Xu et al. show that the lupus-associated polymorphism FcγRIIB-T232 has structural changes of the TM domain that reduces lateral mobility and inhibitory functions.
Abstract
FcγRIIB functions to suppress the activation of immune cells. A single-nucleotide polymorphism in the transmembrane (TM) domain of FcγRIIB, FcγRIIB-T232, is associated with lupus. In this study, we investigated the pathogenic mechanism of FcγRIIB-T232 at both functional and structural levels. Our results showed that FcγRIIB-T232 exhibited significantly reduced lateral mobility compared with FcγRIIB-I232 and was significantly less enriched into the microclusters of immune complexes (ICs) after stimulation. However, if sufficient responding time is given for FcγRIIB-T232 to diffuse and interact with the ICs, FcγRIIB-T232 can restore its inhibitory function. Moreover, substituting the FcγRIIB-T232 TM domain with that of a fast floating CD86 molecule restored both the rapid mobility and the inhibitory function, which further corroborated the importance of fast mobility for FcγRIIB to function. Mechanistically, the crippled lateral mobility of FcγRIIB-T232 can be explained by the structural changes of the TM domain. Both atomistic simulations and nuclear magnetic resonance measurement indicated that the TM helix of FcγRIIB-T232 exhibited a more inclined orientation than that of FcγRIIB-I232, thus resulting in a longer region embedded in the membrane. Therefore, we conclude that the single-residue polymorphism T232 enforces the inclination of the TM domain and thereby reduces the lateral mobility and inhibitory functions of FcγRIIB.
Introduction
Immune cells have developed a sophisticated mechanism to regulate their activations for the purpose of balancing immunoprotection and immunopathology. The receptors for the Fc portion of IgG molecules (FcγRs) well define one of such regulatory strategies. The human immune system contains six types of canonical FcγRs, including FcγRI, FcγRIIA, FcγRIIB, FcγRIIC, FcγRIIIA, and FcγRIIIB, among which FcγRIIB is the only one having an inhibitory function (Smith and Clatworthy, 2010; Nimmerjahn and Ravetch, 2011; Pincetic et al., 2014). Malfunction of FcγRIIB is usually detrimental for the immune system (Niederer et al., 2010; Smith and Clatworthy, 2010; Pincetic et al., 2014). Single-nucleotide polymorphisms (SNPs) of the human FCGR2B gene significantly influence susceptibility to autoimmune diseases (Kyogoku et al., 2002; Niederer et al., 2010; Smith and Clatworthy, 2010). Among all seven nonsynonymous SNPs of FCGR2B, the T-to-C transition in exon 5 (SNP accession no. rs1050501) causes the I232T substitution within the transmembrane (TM) domain of FcγRIIB (FcγRIIB-T232). Strikingly, this polymorphism occurs at a notable frequency with the heterozygous FcγRIIB-T232 polymorphism present in 18–43% of all humans (Niederer et al., 2010; Smith and Clatworthy, 2010). Epidemiological studies revealed the positive association of the homozygous FcγRIIB-T232 polymorphism with systemic lupus erythematosus (SLE) in world-wide populations (Kyogoku et al., 2002; Siriboonrit et al., 2003; Chu et al., 2004; Clatworthy et al., 2007; Niederer et al., 2010; Willcocks et al., 2010). Interestingly, it is also known that homozygosity for this polymorphism is associated with protection against severe malaria, which may contribute to the higher frequency of FcγRIIB-T232 and, hence, SLE in Africans and Southeast Asians (Clatworthy et al., 2007; Willcocks et al., 2010).
Early studies demonstrated that FcγRIIB-T232 polymorphism causes functional loss. Monocytes carrying FcγRIIB-T232 are hyperactivated with enhanced FcγRI-triggered phospholipase D activation and calcium signaling (Floto et al., 2005). B cells carrying FcγRIIB-T232 also become hyperactive, showing abnormally elevated PLCγ2 activation, calcium signaling, and proliferation (Kono et al., 2005). Our early study showed that B cells expressing FcγRIIB-T232 lose the ability to inhibit the oligomerization of B cell receptors (BCRs) upon coligation between BCR and FcγRIIB (Liu et al., 2010c). Our recent live cell–imaging experiments demonstrated that B cells expressing FcγRIIB-T232 fail to inhibit the spatial-temporal colocalization of BCR and CD19 within the B cell immunological synapse (Xu et al., 2014). To our knowledge, FcγRIIB-T232 is the first example that a naturally occurring SNP within the TM domain of a single TM receptor can cripple its normal function. The mechanistic explanation on the function loss of this polymorphism shall be of great interest and importance. Early biochemical studies proposed a model of reduced affinity between FcγRIIB-T232 and lipid rafts to explain the functional effect of this polymorphism (Floto et al., 2005; Kono et al., 2005).
In this study, we investigated the function disruption of FcγRIIB-T232 in a different perspective by combining techniques of high-speed single molecule–tracking, superresolution imaging, Monte Carlo simulations, molecular dynamics (MD) simulations, and nuclear magnetic resonance (NMR) spectroscopy. By live-cell and single-molecule imaging, we first showed that FcγRIIB-T232 diffuses significantly slower than FcγRIIB-I232 on the plasma membrane of laboratory cells and human primary B cells as well as monocytes from lupus patients. Experiments on both BCRs and FcγRIIB receptors on human primary B cells suggested that FcγRIIB diffuses almost three times faster than BCRs. We then proved that rapid lateral mobility is essential for sustaining the inhibitory function of FcγRIIB. In subsequent molecular-level studies, we found that the crippled lateral mobility of FcγRIIB-T232 is possibly caused by changes in conformation and orientation of the TM domain of FcγRIIB-T232. In atomistic simulations, we found that the I232T polymorphism renders the TM helix more bent and more tilted within the membrane. Consistently, NMR measurements suggested that the TM helix of FcγRIIB-T232 is longer than that of FcγRIIB-I232. These structural changes inevitably increase the cross-sectional area of the TM domain and, thus, shall in turn reduce the lateral mobility of FcγRIIB-T232 as predicted by the Stokes-Einstein–like equation (Gambin et al., 2006)–based theoretical calculation. We finally proposed a new “catch me if you can” model to rationalize the function loss of FcγRIIB-T232. FcγRIIB-I232 is a highly mobile plasma membrane receptor that can readily catch the Fc portion of IgG antibodies in immune complexes (ICs) to perform potent inhibitory functions. The I232T polymorphism, however, drastically slows down the lateral diffusion of FcγRIIB-T232 and therefore greatly suppresses its chance to catch the ICs, as indicated from Monte Carlo simulations, which impairs its capability to execute the inhibitory function at the right time in the right place.
Results
FcγRIIB-T232 shows significantly slower lateral mobility than FcγRIIB-I232 in fluorescence recovery after photobleaching (FRAP) assay
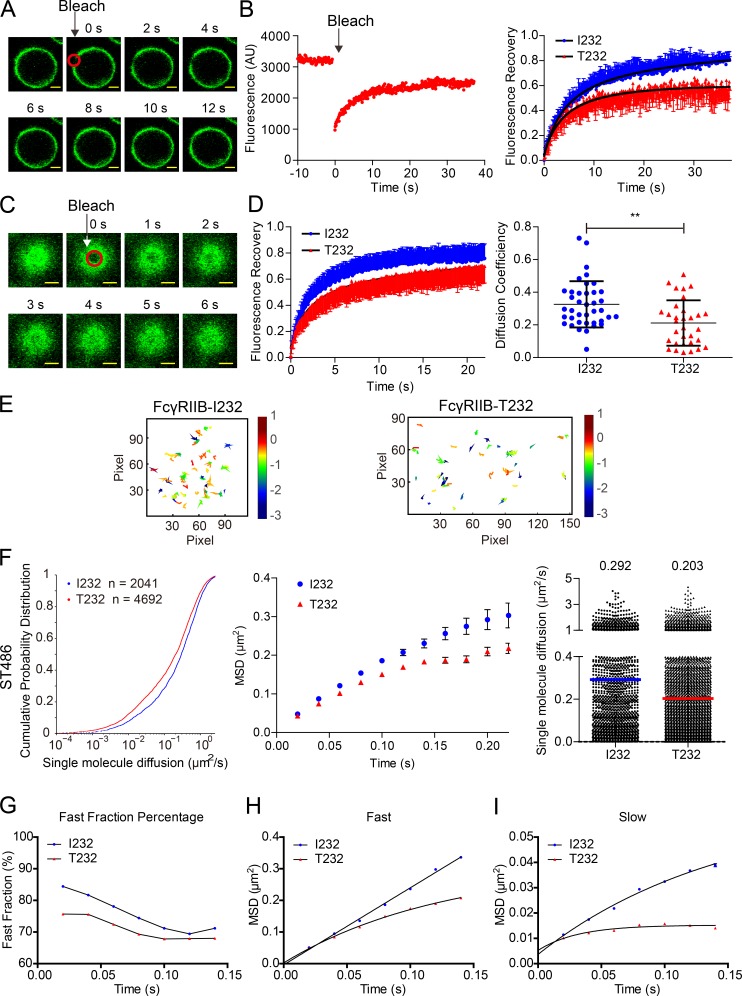
Lateral mobility is one of the key biophysical features of all components of a biomembrane, including membrane proteins, and it is well documented that the TM domain of a receptor protein significantly affects the efficiency of lateral mobility (Fujiwara et al., 2002; Kusumi et al., 2005, 2012; Suzuki et al., 2007; Kasai et al., 2011). Thus, we performed a variety of experiments to compare the diffusional behavior of FcγRIIB-I232 and FcγRIIB-T232 on the plasma membrane. First, we conducted the conventional FRAP experiment to bleach a small region on the equatorial plasma membrane of A20II1.6 B cells that lack endogenous FcγRIIB but express comparable amounts of surface-located FcγRIIB-I232 or FcγRIIB-T232 with YFP as a tag. After photobleaching, we observed that the mean fluorescence intensity of YFP-tagged FcγRIIB is promptly recovered and reaches a plateau in <30 s at room temperature (25°C; Fig. 1, A and B; and Videos 1 and 2). A statistical comparison of the FRAP curves showed that the FRAP recovery kinetics of FcγRIIB-T232 was significantly slower than that of FcγRIIB-I232 (Fig. 1 B). Moreover, after least squares fitting of these two classes of FRAP curves to exponential functions, 69.63 ± 1.42% fast mobile fractions were identified for FcγRIIB-I232, whereas the number was significantly smaller (59.32 ± 0.57%) for FcγRIIB-T232. In addition, the τ1/2 recovery rate was 7.30 ± 1.77 s for FcγRIIB-I232, in comparison with the τ1/2 value of 28.23 ± 2.99 s for FcγRIIB-T232. These analyses demonstrated that FcγRIIB-I232 moved faster than FcγRIIB-T232 in quiescent A20II1.6 B cells.
Figure 1.
FcγRIIB-T232 exhibits significantly slower lateral mobility than FcγRIIB-I232. (A) Representative FRAP images at the indicated time points showing the recovery of FcγRIIB-YFP fluorescence intensity in A20II1.6 B cells. The red circle shows the bleaching area. Bars, 1.5 µm. (B, left) Representative FRAP curve at 25°C experimental condition. (Right) Statistical comparison of the FRAP recovery kinetics from A20II1.6 B cells expressing either FcγRIIB-I232 or FcγRIIB-T232. Error bars represent mean ± SEM from at least 20 cells in three independent experiments. AU, arbitrary units. (C) Representative 2D FRAP images at the indicated time points showing the recovery of FcγRIIB-YFP fluorescence intensity in ST486 B cells. The red circle shows the bleaching area. Bars, 1.5 µm. (D, left) Statistical comparison of FRAP recovery kinetics at 37°C from ST486 B cells expressing either FcγRIIB-I232 or FcγRIIB-T232. Error bars represent mean ± SEM from at least 18 cells in three independent experiments. (Right) Comparison of the diffusion coefficient with each dot representing one measurement for the diffusion coefficient from one cell. Bars represent mean ± SD from at least 18 cells in three independent experiments. Two-tailed Student's t tests were performed with the p-value indicated. **, P < 0.01. (E) Representative trajectories in the whole TIRF imaging time course of either FcγRIIB-I232 or FcγRIIB-T232 on the plasma membrane of ST486 B cells. (F) A series of mathematical comparisons of the Brownian diffusion of FcγRIIB-I232 or FcγRIIB-T232 molecules from ST486 cells in CPD plots (left), MSD plots (middle), or scatter plots (right). Bars represent median value. The p-value in CDP plots is <0.0001 in Kolmogorov-Smirnov tests. (G) PICS analysis of single-molecule TIRF images from either FcγRIIB-I232 or FcγRIIB-T232. (H and I) Two exponential PICS analysis for both the fast (H) and slow (I) fractions of FcγRIIB molecules. (E–I) The results shown are representative of one of at least three independent experiments. See also Videos 1–6.
In conventional FRAP experiments, it is difficult to quantify the absolute Brownian diffusion coefficient from FRAP curves because of the lack of an appropriate mathematical simulation equation. Thus, we performed the two-dimensional (2D) FRAP experiment by adjusting the image plane of a confocal fluorescence microscope to the flat top areas of a cell. Subsequently, we bleached a small circular region and recorded the FRAP curves (Fig. 1, C and D; and Videos 3 and 4). The experiments were performed in human ST486 B cells that lack endogenous FcγRIIB but express comparable amounts of FcγRIIB-I232–YFP or FcγRIIB-T232–YFP. We used ST486 instead of A20II1.6 B cell because ST486 cells were found to easily form a large flat top area at 37°C. Similarly, we observed significantly slower FRAP recovery kinetics in FcγRIIB-T232 than in FcγRIIB-I232 (Fig. 1 D). This configuration allowed us to analyze the 2D FRAP recovery curves with Soumpasis FRAP equations (Soumpasis, 1983) and to quantify the absolute Brownian diffusion coefficients for both FcγRIIB-I232 and FcγRIIB-T232 molecules. The Brownian diffusion coefficient of FcγRIIB-I232 was 0.33 µm2/s, whereas the number of FcγRIIB-T232 was reduced to 0.21 µm2/s (Fig. 1 D). Thus, all FRAP experiments unanimously suggested significantly suppressed FRAP recovery kinetics in FcγRIIB-T232, which further implied its reduced lateral mobility on the plasma membrane of live cells.
Single-molecule imaging showed significantly slower lateral mobility of FcγRIIB-T232 than FcγRIIB-I232
Next, we performed high-resolution high-speed single-molecule imaging experiments using total internal reflection fluorescence (TIRF) microscopy to directly examine the lateral mobility and thus to calculate the Brownian diffusion coefficient of FcγRIIB in quiescent ST486 B cells using a protocol similar to our previous studies to perform single-molecule imaging of BCRs (Liu et al., 2010a,b, 2012). When analyzing the TIRF images of these single FcγRIIB molecules, we first used a method based on single-particle tracking (SPT) to calculate and compare the Brownian diffusion coefficients of FcγRIIB-I232 and FcγRIIB-T232 molecules (Fig. 1 E and Videos 5 and 6). The short-range Brownian diffusion coefficients of 2041 FcγRIIB-I232 and 4692 FcγRIIB-T232 molecules were calculated, and their cumulative probability distribution (CPD) functions were shown for a statistical comparison (Fig. 1 F). In quiescent B cells, only a small fraction (<10%) of FcγRIIB-I232 is immobile as justified by a cutoff diffusion coefficient of 0.01 µm2/s, in contrast to the larger value of >10% for FcγRIIB-T232. After fitting the time-dependent mean square displacement (MSD) data using a confined diffusion model, FcγRIIB-T232 was found to confine to an area of 0.55 ± 0.01 µm2, significantly smaller than FcγRIIB-I232 (0.95 ± 0.03 µm2; Fig. 1 F). In addition, FcγRIIB-I232 also exhibits a faster diffusion coefficient than FcγRIIB-T232 (0.292 vs. 0.203 µm2/s; Fig. 1 F), consistent with the FRAP results (Fig. 1, B and D).
SPT is error prone under conditions of high signal densities and high mobilities that cause particle trajectories to overlap. To avoid these potential errors, we also used a method termed particle image correlation spectroscopy (PICS; Semrau and Schmidt, 2007) to retrieve the nanometer-scale diffusion dynamics from TIRF image sequences of individual diffusing FcγRIIB molecules. Another advantage of PICS is its ability to deliver sufficient statistics where SPT fails to identify enough trajectories that warrant a more in-depth analysis of the mobility behavior. To that end, fast and slow fractions of FcγRIIB can be distinguished through two-exponential fitting of the resulting cumulative probability curves (Semrau and Schmidt, 2007; Xu et al., 2010). The presence of these two fractions was also observed for BCRs and FcγRIIA receptors (Tolar et al., 2009; Liu et al., 2010a,b; Treanor et al., 2010; Jaumouillé et al., 2014), indicating that it can be a general feature for membrane receptors that may be caused by the heterogeneous membrane environments and cytoskeleton regulations. Such refined analyses again confirmed that FcγRIIB-I232 has a larger fast mobile fraction than FcγRIIB-T232 (Fig. 1 G). For both the fast (Fig. 1 H) and slow (Fig. 1 I) mobile factions, FcγRIIB-I232 was confined to an area that is significantly larger than FcγRIIB-T232, consistent with the MSD fitting data above (Fig. 1 F). Thus, PICS analysis not only confirms the faster lateral mobility of FcγRIIB-I232 than FcγRIIB-T232, but also identifies a larger fast mobile fraction of FcγRIIB-I232.
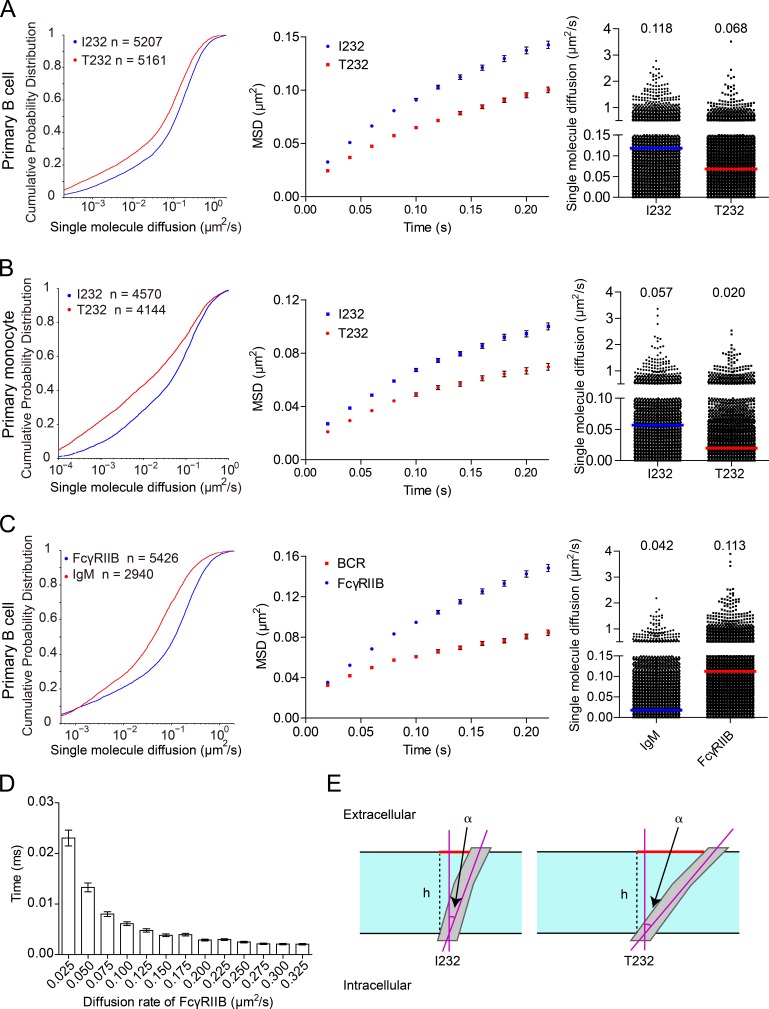
All the experiments in Fig. 1 were performed on B cells by the utilization of an overexpression system with fluorescent protein–tagged FcγRIIB. As a further validation, we examined the lateral mobility of FcγRIIB molecules on primary B cells and monocytes from the peripheral blood of SLE patients homozygous for FcγRIIB-T232 and those carrying FcγRIIB-I232. We used an Alexa Fluor 647–conjugated Fab fragment of a human FcγRIIB-specific antibody (without cross-recognition of FcγRIIA; Rankin et al., 2006) to specifically label FcγRIIB rather than other FcγRs in these immune cells and then quantified their lateral mobility. Indeed, SPT analyses on thousands of FcγRIIB molecules corroborated the statistically significantly slower lateral mobility of FcγRIIB-T232 than FcγRIIB-I232 in both human primary B cells and monocytes (Fig. 2, A and B).
Figure 2.
FcγRIIB-T232 exhibits significantly slower Brownian diffusion than FcγRIIB-I232 in human primary B cells and monocytes. (A and B) Comparison of the Brownian diffusion of FcγRIIB-I232 or FcγRIIB-T232 molecules from either human primary B cells (A) or primary monocytes (B). (C) Comparison of the Brownian diffusion of FcγRIIB and IgM-BCRs molecules in human primary B cells. (A–C) Cells were placed on coverslips tethering anti–MHC-I antibodies in TIRF microscopy imaging. Given are a series of mathematical comparisons in CPD plots (left), MSD plots (middle), and scatter plots (right; bars represent the median value). The p-value in each CDP plots is <0.0001 in Kolmogorov-Smirnov tests. The results shown are representative of one of at least three independent experiments. (D) Monte Carlo simulation of receptors’ binding time. The times for first binding between 625 receptors diffusing at the indicated diffusion rate and 5 random distributed immobile ICs that were confined in a 2.5 × 2.5–µm square were calculated. Error bars represent mean ± SEM from 400× simulation per group that were pooled from at least two independent experiments. (E) In this illustrative diagram, the TM domain is represented as a rectangle colored in gray, whereas the plasma membrane is colored in cyan with the borders indicated by black solid lines. The red solid bars represent the projections of the TM helices within the plane of the plasma membrane, which equal h × tan(α), where h is the thickness of the membrane (denoted as black dashed lines) and α is the tilting angle of a TM helix. The cross-sectional area shall be proportional to tan2(α). The FcγRIIB-I232 TM helix prefers the tilting orientation of 13–18°, whereas the FcγRIIB-T232 TM helix is the most stable at the tilting angle of 35–40° (Fig. 6 J). Therefore, the ratio of cross-sectional area from the FcγRIIB-T232 TM helix to that from FcγRIIB-I232 shall fall in the interval between [tan(35°)/tan(18°)]2 and [tan(40°)/tan(13°)]2, or [4.6, 13.2].
FcγRIIB diffuses faster than BCR in single-molecule imaging assay
The results in Fig. 2 (A and B) indicated that FcγRIIB has higher lateral mobility than published for the BCR (Tolar et al., 2009; Liu et al., 2010a,b; Treanor et al., 2010). To precisely compare the Brownian diffusion coefficients of FcγRIIB and BCR molecules on the same B cells, we performed single-molecule imaging experiments under the same TIRF microscopy imaging conditions for both FcγRIIB and BCR molecules using human peripheral blood primary B cells from healthy individuals bearing FcγRIIB-I232. The SPT analysis of 6,313 IgM-BCRs and 6,896 FcγRIIB-I232 molecules suggested that FcγRIIB diffuses almost three times faster than BCR (0.113 µm 2/s for FcγRIIB vs. 0.042 µm 2/s for BCR; Fig. 2 C).
The high lateral mobility of FcγRIIB is relevant to its potent inhibitory function
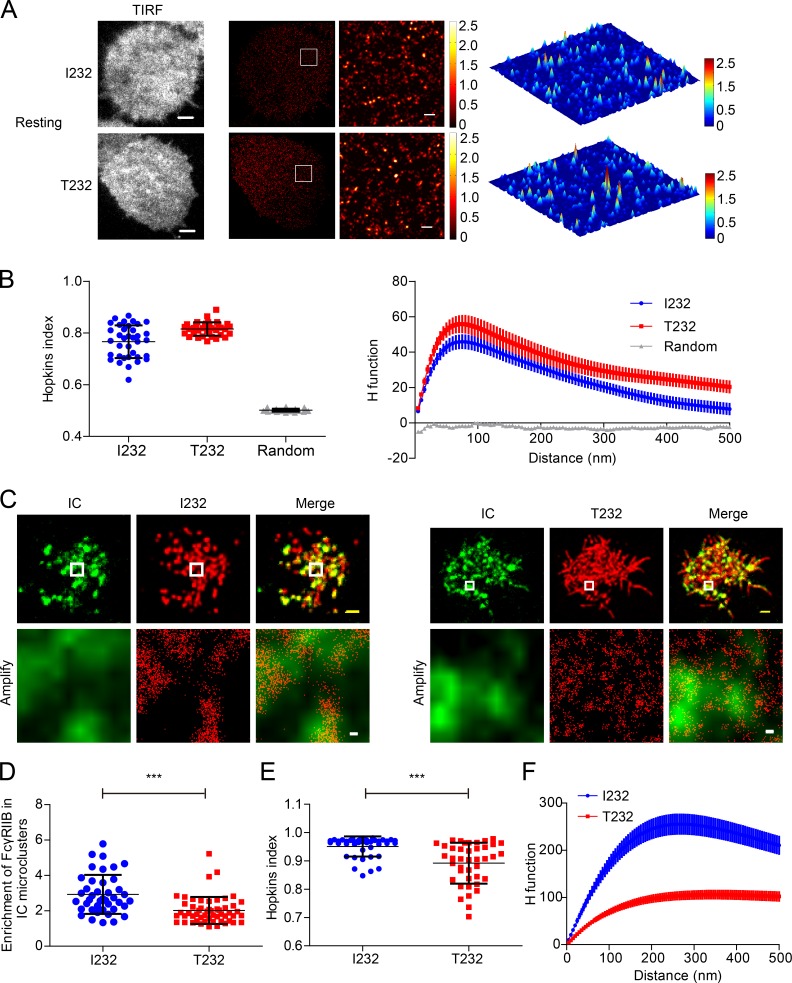
Based on all these data, we hypothesized that the unique high lateral mobility of FcγRIIB might be relevant to its potent inhibitory function. Indeed, Monte Carlo simulation indicated that the high lateral mobility of FcγRIIB can increase its contact possibility with IC microclusters (Fig. 2 D). In contrast, the retarded lateral mobility of FcγRIIB-T232 would substantially reduce its chance to contact IC microclusters (Fig. 2 D). To further validate this hypothesis, we first implemented a superresolution imaging experiment supported by photoactivated localization microscopy (PALM) to visualize the position of both FcγRIIB-I232 and FcγRIIB-T232 molecules on the plasma membrane of A20II1.6 B cells encountering the planar lipid bilayers (PLBs) presenting streptavidin-supported molecules of either biotin-conjugated F(ab’)2 anti–MHC-I as a control or surrogate ICs composed of biotin-conjugated whole IgG anti–mouse Igκ antibodies. After a photoactivatable protein Eos3.2-derived PALM imaging procedure, we found that FcγRIIB-I232 and FcγRIIB-T232 are largely distributed in a comparable manner on the plasma membrane in quiescent B cells (Fig. 3, A and B). When encountering the surrogate ICs inducing BCR and FcγRIIB coligation, FcγRIIB-I232 was found to be significantly enriched within the microcluster of ICs. However, such enrichment is drastically defective in the case of FcγRIIB-T232 (Fig. 3, C and D). Consistently in both Hopkins index (Zhang et al., 2006) and H function analysis, a method derived from Ripley’s K function by calculating the number of FcγRIIB found within a distance of R (Kiskowski et al., 2009), FcγRIIB-I232 can form more large-sized nanoclusters than FcγRIIB-T232 in activated B cells (Fig. 3, E and F). Thus, the results in Fig. 3 suggest that in response to activation by ICs, FcγRIIB-T232 is less enriched in IC microclusters and is therefore less likely to form highly concentrated and large-sized FcγRIIB nanoclusters to execute the inhibitory functions.
Figure 3.
FcγRIIB-T232 is less enriched in IC microclusters in recognition of ICs. (A) A20II1.6 B cell expressing Eos3.2-tagged FcγRIIB-I232 or FcγRIIB-T232 was placed on antigen-free PLBs. Cells were then fixed, imaged, and processed for PALM imaging. (Left) TIRF images. (Middle) PALM images of the same representative cell. (Right) An inset region in the white squares in the PALM images is also shown as a 3D surface plot. Bars: (TIRF image), 3 µm; (superresolution) 300 nm. (B) Given is the quantification of the distribution of FcγRIIB-I232 and FcγRIIB-T232 by Hopkins index (left) and H function (right). Data were pooled from two experiments with a minimum of 14 cells per experiment. Bars represent mean ± SD in Hopkins index (left) and mean ± SEM in H function (right). (C) Similarly as in A, Eos3.2-tagged FcγRIIB-I232 (left)- or FcγRIIB-T232 (right)-expressing B cells were placed on PLBs presenting 20 nM Alexa Fluor 647–conjugated goat IgG anti–mouse Igκ. (Top) Images are conventional TIRF images of IC microclusters and superresolution images of FcγRIIB-I232 (left) or FcγRIIB-T232 (right). Bars, 1.5 µm. (Bottom) Images are the insets in the white squares. Bars, 50 nm. (D) Enrichment of FcγRIIB-I232 and FcγRIIB-T232 molecules within IC microclusters. Data are pooled from at least two independent experiments with a minimum of 12 cells per experiment. Bars represent mean ± SD. A Student's t test was performed with the p-value indicated. (E and F) Given is the quantification of the distribution of FcγRIIB-I232 and FcγRIIB-T232 by Hopkins index (E) and H function (F). Data were pooled from at least two independent experiments with a minimum of 12 cells per experiment. Bars represent mean ± SD (E) or mean ± SEM (F). ***, P < 0.0001.
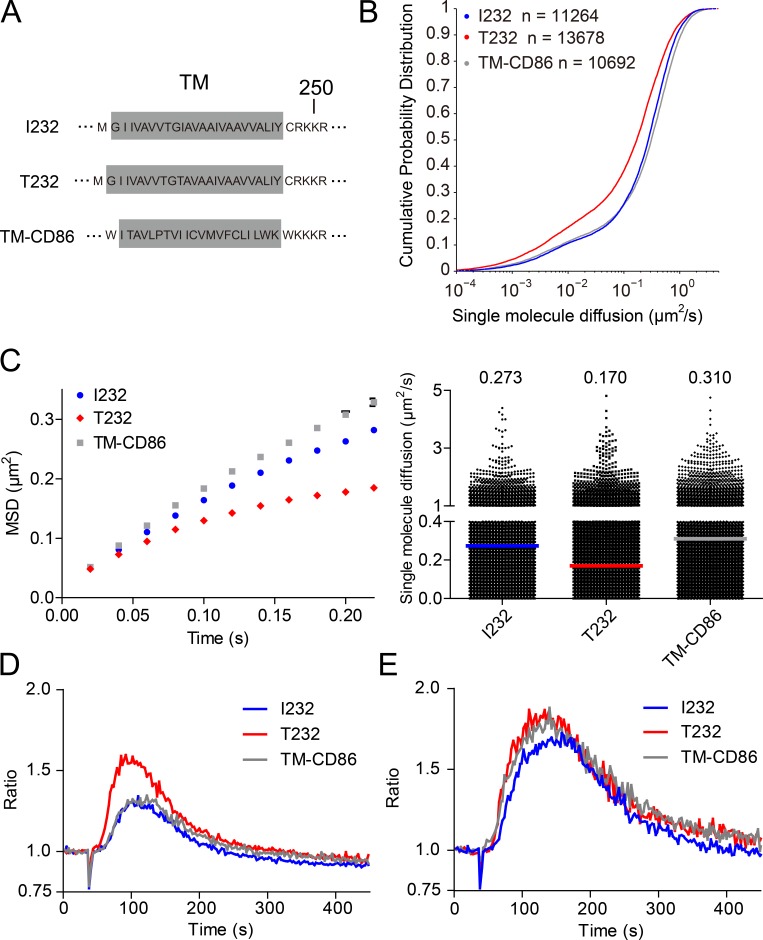
To further check the hypothesis that the high lateral mobility of FcγRIIB-I232 is associated with the potent inhibitory function of FcγRIIB, we used live-cell imaging experiments to evaluate the diffusion coefficient for a chimeric FcγRIIB molecule with the TM domain swapped from a fast floating CD86 molecule (called FcγRIIB-TM-CD86; Fig. 4, A–E). Notably, CD86 is a single TM receptor, very similar to FcγRIIB. In addition, according to the current understanding of plasma membrane biology, the Brownian mobility of a receptor on the plasma membrane is mainly determined by the TM domain of that receptor (Kusumi et al., 2005, 2012). These two points guarantee the feasibility of our swapping experiment. Indeed, single-molecule imaging experiments showed that FcγRIIB-TM-CD86 possesses high lateral mobility at a similar level to FcγRIIB-I232, both of which are significantly faster than FcγRIIB-T232 (Fig. 4, B and C). As a functional test on the potency of the inhibitory function of these three FcγRIIB molecules, we measured Ca2+ mobilization in A20II1.6 B cells expressing comparable amounts of these three FcγRIIB molecules and surface IgG-BCRs (not depicted). We used goat IgG anti–mouse Igκ antibody as the surrogate ICs to induce the coligation of BCR and FcγRIIB. We found that FcγRIIB-I232 and FcγRIIB-TM-CD86 show the most potent inhibitory function against BCR engagement–induced Ca2+ mobilization, whereas FcγRIIB-T232 exhibits a weak inhibitory function (Fig. 4 D). As a system control, F(ab’)2 fragments of goat IgG anti–mouse Igκ, which lack the Fc portion of IgG to interact with FcγRIIB, similarly drive the Ca2+ mobilization in these three types of B cell sublines (Fig. 4 E). Thus, these results support our hypothesis that high mobility of FcγRIIB is essential for its potent inhibitory function.
Figure 4.
The highly motile feature of FcγRIIB is linked to its potent inhibitory function. (A) Shown are the schematic illustrations for the amino acid sequence of the TM domains of FcγRIIB-I232, FcγRIIB-T232, and FcγRIIB-CD86-TM. (B and C) Single-molecule imaging experiments were performed on A20II1.6 B cells expressing FcγRIIB-I232, FcγRIIB-T232, or FcγRIIB-CD86-TM to compare the Brownian motility of these FcγRIIB variants. Given are CPD plots (B), MSD plots (C, left), and scatter plots (C, right), respectively. Bars represent median value. The p-value between FcγRIIB-T232 and FcγRIIB-I232 (or FcγRIIB-CD86-TM) is <0.0001 in Kolmogorov-Smirnov tests. The results shown are representative of one of at least three independent experiments. (D and E) Ratiometric calcium flux in A20II1.6 cells expressing FcγRIIB-I232, FcγRIIB-T232, or FcγRIIB-CD86-TM that were stimulated with 5 µg/ml whole IgG anti–mouse Igκ (D) or F(ab’)2 fragment anti–mouse Igκ (E). The results shown are representative of one of three independent experiments.
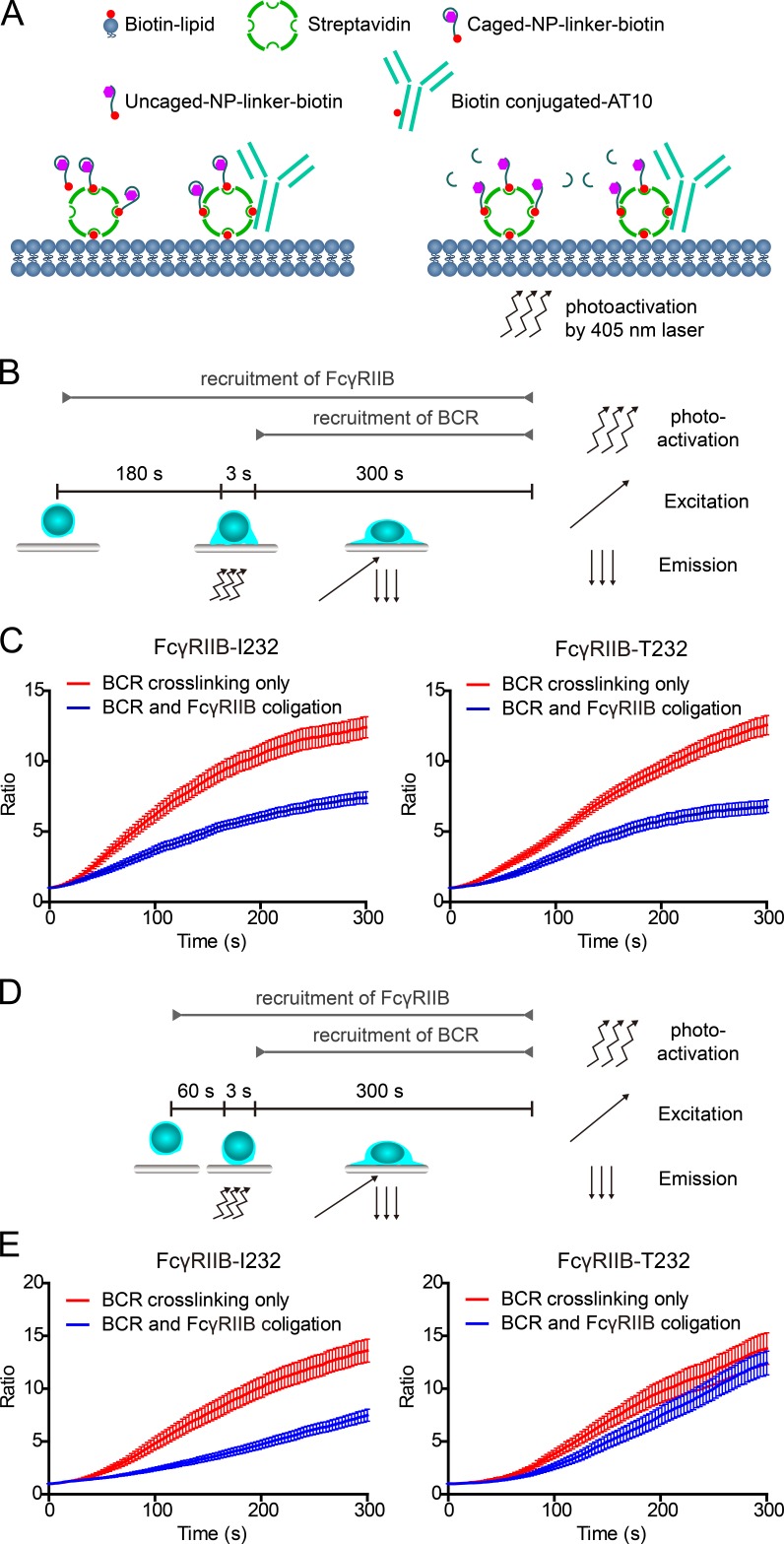
To directly confirm that high lateral mobility is required for the inhibitory functioning of FcγRIIB, we used a photoactivatable antigen system, caged 4-hydroxy-3-nitrophenylacetyl (NP), which was recently developed for the purpose of setting up a controllable trigger point for the recognition of BCRs and antigen (Wang et al., 2016). In such a system, caged NP can only be converted to antigenic form and thus can be recognized by B1-8-BCR–expressing B cells only after photoactivation by 405-nm photons (Fig. 5 A). Before live-cell imaging experiment, we placed B cells expressing NP-specific B1-8-IgM-BCRs on PLBs presenting streptavidin-supported surrogate ICs composed of biotin-conjugated caged NP for the stimulation of B1-8-BCR and biotin-conjugated whole IgG anti-FcγRIIB (clone AT10) for the stimulation of FcγRIIB (schematic representation in Fig. 5 A). Obviously, by using this photoactivatable antigen system, we can purposely give sufficient amount of responding time for FcγRIIB-T232 to diffuse and interact with the ICs before triggering the interaction between caged NP and B1-8-BCR by 405-nm photons (Fig. 5 B). Our hypothesis is that under such experimental conditions, FcγRIIB-T232 might be able to circumvent its defect in lateral mobility and exhibit similar inhibitory power as FcγRIIB-I232. We used 1-, 3-, and 5-min preincubation times (responding time) before we photoactivated caged NP to initiate the recognition between antigen and BCRs (Fig. 5, B and D). Immediately after photoactivation, we examined the synaptic accumulation of B1-8-BCRs by TIRF imaging to quantify and compare the inhibitory functions of FcγRIIB-I232 and FcγRIIB-T232. The results showed that FcγRIIB-T232 restored the capability to inhibit the synaptic accumulation of B1-8-BCRs in comparison with the case of FcγRIIB-I232 when 5- (not depicted) and 3-min but not 1-min responding time was provided for FcγRIIB-T232 in advance (Fig. 5, B–D). Considering the fact that B cells used tens of seconds to land to the surface of PLBs in our experimental system, these results showed that <1-min responding time is not sufficient to restore the inhibitory function of FcγRIIB-T232. Thus, by using the photoactivatable caged-NP system, we further confirmed that the significantly slower lateral mobility of FcγRIIB-T232 than FcγRIIB-I232 accounts for the impaired function of FcγRIIB-T232.
Figure 5.
FcγRIIB-T232 restores the inhibitory function if sufficient responding time is given for FcγRIIB-T232 to interact with the ICs in a photoactivatable antigen system. (A) A schematic diagram depicting the PLBs presenting the surrogate ICs composed of biotin-conjugated caged NP and biotin-conjugated anti-FcγRIIB antibody (AT10) and the action of using 405-nm photos to trigger the interaction between uncaged NP and B1-8-BCRs. (B) A schematic diagram depicting the experimental procedure of using a caged NP–based photoactivatable antigen system to provide sufficient responding time for FcγRIIB-T232 to interact with the ICs. Cells were preincubated on PLBs presenting surrogate ICs for 180 s before the 3-s photoactivation by a 405-nm laser. The accumulation of B1-8-IgM-BCRs into the B cell immunological synapse was examined for 300 s by TIRF microscopy imaging after photoactivation. (C) Statistical quantification of the normalized mean fluorescence intensity to show the synaptic accumulation of B1-8-IgM-BCRs after photoactivation. Error bars represent mean ± SEM of 22 cells from one representative of three independent experiments. (D) Similarly as in B, a 60-s responding time was given. (E) Similarly as in C, statistical quantification is given for the experiments in D. Error bars represent mean ± SEM of 20 cells from one representative of three independent experiments.
The FcγRIIB-T232 TM helix shows aggravated helix bending in MD simulations
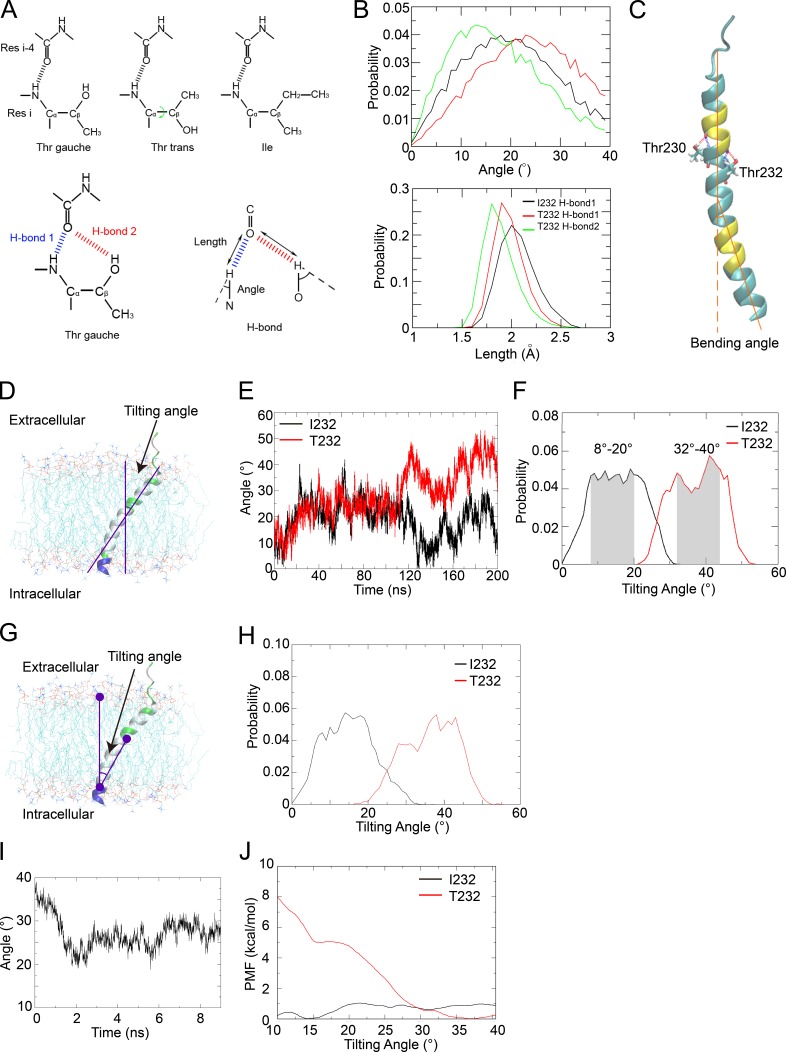
We studied how a single–amino acid change from Ile to Thr within the TM domain substantially slows down the Brownian diffusion of FcγRIIB using MD simulations by particularly focusing on the change in the conformation and orientation of the TM domain caused by the polymorphism. Specifically, we inserted all-atom models for the TM domains of both FcγRIIB-I232 and FcγRIIB-T232 into POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) bilayers (Fig. S1, A and B) and quantitatively compared their conformations and orientations (Fig. S1 B). The TM domain of FcγRIIB-I232 was modeled as a long helix with both termini blocked by chemical groups based on comparative modeling, whereas Ile232 of the simulated structure was mutated to Thr232 to produce the structural model of FcγRIIB-T232 (Fig. S1, A and B; and Videos 7 and 8).
The structural models of both FcγRIIB-I232 and FcγRIIB-T232 TM helices are fairly stable in the membrane (not depicted). Previous studies (Ballesteros et al., 2000; Feldblum and Arkin, 2014) suggested that Thr in the TM helices might slightly bend the helix by preferring gauche conformation of the χ1 torsion angle and thus forming bifurcated hydrogen bonds with residue in the preceding helical turn (Fig. 6 A). As expected, Thr232 in the FcγRIIB-T232 TM helix adopts the gauche conformation in 99.97% of all the simulation snapshots, consistent with bifurcated hydrogen bonds (Fig. 6 A). Comparison of the distributions of hydrogen-bonding parameters (length and angle in Fig. 6 A) for the two candidate hydrogen bonds (denoted as H-bond 1 and H-bond 2 in Fig. 6 A) between residues 232 and 228 shows a shorter distance and a more distorted angle for the backbone hydrogen bond (H-bond 1) in FcγRIIB-T232 than in FcγRIIB-I232 (Fig. 6, A and B), supporting that the bifurcated hydrogen bonds locally bend the helix in FcγRIIB-T232.
Figure 6.
The FcγRIIB-T232 TM helix becomes more bent and prefers a more tilted orientation in the lipid membrane. (A, top) The χ1 angle in the side chain of Thr232 can take either gauche or trans conformations. (Bottom, left) For Thr232, the bifurcated hydrogen bonds (H-bond 1 and H-bond 2) can form only when the χ1 angle takes gauche rather than trans conformation. (Bottom, right) The definitions of the length and angle of the H-bond are illustrated. Res, residue. (B) Comparison of the probability distributions of the angle (top) and length (bottom) of H-bond 1 in the FcγRIIB-I232 (black) and FcγRIIB-T232 (red) systems and H-bond 2 in the FcγRIIB-T232 (green) system. (C) Illustration of the helix bending caused by Thr residues (Thr230 and Thr232 shown in the Licorice representation). The bending angle was estimated by the scalar angle between two vectors representing the directions of both termini of the helix. The terminal residues used to calculate the vectors are colored in yellow. (D) The tilting angle was estimated by the scalar angle between the direction axis of the helix (estimated using the yellow residues) and the norm of the membrane (estimated using the phosphorus atoms). (E) The time series of the tilting angles of the TM helix in 200-ns simulations of the FcγRIIB-I232 (black) and FcγRIIB-T232 (red) systems. (F) Probability distributions of the tilting angles estimated from the last 80 ns in the simulations on the FcγRIIB-I232 (black) and FcγRIIB-T232 (red) TM helices. The favored intervals for the tilting angles are highlighted in shadow. (G and H) Probability distributions of tilting angles (H) estimated using an alternative mathematical method (G), as the scalar angle between the center of phosphorus atoms in the upper lipid leaflet, the center of carbonyl carbon atoms in residues 244–247 that represent the C terminus of the TM helix, and the center of all Cα atoms in residues 224–245 that represent the geometric center of the TM helix. (I) Tilting-angle relaxation of the FcγRIIB TM helix after backward T232I substitution. (J) The free energy profiles against the tilting angle in the FcγRIIB-I232 (black) and FcγRIIB-T232 (red) systems. (A–J) All the MD results have been verified in three independent experiments, using different force fields and different lipid composition. See also Fig. S1 and Videos 7 and 8.
Subsequently, we quantitatively evaluated the extent of bending in both TM helices in the simulation trajectories. The bending angle was estimated as the scalar angle between the directions of both termini of the TM helix (Fig. 6 C). In the FcγRIIB-I232 TM domain, Thr230 that is located half a turn away from the key residue 232 can cause only slight bending in the helix, with a bending angle of 11.14 ± 5.83° in 20,000 structural snapshots. In the FcγRIIB-T232 TM domain, however, the presence of two Thr residues (Thr230 and Thr232) on the opposite sides of the helix significantly raises the bending angle to 13.12 ± 4.39° in 20,000 snapshots (P < 1 × 10−7). Therefore, the residue substitution of I232T aggravates the bending of the FcγRIIB TM helix.
The FcγRIIB-T232 TM helix prefers a more tilted orientation in the lipid membrane in MD simulations
Next, we compared the orientations of the TM domains for FcγRIIB-I232 and FcγRIIB-T232. The TM helices are completely perpendicular to the membrane before simulation. After sufficient equilibration (200 ns), the tilting angle of the FcγRIIB-I232 TM helix converges to 8–20° in the membrane (Fig. 6, D–F; and Video 7), generally consistent with the observation (8°) by Vostrikov et al. (2010) on the RGWALP helix in the dioleoylphosphocholine membrane, considering that the difference of eight residues in TM length could roughly account for the distinction in the observed tilting angles of 4–12° (similar to the effect of shortening in lipid tails by one to two CH2 groups in both leaflets in the work of Vostrikov et al.).
Strikingly, the FcγRIIB-T232 TM helix finally stabilizes at a significantly more tilted posture (32–44°) after sufficient equilibration (Fig. 6, E and F; and Video 8). Statistical analysis on the tilting angles evaluated from the last 80-ns convergent trajectories (15.49 ± 6.62° vs. 37.40 ± 6.55°), indicating a significant difference (P < 1 × 10−10) in the orientation of the TM domain (Fig. 6 F). Notably, the marked difference in orientations is independent of the methods chosen for the angle estimation (see the results derived from an alternative mathematical definition of the bending angle in Fig. 6, G and H). In a further validation, a structure of the FcγRIIB-T232 TM helix tilted at 40° was taken as the initial structure, and a back amino acid change was performed to restore the I232 sequence. As expected, the TM helix gradually changed its orientation from 40° to the interval of 20–30° (Fig. 6 I).
The simulation results indicate that the FcγRIIB-T232 TM helix prefers a more inclined orientation than FcγRIIB-I232. To rigorously validate this hypothesis, we calculated the free energy profiles of both TM helices against tilting angles in the POPC membrane using the adaptive biasing force (ABF) method (Rodriguez-Gomez et al., 2004; Darve et al., 2008). To simplify the free energy calculation, the tilting angle was calculated using the alternative definition shown in Fig. 6 G. The free energy curves suggested that the FcγRIIB-I232 TM helix prefers a tilting orientation of 13–18° (Fig. 6, H and J), consistent with the equilibrium simulation. In contrast, the FcγRIIB-T232 TM helix exhibits two favored tilting orientations, one at ∼15° and the other at 35–40° (Fig. 6 J). Notably, the latter orientation is 5 kcal/mol more favorable than the former one (Fig. 6 J), which explains the further inclination of this helix observed in the equilibrium simulation, as compared with FcγRIIB-I232.
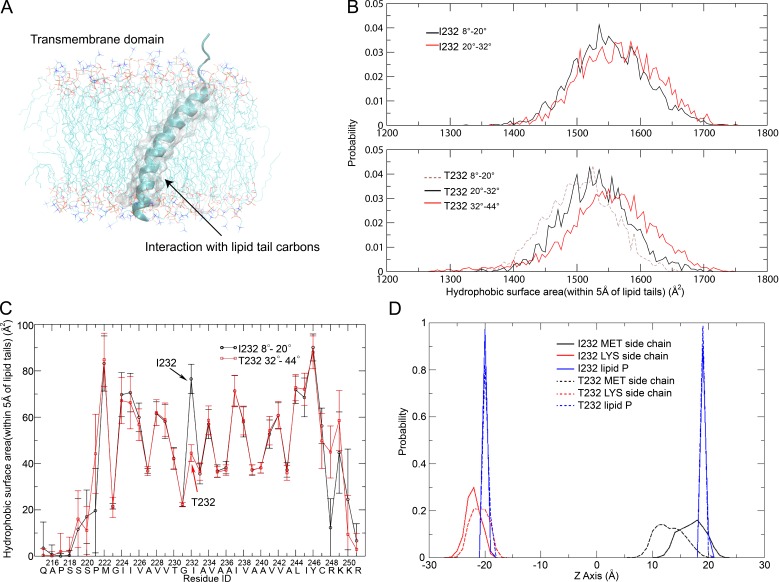
The degree of helix inclination is adjusted to maintain hydrophobic interaction between the TM domain and lipids
To explore the molecular mechanism of the distinct orientation preferences of FcγRIIB-I232 and FcγRIIB-T232, we evaluated the hydrophobic interactions between the TM helices and membrane lipids by calculating the surface area of nonpolar atoms in the helix that physically interact with the lipid carbon atoms (Fig. 7 A). The FcγRIIB-T232 TM helix exhibits a small hydrophobic interacting surface area when the helix tilts at 8–20° but shows markedly elevated interaction accompanying further inclination (Fig. 7 B). In particular, at the tilting orientations of 20–32° and 32–44°, its values agree with the ones of the FcγRIIB-I232 TM helix tilted at 8–20° and 20–32°, respectively (Fig. 7 B). These calculations imply that the FcγRIIB-T232 TM helix has to take a more tilted orientation to maintain adequate TM–lipid interaction or, in other words, to reduce any energetically unfavorable hydrophobic mismatch. The overall hydrophobic interacting surface area was then decomposed to the contributions of every single residue (Fig. 7 C). As expected, Thr232 weakens the TM–lipid hydrophobic interaction in the FcγRIIB-T232 TM helix, as compared with Ile232 in the FcγRIIB-I232 TM helix, possibly because Thr232 introduces a hydrophilic hydroxyl group within the membrane lipids (Fig. 7 C). Such a loss in hydrophobic interaction, however, can be compensated by the burial of more flanking residues in the membrane when the FcγRIIB-T232 TM helix becomes more inclined.
Figure 7.
The degree of helix inclination is adjusted to maintain the hydrophobic interaction between the TM domain and lipids. (A) Illustration of the protein–lipid hydrophobic interaction, which was evaluated by the surface area of the nonpolar atoms (shown in gray and transparent surface representation) in TM helices that fall within 5 Å of the carbon atoms in lipid tails. (B) Distributions of hydrophobic surface area for FcγRIIB-I232 and FcγRIIB-T232 TM helices at various tilting angles (8–20°, 20–32°, and 32–44°). (C) Decomposition of the hydrophobic surface area to the contribution of each residue for the FcγRIIB-I232 TM helix tilting by 8–20° (black) and the FcγRIIB-T232 TM helix tilting by 32–44° (red). Error bar represent the mean ± SD. (D) Probability distributions of positions of the side groups of Met222 (MET; black) and Lys249 (LYS; red) along the axis perpendicular to the membrane surface in the FcγRIIB-I232 (continuous lines) and FcγRIIB-T232 (dashed lines) systems, respectively. The borders of the membrane bilayers are indicated by the same distributions of phosphorus atoms (blue) in the phospholipid molecules located in the two leaflets. (A–D) All the MD results have been verified in three independent experiments, using different force fields and different lipid composition.
In summary, the residue substitution of I232T impairs the favorable hydrophobic interaction between the TM helix and membrane lipids, which is compensated in an alternative approach by increasing the tilting angle of FcγRIIB-T232. Considering the strong membrane localization effect of the positively charged residues (RKKR from residues 248 to 251) at the C terminus of the TM helix (Fig. 4 A), residues in the N-terminal juxtamembrane region are expected to become buried in the membrane, accompanying the inclination of the FcγRIIB-T232 TM helix. Accordingly, we indeed observed a drastic difference in the position of the N-terminal Met222 between FcγRIIB-I232 and FcγRIIB-T232 (Fig. 7 D).
FcγRIIB-T232 shows a longer TM helix than FcγRIIB-I232 as measured by NMR spectroscopy
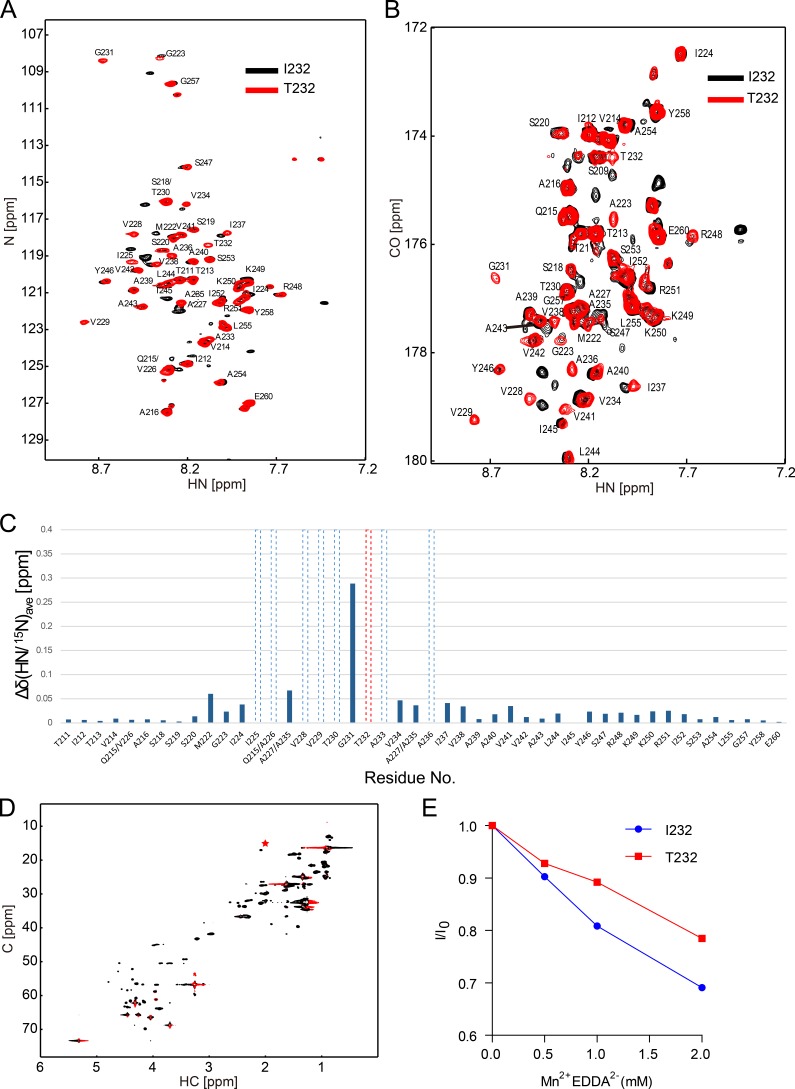
To further validate the simulation results, we assessed the structural differences of the FcγRIIB-I232 and FcγRIIB-T232 TM domains by NMR spectroscopy. The TM peptides (Pro209 to Glu260) of FcγRIIB-I232 or FcγRIIB-T232 were purified and reconstituted into a lipid bicelle system with a symmetric long-chain phospholipid bilayer rimed by short-chain detergents (Fig. S2). Both the 1H-15N heteronuclear single quantum coherence (HSQC) and 2D version of HNCO were performed to accurately quantify the backbone conformations of the TM peptides of FcγRIIB-I232 and FcγRIIB-T232 (Fig. 8). The well-dispersed 1H-15N HSQC spectra indicated that the peptides were reconstituted successfully into the bicelle (Fig. 8, A and B). The results from both methods demonstrated that >10 residues of the FcγRIIB-T232 TM domain had different chemical environments from that of the FcγRIIB-I232 TM domain. These perturbed residues are mainly located at the N terminus of the TM domain preceding the polymorphism, strongly suggesting that the structure of this region is significantly affected by the I232T polymorphism (Fig. 8 C). This observation agrees well with MD simulation.
Figure 8.
The substitution of Ile232 to Thr232 induced substantial conformational changes in the TM domain of FcγRIIB as measured by solution NMR. (A and B) Overlay of 1H-15N HSQC spectra (A) and 2D version of HNCO spectra of FcγRIIB-I232 and FcγRIIB-T232 (B). Spectra were recorded at 27°C and an 1H frequency of 600 MHz using 0.1 mM 13C, 15N–labeled FcγRIIB-I232 or FcγRIIB-T232 reconstituted in a bicelle composed of 40 mM DHPC and 12 mM POPC in 20 mM Bis-Tris, pH 6.7, and 10% D2O solution. The resonance signals of FcγRIIB-T232 were assigned by a set of triple resonance experiments and were labeled in the HSQC and HNCO spectras. ppm, parts per million. (C) The averaged chemical shift difference of the amide signals between FcγRIIB-I232 and FcγRIIB-T232 in the HSQC spectra. The polymorphism is shown by red dashed lines. The residues that had big chemical shift changes are shown by the dashed lines. (D) 1H-13C HSQC was recorded at 27°C and an 1H frequency of 900 MHz. The methyl signal (CεHε3) of the M222 sidechain is marked with a red asterisk. (E) PRE effects on the methyl signal of the M222 sidechain upon titration of Mn2+EDDA2−. The methyl signals of M222 in the presence of Mn2+EDDA2− were normalized on the signal in the absence of the PRE reagent. (A–E) The results shown are one representative of at least two independent experiments. See also Fig. S2.
The MD simulations strongly suggested that the TM helix of FcγRIIB-T232 preferred a more tilted orientation in the membrane bilayer than that of FcγRIIB-I232, which could lead to a longer membrane-embedded region of FcγRIIB-T232 (Fig. 6 F and Fig. 7 D). To validate this proposition, we applied a NMR-based paramagnetic relaxation enhancement (PRE) experiment to study the length difference of the TM domain of FcγRIIB-I232 and FcγRIIB-T232. We chose a solvent PRE reagent Mn2+EDDA2− (ethylenediamine-N, N’-diacetic acid; EDDA2− chelated Mn2+) that has been used for defining the border of the TM region in various studies and has been reported to have no biasing effect to polar residues and lipid functional groups (Lau et al., 2008; Claridge et al., 2013). Theoretically, the membrane-embedded residues of the TM domain should be insensitive to Mn2+EDDA2−, whereas the solvent-exposed residues of the extracellular and intracellular domains should be sensitive. According to the MD simulations, the FcγRIIB-T232 TM domain is more tilted, leading to the deeper membrane embedment of Met222 that is otherwise more solvent exposed in the case of FcγRIIB-I232 (Fig. 7 D). Because the methyl (CεHε3) signal of the Met222 side chain is well separated from other signals in the 1H-13C HSQC spectrum (Fig. 8 D), we measured the signal reduction of Met222-CH3 of either FcγRIIB-I232 or FcγRIIB-T232 induced by the addition of Mn2+EDDA2−. A 1H-13C HSQC–based PRE titration experiment showed that the intensity of the Met222-CH3 of both proteins decreased in correspondence to the increase of the concentration of Mn2+EDDA2−. However, the Met222-CH3 signal of FcγRIIB-T232 decreased at a significantly slower rate than that of FcγRIIB-I232 (Fig. 8 E), indicating the better membrane embedment of Met222 in FcγRIIB-T232. The PRE data agree well with the HSQC/HNCO data, showing that the structure of the N terminus of the TM domain is more affected by the I232T polymorphism.
Tilted posture of the TM helix of FcγRIIB-T232 predicts its slower lateral mobility and fewer collisions with ICs than FcγRIIB-I232
MD simulations suggested that the FcγRIIB-T232 TM helix prefers a more tilted orientation than FcγRIIB-I232 (Figs. 6 and 7). NMR measurement on the recombinant peptide of the TM helix also supported such a conclusion (Fig. 8). The current understanding in plasma membrane biology shows that the Brownian diffusion coefficient of a given TM receptor on the plasma membrane is inversely proportional to the size of the cross-sectional area of its TM domain (Kusumi et al., 2005, 2012; Gambin et al., 2006). For a single TM receptor like FcγRIIB, an inclined TM helix of FcγRIIB-T232 with the tilting angle of 35–40° (Fig. 6 F) will inevitably have an ∼4.6–13.2-fold higher cross-sectional area within the plasma membrane (Fig. 2 E) than the relatively perpendicular FcγRIIB-I232 TM helix with the tilting angle of only 13–18° (Fig. 6 F). The Brownian diffusion coefficients of FcγRIIB-I232 and FcγRIIB-T232 are therefore supposed to differ significantly on the plasma membrane. Thus, the theoretical prediction based on the heuristic Stokes-Einstein–like equation (Gambin et al., 2006) and the changes in conformation and orientation of the TM domain of FcγRIIB-T232 observed from both in silico MD simulations and experimental NMR studies are coherently consistent with the in vivo experimental observations in live immune cells, showing that FcγRIIB-T232 exhibits significantly slower lateral mobility than FcγRIIB-I232 as extensively demonstrated in Figs. 1 and 2. The difference between the lateral mobility of FcγRIIB-I232 and FcγRIIB-T232 implied that FcγRIIB-I232 may be more likely than FcγRIIB-T232 to collide with the ICs and hence to form microclusters, which explains its stronger inhibition. Indeed, Monte Carlo simulations indicated that the collision rate increased as the diffusion coefficient was ramped up, predicating that slowly mobile FcγRIIB-T232 were significantly less likely to collide with the ICs than the highly mobile FcγRIIB (Fig. 2 D).
Discussion
In this study, we mechanistically investigated the functional and structural disruption of FcγRIIB-T232. One of the primary findings of our study is that the I232T polymorphism causes significant changes in both conformations and orientations of the TM domain of FcγRIIB. In contrast, the I232T polymorphism aggravates the bending of the TM helix by engaging the Thr232 side chain to form bifurcated hydrogen bonds, consistent with previous studies showing that Ser and Thr present in TM helices frequently cause helix bending (Ballesteros et al., 2000; Feldblum and Arkin, 2014). However, the I232T polymorphism triggers the TM domain of FcγRIIB to adopt a significantly more inclined posture. Although early studies on the WALP (peptides composed of multiple Trp, Ala, and Leu residues) family helix using NMR spectroscopy (Vostrikov et al., 2010) showed that the TM helix can automatically adjust its degree of inclination to adapt the changes in lipid composition, to our knowledge, it has never been reported that a single-residue substitution can cause the substantial change of tilting the angle of a TM helix (from ∼15° to ∼40°) when all the other conditions (e.g., the lipid composition) are identical. Helix bending and inclination can jointly increase the cross-sectional area of the TM domain, which provides the molecular-level explanation for the reduction in the lateral mobility induced by this polymorphism. Moreover, the simulation results were successfully confirmed by NMR measurements: (a) the observation of more significant perturbation on the microenvironment for the N-terminal residues than C-terminal ones is consistent with the prediction based on simulation, considering that helix bending occurs at the N-terminal half and that, during the helix inclination, the positively charges residues (KRRK) restrict the movement of C-terminal residues by favorably interacting with the negatively charged lipid head group, and (b) NMR-based PRE measurement also captured deeper membrane burial of Met222 in FcγRIIB-T232 than in FcγRIIB-I232, providing direct experimental evidence for the simulation observation of enhanced inclination of the TM helix induced by the I232T polymorphism. Our further mechanistic investigation implied that the TM helix of FcγRIIB-T232 uses a strategy of enhancing inclination to maintain hydrophobic interaction between the TM domain and lipids because the Ile-to-Thr polymorphism introduces a hydrophilic hydroxyl group within the membrane bilayer. We proposed that the helix inclination could drag juxtamembrane residues into the membrane to compensate the loss of hydrophobic interaction caused by the introduction of a polar group. This proposition is consistent with the well-accepted perspective that the TM helices of integral membrane proteins shall be perfectly oriented for the purpose of maintaining optimal interactions with the lipids (Strandberg et al., 2004; Vostrikov et al., 2010).
The second key finding of this study is that the structural changes in the TM domains of FcγRIIB-T232 dramatically impairs its Brownian lateral mobility on the plasma membrane, and as a consequence, all these influences help explain the functional disruption of FcγRIIB-T232 compared with the case of FcγRIIB-I232. TM receptors on the plasma membrane follow unceasing lateral motion (or termed as Brownian diffusion in a 2D surface; Kusumi et al., 2005, 2012). The lateral mobility allows a receptor to frequently communicate with other molecules including ligands in the extracellular space, co-receptors on the membrane, and downstream signaling molecules in the cytosol. Thus, the linkage between this biophysical property and the function of membrane receptors has attracted more and more attention in biological studies (Fujiwara et al., 2002; Kusumi et al., 2005, 2012; Suzuki et al., 2007; Kasai et al., 2011; Jaumouillé et al., 2014). Recent advances in membrane biology have demonstrated that the plasma membrane is compartmentalized into hundreds of nanometer-sized microdomains by a membrane-skeleton fence and an anchored TM-protein picket (Kusumi et al., 2005, 2012). Such a comprehensive view determines that the Brownian diffusion coefficient of a given receptor on the plasma membrane shall be inversely proportional to the size of the cross-sectional area of its TM domain. Indeed, the empirical study by Gambin et al. (2006) summarized a heuristic Stokes-Einstein–like equation to mathematically quantify influence of the dimension of the cross-sectional area of the TM domain of a receptor on its lateral mobility (D ∝ 1/R). Here, we consistently found that FcγRIIB-T232 performed much slower Brownian diffusion than FcγRIIB-I232. These results are consistent with the general understanding of the current plasma membrane biology (Kusumi et al., 2005, 2012; Gambin et al., 2006), considering that the TM helix of FcγRIIB-T232 tilting by 35–40° is theoretically predicted to have ∼4.6–13.2-fold more cross-sectional area within the plasma membrane than that of FcγRIIB-I232 with tilting by 13–18° (Fig. 6 F and Fig. 2 E). Thus, these live-cell imaging experiments validated the calculations from in silico MD simulations and the measurements from experimental NMR experiments.
The third and the most important finding of this study is that reduction in the lateral mobility of FcγRIIB-T232 crucially correlates with its function loss. FcγRI and BCR are two of the main activating immune receptors subjected to the inhibition of FcγRIIB. According to the literature, they show strong affinities with the target ligands. The reported dissociation constant between FcγRI and the Fc portion of IgG1 is 7–9 × 10−9 M (Lu et al., 2015). The dissociation constant between BCR and the antigen would be within a wide range from 10−6 to 10−10 M (Batista and Neuberger, 2000). In contrast, human FcγRIIB is well known to recognize the Fc portion of human IgG antibody with weak affinity (the dissociation constant ≈ 0.9–1 × 10−6 M; Mimura et al., 2001). Therefore, when FcγRI or BCR molecules on immune cells (e.g., monocytes for FcγRI and B cells for BCR) encounter the antigen and IgG antibody ICs, these two activating receptors instantly bind with ICs to form IC microclusters, thus initiating the signaling cascades. Defeated in the initial binding competition, FcγRIIB has to diffuse in a random manner until it collides with the Fc portion of clustered IgG with avidity effects in preformed IC microclusters, which finally enriches these molecules to initiate their inhibitory function. The high mobility of FcγRIIB is thus essential for this process, and the faster floating FcγRIIB-I232 is more likely to collide with the Fc portion of the IgG antibodies in ICs through random diffusion than the slower FcγRIIB-T232. As a consequence, FcγRIIB-T232 may fail to perform the inhibitory function because of the lack of sufficient collision between FcγRIIB-T232 and the Fc portion of IgG. Our hypothesis that the high lateral mobility of FcγRIIB-I232 is associated with the potent inhibitory function of FcγRIIB is supported by the experiments using photoactivatable antigen caged NP. In those experiments, caged NP can only be recognized by B1-8-BCR–expressing B cells upon photoactivation. Indeed, if sufficient amount of responding time was provided in advance for FcγRIIB-T232 to diffuse and interact with the ICs before triggering the interaction between caged NP and B1-8-BCR, FcγRIIB-T232 could inhibit the recruitment of B1-8-BCRs into the B cell immunological synapse as potently as FcγRIIB-I232.
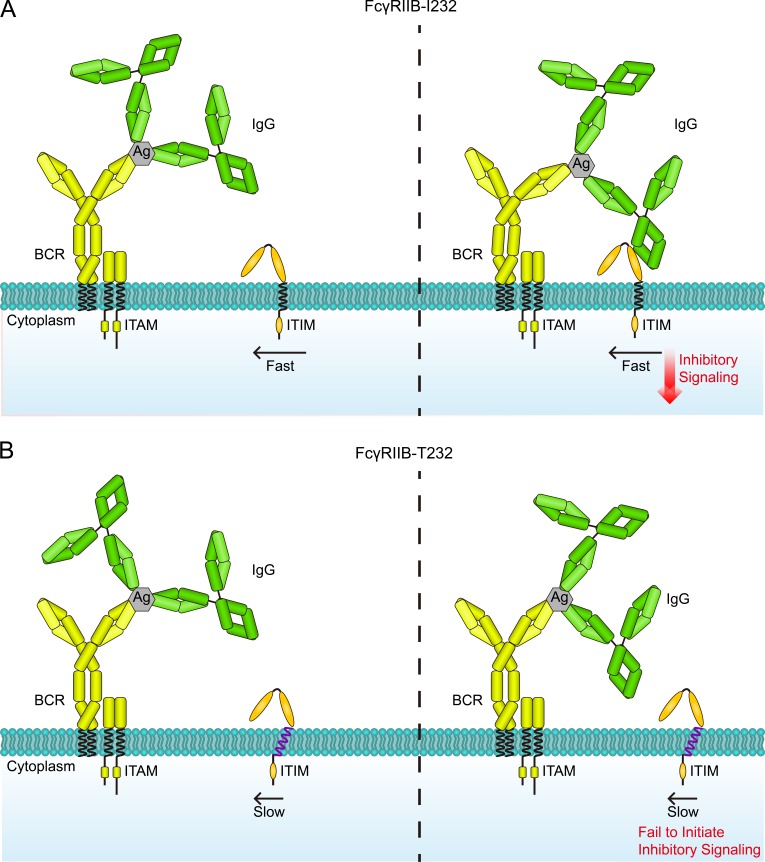
Based on all these data and thoughts, we suggest the catch me if you can model as a new mechanistic explanation of how a single–amino acid change from Ile232 to Thr232 in the TM domain results in functional disruption of the lupus-associated FcγRIIB-T232 polymorphism (Fig. 9, A and B). Our model is originally inspired by the observations that FcγRIIB-T232 showed impaired accumulation in the synapse at the early time point after activation (Xu et al., 2014) and is later supported by the observation in this study that a chimeric FcγRIIB with the TM domain replaced by that of a fast floating molecule CD86 restored the inhibitory function and the observation that FcγRIIB-T232 could circumvent its defect in lateral mobility and efficiently execute its inhibitory function as long as a sufficient amount of responding time is provided for FcγRIIB-T232 to interact with the ICs. Thus, the inhibitory function of FcγRIIB is strikingly found to depend on its lateral mobility, a property that is known to be mainly determined by the TM domain of the receptor rather than by the extracellular or cytoplasmic domains (Kusumi et al., 2005, 2012). Notably, FcγRIIB is well known as a monomeric TM receptor that does not function through forming complexes with other plasma membrane receptors (Jaumouillé et al., 2014), a fact that lays the foundation for our prediction on lateral mobility based on a cross-sectional area of the TM domain. Our model is also supported by the PALM-based superresolution imaging and Monte Carlo simulation analyses in this study that the lack of sufficient lateral mobility of FcγRIIB-T232 hinders its successful collision and enrichment in the IC microclusters and the subsequent assembly of larger-sized FcγRIIB nanoclusters in response to antigen and IgG antibody ICs. Indeed, the formation of potent FcγRIIB microclusters and their concomitant colocalization with BCR microclusters are prerequisites for FcγRIIB to block B cell activation (Liu et al., 2010c; Xu et al., 2014). Early studies reported that the function loss of FcγRIIB-T232 arises from its reduced affinity with the lipid rafts composed of sphingolipids and cholesterols within the plasma membrane upon IC activation (Floto et al., 2005; Kono et al., 2005). Because the lipid rafts are usually coincident with the microdomains to which immune receptors (e.g., BCR and FcγRI) would be translocated to in response to recognition of ICs, they would also be the places that FcγRIIB tries to reach through random diffusion. In this perspective, our catch me if you can model does not contradict this theory. Instead, we propose that our catch me if you can model could help explain the lack of stability of FcγRIIB-T232 within the microdomains enriched by sphingolipids and cholesterols.
Figure 9.
A catch me if you can model to explain the function loss of FcγRIIB caused by the lupus-associated polymorphism I232T. (A and B) When B cells encounter the antigen (Ag) and IgG antibody ICs, BCR first interacts with antigens favorably to form BCR-IC microclusters, so as to initiate the BCR signaling cascades. Because of the weak affinity between FcγRIIB and the Fc portion of human IgG antibody, only the highly mobile FcγRIIB-I232 (A) could catch the IgG Fc portion in the IC microclusters through random diffusion, and its subsequent enrichment allows proper functioning by initiation of its inhibitory signaling. In contrast, the I232T polymorphism significantly impairs the mobility of FcγRIIB-T232 (B), which causes function loss by substantially reducing its chance to collide with the IgG Fc portion in the IC microclusters. ITAM, immunoreceptor tyrosine-based activation motif; ITIM, immunoreceptor tyrosine-based inhibition motif.
Conclusively, in the catch me if you can model, the I232T polymorphism enforces the inclination of the TM domain and thereby reduces the lateral mobility of FcγRIIB-T232, which is critical for FcγRIIB molecules to catch the Fc portion of IgG antibodies in the IC microclusters to execute inhibitory function at the right time in the right place.
Materials and methods
Cells, antigens, antibodies, and chemical reagents
A20II1.6, ST486, and THP-1 cell lines were gifts for laboratory studies from S.K. Pierce (National Institute of Allergy and Infectious Diseases, Bethesda, MD), all of which were originally purchased from ATCC. 293T was purchased from cellbank (Chinese Academy of Sciences). The biotin-conjugated goat F(ab’)2 anti–mouse IgG Fc portion, biotin-conjugated goat anti–mouse IgG Fc portion, goat anti–anti–mouse Igκ, goat anti–human IgM + IgG (H+L), and biotin-conjugated goat F(ab’)2 anti–human IgM + IgG (H+L) were purchased from Jackson ImmunoResearch Laboratories, Inc. Anti–MHC-I (H-2Db), FITC-conjugated mouse anti–human CD14 and FITC-conjugated mouse anti–human CD19 was purchased from eBioscience. A human FcγRIIB (no cross-reaction to FcγRIIA)-specific mAb and mouse–human chimeric ch2B6-N297Q (Rankin et al., 2006) were a gift from S. Koenig and T.J. Mayer (Macrogenics, Rockville, MD). The biotinylation of antibodies was performed using EZ-Link Sulfo-NHS-LC-Biotin kits (Thermo Fisher Scientific) following manufacturer’s protocols. Conjugations of antibodies with Alexa Fluor 568 or 647 were performed using Alexa Fluor mAb labeling kits (Molecular Probes) following the manufacturer’s protocols. The digestion of antibodies into Fab or F(ab’)2 fragments was done using Fab or F(ab’)2 micro preparation kits following a protocol in our published studies (Li et al., 2004; Liu et al., 2010a,b,c, 2012). Primary B cells and monocytes from the peripheral blood of SLE patients were collected from participant patients enrolled in the Department of Rheumatology and Immunology, Peking University People’s Hospital. All participants signed informed consent to donate their blood samples for research. The human experimental guidelines were approved by the Medical Ethics Committee of Peking University People’s Hospital (approval no. 2014PHB116-01).
Plasmid construction and cell transfection
The pMSCVpuro plasmids containing human FcγRIIB WT or I232T were provided by Z.I. Honda (Tokyo University, Tokyo, Japan; Kono et al., 2005). The mEos3.2 cDNA was provided by P. Xu (Chinese Academy of Sciences, Beijing, China). Following Gibson assembly protocols (Gibson et al., 2009), we subcloned FcγRIIB cDNA into a pEYFP-N1 plasmid or pHAGE plasmid containing mEos3.2 cDNA. Similarly, the TM domain of FcγRIIB was swapped with the TM domain of CD86 following Gibson assembly protocols. Stable A20II1.6 cell sublines expressing FcγRIIB-I232, FcγRIIB-T232, or FcγRIIB-CD86-TM were acquired by electroporation-based transfection, G418 (1 mg/ml) selection, and flow cytometry–based cell sorting. Multiple rounds of cell sorting were used to get cell sublines with similar expression level of FcγRIIB. Similarly, stable ST486 expressing a comparable amount of FcγRIIB-I232 or FcγRIIB-T232 was acquired by three vector (pHAGE, psPAX2, and pMD2.G)–based lentivirus infection, followed by multiple rounds of cell sorting.
Modeling the TM domain and the equilibrium MD simulations
The sequence boundary of the TM domain of FcγRIIB was predicted using the TMHMM server (Krogh et al., 2001). The representative structure for the TM domain (residues 215–251) was then modeled by comparative modeling using the program MODELLER 9.10 (Šali and Blundell, 1993; Fiser and Sali, 2003), with the N and C termini blocked by acetyl and N-methyl-amide groups, respectively. In the simulations, the modeled TM domain structure was inserted into a lipid bilayer composed of 101 POPC molecules. The system was subsequently solvated by TIP3P water (6,393 molecules) and was neutralized by 150 mM NaCl. The polymorphism was achieved using the Mutator plugin of Visual Molecular Dynamics (version 1.9.1; Šali and Blundell, 1993). All simulations were performed by the Nanoscale Molecular Dynamics program (version 2.9; Phillips et al., 2005) using the CHARMM36 force field with the CMAP correction (MacKerell et al., 1998). The simulations were conducted in an isothermal-isobaric ensemble, with the pressure and temperature held at 1 atm and 310 K using a Langevin thermostat and Nose-Hoover Langevin piston, respectively (Martyna et al., 1994; Feller et al., 1995). The van der Waals interactions were cutoff at 12 Å with a smooth switch at 10 Å. The electrostatic interactions were evaluated using the particle mesh Eward method with periodic boundary conditions applied in all directions (Essmann et al., 1995).
All systems were first preequilibrated in the following four steps: (1) The system was minimized by 1,000 steps and equilibrated for 0.5 ns with all atoms fixed except the lipid tails; (2) The protein atoms were restrained at the initial position by a force constant of 1 kcal/mol/Å2, and the system was equilibrated for 0.5 ns after 1,000 steps of minimization; (3) all constraints were removed, and the system was equilibrated for 0.5 ns; and (4) the system was equilibrated for an additional 0.5 ns with the area of the membrane held as a constant. After the standard preequilibration, all systems were further preequilibrated for 20 ns with a time step of 1 fs.
In the following productive simulations, the time step was switched to 2 fs, and the productive simulations were conducted for 200 ns, with the snapshots in the last 80 ns adopted to evaluate the probability distributions for the tilting angle of the TM domain. In a control simulation in which a snapshot of FcγRIIB-T232 at 200 ns was mutated back to FcγRIIB-I232 to observe the responsive changes in the tilting angle, the productive simulations were performed for 9 ns after 3 ns of preequilibration. In the simulations, the tilting angle was calculated as the scalar angle between two axes: the mean normal vectors of the outer and inner lipid leaflets (defined by all phosphorus atoms) as well as the central axis of the TM helix (defined by all backbone atoms of residues 224–245).
ABF simulation
The ABF method was used to evaluate the potential of mean force (PMF; Darve and Pohorille, 2001; Darve et al., 2002, 2008; Rodriguez-Gomez et al., 2004). The C terminus of the TM domain (the Cα atom of Arg248) was restrained in the x-y plane (parallel to the membrane surface) using a flat-bottom potential (no penalties within 0.3 Å around the center position in both the x and y directions) with a force constant of 100 kcal/mol/Å2. The reaction coordinate was chosen as the scalar angle of three groups: the center of phosphorus atoms in upper lipid leaflet, the center of carbonyl carbon atoms in residues 244–247 that represent the C terminus of the TM helix, and the center of all Cα atoms in residues 224–245 that represent the geometric center of the TM helix. The reaction coordinate was divided into six windows with each one spanning 5.5°. The neighboring windows had an overlap of 0.5° to guarantee smooth splicing. A force constant of 100 kcal/mol/Å2 was used to retrain the tilting angle within each window. For the FcγRIIB-I232 and FcγRIIB-T232 systems, the initial structure for each window was chosen from the equilibrium simulations. Before the simulation, all systems were translated so that the Cα atom of Arg248 was positioned at the origin. All systems were then preequilibrated for >1 ns before the ABF calculation. The PMF was calculated with a width of 0.1° in each window, and 1,000 samples were used to preestimate the biasing force in each bin. The time step of 1 fs was used in all ABF simulations. The PMF profiles were updated every 4 ns. The calculation was believed as convergent both when the two latest PMF profiles differed by <0.5 kcal/mol and when the maximum and minimum sampling counts within each window differed by less than fivefold in the last 4-ns simulations. The overall PMF profiles were finally derived by splicing the curves from all windows.
FcγRIIB TM peptide expression and purification
FcγRIIB TM WT and I232T peptides were expressed as a TrpLE fusion protein in Escherichia coli BL21 (DE3) cells. C247S substitution was incorporated to avoid peptide cross-linking. The fusion protein was expressed in inclusion bodies, which were dissolved in wash buffer containing 50 mM Tris-HCl, pH 8.0, 6 M guanidine hydrochloride, 200 mM NaCl, and 1% Triton X-100, and then applied onto a Ni-nitrilotriacetic acid affinity column (QIAGEN). The FcγRIIB TM peptide was cleaved from the fusion protein at the Asp-Pro site in 10% formic acid containing 6 M guanidine-HCl. The digest was dialyzed to water, lyophilized, and loaded on a column (300SB-C3; ZORBAX) in 70% TFA. A linear gradient of 40–100% buffer B (acetonitrile and 0.2% TFA) in 60 min was run to purify the target peptide.
NMR sample preparation
FcγRIIB TM peptide was reconstituted in POPC bicelle solution containing 400 mM DHPC (1, 2-dihexanoyl-sn-glycero-3-phosphocholine) and 120 mM POPC (Q = 0.3) by repeated vortex, freezing, and thawing until a clear solution was obtained. The bicelles of this q-value have been reported to form magnetically isotropic units that exhibit planar geometry (Prosser et al., 2006) and are well suited for the study of single-span TM proteins (Lau et al., 2008). Bis-Tris buffer solution, pH 6.7, D2O, and H2O were then added into the sample. The NMR sample for chemical shift assignments contained 0.3 mM 13C, 15N-labeled TM domain peptide of FcγRIIB, 20 mM Bis-Tris, pH 6.7, 200 mM DHPC, 60 mM POPC, and 10% D2O/90% H2O.
NMR spectroscopy
NMR experiments were conducted at 27°C on ASC 600 and 800 MHz (Agilent Technologies) and AVANCE III 900 MHz (Bruker) spectrometers equipped with cryogenic probes. Sequence-specific assignment of the backbone chemical shifts was accomplished using triple resonance experiments, including HNCA, HNCACB, CBCA(CO)NH, HNCO, and 15N-edited NOESY-HSQC with a mixing time of 120 ms. In PRE experiments, a 2D 1H-13C HSQC spectrum was recorded with the addition of 0, 0.5, 1, and 2 mM of Mn2+EDDA2−. Mn2+EDDA2− was freshly prepared as previously described (Lau et al., 2008). The intensity of each spectrum was analyzed by NMR View (Delaglio et al., 1995) and KUJIRA (Kobayashi et al., 2007), and then the intensity ratio without Mn2+EDDA2− (I/I0) was calculated for quantifying the signal broadening caused by PRE. The chemical shift difference per residue of FcγRIIB-I232 and FcγRIIB-T232 in the HSQC spectra was calculated as Δδ(HN/15N) = [(δHN)2 + (0.154 × δN)2]1/2 (Ayed et al., 2001).
FRAP measurements
A20II1.6 or ST486 B cells expressing YFP tag were cultured on the glass of Lab-Tek chambers (Thermo Fisher Scientific). Subsequently FRAP measurements were performed at the indicated temperature on a confocal microscope (LSM510; ZESIS) using a 100× oil objective and the 514-nm line of a Kr/Ar laser. Photobleaching was performed by illuminating the region of interest only with the 514-nm laser at 100% power. Recovery of the fluorescence intensity was then recorded at an imaging rate of 93 ms per frame. For each set of conditions, >12 cells were collected and averaged before analysis. FRAP datasets were analyzed by nonlinear least squares fitting to the Soumpasis equation (Soumpasis, 1983):
Preparation of antigen-containing PLBs
PLBs were prepared following our published protocol (Liu et al., 2010a,b; Wan and Liu, 2012), which biotinylated antibodies against BCR, and FcγRIIB was attached through streptavidin to the planar fluid lipid bilayers. In brief, biotin liposomes were prepared by sonication of 1,2-dioleoyl-sn-glycero-3-phosphocholine and 1,2-dioleoyl-sn-glycero-3-sphosphoethanolamine-cap-biotin (Avanti Polar Lipids, Inc.) in a 25:1 molar ratio in PBS at a lipid concentration of 5 mM. Lab-Tek chambers (Thermo Fisher Scientific) were prepared with replaced nanostrip-washed coverslips. The coverslips were incubated with 0.1 mM biotin liposomes in PBS for 20 min. After washing with 10 ml PBS, the PLB was incubated with 30 nM streptavidin for 15 min, and excessive streptavidin was removed by washing with 10 ml PBS. The streptavidin-containing PLBs were incubated for 15 min with 20 nM biotinylated F(ab’)2 anti–MHC-I. The excessive antigen was removed by washing. Next, PLB was blocked with 5% BSA in PBS at 37°C for 30 min and washed thoroughly before usage.
For caged NP experiments, the nanostrip-washed coverslips were incubated with 0.15 mM biotin liposomes in PBS for 20 min. After washing with 10 ml PBS, the PLB was incubated with 40 nM streptavidin for 15 min, and excessive streptavidin was removed by washing with 10 ml PBS. The following steps were performed in a dark room for the protection of caged-NP molecules. The streptavidin-containing PLBs were incubated for 15 min with 100 nM biotinylated caged NP and 20 nM biotinylated anti-FcγRIIB mAb (clone AT10). The excessive molecules were removed by washing. Next, PLB was blocked with 5% BSA in PBS at 37°C for 30 min and washed thoroughly before usage.
Single-molecule imaging of FcγRIIB
To evaluate the Brownian motility of FcγRIIB variants with different TM domains, A20II1.6 B cells expressing YFP (or mEos3.2)-tagged FcγRIIB-I232, FcγRIIB-T232, or FcγRIIB-CD86-TM were imaged following a YFP-based single-molecule imaging protocol (Xu et al., 2010). To image the Brownian motility of FcγRIIB on primary cells, B cells or monocytes from human peripheral blood mononuclear cells were labeled with an Alexa Fluor 647–conjugated Fab fragment of human FcγRIIB-specific mAb, ch2B6-N297Q, in combination with either FITC-conjugated anti-CD19 or FITC-conjugated anti-CD14. The Brownian motility of IgM-BCR was performed by labeling the primary B cells with an Alexa Fluor 647–conjugated Fab fragment goat anti–human IgM Fc5μ. The cells were generally suspended in PBS with 1% FBS and were placed on coverslips that were coated with anti–MHC-I antibodies. Unless specifically indicated, the single-molecule images were acquired by Olympus IX-81–based TIRF microscopy at 30°C with 5% CO2 supplies following our published protocols (Tolar et al., 2009). In brief, a subregion including an intact cell contact area in the electron-multiplying charged-coupled device chip (512 × 512 pixels) was used to achieve a recording rate of 20 ms per frame. Single FcγRIIB or BCRs were captured on 200 frames in streamline acquisition mode. Single-molecule tracking was performed using Matlab (Mathworks) code following our published protocols (Crocker and Grier, 1996; Douglass and Vale, 2008). MSD and short-range diffusion coefficients for each molecule trajectory (D0; based on time intervals of 20 ms) were calculated from positional coordinates following our published protocols (Douglass and Vale, 2008). The MSD plot was mathematically fitted into a confined diffusion model by an exponential function to acquire the size of the confined area. PICS analysis was performed as described in detail in the Supplemental materials section of Xu et al. (2010).
PALM imaging of FcγRIIB on the plasma membrane of B cells
To acquire the PALM images of FcγRIIB at the quiescent state, A20II1.6 B cells expressing mEos3.2-tagged FcγRIIB-I232 or FγRIIB-I232T were placed on PLBs tethering biotin-conjugated F(ab’)2 anti–mouse MHC-1 at 37°C with 5% CO2 supplies for 10 min before fixation with 4% paraformaldehyde. To acquire the PALM images of FcγRIIB at the activated state, A20II1.6 B cells expressing mEos3.2-tagged FcγRIIB-I232 or FγRIIB-I232T were placed on PLBs tethering biotin-conjugated Alexa Fluor 647 goat IgG anti–mouse Igκ (SouthernBiotech) against both BCR and FcγRIIB. TIRF images were captured using a microscope (IX-81; Olympus) equipped with a TIRF port, electron-multiplying charged-coupled device camera (iXon + DU-897D; Andor Technology), a 100× 1.49 NA objective TIRF lens (Olympus), and 405-, 488-, 561-, and 647-nm lasers (sapphire laser; Coherent). A continuous 405-nm laser with a 561-nm laser irradiation was used to convert the mEos3.2 molecules. Acquisition was controlled by Metamorph software (Molecular Devices) and with an exposure time of 30 ms.
In the analyses of PALM images, peaks in single-molecule images were first identified by using Insight3 software that was provided by X. Zhuang (Harvard University, Cambridge, MA) and B. Huang (University of California, San Francisco, San Francisco, CA). Drift correction and superresolution image reconstruction were done using custom-written Matlab scripts from Y. Sun (Peking University, Beijing, China). Ripley’s K-derived H function (Kiskowski et al., 2009), quantification of the enrichment of FcγRIIB inside immune-complex microclusters, nanocluster size, and the number of nanoclusters were processed according to protocols previously described (Zhang et al., 2006; Owen et al., 2010; Mattila et al., 2013).
Calcium influx measurement by flow cytometry
A20II1.6 expressing the same amount of YFP-tagged FcγRIIB-I232, FcγRIIB-T232, or FcγRIIB-TM-CD86 was used for intercellular calcium influx measurement. In brief, the cells were incubated with a ratiometric indicator INDO-1 following the instructions and then suspended in HBSS buffer with 1% FBS and 1 mM Ca2+. Intracellular calcium flux was measured by flow cytometry (LSRII; BD) stimulating with 5 µg/ml goat IgG or F(ab’)2 anti–mouse Igκ and analyzed by FlowJo software (Tree Star).
Calculation for the ratio of the cross-sectional area from the FcγRIIB-T232 TM helix to that from FcγRIIB-I232
As depicted in the illustrative diagram in Fig. 2 E, the TM domain of FcγRIIB is represented as a rectangle colored in gray, whereas the plasma membrane is colored in cyan with the borders indicated by black solid lines. The red solid bars represent the projections of the TM helices within the plane of the plasma membrane, which are equal to h × tan(α), where h is the thickness of the membrane (denoted as black dashed lines) and α is the tilting angle of a TM helix. By assuming that the TM helices are freely rotatable within the membrane and by neglecting the difference between the thickness of the membrane in the FcγRIIB-I232 and FcγRIIB-T232 systems, the cross-sectional area is supposed to be proportional to [h × tan(α)]2. According to the free energy calculations in this study, the FcγRIIB-I232 TM helix prefers the tilting orientation of 13–18°, whereas the FcγRIIB-T232 TM helix is the most stable at the tilting angle of 35–40° (Fig. 6 J). Therefore, the ratio of cross-sectional area from the FcγRIIB-T232 TM helix to that from the FcγRIIB-I232 shall fall in the interval between [tan(35°)/tan(18°)]2 and [tan(40°)/tan(13°)]2 or [4.6, 13.2].
Online supplemental material
Fig. S1 shows the MD simulation strategy for the TM domain helix of FcγRIIB-I232 or FcγRIIB-T232 in the POPC bilayer. Fig. S2 shows the preparation of TM peptides for the NMR spectrum. Videos 1 and 2 are representative videos indicating the dynamic recovery of FcγRIIB-I232 or FcγRIIB-T232 within the photobleached area on the plasma membrane of A20II1.6 B cells. Videos 3 and 4 are representative videos indicating the dynamic recovery of FcγRIIB-I232 or FcγRIIB-T232 within the photobleached area on the top flat area of the plasma membrane of ST486 human B cells. Videos 5 and 6 are representative videos indicating the SPT of FcγRIIB-I232 or FcγRIIB-T232 molecules on the plasma membrane of ST486 human B cells. Videos 7 and 8 are representative videos of the trajectory of FcγRIIB-I232 or FcγRIIB-T232 TM helices in POPC.
Supplementary Material
Acknowledgments
We thank Dr. Susan K. Pierce, Dr. Scott Koenig, Dr. Timothy J. Mayer, Dr. Pingyong Xu, Dr. Z.I. Honda, Dr. Xiaowei Zhuang, and Dr. Bo Huang for experimental materials and image analyzing tools. We thank Dr. Kenneth G. C. Smith (Cambridge Institute for Medical Research and the Department of Medicine, University of Cambridge, Cambridge, England, UK) and Dr. Hai Qi (School of Medicine, Tsinghua University, Beijing, China) for helpful discussions.
W. Liu is supported by funds from the Chinese Ministry of Science and Technology (MOST; 2014CB542500 and 2014AA020527), National Natural Science Foundation of China (NSFC; 81361120384, 31270913, 81621002, and 81422020), One-Thousand-Youth-Talents program of the Chinese Central Government (2069999-3), and Tsinghua University Initiative Scientific Research Program (20131089279). L. Xu is funded by NSFC grants (31501140) and a postdoctoral fellowship from the Center for Life Sciences of Tsinghua University. H. Gong is funded by NSFC grants (31470033 and 31321062). Chenqi Xu is funded by Chinese Academy of Sciences grants (Strategic Priority Research Program grant nos. XDB08020100 and KSCX2-EW-J-11), NSFC grants (31370860 and 31425009), and MOST (2011CB910901 and 2012CB910804). H. Li is funded by MOST (2014CB541903) and NSFC (31470734).
The authors declare no competing financial interests.
Author contributions: W. Liu conceived the project. L. Xu, Chenguang Xu, and X. Gu performed biochemical and imaging experiments. Chenguang Xu, L. Xu, H. Zhang, Y. Fang, and Z. Shen performed superresolution imaging experiments. H. Gong and M. Xia performed the MD simulation and analyzed the data. Chenqi Xu, H. Li, and J. Guo performed and analyzed NMR experiments. X. Sun, Z. Li, L. Xu, and Z. Hu performed the epidemiological experiments and analyzed the clinical data. Y. Sun and B. Xue helped with the superresolution analyses. J. Wang helped with photoactivatable antigen experiments. H. Xie and H. Zhang helped with Monte Carlo simulations of collision. J. Yi, T. Meckel, and Y.-H. Chen provided help in sample preparations and data analyses. W. Liu, H. Gong, and Chenqi Xu wrote the manuscript.
Footnotes
Abbreviations used:
- 2D
- two-dimensional
- ABF
- adaptive biasing force
- BCR
- B cell receptor
- CPD
- cumulative probability distribution
- EDDA
- ethylenediamine-N, N’-diacetic acid
- FRAP
- fluorescence recovery after photobleaching
- HSQC
- heteronuclear single quantum coherence
- IC
- immune complex
- MD
- molecular dynamics
- MSD
- mean square displacement
- NMR
- nuclear magnetic resonance
- NP
- 4-hydroxy-3-nitrophenylacetyl
- PALM
- photoactivated localization microscopy
- PICS
- particle image correlation spectroscopy
- PLB
- planar lipid bilayer
- PMF
- potential of mean force
- POPC
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- PRE
- paramagnetic relaxation enhancement
- SLE
- systemic lupus erythematosus
- SNP
- single-nucleotide polymorphism
- SPT
- single-particle tracking
- TIRF
- total internal reflection fluorescence
- TM
- transmembrane
References
- Ayed A., Mulder F.A., Yi G.S., Lu Y., Kay L.E., and Arrowsmith C.H.. 2001. Latent and active p53 are identical in conformation. Nat. Struct. Biol. 8:756–760. 10.1038/nsb0901-756 [DOI] [PubMed] [Google Scholar]
- Ballesteros J.A., Deupi X., Olivella M., Haaksma E.E., and Pardo L.. 2000. Serine and threonine residues bend α-helices in the χ1 = g− conformation. Biophys. J. 79:2754–2760. 10.1016/S0006-3495(00)76514-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista F.D., and Neuberger M.S.. 2000. B cells extract and present immobilized antigen: implications for affinity discrimination. EMBO J. 19:513–520. 10.1093/emboj/19.4.513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Z.T., Tsuchiya N., Kyogoku C., Ohashi J., Qian Y.P., Xu S.B., Mao C.Z., Chu J.Y., and Tokunaga K.. 2004. Association of Fcγ receptor IIb polymorphism with susceptibility to systemic lupus erythematosus in Chinese: a common susceptibility gene in the Asian populations. Tissue Antigens. 63:21–27. 10.1111/j.1399-0039.2004.00142.x [DOI] [PubMed] [Google Scholar]
- Claridge J.K., Aittoniemi J., Cooper D.M., and Schnell J.R.. 2013. Isotropic bicelles stabilize the juxtamembrane region of the influenza M2 protein for solution NMR studies. Biochemistry. 52:8420–8429. 10.1021/bi401035m [DOI] [PubMed] [Google Scholar]
- Clatworthy M.R., Willcocks L., Urban B., Langhorne J., Williams T.N., Peshu N., Watkins N.A., Floto R.A., and Smith K.G.. 2007. Systemic lupus erythematosus-associated defects in the inhibitory receptor FcγRIIb reduce susceptibility to malaria. Proc. Natl. Acad. Sci. USA. 104:7169–7174. 10.1073/pnas.0608889104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker J.C., and Grier D.G.. 1996. Methods of digital video microscopy for colloidal studies. J. Colloid Interface Sci. 179:298–310. 10.1006/jcis.1996.0217 [DOI] [Google Scholar]
- Darve E., and Pohorille A.. 2001. Calculating free energies using average force. J. Chem. Phys. 115:9169–9183. 10.1063/1.1410978 [DOI] [Google Scholar]
- Darve E., Wilson M.A., and Pohorille A.. 2002. Calculating free energies using a scaled-force molecular dynamics algorithm. Mol. Simul. 28:113–144. 10.1080/08927020211975 [DOI] [Google Scholar]
- Darve E., Rodríguez-Gómez D., and Pohorille A.. 2008. Adaptive biasing force method for scalar and vector free energy calculations. J. Chem. Phys. 128:144120 10.1063/1.2829861 [DOI] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., and Bax A.. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 6:277–293. 10.1007/BF00197809 [DOI] [PubMed] [Google Scholar]
- Douglass A.D., and Vale R.D.. 2008. Single-molecule imaging of fluorescent proteins. Methods Cell Biol. 85:113–125. 10.1016/S0091-679X(08)85006-6 [DOI] [PubMed] [Google Scholar]
- Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., and Pedersen L.G.. 1995. A smooth particle mesh Ewald method. J. Chem. Phys. 103:8577–8593. 10.1063/1.470117 [DOI] [Google Scholar]
- Feldblum E.S., and Arkin I.T.. 2014. Strength of a bifurcated H bond. Proc. Natl. Acad. Sci. USA. 111:4085–4090. 10.1073/pnas.1319827111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller S.E., Zhang Y., Pastor R.W., and Brooks B.R.. 1995. Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 103:4613–4621. 10.1063/1.470648 [DOI] [Google Scholar]
- Fiser A., and Sali A.. 2003. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 374:461–491. 10.1016/S0076-6879(03)74020-8 [DOI] [PubMed] [Google Scholar]
- Floto R.A., Clatworthy M.R., Heilbronn K.R., Rosner D.R., MacAry P.A., Rankin A., Lehner P.J., Ouwehand W.H., Allen J.M., Watkins N.A., and Smith K.G.. 2005. Loss of function of a lupus-associated FcγRIIb polymorphism through exclusion from lipid rafts. Nat. Med. 11:1056–1058. 10.1038/nm1288 [DOI] [PubMed] [Google Scholar]
- Fujiwara T., Ritchie K., Murakoshi H., Jacobson K., and Kusumi A.. 2002. Phospholipids undergo hop diffusion in compartmentalized cell membrane. J. Cell Biol. 157:1071–1081. 10.1083/jcb.200202050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambin Y., Lopez-Esparza R., Reffay M., Sierecki E., Gov N.S., Genest M., Hodges R.S., and Urbach W.. 2006. Lateral mobility of proteins in liquid membranes revisited. Proc. Natl. Acad. Sci. USA. 103:2098–2102. 10.1073/pnas.0511026103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A. III, and Smith H.O.. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 6:343–345. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- Jaumouillé V., Farkash Y., Jaqaman K., Das R., Lowell C.A., and Grinstein S.. 2014. Actin cytoskeleton reorganization by Syk regulates Fcγ receptor responsiveness by increasing its lateral mobility and clustering. Dev. Cell. 29:534–546. 10.1016/j.devcel.2014.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai R.S., Suzuki K.G., Prossnitz E.R., Koyama-Honda I., Nakada C., Fujiwara T.K., and Kusumi A.. 2011. Full characterization of GPCR monomer–dimer dynamic equilibrium by single molecule imaging. J. Cell Biol. 192:463–480. 10.1083/jcb.201009128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiskowski M.A., Hancock J.F., and Kenworthy A.K.. 2009. On the use of Ripley’s K-function and its derivatives to analyze domain size. Biophys. J. 97:1095–1103. 10.1016/j.bpj.2009.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi N., Iwahara J., Koshiba S., Tomizawa T., Tochio N., Güntert P., Kigawa T., and Yokoyama S.. 2007. KUJIRA, a package of integrated modules for systematic and interactive analysis of NMR data directed to high-throughput NMR structure studies. J. Biomol. NMR. 39:31–52. 10.1007/s10858-007-9175-5 [DOI] [PubMed] [Google Scholar]
- Kono H., Kyogoku C., Suzuki T., Tsuchiya N., Honda H., Yamamoto K., Tokunaga K., and Honda Z.. 2005. FcγRIIB Ile232Thr transmembrane polymorphism associated with human systemic lupus erythematosus decreases affinity to lipid rafts and attenuates inhibitory effects on B cell receptor signaling. Hum. Mol. Genet. 14:2881–2892. 10.1093/hmg/ddi320 [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., and Sonnhammer E.L.L.. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580. 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- Kusumi A., Nakada C., Ritchie K., Murase K., Suzuki K., Murakoshi H., Kasai R.S., Kondo J., and Fujiwara T.. 2005. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: high-speed single-molecule tracking of membrane molecules. Annu. Rev. Biophys. Biomol. Struct. 34:351–378. 10.1146/annurev.biophys.34.040204.144637 [DOI] [PubMed] [Google Scholar]
- Kusumi A., Fujiwara T.K., Chadda R., Xie M., Tsunoyama T.A., Kalay Z., Kasai R.S., and Suzuki K.G.. 2012. Dynamic organizing principles of the plasma membrane that regulate signal transduction: commemorating the fortieth anniversary of Singer and Nicolson’s fluid-mosaic model. Annu. Rev. Cell Dev. Biol. 28:215–250. 10.1146/annurev-cellbio-100809-151736 [DOI] [PubMed] [Google Scholar]
- Kyogoku C., Dijstelbloem H.M., Tsuchiya N., Hatta Y., Kato H., Yamaguchi A., Fukazawa T., Jansen M.D., Hashimoto H., van de Winkel J.G.J., et al. 2002. Fcγ receptor gene polymorphisms in Japanese patients with systemic lupus erythematosus: Contribution of FCGR2B to genetic susceptibility. Arthritis Rheum. 46:1242–1254. 10.1002/art.10257 [DOI] [PubMed] [Google Scholar]
- Lau T.L., Partridge A.W., Ginsberg M.H., and Ulmer T.S.. 2008. Structure of the integrin β3 transmembrane segment in phospholipid bicelles and detergent micelles. Biochemistry. 47:4008–4016. 10.1021/bi800107a [DOI] [PubMed] [Google Scholar]
- Li Q., Lau A., Morris T.J., Guo L., Fordyce C.B., and Stanley E.F.. 2004. A syntaxin 1, Gαo, and N-type calcium channel complex at a presynaptic nerve terminal: analysis by quantitative immunocolocalization. J. Neurosci. 24:4070–4081. 10.1523/JNEUROSCI.0346-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Meckel T., Tolar P., Sohn H.W., and Pierce S.K.. 2010a Antigen affinity discrimination is an intrinsic function of the B cell receptor. J. Exp. Med. 207:1095–1111. 10.1084/jem.20092123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Meckel T., Tolar P., Sohn H.W., and Pierce S.K.. 2010b Intrinsic properties of immunoglobulin IgG1 isotype-switched B cell receptors promote microclustering and the initiation of signaling. Immunity. 32:778–789. 10.1016/j.immuni.2010.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Won Sohn H., Tolar P., Meckel T., and Pierce S.K.. 2010c Antigen-induced oligomerization of the B cell receptor is an early target of FcγRIIB inhibition. J. Immunol. 184:1977–1989. 10.4049/jimmunol.0902334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Chen E., Zhao X.W., Wan Z.P., Gao Y.R., Davey A., Huang E., Zhang L., Crocetti J., Sandoval G., et al. 2012. The scaffolding protein synapse-associated protein 97 is required for enhanced signaling through isotype-switched IgG memory B cell receptors. Sci. Signal. 5:ra54 10.1126/scisignal.2002820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Chu J., Zou Z., Hamacher N.B., Rixon M.W., and Sun P.D.. 2015. Structure of FcγRI in complex with Fc reveals the importance of glycan recognition for high-affinity IgG binding. Proc. Natl. Acad. Sci. USA. 112:833–838. 10.1073/pnas.1418812112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKerell A.D., Bashford D., Bellott M., Dunbrack R.L., Evanseck J.D., Field M.J., Fischer S., Gao J., Guo H., Ha S., et al. 1998. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 102:3586–3616. 10.1021/jp973084f [DOI] [PubMed] [Google Scholar]
- Martyna G.J., Tobias D.J., and Klein M.L.. 1994. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 101:4177–4189. 10.1063/1.467468 [DOI] [Google Scholar]
- Mattila P.K., Feest C., Depoil D., Treanor B., Montaner B., Otipoby K.L., Carter R., Justement L.B., Bruckbauer A., and Batista F.D.. 2013. The actin and tetraspanin networks organize receptor nanoclusters to regulate B cell receptor-mediated signaling. Immunity. 38:461–474. 10.1016/j.immuni.2012.11.019 [DOI] [PubMed] [Google Scholar]
- Mimura Y., Sondermann P., Ghirlando R., Lund J., Young S.P., Goodall M., and Jefferis R.. 2001. Role of oligosaccharide residues of IgG1-Fc in FcγRIIb binding. J. Biol. Chem. 276:45539–45547. 10.1074/jbc.M107478200 [DOI] [PubMed] [Google Scholar]
- Niederer H.A., Clatworthy M.R., Willcocks L.C., and Smith K.G.. 2010. FcγRIIB, FcγRIIIB, and systemic lupus erythematosus. Ann. N. Y. Acad. Sci. 1183:69–88. 10.1111/j.1749-6632.2009.05132.x [DOI] [PubMed] [Google Scholar]
- Nimmerjahn F., and Ravetch J.V.. 2011. FcγRs in health and disease. Curr. Top. Microbiol. Immunol. 350:105–125. [DOI] [PubMed] [Google Scholar]
- Owen D.M., Rentero C., Rossy J., Magenau A., Williamson D., Rodriguez M., and Gaus K.. 2010. PALM imaging and cluster analysis of protein heterogeneity at the cell surface. J. Biophotonics. 3:446–454. 10.1002/jbio.200900089 [DOI] [PubMed] [Google Scholar]
- Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kalé L., and Schulten K.. 2005. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26:1781–1802. 10.1002/jcc.20289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pincetic A., Bournazos S., DiLillo D.J., Maamary J., Wang T.T., Dahan R., Fiebiger B.M., and Ravetch J.V.. 2014. Type I and type II Fc receptors regulate innate and adaptive immunity. Nat. Immunol. 15:707–716. 10.1038/ni.2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser R.S., Evanics F., Kitevski J.L., and Al-Abdul-Wahid M.S.. 2006. Current applications of bicelles in NMR studies of membrane-associated amphiphiles and proteins. Biochemistry. 45:8453–8465. 10.1021/bi060615u [DOI] [PubMed] [Google Scholar]
- Rankin C.T., Veri M.C., Gorlatov S., Tuaillon N., Burke S., Huang L., Inzunza H.D., Li H., Thomas S., Johnson S., et al. 2006. CD32B, the human inhibitory Fc-γ receptor IIB, as a target for monoclonal antibody therapy of B-cell lymphoma. Blood. 108:2384–2391. 10.1182/blood-2006-05-020602 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Gomez D., Darve E., and Pohorille A.. 2004. Assessing the efficiency of free energy calculation methods. J. Chem. Phys. 120:3563–3578. 10.1063/1.1642607 [DOI] [PubMed] [Google Scholar]
- Šali A., and Blundell T.L.. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779–815. 10.1006/jmbi.1993.1626 [DOI] [PubMed] [Google Scholar]
- Semrau S., and Schmidt T.. 2007. Particle image correlation spectroscopy (PICS): retrieving nanometer-scale correlations from high-density single-molecule position data. Biophys. J. 92:613–621. 10.1529/biophysj.106.092577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siriboonrit U., Tsuchiya N., Sirikong M., Kyogoku C., Bejrachandra S., Suthipinittharm P., Luangtrakool K., Srinak D., Thongpradit R., Fujiwara K., et al. 2003. Association of Fcγ receptor IIb and IIIb polymorphisms with susceptibility to systemic lupus erythematosus in Thais. Tissue Antigens. 61:374–383. 10.1034/j.1399-0039.2003.00047.x [DOI] [PubMed] [Google Scholar]
- Smith K.G., and Clatworthy M.R.. 2010. FcγRIIB in autoimmunity and infection: evolutionary and therapeutic implications. Nat. Rev. Immunol. 10:328–343. 10.1038/nri2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soumpasis D.M. 1983. Theoretical analysis of fluorescence photobleaching recovery experiments. Biophys. J. 41:95–97. 10.1016/S0006-3495(83)84410-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strandberg E., Ozdirekcan S., Rijkers D.T., van der Wel P.C., Koeppe R.E. II, Liskamp R.M., and Killian J.A.. 2004. Tilt angles of transmembrane model peptides in oriented and non-oriented lipid bilayers as determined by 2H solid-state NMR. Biophys. J. 86:3709–3721. 10.1529/biophysj.103.035402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K.G., Fujiwara T.K., Sanematsu F., Iino R., Edidin M., and Kusumi A.. 2007. GPI-anchored receptor clusters transiently recruit Lyn and Gα for temporary cluster immobilization and Lyn activation: single-molecule tracking study 1. J. Cell Biol. 177:717–730. 10.1083/jcb.200609174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolar P., Hanna J., Krueger P.D., and Pierce S.K.. 2009. The constant region of the membrane immunoglobulin mediates B cell-receptor clustering and signaling in response to membrane antigens. Immunity. 30:44–55. 10.1016/j.immuni.2008.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treanor B., Depoil D., Gonzalez-Granja A., Barral P., Weber M., Dushek O., Bruckbauer A., and Batista F.D.. 2010. The membrane skeleton controls diffusion dynamics and signaling through the B cell receptor. Immunity. 32:187–199. 10.1016/j.immuni.2009.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vostrikov V.V., Daily A.E., Greathouse D.V., and Koeppe R.E. II. 2010. Charged or aromatic anchor residue dependence of transmembrane peptide tilt. J. Biol. Chem. 285:31723–31730. 10.1074/jbc.M110.152470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Z., and Liu W.. 2012. The growth of B cell receptor microcluster is a universal response of B cells encountering antigens with different motion features. Protein Cell. 3:545–558. 10.1007/s13238-012-2054-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Tang S., Wan Z., Gao Y., Cao Y., Yi J., Si Y., Zhang H., Liu L., and Liu W.. 2016. Utilization of a photoactivatable antigen system to examine B-cell probing termination and the B-cell receptor sorting mechanisms during B-cell activation. Proc. Natl. Acad. Sci. USA. 113:E558–E567. 10.1073/pnas.1517612113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willcocks L.C., Carr E.J., Niederer H.A., Rayner T.F., Williams T.N., Yang W., Scott J.A., Urban B.C., Peshu N., Vyse T.J., et al. 2010. A defunctioning polymorphism in FCGR2B is associated with protection against malaria but susceptibility to systemic lupus erythematosus. Proc. Natl. Acad. Sci. USA. 107:7881–7885. 10.1073/pnas.0915133107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Li G., Wang J., Fan Y., Wan Z., Zhang S., Shaheen S., Li J., Wang L., Yue C., et al. 2014. Through an ITIM-independent mechanism the FcγRIIB blocks B cell activation by disrupting the colocalized microclustering of the B cell receptor and CD19. J. Immunol. 192:5179–5191. 10.4049/jimmunol.1400101 [DOI] [PubMed] [Google Scholar]
- Xu X., Meckel T., Brzostowski J.A., Yan J., Meier-Schellersheim M., and Jin T.. 2010. Coupling mechanism of a GPCR and a heterotrimeric G protein during chemoattractant gradient sensing in Dictyostelium. Sci. Signal. 3:ra71 10.1126/scisignal.2000980 [DOI] [PubMed] [Google Scholar]
- Zhang J., Leiderman K., Pfeiffer J.R., Wilson B.S., Oliver J.M., and Steinberg S.L.. 2006. Characterizing the topography of membrane receptors and signaling molecules from spatial patterns obtained using nanometer-scale electron-dense probes and electron microscopy. Micron. 37:14–34. 10.1016/j.micron.2005.03.014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.