Moore and colleagues present new strains of optically clear immune-deficient zebrafish that allow for dynamic imaging of regeneration and tumor progression at single-cell resolution in live animals.
Abstract
Cell transplantation into immunodeficient mice has revolutionized our understanding of regeneration, stem cell self-renewal, and cancer; yet models for direct imaging of engrafted cells has been limited. Here, we characterize zebrafish with mutations in recombination activating gene 2 (rag2), DNA-dependent protein kinase (prkdc), and janus kinase 3 (jak3). Histology, RNA sequencing, and single-cell transcriptional profiling of blood showed that rag2 hypomorphic mutant zebrafish lack T cells, whereas prkdc deficiency results in loss of mature T and B cells and jak3 in T and putative Natural Killer cells. Although all mutant lines engraft fluorescently labeled normal and malignant cells, only the prkdc mutant fish reproduced as homozygotes and also survived injury after cell transplantation. Engraftment into optically clear casper, prkdc-mutant zebrafish facilitated dynamic live cell imaging of muscle regeneration, repopulation of muscle stem cells within their endogenous niche, and muscle fiber fusion at single-cell resolution. Serial imaging approaches also uncovered stochasticity in fluorescently labeled leukemia regrowth after competitive cell transplantation into prkdc mutant fish, providing refined models to assess clonal dominance and progression in the zebrafish. Our experiments provide an optimized and facile transplantation model, the casper, prkdc mutant zebrafish, for efficient engraftment and direct visualization of fluorescently labeled normal and malignant cells at single-cell resolution.
Introduction
Allogeneic cell transplantation into mice has advanced our understanding of stem cell self-renewal, regeneration, and cancer. For example, hematopoietic stem cells were purified and then assessed for long-term allogeneic engraftment studies (Spangrude et al., 1988), and muscle satellite cells were identified for possible therapy for regenerative muscle disorders (Cerletti et al., 2008). In the setting of cancer, allogeneic cell transplantation studies have been integral for assessing tumorigenicity (Curtis et al., 2010; Hettmer et al., 2011) and metastatic cancer growth (Mito et al., 2009). The generation of immune-compromised genetic models like DNA-activated catalytic peptide (Prkdc, SCID) knockout mice have now become a standard model for assessing tissue engraftment potential (Ito et al., 2012). Prkdc mutant mice have impaired nonhomologous end joining (NHEJ) DNA repair, preventing V(D)J receptor recombination and, subsequently, the production of mature T and B cells (Bosma et al., 1983; Blunt et al., 1995). When Prkdc loss is complexed with the interleukin receptor 2 gamma chain (Il2rγ)–null mutation, which ablates NK cell function, cellular engraftment is vastly improved (Ito et al., 2002). Similarly, compound Prkdc; Janus kinase 3 (Jak3)–deficient mice lack both adaptive and NK cell immunity (Okada et al., 2008; Sato et al., 2010). Both models are capable of extremely high allogeneic engraftment rates and have been used in xenograft transplantation assays to assess cancer progression, metastasis, and therapy responses in vivo (Ito et al., 2012). Despite the utility of these models for engrafting non–immune-matched tissues, murine models are expensive and require complex imaging modalities for direct in vivo imaging of engrafted tissue, making single-cell visualization difficult (Ellenbroek and van Rheenen, 2014).
Zebrafish are a powerful experimental model for cell transplantation studies. For example, normal and malignant tissues can be engrafted into γ-irradiated (Traver et al., 2004), dexamethasone-treated (Stoletov et al., 2007) or syngeneic adult recipients (Mizgireuv and Revskoy, 2006; Smith et al., 2010). Development of optically clear casper zebrafish has further optimized the direct visualization of fluorescently labeled cells into engrafted animals (White et al., 2008; Feng et al., 2010; Heilmann et al., 2015; Li et al., 2015; Tang et al., 2016). Despite the proven utility of these approaches, chemical and γ-irradiation ablation of the immune system is only temporary, preventing durable long-term engraftment of tissues (Stoletov et al., 2007; Smith et al., 2010). Moreover, transplantation into syngeneic animals is limited to donor cells derived from these same isogenic lines, preventing the wider adoption of these models (Mizgireuv and Revskoy, 2006; Mizgirev and Revskoy, 2010; Smith et al., 2010). To begin to address these limitations, we have recently developed rag2E450fs homozygous mutant zebrafish that can engraft allogeneic tissues from multiple donor strains (Tang et al., 2014, 2016). Though the rag2E450fs model is an important conceptual advance in zebrafish transplant technology, the model is not optimal. For example, homozygous casper, rag2E450fs mutant zebrafish do not breed and the line must be maintained through heterozygous in-crossing (Tang et al., 2016). Because the rag2E450fs mutation is hypomorphic, these fish only lack T cells and have variable B cell defects that differ greatly between fish, likely impacting engraftment potential within individual animals (Tang et al., 2014). Finally, these animals develop gill inflammation and likely autoimmunity, which would be predicted based on the similarity of rag2E450fs truncation allele with human mutations that cause Omen’s syndrome and result in variable immune deficiency, autoimmunity, and inflammation (Santagata et al., 2000; Tang et al., 2014). Therefore, the development of new immune-comprised zebrafish models will be required to advance transplant biology in the zebrafish.
Here, we develop new immune-deficient zebrafish models that are optically clear and have more complete immune deficiencies that affect T, B, and presumptive NK cells. The prkdc and jak3 mutant fish are similar to transplant models currently used in the mouse, yet provide new opportunities to dynamically visualize engraftment at single-cell resolution and answer important questions in muscle regeneration and tumor cell heterogeneity. These new zebrafish lines, especially the prkdc-null, casper zebrafish, will transform our ability for direct, live animal imaging of self-renewal, cell state transitions, regeneration, and the hallmarks of cancer at single-cell resolution in the allogeneic transplantation setting.
Results and discussion
Generation and cellular characterization of immune-compromised zebrafish models
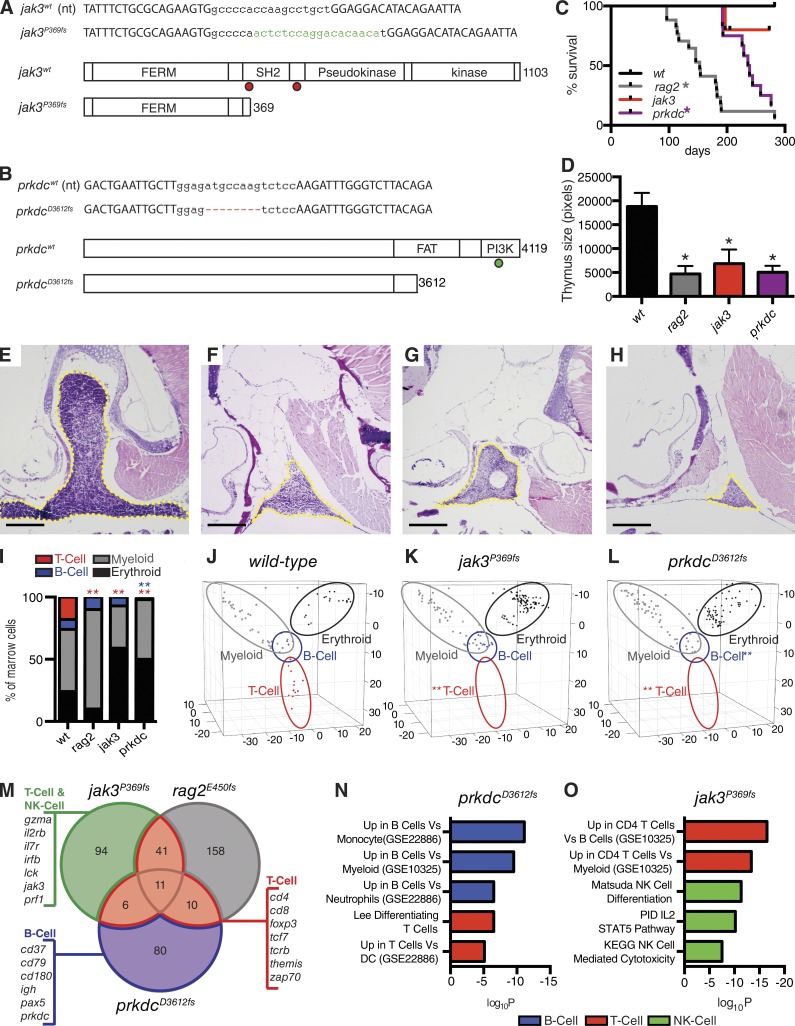
In a concerted effort to expand available immune compromised zebrafish models that exhibit differential immune deficiencies and have elevated engraftment potential, we generated zebrafish with truncating mutations in the jak3 and prkdc genes (Fig. 1, A and B) using transcription activator-like effector nucleases (TALENs; Dahlem et al., 2012; Moore et al., 2012). A jak3 mutant line was identified that harbored a 10-nt deletion and an 18-nt addition, resulting in a frame shift at proline residue 369 and leading to a premature stop codon. The resulting truncated jak3P369fs protein is predicted to lack the SH2 and kinase domains, and is therefore likely functionally null (Fig. 1 A, jak3P369fs mutants). Similar truncation mutations in humans are found at amino acid residues 408 and 565, leading to JAK3 loss-of-function and reductions in T and NK cells (Russell et al., 1995). We also identified prkdc mutant zebrafish that harbor an 8-nt deletion, causing a frame shift at aspartic acid residue 3612 and resulting in the formation of a premature stop codon (Fig. 1 B, prkdcD3612fs mutants). Similar mutations have been reported in SCID mice and lead to the production of a truncated protein that lacks catalytic activity (Blunt et al., 1996). Because PRKDC is required for NHEJ DNA repair, mice with truncation mutations in PRKDC fail to efficiently recombine T and B cell immune receptors, and thus, lack functionally mature T and B cells (Bosma et al., 1983; Blunt et al., 1995). Although we predict that both the jak3P369fs and prkdcD3612fs alleles result in truncated protein products, we cannot exclude the possibility that loss-of-function phenotypes could be caused in part by nonsense-mediated mRNA decay. The jak3P369fs and prkdcD3612fs mutant zebrafish were outcrossed twice into the casper background to minimize the likelihood of carrying off-target mutations and to take advantage of the superior imaging accessibility afforded by this optically clear zebrafish background (White et al., 2008).
Figure 1.
Characterization of rag2E450fs, jak3P369fs, and prkdcD3612fs mutant zebrafish. (A and B) Genetic mutations created for jak3 and prkdc. Capital letters show TALEN-binding sites. Green lowercase letters denote additions and red hyphens represent deletions. Dots indicate truncation mutations found in human (red) and mouse (green) SCID. (C) Kaplan-Meier survival analysis for wild-type and mutant zebrafish lines (rag2E450fs, n = 17; jak3P369fs, n = 12; prkdcD3612fs, n = 12; casper wild-type, n = 12; *, P < 0.0001 survival compared with wild-type survival, Log-rank test). (D) Quantification of thymus size in mutant and wild-type fish (*, P < 0.028, Student’s t test; n ≥ 5 animals per group analyzed). (E–H) Hematoxylin and eosin–stained sections of thymus from 90-d-old wild-type (E), rag2E450fs (F), jak3P369fs (G), and prkdcD3612fs (H) mutant zebrafish; yellow dashed lines denote thymus (n ≥ 5 animals per group analyzed). Bars, 25 µm. (I–L) Quantization of blood cells contained in the marrow as assessed by Fluidigm single-cell quantitative PCR (I) and visualized by principle component analysis (J–L). Significant reductions in T cells are denoted by red asterisks (P = 0.0001) and B cells by blue asterisk (P = 0.001; Fisher’s exact test, n = 72, 138, and 145 cells analyzed in wild-type, jak3P369fs, and prkdcD3612fs mutant fish, respectively; cells were combined from the marrow of two fish in each analysis). Blood cell lineages are denoted by open ovals in J–L. (M) Venn diagram showing overlap of selected down-regulated genes identified after RNA sequencing of the whole kidney marrow of jak3P369fs, prkdcD3612fs, and rag2E450fs mutants (n = 3 fish analyzed/genotype). Gene expression is relative to wild-type marrow (>2-fold change; FDR < 0.05, Benjamini-Hochberg test). (N and O) Molecular pathways down-regulated in prkdcD3612fs (N) and jak3P369fs (O) mutant marrow.GSEAsig FDR ≤ 6.33 × 10−4 for each analysis.
Mutant zebrafish lines were assessed for differences in survival and fecundity. Rag2E450fs, jak3P369fs, and prkdcD3612fs casper heterozygous fish were individually in-crossed. Resulting progeny survived in Mendelian ratios without morphological defects to 3 mo of age (Fig. 1 C). However, after routine tail fin clip genotyping at 3 mo of age, all jak3P369fs homozygous mutants died within just 1–2 d (n = 44 animals), whereas all wild-type, rag2E450fs, and prkdcD3612fs fish survived well after this procedure (wild-type, n = 70; rag2E450fs, n = 82; prkdcD3612fs, n = 71). To increase survival of jak3P369fs homozygous fish, we refined a method for genotyping using DNA obtained from one to three scales isolated from the lateral flank of anesthetized fish. This approach led to less injury and increased survival rates to ∼95% in jak3P369fs homozygous mutant animals after genotyping (n = 50 of 53). All three homozygous mutant lines were assayed for long-term survival after genotyping. The jak3P369fs homozygous mutant fish survived well for up to 1 yr after scale genotyping, whereas rag2E450fs and prkdcD3612fs mutant fish had elevated morbidity after tail fin clip genotyping, peaking at 6–9 mo of age (Fig. 1 C). We observed no qualitative difference in longevity after fin clip or scale genotyping for rag2E450fs and prkdcD3612fs mutants. Immune-deficient mice also have elevated mortality when compared with normal mice, similar to what is reported for our mutant zebrafish. Pairwise matings revealed that only the prkdcD3612fs mutant zebrafish could reproduce and generate viable homozygous progeny (n = 18 of 24 matings). In contrast, homozygous rag2E450fs and jak3P369fs fish in the casper background failed to reproduce (n > 25 matings/line).
To better understand the immune phenotypes of homozygous mutant zebrafish, we next performed a detailed cellular and molecular characterization of blood cell development. Rag2, Jak3, and Prkdc mutant mice and humans have common defects in thymic T cell maturation (Shinkai et al., 1992; Blunt et al., 1995; Nosaka et al., 1995; Kalman et al., 2004). As expected, all three mutant zebrafish lines had significantly smaller thymi, along with a concomitant reduction in T lymphocytes when compared with both wild-type and heterozygous siblings (Fig. 1, D–H). We next assessed mutant zebrafish for deficiencies in blood cell lineages found in the kidney marrow, the site of hematopoiesis in adult zebrafish. Using a recently developed Fluidigm microfluidics system that can accurately quantify blood cell defects based on single cell transcriptional analysis using lineage-specific quantitative PCR primers (Moore et al., 2016), we have previously demonstrated that immune deficiency observed in rag2E450fs mutants are caused by the lack of T lymphocytes with no effect on overall numbers of B cells (Tang et al., 2014). Here, we demonstrate that jak3P369fs and prkdcD3612fs mutants also have a loss of mature T cells in the marrow (Fig. 1, I–L; P < 0.0001). Single-cell transcriptional analysis also revealed that prkdcD3612fs mutant fish lacked mature B cells (Fig. 1, I and L; P < 0.001), whereas B cells were present in both the rag2E450fs and jak3P369fs mutant marrow (Fig. 1, I and K). These data are consistent with mouse and human studies, where PRKDC mutations result in ablation of T and B cells (Bosma et al., 1983; Sipley et al., 1995). Gene expression analysis, hierarchical clustering, and principle component analysis (PCA) independently confirmed blood cell defects in all three mutant lines (Fig. 1, I–L).
JAK3 deficiency results in impaired T and NK cell development in both mouse and human patients (Baird et al., 1998; Notarangelo et al., 2001). JAK3 is a critical downstream regulator of IL2Rγ, is required in both T and NK cells to regulate STAT5 signaling and is required for maturation of these cells (Lin and Leonard, 2000). Single-cell Fluidigm transcriptional analysis revealed that presumptive NK cells comprise <1% of wild-type whole kidney marrow cells, and thus are difficult to quantify using this approach (Moore et al., 2016). We therefore performed RNA sequencing analysis on the whole kidney marrow of 3-mo-old wild-type and homozygous mutant zebrafish to assess loss of well-known T, B, and NK cell marker gene expression. This analysis identified lineage-restricted genes that were specifically lost in each mutant line. For example, T cell marker genes (lck, cd4, cd8, foxp3, tcrb, and zap70), B cell marker genes (pax5, cd37, cd79, and igh), and presumptive markers for NK cells (prf1 homologues, gzma, il2rb, irfb, and jak3) were down-regulated in specific mutants (Fig. 1 M and Table S1). Down-regulated genes found in each mutant line were compared for overlap with gene sets found in the Molecular Signatures Database (MsigDB; Subramanian et al., 2005), providing independent and unbiased assessment of loss of specific blood cell types (Fig. 1, N and O; and Tables S2–S4). This analysis confirmed T cell losses in rag2E450fs mutant fish (Tables S1 and S4) and losses of both T and B cells in prkdcD3612fs mutant fish (Fig. 1 N and Tables S2). In contrast, jak3P369fs mutant fish specifically lacked T and NK cell signatures and had impaired Jak–Stat5 signaling (Fig. 1 O and Table S3). Expression of NK-lysins were not differentially regulated in the marrow of either prkdcD3612fs or jak3P369fs mutant fish, suggesting that NK-like cells identified by our group may be retained in these animals (Tables S2 and S3). In sum, our analysis uncovered a striking loss of expected gene signatures in each mutant, with a majority of the most significantly associated gene sets defining lineage-restricted and functional gene pathways associated with T, B, and NK cell defects (Tables S2–S4). Collectively, our studies show a remarkable similarity in the immune cell defects present in immune-deficient zebrafish and those found in mouse and humans, underscoring the high conservation of gene programs that regulate T, B, and NK cell development and their function throughout vertebrate evolution.
Functional characterization of prkdcD3612fs homozygous mutant zebrafish
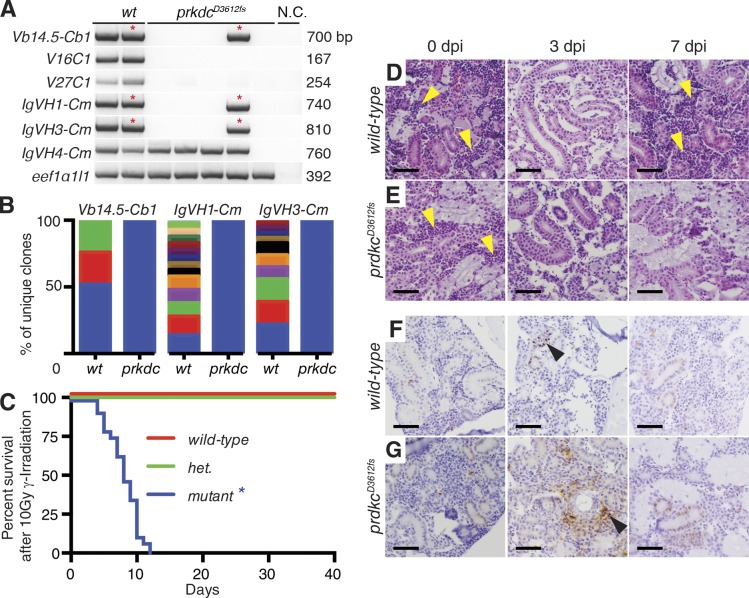
PRKDC is an integral component of the DNA repair complex and is specifically required for NHEJ. Subsequently, Prkdc-null mice have a reduced immune receptor repertoire resulting from failed V(D)J receptor recombination (Blunt et al., 1995). To characterize the impact of the prkdcD3612fs mutation on V(D)J recombination, we assayed t cell receptor β (tcrb) and immunoglobulin heavy chain (igm) rearrangements from blood cells isolated from the whole kidney marrow. prkdcD3612fs mutant zebrafish exhibited near complete absence of tcrb and igm rearrangements when compared with wild-type fish (Fig. 2 A). For those rare instances where recombination events were detected in mutant prkdcD3612fs fish, rearrangement products were sequenced (n = 3 out of 13 animals total; Table S5). Sequencing revealed a striking loss of immune cell repertoire in prkdcD3612fs mutant fish, with each band representing only a single clone. In contrast, a wide-range of recombined receptor rearrangements were detected from amplified products isolated from wild-type fish (Fig. 2 B and Table S5). Similar results have been reported for PRKDC-deficient mice, with these animals having a leaky SCID phenotype resulting in the production of rare B and T cell clones and detectable serum IgG levels (Custer et al., 1985). Collectively, these results confirm the molecular defect accompanying the loss of T and B cells in prkdcD3612fs mutant fish is caused by impaired receptor recombination.
Figure 2.
prkdcD3612fs mutants have defective V(D)J receptor recombination and develop anemia after low-dose γ-irradiation. (A) PCR analysis for tcrb and igm rearrangement of whole kidney marrow cells from wild-type (wt) and prkdcD3612fs zebrafish. N.C., negative control. tcrb transcripts specific to the variable (V) and constant (C) regions are noted with band size to the right. Analysis of eef1a1l1 ensured equal loading and was a positive control. (B) Percentage of different immune receptor recombined clones found in the asterisk-labeled bands in A (n > 19 individual clones sequenced). Each color represents a different clone found in the marrow. (C) Kaplan-Meier survival analysis for wild-type, heterozygous (het.), and prkdcD3612fs mutant zebrafish lines after 10 Gy γ-irradiation (wild-type, n = 29; het, n = 29; prkdcD3612fs, n = 28; *, P < 0.0001 comparing prkdcD3612fs to wild-type survival, Log-rank test). (D and E) Histological section of wild-type (D) and prkdcD3612fs mutant (E) whole kidney marrow at the indicated times after 10 Gy γ-irradiation (n ≥ 6 animals assessed/genotype). dpi, days post irradiation. Yellow arrowheads denote nests of hematopoietic cells. (F and G) Activated caspase3 staining of wild-type (F) and prkdcD3612fs mutant (G). Black arrowheads denote apoptotic blood cells. Bars, 50 µm.
Mice with PRKDC deficiency have impaired DNA repair and are exquisitely sensitive to γ-irradiation. Specifically, mice deficient in PRKDC die after irradiation as a result of failed DNA repair, loss of blood stem and progenitor cells, and eventual anemia (Biedermann et al., 1991). Thus, we assessed if prkdcD3612fs mutant fish were more sensitive to ionizing radiation-induced DNA damage. 3-mo-old wild-type and prkdcD3612fs mutant zebrafish were exposed to a sublethal dose of γ-irradiation (10 Gy; Traver et al., 2004). By 12 d after irradiation, all wild-type fish and heterozygous mutant fish were alive (n = 29 fish/genotype). In contrast, homozygous mutant prkdcD3612fs animals developed progressive pallor and died within 12 d after γ-irradiation (n = 28; P < 0.0001, Log-rank statistic; Fig. 2 C). Histological analysis revealed that both prkdcD3612fs and wild-type fish had reduced whole kidney marrow cellularity by 3 d after irradiation (Fig. 2, D and E). PrkdcD3612fs mutant animals continued to have reduced marrow cellularity past 3 d after irradiation, resulting in elevated apoptosis of hematopoietic cells when assessed by activated Caspase-3 staining. These fish developed fatal anemia by 10 d after irradiation (Fig. 2, E and G). In contrast, wild-type fish recovered marrow cells by 7 d after irradiation (Fig. 2, D and F). We conclude that blood cells from prkdcD3612fs mutant zebrafish are defective in NHEJ, preventing both immune receptor rearrangement and DNA repair after γ-irradiation.
Immune-compromised zebrafish models accept allogeneic transplants
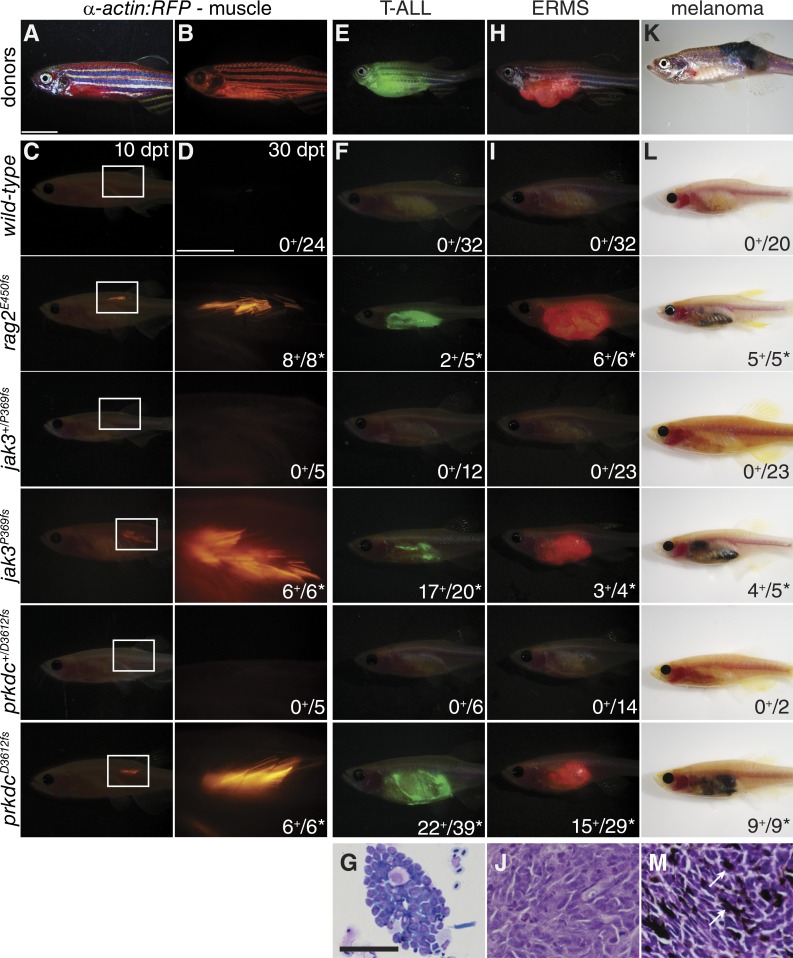
Given that homozygous rag2E450fs, jak3P369fs, and prkdcD3612fs mutant fish have variable immune deficiencies, we next wanted to assess each line for the ability to engraft allogeneic cells isolated from a wide range of tissues and genetic backgrounds. We first assessed engraftment of α-actin-RFP–labeled zebrafish muscle. As expected, neither wild-type nor heterozygous mutant zebrafish engrafted fluorescently labeled muscle cells when implanted into the dorsal musculature (Fig. 3, A–D; and Table S6). In contrast, fluorescent muscle engraftment was robust in all three mutant lines tested, with durable engraftment lasting beyond 30 d post transplantation (dpt; Fig. 3, C and D; and Table S6). Importantly, engraftment into these models did not require matching at the MHC or immune suppression with γ-irradiation or dexamethasone.
Figure 3.
Immune-deficient zebrafish efficiently engraft fluorescently labeled normal and malignant tissues. (A and B) Brightfield and fluorescent images of α-actin-RFP transgenic donor fish, respectively. (C and D) Merged fluorescence and brightfield images of α-actin-RFP muscle transplanted wild-type and mutant fish at 10 dpt (C, genotypes indicated) and a magnified view of the boxed region at 30 dpt (D). Merged (E, F, H, and I) and brightfield (K and L) images of donors and recipient mutant zebrafish with tumors. (E–G) GFP-labeled Myc-induced T-ALL arising in donor CG1-strain fish. (H–J) mCherry-labeled ERMS from donor CG1-strain fish. (K–M) BRAFV600E-induced melanoma arising in p53-deficient nacre-strain donor fish. Tumor engraftment shown at 30 dpt in indicated genotypes. (G, J, and M) Representative tumor histology from prkdcD3612fs animals by cytospin (G) and sectioning (J and M). Arrows show pigmented melanoma cells in M. Number of animals with successful engraftment are shown in the bottom right corner of images. *, P < 0.05, Fisher Exact test comparing engraftment of tumors into mutant versus wild-type fish. Bars: (A–C, E, F, H, K, and L) 1 cm; (D) 0.25 cm; (G, J, and K) 50 µm.
We next wanted to assess rag2E450fs, jak3P369fs, and prkdcD3612fs mutant fish for their utility in engrafting cancer cells. As expected, homozygous mutant fish efficiently engrafted a wide array of fluorescently labeled cancers arising in various backgrounds (Fig. 3, E–M; and Table S6). These included Myc-induced T cell acute lymphoblastic leukemia (T-ALL; Fig. 3, E, F, and G; Langenau et al., 2003; Blackburn et al., 2014), kRASG12D-induced embryonal rhabdomyosarcoma (ERMS; Fig. 3, H, I, and J; Langenau et al., 2007; Ignatius et al., 2012), BRAFV600E, p53-deficient melanoma (Fig. 3, K, L, and M; Patton et al., 2005; Ceol et al., 2011), and neuroblastomas (Fig. S1; Zhu et al., 2012). Importantly, engrafted tumors exhibited similar histology as donor tumors (Fig. 3, G, J, and M; and Fig. S2). In addition, T-ALL and ERMS could be serially transplanted from engrafted recipient fish into new prkdcD3612fs homozygous hosts (Fig. S6). As may have been expected based on our injury studies, homozygous jak3P369fs mutant fish did not tolerate intraperitoneal transplantation of tumor cells, with 51 of 87 homozygous jak3P369f mutant fish dying immediately after the procedure (Fig. S6). In contrast, engraftment of tumor cells into the peritoneum of either rag2E450fs or prkdcD3612fs mutant fish was well tolerated (Fig. S6). Collectively, these results show that all three zebrafish mutant lines can accept allogeneic cells from unrelated donor animals without preconditioning, confirming both immunodeficiency and utility as a cellular transplantation model.
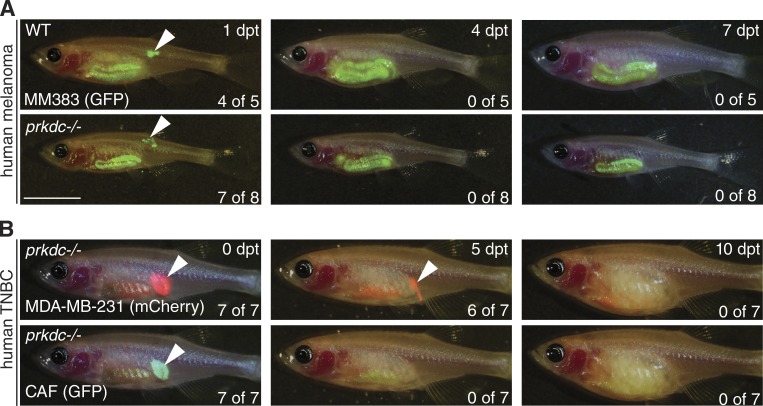
Given the utility of prkdcD3612fs mutant zebrafish for engrafting allogeneic tissues and cancers, we next assessed their ability to accept mouse and human tumor xenografts. Despite optimizing long-term survival of mutant fish at up to 36°C and observing initial implantation of fluorescently labeled tumor cells at the site of injection, human melanoma, triple-negative breast cancer, and pancreatic adenocarcinoma cells all regressed and were rejected by the immune system by 7 dpt (n = 36; Fig. 4 and Fig. S7). Similar results were observed for engraftment of B16 mouse melanoma cells (n = 4; Fig. S7). Engraftment was not enhanced by addition of Matrigel or implantation of tumor cells with cancer-associated fibroblasts. We believe the failure to robustly engraft human and mouse cells likely reflects rejection by innate immune cells retained in prkdcD3612fs mutant fish. For example, we have recently identified a novel population of NK-lysin+ blood cells that likely define an innate NK-like cell that is uniquely found in zebrafish (Moore et al., 2016). These and other innate immune cells are retained in prkdcD3612fs mutant fish and may elicit rejection of mouse and human xenografts. These data are expected based on mouse studies where NK and innate immune cells are retained in PRKDC-null mice (Hackett et al., 1986). We posit that additional compound mutant lines that ablate both the innate and adaptive immune systems will be required to achieve robust and sustained xenograft cell transplantation into adult zebrafish. Moreover, it is also likely that refined cell transplantation approaches and delivery of human growth factors will be required for optimized xenograft growth in vivo.
Figure 4.
Human cancer xenografts are rejected in prkdcD3612fs mutant zebrafish. (A and B) Merged brightfield and epifluorescence images of human tumor cells transplanted into immune-compromised zebrafish. (A) Wild-type (top) and prkdcD3612fs homozygous mutant zebrafish (bottom) transplanted with 6 × 104 human melanoma cells (MM383, GFP-labeled) and imaged at the indicated time points after transplantation. (B) PrkdcD3612fs homozygous mutant zebrafish transplanted with 7.8 × 104 human triple negative breast cancer (TNBC) cell line (MDA-MB-231, mCherry-labeled) with equal number of matched cancer-associated fibroblasts (CAF; GFP-labeled). Fish were imaged at the indicated time points. Arrows denote site of injection for tumor cell transplantation. Number of animals with successful engraftment is shown in the bottom right corner of images. Note autofluorescence in the gastrointestinal tract of recipient zebrafish was caused by diet. Bar, 1 cm.
Imaging engraftment at single-cell resolution in homozygous prkdcD3612fs zebrafish
Allogeneic transplantation studies provide novel functional assays to assess stem cell self-renewal (Spangrude et al., 1988), tumorigenenicity of specific cell types (Curtis et al., 2010; Hettmer et al., 2011), and powerful models for performing in vivo drug efficacy studies for regeneration and cancer (Udina et al., 2004; Wang et al., 2011). Our analysis has uncovered that prkdcD3612fs mutant zebrafish are likely the best engraftment model for allogeneic transplantation of the three immune-deficient zebrafish lines presented here. This comes from analysis of high fecundity as homozygous mutants, ability to survive after injury and transplantation, and lack of both T and B cells. Despite the well-established allogeneic transplantation models in mice that are commonly used to assess important questions in regenerative medicine and cancer, it has been difficult to image engraftment of cells at single-cell resolution. Building on the optical transparency of the prkdcD3612fs (casper) mutants, we next wanted to assess the model for the ability to visualize fluorescent cell engraftment at single-cell resolution in live animals, answering important and fundamental questions in muscle regeneration and in defining roles for clonal competition in regulating progression in leukemia.
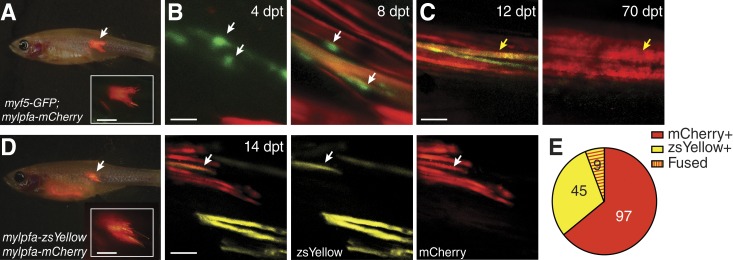
Initially, we visualized muscle cell heterogeneity, repopulation of individual satellite cells into their endogenous niche, and timing of regeneration after engraftment. Muscle cells were isolated from 60-d-old myf5-GFP, mylpfa-mCherry double-transgenic fish and engrafted into the dorsal musculature of prkdcD3612fs mutant zebrafish (Fig. 5 A). Serial imaging of engrafted fish revealed two prominent modes of fiber regeneration. For example, 86.7% of regenerating fibers expressed only the mylpfa-mCherry transgene between 4 and 12 dpt, suggesting that differentiated myosin-expressing myoblasts resident in donor cell preparation drove regeneration (n = 189 of 218 fibers; n = 14 animals analyzed). In some instances, single myf5-GFP+ progenitor cells engrafted into recipient fish, taking up residence adjacent injured muscle, and then driving regeneration of fibers (Fig. 5 B). When combined with our observation that ∼14% of early forming regenerative fibers expressed both mCherry and GFP (Fig. 5 C, left), we conclude that myf5-GFP+ progenitor cells contribute to a small fraction of regenerative fibers, with persistence of GFP being seen in early differentiated fibers but lost with time (Fig. 5 C, right). These same fully regenerated fibers also largely lacked myf5-GFP+ progenitor cells adjacent fibers, suggesting that most donor cells differentiate into fusion-competent myoblasts and contribute to fiber regeneration without replenishing the satellite cells. These data are consistent with reports that satellite and muscle precursor cells become active after isolation from muscle and have limited regenerative capacity after engraftment into recipient mice (Montarras et al., 2005). Our data provide evidence that muscle regeneration after injury follows in a highly coordinated and hierarchically organized differentiation cascade, with dormant progenitor cells from the donor population becoming reactivated, situating themselves near the damaged tissue of target, and reinitiating muscle developmental programs to reconstruct nascent muscle fibers. The ability to visualize this process in an organism known for its facile large-scale in vivo drug screening will likely provide unprecedented opportunities to discover therapeutics that restore and enhance muscle regeneration, with implications in clinical intervention of muscular dystrophy and exercise-related muscle injuries.
Figure 5.
Visualizing fluorescent muscle cell regeneration at single-cell resolution in engrafted prkdcD3612fs mutant zebrafish. (A) Brightfield and fluorescent merged image of a prkdcD3612fs mutant fish engrafted with myf5-GFP; mylpfa-mCherry double transgenic skeletal muscle cells at 8 dpt. (B) Confocal images of the same engrafted animal at 4 and 8 dpt. myf5-GFP+ muscle progenitor cells (white arrows) appear by 4 dpt and are positioned adjacent to a double-transgenic GFP+ and mCherry+ fiber by 8 dpt. (C) Merged confocal images of a myf5-GFP; mylpfa-mCherry muscle fibers at 12 dpt (left); the fibers eventually expressed only mCherry by 70 dpt (yellow arrow). (D and E) prkdcD3612fs engrafted zebrafish with muscle cells isolated from mylpfa-zsYellow and mylpfa-mCherry transgenic donors. Whole animal image (left) with site of engraftment noted by a white arrow and high magnification confocal images (right, merged and single fluorescent confocal stacks are shown). Double fluorescently labeled muscle fiber denoted by white arrow. (E) Quantification of single and double fluorescently positive fibers in all animals analyzed (n = 13). Bars: (A and D, inset) 2 mm; (B) 50 µm; (C and D, rightmost panel) 200 µm.
We next wanted to dynamically visualize the relative contribution of engrafted precursors and myoblasts to fiber fusion and creation. Here, we performed competitive engraftment studies using muscle cells isolated from juvenile (60-d-old) mylpfa-mCherry and mylpfa-zsYellow transgenic zebrafish donors and injected equal numbers of muscle cells of each color into the dorsal musculature of prkdcD3612fs mutant zebrafish (104 cells per color/fish; Fig. 5 D). After engraftment for 8–14 d, we observed that most reconstituted fibers expressed a single fluorescent protein, with myoblast fusion contributing to regeneration in only 9 of 151 fibers analyzed (n = 14 animals; Fig. 5 E). These data suggest a model of muscle repair whereby single myoblasts fuse with existing injured fibers to restore muscle integrity. This model is supported by studies in mice and flies, but has not been proven at single-cell resolution in live animals over time (Chargé and Rudnicki, 2004; Abmayr and Pavlath, 2012). Alternatively, it is also possible that single muscle stem cells reinitiate muscle fiber formation from first expanding a myoblast population locally, with these cells ultimately dominating the regeneration process and creating new muscle fibers in the zebrafish, akin to what has been recently described for embryonic muscle regeneration in zebrafish (Gurevich et al., 2016). In total, this work has provided new insights and novel experimental models for directly imaging regeneration in live animals.
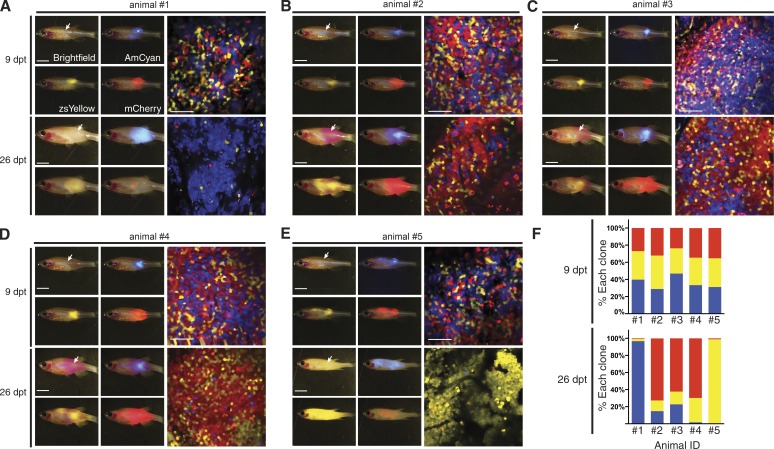
It is well known that human T-ALLs are oligoclonal at diagnosis; however, progression and relapse are often driven by emergence of an underrepresented clone contained within the primary leukemia (Mullighan et al., 2008). The role that stochasticity has in governing emergence of clonal dominance has gone unstudied, largely because this process has been difficult to study experimentally because of the lack of robust models and the inability to serially assess animals over time. To visualize this process in live fish, we mixed equal numbers of AmCyan, zsYellow, and mCherry-labeled T-ALL with similar growth kinetics and leukemia-propagating cell frequencies when grown individually in syngeneic fish (Blackburn et al., 2014). These cell mixtures were injected into the dorsal musculature of optically clear recipient prkdcD3612fs mutants (3.3 × 104 of each clone, 1 × 105 total cells per recipient animal). Engrafted fish were imaged by confocal microscopy at 9 and 26 dpt. At 9 dpt, the three clones were present in roughly equal numbers (Fig. 6, A–F, top; total of five fish analyzed); however, by 26 dpt, leukemias had become dominated by a single fluorescent clone (Fig. 6, A–F, bottom). Remarkably, the same clone did not dominate in individual engrafted fish (Fig. 6, A–F, bottom), supporting a model that clonal dominance can emerge as a consequence of neutral stochastic drift. Similar clonal drift and stochastic emergence of dominant clones has been documented in intestinal crypt stem cells (Snippert et al., 2010) and mouse intestinal adenomas (Schepers et al., 2012). Our results show that clonal dominance can evolve stochastically in rapidly growing leukemias that have similar growth rates and self-renewal/stem cell activity, suggesting differential activity of individual leukemia-propagating cells in driving growth over time. Collectively, our work provides unprecedented access to dynamically visualize clonal dominance, and cancer progression at single-cell resolution in engrafted prkdcD3612fs (casper) mutant zebrafish, which will be broadly useful for assessing clonal evolution and progression in a wide array of nonmalignant tissues and cancers.
Figure 6.
Imaging the emergence of clonal dominance in Myc-induced T-ALL at single-cell resolution in engrafted prkdcD3612fs mutant zebrafish. (A–E) Equal numbers of fluorescently labeled T-ALL cells were engrafted into the dorsal musculature of prkdcD3612fs (casper) mutant fish (1 × 105 cells/clone). Images were taken at 9 dpt (top) and 26 dpt (bottom). Whole animal imaging showing brightfield and epifluorescence in three color channels (left, site of injection indicated by white arrows) and confocal images for engraftment at site of injection (right). (F) Quantification of relative T-ALL clone proportions found within individual animals at the site of injection. Bars: (epifluorescence images) 5 mm; (confocal images) 50 µm.
Our work has detailed the remarkable genetic and functional conservation of T, B, and NK cell differentiation programs between zebrafish, mice, and humans. Moreover, our work has provided a novel, optimized, and facile allogeneic transplantation model, the casper, prkdcD3612fs mutant fish, for efficient engraftment and direct visualization of fluorescently labeled normal and malignant cells at single-cell resolution. This model will be invaluable for the dynamic live-cell imaging of regeneration and the hallmarks of cancer at single-cell resolution, including tumor heterogeneity, metastasis, neovascularization, and clonal evolution.
Materials and methods
Creation of jak3P369fs and prkdcD3612fs homozygous mutant zebrafish
All zebrafish experiments were completed with IACUC approval from the Massachusetts General Hospital (2011N000127). Jak3P369fs mutant zebrafish were made using TALENs and previously described injection strategies (Moore et al., 2012). The prkdcD3612fs mutant was made using TALENs made by the University of UTAH Health and Sciences Genetic Mutation and Detection Core as previously described (Dahlem et al., 2012). Specifically, RNA was prepared for each TALEN arm and microinjected into AB or casper zebrafish. F0-injected animals were raised to adulthood, and single male-by-female matings performed. 12 resultant progeny from each cross were arrayed into 96-well plates and genomic DNA was extracted. PCR amplification of the target sites was performed, and amplicons were sent for Sanger sequencing. From this analysis, one mutant line for each gene was identified. F1 progeny were subsequently raised to adulthood, and heterozygous fish were identified by genotyping. Heterozygous mutant AB fish were bred to casper zebrafish (roy−/−; mitfa−/−; White et al., 2008), resultant progeny were genotyped, and heterozygous mutants were identified and in-crossed to produce triple homozygous animals.
prkdcD3612fs genotyping
The prkdcD3612fs allele introduces a de novo HinfI site, allowing for restriction enzyme–mediated identification of the mutant allele. Specifically, adult 2–4-mo-old fish were tail fin clipped, and genomic DNA was prepared using the modified HotSHOT method by placing the clipped fin in 50 µl of 50 mM NaOH, boiling at 95°C for 20 min, and neutralizing with 2.5 µl 1 M Tris buffer (pH 8.0; Meeker et al., 2007). 2.5 µl of genomic DNA were used in a standard 25-µl volume PCR using Taq DNA polymerase (New England BioLabs) with forward and reverse primers (Table S8) to produce a 207-bp PCR amplicon. The PCR cycle parameters were (i) denaturation, 94°C for 30 s; (ii) annealing, 53°C for 30 s; and (iii) elongation, 68°C for 25 s; these steps were repeated for 35 cycles. For enzymatic digestion, 15 µl of nonpurified PCR reaction were combined with 0.3 µl HinfI (New England Biolabs)+ 2 µl 10× NEB buffer CutSmart (New England Biolabs) plus 2.7 µl water. The 20-µl reaction was incubated at 37°C for 1–2 h. Enzymatic digestion was visualized by electrophoresis on a 2.5% Tris-acetate-EDTA (TAE) agarose gel containing ethidium bromide or Qiaxcel Screening cartridge (QIAGEN). After HinfI digestion, wild-type fish produced two bands at 183 and 24 bp. Heterozygous rag2E450fs fish produced four bands at 183, 107, 68, and 24 bp as a result of digestion of the mutant allele. Homozygous mutant fish produced bands at 107, 68, and 24 bp (Fig. S6).
jak3P369fs genotyping
The jak3P369fs allele removes a natural EcoNI site found within the wild-type site, allowing for restriction enzyme–mediated identification of the mutant allele. Specifically, anesthetized 2–4-mo-old fish had 1–3 scales removed using fine forceps. Scales were placed in 25 µl 50 mM NaOH solution, boiled at 95°C for 20 min, and neutralized with 2.5 µl 1 M Tris buffer (pH 8.0; Meeker et al., 2007). 2.5 µl of genomic DNA were used in a standard 25-µl vol PCR using Taq DNA polymerase (New England BioLabs) with forward primer and reverse primer (Table S8). The PCR cycle parameters were (i) denaturation, 94°C for 30 s; (ii) annealing, 68°C (−0.4°C/Cycle) for 30 s; and (iii) elongation, 68°C for 45 s, repeated for 35 cycles. For enzymatic digestion, 15 µl of nonpurified PCR reaction were combined with 0.3 µl EcoNI (New England Biolabs) plus 2 µl 10× NEB buffer CutSmart (New England Biolabs) plus 2.7 µl water. The 20-µl reaction was incubated at 37°C for ≥2 h. Enzymatic digestion was visualized by electrophoresis on a 2.5% TAE agarose gel containing ethidium bromide or Qiaxcel Screening cartridge (QIAGEN). After EcoNI digestion, wild-type fish produced two bands at 111 and 110 bp. Heterozygous jak3P369fs fish produced three bands at 229, 111, and 110 bp as a result of digestion of the mutant allele. Homozygous mutant fish are undigested and produce one band at 229 bp (Fig. S3).
Thymus histology and size quantification
Thymus histology was performed as previously described (Tang et al., 2014). In brief, 3-mo-old fish were sacrificed and fixed in 4% paraformaldehyde, embedded in paraffin, and step sectioned. Slides were stained with hematoxylin and eosin by the Specialized Histopathology Services at Massachusetts General Hospital. Thymus sections were imaged at 200× magnification using an Olympus BX41 compound microscope. Images were imported into Fiji (ImageJ; National Institutes of Health) and the thymus was traced using the Freehand Selection tool and area quantified. The mean area of thymi (n ≥ 5) where graphed and SEM indicated. Significance was calculated by Student’s t test.
Receptor rearrangements
RNA was extracted from whole kidney marrow and made into cDNA using SuperScript III Reverse transcription (Invitrogen). tcrb and igm targets were amplified using nested PCR primers (Table S7) as previously described (Tang et al., 2014). The first PCR used 2 ng cDNA and was performed using a standard 25-µl reaction with Choice TaqBlue Mastermix (Denville Scientific). PCR cycle parameters were (i) denaturation, 94°C for 45 s; (ii) annealing, 56°C for 30 s; and (iii) elongation, 72°C for 1 min; these steps were repeated for 35 cycles. In the nested PCR, 1 µl of the first PCR was combined in a 25 µl reaction using the same polymerase and the inner set of primers. The cycling parameters are identical to the first reaction, except repeated for 25 cycles. The final products of this nested PCR reaction are visualized on a 2% TAE agarose gel containing ethidium bromide. Gel bands were excised and DNA isolated using Zymoclean Gel DNA recovery kit (Zymo Research). DNA was cloned into the pGEM-T vector (Promega) using the manufactures protocol, transformed into DH5α cells, selected on Carbenicillin, and then sequenced.
prkdcD3612fs irradiation
3-mo-old fish were irradiated with10-Gy γ-irradiation (Cs137) in 60-mm dishes, sacrificed at the indicated time points, fixed in 4% paraformaldehyde, embedded, sectioned and HE stained as above. Adjacent slides were assessed by immunohistochemistry for cleaved Caspase-3 (Cell Signaling Technology). Kidney sections where imaged at 400× on an Olympus BX41 compound microscope.
Whole kidney marrow gene expression as assessed by RNA sequencing
Wild-type, rag2E450fs, jak3P369fs, and prkdcD3612fs kidneys were dissected and placed into 350 µl of QIAGEN RLT buffer (supplemented with 1% 2-Mercaptoethanol; Bio-Rad Laboratories). Whole RNA was isolated using the RNAeasy Mini kit (QIAGEN) following the manufacturer’s recommended protocol, including on-column DNA digestion with RNase-free DNase (QIAGEN). Total RNA isolated from three animals for each genotype was subjected to rRNA depletion using RiboZero kit (Illumina), followed by NGS library construction using NEBNext Ultra Directional RNA Library Prep kit for Illumina (New England Biolabs) using 15 cycles of PCR amplification. Experiments were performed in triplicate for wild-type, rag2, prkdc, and jak3 mutants. Sequencing was performed on an Illumina HiSeq 2500 instrument, and reads were mapped to the Danio rerio reference genome (Zv9 build) using STAR (Dobin et al., 2013), resulting in a mean of 18 million aligned pairs of 50-bp reads per sample. Read counts over transcripts were calculated using HTSeq v.0.6.0 (Anders et al., 2015) based on the most current Ensembl annotation file for Zv9. Differential expression analysis was performed using EdgeR package (version 3.8.6; Robinson et al., 2010) based on the criteria of >2-fold change in expression value and false discovery rates (FDR; Benjamini-Hochberg Test) <0.05.
Whole kidney marrow single-cell isolation and quantitative PCR using the Fluidigm BioMark HD platform
Cell extraction, sorting, gene expression reactions, quantification, and analysis were performed as previously described (Moore et al., 2016). Specifically, whole kidney marrow was isolated from 2.5–3-mo-old jak3P369fs and prkdcD3612fs homozygous mutants, placed in 5% FBS/1xPBS, triturated by pipet, and filtered through 35-mm cell strainer. Single-cells were sorted into 96-well plates using FACSAria Fusion Cell Sorter (BD), with each well containing 5 µl of lysis buffer. Plates were heated at 65°C for 90 s to lyse cell membranes. cDNA was made by the addition of Fluidigm Reverse Transcription Mix (Fluidigm). Next, samples were preamplified using gene target specific nested outer primers (166 nM; primers used and genes analyzed in Moore et al., 2016) and 10x PreAmp Master Mix (Fluidigm). Unincorporated primers were removed by exonuclease treatment (New England Biolabs). Samples were then diluted by the addition of 36 µl of 1xTE. Each well now contained 50 µl of preamplified cDNA that was ready for qPCR using the Fluidigm BioMark HD. Samples were prepared for qPCR by combining 3 µl of preamplified cDNA sample with 3.5 µl 2× SSo Fast EvaGreen Supermix with low ROX (Bio-Rad Laboratories), 0.35 µl 20× DNA Binding Dye Sample Loading Reagent (Fluidigm), and 0.15 µl water. After priming, the 96.96 Dynamic Array Chip for Gene Expression (Fluidigm), 5 µl of 96 separate inner primer pairs (5 µM) and 5-µl samples were loaded onto the Dynamic Array Chip and mixed in the Fluidigm IFC controller HX. Dynamic Array Chip was then loaded into the Fluidigm BioMark HD following the manufacturer’s protocol. Thermal cycling was completed using protocol GE 96 × 96 PCR melt v2.pcl.
Statistical analysis and display of single-cell data
Ct values were recovered from the BioMark HD. The quality threshold was set to 0.65, and a linear derivative as baseline correction was used. Using SINGuLAR, a limit of detection was set to the Ct of 28; expression was calculated using default settings. Sample wells that failed to express housekeeping genes (eef1a1l1 and actb1) or other lineage-specific markers were deemed to lack a cell and were eliminated from further analysis. Unsupervised hierarchical clustering was performed using SINGuLAR software from Fluidigm. jak3P369fs and prkdcD3612fs single-cell samples were combined and compared with the datasets generated previously (Moore et al., 2016) and lineage assignments were made following hierarchical clustering. The PCA hierarchical clustering, three-dimensional PCA, was also generated using the SINGuLAR R package, with limit of detection set to the Ct of 28.
Muscle and tumor cell transplantation in zebrafish
Muscle and tumor cell isolation and transplantation were performed as previously described (Tang et al., 2014; Tenente et al., 2014). In short, tissues were isolated from donor fish following Tricaine (Western Chemical) overdose. Tumors were excised from dissected fish and placed into 500 µl of 0.9x PBS + 5% FBS on a 10-cm Petri dish. Single-cell suspensions were obtained by maceration with a razor blade, followed by manual pipetting to disassociate cell clumps. Cells were filtered through a 40-µm filter (Falcon), centrifuged at 1,000 g for 10 min, and resuspended to the desired volume. For zebrafish tumor cell transplantation, 5-µl suspension containing 105–106 tumor cells were injected into the peritoneal cavity of each recipient fish using a 26s Hamilton 80366 syringe. For high-resolution confocal imaging of muscle and T-ALL engraftment, 1 µl cell suspension containing desired amount of donor cells were injected intramuscularly into the anesthetized recipient fish using a 33-gauge needle (Hamilton). Cellular engraftment was assessed at 10, 20, and 30 dpt by epifluorescence microscopy (Fig. 3). Recipient fish were sacrificed when moribund or at ≥30 dpt. If animals failed to engraft disease, they were then subjected to either (i) fixation in 4% paraformaldehyde for sectioning or (ii) isolation of peripheral blood for cytospins and histological examination.
Confocal imaging of multicolored cellular engraftment in recipient zebrafish
Imaging was performed using an inverted LSM 710 confocal microscope (Zeiss). Recipient prkdcD3612fs and jak3P369fs mutant zebrafish were anesthetized using 168 mg/l Tricaine (Western Chemical) before being placed onto a 12-mm glass-bottom dish (Thermo Fisher Scientific) with a small amount of the Tricaine solution to allow slow breathing. Images were taken under the acquisition mode using color combinations including AmCyan (excitation, 458 nm; emission, 472–508 nm), zsYellow (excitation, 514 nm; emission, 521–547 nm), and mCherry (excitation, 561 nm; emission, 602–623 nm). The Best Signal option in the Smart Setup was used to achieve strict separation of different color channels. Laser intensity percentage was set with respect to the intensity of each fluorescent marker used, between 10 and 18%. For imaging of muscle fiber regrowth, 100× magnification was achieved with a 10× objective (NA, 0.45; coverglass thickness, 0.17 mm; working distance, 2.0 mm), and 200× magnification with a 20× objective (NA, 0.8; coverglass thickness, 0.17 mm; working distance, 0.55 mm). For T-ALL competition, 400× magnification was achieved with a 40× water emersion objective (NA, 1.3; coverglass thickness, 0.14–0.19 mm; working distance, 0.62 mm for 0.17 mm coverglass). The proportion of T-ALL clones was quantified in the Fiji (ImageJ) software by measuring the area covered by each fluorescent color, which accurately estimate fluorescence cell number (Tang et al., 2016).
Online supplemental material
Fig. S1 shows immune-deficient zebrafish engraft fluorescently labeled neuroblastoma. Fig. S2 shows histology of tumors engrafted in immune-deficient zebrafish. Fig. S3 shows genotyping strategies for jak3P369fs and prkdcD3612fs mutant zebrafish. Tables S1–S8 are available as Excel files. Table S1 lists genes down-regulated in mutant whole kidney marrow compared with wild-type by RNA-Seq. Table S2 lists gene expression signatures that are deregulated in the marrow of prkdcD3612fs mutant zebrafish. Table S3 lists gene expression signatures that are deregulated in the marrow of jak3P369fs mutant zebrafish. Table S4 lists gene expression signatures that are deregulated in the marrow of rag2E450fs mutant zebrafish. Table S5 lists sequence analysis for Igh-V3 V(D)J recombined clones in wild-type and prkdcD3612fs mutant zebrafish. Table S6 lists results of allortransplantation into rag2E450fs, jakP369fs and prkdcD3612fs mutant zebrafishTable S7 lists results of xenotransplantation into prkdcD3612fs mutant zebrafish. Table S8 lists PCR primers.
Supplementary Material
Acknowledgments
We thank Drs. Daniel Haber and Matteo Ligorio for helpful advice and cell lines.
This work was supported by Alex’s Lemonade Stand Foundation (D.M. Langenau), the Live Like Bella Foundation for Childhood Cancer (D.M. Langenau), American Cancer Society (D.M. Langenau), the Massachusetts General Hospital (MGH) Howard Goodman Fellowship (D.M. Langenau), and National Institutes of Health grants R24OD016761, R01CA154923, and U54CA168512. Q. Tang is funded by the China Scholarship Council. We thank the MGH Next Generation Sequencing Core, MGH Specialized Histopathology Services, and the Dana-Farber/Harvard Cancer Center (P30 CA06516), MGH Cancer Center/Molecular Pathology Confocal Core, the MGH Pathology Flow and Image Cytometry Research Core which obtained support from the NIH Shared Instrumentation program with 1S10OD012027-01A1, 1S10OD016372-01, S10RR020936-01, and 1S10RR023440-01A1, and the University of Utah Health and Sciences Genetic Mutation and Detection Core.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- dpt
- days post-transplantation
- ERMS
- embryonal rhabdomyosarcoma
- NHEJ
- nonhomologous end joining
- PCA
- principal component analysis
- TAE
- Tris-acetate-EDTA
- TALEN
- transcription activator-like effector nuclease
- T-ALL
- T cell acute lymphoblastic leukemia
References
- Abmayr S.M., and Pavlath G.K.. 2012. Myoblast fusion: lessons from flies and mice. Development. 139:641–656. 10.1242/dev.068353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., and Huber W.. 2015. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 31:166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird A.M., Thomis D.C., and Berg L.J.. 1998. T cell development and activation in Jak3-deficient mice. J. Leukoc. Biol. 63:669–677. [DOI] [PubMed] [Google Scholar]
- Biedermann K.A., Sun J.R., Giaccia A.J., Tosto L.M., and Brown J.M.. 1991. scid mutation in mice confers hypersensitivity to ionizing radiation and a deficiency in DNA double-strand break repair. Proc. Natl. Acad. Sci. USA. 88:1394–1397. 10.1073/pnas.88.4.1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn J.S., Liu S., Wilder J.L., Dobrinski K.P., Lobbardi R., Moore F.E., Martinez S.A., Chen E.Y., Lee C., and Langenau D.M.. 2014. Clonal evolution enhances leukemia-propagating cell frequency in T cell acute lymphoblastic leukemia through Akt/mTORC1 pathway activation. Cancer Cell. 25:366–378. 10.1016/j.ccr.2014.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blunt T., Finnie N.J., Taccioli G.E., Smith G.C., Demengeot J., Gottlieb T.M., Mizuta R., Varghese A.J., Alt F.W., Jeggo P.A., and Jackson S.P.. 1995. Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell. 80:813–823. 10.1016/0092-8674(95)90360-7 [DOI] [PubMed] [Google Scholar]
- Blunt T., Gell D., Fox M., Taccioli G.E., Lehmann A.R., Jackson S.P., and Jeggo P.A.. 1996. Identification of a nonsense mutation in the carboxyl-terminal region of DNA-dependent protein kinase catalytic subunit in the scid mouse. Proc. Natl. Acad. Sci. USA. 93:10285–10290. 10.1073/pnas.93.19.10285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosma G.C., Custer R.P., and Bosma M.J.. 1983. A severe combined immunodeficiency mutation in the mouse. Nature. 301:527–530. 10.1038/301527a0 [DOI] [PubMed] [Google Scholar]
- Ceol C.J., Houvras Y., Jane-Valbuena J., Bilodeau S., Orlando D.A., Battisti V., Fritsch L., Lin W.M., Hollmann T.J., Ferré F., et al. 2011. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 471:513–517. 10.1038/nature09806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerletti M., Jurga S., Witczak C.A., Hirshman M.F., Shadrach J.L., Goodyear L.J., and Wagers A.J.. 2008. Highly efficient, functional engraftment of skeletal muscle stem cells in dystrophic muscles. Cell. 134:37–47. 10.1016/j.cell.2008.05.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chargé S.B., and Rudnicki M.A.. 2004. Cellular and molecular regulation of muscle regeneration. Physiol. Rev. 84:209–238. 10.1152/physrev.00019.2003 [DOI] [PubMed] [Google Scholar]
- Curtis S.J., Sinkevicius K.W., Li D., Lau A.N., Roach R.R., Zamponi R., Woolfenden A.E., Kirsch D.G., Wong K.K., and Kim C.F.. 2010. Primary tumor genotype is an important determinant in identification of lung cancer propagating cells. Cell Stem Cell. 7:127–133. 10.1016/j.stem.2010.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custer R.P., Bosma G.C., and Bosma M.J.. 1985. Severe combined immunodeficiency (SCID) in the mouse. Pathology, reconstitution, neoplasms. Am. J. Pathol. 120:464–477. [PMC free article] [PubMed] [Google Scholar]
- Dahlem T.J., Hoshijima K., Jurynec M.J., Gunther D., Starker C.G., Locke A.S., Weis A.M., Voytas D.F., and Grunwald D.J.. 2012. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet. 8:e1002861 10.1371/journal.pgen.1002861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenbroek S.I., and van Rheenen J.. 2014. Imaging hallmarks of cancer in living mice. Nat. Rev. Cancer. 14:406–418. 10.1038/nrc3742 [DOI] [PubMed] [Google Scholar]
- Feng H., Stachura D.L., White R.M., Gutierrez A., Zhang L., Sanda T., Jette C.A., Testa J.R., Neuberg D.S., Langenau D.M., et al. 2010. T-lymphoblastic lymphoma cells express high levels of BCL2, S1P1, and ICAM1, leading to a blockade of tumor cell intravasation. Cancer Cell. 18:353–366. 10.1016/j.ccr.2010.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich D.B., Nguyen P.D., Siegel A.L., Ehrlich O.V., Sonntag C., Phan J.M., Berger S., Ratnayake D., Hersey L., Berger J., et al. 2016. Asymmetric division of clonal muscle stem cells coordinates muscle regeneration in vivo. Science. 353:aad9969 10.1126/science.aad9969 [DOI] [PubMed] [Google Scholar]
- Hackett J. Jr., Bosma G.C., Bosma M.J., Bennett M., and Kumar V.. 1986. Transplantable progenitors of natural killer cells are distinct from those of T and B lymphocytes. Proc. Natl. Acad. Sci. USA. 83:3427–3431. 10.1073/pnas.83.10.3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann S., Ratnakumar K., Langdon E.M., Kansler E.R., Kim I.S., Campbell N.R., Perry E.B., McMahon A.J., Kaufman C.K., van Rooijen E., et al. 2015. A quantitative system for studying metastasis using transparent zebrafish. Cancer Res. 75:4272–4282. 10.1158/0008-5472.CAN-14-3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettmer S., Liu J., Miller C.M., Lindsay M.C., Sparks C.A., Guertin D.A., Bronson R.T., Langenau D.M., and Wagers A.J.. 2011. Sarcomas induced in discrete subsets of prospectively isolated skeletal muscle cells. Proc. Natl. Acad. Sci. USA. 108:20002–20007. 10.1073/pnas.1111733108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatius M.S., Chen E., Elpek N.M., Fuller A.Z., Tenente I.M., Clagg R., Liu S., Blackburn J.S., Linardic C.M., Rosenberg A.E., et al. 2012. In vivo imaging of tumor-propagating cells, regional tumor heterogeneity, and dynamic cell movements in embryonal rhabdomyosarcoma. Cancer Cell. 21:680–693. 10.1016/j.ccr.2012.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M., Hiramatsu H., Kobayashi K., Suzue K., Kawahata M., Hioki K., Ueyama Y., Koyanagi Y., Sugamura K., Tsuji K., et al. 2002. NOD/SCID/gamma(c)(null) mouse: an excellent recipient mouse model for engraftment of human cells. Blood. 100:3175–3182. 10.1182/blood-2001-12-0207 [DOI] [PubMed] [Google Scholar]
- Ito R., Takahashi T., Katano I., and Ito M.. 2012. Current advances in humanized mouse models. Cell. Mol. Immunol. 9:208–214. 10.1038/cmi.2012.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalman L., Lindegren M.L., Kobrynski L., Vogt R., Hannon H., Howard J.T., and Buckley R.. 2004. Mutations in genes required for T-cell development: IL7R, CD45, IL2RG, JAK3, RAG1, RAG2, ARTEMIS, and ADA and severe combined immunodeficiency: HuGE review. Genet. Med. 6:16–26. 10.1097/01.GIM.0000105752.80592.A3 [DOI] [PubMed] [Google Scholar]
- Langenau D.M., Traver D., Ferrando A.A., Kutok J.L., Aster J.C., Kanki J.P., Lin S., Prochownik E., Trede N.S., Zon L.I., and Look A.T.. 2003. Myc-induced T cell leukemia in transgenic zebrafish. Science. 299:887–890. 10.1126/science.1080280 [DOI] [PubMed] [Google Scholar]
- Langenau D.M., Keefe M.D., Storer N.Y., Guyon J.R., Kutok J.L., Le X., Goessling W., Neuberg D.S., Kunkel L.M., and Zon L.I.. 2007. Effects of RAS on the genesis of embryonal rhabdomyosarcoma. Genes Dev. 21:1382–1395. 10.1101/gad.1545007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Lahvic J.L., Binder V., Pugach E.K., Riley E.B., Tamplin O.J., Panigrahy D., Bowman T.V., Barrett F.G., Heffner G.C., et al. 2015. Epoxyeicosatrienoic acids enhance embryonic haematopoiesis and adult marrow engraftment. Nature. 523:468–471. 10.1038/nature14569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.X., and Leonard W.J.. 2000. The role of Stat5a and Stat5b in signaling by IL-2 family cytokines. Oncogene. 19:2566–2576. 10.1038/sj.onc.1203523 [DOI] [PubMed] [Google Scholar]
- Meeker N.D., Hutchinson S.A., Ho L., and Trede N.S.. 2007. Method for isolation of PCR-ready genomic DNA from zebrafish tissues. Biotechniques. 43:610–614. 10.2144/000112619 [DOI] [PubMed] [Google Scholar]
- Mito J.K., Riedel R.F., Dodd L., Lahat G., Lazar A.J., Dodd R.D., Stangenberg L., Eward W.C., Hornicek F.J., Yoon S.S., et al. 2009. Cross species genomic analysis identifies a mouse model as undifferentiated pleomorphic sarcoma/malignant fibrous histiocytoma. PLoS One. 4:e8075 10.1371/journal.pone.0008075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizgireuv I.V., and Revskoy S.Y.. 2006. Transplantable tumor lines generated in clonal zebrafish. Cancer Res. 66:3120–3125. 10.1158/0008-5472.CAN-05-3800 [DOI] [PubMed] [Google Scholar]
- Mizgirev I.V., and Revskoy S.. 2010. A new zebrafish model for experimental leukemia therapy. Cancer Biol. Ther. 9:895–902. 10.4161/cbt.9.11.11667 [DOI] [PubMed] [Google Scholar]
- Montarras D., Morgan J., Collins C., Relaix F., Zaffran S., Cumano A., Partridge T., and Buckingham M.. 2005. Direct isolation of satellite cells for skeletal muscle regeneration. Science. 309:2064–2067. 10.1126/science.1114758 [DOI] [PubMed] [Google Scholar]
- Moore F.E., Reyon D., Sander J.D., Martinez S.A., Blackburn J.S., Khayter C., Ramirez C.L., Joung J.K., and Langenau D.M.. 2012. Improved somatic mutagenesis in zebrafish using transcription activator-like effector nucleases (TALENs). PLoS One. 7:e37877 10.1371/journal.pone.0037877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore F.E., Garcia E.G., Lobbardi R., Jain E., Tang Q., Moore J.C., Cortes M., Molodtsov A., Kasheta M., Luo C.C., et al. 2016. Single-cell transcriptional analysis of normal, aberrant, and malignant hematopoiesis in zebrafish. J. Exp. Med. 213:979–992. 10.1084/jem.20152013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan C.G., Phillips L.A., Su X., Ma J., Miller C.B., Shurtleff S.A., and Downing J.R.. 2008. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 322:1377–1380. 10.1126/science.1164266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosaka T., van Deursen J.M., Tripp R.A., Thierfelder W.E., Witthuhn B.A., McMickle A.P., Doherty P.C., Grosveld G.C., and Ihle J.N.. 1995. Defective lymphoid development in mice lacking Jak3. Science. 270:800–802. 10.1126/science.270.5237.800 [DOI] [PubMed] [Google Scholar]
- Notarangelo L.D., Mella P., Jones A., de Saint Basile G., Savoldi G., Cranston T., Vihinen M., and Schumacher R.F.. 2001. Mutations in severe combined immune deficiency (SCID) due to JAK3 deficiency. Hum. Mutat. 18:255–263. 10.1002/humu.1188 [DOI] [PubMed] [Google Scholar]
- Okada S., Harada H., Ito T., Saito T., and Suzu S.. 2008. Early development of human hematopoietic and acquired immune systems in new born NOD/Scid/Jak3null mice intrahepatic engrafted with cord blood-derived CD34+ cells. Int. J. Hematol. 88:476–482. 10.1007/s12185-008-0215-z [DOI] [PubMed] [Google Scholar]
- Patton E.E., Widlund H.R., Kutok J.L., Kopani K.R., Amatruda J.F., Murphey R.D., Berghmans S., Mayhall E.A., Traver D., Fletcher C.D., et al. 2005. BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr. Biol. 15:249–254. 10.1016/j.cub.2005.01.031 [DOI] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., and Smyth G.K.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell S.M., Tayebi N., Nakajima H., Riedy M.C., Roberts J.L., Aman M.J., Migone T.S., Noguchi M., Markert M.L., Buckley R.H., et al. 1995. Mutation of Jak3 in a patient with SCID: essential role of Jak3 in lymphoid development. Science. 270:797–800. 10.1126/science.270.5237.797 [DOI] [PubMed] [Google Scholar]
- Santagata S., Villa A., Sobacchi C., Cortes P., and Vezzoni P.. 2000. The genetic and biochemical basis of Omenn syndrome. Immunol. Rev. 178:64–74. 10.1034/j.1600-065X.2000.17818.x [DOI] [PubMed] [Google Scholar]
- Sato Y., Takata H., Kobayashi N., Nagata S., Nakagata N., Ueno T., and Takiguchi M.. 2010. Failure of effector function of human CD8+ T Cells in NOD/SCID/JAK3−/− immunodeficient mice transplanted with human CD34+ hematopoietic stem cells. PLoS One. 5:5 10.1371/journal.pone.0013109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers A.G., Snippert H.J., Stange D.E., van den Born M., van Es J.H., van de Wetering M., and Clevers H.. 2012. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 337:730–735. 10.1126/science.1224676 [DOI] [PubMed] [Google Scholar]
- Shinkai Y., Rathbun G., Lam K.P., Oltz E.M., Stewart V., Mendelsohn M., Charron J., Datta M., Young F., Stall A.M., et al. 1992. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 68:855–867. 10.1016/0092-8674(92)90029-C [DOI] [PubMed] [Google Scholar]
- Sipley J.D., Menninger J.C., Hartley K.O., Ward D.C., Jackson S.P., and Anderson C.W.. 1995. Gene for the catalytic subunit of the human DNA-activated protein kinase maps to the site of the XRCC7 gene on chromosome 8. Proc. Natl. Acad. Sci. USA. 92:7515–7519. 10.1073/pnas.92.16.7515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.C., Raimondi A.R., Salthouse C.D., Ignatius M.S., Blackburn J.S., Mizgirev I.V., Storer N.Y., de Jong J.L., Chen A.T., Zhou Y., et al. 2010. High-throughput cell transplantation establishes that tumor-initiating cells are abundant in zebrafish T-cell acute lymphoblastic leukemia. Blood. 115:3296–3303. 10.1182/blood-2009-10-246488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippert H.J., van der Flier L.G., Sato T., van Es J.H., van den Born M., Kroon-Veenboer C., Barker N., Klein A.M., van Rheenen J., Simons B.D., and Clevers H.. 2010. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 143:134–144. 10.1016/j.cell.2010.09.016 [DOI] [PubMed] [Google Scholar]
- Spangrude G.J., Heimfeld S., and Weissman I.L.. 1988. Purification and characterization of mouse hematopoietic stem cells. Science. 241:58–62. 10.1126/science.2898810 [DOI] [PubMed] [Google Scholar]
- Stoletov K., Montel V., Lester R.D., Gonias S.L., and Klemke R.. 2007. High-resolution imaging of the dynamic tumor cell vascular interface in transparent zebrafish. Proc. Natl. Acad. Sci. USA. 104:17406–17411. 10.1073/pnas.0703446104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q., Abdelfattah N.S., Blackburn J.S., Moore J.C., Martinez S.A., Moore F.E., Lobbardi R., Tenente I.M., Ignatius M.S., Berman J.N., et al. 2014. Optimized cell transplantation using adult rag2 mutant zebrafish. Nat. Methods. 11:821–824. 10.1038/nmeth.3031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q., Moore J.C., Ignatius M.S., Tenente I.M., Hayes M.N., Garcia E.G., Torres Yordán N., Bourque C., He S., Blackburn J.S., et al. 2016. Imaging tumour cell heterogeneity following cell transplantation into optically clear immune-deficient zebrafish. Nat. Commun. 7:10358 10.1038/ncomms10358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenente I.M., Tang Q., Moore J.C., and Langenau D.M.. 2014. Normal and malignant muscle cell transplantation into immune compromised adult zebrafish. J. Vis. Exp. 10.3791/52597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traver D., Winzeler A., Stern H.M., Mayhall E.A., Langenau D.M., Kutok J.L., Look A.T., and Zon L.I.. 2004. Effects of lethal irradiation in zebrafish and rescue by hematopoietic cell transplantation. Blood. 104:1298–1305. 10.1182/blood-2004-01-0100 [DOI] [PubMed] [Google Scholar]
- Udina E., Rodríguez F.J., Verdú E., Espejo M., Gold B.G., and Navarro X.. 2004. FK506 enhances regeneration of axons across long peripheral nerve gaps repaired with collagen guides seeded with allogeneic Schwann cells. Glia. 47:120–129. 10.1002/glia.20025 [DOI] [PubMed] [Google Scholar]
- Wang G.Y., So P.L., Wang L., Libove E., Wang J., and Epstein E.H. Jr. 2011. Establishment of murine basal cell carcinoma allografts: a potential model for preclinical drug testing and for molecular analysis. J. Invest. Dermatol. 131:2298–2305. 10.1038/jid.2011.204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R.M., Sessa A., Burke C., Bowman T., LeBlanc J., Ceol C., Bourque C., Dovey M., Goessling W., Burns C.E., and Zon L.I.. 2008. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell. 2:183–189. 10.1016/j.stem.2007.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Lee J.S., Guo F., Shin J., Perez-Atayde A.R., Kutok J.L., Rodig S.J., Neuberg D.S., Helman D., Feng H., et al. 2012. Activated ALK collaborates with MYCN in neuroblastoma pathogenesis. Cancer Cell. 21:362–373. 10.1016/j.ccr.2012.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.