Abstract
Chromosome 7q11.23 duplication syndrome is a well-recognised syndrome which involves the duplication of the same genes located in the Williams-Beuren critical region. However, in 2010, 4 patients were reported with a microduplication only in the HIP1 and YWHAG genes. We refer to this as a distal 7q11.23 duplication (dup7q11.23D). Here, we report the fifth de novo patient with dup7q11.23D, whose symptoms may be explained by YWHAG overexpression as was demonstrated recently in mice and obese patients. Finally, further studies will be necessary to delineate this emerging microduplication syndrome.
Key Words: Chromosome 7q11.23 region, Development delay, HIP1, Williams-Beuren duplication syndrome, YWHAG
The chromosome 7q11.23 region is widely recognised because it contains the critical genes leading to Williams-Beuren microdeletion (MIM 194050) and microduplication (MIM 609757) syndromes [Zarate et al., 2014; Morris et al., 2015]. It is flanked by 3 main clusters of low-copy repeats (LCR) named proximal (LCR-P), central (LCR-C), and distal (LCR-D), and it encompasses many and highly transcribed genes. Indeed, its gene density and transcriptional rate are significantly higher when compared with other segments of chromosome 7 [Ebert et al., 2014]. To date, the genes between LCR-P and LCR-C have been marked as causative of these 2 disorders, mainly ELN, GTF2I, and GTF2IRD1 [Antonell et al., 2010; Zarate et al., 2014]. Subsequently, in 2010, the deletion of the distal portion (between LCR-C and LCR-D) was identified as a different entity, named distal chromosome 7q11.23 deletion (MIM 613729). The haploinsufficiency of the genes HIP and YWHAG was suggested to be critical for the manifestations of that microdeletion syndrome [Ramocki et al., 2010]. Although 4 patients with a duplication of HIP or HIP/YWHAG were also described by Ramocki et al. [2010], which did not comprise the genes between LCR-P and LCR-C, there was no evidence of the relevance of the overexpression of these genes. However, Cornell et al. [2016] recently provided strong evidence that the duplication of YWHAG causes neuronal migration delay in the developing cerebral cortex of mice, in a similar manner as YWHAG knockdown does. Here, we report the fifth patient with a distal 7q11.23 duplication (dup7q11.23D) and show the manifestations which could characterise this emerging microduplication syndrome.
Patient and Methods
Clinical Report
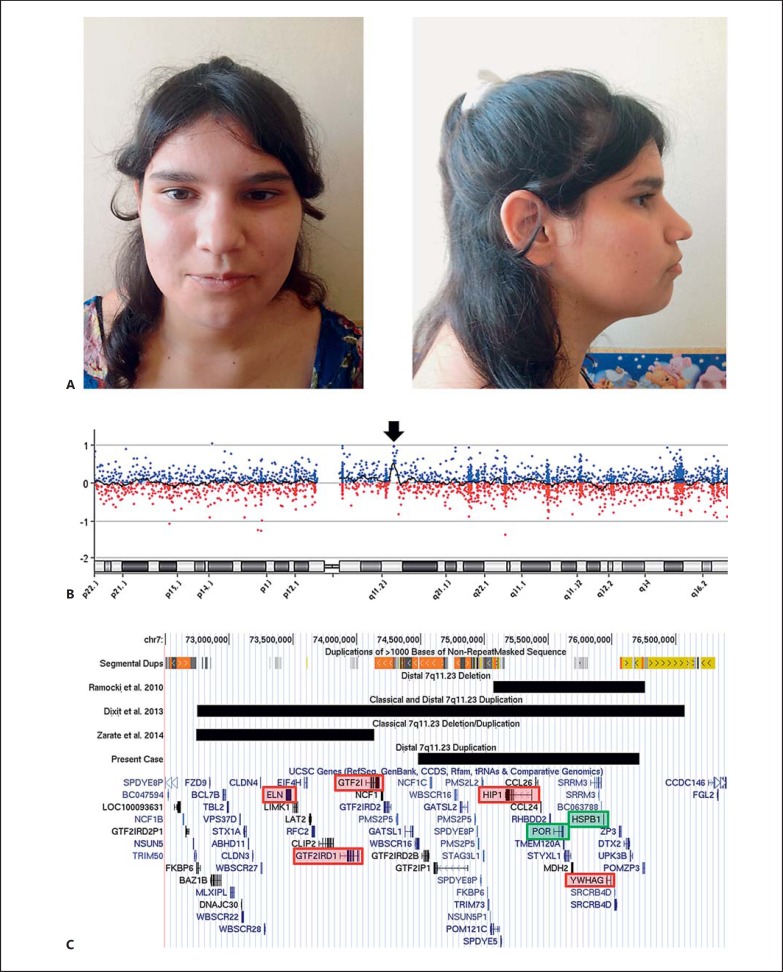
A 16-year-old female patient was referred to our centre from a psychiatric unit with a suspicion of Prader-Willi syndrome due to obesity, mild intellectual delay, aggressiveness as well as attention deficit hyperactivity, anxious and impulsive control disorders. She is the second child of nonconsanguineous healthy parents; her mother has a lower IQ and a normal physical phenotype. The older brother was diagnosed with a bipolar disorder; he was not willing to participate in this study and refused any genetic analyses. Our patient had a normal perinatal history but subsequently showed global development delay as reported by the parents. Neurological evaluations revealed a normal EEG and brain CT. Since 1 year of age, she manifested obesity, hyperphagia and insulin resistance, which has been managed with metformin. Currently, she is being treated with anxiolytic and antipsychotic drugs as well as mood stabilizers. At physical examination, her height was 165 cm (64th percentile) and her weight was 76.5 kg. Thus, her BMI was in the obese range at 28.1 (96th percentile). She showed mild facial dysmorphisms such as a broad face, straight eyebrows, deep-set eyes, a protruding nasal tip, a high-arched palate as well as a mild systolic heart murmur (fig. 1A). The echocardiography did not reveal abnormalities in the heart structure. The girl's developmental milestones, IQ scores of the family, and patient's growth curves and insulin levels were not available.
Fig. 1.
Clinical and molecular findings of our patient with dup7q11.23D. A Facial characteristics of the patient, which are subtle in general. B Array-CGH finding in 7q11.23, which clearly demonstrates the duplication (arrow). C Schematic visualisation of dup7q11.23D and its relationship with other pathogenic deletions/duplications reported in the same region, its LCRs (or segmental duplications), and the compromised genes, using the GRCh37/hg19 genome assembly. The genes in the red boxes are critical for the classical 7q11.23 deletion/duplication (ELN, GTF2I, and GTF2IRD1) and distal 7q11.23 deletion/duplication (HIP1 and YWHAG) as described by Zarate et al. [2014] and Ramocki et al. [2010], respectively. The genes in the green boxes were duplicated using the MLPA P029 WBS probemix.
Methods
Blood samples (5 ml) were collected in ethylenediaminetetraacetic acid tubes for DNA extraction and in heparin tubes for chromosome analysis. DNA was extracted from the patient and her parents using the Wizard Genomic DNA Purification Kit, according to the manufacturer's protocols (Promega, Madison, Wis., USA). Only the patient underwent conventional chromosome analysis and SNRPN methylation studies via methylation-sensitive PCR analysis (MS-PCR), whereas the girl and her parents were examined using chromosome microarray and MLPA analyses. The metaphase chromosome spreads were obtained from patient lymphocytes using conventional methods, and GTG-banded chromosomes were analysed at the 550-band level. MS-PCR was carried out as recommended by Kubota et al. [1997] for studying the suspicion of Prader-Willi syndrome. Subsequently, chromosomal microarray analysis was performed using the Agilent 8x60K, ISCA design platform (Agilent Technologies, Santa Clara, Calif., USA), and all procedures were carried out as instructed by the manufacturer. The analysis was done using the Agilent CytoGenomics Software and its algorithm ADM-2. Finally, we performed MLPA to confirm the findings with 250 ng of genomic DNA using the SALSA P029 Williams-Beuren Syndrome probemix (MRC-Holland, Amsterdam, The Netherlands), following the manufacturer's instructions. The amplified probes were detected by an automated capillary system (ABI PRISM 310, Applied Biosystems, Tokyo, Japan), and the results were analysed using the software Coffalyser (MRC-Holland), which were standardised to the normal controls. Genes with MLPA ratios <0.7 were defined as deletions, ratios ≥0.7 and ≤1.3 were considered normal, whereas all ratios >1.3 were defined as duplications.
Results
Initial evaluations showed both a normal karyotype and Prader-Willi syndrome MS-PCR analysis in the patient. The chromosome microarray technique revealed a duplication of 1.7 Mb in the 7q11.23 region that did not compromise ELN but a more distal portion (mean log2 ratio = 0.55; 74,481,481-76,214,077 bp; genome assembly GRCh37/hg19; fig. 1B). This CNV is restricted by segmental duplications of more than 98% of similarity, which encompasses more than 30 UCSC genes, including HIP1 and YWHAG (fig. 1C). This distal duplication was confirmed by MLPA for Williams-Beuren syndrome, which revealed a mean ratio >1.35 only for the 5 probes located at genes POR and HSPB1 (fig. 1C). Both parents were examined with both techniques and had normal results (data not shown).
Discussion
As seen in table 1, it seems that dup7q11.23D exhibits a predominantly neuropsychiatric phenotype with a few dysmorphic characteristics. Interestingly, the 3 patients with a duplication of YWHAG manifested attention deficit hyperactivity disorder and aggressiveness, whereas the 2 patients with involvement of only HIP1 had central nervous system abnormalities, including a neoplasm. The duplication of YWHAG causes a delayed migration of pyramidal neurons to external layers of the cerebral cortex in different stages of brain development in mice. This is explained by a deficit in the locomotion stage of neuronal migration and not by an altered neurogenesis, which may reflect pathologic changes on microtubule dynamics [Cornell et al., 2016]. Importantly, this aberration was extremely similar to that seen in foetal mice brains when the YWHAG gene was knocked down, which reflects that a correct dose of the protein product of this gene is necessary for a normal brain development [Cornell et al., 2016]. Furthermore, the obesity seen in our patient may also be explained by the overexpression of this gene as was demonstrated by Capobianco et al. [2012] because this gene is also involved in both cellular insulin-mediated glucose transport and lipid metabolism. Concerning HIP1, it has been demonstrated that its protein is overexpressed in brain cancers [Bradley et al., 2007], which may clarify why patient 2 developed a spinal cord schwannoma. Nevertheless, it is unclear how the HIP1 duplication can contribute to the neuropsychiatric phenotype, albeit the gene dose of this part of the 7q11.23 region may be significantly important for neural development [Cornell et al., 2016].
Table 1.
Characteristics and patients
| Characteristics | Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 |
|---|---|---|---|---|---|
| Gender | male | male | male | female | Female |
| Age at diagnosis | 3 months | 34 years | 16 years | 5 years | 16 years |
| Mode of inheritance | paternal | ND | ND | ND | de novo |
| Critical genes compromised | HIP1 | HIP1 | HIP1, YWHAG | HIP1, YWHAG | HIP1, YWHAG |
| Growth parameters | |||||
| Weight | ND | ND | ND | ND | p96 |
| Stature | ND | ND | ND | ND | p64 |
| Head circumference | ND | ND | ND | ND | p40–50 |
| Birth weight | ND | ND | ND | ND | Normal |
| Major congenital/structural anomalies | ND | ND | ND | ND | - |
| Medical diseases | ND | ND | ND | ND | + |
| Physical manifestations | |||||
| Broad face | ND | ND | ND | ND | + |
| Deep-set eyes | ND | ND | ND | ND | + |
| Straight eyebrows | ND | ND | ND | ND | + |
| Other minor corporal anomalies | ND | ND | ND | ND | + |
| Neuropsychiatric characteristics | |||||
| Speech delay/disorder | ND | ND | ND | + | + |
| Motor development delay/disorder | ND | ND | ND | ND | + |
| Behavioural problems (aggressiveness) | ND | ND | + | + | + |
| Intellectual disability | ND | ND | ND | ND | + |
| Attention deficit hyperactivity disorder | ND | ND | + | + | + |
| Central nervous system anomalies | Chiari III malformation, hydrocephalus | spinal cord schwannoma | ND | ND | – |
| Other neuropsychiatric problems | ND | ND | bipolar disorder | ND | anxious and impulsive control Disorders |
+ = Present; – = absent; ND = not determined/described; p = percentile. Patients 1–4 from Ramocki et al. [2010] and patient 5 from this study.
Although it is premature to compare our case with the Williams-Beuren duplication syndrome and considering the whole clinical data of our patient and of those patients reported by Ramocki et al. [2010] were not available and well delineated, this microduplication may involve less congenital malformations and dysmorphic features when compared with the Williams-Beuren duplication [Morris et al., 2015]. This could explain why some patients with a duplication of the entire 7q11.23 region (fig. 1C) did not manifest a more severe phenotype when compared with those patients with a shorter duplication [Berg et al., 2007; Dixit et al., 2013].
Finally, and as for Williams-Beuren deletion/duplication, the nonallelic homologous recombination is the most plausible mechanism which explains the emergence of dup7q11.23D due to the high similarity of the flanking LCRs. This mechanism may also explain the inversion of this region, which is a predisposing factor for both Williams-Beuren syndrome and the 7q11.23 duplication [Morris et al., 2015]. Albeit the mother should be tested in the future for this rearrangement and her son's genetic status is unknown, we could hypothesise that the mother carries this inversion and the girl's brother also inherited this duplication or a similar one. Moreover, the mother does not have conspicuous facial features, which is concordant with the fact that the 7q11.23 inversion may not involve severe clinical symptoms [Tam et al., 2008]. However, it has been recently demonstrated that the disruption of the ‘topologically associating domain’, i.e., distant and separated regions of the genome that share the same enhancers, promoters, and transcriptional machinery by chromosomal rearrangements, can cause gene misexpression and disease [Lupiáñez et al., 2016]. Thus, this situation could explain the clinical suspicion of a lower IQ but a normal physical phenotype in the mother. Finally, we encourage other clinicians to report patients with similar amplifications in order to delineate and establish clinical guidelines for long-term care management.
Statement of Ethics
The authors have no ethical conflicts to disclose.
Disclosure Statement
The authors have no conflicts of interest to declare.
Acknowledgement
We would like to thank the patient and her family for their collaboration.
References
- 1.Antonell A, Del Campo M, Magano LF, Kaufmann L, De la Iglesia JM, et al. Partial 7q11.23 deletions further implicate GTF2I and GTF2IRD1 as the main genes responsible for the Williams-Beuren syndrome neurocognitive profile. J Med Genet. 2010;47:312–320. doi: 10.1136/jmg.2009.071712. [DOI] [PubMed] [Google Scholar]
- 2.Berg JS, Brunetti-Pierri N, Peters SU, Kang SH, Fong CT, et al. Speech delay and autism spectrum behaviors are frequently associated with duplication of the 7q11.23 Williams-Beuren syndrome region. Genet Med. 2007;9:427–441. doi: 10.1097/gim.0b013e3180986192. [DOI] [PubMed] [Google Scholar]
- 3.Bradley SV, Holland EC, Liu GY, Thomas D, Hyun TS, Ross TS. Huntingtin interacting protein 1 is a novel brain tumor marker that associates with epidermal growth factor receptor. Cancer Res. 2007;67:3609–3615. doi: 10.1158/0008-5472.CAN-06-4803. [DOI] [PubMed] [Google Scholar]
- 4.Capobianco V, Nardelli C, Ferrigno M, Iaffaldano L, Pilone V, et al. miRNA and protein expression profiles of visceral adipose tissue reveal miR-141/YWHAG and miR-520e/RAB11A as two potential miRNA/protein target pairs associated with severe obesity. J Proteome Res. 2012;11:3358–3369. doi: 10.1021/pr300152z. [DOI] [PubMed] [Google Scholar]
- 5.Cornell B, Wachi T, Zhukarev V, Toyo-Oka K. Overexpression of the 14-3-3gamma protein in embryonic mice results in neuronal migration delay in the developing cerebral cortex. Neurosci Lett. 2016;628:40–46. doi: 10.1016/j.neulet.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 6.Dixit A, McKee S, Mansour S, Mehta SG, Tanteles GA, et al. 7q11.23 Microduplication: a recognizable phenotype. Clin Genet. 2013;83:155–161. doi: 10.1111/j.1399-0004.2012.01862.x. [DOI] [PubMed] [Google Scholar]
- 7.Ebert G, Steininger A, Weißmann R, Boldt V, Lind-Thomsen A, et al. Distribution of segmental duplications in the context of higher order chromatin organisation of human chromosome 7. BMC Genomics. 2014;15:537. doi: 10.1186/1471-2164-15-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kubota T, Das S, Christian SL, Baylin SB, Herman JG, Ledbetter DH. Methylation-specific PCR simplifies imprinting analysis. Nat Genet. 1997;16:16–17. doi: 10.1038/ng0597-15. [DOI] [PubMed] [Google Scholar]
- 9.Lupiáñez DG, Spielmann M, Mundlos S. Breaking TADs: how alterations of chromatin domains result in disease. Trends Genet. 2016;32:225–237. doi: 10.1016/j.tig.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Morris CA, Mervis CB, Paciorkowski AP, Abdul-Rahman O, Dugan SL, et al. 7q11.23 duplication syndrome: physical characteristics and natural history. Am J Med Genet A. 2015;167A:2916–2935. doi: 10.1002/ajmg.a.37340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramocki MB, Bartnik M, Szafranski P, Kołodziejska KE, Xia Z, et al. Recurrent distal 7q11.23 deletion including HIP1 and YWHAG identified in patients with intellectual disabilities, epilepsy, and neurobehavioral problems. Am J Hum Genet. 2010;87:857–865. doi: 10.1016/j.ajhg.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tam E, Young EJ, Morris CA, Marshall CR, Loo W, et al. The common inversion of the Williams-Beuren syndrome region at 7q11.23 does not cause clinical symptoms. Am J Med Genet A. 2008;146A:1797–1806. doi: 10.1002/ajmg.a.32360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zarate YA, Lepard T, Sellars E, Kaylor JA, Alfaro MP, et al. Cardiovascular and genitourinary anomalies in patients with duplications within the Williams syndrome critical region: phenotypic expansion and review of the literature. Am J Med Genet A. 2014;164A:1998–2002. doi: 10.1002/ajmg.a.36601. [DOI] [PubMed] [Google Scholar]