutically important secondary metabolites such as therapeutic enzymes, antibiotics, immunosuppressants, anti tumour agents and vitamins (Watve et al., 2001). Actinomycetes are usually Gram-positive bacteria containing a percentage of guanine, cytosine higher than 55%, and most of actinomycetes produce mycelia. The actinomycetes are very interesting due to their ability to synthesize biologically active metabolites with diverse chemical molecules. (Valan Arasu et al., 2008). Streptomycetes especially the genus Streptomyces were very potent producers of secondary metabolites including antibacterial enzymes and toxins (Ren et al., 2010). Major types of antibiotics produced by Streptomyces are aminoglycosides, anthracyclines, glycopeptides, b-lactams, macrolides, nucleosides, peptides, polyenes, polyethers, and antetracyclines (Miyadoh, 1993). The widespread use of antibiotics in medicine plays a significant role in the emergence of resistant bacteria and specifically, misuse and/or overuse of antibiotics are the major causes (Mathew et al., 2007, Ferber, 2002, Goossens et al., 2005).
The actinomycete class produces novel secondary metabolites, and sometimes very unique metabolites which significantly show biological activities with low toxicity (Berdy, 2005; Kurtböke, 2012). Most of the antimicrobial compounds have been isolated and characterized from actinomycete group including anthracyclines, aminoglycosides, macrolides, glycopeptides, nucleosides, beta-lactams, peptides, polyketides, tetracyclines and actinomycins by Berdy (2005).
Number of antibiotics are isolated from extracellular of microbes which are diffused in culture media (Bode et al., 2002, Charoensopharat et al., 2008). The present studies are aimed to isolate the novel molecule from Streptomyces sp. ERI-26 and tested against pathogenic bacteria and fungi.
2. Materials and methods
2.1. Isolation and identification
The isolation and identification of selected Streptomyces isolate. ERI-26 have been reported in our earlier article (Valan Arasu et al., 2008).
2.2. Chemical
The pure organic solvents were used in this experiment. Hexane, ethyl acetate and methanol solvents were purchased from Ranbaxy laboratory, Saidapet, Chennai-15. The Silica gel was purchased from Merck (NJ, USA), which is used for thin layer chromatography (TLC) and column chromatography. All other chemicals were pure grade.
2.3. Extract preparation and isolation of active fraction by column chromatography
The isolate Streptomyces sp. isolate ERI-26 was grown in MNGA agar media and kept for incubation for 6 days. After the incubation period the culture with agar media was put into 500 ml of methanol in a conical flask. The content was allowed to diffuse the compounds in methanol. After that it was subjected to centrifuge (4000 rpm, 10 min) and extract was taken in methanol. The methanol phase was evaporated using vacuum rotary evaporator. Finally crude extract was obtained. The extract 5.25 g was subjected to fractionation using silica gel column chromatography. Totally 12 fractions were collected using the solvent system of chloroform, ethylacetate and methanol and their mixture. The active fraction 5 was taken for further isolation of active compound.
2.4. Reversed-phase high performance liquid chromatography (RPHPLC) purification
The 5th fraction yielded 5 g. The fraction 5 was further purified with the help of preparative HPLC with an isocratic elution capability, ultraviolet spectrophotometer as detector and an auto sampler (Waters Alliance System). The purification of 5th fraction in preparative HPLC the solvent system were used: acetonitrile and aqueous acetic acid (15:85, v/v). The flow-rate was 3 ml/min and sample injection volume was 100 μl. The fraction was monitored on the screen also at 254 nm and the peak fraction was carefully collected. The fraction 5 was purified as two fractions. The second fraction B was taken for further structure elucidation which is shown in HPLC 99.14%.
2.5. Identification and characterization of anthraquinone by spectroscopic method
Fraction B was obtained from fraction 5 using preparative HPLC method. Fraction B was submitted to spectroscopic analysis. The 1H NMR (300 MHz), (AL-300 JEOL) spectra were measured and 13C NMR, AL-300 JEOL spectra were measured on a (75.45 MHz). The mass spectrum of the isolated compound (ESI-MS-JEOL instrument) was taken, IR spectrum of the isolated compound was taken from Shimadzu by KBr pellet method.
2.6. Tested microorganisms
The present study was antimicrobial screening of isolated compound B against following pathogenic microbes; Bacteria: Enterococcus faecalis ATCC 29212, Bacillus subtilis MTCC 441, Staphylococcus aureus ATCC 25923, Escherichia coli ATCC 25922, Staphylococcus epidermidis MTCC 3615, Klebsiella pneumoniae ATCC 15380, Pseudomonas aeruginosa ATCC 27853, Proteus vulgaris MTCC 1771. Fungi: Trichophyton rubrum MTCC 296, Trichophyton mentagrophytes 66/01, T. rubrum 57/01, Trichophyton simii 110/02, Aspergillus niger MTCC 1344, Epidermophyton floccosum 73/01, Aspergillus flavus, Curvularia lunata 46/0, Botrytis cinerea, Candida albicans MTCC 227 and Magnaporthe grisea. The tested microbes were obtained from the Institute of Microbial Technology (IMTECH), Chandigarh, India.
2.7. Preparation of inoculums
The preparation of inoculums was followed by standard protocol. The bacterial cells were grown in Mueller Hinton Broth (Himedia) for 24 h at 37 °C. These cell suspensions were diluted with sterile MHB to provide initial cell counts of about 104 CFU/ml. The fungal culture was grown in Sabouraud Dextrose Agar (SDA) slants in suitable temperature at 28 °C for 3 days. After incubation period the spores were collected from slants using sterile DD water and homogenized. The yeast inoculums were prepared by using Sabouraud Dextrose Broth (SDB) at 28 °C for 48 h.
2.8. Minimum inhibitory concentration
The minimum inhibitory concentration of the isolated compound tested against microbes using standard method for bacteria (Duraipandiyan and Ignacimuthu, 2009), fungi (CLSI, 2008) and yeasts (NCCLS, 2002). The isolated compound dissolved in suitable solvents. The concentration of the tested compound was 250 μg/ml. This was serially diluted twofold. The bacterial 100 μl of inoculums (108 CFU/ml) was inoculated in each well and 104 spore/ml of fungi, respectively. The Fluconazole for fungi and Ciprofloxacin for bacteria were included in the assays as positive controls. The plates were incubated in 24, 48 or 72 h at 27 °C for fungi, up to 9 days for dermatophytes. The bacterial plates were incubated for 24 h at 37 °C. After the incubation period the Minimum inhibitory concentration of the tested compound was observed and the lowest extract concentration was noted , showing no visible fungal growth after incubation time.
3. Results and discussion
3.1. Isolation and purification
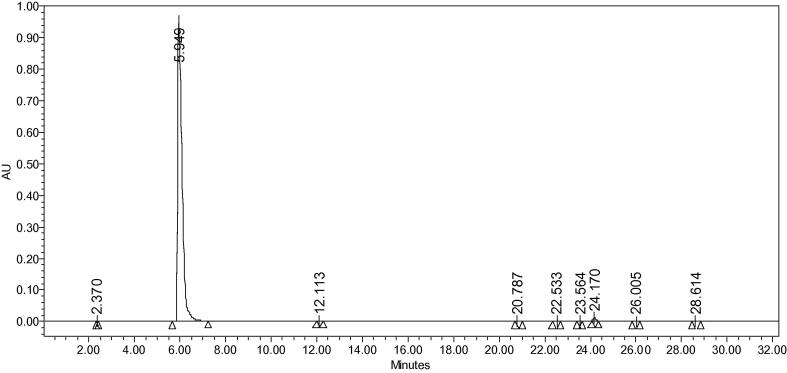
Streptomyces isolate (ERI-26) methanol extract was subjected to chromatography separation. Totally 12 fractions were collected and identified by the screening method, it was reported in our previous article (Valan Arasu et al., 2008). The active fraction 5 was further separated and purified using semi-preparative HPLC. This fraction gave two fractions. Both the fractions purified and showed activity against tested microbes. The fraction A was already published (Duraipandiyan et al., 2014). The fraction B was presently reported. The pure compound was purified by using preparative HPLC. The purified compound was eluted at 5.94 min yielding 95 mg, purity was 99.14% in HPLC (Fig. 1).
Figure 1.
HPLC chromatogram of compound 6,61-bis (1,5,7-trihydroxy-3-hydroxymethylanthraquinone).
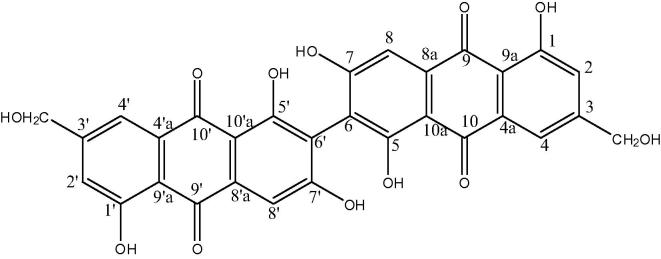
3.2. Structure elucidation of compound 6,61-bis (1,5,7-trihydroxy-3-hydroxymethylanthraquinone)
Further elution of the column with the same solvent mixture gave compound which was crystallized as yellow orange crystals from acetone (yield 82 mg, mp.293°). The compound answered for phenol and also for quinone. TLC of the isolated compound showed single spot with ethyl acetate: methanol: acetic acid 3:2:02 as developing system, (Rf = 0.5) turning pink on exposure to ammonia vapour. Its molecular formula is C30H18O12.
UV λmax MeOH nm: 222,246,272,316,439. IR γmax KBR cm−1: 3429 (hydroxyl), 2924, 2853, 1630 (chelated quinone carbonyl), 1609, 1570, 1450, 1386, 1363, 1421, 1257, 1224, 1100, 1082, 1063, 878, 761, 731. 1H NMR (δ, DMSO-d6, 300 MHz): 7.27 (2H, brs, H-2 and H-2′), 7.65 (2H,brs, H4 and H-4′), 7.32 (2H,brs, H-8 and H-8′), 12.22 and 12.76 (2H each, brs, 4× chelated OH), 11.86 (2H, brs, C-7OH and C-7′ OH) 4.60 (4H, brs, 2× CH2OH). 13C NMR (δ, DMSO-d6, 75 MHz): 159.52 (C-1 and C-1′) 120.89 (C-2 and C-2′), 153.30 (C3 and C-3′), 117.22 (C-4 and C-4′) 132.22 (C-4a and C-4′a), 161.45 (C-5 and C-5′), 112.86 (C-6 and C-6′), 160.07 (C-7 and C-7′), 109.42 (C-8 and C8′), 132.83 (C-8a and C-8′a), 189.86 (C-9 and C-9′), 114. 09 (C-9a and C-9′a), 181.01 (C-10 and C-10′), 107.77 (C-10a and C-10′a), 61.99 (2× CH2OH). EI-MS (m/z): 551 [M-H2O-H]+. The above spectral data showed the compound B to be 6,61-bis (1,5,7-trihydroxy-3-hydroxymethylanthraquinone) which was novel (Fig. 2).
Figure 2.
Structure of the isolated compound 6,61-bis (1,5,7-trihydroxy-3-hydroxylmethylanthraquinone).
3.3. Antimicrobial activities of compound
The compound 6,61-bis (1,5,7-trihydroxy-3-hydroxymethylanthraquinone) was tested against bacteria and fungi. Minimum inhibitory concentration values for the compound are reported in Table 1. The compound (6,61-bis (1,5,7-trihydroxy-3-hydroxymethylanthraquinone) inhibited the growth of S. aureus at 62.5 μg/ml, S. epidermidis at 15.62 μg/ml, B. subtilis at 62.5 μg/ml, E. faecalis at 62.5 μg/ml and P. aeruginosa at 150 μg/ml. Fungi: T. mentagrophytes at 15.62 μg/ml, E. floccosum at 125 μg/ml, T. simii at 15.62 μg/ml, T. rubrum296 at 62.5 μg/ml, T. rubrum 57 at 7.81 μg/ml, A. niger at 7.81 μg/ml, A. flavus at 3.90 μg/ml, B. cinerea 3.90 μg/ml and M. grisea at 15.62 μg/ml. (Table 1). The significant MIC values of tested anthraquinone showed against A. flavus and B. cinerea.
Table 1.
Minimum inhibitory concentrations of compound (6,61-bis (1,5,7-trihydroxy-3-hydroxylmethylanthraquinone) tested microbes.
| Tested organisms | Tested compounds standard |
|
|---|---|---|
| Bacteria | C | Streptomycin |
| Staphylococcus aureus | 62.5 | <0.78 |
| Staphylococcus epidermidis | 15.62 | 6.25 |
| Bacillus subtilis | 62.5 | <0.78 |
| Pseudomonas aeruginosa | 150 | 25 |
| Escherichia coli | >250 | 6.25 |
| Klebsiella pneumoniae | >250 | <0.78 |
| Enterococcus faecalis | 62.5 | 6.25 |
| Proteus vulgaris | >250 | 25 |
| Fungi | Fluconazole | |
| Trichophyton mentagrophytes | 15.62 | 25 |
| Epidermophyton floccosum | 125 | 12.5 |
| Trichophyton rubrum 296 | 62.5 | <12.5 |
| Trichophyton rubrum 57/01 | 7.81 | 25 |
| Trichophyton simii | 15.62 | <12.5 |
| Curvularia lunata | >250 | <12.5 |
| Aspergiller flavus | 3.90 | 25 |
| Magnaporthe grisea | 15.62 | >100 |
| Botrytis cinerea | 3.90 | 25 |
| Aspergillus niger | 7.81 | 100 |
| Candida albicans | 125 | >100 |
Compound: 6,61-bis (1,5,7-trihydroxy-3-hydroxylmethylanthraquinone).
Standard: Streptomycin (antibacterial agent), Fluconazole (antifungal agent).
Above isolated compound spectral data showed anthraquinone skeleton. The spectral data closely resembled to that of citreorosein (ω-hydroxyl emodin) an anthraquinone isolated from plants and fungi (Fujimoto et al., 2004). Compound was isolated as yellow orange crystals from acetone (m.p. 293°); it showed the molecular formula as C30H18O12 based on mass, 1H and 13C NMR (DEPT) spectral data. It gave positive ferric reaction for phenol and also answered for quinone by giving bluish pink colour with alcoholic NaOH. The UV spectrum showed maxima at 222,246,272,316 and 439. The IR spectrum similarly showed the presence of hydroxyl (3429 cm−1) and only chelated quinone carbonyl (1630 cm−1). The small peak corresponding to unchelated quinone carbonyl which appears around 1664 cm−1 in anthraquinones (Thomason, 1977) was absent.
Compound was shown to be dimer of compound (Duraipandiyan et al., 2014), linked at C-6 by its 1H and 13C NMR spectra. The two meta linked aromatic proton H-2 and H-4 adjacent to the hydroxyl methyl group appeared as unresolved broad singlet as usual at δ 7.27 and 7.65. H-8 appeared as singlet δ 7.32. The methylene proton of the hydroxyl methyl group at C-3 appeared as broad singlet at δ 4.60. The chelated hydroxyl groups appeared at δ 12.22 and 12.76. The unchelated hydroxyl groups at C-7 appeared at δ 11.86. In the 13C NMR spectrum the linkage of C-6 and C-61 in the dimeric structure of isolated compound is evident since C-6 appeared as a singlet in compound and moved down field to δ 112.86 from δ 107.88. It is also found that C-5 and C-7 undergo a slight upfield shift.
Isolated compound 6,61-bis (1,5,7-trihydroxy-3-hydroxymethylanthraquinone) is new anthraquinone hitherto unreported from fungi, insects, plants and microbes. The oxynation pattern of the compound does not relate to anthraquinones from the plant, microbes and insect. It is interesting to observe however that recently many reports showed that plant anthraquinones have been reported from Streptomycetes (Cui et al., 2008). Many researchers have reported anthraquinones from actinomycetes. For instance, Cui et al.(2006) have reported that two anthraquinones (Aloesaponarin II and 1,6-dihydroxy-8-hydroxymethylanthraquinone) were isolated from marine Actinomycete isolate M097.
Two novel anthraquinones, were isolated from culture of Micromonospora sp. named a as lupinacidins A (Ia) and B (Ib), are isolated and Lupinacidins show significant inhibitory effects on the invasion of murine colon 26-L5 carcinoma cells without inhibiting cell growth (Igarashi et al., 2007).
Increasing emergence of resistant pathogens emphasizes the need for new and effective antimicrobials. In this investigation, many actinomycete isolates are competent of producing antimicrobial compounds, active against various infectious diseases causing resistant bacteria (Yoo et al., 2007, Sohng et al., 2008, Mellouli et al., 2003). The isolated compound from Streptomyces sp. ERI 26 inhibited the growth of P. aeruginosa at 150 μg /ml. The same organism P. aeruginosa was inhibited by the Streptomyces sp.KH003 at 50 mg /ml of crude extracts. Seung Sik Cho et al., 2012, reported newly isolated Streptomyces sp. CS392 producing antimicrobial compounds which were active against pathogenic microbes.
Previously we have reported an anthraquinone 2,3-dihydroxy-9,10-anthraquinone isolated from Streptomyces galbus ERINLG-127 ethyl acetate extract which showed good antimicrobial activity against tested bacteria and fungi. The compound showed significant MIC values of 12.5 μg/mL against P. aeruginosa, Salmonella typhimurium, K. pneumoniae (ESBL-3894), K. pneumoniae (ESBL-3971), and S. aureus (MRSA) (Balachandran et al., 2014).
Poumale et al. (2006) reported that new anthraquinone isolated from marine Streptomyces sp. which is named as 8-hydroxy-3-methoxy-1-propylanthraquinone and 3,8-dihydroxy-1-propylanthraquinone and the compound showed activity against bacteria at a concentration of 40 μg/ml.
4. Conclusion
The present report is about the isolated novel antimicrobial anthraquinone from Streptomyces sp. ERI 26, compound 6,61-bis (1,5,7-trihydroxy-3-hydroxymethylanthraquinone showed good antimicrobial activity.
Acknowledgement
The Project was fully financially supported by King Saud University, through Vice Deanship of Research Chairs.
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.sjbs.2016.02.008.
Appendix A. Supplementary data
References
- Balachandran C., Arun Y., Duraipandiyan V., Ignacimuthu S., Balakrishna K., Al-Dhabi N.A. Antimicrobial and cytotoxicity properties of 2,3-dihydroxy-9,10-anthraquinone Isolated from Streptomyces galbus (ERINLG-127) Appl. Biochem. Biotechnol. 2014;172:3513–3528. doi: 10.1007/s12010-014-0783-8. [DOI] [PubMed] [Google Scholar]
- Berdy J. Bioactive microbial metabolites. J. Antibiot. (Tokyo) 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- Bode H.B., Bethe B., Höfs R., Zeeck A. Big effects from small changes: possible ways to explore nature’s chemical diversity. Chem. Biol. Chem. 2002;3:619–627. doi: 10.1002/1439-7633(20020703)3:7<619::AID-CBIC619>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Charoensopharat K., Thummbenjapone P., Sirithorn P., Thammasirirak S. Antibacterial substance produced by Streptomyces sp. No 87. Afr. J. Biotechnol. 2008;7:1362–1368. [Google Scholar]
- Cho S.S., Choi Y.H., Simkhada J., Mander P., Park D.J., Yoo J.C. A newly isolated Streptomyces sp. CS392 producing three antimicrobial compounds. Bioprocess Biosyst. Eng. 2012;35:247–254. doi: 10.1007/s00449-011-0599-7. [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute., (CLSI) 2008. Reference method for Broth dilution antifungal susceptibility testing of filamentous fungi; Approved standard second edition. CLSI document M38–A2 (ISBN 1-56238-668-9). Clinical and Laboratory Standards Institute, 940, West valley Road, Suite 1400, Wayne, Pennsylvania 19087-1898 USA.
- Cui H., Shaaban K.A., Qin S. Two anthraquinone compounds from a marine actinomycete isolate M097 isolated from Jiaozhou Bay. World J. Microbiol. Biotechnol. 2006;22:1377–1379. [Google Scholar]
- Cui H.X., Shaaban A.K., Schiebel M., Qin S., Laatsch H. New antibiotic with typical plant anthraquinone structure obtained studying terrestrial and marine Streptomycetes. W. J. Microbiol. Biotechnol. 2008;24:419–421. [Google Scholar]
- Duraipandiyan V., Ignacimuthu S. Antibacterial and Antifungal activity of Flintersine isolated from the traditional medicinal plant, Toddalia asiatica (L.) Lam. J. Ethnopharmacol. 2009;123:494–498. doi: 10.1016/j.jep.2009.02.020. [DOI] [PubMed] [Google Scholar]
- Duraipandiyan V., AL-Dhabi N.A., Balachandran C., Karunai Raj M., Valan Arasu M., Ignacimuthu M. Novel 1,5,7-trihydroxy-3-hydroxy methyl anthraquinone isolated from terrestrial streptomyces sp. (ERI-26) with antimicrobial and molecular docking studies. Appl. Biochem. Biotechnol. 2014;174:1784–1794. doi: 10.1007/s12010-014-1157-y. [DOI] [PubMed] [Google Scholar]
- Ferber D. Antibiotic resistance. Livestock feed ban preserves drugs’ power. Science. 2002;295:27–28. doi: 10.1126/science.295.5552.27a. [DOI] [PubMed] [Google Scholar]
- Fujimoto H., Nakamura E., Okuyama E., Ishibashi M. Six immunosuppressive features from Ascomycete Zopfiella longicaudata found in screening study monitored by immunomodulatory activity. Chem. Pharm. Bull. 2004;52:1065–1068. doi: 10.1248/cpb.52.1005. [DOI] [PubMed] [Google Scholar]
- Goossens H., Ferech M., Vander Stichele R., Elseviers M. Outpatient antibiotic use in Europe and association with resistance: a cross-national database study. Lancet. 2005;365:579–587. doi: 10.1016/S0140-6736(05)17907-0. [DOI] [PubMed] [Google Scholar]
- Igarashi Y., Trujillo M.E., Martinez-molina E., Yanase S., Miyanaga S., Obata T., Sakurai H., Saiki I., Fujita T., Furumai T. Antitumor Anthraquinones from an Endophytic Actinomycete Micromonospora lupini sp. Bioorg. Med. Chem. Lett. 2007;17:3702–3705. doi: 10.1016/j.bmcl.2007.04.039. [DOI] [PubMed] [Google Scholar]
- Kurtböke D.I. Biodiscovery from rare actinomycetes: an eco-taxonomical perspective. Appl. Microbiol. Biotechnol. 2012;93:1843–1852. doi: 10.1007/s00253-012-3898-2. [DOI] [PubMed] [Google Scholar]
- Mathew A.G., Cissell R., Liamthong S. Antibiotic resistance in bacteria associated with food animals: a United States perspective of livestock production. Foodborne Pathog. Dis. 2007;4:115–133. doi: 10.1089/fpd.2006.0066. [DOI] [PubMed] [Google Scholar]
- Mellouli L., Ben Ameur-Mehdi R., Sioud S., Salem M., Bejar S. Isolation, purification and partial characterization of antibacterial activities produced by a newly isolated Streptomyces sp. US24 strain. Res. Microbiol. 2003;154:345–352. doi: 10.1016/S0923-2508(03)00077-9. [DOI] [PubMed] [Google Scholar]
- Miyadoh S. Research on antibiotic screening in Japan over the last decade: a producing microorganisms approach. Actinomycetologica. 1993;9:100–106. [Google Scholar]
- NCCLS, 2002. M27–A2. In National Committee for Clinical Laboratory Standards. Reference method for broth dilution antifungal susceptibility testing of yeasts: proposed standard.
- Poumale H.M.P., Ngadjui B.T., Helmke E., Laatsch H. New anthraquinones from a marine streptomyces sp. – isolation, structure determination and biological activities. Z. Naturforsch. 2006;61b:1450–1454. [Google Scholar]
- Ren H., Zhang P., Liu C., Xue Y., Lian B. The potential use of bacterium strain R219 for controlling of the bloom-forming cyanobacteria in freshwater lake. W. J. Microbiol. Biotechnol. 2010;26:465–472. [Google Scholar]
- Sohng J.K., Yamaguchi T., Seong C.N., Baik K.S., Park S.C., Lee H.J., Jang S.Y., Simkhada J.R., Yoo J.C. Production, isolation and biological activity of nargenicin from Nocardia sp. CS682. Arch. Pharm. Res. 2008;31:1339–1345. doi: 10.1007/s12272-001-2115-0. [DOI] [PubMed] [Google Scholar]
- Spizek J., Novotna J., Rezanka T., Demain L.A. Do we need new antibiotics? the search for new targets. J. Ind. Microbiol. Biotechnol.2010;37:1241. doi: 10.1007/s10295-010-0849-8. [DOI] [PubMed] [Google Scholar]
- Thomason R.H. Academic press; London: 1977. Naturally Occurring Quinones. pp. 67. [Google Scholar]
- Valan Arasu M., Duraipandiyan V., Agastian P., Ignacimuthu S. Antimicrobial activity of Streptomyces spp. ERI-26 recovered from Western Ghats of Tamil Nadu. J. Mycol. Med.2008;18:147–153. [Google Scholar]
- Watve M.G., Tickoo R., Jog M.M., Bhole B.D. How many antibiotics are produced by the genus Streptomyces? Arch. Microbiol. 2001;176:386–390. doi: 10.1007/s002030100345. [DOI] [PubMed] [Google Scholar]
- Yoo J.C., Kim J.H., Ha J.W., Park N.S., Sohng J.K., Lee J.W., Park S.C., Kim M.S., Seong C.N. Production and biological activity of laidlomycin, anti-MRSA/VRE antibiotic from Streptomyces sp. CS684. J. Microbiol. 2007;45:6–10. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.