Abstract
Cyclin A2 activates the cyclin-dependent kinases Cdk1 and Cdk2 and is expressed at elevated levels from S phase until early mitosis. We found that mutant mice that cannot elevate cyclin A2 are chromosomally unstable and tumor-prone. Underlying the chromosomal instability is a failure to up-regulate the meiotic recombination 11 (Mre11) nuclease in S phase, which leads to impaired resolution of stalled replication forks, insufficient repair of double-stranded DNA breaks, and improper segregation of sister chromosomes. Unexpectedly, cyclin A2 controlled Mre11 abundance through a C-terminal RNA binding domain that selectively and directly binds Mre11 transcripts to mediate polysome loading and translation.These data reveal cyclin A2 as a mechanistically diverse regulator of DNA replication combining multifaceted kinase-dependent functions with a kinase-independent, RNA binding–dependent role that ensures adequate repair of common replication errors.
Cyclin A2 is a core cell cycle regulator that activates Cdk1 and Cdk2. Cyclin A2 levels increase upon S phase entry and remain high until its proteasome-dependent destruction in prometaphase (1). Cyclin A2–Cdk complexes that assemble at the onset of S phase drive chromosome duplication through phosphorylation of key DNA replication factors. Subsequently, cyclin A2–Cdk activity initiates mitosis by phosphorylating and inactivating the protein kinase Wee1, resulting in activation and nuclear localization of cyclin B1–Cdk1. During early mitosis, cyclin A2 is implicated in mitotic spindle anchoring (2) and correction of aberrant kinetochore-microtubule attachments (3). Furthermore, in a Cdk-independent manner, cyclin A2 regulates RhoA and RhoC, two guanosine triphosphatases implicated in cell morphogenesis, adhesion, and migration (4).
In mice, cyclin A2 is essential for early embryogenesis, limiting investigations of its biological functions (5). Studies of conditional knockout mice revealed that cyclin A2 is essential for cell cycle progression of certain cell types, including pluripotent and hematopoietic stem cells and select neuronal progenitors, yet is dispensable in fibroblasts because of redundancy with cyclin E in this cell type (6). To uncover novel biological processes that critically depend on a full complement of cyclin A2, we used a combination of knockout (Ccna2−) and hypomorphic (Ccna2H) alleles to markedly down-regulate cyclin A2 expression in mice without overtly affecting embryogenesis or postnatal development. Ccna2H was created by targeted insertion of a neomycin resistance cassette into Ccna2 intron 2; Ccna2− was created by gene trap mutagenesis (fig. S1, A to D). Ccna2−/H mice showed markedly reduced levels of cyclin A2 protein in tissues with a high mitotic index, including small intestine, bone marrow, and spleen (Fig. 1A). In contrast, cyclin A2 levels appeared normal in tissues with few cycling cells, such as brain, liver, and lung. However, actively cycling cultured lung epithelial cells from Ccna2−/H mice had lower amounts of cyclin A2 relative to the wild type (Fig. 1A). Analysis of mouse embryonic fibroblasts (MEFs) confirmed this, with Ccna2−/H MEFs expressing only ~25% of normal cyclin A2 levels (Fig. 1A and fig. S1E). In Ccna2+/+ MEFs, cyclin A2 expression typically started to increase during G1 and peaked from G2 until prophase (fig. S2). Cyclin A2 levels remained low in Ccna2−/H MEFs throughout the cell cycle, resulting in markedly reduced Cdk activity during S and G2 phase (fig. S3). Despite these abnormalities, Ccna2−/H MEFs showed a normal cell cycle profile (fig. S4).
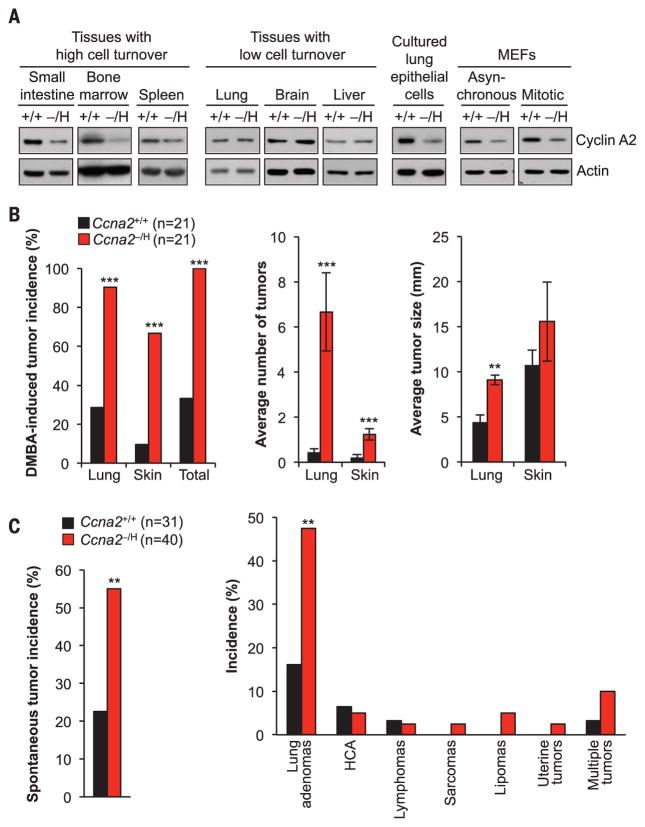
Fig. 1. Cyclin A2 insufficiency drives tumorigenesis.
(A) Immunoblot analysis of cyclin A2 in tissues and cultured primary cells of Ccna2+/+ (+/+) and Ccna2−/H (−/H) mice. Actin was used as a loading control. (B) Incidence, multiplicity, and size of DMBA-induced tumors in 5-month-old mice. (C) Overall and tissue-specific incidences of spontaneous tumors in 16-month-old mice. HCA, hepatocellular adenomas. ** P < 0.01, ***P < 0.001.
Both cyclin A2 deficiency and overabundance have been observed in human tumors and predict poor clinical outcome (7), but whether and how cyclin A2 deregulation drives malignant growth is unknown. To determine whether reduced cyclin A2 expression contributes to tumorigenesis, we treated Ccna2+/+ and Ccna2−/H mice with 7,12-dimethylbenz(a)anthracene (DMBA), a carcinogen that predisposes mice to lung adenomas and skin papillomas (8). Ccna2−/H mice showed a marked increase in tumor incidence and multiplicity in both lung and skin (Fig. 1B). Furthermore, lung adenomas of Ccna2−/H mice were larger in size. Ccna2−/H mice were also more susceptible to spontaneous tumors, particularly lung adenomas (Fig. 1C). Collectively, these data establish that cyclin A2 insufficiency promotes neoplastic transformation.
To identify the underlying defects, we screened for chromosomal instability, a hallmark of human malignancies. Splenocytes and MEFs of Ccna2−/H mice had increased aneuploidy (fig. S5A). Ccna2−/H MEFs showed predisposition to two types of chromosome segregation errors: chromatin bridges and lagging chromosomes (Fig. 2A). The latter are the result of merotelic attachment, a microtubule-kinetochore malattachment caused by spindle defects, including defects in attachment error correction, microtubule dynamics, mitotic timing, centrosome disjunction, and centrosome movement (9). We systematically screened Ccna2−/H MEFs for lagging chromosomes, and by measuring the separation between centrosomes in G2 and prophase, we found that the movement of sister centrosomes to opposite poles was impaired (Fig. 2B and fig. S5, B to F). Cells with delayed centrosome separation form asymmetrical spindles and lagging chromosomes at increased rates (10). Consistent with this, spindle geometry defects occurred at high frequency in Ccna2−/H MEFs (Fig. 2C). Furthermore, lagging chromosomes were more prevalent in Ccna2−/H MEFs with asymmetrical as opposed to symmetrical spindles (fig. S5, G and H).
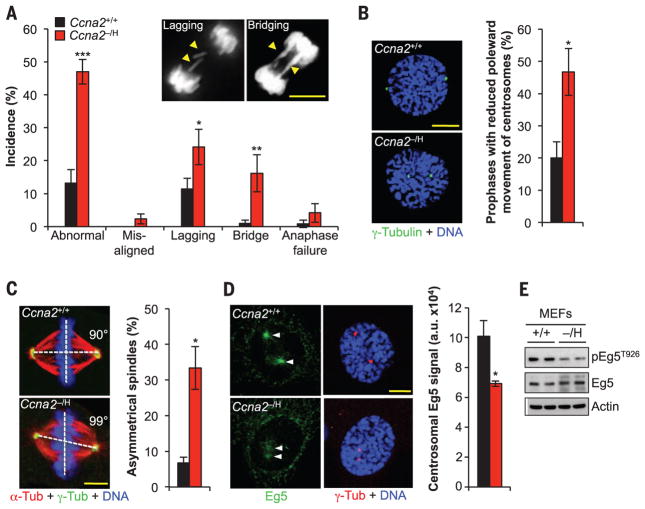
Fig. 2. Cyclin A2–Cdk1 regulates spindle pole movement by targeting Eg5 to centrosomes.
(A) Live-cell imaging analysis of chromosome mis-segregations in MEFs expressing histone H2B tagged with a monomeric form of red fluorescent protein (H2B-mRFP). Inset shows examples of Ccna2−/H MEFs with chromosome lagging or bridging. (B) Left: Images of MEFs in prophase stained for γ-tubulin and DNA. Right: Incidence of prophases with delayed centrosome movement. (C) Left: Maximum-intensity projection images of immunostained metaphases. Right: Incidence of cells with abnormal spindle geometry. (D) Left: Images of prophase MEFs immunostained for Eg5 and γ-tubulin. Right: Quantification of Eg5 signal at centrosomes in prophase. (E) Immunoblot analysis of phosphorylated Eg5T926 and total Eg5 in asynchronously cultured MEFs. Actin was used as loading control. DNA in (B) to (D) was visualized with Hoechst. Scale bars, 10 μm. Data are means ± SEM [N = 6 independent MEF lines per genotype in (A), and N = 3 in (B) to (D); 10 cells per line were analyzed in (B) to (D)]. *P < 0.05, **P < 0.01, ***P < 0.001 (unpaired t test).
The motor protein Eg5 accumulates at centrosomes in prophase to drive centrosome movement (11). Loading of Eg5 was markedly reduced in Ccna2−/H MEFs, as was centrosomal accumulation of cyclin A2 (Fig. 2D and fig. S5I). Centrosome targeting of Eg5 is dependent on Cdk-mediated phosphorylation of Thr926 (T926) (11). T926 phosphorylation was reduced in Ccna2−/H MEFs despite normal expression of Eg5 and alternative Cdk1 and Cdk2 partners, including cyclins B and E (Fig. 2E and fig. S5J). Restoration of cyclin A2 expression in Ccna2−/H MEFs normalized T926 phosphorylation and corrected centrosomal loading of Eg5, centrosome movement, and spindle geometry (fig. S5, K to N). Collectively, these results uncover a nonredundant catalytic role of cyclin A2–Cdk in targeting Eg5 to centrosomal microtubules for proper centrosome movement and spindle formation.
Chromatin bridges, the other chromosome segregation defect observed in Ccna2−/H MEFs, can be caused by defects in DNA replication, DNA decatenation, or cohesin cleavage (12, 13). Ccna2−/H MEFs properly targeted topoisomerase IIα to inner centromeric regions early in mitosis and had normal separase activity (fig. S6, A and B), which suggests that bridge formation was unlikely to result from decatenation or cohesin cleavage defects. In DNA fiber assays, replication forks of Ccna2−/H cells progressed at reduced rates and stalled more frequently than did Ccna2+/+ cells (Fig. 3A and fig. S6, C to G). Phosphorylation of checkpoint kinase 1 (Chk1) at Ser345 (S345), a marker of replication stress (14), was elevated in Ccna2−/H MEFs (Fig. 3B). Furthermore, the efficiency with which hydroxyureastalled replication forks of Ccna2−/H MEFs restarted was markedly reduced (fig. S6H). Unreplicated single-stranded DNA (ssDNA) at stalled replication forks is vulnerable to breakage (14). Visualization of DNA damage repair foci by staining for the presence of the DNA repair proteins γH2AX and 53BP1 demonstrated that Ccna2−/H cells accumulate more double-stranded DNA breaks (DSBs; fig. S7, A to F). Furthermore, radiation-induced DSBs were repaired with reduced efficiency in Ccna2−/H MEFs (fig. S7, G and H). Rad51, a key component of DSB repair (15), failed to accumulate at radiation-induced DSBs in a large proportion of Ccna2−/H MEFs (fig. S7, I to L). Rad51 accumulation at DSBs is dependent on end resection, a key step in the repair process that is measured by staining for RPA2, a repair protein that stabilizes ssDNA intermediates (15). Analysis of RPA2 and γH2AX at 8 hours after γ-irradiation revealed that RPA2 localization at DSBs was markedly reduced in Ccna2−/H MEFs (fig. S7, M and N).
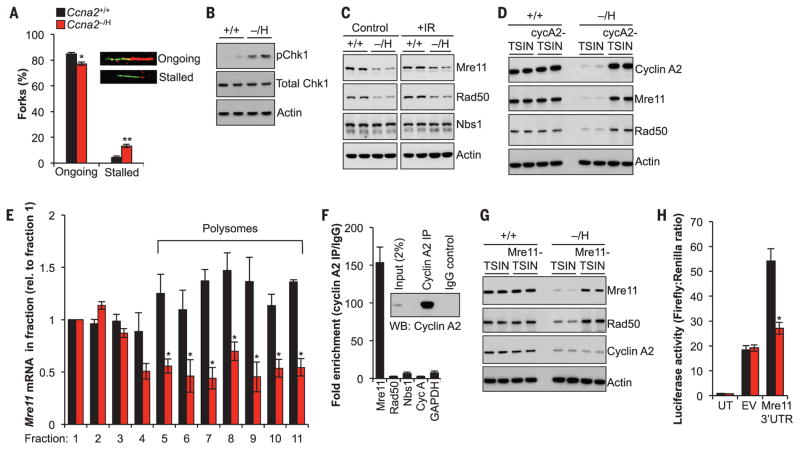
Fig. 3. Cyclin A2 interacts with Mre11 mRNA to regulate its translation.
(A) Percentages and examples of ongoing and stalled replication forks in MEFs. Inset shows examples of fibers. (B) Immunoblot analysis for Chk1 phosphorylation at S345. (C) Immunoblots of control and γ-irradiated (IR) MEF lysates probed for the indicated proteins. (D) Immunoblot analysis of asynchronously cultured MEFs transduced with control (TSIN) or Ccna2-containing (cycA2-TSIN) lentiviruses. (E) Quantification of Mre11 mRNA in polysome fractions of MEFs. (F) Cyclin A2 RNA immunoprecipitation (IP) in MEFs followed by RT-qPCR analysis for the indicated genes. IgG, immunoglobulin G; WB, Western blot. (G) As (D) but using control (TSIN) and Mre11-containing lentivirus. (H) Luciferase activity in cells transfected with 3′UTR of Mre11 mRNA. UT, untransfected; EV, empty vector. Actin was used as loading control in (B), (C), (D), and (G). Data in (A), (E), (F), and (H) are means ± SEM (N = 3 independent MEF lines per genotype). *P < 0.05, **P < 0.01 (unpaired t test).
The MRN complex, composed of Mre11, Rad50, and Nbs1, plays a central role in both replication fork restart and DSB repair (15). Western blot analysis revealed that levels of Mre11 and Rad50 protein, but not Nbs1 protein, were consistently reduced in both irradiated and nonirradiated Ccna2−/H MEFs (Fig. 3C). Levels of Mre11 and Rad50 were also low in Ccna2−/H tumors, although Cdk2 activity appeared normal (fig. S8). Analysis of cell cycle–dependent expression showed that both Mre11 and Rad50 levels peaked in Ccna2+/+ MEFs during S and G2 phase, similar to cyclin A2 expression (figs. S9 and S10). Expression of Mre11 and Rad50 in S and G2 phases was substantially lower in Ccna2−/H MEFs. Ectopic expression of cyclin A2 restored both Mre11 and Rad50 protein levels (Fig. 3D). Mre11 and Rad50 mRNA levels and ratios of nuclear to cytoplasmic distribution in Ccna2−/H MEFs were similar to those of Ccna2+/+ MEFs (fig. S11, A to C), indicating that reduced expression of Mre11 and Rad50 was not due to altered transcription or defective nuclear export. Furthermore, proteasome degradation was not responsible, as treatment of Ccna2−/H MEFs with the proteasome inhibitor MG132 did not alter Mre11 and Rad50 levels (fig. S11D).
However, Mre11 and Rad50 mRNA abundance in polysomes was reduced in Ccna2−/H MEFs, suggesting reduced translation (Fig. 3E and fig. S12, A to C). To determine whether this was a direct effect, we immunoprecipitated cyclin A2 and analyzed it for the presence of Mre11, Rad50 transcripts, and several control transcripts by quantitative reverse transcription polymerase chain reaction (RT-qPCR). Intriguingly, cyclin A2 selectively precipitated Mre11 transcripts in Ccna2+/+ MEFs (Fig. 3F). Similar results were obtained using human primary fibroblasts and HeLa cells (fig. S12D). Cyclin A2 knockdown in these cells resulted in reduced MRE11 and RAD50 protein levels, consistent with results in Ccna2−/H MEFs (fig. S12E). Cyclin A1 had no Mre11 RNA binding ability (fig. S12, F and G). Two observations suggested that cyclin A2 regulation of Mre11 and Rad50 was Cdk-independent: Neither Cdk2 nor Cdk1 immunoprecipitation enriched Mre11 transcripts comparably to cyclin A2 immunoprecipitation (fig. S12H), and inhibition of Cdk2 activity in wild-type MEFs by roscovitine had no impact on Mre11 levels (fig. S12I). Mre11 knockdown resulted in low Rad50 protein levels, but Rad50 depletion did not affect Mre11, indicating that Rad50 abundance is Mre11-dependent (fig. S13, A and B). Ectopic expression of Mre11 in Ccna2−/H MEFs normalized both Mre11 and Rad50 protein levels but had no impact on cyclin A2 levels (Fig. 3G). Coincidentally, polysome association of Rad50 mRNA was restored (fig. S13C), reinforcing the idea that Rad50 translation is Mre11-dependent, although Mre11 immunoprecipitates did not contain Rad50 transcripts (fig. S13D). Ectopic expression of Mre11 in Ccna2−/H cells rescued DSBs and chromatin bridges, indicating that Mre11 deficiency is the driver of these chromosomal phenotypes (fig. S13, E and F). Expression of CtIP, a binding partner of the MRN complex implicated in end resection, was subnormal in Ccna2−/H MEFs through a Cdk2-dependent, Mre11-independent mechanism and therefore is unlikely to contribute to the chromosomal phenotype of Ccna2−/H cells (fig. S14).
RNA sequencing analysis of cyclin A2, E2, and B1 immunoprecipitates demonstrated that cyclin A2 exclusively binds Mre11 transcripts and that cyclins E2 and B1 have no RNA binding ability (tables S1 to S3). Mre11 RNA sequence reads exclusively mapped to the 3′ untranslated region (3′UTR), indicating that this region contains the cyclin A2 binding site (fig. S15A). When introduced downstream of a luciferase reporter, the Mre11 3′UTR enhanced luciferase activity in Ccna2+/+ MEFs but not in Ccna2−/H MEFs (Fig. 3H). Ectopic expression of cyclin A2 in Ccna2−/H MEFs corrected this deficiency (fig. S15B).
Expression of hemagglutinin (HA)–tagged cyclin A2 deletion mutants in MEFs revealed that residues 302 to 432 are necessary and sufficient for Mre11 transcript binding (Fig. 4A). This fragment, which lacks Cdk-binding ability, restored Mre11 and Rad50 expression and prevented DSB formation in Ccna2−/H MEFs (Fig. 4B and fig. S16A) but did not correct Cdk-dependent phenotypes such as Eg5 loading, centrosome movement, spindle geometry, and chromosome lagging (fig. S16, B to F). Conversely, expression of amino acids 1 to 301, a fragment that binds to Cdk, corrected the latter defects but failed to restore Mre11 and Rad50 expression and DSB repair (fig. S16, G to K). Using deletion mutagenesis, we identified two subregions within the Mre11 3′UTR, a 150-nucleotide (nt) segment at the 5′ end and a 34-nt segment at the 3′ end, as potential cyclin A2 binding sites (Fig. 4C). Recombinant glutathione S-transferase (GST)–cyclin A2302–432 directly bound a radiolabeled RNA probe spanning the last 32 nt of the Mre11 3′UTR in an electrophoretic mobility shift assay, but not a scrambled 32-nt oligomer (32-mer) (Fig. 4D and fig. S17, A and B). Neither GST–cyclin A21–209 nor GST–cyclin A2302–432(4M), which carries mutations in four putative ribonucleic acid contact sites required for restoration of Mre11 and Rad50 expression in Ccna2−/H MEFs by ectopic expression of cyclin A2 (fig. S17, B to E), was able to bind the Mre11 RNA probe (Fig. 4D).
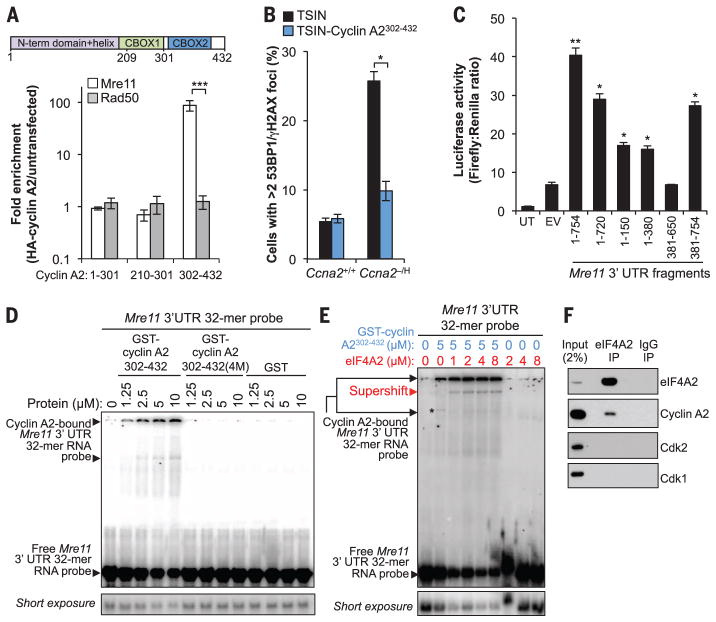
Fig. 4. The cyclin A2 C terminus directly binds to a conserved sequence in the Mre11 3′UTR.
(A) RNA immunoprecipitation in wild-type MEFs expressing the indicated HA-tagged cyclin A2 deletion mutants. RNAs coprecipitating with HA–cyclin A2 mutants were analyzed by RT-qPCR. (B) Quantification of cyclin A2302–432–expressing cells with two or more γH2AX- and 53BP1-colocalized foci. (C) Luciferase activity in MEFs transfected with different regions of the Mre11 3′UTR. (D) Representative native gel shifts showing a specific interaction between recombinant cyclin A2302–432 and the Mre11 3′UTR conserved sequence. (E) Same as (D) but in the presence of recombinant human eIF4A2. (F) Western blot analysis of eIF4A2-coprecipitating proteins in wild-type MEFs. Data in (A) to (C) are means ± SEM (N = 3 independent MEF lines per genotype). *P < 0.05, **P < 0.01, ***P < 0.001 (unpaired t test).
Translation initiation factor eIF4A2, a previously documented putative cyclin A2 interactor (16), supershifted GST–cyclin A2302–432–bound Mre11 RNA complexes but did not bind the probe in the absence of cyclin A2 (Fig. 4E). eIF4A2, but not Cdk1 or Cdk2, coimmunoprecipitated cyclin A2 under physiological conditions, indicating that cyclin A2 bound to Mre11 mRNA is free of Cdk binding partners (Fig. 4F). Cyclin A2 interacted with eIF4A2 and Mre11 RNA through a common domain (fig. S17F). Finally, using the CRISPR/Cas9 technique, we generated mice homozygously lacking the last 47 nt of the Mre11 3′UTR (designated Mre11ΔA2/ΔA2 MEFs), which were viable and overtly normal (fig. S18). Mre11ΔA2/ΔA2 MEFs had normal amounts of Mre11 mRNA, but the transcripts showed markedly reduced polysome association, resulting in low Mre11 protein levels (fig. S19, A to D). Mre11ΔA2/ΔA2 MEFs entirely phenocopied Ccna2−/H MEFs, except for phenotypes related to phosphorylation of Eg5 mediated by cyclin A2–Cdk (fig. S19, E to K, and fig. S20).
Our work exposed two novel cyclin A2 functions. First, we identified a role for the cyclin A2 N terminus in the formation of bipolar mitotic spindles by regulating Eg5 loading onto centrosomes through phosphorylation of T926. Second, the C terminus of cyclin A2, which has low sequence conservation among cyclin family members, directly binds to a conserved region in the 3′UTR of Mre11 transcripts to promote their translation, presumably through an interaction with eIF4A2. In doing so, cyclin A2 coordinates its well-established Cdk-dependent functions in the initiation and progression of DNA synthesis (16) while simultaneously reinforcing stabilization of the key machinery responsible for repairing DNA lesions caused by replication errors. Cyclins have hitherto not been implicated in RNA binding. The observation that cyclin A2 seemingly interacts with just a single transcript, Mre11 mRNA, was unexpected and appears to be a unique feature among RNA binding proteins.
Supplementary Material
Acknowledgments
We thank W. Zhou, M. Li, F. Jin, and X. Wang for assistance and D. Compton for providing photoactivatable GFP-tagged α-tubulin. Supported by NIH grants CA126828 and CA168709 (J.M.v.D.) and CA166025 (L.J.M.).
Footnotes
REFERENCES AND NOTES
- 1.Hochegger H, Takeda S, Hunt T. Nat Rev Mol Cell Biol. 2008;9:910–916. doi: 10.1038/nrm2510. [DOI] [PubMed] [Google Scholar]
- 2.Beamish H, et al. J Biol Chem. 2009;284:29015–29023. doi: 10.1074/jbc.M109.042820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kabeche L, Compton DA. Nature. 2013;502:110–113. doi: 10.1038/nature12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arsic N, et al. J Cell Biol. 2012;196:147–162. doi: 10.1083/jcb.201102085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murphy M, et al. Nat Genet. 1997;15:83–86. doi: 10.1038/ng0197-83. [DOI] [PubMed] [Google Scholar]
- 6.Kalaszczynska I, et al. Cell. 2009;138:352–365. doi: 10.1016/j.cell.2009.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yam CH, Fung TK, Poon RY. Cell Mol Life Sci. 2002;59:1317–1326. doi: 10.1007/s00018-002-8510-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Serrano M, et al. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 9.Naylor RM, Jeganathan KB, Cao X, van Deursen JM. J Clin Invest. 2016;126:543–559. doi: 10.1172/JCI82277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Ree JH, Nam HJ, Jeganathan KB, Kanakkanthara A, van Deursen JM. Nat Cell Biol. 2016;18:814–821. doi: 10.1038/ncb3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slangy A, et al. Cell. 1995;83:1159–1169. doi: 10.1016/0092-8674(95)90142-6. [DOI] [PubMed] [Google Scholar]
- 12.Nam HJ, van Deursen JM. Nat Cell Biol. 2014;16:538–549. doi: 10.1038/ncb2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burrell RA, et al. Nature. 2013;494:492–496. doi: 10.1038/nature11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeman MK, Cimprich KA. Nat Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stracker TH, Petrini JH. Nat Rev Mol Cell Biol. 2011;12:90–103. doi: 10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pagliuca FW, et al. Mol Cell. 2011;43:406–417. doi: 10.1016/j.molcel.2011.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.