LUBAC components interact with the TLR3 signaling cascade at different levels, thereby tightly controlling TLR3-mediated innate immunity.
Abstract
The linear ubiquitin chain assembly complex (LUBAC), consisting of SHANK-associated RH-domain–interacting protein (SHARPIN), heme-oxidized IRP2 ubiquitin ligase-1 (HOIL-1), and HOIL-1–interacting protein (HOIP), is a critical regulator of inflammation and immunity. This is highlighted by the fact that patients with perturbed linear ubiquitination caused by mutations in the Hoip or Hoil-1 genes, resulting in knockouts of these proteins, may simultaneously suffer from immunodeficiency and autoinflammation. TLR3 plays a crucial, albeit controversial, role in viral infection and tissue damage. We identify a pivotal role of LUBAC in TLR3 signaling and discover a functional interaction between LUBAC components and TLR3 as crucial for immunity to influenza A virus infection. On the biochemical level, we identify LUBAC components as interacting with the TLR3-signaling complex (SC), thereby enabling TLR3-mediated gene activation. Absence of LUBAC components increases formation of a previously unrecognized TLR3-induced death-inducing SC, leading to enhanced cell death. Intriguingly, excessive TLR3-mediated cell death, induced by double-stranded RNA present in the skin of SHARPIN-deficient chronic proliferative dermatitis mice (cpdm), is a major contributor to their autoinflammatory skin phenotype, as genetic coablation of Tlr3 substantially ameliorated cpdm dermatitis. Thus, LUBAC components control TLR3-mediated innate immunity, thereby preventing development of immunodeficiency and autoinflammation.
Introduction
Influenza viruses belong to the family Orthomyxoviridae and cause millions of cases of severe illness and thousands of deaths per year.
We and others recently discovered the linear ubiquitin chain assembly complex (LUBAC) to be a critical regulator of innate immune signaling and inflammation (Walczak et al., 2012). The tripartite LUBAC is comprised of the SHANK-associated RH-domain–interacting protein (SHARPIN), heme-oxidized IRP2 ubiquitin ligase-1 (HOIL-1), and HOIL-1–interacting protein (HOIP; Gerlach et al., 2011; Ikeda et al., 2011; Tokunaga et al., 2011). To date, LUBAC is the only complex known to generate N- to C-terminal—also referred to as linear—ubiquitin linkages under native conditions (Kirisako et al., 2006).
SHARPIN-deficient mice suffer from severe chronic skin inflammation and several other organ dysfunctions (HogenEsch et al., 1993). Because of their overt skin phenotype, they are also known as chronic proliferative dermatitis mice (cpdm; HogenEsch et al., 1993). We recently discovered that absence of SHARPIN results in a cell-death–favoring dysregulation of TNF signaling and that dermatitis of SHARPIN-deficient mice was completely prevented by TNF deficiency, implying that in the absence of SHARPIN, aberrant TNF-induced cell death causes cpdm dermatitis (Gerlach et al., 2011). Subsequently, we and others provided genetic proof for this mechanism, as genetic ablation of essential components of the TNFR1-induced cell death pathway prevented cpdm dermatitis (Kumari et al., 2014; Rickard et al., 2014).
Mice lacking HOIL-1 have been reported to present with no overt phenotype (Tokunaga et al., 2009) whereas absence of HOIP, the central LUBAC component, results in lethality of developing mouse embryos at day 10.5 of embryonic development (Peltzer et al., 2014). Linear ubiquitination has further been implicated in prevention of immunodeficiency and autoinflammation, as patients with mutations in HOIL-1 or HOIP present with recurrent bacterial infections and, concomitantly, with hyperinflammation (Boisson et al., 2012, 2015).
Members of the TLR family are crucial regulators of inflammation and become activated by conserved pathogen-associated molecular patterns (PAMPs) from bacteria, viruses, and fungi (Akira et al., 2006). Equally, endogenous molecules, such as high mobility group protein B1, mRNA, or DNA, can act as danger signals, or damage-associated molecular patterns (DAMPs), by activating TLRs after their release from damaged cells (Rifkin et al., 2005). TLR3, a member of the TLR family involved in sensing of both viral infection and tissue damage, is activated by double-stranded (ds) RNA, which is either generated by viruses during their replication cycle acting as a PAMP (Alexopoulou et al., 2001) or released from damaged cells as a DAMP (Cavassani et al., 2008; Bernard et al., 2012). TLR3 is a type I transmembrane protein and localized in the cell’s endosomal compartment (Matsumoto et al., 2014). Ligation of TLR3 by dsRNA results in formation of a TLR3-signaling complex (TLR3-SC) across the endosomal membrane. This complex activates the following different signaling outputs: (i) activation of NF-κB and MAPK (Meylan et al., 2004); (ii) induction of type I IFNs (Fitzgerald et al., 2003); and (iii) cell death (Feoktistova et al., 2011; Estornes et al., 2012). Apart from TLR3, the cytosolic receptors retinoic acid inducible gene I (RIG-I) and melanoma differentiation-associated protein 5 (MDA5) are known to sense dsRNA (Takeuchi and Akira, 2009).
Whereas it is clear that TLR3 is involved in the host response to viral infection, its precise role remains rather poorly defined (Perales-Linares and Navas-Martin, 2013). Patients deficient in TLR3 and downstream signaling molecules, i.e., TIR-domain–containing adapter inducing IFN-β (TRIF), TNFR-associated factor (TRAF) 3, TANK-binding kinase (TBK) 1, or IFN regulatory factor (IRF) 3, have been identified as being highly susceptible to HSV 1 encephalitis (Zhang et al., 2007, 2013; Pérez de Diego et al., 2010; Sancho-Shimizu et al., 2011; Herman et al., 2012; Andersen et al., 2015). A missense mutation in the Tlr3 gene was identified in a patient with influenza A virus (IAV)–associated encephalopathy (Hidaka et al., 2006), and TLR3 polymorphisms have been associated with development of pneumonia in children infected with the H1N1/2009 pandemic strain of IAV (Esposito et al., 2012). In contrast, TLR3 deficiency was proposed to protect mice from IAV-induced lethal hyperinflammation (Le Goffic et al., 2006).
In addition to TLR3′s complex function in sensing viral infections, its role in tissue damage is also not fully understood. Whereas wound healing and skin regeneration was shown to critically depend on TLR3-mediated inflammation (Lai et al., 2009; Nelson et al., 2015), TLR3 has also been shown to mediate deleterious effects of tissue damage (Cavassani et al., 2008; Bernard et al., 2012). Thus, whereas TLR3 is a crucial mediator of various physiological processes ensuring host-protective immunity and proper wound healing, in both cases the outcome of TLR3 signaling can also be deleterious. Hence, the TLR3 gene-activatory and cell-death–inducing signaling outputs need to be precisely balanced and tightly controlled to achieve an adequate physiological response. In situations in which this is not the case and the physiological response is perturbed, TLR3 activation can cause morbidity or even mortality. It is therefore important to fully understand how TLR3 signaling is regulated, i.e., which factors are responsible for achieving an appropriate response to TLR3 stimulation.
Based on the aforementioned data, we set out to investigate a possible functional interplay between LUBAC components and TLR3 at the physiological, pathological, and mechanistic biochemical levels.
Results
Adequate host response against IAV infection requires SHARPIN and TLR3
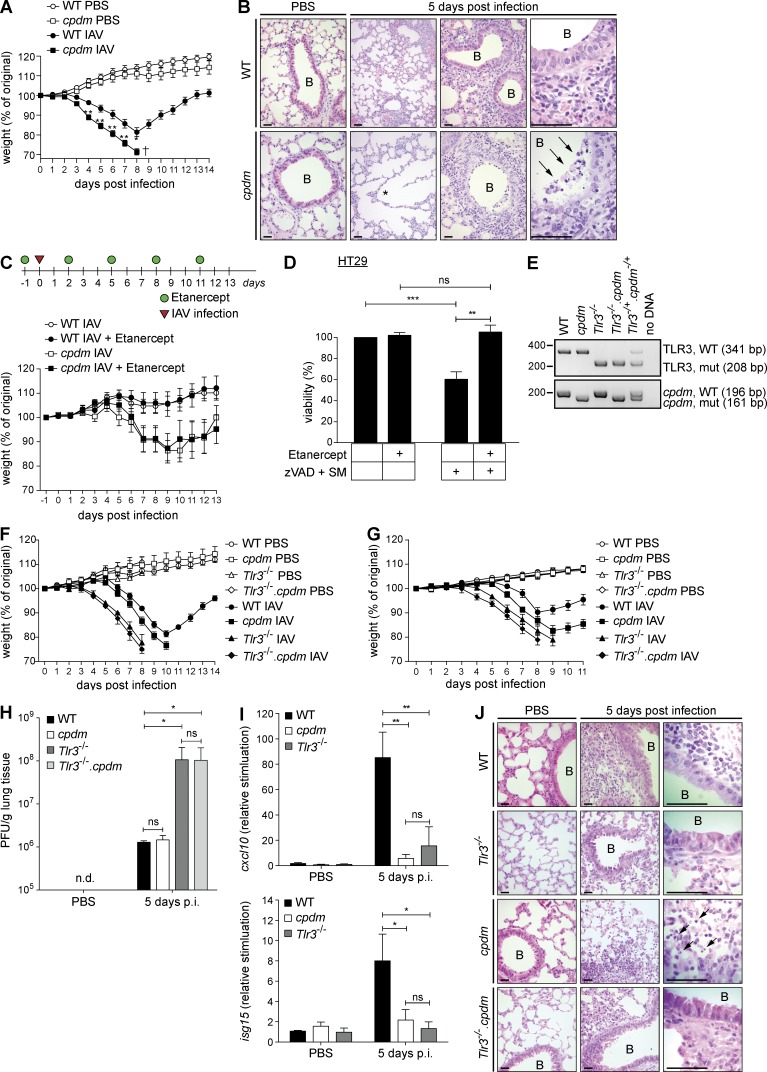
Patients with defects in linear ubiquitination caused by mutations in Hoil-1 or Hoip genes resulting in deficiency for the respectively encoded proteins are prone to various infections (Boisson et al., 2012, 2015). As linear ubiquitination appears to be essential to maintain host protection during the course of an infection, we hypothesized that absence of SHARPIN, the third LUBAC component, may influence disease outcome after infection by IAV. We therefore infected age- and sex-matched WT and SHARPIN-deficient cpdm mice with IAV and monitored their weight for 14 consecutive days after infection. Strikingly, cpdm mice were substantially more susceptible to IAV infection than WT littermates (Fig. 1 A). Histological analysis revealed significant presence of dead cells and evidence of alveolar damage in the lungs of infected cpdm mice, whereas very few dead cells could be detected in the lungs of infected WT mice (Fig. 1 B).
Figure 1.
Adequate host response against IAV infection requires SHARPIN and TLR3. (A) Weight of cpdm mice and WT littermate controls (n ≥ 6 mice per group) infected i.n. with 103 PFU IAV was monitored for 14 consecutive days after infection. (B) Representative H&E stainings of lungs of cpdm mice and WT littermate controls noninfected (PBS) or infected i.n. with IAV are shown (lungs of n ≥ 3 mice per genotype and group were analyzed). Alveolar damage and dead/dying cells are indicated by asterisk and arrows, respectively. Bars, 100 µm. B, bronchiole. (C) Treatment schedule. cpdm mice and WT littermate controls (n ≥ 9 mice per group) were injected i.p. with Etanercept or Pentaglobin as control at a dose of 0.5 mg/mouse 1 d before infection with 50 PFU IAV, and then every third day. Weight was monitored for 13 consecutive days after infection. (D) HT29 cells were incubated with 50 µg/ml Etanercept before stimulation with 100 nM SM and 20 µM zVAD. Cell viability was assessed after 48 h. Data are presented as means ± SD of three independent experiments performed in triplicates. (E) Representative PCR genotyping of experimental mice is shown. (F and G) Weight of WT, cpdm, Tlr3−/−, and Tlr3−/−.cpdm mice (n ≥ 5 mice per group) infected i.n. with 300 PFU IAV (F) or 100 PFU IAV (G) was monitored for 11 consecutive days after infection. (H and I) IAV lung titers (H) and induction of cxcl10 and isg15 by RT-PCR (I) were determined at day 5 after infection (lungs of n ≥ 3 mice per genotype and group were analyzed). (J) Representative H&E stainings of lungs of mice of indicated genotypes (n ≥ 3 per genotype and group were analyzed) at day 5 after infection or noninfection (PBS) are shown. Dead/dying cells are indicated by arrows. Bars, 100 µm. B, bronchiole. Values are plotted as means ± SEM in A, C, F, and G and as means ± SD in D, H, and I. n.d., nondetectable; p.i., post infection. *, P < 0.05; **, P < 0.01; ***, P < 0.001, unpaired Student’s t test.
IAV is known to induce excessive inflammation via production of proinflammatory mediators, including TNF (Szretter et al., 2007), and SHARPIN deficiency has been shown to sensitize cells to TNF-induced death (Gerlach et al., 2011; Kumari et al., 2014; Rickard et al., 2014). To test whether TNF, produced in response to infection, could be responsible for the observed aberrant cell death in lungs of cpdm mice and thereby contribute to their IAV-induced lethality, we treated IAV-infected WT and cpdm mice with the TNF blocker Etanercept (a soluble TNFR2-Fc fusion protein) or vehicle control (Fig. 1 C). Again, cpdm mice lost significantly more weight than WT mice but, importantly, this additional weight loss was not prevented by TNF inhibition (Fig. 1 C) despite autocrine TNF-mediated cell death, induced by caspase inhibitor and Smac Mimetic (SM; He et al., 2009), being effectively inhibited by Etanercept in vitro (Fig. 1 D). Hence, TNF is not responsible for induction of aberrant cell death and pathology in IAV-infected cpdm mice.
The presence of TLR3 has been described to be detrimental during IAV infection (Le Goffic et al., 2006). Because IAV-induced death of cpdm mice was independent of TNF, we addressed whether the presence of TLR3 may be causative for their enhanced susceptibility. We reasoned that, if this were the case, then absence of TLR3 should prevent lethality. However, when we challenged WT, cpdm, Tlr3−/−, and Tlr3−/−.cpdm mice (Fig. 1 E) with IAV, we found that TLR3 deficiency failed to protect cpdm mice from IAV-induced lethality. In fact, both, Tlr3−/− and Tlr3−/−.cpdm mice were even more susceptible to IAV infection than cpdm mice at two different viral doses (Fig. 1, F and G).
The fact that Tlr3−/−.cpdm mice succumbed with similar kinetics to IAV infection as Tlr3−/− mice suggests that SHARPIN fulfills its antiviral role by acting downstream of TLR3. This conclusion received additional support from our analysis of viral titers in the lungs of the respective mouse strains after infection. Whereas TLR3 deficiency resulted in a drastic increase in viral load, in line with a previous publication (Le Goffic et al., 2006), SHARPIN deficiency did not interfere with appropriate control of viral propagation as lungs of infected cpdm mice displayed viral loads equal to those of lungs of infected WT mice (Fig. 1 H). Yet, when TLR3 and SHARPIN were coablated, viral titers were as high as in the sole absence of TLR3 (Fig. 1 H). Interestingly, ablation of TLR3 or SHARPIN alone resulted in reduced chemokine and interferon-stimulated gene production in the lungs after IAV infection as compared with WT mice (Fig. 1 I). Thus, whereas in the absence of SHARPIN, TLR3 can still control viral replication, gene-activatory signaling requires the presence of SHARPIN and TLR3. Intriguingly, cell death levels were increased in the lungs of IAV-infected cpdm mice as compared with those in WT, Tlr3−/−, and Tlr3−/−.cpdm mice (Fig. 1 J). These results indicate that the increase in cell death upon infection with IAV in the lungs of SHARPIN-deficient mice is mediated by TLR3. Thus, both TLR3 and SHARPIN are required to fight infection with IAV.
LUBAC components are required for TLR3-mediated gene activation
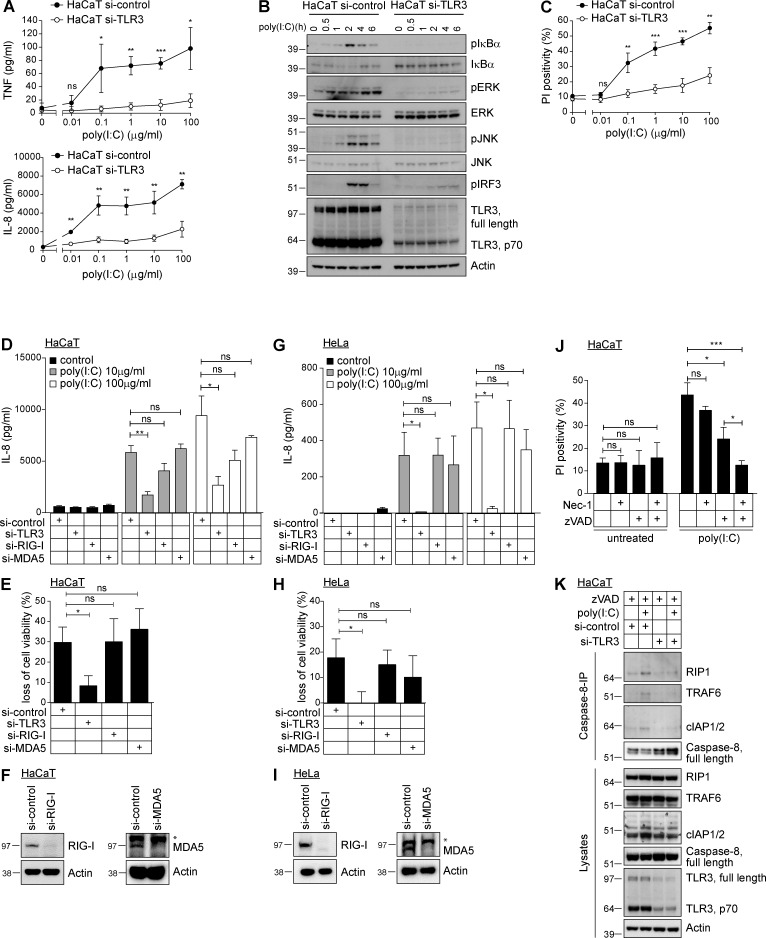
As the results obtained with Tlr3−/− and cpdm mice in the IAV infection model suggested LUBAC involvement in TLR3 signaling, we next aimed to define the biochemical basis for this functional interaction. To study signaling downstream of TLR3, a cellular system was required in which signaling induced by administration of the synthetic analogue of dsRNA, known as poly(I:C), was dependent on TLR3 and not on the cytosolic sensors of dsRNA, RIG-I and MDA5 (Takeuchi and Akira, 2009). To test this, we stimulated the human keratinocyte line HaCaT with and without RNAi-suppressed TLR3 expression with poly(I:C). This stimulation resulted in cytokine/chemokine secretion (Fig. 2 A) and activation of NF-κB, MAPK, and IRF3 (Fig. 2 B) only in TLR3-expressing and not in TLR3-suppressed HaCaT cells. Also cell death induction by poly(I:C) critically depended on the presence of TLR3 (Fig. 2 C). High concentrations of poly(I:C) still resulted in cytokine/chemokine secretion and cell death induction, albeit at substantially reduced levels. To evaluate whether this residual activation was mediated by stimulation of other dsRNA-sensing receptors, such as RIG-I or MDA5, we performed RNAi-mediated knockdown of TLR3, RIG-I, and MDA5 in parallel in HaCaT (Fig. 2, D–F) and HeLa cells (Fig. 2, G–I). Despite efficient knockdown of both receptors on the protein level (Fig. 2, F and I), neither RNAi targeting of RIG-I nor MDA5 significantly reduced poly(I:C)-induced chemokine secretion or cell death, whereas that of TLR3 did (Fig. 2, D and E and G and H). Using caspase inhibitors and inhibitors of the kinase activity of receptor-interacting protein (RIP) 1, we next determined that poly(I:C)-induced cell death was mainly apoptotic and that caspase inhibition partially converted this death to necroptosis (Fig. 2 J). Caspase-8 was previously shown to be a component of a signaling complex that forms upon stimulation with poly(I:C) (Feoktistova et al., 2011; Estornes et al., 2012). We confirmed this by performing Caspase-8 immunoprecipitation (IP), which revealed that RIP1, TRAF6, and cellular inhibitor of apoptosis proteins (cIAPs) 1 and 2 associate with Caspase-8 in a poly(I:C) stimulation-dependent manner. Again, this association was entirely dependent on the presence of TLR3 (Fig. 2 K). In summary, we can conclude that in the cell lines used, poly(I:C) achieves its activity by stimulating TLR3.
Figure 2.
TLR3 presence is required to mediate poly(I:C)-induced signaling. (A) HaCaT cells transfected with control or TLR3 RNAi were stimulated with poly(I:C) as indicated and concentrations of TNF and IL-8 were determined by ELISA. (B) HaCaT cells transfected with control or TLR3 RNAi were stimulated with 5 µg/ml poly(I:C) as indicated and subjected to analysis by immunoblotting. (C) HaCaT cells transfected with control or TLR3 RNAi were stimulated with poly(I:C) as indicated and analyzed for propidium iodide positivity by FACS after 24 h. (D–I) HaCaT cells (D–F) or HeLa cells (G–I) transfected with control siRNA or siRNA targeting TLR3, RIG-I, or MDA5 were stimulated with 10 or 100 µg/ml poly(I:C). After 24 h, IL-8 secretion was determined by ELISA (D and G) and loss of cell viability was assessed in parallel (E and H). Knockdown efficiency of MDA5 and RIG-I was determined by immunoblotting (F and I). (J) HaCaT cells were preincubated for 1 h with 10 µM 7-Cl-O-Nec-1 (Nec-1), 20 µM zVAD (zVAD), or a combination thereof before stimulation with 50 µg/ml poly(I:C). Propidium iodide positivity was determined by FACS 24 h later. (K) HaCaT cells transfected with control or TLR3 siRNA were stimulated with 20 µg/ml poly(I:C) in the presence of 20 µM zVAD. Caspase-8 was immunoprecipitated and coimmunoprecipitated proteins were analyzed by Western blot. All values are means ± SD of at least three independent experiments performed in triplicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001, unpaired Student’s t test. Western blots are representative of at least two independent experiments. Nonspecific bands are indicated by asterisk.
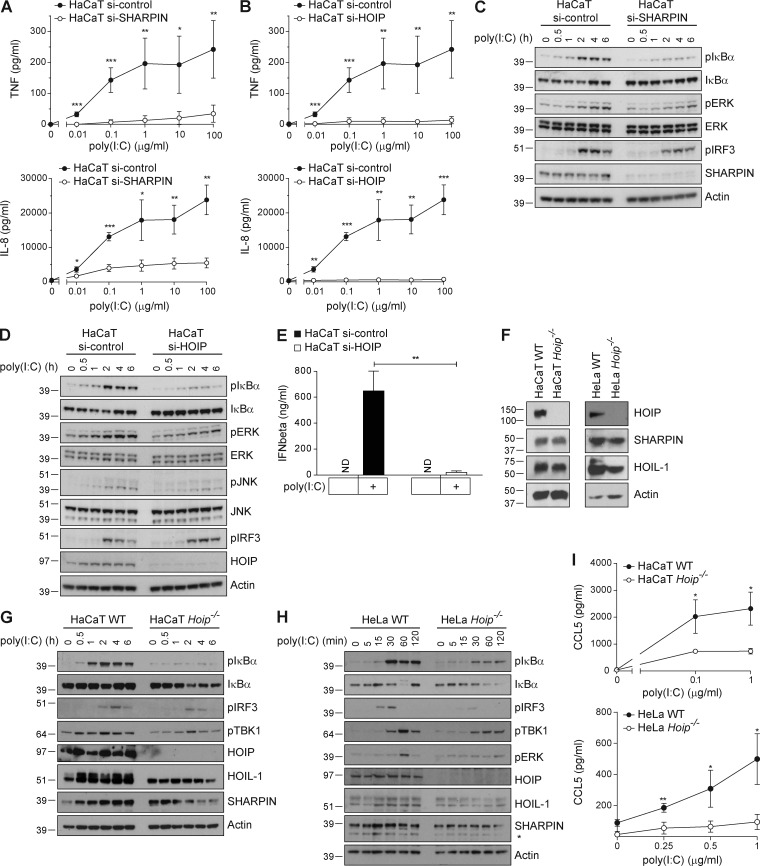
Having established that all known TLR3 signaling outputs were induced by poly(I:C) stimulation in a TLR3-dependent manner, we next sought to evaluate the role of LUBAC components in enabling or regulating these signaling outputs. We first studied whether absence of LUBAC components altered TLR3-induced gene activation. RNAi-mediated knockdown of SHARPIN (Fig. 3 A) or HOIP (Fig. 3 B) significantly reduced poly(I:C)-induced secretion of IL-8 and TNF. Silencing of SHARPIN or HOIP in HaCaT cells diminished poly(I:C)-induced NF-κB and MAPK activation, whereas activation of IRF3 remained largely unaffected (Fig. 3, C and D). Interestingly, even though phosphorylation of IRF3, a prerequisite of IFN gene expression, was largely unaffected in these cells, RNAi-mediated knockdown of HOIP in HaCaT cells abolished poly(I:C)-induced IFN-β secretion (Fig. 3 E).
Figure 3.
LUBAC components are required for TLR3-mediated gene activation. (A and B) HaCaT cells transfected with control or siRNA targeting SHARPIN (A) or HOIP (B) were stimulated with poly(I:C) as indicated. Cell supernatants were collected after 24 h of stimulation and concentration of TNF and IL-8 was determined by ELISA. (C and D) HaCaT cells transfected with control or RNAi targeting SHARPIN (C) or HOIP (D) were stimulated with 5 µg/ml poly(I:C) as indicated and subjected to analysis by immunoblotting. (E) HaCaT cells transfected with nontargeting or HOIP RNAi were stimulated with 10 µg/ml poly(I:C). Cell supernatants were collected after 4 h of stimulation and analyzed for concentration of IFN-β by ELISA. (F) HaCaT Hoip WT and Hoip−/− cells and HeLa Hoip WT and Hoip−/− cells were generated, and the absence of HOIP protein was confirmed by Western blotting. Protein levels of SHARPIN and HOIL-1 were determined in parallel. Actin served as loading control. (G and H) HaCaT Hoip WT and Hoip−/− cells (G) or HeLa Hoip WT and Hoip−/− cells (H) were stimulated with 5 µg/ml poly(I:C) as indicated and subjected to analysis by immunoblotting. (I) HaCaT and HeLa Hoip WT and Hoip−/− cells were stimulated with poly(I:C) as indicated. Cell supernatants were collected 24 h later and analyzed for concentration of CCL5 by ELISA. All values are means ± SD of at least three independent experiments performed in triplicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001, unpaired Student’s t test. Western blots are representative of at least two independent experiments. Nonspecific bands are indicated by asterisk.
We next generated knockout cell lines for Hoip (Fig. 3 F) to analyze gene-activatory signaling in WT and HOIP-deficient HaCaT and HeLa cells after stimulation with poly(I:C). Absence of HOIP consistently resulted in diminished NF-κB and ERK activation and consequent reduction in chemokine secretion in both cell lines (Fig. 3, G–I). Of note, IRF3 phosphorylation was strongly reduced in HOIP-deficient HeLa cells, but not in HOIP-deficient HaCaT cells, indicating that LUBAC may differentially regulate IRF3 phosphorylation in a cell type–specific manner (Fig. 3, G and H).
LUBAC components limit TLR3-induced cell death
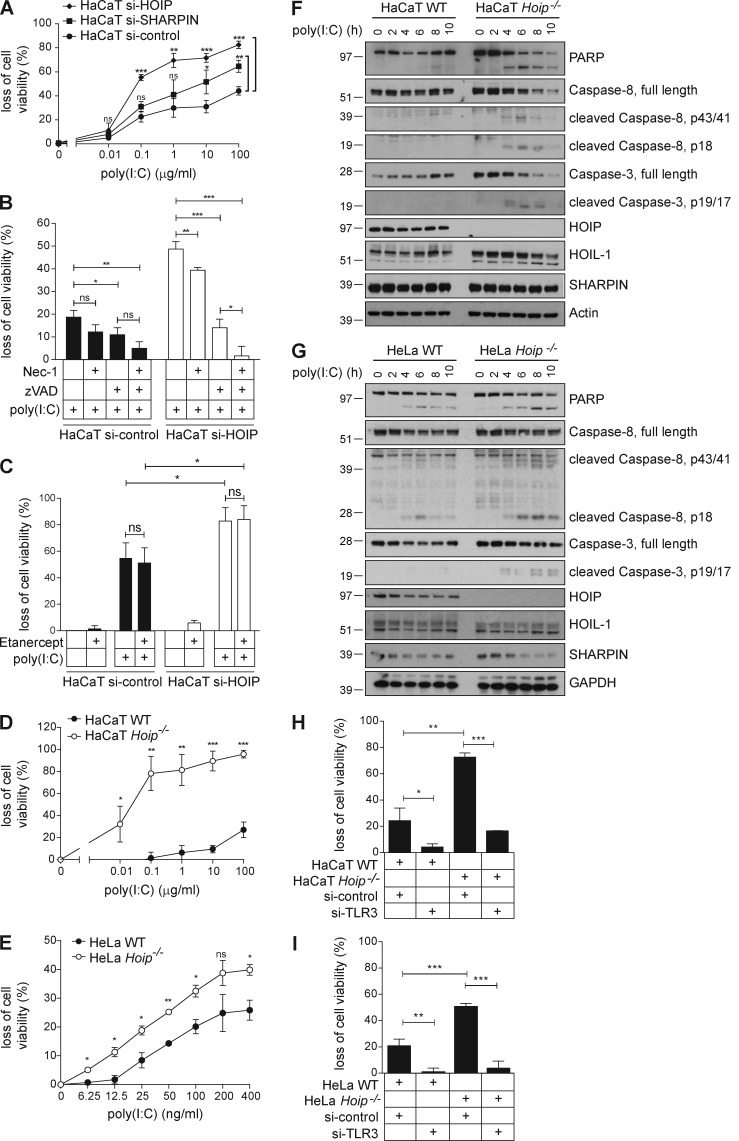
When analyzing whether TLR3-mediated cell death was regulated by LUBAC components, we found that silencing of either SHARPIN or HOIP sensitized cells to TLR3-mediated death with absence of HOIP as the central LUBAC component having the most pronounced effect (Fig. 4 A). Silencing of HOIP did not alter the type of cell death induced by poly(I:C), as it remained largely apoptotic (Fig. 4 B). Because poly(I:C) stimulation of HaCaT cells induced secretion of TNF (Fig. 2 A), we next addressed whether autocrine TNF might be responsible for the observed cell death after TLR3 stimulation by poly(I:C) (Haas et al., 2009; Gerlach et al., 2011). However, in line with our in vivo results on IAV infection (Fig. 1 C), this was not the case. TNF inhibition did not prevent poly(I:C)-induced death of HaCaT cells, neither in the presence nor absence of LUBAC components (Fig. 4 C), whereas SM/zVAD-induced death of HT-29 cells, which is known to be mediated by autocrine TNF (He et al., 2009), was blocked (Fig. 1 D). Both HOIP-deficient HaCaT and HeLa cells were sensitized to poly(I:C)-induced death (Fig. 4, D and E). Further analysis revealed that hallmarks of apoptosis, including cleavage of Caspase-3 and -8 and poly(ADP-ribose) polymerase, were increased in the absence of HOIP (Fig. 4, F and G). Importantly, independent of HOIP being absent or present, poly(I:C)-induced cell death was strictly dependent on the presence of TLR3, as RNAi-mediated knockdown of TLR3 prevented cell death in both HaCaT and HeLa cells (Fig. 4, H and I).
Figure 4.
LUBAC components limit TLR3-induced cell death. (A) HaCaT cells transfected with control, SHARPIN, or HOIP siRNA were stimulated with poly(I:C) as indicated and loss of cell viability was determined after 24 h. (B) HaCaT cells transfected with control or HOIP siRNA were incubated with 10 µM 7-Cl-O-Nec-1 (Nec-1), 20 µM zVAD (zVAD), or a combination thereof 1 h before stimulation with 1 µg/ml poly(I:C). Loss of cell viability was assessed after 24 h. (C) HaCaT cells transfected with control or HOIP siRNA were incubated in the presence or absence of 50 µg/ml Etanercept 2 h before stimulation with 100 µg/ml poly(I:C). Loss of cell viability was determined after 24 h. (D and E) HaCaT Hoip WT and Hoip−/− cells (D) and HeLa Hoip WT and Hoip−/− cells (E) were stimulated with poly(I:C) as indicated. Loss of cell viability was assessed after 24 h. (F and G) HaCaT Hoip WT and Hoip−/− cells (F) or HeLa Hoip WT and Hoip−/− cells (G) were stimulated with 5 µg/ml poly(I:C) as indicated and subjected to further analysis by immunoblotting. (H) HaCaT Hoip WT and Hoip−/− cells were transfected with control RNAi or RNAi targeting TLR3. Cells were stimulated with 1 µg/ml and loss of cell viability was assessed after 24 h. (I) HeLa Hoip WT and Hoip−/− cells were transfected with control or TLR3 siRNA. Cells were stimulated with 100 µg/ml and loss of cell viability was assessed after 24 h. All values are presented as means ± SD of at least three independent experiments performed in triplicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001, unpaired Student’s t test. Images are representative of at least two independent experiments.
LUBAC components form part of the TLR3-SC and prevent TLR3-induced DISC formation
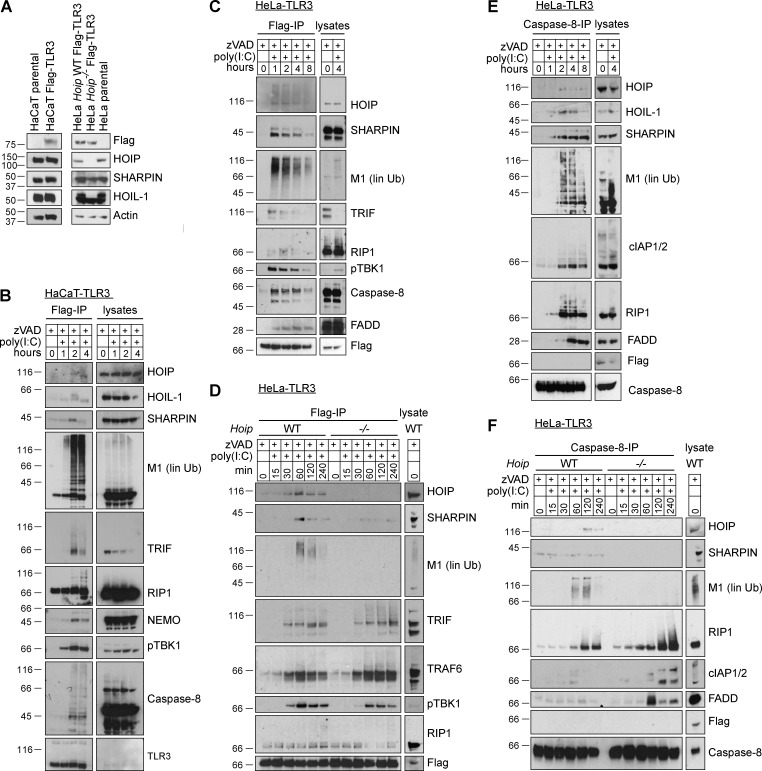
Having identified LUBAC components as crucial for balancing TLR3-induced signaling outputs, we next determined whether they achieve this by forming part of the TLR3-SC induced by poly(I:C) stimulation. To compensate for the fact that the native TLR3-SC is of low abundance and therefore difficult to analyze after IP, we generated HeLa and HaCaT cells stably expressing Flag-tagged TLR3 at intermediate levels (HaCaT-TLR3 and HeLa-TLR3 cells; Fig. 5 A) so that activation of TLR3 signaling still requires ligand-induced receptor cross-linking. After stimulation of these cells with poly(I:C), we isolated the TLR3-SC by anti-Flag-IP and evaluated it for the presence of various factors, including LUBAC components (Fig. 5, B and C). This revealed the recruitment of LUBAC components to TLR3 upon stimulation with poly(I:C), along with components previously implicated in TLR3 signaling, such as TRIF, RIP1, TBK1, Fas-associated protein with death domain (FADD), and Caspase-8. Importantly, recruitment of LUBAC components results in generation of linear ubiquitin chains within this complex, as demonstrated by the presence of linear ubiquitin, also referred to as Met1 or M1 linkages, after poly(I:C) stimulation in the TLR3-SC obtained from both HaCaT and HeLa cells (Fig. 5, B and C).
Figure 5.
LUBAC components form part of the TLR3-SC and prevent TLR3-induced DISC formation. (A) HaCaT parental and Flag-tagged TLR3-expressing cells and HeLa parental and Flag-tagged TLR3-expressing HeLa Hoip WT and Hoip−/− cells were lysed, and levels of TLR3 (by staining for Flag), HOIP, HOIL-1, and SHARPIN were determined by immunoblotting. Actin served as loading control. (B) HaCaT-TLR3 cells were stimulated with 20 µg/ml poly(I:C) in the presence of 20 µM zVAD as indicated. Cells were subsequently lysed and TLR3 was immunoprecipitated via Flag and coimmunoprecipitated proteins were analyzed by immunoblotting. Levels of Flag-tagged TLR3 were determined by staining for TLR3. (C) HeLa cells stably expressing Flag-TLR3 were incubated in the presence of 20 µM zVAD before stimulation with 20 µg/ml poly(I:C) as indicated. TLR3 was immunoprecipitated via its Flag-tag and analysis of coimmunoprecipitated proteins by immunoblotting was performed as indicated. (D) HeLa Hoip WT and Hoip−/− cells stably expressing Flag-TLR3 were stimulated with 20 µg/ml poly(I:C) in the presence of 20 µM zVAD and TLR3 was immunoprecipitated via Flag and coimmunoprecipitated proteins were analyzed by immunoblotting. (E) Caspase-8-IP was performed after lysates were depleted of the TLR3-SC by Flag-IP (C), and coimmunoprecipitated proteins were analyzed by immunoblotting. (F) Caspase-8-IP was performed after lysates were depleted of the TLR3-SC by Flag-IP (shown in D) and coimmunoprecipitated proteins were subjected to analysis by immunoblotting. Images are representative of at least three independent experiments. M1, linear ubiquitin linkages.
Having identified LUBAC as a novel component of the TLR3-SC, we next analyzed how the presence of LUBAC components determines composition of this complex (Fig. 5 D). This analysis revealed that, apart from preventing recruitment of SHARPIN and HOIL-1 and formation of M1 linkages within the TLR3-SC, composition of this complex precipitated from HOIP-deficient HeLa cells did not appear to be majorly altered compared with the TLR3-SC precipitated from HeLa cells expressing HOIP (Fig. 5 D). Given that TNF induces cell death via a second complex that has been proposed to emerge from complex I of TNFR1 (Micheau and Tschopp, 2003), we hypothesized that TLR3-mediated cell death might also be executed from a second signaling platform and that composition of such a TLR3-dependent secondary complex might be altered by the absence of LUBAC components, thereby providing a possible explanation as to how LUBAC prevents cell death. To test this hypothesis, we performed Caspase-8-IP after lysates were depleted of the TLR3-SC by Flag-IP shown in Fig. 5 C and probed for associated proteins implicated in cell death signaling (Fig. 5 E). This experiment indeed revealed the existence of a second signaling platform. Importantly, in spite of being triggered by activation of TLR3, this complex, which we will refer to as TLR3-induced death-inducing signaling complex (DISC), was devoid of the receptor (Fig. 5 E). The TLR3-induced DISC contains LUBAC, RIP1, FADD, cIAP1/2, and Caspase-8 (Fig. 5 E). In addition, we subjected lysates of HeLa cells proficient and deficient in HOIP from the experiment in Fig. 5 D to further analysis of the TLR3-induced DISC by performing Caspase-8-IP after the TLR3-SC was immunoprecipitated via Flag. Formation of the TLR3-induced DISC was markedly enhanced in the absence of HOIP as association of RIP1, cIAP1/2, and FADD with Caspase-8 was substantially increased in HOIP-deficient cells, whereas LUBAC components and M1 linkages were also absent from this complex (Fig. 5 F). Together, these results demonstrate that absence of LUBAC and linear ubiquitin from the TLR3-SC results in an aberrant increase in formation of the TLR3-induced DISC.
LUBAC components regulate TLR3 signaling induced by poly(I:C) ex vivo and in vivo
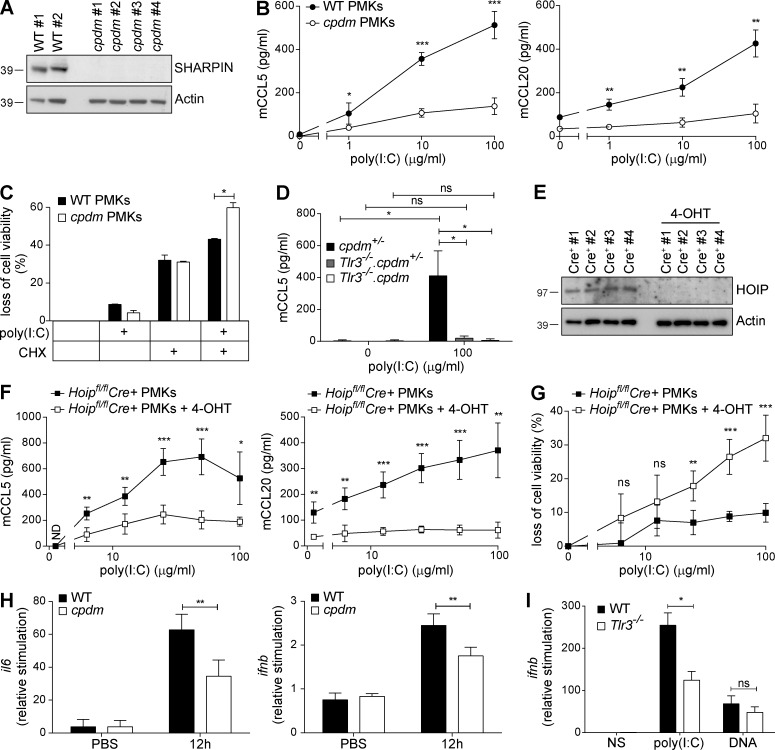
Having identified the presence of LUBAC components in the TLR3-SC, and in the newly described TLR3-induced DISC, as crucial for a balanced TLR3 signaling output in vitro in cell lines, we next investigated the role of LUBAC components in TLR3 signaling in primary cells. As TLR3 signaling has been shown to be involved in skin homeostasis (Lai et al., 2009; Nelson et al., 2015), we used primary murine keratinocytes (PMKs) isolated from newborn WT versus cpdm mice as the primary cell model to assess the role of LUBAC components in TLR3 signaling ex vivo. After determining that WT PMKs express SHARPIN protein, whereas cpdm–derived PMKs do not (Fig. 6 A), we compared chemokine secretion upon poly(I:C) stimulation from cpdm- and WT-derived PMKs. In line with the in vitro results obtained in cell lines, cpdm PMKs were found to secrete significantly less chemokines than WT PMKs when stimulated with poly(I:C) (Fig. 6 B). Upon stimulation with poly(I:C), cpdm PMKs were significantly sensitized to poly(I:C)-induced cell death (Fig. 6 C). Poly(I:C)-induced chemokine secretion was strictly dependent on presence of TLR3, independent of whether SHARPIN was absent or present (Fig. 6 D). Moreover, PMKs with inducible deletion of HOIP (Fig. 6 E) exhibited reduced chemokine secretion (Fig. 6 F) along with a significant increase in poly(I:C)-induced cell death (Fig. 6 G), demonstrating that HOIP limits TLR3-mediated death of PMKs ex vivo. These experiments were performed using PMKs from 4-hydroxy-tamoxifen (4-OHT)-inducible Hoipfl/fl Cre+ mice.
Figure 6.
LUBAC components regulate TLR3 signaling induced by poly(I:C) ex vivo and in vivo. (A) SHARPIN protein expression of PMKs from WT mice (n = 2) and cpdm (n = 4) littermates was analyzed by Western blot. Actin served as loading control. (B) PMKs isolated from newborn WT (n = 2) and cpdm (n = 4) littermates were stimulated by poly(I:C) as indicated. Concentrations of murine CCL5 (mCCL5) and murine CCL20 (mCCL20) in the supernatants were determined by ELISA after 24 h of stimulation. One representative of two independent experiments performed in triplicates is shown. (C) PMKs from WT mice (n = 2) and cpdm littermates (n = 2) were isolated at the age of 4 wk and cells were stimulated with 50 µg/ml poly(I:C) in absence versus presence of 250 ng/ml CHX. Loss of cell viability was determined after 48 h of stimulation. One representative of two independent experiments performed in triplicates is shown. (D) PMKs from cpdm+/− (n = 3), Tlr3−/−.cpdm+/− (n = 3), and Tlr3−/−.cpdm−/− (n = 3) littermates isolated at the age of 4 wk were stimulated with 100 µg/ml poly(I:C) and concentration of mCCL5 was determined in the supernatant after 24 h by ELISA. (E) HOIP protein expression of PMKs isolated from one litter of newborn Cre+ Hoipfl/fl mice (n = 4) cultured in absence versus presence of 1 µM 4-OHT for at least 72 h was analyzed by Western blot. Actin served as loading control. (F) PMKs with 4-OHT–inducible deletion of HOIP from Cre+ Hoipfl/fl newborn littermates (n = 4) were stimulated with poly(I:C) as indicated. Concentrations of mCCL5 and mCCL20 were determined by ELISA after 24 h of stimulation. One representative of two independent experiments performed in triplicates is shown. (G) Loss of cell viability was determined in parallel. (H) Naked poly(I:C) was injected into the ear pinnae of WT and cpdm littermates (n = 5) and induction of il6 and ifnb mRNA was assessed by RT-PCR. (I) Naked poly(I:C) was injected into the ear pinnae of WT and Tlr3−/− littermates (n = 5) and ifnb mRNA level was assessed by RT-PCR. Values are plotted as means ± SD for the indicated number of mice. *, P < 0.05; **, P < 0.01; ***, P < 0.001, unpaired Student’s t test. Images are representative of at least two independent experiments.
We next evaluated whether absence of SHARPIN influences TLR3 signaling in response to poly(I:C) in vivo. For this purpose, we injected poly(I:C) into the ear pinnae of WT and cpdm mice. In line with our ex vivo results, cytokine and IFN-β mRNA induction was significantly suppressed in the ear pinnae of cpdm mice (Fig. 6 H). Intradermal stimulation with poly(I:C) is dependent on the presence of TLR3, as injection of poly(I:C) into the ear pinnae of TLR3-deficient mice resulted in significantly reduced IFN-β mRNA induction (Fig. 6 I).
TLR3 deficiency ameliorates cpdm dermatitis
In this study, we identified a crucial role for LUBAC components in TLR3-mediated host protection against IAV infection. However, TLR3 has also been implicated in the sensing of tissue damage (Brentano et al., 2005; Cavassani et al., 2008; Bernard et al., 2012). Intriguingly, cpdm mice were previously shown to display increased cell death in the skin even before onset of inflammation (HogenEsch et al., 1993) and we subsequently showed that TNF is the trigger of this early cell death (Gerlach et al., 2011; Kumari et al., 2014; Rickard et al., 2014). This, together with our discovery that the presence of LUBAC components prevents TLR3-induced DISC formation and, consequently, TLR3-induced cell death, led us to develop the following hypothesis: release of dsRNA as a DAMP from TNF-killed keratinocytes in the skin of cpdm mice could trigger further release of dsRNA, resulting in stimulation of TLR3, which, in the absence of SHARPIN, would further increase cell death.
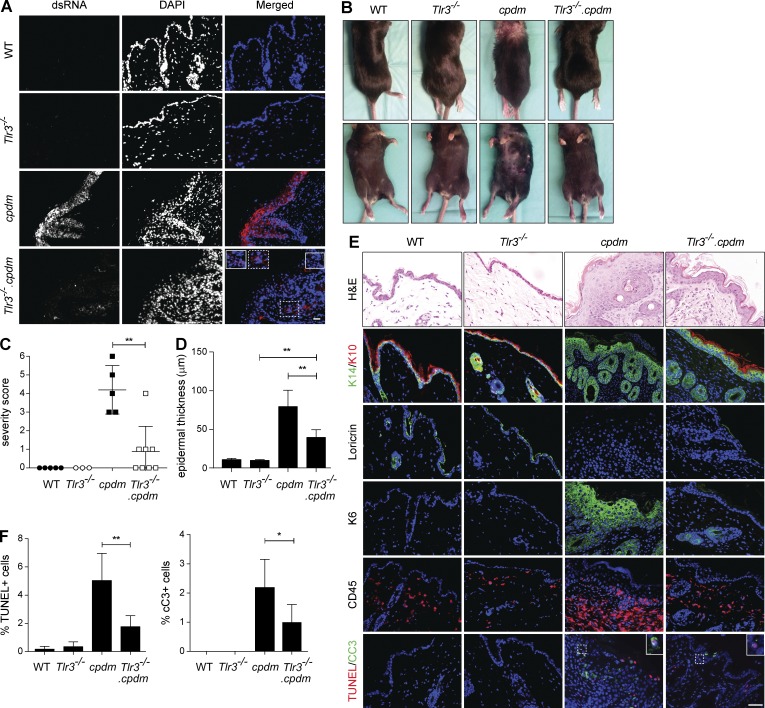
To test this hypothesis, we first analyzed cpdm skin sections for the presence of dsRNA. Staining with a dsRNA-specific antibody showed that dsRNA is indeed highly abundant in cpdm, whereas it is virtually absent from WT skin (Fig. 7 A). To address whether deregulated TLR3 signaling in the absence of SHARPIN might serve as an inflammation accelerant by triggering additional release of DAMPs from cells dying as a consequence of TLR3 ligation, we next assessed dsRNA presence in the skin of Tlr3−/− and Tlr3−/−.cpdm mice (Fig. 7 A). Interestingly, release of dsRNA, although clearly detectable, was significantly diminished in Tlr3−/−.cpdm skin, indicating that TLR3 stimulation indeed exacerbates DAMP release (Fig. 7 A). These results imply that, in the skin of cpdm mice, TLR3 serves as both damage sensor and amplifier of inflammation.
Figure 7.
TLR3 deficiency ameliorates cpdm dermatitis. (A) Representative images of dsRNA staining (J2) in the skin of 8-wk-old WT, Tlr3−/−, cpdm, and Tlr3−/−.cpdm littermates are shown (two independent experiments, n = 3 per genotype). (B) Representative pictures of 8-wk-old littermates of indicated genotypes are shown. (C) Severity scoring of dermatitis was assessed at the age of 8 wk. Dots represent scoring of individual mice ± SD. (D) Quantification of epidermal thickness of mice of indicated genotype (n = 3 per genotype) is shown. (E) Representative H&E, keratin 14/10, loricrin, keratin 6, CD45, and TUNEL/cleaved Caspase-3 stainings of skin sections of 8-wk-old WT, Tlr3−/−, cpdm, and Tlr3−/−.cpdm littermates are shown (three independent experiments, n ≥ 3 per genotype). Bar, 50 µm. (F) Quantification of TUNEL-positive cells and cleaved Caspase-3-positive cells in the skin sections of indicated genotypes is shown (n ≥ 3 per genotype). *, P < 0.05, **, P < 0.01, unpaired Student’s t test.
To test whether deregulation of TLR3 signaling in the absence of SHARPIN indeed contributes to cpdm pathogenesis, we coablated the Tlr3 gene and evaluated whether this resulted in amelioration of dermatitis induced by SHARPIN deficiency. Strikingly, this was the case, as TLR3 deficiency markedly attenuated skin inflammation in cpdm mice (Fig. 7 B, C). Genetic coablation of Tlr3 also significantly reduced epidermal thickness in cpdm mice (Fig. 7 D). Furthermore, skin sections of Tlr3−/−.cpdm mice were less inflamed and displayed less hyperkeratosis and parakeratosis than those of cpdm mice (Fig. 7 E).
Further characterization of these skin sections revealed that keratin 10, normally expressed in terminally differentiating epidermal keratinocytes, is expressed in WT and Tlr3−/− mice but absent from cpdm skin. Interestingly, co-deficiency in TLR3 restored keratin 10 expression (Fig. 7 E). Expression of loricrin, an additional marker of terminally differentiated epidermal cells, was found in WT and Tlr3−/− mice. Whereas cpdm skin lacked expression of loricrin, it was present in the skin of Tlr3−/−.cpdm mice (Fig. 7 E). Interestingly, keratin 6, a marker expressed by keratinocytes that undergo rapid turnover and thus serve as an indicator of proliferation, was highly expressed in cpdm keratinocytes, whereas almost no keratin 6 expression was detectable in Tlr3−/−.cpdm skin (Fig. 7 E). Staining of skin sections for CD45 revealed that immune cell infiltration of cpdm skin was also markedly reduced in the absence of TLR3 (Fig. 7 E).
We next assessed the extent of cell death in the skin of the different mice. Although TUNEL and cleaved Caspase-3 positivity was detectable in both cpdm and Tlr3−/−.cpdm skin (Fig. 7 E), quantification revealed that coablation of TLR3 resulted in disappearance of the majority of cell death in the skin of cpdm mice (Fig. 7 F).
Discussion
In this study, we discover an intricate mechanistic interaction between LUBAC components and activated TLR3 at the biochemical level, with profound immunological consequences on host response to viral infection and development of autoinflammation. The role of TLR3 in innate sensing of IAV is not entirely clear (Perales-Linares and Navas-Martin, 2013). A missense mutation in the Tlr3 gene was identified in a patient with IAV-associated encephalopathy (Hidaka et al., 2006), and children with TLR3 polymorphisms have an increased risk of pneumonia when infected by the pandemic A/H1N1/2009 influenza virus (Esposito et al., 2012). Le Goffic et al. (2006) reported that TLR3 deficiency protects mice against H3N2 IAV infection despite elevated viral titers and diminished chemokine production in the lungs of TLR3-deficient mice during infection, although this was later suggested to be strain specific (Leung et al., 2014). Here, we establish further that the presence of TLR3 is crucial for the initiation of an adequate antiviral immune response to infection with the IAV strain A/Puerto Rico/8/1934 (H1N1).
We show that both TLR3 and SHARPIN are required to initiate an adequate host response to IAV infection. TLR3 is indispensable for IAV-induced gene activation and control of viral load, as viral particles replicate uncontrolled in its absence. SHARPIN maintains gene activation and prevents excessive cell death upon infection with IAV. In the absence of both TLR3 and SHARPIN, no control of viral load is achieved, no gene activation initiated, and no cell death observed. This shows that TLR3 acts upstream of SHARPIN and, importantly, that the increased cell death in response to IAV infection in SHARPIN-deficient mice is indeed caused by triggering of TLR3. Interestingly, although SHARPIN is required to control TLR3-mediated signaling outputs, viral control is not altered in SHARPIN-deficient as compared with WT mice. One plausible explanation for these findings could be that in the absence of SHARPIN, diminished TLR3-induced gene activation would result in an increase of viral replication but that this is limited by increased TLR3-mediated cell death in the lungs of SHARPIN-deficient mice. Thus, sensitization to TLR3-mediated cell death in the absence of SHARPIN may be beneficial to the extent that it enables control of viral replication, as infected and possibly also bystander cells are more prone to die. However, when the initial viral dose is too high, damage to the lungs of SHARPIN-deficient mice, possibly caused by excessive TLR3-mediated cell death, is incompatible with survival and results in diminished disease tolerance. Thus, whereas at lower viral doses TLR3-induced cell death may contribute to antiviral immunity, when the viral titer surpasses a certain limit, this cell death may become harmful and eventually lethal.
On the biochemical level, we identify LUBAC as a novel component of the dsRNA-induced TLR3-SC. LUBAC components are required in this complex for enabling TLR3-induced gene activation and subsequent secretion of chemo- and cytokines. In addition, IFN-β secretion critically depended on the presence of HOIP. Interestingly, and contrary to the role of LUBAC components in TLR3-mediated type I IFN production identified in this study, it has been claimed that LUBAC negatively regulates virus-induced type I IFN production via degradation of tripartite motif-containing protein 25 in the RIG-I–signaling pathway (Inn et al., 2011). Type I IFN production in RIG-I signaling is mediated by IRF3. IRF3 has recently been shown to be involved in RIG-I–mediated induction of apoptosis after viral infection. Interestingly, this process was shown to require linear ubiquitination of IRF3 (Chattopadhyay et al., 2016), suggesting opposing roles for LUBAC in RIG-I– versus TLR3-mediated apoptosis, as we demonstrate that LUBAC prevents aberrant TLR3-induced cell death without regulating the type of death. Absence of LUBAC components therefore perturbs the balance of TLR3′s signaling outputs by reducing gene activation and enhancing cell death. TLR3-SC analysis revealed recruitment of various endogenous proteins to the TLR3-SC, including the three LUBAC components. Mechanistically, we show that ligation of TLR3 results in formation of the TLR3-SC, leading to gene activation in a LUBAC component–dependent manner. Subsequently, a previously unrecognized TLR3-induced DISC, which is devoid of TLR3, forms and triggers cell death. In HOIP-deficient cells, formation of this DISC, and consequently TLR3-induced cell death, is substantially enhanced. This provides the molecular explanation of how LUBAC limits cell death induction by TLR3. We propose that the TLR3-induced DISC forms subsequently to the TLR3-SC by dissociation of part of the TLR3-SC from the receptor over time. This proposed model is supported by the observations that the kinetics of formation of the TLR3-SC precedes that of the TLR3-induced DISC and that TLR3 is absent from the TLR3-induced DISC. Notably, because of this sequence of events TLR3 signaling appears to parallel TNFR1 signaling in which complex II, originating from the TNFR1-SC, which is known to induce gene activation, was identified to induce cell death (Micheau and Tschopp, 2003). However, although the data provide compelling support for a transition from complex I to complex II in TLR3 signaling, it cannot be excluded that these complexes form sequentially, yet independently, of each other. Thus, as for TNFR1 signaling, formal proof for provenience of complex II from complex I remains to be provided.
Related to this matter, poly(I:C)-induced cell death was previously shown to be mediated by a platform containing Caspase-8, RIP1, cIAPs, TRIF, and TLR3 (Estornes et al., 2012; Weiss et al., 2013). In addition, cIAPs have been identified to prevent formation of a poly(I:C)-induced signaling platform termed Ripoptosome consisting of RIP1 as its core component, Caspase-8, and TRIF (Feoktistova et al., 2011). These studies isolated the poly(I:C)-induced signaling complex by Caspase-8-IP. We show here that Caspase-8 is only transiently associated with TLR3 and that it dissociates from the receptor over time to form part of the TLR3-induced DISC. Our results demonstrate the stimulation- and HOIP-dependent presence of linear ubiquitin linkages in both, the TLR3-SC and the TLR3-induced DISC. It remains to be determined, however, which of these complexes’ individual components are modified by M1-linked ubiquitin.
Recently, cIAP2 was identified as crucial for protection against IAV-induced host lethality (Rodrigue-Gervais et al., 2014). In this study, the authors proposed that IAV infection results in secretion of TRAIL and CD95L, which were at least partially responsible for cell death induction of lung epithelial cells in the absence of cIAP2. Because we identified cIAPs to form part of the TLR3-induced DISC, we postulate that deregulated TLR3 signaling, with a signaling output shifted toward cell death in the absence of cIAP2, is likely to contribute to IAV-induced lethality in this scenario.
TLR3 has recently been implicated in skin regeneration and wound healing by serving as DAMP sensor (Lai et al., 2009; Nelson et al., 2015), in addition to serving as a PAMP sensor. Here, we show that, in the absence of SHARPIN, deregulated TLR3 signaling contributes to the pathogenesis of cpdm dermatitis, as genetic ablation of TLR3 significantly ameliorates dermatitis of SHARPIN-deficient mice. We demonstrate that dsRNA is highly abundant in inflamed skin of cpdm mice, whereas there is significantly less dsRNA in Tlr3−/−.cpdm skin, suggesting that the presence of TLR3 serves as an accelerant of the disease caused by absence of SHARPIN. Importantly, TNF deficiency completely prevented inflammation in cpdm mice (Gerlach et al., 2011), whereas genetic ablation of Tlr3, though exerting a substantial effect, only ameliorated it. We therefore propose a model according to which absence of SHARPIN results in TNF-induced death of keratinocytes (Gerlach et al., 2011), inducing an initial release of different DAMPs, including dsRNA, in turn resulting in activation of TLR3, which, in the absence of SHARPIN, triggers further release of DAMPs, including more dsRNA, because of a further increase in aberrant cell death. As TNF-induced cell death results in release of all different kinds of DAMPs, it is likely that apart from TLR3, other damage sensors also play a role in fueling the inflammation in SHARPIN-deficient mice, which explains why absence of TLR3, though substantially ameliorating cpdm skin inflammation, fails to completely rescue the phenotype in contrast to TNF deficiency. According to this model, this sequence of events ultimately results in a self-perpetuating, TLR3/dsRNA-driven cycle of chronic inflammation and tissue malfunctioning in the skin of cpdm mice. It is tempting to speculate that the disease etiology in cpdm mice proposed by this model may provide an explanation as to why certain autoimmune patients do not respond to anti-TNF therapy, even though TNF could still be the initial trigger.
In this study, we demonstrate that SHARPIN is involved in host protection against IAV infection by serving a crucial role in TLR3 signaling and that TLR3 contributes to the autoinflammatory skin phenotype that characterizes SHARPIN-deficient cpdm mice. Interestingly, this co-occurrence of impaired host defense and autoinflammation caused by deregulation of TLR3 signaling in mice deficient in SHARPIN, is reminiscent of the severe pathological syndrome observed in some of the patients with knockout mutations in either of the other two LUBAC components, HOIP and HOIL-1 (Boisson et al., 2012, 2015). The autoinflammatory phenotype of SHARPIN-deficient mice is, however, characterized by severe skin inflammation, whereas the described patients deficient in HOIL-1 or HOIP appear not to suffer from inflammatory skin symptoms. Notably, human patients with deficiency in SHARPIN have not yet been described. Two independent studies identified truncating mutations in HOIL-1 to be causative for a phenotype encompassing muscular weakness, progressive cardiomyopathy, and signs of amylopectinosis; however, none of the patients suffered from severe immunodeficiency or overt hyperinflammation (Nilsson et al., 2013; Wang et al., 2013). The authors speculated that this discrepancy to the LUBAC-deficient patients described by Boisson et al. (Boisson et al., 2012, 2015) may result from different sites of the mutations. Variability seen in the phenotype of patients with the same genetic disorder may also come from the human virome, as suggested by MacDuff et al. (2015), who showed that chronic viral infection results in activation of the innate immune system thereby complementing immunodeficiency of mice deficient in e.g., HOIL-1 to bacterial infections. To answer the question whether the immunodeficient and autoinflammatory phenotypes of HOIP/HOIL-1–deficient patients are related to, or completely independent of, the immunodeficient and autoinflammatory phenotypes of SHARPIN-deficient mice will require further analysis. Based on our results, we suggest that deregulated TLR3 signaling may contribute to the immunodeficiency and autoinflammation observed in patients with LUBAC deficiency, and, although none of the patients with LUBAC deficiency identified so far suffered from IAV infection, we propose that outcome of TLR3-triggering viral infections may be worse in patients with deficiency in LUBAC components.
Materials and methods
Reagents
The following antibodies were used: α-pIκBα (IgG1), α-IκBα (rabbit), α-pERK (rabbit), α-ERK (rabbit), α-pJNK (rabbit), α-JNK (rabbit), α-pIRF3 (Ser396; rabbit), and α-pTBK1 (rabbit) were purchased from Cell Signaling Technology; α-actin (IgG1), α-FLAG (IgG1), and α-human-HOIP (rabbit) were obtained from Sigma-Aldrich; α-mouse-HOIP (rabbit) was custom-made by Thermo Fisher Scientific; α-MDA5 (rabbit) was purchased from Thermo Fisher Scientific; α-HOIL-1 (IgG2a) and α-SHARPIN (IgG1) are previously described (Haas et al., 2009; Gerlach et al., 2011); α-SHARPIN (rabbit) was purchased from Proteintech; α-Caspase-8 (C15; IgG2b), α-cFLIP (NF6; IgG1), and α-FADD (IgG1) were obtained from Enzo Life Sciences; α-TRAF6 (C-term; rabbit) was purchased from Epitomics; α-PARP (IgG1) and α-RIP1 (IgG2a) were purchased from BD; α-Caspase-3 (goat), α-HOIP (IgG1), and α-cIAP1/2 (IgG2a) were obtained from R&D Systems; α-GAPDH (IgG1) was purchased from Abcam; α-linear Ubiquitin (1E3; rabbit) was obtained from Merck; α-RIG-I (rabbit) was obtained from IBL; and α-IKKγ (FL419; rabbit) was purchased from Santa Cruz Biotechnology, Inc.
α-Caspase-8 (goat; Santa Cruz Biotechnology, Inc.) and α-FLAG (IgG1; Sigma-Aldrich) were used for IP experiments. α-J2 (English & Scientific Consulting Kft.), α-K14, α-K10, α-K6, and α-loricirin (Covance), α-cleaved Caspase-3 (Cell Signaling Technology), and α-CD45 (BD) were used for immunofluorescence. α-TLR3 (IgG1) was provided by S. Lebecque (Centre de Recherche en Cancérologie de Lyon, INSERM Unité Mixte de Recherche 1052/CNRS 5286, Centre Léon Bérard, Lyon, France).
Secondary antibodies for Western blot were purchased from Southern Biotech. Alexa Fluor 488 and 594 goat α-rabbit IgG were purchased from Invitrogen, and goat α-rat HRP was obtained from Cambridge Bioscience.
Poly(I:C) HMW was purchased from InvivoGen; z-VAD(OMe)-FMK was obtained from Abcam; and 7-Cl-O-Nec-1 was obtained from Millipore. Smac Mimetic 083 (SM083) was provided by M. Bolognesi (Protein Biochemistry Unit, Department of Biosciences, University of Milano, Milano, Italy). Etanercept (Enbrel) was purchased from Pfizer, and Pentaglobin was obtained from Biotest.
Primers
The following primers were used: il6, forward, 5′-GTAGCTATGGTACTCCAGAAGAC-3′, and reverse 5′-ACGATGATGCACTTGCAGAA-3′; ifnb, forward, 5′-CATCAACTATAAGCAGCTCCA-3′, and reverse, 5′-TTCAAGTGGAGAGCAGTTGAG-3′; cxcl10, forward, 5′-ACTGCATCCATATCGATGAC-3′, and reverse, 5′-TTCATCGTGGCAATGATCTC-3′; hprt, forward 5′-GTTGGATACAGGCCAGACTTTGTTG-3′, and reverse, 5′-GATTCAACTTGCGCTCATCTTAGGC-3′; and isg15, forward, 5′-GCAAGCAGCCAGAAGCAGACTCC-3′ and reverse, 5′-CGGACACCAGGAAATCGTTACCCC-3′.
Culture of cell lines
The human immortalized keratinocyte line HaCaT, the human colon adenocarcinoma grade II cell line HT29, and the cervical cancer cell line HeLa were maintained in DMEM supplemented with 10% FCS. Parental cell lines were tested on a regular basis and determined to be mycoplasma free using MycoAlert Mycoplasma Detection kit (Lonza).
Retroviral transduction of cells
For tagging of exogenously expressed proteins, we used a C-terminal Tandem Affinity Purification-tag consisting of 2x Strep-tag II sequence followed by a PreScission cleavage site and 1x Flag-tag. Coding sequences for TLR3 fused at the C terminus to a Tandem Affinity Purification-tag were inserted into the retroviral murine stem cell virus vector, followed by an internal ribosome entry site and the open reading frame of enhanced green fluorescent protein. These vectors were transfected using Lipofectamine 2000 in Phoenix cells cultured in DMEM supplemented with 10% FCS. 1 d after transfection, the medium was replaced and subsequently collected at day three. Viral supernatants were passed through a 0.45-µm filter and added to HaCaT or HeLa cells in the presence of Polybrene at 6 µg/ml before cells were subjected to spin-infection (2,500 rpm, 45 min, 30°C). EGFP-positive cells were isolated using MoFlo FACS (Beckman Coulter) to >95% purity.
Generation of Hoip−/− cell lines
For production of Hoip−/− HaCaT and HeLa cells, LentiCRISPR version 2 vector (F. Zhang, Broad Institute of MIT and Harvard, Cambridge, MA; plasmid #52961; Addgene; Sanjana et al., 2014) targeting the following sequence was used: 5′-CGAGATGTGCTGCGATTATA-3′.
Lentiviral particles were produced in HEK293FT cells upon co-transfection of LentiCRISPR vector together with packaging vectors psPAX2 and pMD2G. Cells were infected as in the case of retroviral transduction and subjected to puromycin (2 µg/ml) selection for 1 wk. Single-cell cloning was achieved by limiting dilution, and HOIP-deficient cells were validated by Western blotting.
Cell viability and cell death assays
Cell viability was determined using Cell Titer Glo assay (Promega) according to the manufacturer’s instructions. Cell death was determined by percentage of propidium iodide–positive cells.
Western blot analysis
Cells were washed twice with ice-cold PBS before lysis in lysis buffer containing 30 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM EDTA, 2 mM KCl, 10% Glycerol, 1% Triton X-100, 1× complete EDTA-free protease-inhibitor mix (Roche), and 1× phosphatase-inhibitor cocktail 2 (Sigma-Aldrich). Protein concentration of lysates was determined using BCA protein assay (Thermo Fisher Scientific). Lysates were denatured in reducing sample buffer at 95°C for 10 min before separation by SDS-PAGE (NuPAGE), and analyzed by Western blotting. Membranes were incubated with primary antibodies at 4°C overnight or for 1 h at room temperature. Washing of membranes was performed in 1xPBS containing 0.05% Tween-20 (Sigma-Aldrich) for 3 × 10 min before incubation with the secondary antibody for 1 h at room temperature. Membranes were stripped with stripping buffer containing 50 mM glycine, pH 2.3, before reprobing with the next primary antibody.
IP
Cells were seeded at 0.5–107 cells in 150 cm2 dishes. The next day, cells were stimulated as indicated in DMEM without FCS. Cells were washed twice with ice-cold PBS and harvested in 0.5 ml lysis buffer containing 100 mM NaCl, 40 mM Tris-HCl, pH 7.5, 1 mM CaCl2, 1 mM MgCl2, and 1× complete EDTA-free protease-inhibitor mix supplemented with 0.5% Triton X-100. Cells were lysed for 20 min by gentle rocking at 4°C. Lysates were subsequently cleared by centrifugation at 13,000 rpm for 10 min and transferred to a new Eppendorf tube. The remaining pellets were resuspended in 0.5 ml of lysis buffer supplemented with 1% Triton X-100 and 0.1% SDS and subjected to sonification before centrifugation at 13,000 rpm for 20 min. The cleared lysate was added to the lysate of centrifugation step 1. 30 µl of lysates were subsequently stored at −20°C for analysis of whole-cell lysates. For IP of Flag, 10 µl of M2 beads (Sigma-Aldrich) were added to each sample. For IP of Caspase-8, 10 µl Protein-G beads that were precoupled to Caspase-8 antibody were added to each sample. Samples were incubated at 4°C for 8–24 h. For IP of Caspase-8 after IP of Flag, lysates were cleared by centrifugation from M2 beads, and Protein-G beads that were precoupled to Caspase-8 were added to each sample and incubated for 8–24 h at 4°C. Samples were washed at least five times with lysis buffer containing 1% Triton X-100. After the last centrifugation step, beads were sucked dry and resuspended in 2x reducing sample buffer, boiled for 10 min at 95°C, and separated by SDS-PAGE before analysis by Western blotting.
ELISA
Cells were stimulated as indicated for 4 h (IFN-β) or 24 h (IL-8, TNF, CCL5, and CCL20). Cell supernatants were subsequently collected and stored at −20°C until further analysis. DuoSet ELISA for human α-IL-8, human α-TNF, human and murine α-CCL5, and murine α-CCL20 were purchased from R&D Systems. Human IFN-β was determined using the VeriKine human IFN-β ELISA kit (PBL Assay Science). The respective ELISA was performed according to the manufacturer's instructions.
RNA interference
Cells were plated in 6-well plates at a seeding density of 1.2 × 105 cells/well before transfection by the respective siRNAs using Lipofectamine 2000 transfection reagent (Invitrogen) for HaCaT or using DharmaFECT transfection reagent (Dharmacon) for HeLa according to the manufacturer's instructions. siRNA sequences targeting the gene of interest were purchased from Dharmacon (Non-Targeting siRNA [sequence #3], human HOIP siRNA [sequence #4], human SHARPIN siRNA [ON-TARGETplus SMARTpool], human HOIL-1 siRNA [ON-TARGETplus SMARTpool], human TLR3 siRNA [ON-TARGETplus Set of 4 Upgrade {sequences #5–#8}], human RIG-I siRNA [ON-TARGETplus Set of 4 Upgrade {sequences #5–#8}], and human MDA5 siRNA [ON-TARGETplus Set of 4 Upgrade {sequences #5–#8}]). Cells were reseeded 48 h after transfection and subjected to further experimental analysis 24 h later.
Mice
Tlr3−/− (9675, B6N.129S1-Tlr3tm1Flv/J), cpdm (7599, C57BL/KaLawRij-Sharpincpdm/RijSunJ), and Tamoxifen-inducible Cre mice (4682, B6.Cg-Tg(CAG-cre/Esr1*)5Amc/J) were obtained from The Jackson Laboratory. HOIP-floxed mice were generated as previously described (Peltzer et al., 2014). HOIP-floxed mice were crossed with transgenic mice expressing the Tamoxifen-inducible loxP-deleter Cre recombinase to generate Tamoxifen-inducible HOIP-deficient mice. All mice were crossed for at least five generations before the experimental studies. All mice were typed by PCR analysis. Colonies were fed ad libitum. All animal experiments were conducted under an appropriate UK project license in accordance with the regulations of UK home office for animal welfare according to the ASPA (Animals [Scientific Procedures] Act 1986).
Isolation and culture of PMKs from newborn mice
PMKs were isolated according to the protocol established and described in detail by Lichti et al. (2008) from newborn littermates at the age of 0–2 d. In summary, newborn mice were humanely killed and their skin was floated on trypsin (Cellgro) at 4°C overnight. The next day, epidermis was separated from dermis and PMKs were isolated using high-calcium medium (HiCa medium, 1.3 mM Ca2+). After isolation, PMKs were counted and seeded at equal numbers in low-calcium medium (LoCa medium, 0.05 mM Ca2+). 24 h after plating, dead cells were removed by washing once with DPBS (Invitrogen). Fresh LoCa medium was subsequently added. Medium was changed every other day until cells reached 70–90% confluency, usually between day 3 and 5 after plating. For PMKs from Tamoxifen-inducible Cre HOIPfl/fl newborn littermates, 4-OHT (Sigma-Aldrich) was added to the medium at a concentration of 1 µM directly after isolation for at least 72 h.
Isolation and culture of PMKs from tails of adult mice
Tails of littermates were wiped with 70% Ethanol, and then removed at base and carefully incised using a scalpel to make a longitudinal incision down the tail; skin was subsequently removed from the tails using tweezers. Skin was then incubated overnight at 4°C in keratinocyte serum free media (from Invitrogen) with dispase II at a final concentration of 2.1 U/ml to cleave epidermal–dermal junctions. The next day, epidermis was separated from dermis. Epidermis was then incubated for 15 min in 2 ml TrypLE express trypsin (from Invitrogen) to isolate keratinocytes. After addition of medium, cells were centrifuged at 600 rpm for 10 min. Supernatant was removed and the pellet was resuspended in medium supplemented with bovine pituitary extract and epidermal growth factor, including gentamicin at 50 µg/ml (Sigma-Aldrich). Floating dead cells were removed the next day. Keratinocytes were then grown to 70–90% confluency until further experiments were performed, usually after 3–5 additional days in culture.
Histology
Skin or lungs were removed, fixed in 10% formalin, and then transferred to 70% ethanol. Paraffin embedding, preparation of sections and H&E staining of tissues were performed by histological services at the National Heart & Lung Institute (London, England, UK) or the Department of Pathology, University of Cambridge (Cambridge, England, UK).
Immunofluorescence
Skin sections were de-waxed and rehydrated by passing the slides through xylene and descending grades of alcohol, then rinsed in water. Antigen retrieval was performed by immersing sections in 10 mM sodium citrate buffer, pH 6.0. Sections were washed and then blocked for at least 30 min with TBS 1x, pH 7.6, containing Tween-20 0.5% and BSA 0.2% in a humid chamber. For CD45 staining, Retrievagen A (BD) was used for antigen retrieval. Slides were incubated with 100 µl of the primary antibody overnight at 4°C. The following antibodies were used: α-K14 (Covance), α-K10 (Covance), α-K6 (Covance), α-loricirin (Covance), and α-CD45 (BD) using the following dilutions: K14 1:1000, K10 1:500, K6 1:500, loricrin 1:500, and CD45 1:20. On the day after, slides were washed and subsequently incubated with the secondary antibody Alexa Fluor 488 (Invitrogen) or 594 goat α-rabbit IgG (Invitrogen) or goat α-rat HRP (Cambridge Bioscience) at room temperature for 1 h at a dilution of 1:500. Where HRP antibody was applied, the TSA Plus Cyanine 3 System (Perkin Elmer) was applied according to the manufacturer’s instructions. Sections were extensively washed and counterstained with 1:2,000 DAPI (Roche) to visualize the nuclei. Sections were mounted with StayBrite Hardset Mounting Medium (Biotium Inc.) and analyzed by fluorescent microscopy. At least 10 different images (40×) for each slide were acquired.
dsRNA staining with J2 antibody
Slide deparaffinization was performed by incubating the slides at 60°C. Slides were subsequently immersed in histoclear, followed by incubation in decreasing concentrations of ethanol and subsequent washing of the slides in water. Antigen retrieval was performed using EDTA buffer (1 mM EDTA, pH 8.0). Subsequent treatment of slides with H2O2 3% for 10 min was performed to block endogenous peroxidases. Blocking of unspecific sites was performed using TBS 1x, pH 7.6, containing Tween 20 0.05%, BSA 0.2%, and goat serum 10%. Staining with the J2 antibody (English & Scientific Consulting Kft.) was performed diluting the antibody in TBS 1x, pH 7.6, containing Tween 20 0.05%, BSA 0.2%, and goat serum 10% to a final concentration of 1 µg/ml for 2 h. Afterward, slides were incubated with the secondary antibody α-mouse IgG2a-HRP (1:1,000) for 1 h at room temperature. TSA amplification system Cy3 (Promega) was added. Finally, nuclei were stained using DAPI dye 1:1,000. Slides were subsequently mounted with 10 µl Everbright mounting medium and sealed with a coverslip. At least 10 different images (40×) for each slide were acquired.
TUNEL and cleaved Caspase-3 staining
Sections were immersed in 10 mM sodium citrate buffer, pH 6.0, for antigen retrieval. Slides were washed with water and blocked with 1x PBS containing 10% goat serum at room temperature for 20 min. Subsequently, slides were incubated with anti-cleaved Caspase-3 (Cell Signaling Technology) diluted 1:100 in 1.5% goat serum in PBS 1x at room temperature for 1 h. After washing, sections were incubated with rabbit biotinylated antibody (Vector) 1:200 diluted in 1.5% goat serum in PBS 1x at room temperature for 45 min. Slides were washed and incubated with streptavidin conjugate Cy2 (Jackson ImmunoResearch Laboratories) diluted 1:200 in 1.5% goat serum in PBS 1x and counterstained with DAPI 1:1,000 at room temperature. Next, slides were washed and ApopTag Red In Situ Apoptosis Detection (Merch Millipore) protocol was applied according to the manufacturer’s instructions. Slides were mounted with StayBrite Hardset Mounting Medium (Biotium Inc.) and analyzed by fluorescent microscopy. At least 10 different images (40×) for each slide were acquired and quantification was performed with ImageJ Software (National Institutes of Health) on monochrome images counting the DAPI-positive cells in the epidermis and cells positive for the specific staining within the epidermis.
Epidermal thickness quantification
The epidermal thickness was measured in five different positions per field and for at least 10 different fields per mouse. Quantification was performed using ImageJ Software.
In vivo RNA stimulation
1 µg poly(I:C) in 10 µl PBS was injected i.d. into the ear pinnae of age- and sex-matched littermates of the indicated genotypes (n ≥ 5 per time point), and induction of innate immune transcriptional responses was assessed by RT-PCR analysis of extracted RNA after 12 h. Data are presented as fold-induction relative to the hprt gene.
IAV infection
IAV strain A/PR/8 was grown and titrated on Madin Darby canine kidney cells. For weight loss experiments, age- and sex-matched WT, cpdm, Tlr3−/−, and Tlr3−/−.cpdm littermates (4–5 wk old) were anaesthetized and infected i.n. with the indicated dose of IAV, and their weights were monitored daily. Etanercept (or Pentaglobin as mock control) was administered at a concentration of 0.5 mg/mouse/injection. Injections were started 1 d before the infection and then every third day for 2 wk. For analysis of lung pathology, age- and sex-matched littermates (n ≥ 10 per group; 4–5 wk old) were infected i.n. with IAV and lung tissue was harvested at the indicated times after infection and fixed in formalin for histological processing or frozen for further analysis. For RT-PCR, frozen lung tissue was ground under liquid nitrogen and total cellular RNA was extracted using an RNeasy kit (QIAGEN). cDNA synthesis was performed with Superscript III Reverse transcription (Life Technologies) using 500 ng of template RNA. RT-PCR was performed on a 7900HT series thermocycler (Life Technologies) with Fast SYBR Green Master Mix (Life Technologies). Hprt was used as the reference gene in all assays. Data were analyzed with RQ manager 1.2 software (Life Technologies). For IAV titrations, serial dilutions of lung homogenates of known mass were incubated on monolayers of Madin Darby canine kidney cells for 1 h before being overlayed with medium containing 0.5% agarose and 0.1 µg/ml TPCK-trypsin (Worthington Biochemical Corporation). Plaques were counted 3–4 d later and data are presented as virus titer per milligram of lung tissue. For histological analysis, lungs were fixed in 10% formalin before being transferred into 20% ethanol before paraffin embedding. Hematoxylin and eosin (H&E) staining was performed and at least 10 different images (40×) for each slide were acquired for analysis.
Severity of dermatitis
Severity of dermatitis of cpdm mice and WT littermate controls was assessed at the age of 8 wk on six different body parts (head, neck, anterior trunk, posterior trunk, forelegs, and hind legs) in a blinded manner. The score was obtained by grading each of the six body parts lesion absent (0 points) or lesion detectable (1 point), accounting to a maximum of six points/mouse.
Microscope image acquisition
All microscopic images were acquired using an Axio Imager A1 microscope (EC Plan-neofluar Air; magnification 40×; NA, 0.75; ZEISS) at room temperature (20°C). DAPI, GFP, Cy3, and Alexa Fluor 568 were used as fluorochromes. Images were taken with an Axiocam (ZEISS) Mrm rev:3.0 –12 bit monochrome 2/3”CCD camera. Axiovision 4.8.1 (ZEISS) was used as acquisition software. Images were further processed using ImageJ and Photoshop (Adobe).
Statistical analysis
Data were analyzed using Prism 6 software (GraphPad Software). Statistical significance between groups was determined using unpaired Student’s t test. P < 0.05 was considered significant. *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
Acknowledgments
We thank S. Lebecque and Y. Estornes for providing reagents; K. Roberts (Trinity College Dublin) and Martin Leverkus for help and advice; and Helena Draberova, Aida Sarr, and Dina Hochhauser for excellent technical assistance.
This work was funded by a Wellcome Trust Senior Investigator award (096831/Z/11/Z; and grant 090315 to H. Ren) and an European Research Council advanced grant (294880; H. Walczak). J. Zinngrebe received support from the Boehringer Ingelheim Fonds and N. Peltzer received funds from the Swiss National Science Foundation. B.J. Ferguson is supported by an Isaac Newton Trust/Wellcome Trust ISSF/University of Cambridge research grant. B. Dome received support from the Hungarian Scientific Research Fund (OTKA-K108465).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- 4-OHT
- 4-hydroxy-tamoxifen
- cIAP
- cellular inhibitor of apoptosis protein
- cpdm
- chronic proliferative dermatitis mice
- DAMP
- damage-associated molecular pattern
- DISC
- death-inducing signaling complex
- ds
- double stranded
- FADD
- Fas-associated protein with death domain
- HOIL-1
- heme-oxidized IRP2 ubiquitin ligase-1
- HOIP
- HOIL-1–interacting protein
- IAV
- influenza A virus
- IP
- immunoprecipitation
- IRF
- IFN regulatory factor
- LUBAC
- linear ubiquitin chain assembly complex
- MDA5
- melanoma differentiation-associated protein 5
- PAMP
- pathogen-associated molecular pattern
- PMK
- primary murine keratinocyte
- RIG-I
- retinoic acid inducible gene I
- RIP
- receptor-interacting protein
- SC
- signaling complex
- SHARPIN
- SHANK-associated RH-domain-interacting protein
- SM
- Smac Mimetic
- TBK
- TANK-binding kinase
- TRAF
- TNFR-associated factor
- TRIF
- TIR-domain–containing adapter inducing IFN-β
References
- Akira S., Uematsu S., and Takeuchi O.. 2006. Pathogen recognition and innate immunity. Cell. 124:783–801. 10.1016/j.cell.2006.02.015 [DOI] [PubMed] [Google Scholar]
- Alexopoulou L., Holt A.C., Medzhitov R., and Flavell R.A.. 2001. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 413:732–738. 10.1038/35099560 [DOI] [PubMed] [Google Scholar]
- Andersen L.L., Mørk N., Reinert L.S., Kofod-Olsen E., Narita R., Jørgensen S.E., Skipper K.A., Höning K., Gad H.H., Østergaard L., et al. 2015. Functional IRF3 deficiency in a patient with herpes simplex encephalitis. J. Exp. Med. 212:1371–1379. 10.1084/jem.20142274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard J.J., Cowing-Zitron C., Nakatsuji T., Muehleisen B., Muto J., Borkowski A.W., Martinez L., Greidinger E.L., Yu B.D., and Gallo R.L.. 2012. Ultraviolet radiation damages self noncoding RNA and is detected by TLR3. Nat. Med. 18:1286–1290. 10.1038/nm.2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson B., Laplantine E., Prando C., Giliani S., Israelsson E., Xu Z., Abhyankar A., Israël L., Trevejo-Nunez G., Bogunovic D., et al. 2012. Immunodeficiency, autoinflammation and amylopectinosis in humans with inherited HOIL-1 and LUBAC deficiency. Nat. Immunol. 13:1178–1186. 10.1038/ni.2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson B., Laplantine E., Dobbs K., Cobat A., Tarantino N., Hazen M., Lidov H.G., Hopkins G., Du L., Belkadi A., et al. 2015. Human HOIP and LUBAC deficiency underlies autoinflammation, immunodeficiency, amylopectinosis, and lymphangiectasia. J. Exp. Med. 212:939–951. 10.1084/jem.20141130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brentano F., Kyburz D., Schorr O., Gay R., and Gay S.. 2005. The role of Toll-like receptor signalling in the pathogenesis of arthritis. Cell. Immunol. 233:90–96. 10.1016/j.cellimm.2005.04.018 [DOI] [PubMed] [Google Scholar]
- Cavassani K.A., Ishii M., Wen H., Schaller M.A., Lincoln P.M., Lukacs N.W., Hogaboam C.M., and Kunkel S.L.. 2008. TLR3 is an endogenous sensor of tissue necrosis during acute inflammatory events. J. Exp. Med. 205:2609–2621. 10.1084/jem.20081370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S., Kuzmanovic T., Zhang Y., Wetzel J.L., and Sen G.C.. 2016. Ubiquitination of the Transcription Factor IRF-3 Activates RIPA, the Apoptotic Pathway that Protects Mice from Viral Pathogenesis. Immunity. 44:1151–1161. 10.1016/j.immuni.2016.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito S., Molteni C.G., Giliani S., Mazza C., Scala A., Tagliaferri L., Pelucchi C., Fossali E., Plebani A., and Principi N.. 2012. Toll-like receptor 3 gene polymorphisms and severity of pandemic A/H1N1/2009 influenza in otherwise healthy children. Virol. J. 9:270 10.1186/1743-422X-9-270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estornes Y., Toscano F., Virard F., Jacquemin G., Pierrot A., Vanbervliet B., Bonnin M., Lalaoui N., Mercier-Gouy P., Pachéco Y., et al. 2012. dsRNA induces apoptosis through an atypical death complex associating TLR3 to caspase-8. Cell Death Differ. 19:1482–1494. 10.1038/cdd.2012.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feoktistova M., Geserick P., Kellert B., Dimitrova D.P., Langlais C., Hupe M., Cain K., MacFarlane M., Häcker G., and Leverkus M.. 2011. cIAPs block Ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Mol. Cell. 43:449–463. 10.1016/j.molcel.2011.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald K.A., McWhirter S.M., Faia K.L., Rowe D.C., Latz E., Golenbock D.T., Coyle A.J., Liao S.M., and Maniatis T.. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491–496. 10.1038/ni921 [DOI] [PubMed] [Google Scholar]
- Gerlach B., Cordier S.M., Schmukle A.C., Emmerich C.H., Rieser E., Haas T.L., Webb A.I., Rickard J.A., Anderton H., Wong W.W., et al. 2011. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature. 471:591–596. 10.1038/nature09816 [DOI] [PubMed] [Google Scholar]
- Haas T.L., Emmerich C.H., Gerlach B., Schmukle A.C., Cordier S.M., Rieser E., Feltham R., Vince J., Warnken U., Wenger T., et al. 2009. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol. Cell. 36:831–844. 10.1016/j.molcel.2009.10.013 [DOI] [PubMed] [Google Scholar]
- He S., Wang L., Miao L., Wang T., Du F., Zhao L., and Wang X.. 2009. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell. 137:1100–1111. 10.1016/j.cell.2009.05.021 [DOI] [PubMed] [Google Scholar]
- Herman M., Ciancanelli M., Ou Y.H., Lorenzo L., Klaudel-Dreszler M., Pauwels E., Sancho-Shimizu V., Pérez de Diego R., Abhyankar A., Israelsson E., et al. 2012. Heterozygous TBK1 mutations impair TLR3 immunity and underlie herpes simplex encephalitis of childhood. J. Exp. Med. 209:1567–1582. 10.1084/jem.20111316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidaka F., Matsuo S., Muta T., Takeshige K., Mizukami T., and Nunoi H.. 2006. A missense mutation of the Toll-like receptor 3 gene in a patient with influenza-associated encephalopathy. Clin. Immunol. 119:188–194. 10.1016/j.clim.2006.01.005 [DOI] [PubMed] [Google Scholar]
- HogenEsch H., Gijbels M.J., Offerman E., van Hooft J., van Bekkum D.W., and Zurcher C.. 1993. A spontaneous mutation characterized by chronic proliferative dermatitis in C57BL mice. Am. J. Pathol. 143:972–982. [PMC free article] [PubMed] [Google Scholar]
- Ikeda F., Deribe Y.L., Skånland S.S., Stieglitz B., Grabbe C., Franz-Wachtel M., van Wijk S.J., Goswami P., Nagy V., Terzic J., et al. 2011. SHARPIN forms a linear ubiquitin ligase complex regulating NF-κB activity and apoptosis. Nature. 471:637–641. 10.1038/nature09814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inn K.S., Gack M.U., Tokunaga F., Shi M., Wong L.Y., Iwai K., and Jung J.U.. 2011. Linear ubiquitin assembly complex negatively regulates RIG-I- and TRIM25-mediated type I interferon induction. Mol. Cell. 41:354–365. 10.1016/j.molcel.2010.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T., Kamei K., Murata S., Kato M., Fukumoto H., Kanie M., Sano S., Tokunaga F., Tanaka K., and Iwai K.. 2006. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 25:4877–4887. 10.1038/sj.emboj.7601360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari S., Redouane Y., Lopez-Mosqueda J., Shiraishi R., Romanowska M., Lutzmayer S., Kuiper J., Martinez C., Dikic I., Pasparakis M., and Ikeda F.. 2014. Sharpin prevents skin inflammation by inhibiting TNFR1-induced keratinocyte apoptosis. eLife. 3:3 10.7554/eLife.03422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y., Di Nardo A., Nakatsuji T., Leichtle A., Yang Y., Cogen A.L., Wu Z.R., Hooper L.V., Schmidt R.R., von Aulock S., et al. 2009. Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nat. Med. 15:1377–1382. 10.1038/nm.2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Goffic R., Balloy V., Lagranderie M., Alexopoulou L., Escriou N., Flavell R., Chignard M., and Si-Tahar M.. 2006. Detrimental contribution of the Toll-like receptor (TLR)3 to influenza A virus-induced acute pneumonia. PLoS Pathog. 2:e53 10.1371/journal.ppat.0020053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung Y.H., Nicholls J.M., Ho C.K., Sia S.F., Mok C.K., Valkenburg S.A., Cheung P., Hui K.P., Chan R.W., Guan Y., et al. 2014. Highly pathogenic avian influenza A H5N1 and pandemic H1N1 virus infections have different phenotypes in Toll-like receptor 3 knockout mice. J. Gen. Virol. 95:1870–1879. 10.1099/vir.0.066258-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichti U., Anders J., and Yuspa S.H.. 2008. Isolation and short-term culture of primary keratinocytes, hair follicle populations and dermal cells from newborn mice and keratinocytes from adult mice for in vitro analysis and for grafting to immunodeficient mice. Nat. Protoc. 3:799–810. 10.1038/nprot.2008.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDuff D.A., Reese T.A., Kimmey J.M., Weiss L.A., Song C., Zhang X., Kambal A., Duan E., Carrero J.A., Boisson B., et al. 2015. Phenotypic complementation of genetic immunodeficiency by chronic herpesvirus infection. eLife. 4:4 10.7554/eLife.04494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M., Funami K., Tatematsu M., Azuma M., and Seya T.. 2014. Assessment of the Toll-like receptor 3 pathway in endosomal signaling. Methods Enzymol. 535:149–165. 10.1016/B978-0-12-397925-4.00010-9 [DOI] [PubMed] [Google Scholar]
- Meylan E., Burns K., Hofmann K., Blancheteau V., Martinon F., Kelliher M., and Tschopp J.. 2004. RIP1 is an essential mediator of Toll-like receptor 3-induced NF-kappa B activation. Nat. Immunol. 5:503–507. 10.1038/ni1061 [DOI] [PubMed] [Google Scholar]
- Micheau O., and Tschopp J.. 2003. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 114:181–190. 10.1016/S0092-8674(03)00521-X [DOI] [PubMed] [Google Scholar]
- Nelson A.M., Reddy S.K., Ratliff T.S., Hossain M.Z., Katseff A.S., Zhu A.S., Chang E., Resnik S.R., Page C., Kim D., et al. 2015. dsRNA Released by Tissue Damage Activates TLR3 to Drive Skin Regeneration. Cell Stem Cell. 17:139–151. 10.1016/j.stem.2015.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson J., Schoser B., Laforet P., Kalev O., Lindberg C., Romero N.B., Dávila López M., Akman H.O., Wahbi K., Iglseder S., et al. 2013. Polyglucosan body myopathy caused by defective ubiquitin ligase RBCK1. Ann. Neurol. 74:914–919. 10.1002/ana.23963 [DOI] [PubMed] [Google Scholar]
- Peltzer N., Rieser E., Taraborrelli L., Draber P., Darding M., Pernaute B., Shimizu Y., Sarr A., Draberova H., Montinaro A., et al. 2014. HOIP deficiency causes embryonic lethality by aberrant TNFR1-mediated endothelial cell death. Cell Reports. 9:153–165. 10.1016/j.celrep.2014.08.066 [DOI] [PubMed] [Google Scholar]
- Perales-Linares R., and Navas-Martin S.. 2013. Toll-like receptor 3 in viral pathogenesis: friend or foe? Immunology. 140:153–167. 10.1111/imm.12143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez de Diego R., Sancho-Shimizu V., Lorenzo L., Puel A., Plancoulaine S., Picard C., Herman M., Cardon A., Durandy A., Bustamante J., et al. 2010. Human TRAF3 adaptor molecule deficiency leads to impaired Toll-like receptor 3 response and susceptibility to herpes simplex encephalitis. Immunity. 33:400–411. 10.1016/j.immuni.2010.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickard J.A., Anderton H., Etemadi N., Nachbur U., Darding M., Peltzer N., Lalaoui N., Lawlor K.E., Vanyai H., Hall C., et al. 2014. TNFR1-dependent cell death drives inflammation in Sharpin-deficient mice. eLife. 3:3 10.7554/eLife.03464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifkin I.R., Leadbetter E.A., Busconi L., Viglianti G., and Marshak-Rothstein A.. 2005. Toll-like receptors, endogenous ligands, and systemic autoimmune disease. Immunol. Rev. 204:27–42. 10.1111/j.0105-2896.2005.00239.x [DOI] [PubMed] [Google Scholar]
- Rodrigue-Gervais I.G., Labbé K., Dagenais M., Dupaul-Chicoine J., Champagne C., Morizot A., Skeldon A., Brincks E.L., Vidal S.M., Griffith T.S., and Saleh M.. 2014. Cellular inhibitor of apoptosis protein cIAP2 protects against pulmonary tissue necrosis during influenza virus infection to promote host survival. Cell Host Microbe. 15:23–35. 10.1016/j.chom.2013.12.003 [DOI] [PubMed] [Google Scholar]
- Sancho-Shimizu V., Pérez de Diego R., Lorenzo L., Halwani R., Alangari A., Israelsson E., Fabrega S., Cardon A., Maluenda J., Tatematsu M., et al. 2011. Herpes simplex encephalitis in children with autosomal recessive and dominant TRIF deficiency. J. Clin. Invest. 121:4889–4902. 10.1172/JCI59259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana N.E., Shalem O., and Zhang F.. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 11:783–784. 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szretter K.J., Gangappa S., Lu X., Smith C., Shieh W.J., Zaki S.R., Sambhara S., Tumpey T.M., and Katz J.M.. 2007. Role of host cytokine responses in the pathogenesis of avian H5N1 influenza viruses in mice. J. Virol. 81:2736–2744. 10.1128/JVI.02336-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O., and Akira S.. 2009. Innate immunity to virus infection. Immunol. Rev. 227:75–86. 10.1111/j.1600-065X.2008.00737.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga F., Sakata S., Saeki Y., Satomi Y., Kirisako T., Kamei K., Nakagawa T., Kato M., Murata S., Yamaoka S., et al. 2009. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat. Cell Biol. 11:123–132. 10.1038/ncb1821 [DOI] [PubMed] [Google Scholar]
- Tokunaga F., Nakagawa T., Nakahara M., Saeki Y., Taniguchi M., Sakata S., Tanaka K., Nakano H., and Iwai K.. 2011. SHARPIN is a component of the NF-κB-activating linear ubiquitin chain assembly complex. Nature. 471:633–636. 10.1038/nature09815 [DOI] [PubMed] [Google Scholar]
- Walczak H., Iwai K., and Dikic I.. 2012. Generation and physiological roles of linear ubiquitin chains. BMC Biol. 10:23 10.1186/1741-7007-10-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Kim C., Bradfield J., Guo Y., Toskala E., Otieno F.G., Hou C., Thomas K., Cardinale C., Lyon G.J., et al. 2013. Whole-genome DNA/RNA sequencing identifies truncating mutations in RBCK1 in a novel Mendelian disease with neuromuscular and cardiac involvement. Genome Med. 5:67 10.1186/gm471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R., Sachet M., Zinngrebe J., Aschacher T., Krainer M., Hegedus B., Walczak H., and Bergmann M.. 2013. IL-24 sensitizes tumor cells to TLR3-mediated apoptosis. Cell Death Differ. 20:823–833. 10.1038/cdd.2013.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.Y., Jouanguy E., Ugolini S., Smahi A., Elain G., Romero P., Segal D., Sancho-Shimizu V., Lorenzo L., Puel A., et al. 2007. TLR3 deficiency in patients with herpes simplex encephalitis. Science. 317:1522–1527. 10.1126/science.1139522 [DOI] [PubMed] [Google Scholar]
- Zhang S.Y., Herman M., Ciancanelli M.J., Pérez de Diego R., Sancho-Shimizu V., Abel L., and Casanova J.L.. 2013. TLR3 immunity to infection in mice and humans. Curr. Opin. Immunol. 25:19–33. 10.1016/j.coi.2012.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]