Nabekura and Lanier demonstrate that two distinct long-lived NK cell subsets with different functional properties differentiate during mouse model of cytomegalovirus (MCMV) infection. NK cells expressing the MCMV-specific Ly49H receptor differentiated into memory NK cells and Ly49H− NK cells differentiated into cytokine-activated NK cells. Memory NK cells show enhanced effector function, whereas cytokine-activated NK cells persisted better in an MCMV-free environment.
Abstract
Natural killer (NK) cells provide important host defense and can generate long-lived memory NK cells. Here, by using novel transgenic mice carrying inducible Cre expressed under the control of Ncr1 gene, we demonstrated that two distinct long-lived NK cell subsets differentiate in a mouse model of cytomegalovirus (MCMV) infection. NK cells expressing the MCMV-specific Ly49H receptor differentiated into memory NK cells by an activating signaling through Ly49H and Ly49H− NK cells differentiated into cytokine-activated NK cells by exposure to inflammatory cytokines during infection. Interleukin-12 is indispensable for optimal generation of both antigen-specific memory NK cells and cytokine-activated NK cells. MCMV-specific memory NK cells show enhanced effector function and augmented antitumor activity in vivo as compared with cytokine-activated NK cells, whereas cytokine-activated NK cells exhibited a more robust response to IL-15 and persisted better in an MCMV-free environment. These findings reveal that NK cells are capable of differentiation into distinct long-lived subsets with different functional properties.
Introduction
NK cells recognize and eliminate virus-infected cells and transformed cells by using a repertoire of NK cell receptors that regulates their activation and effector functions (Lanier, 2005). NK cells were traditionally classified as innate immune cells that respond rapidly against pathogens, but were considered short-lived and unable to differentiate into long-lived memory cells. Accumulating evidence, however, demonstrates that NK cells possess adaptive immune features, which include antigen-specific clonal expansion and differentiation into self-renewing, long-lived memory NK cells (O’Leary et al., 2006; Sun et al., 2009, 2010; Min-Oo et al., 2013). In mouse models, NK cells that are activated by pathogens, haptens, alloantigens, or a combination of cytokines, and are subsequently capable of differentiating into memory or memory-like NK cells with augmented effector functions in response to a variety of secondary stimulations as compared with naive NK cells (O’Leary et al., 2006; Cooper et al., 2009; Sun et al., 2009; Nabekura and Lanier, 2014).
We have demonstrated that mouse NK cells expressing the activating Ly49H receptor, which specifically recognizes the m157 mouse cytomegalovirus (MCMV) glycoprotein on the infected cells (Arase et al., 2002; Smith et al., 2002), undergo activation, a robust expansion, contraction, differentiation into a long-lived memory subset with enhanced effector functions, and persistence for several months after MCMV infection in a manner similar to antigen-specific T cells (Sun et al., 2009, 2010). These MCMV-primed memory Ly49H+ NK cells are capable of mounting a secondary antigen-specific recall response and provide effective host protection against rechallenge with MCMV infection (Sun et al., 2009). We have also demonstrated that mouse NK cells bearing the activating Ly49D receptor, which is specific for H-2Dd, preferentially expand and differentiate into memory NK cells when challenged with allogeneic H-2Dd–expressing cells in the context of an inflammatory environment (Nabekura and Lanier, 2014). Like Ly49H+ NK cells generated during MCMV infection, alloantigen-primed Ly49D+ NK cells exert enhanced effector functions and proliferate in response to a secondary alloantigen stimulation (Nabekura and Lanier, 2014). These activating receptor ligand–driven memory NK cell subsets share a memory immunophenotype (KLRG1highLy6C+DNAM-1lowCD11b+CD27−), mount a secondary response when rechallenged with the same antigenic stimulation, and demonstrate augmented effector functions in vitro. The generation of memory Ly49D+ and Ly49H+ NK cells requires signals transmitted by the immunoreceptor tyrosine-based activation motif (ITAM)–containing DAP12 adapter and by the proinflammatory cytokine IL-12 (Sun et al., 2009, 2012; Nabekura and Lanier, 2014). In humans, the activating CD94-NKG2C receptor has been implicated in the NK cell response to human cytomegalovirus (HCMV). The existence of human memory NK cells is supported by an increased frequency of CD94-NKG2ChighCD57+ NK cells in HCMV-seropositive human subjects as compared with HCMV-seronegative individuals (Lopez-Vergès et al., 2011). Similar to the response of Ly49H+ NK cells during MCMV infection in mice, human NK cells bearing NKG2C expand during acute HCMV infection, up-regulate expression of NKG2C and the maturation marker CD57, and exert enhanced IFN-γ production in response to target cells (Lopez-Vergès et al., 2011; Foley et al., 2012b). These HCMV-induced CD94-NKG2ChighCD57+ NK cells show long-term persistence in HCMV-seropositive individuals and preferentially proliferate in response to reactivation of HCMV in allogeneic hematopoietic cell and organ transplant recipients (Gumá et al., 2004; Lopez-Vergès et al., 2011; Foley et al., 2012a,b).
Activation of NK cells by cytokines alone in vitro leads to the generation of NK cells with memory-like properties (Cooper et al., 2009). Mouse and human NK cells preactivated with a combination of IL-12, IL-15, and IL-18 exhibit enhanced IFN-γ production, but not cytotoxicity, upon restimulation with IL-12 and IL-15 or activating receptor ligation as compared with NK cells pretreated with IL-15 alone (Cooper et al., 2009; Romee et al., 2012). These cytokine-induced memory-like NK cells highly express IL-2Rα, and they persist and demonstrate sustained IFN-γ production in vivo when they are adoptively transferred into lymphopenic recipient mice (Cooper et al., 2009; Ni et al., 2012; Romee et al., 2012; Leong et al., 2014). Although the phenotype of mouse cytokine-induced memory-like NK cells is not well defined (Cooper et al., 2009; Ni et al., 2012), human cytokine-induced memory-like NK cells exhibit a distinct phenotype compared with naive NK cells, including high expression of CD94, NKG2A, CD69, and NKp46 (Romee et al., 2012). It is unknown whether the in vitro cytokine–induced memory-like NK cells are representative of NK cells generated in physiological situations in vivo, for example during infection or in other inflammatory environments. A prior study reported that mouse NK cells do not undergo proliferation and maturation in peripheral tissues during infection with influenza virus (van Helden et al., 2012). However, adoptively transferred NK cells from influenza virus–infected mice survive longer than naive NK cells from uninfected mice and undergo homeostatic proliferation in naive recipient mice (van Helden et al., 2012). NK cells from respiratory syncytial virus (RSV)–infected mice have the same properties (van Helden et al., 2012), suggesting that these long-lived NK cells with proliferative activity are generated not in response to virus-specific antigens but rather may be caused by exposure to inflammatory cytokines during these viral infections. Unlike the CD94-NKG2ChighCD57+ memory NK cells in HCMV-seropositive humans, the in vivo counterparts of in vitro–generated cytokine-induced memory-like NK cells in humans have not been reported. In the present study, by using novel bacterial artificial chromosome (BAC) transgenic mice carrying an inducible Cre recombinase that is expressed under the control of the mouse Ncr1 gene, we addressed whether NK cells differentiate into distinct subsets of long-lived antigen-specific and non–antigen-specific NK cells with different functional properties and fates in a model of MCMV infection.
Results
Generation of NKp46-CreERT2 transgenic mice
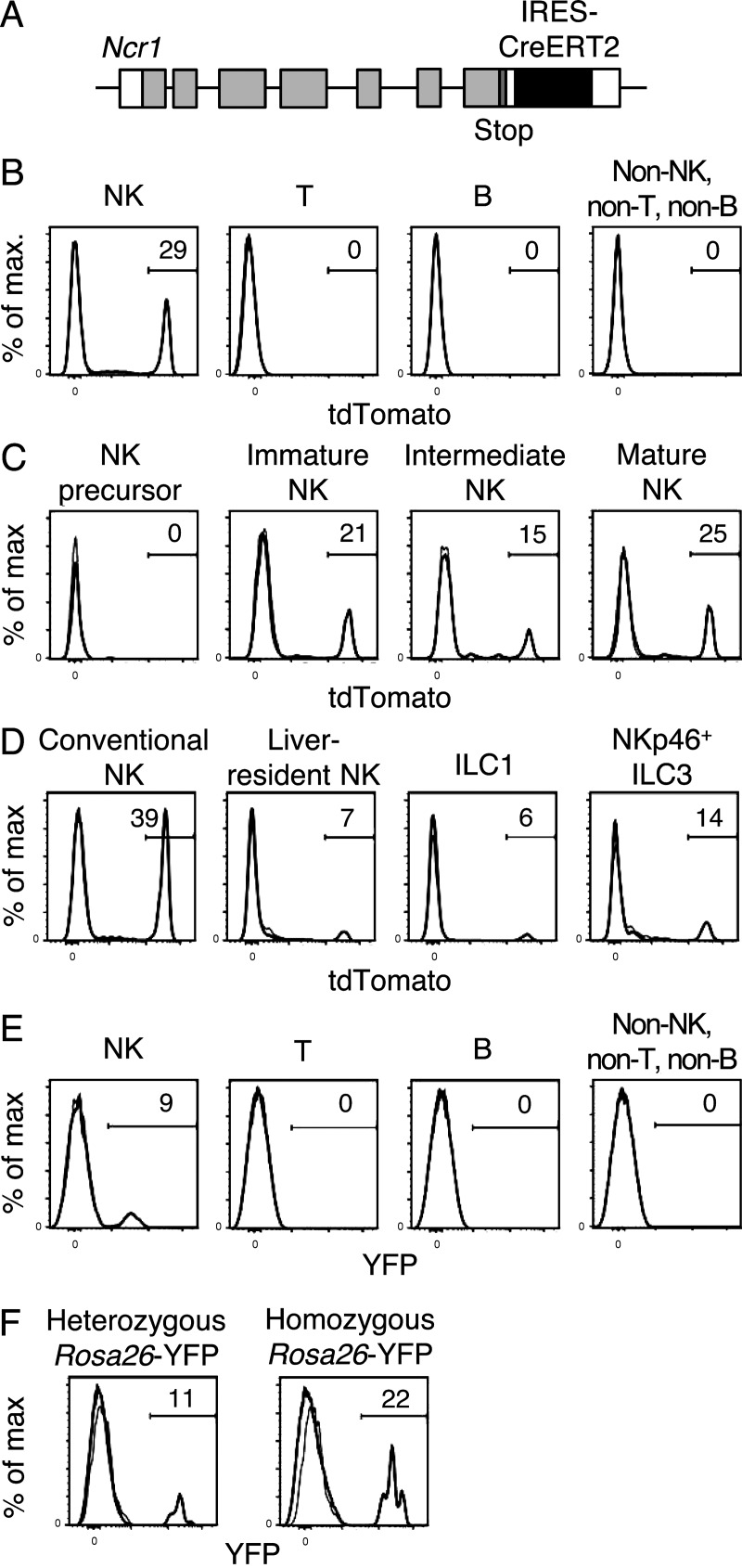
Our prior studies have used transfer of MCMV-specific Ly49H+ NK cells into Ly49H-deficient or DAP12-deficient mice, which lack functionally competent Ly49H+ NK cells, to track their fate during MCMV infection (Sun et al., 2009; Nabekura et al., 2014). A technical limitation of the transfer method is the detection of the small number of donor Ly49H+ NK cells in recipient mice over the course of MCMV infection. Furthermore, the differentiation and fate of Ly49H− NK cells in response to inflammatory cytokines during MCMV infection remains to be addressed. To address these issues, we developed a transgenic mouse carrying the CreERT2 recombinase expressed under the control of the mouse Ncr1 gene, which is encoded in a bacterial artificial chromosome (NKp46-CreERT2 Tg mice; Fig. 1 A). NKp46-CreERT2 Tg mice were intercrossed with Rosa26-tdTomato C57BL/6 (B6) mice (their Rosa26 loci are derived from 129S6 chromosome 6 containing the 129S6 NK gene cluster, which lacks the Klra8 gene encoding Ly49H) or with Rosa26-YFP B6 mice (their Rosa26 loci are derived from B6 chromosome 6 containing the B6 NK gene cluster with the Klra8 gene). These mice express tdTomato or YFP, respectively, upon CreERT2-mediated excision of the loxP-floxed stop codon in the Rosa26 loci only after tamoxifen administration. These NKp46-CreERT2 Tg mice with the Rosa26-tdTomato or Rosa26-YFP allele were obtained at Mendelian frequencies, developed normally with fertility, and exhibited no significant variations in frequencies of immune cell subsets, including NK cells, in the lymphoid tissues compared with WT littermate controls without the BAC construct (unpublished data). NK cells in these NKp46-CreERT2 Tg mice and WT littermates expressed equivalent amounts of NKp46 on their cell surfaces (unpublished data). NK cells were identified as DX5+NKp46+TCRβ−B220− lymphocytes in the spleen, blood, liver, and bone marrow. 20–40% of NK cells in the spleens of NKp46-CreERT2 Tg mice carrying a heterozygous Rosa26-tdTomato allele were positive for tdTomato after oral administration of tamoxifen (Fig. 1 B and not depicted). T cells, B cells, and non-T, non-B, non-NK cells (i.e., predominantly myeloid cells) did not express tdTomato (Fig. 1 B) Consistent with a prior study using a NKp46-iCre knock-in mouse to fate map NK cells expressing NKp46 (Narni-Mancinelli et al., 2011), immature NK cells and mature NK cells, but not precursor NK cells, in the bone marrow expressed tdTomato after tamoxifen administration (Fig. 1 C). NK cells express the pan-NK cell marker DX5, whereas liver-resident NK cells (also referred to as innate lymphoid cell 1 [ILC1]) do not express DX5. Additionally, some other ILC subsets, e.g., some ILC3, express NKp46 (Peng et al., 2013; Serafini et al., 2015). After tamoxifen treatment, ILC1 and NKp46+ ILC3 were positive for tdTomato, but at a lower frequency than NK cells in the liver (Fig. 1 D). NKp46-CreERT2 Tg mice were intercrossed with Rosa26-YFP B6 mice bearing the B6-derived NK gene cluster, including the Klra8 gene on chromosome 6, which encodes Ly49H. 10–20% of splenic NK cells in NKp46-CreERT2 Tg x Rosa26-YFP mice expressed YFP after tamoxifen administration (Fig. 1, E and F). After tamoxifen treatment, we observed the same pattern of expression of YFP and tdTomato in NKp46+ cells, including NK cells, liver-resident ILC1, and NKp46+ ILC3, in mice having the Rosa26-YFP or Rosa26-dtTomato alleles (unpublished data).
Figure 1.
NK cells in NKp46-CreERT2 Tg mice carrying the Rosa26-tdTomato allele or the Rosa26-YFP alleles express tdTomato or YFP after tamoxifen administration. (A) Schematic representation of the strategy used to generate NKp46-CreERT2 Tg mice. An IRES-CreERT2 cassette was inserted into the 3′ untranslated region of exon 7 of the mouse Ncr1 gene on mouse chromosome 7 in a C57BL/6 BAC. (B–D) NKp46-CreERT2 Tg mice with a heterozygous Rosa26-tdTomato allele were treated with tamoxifen for 5 consecutive days and immune cells were analyzed after the last tamoxifen administration. (B) Expression of tdTomato in NK cells (DX5+NKp46+TCRβ−B220−); T cells (NKp46−TCRβ+B220−); B cells (NKp46−TCRβ−B220+); and non-NK, non-T, and non-B cells (NKp46−TCRβ−B220−, predominantly myeloid cells) in the spleen. (C) Expression of tdTomato in NK precursor cells (TCRβ−CD19−CD11b−DX5−NKp46−CD122+) and immature (CD11b−CD27+), intermediate (CD11b+CD27+), and mature (CD11b+CD27+) NK cells (TCRβ−CD19−DX5+NKp46+) in the bone marrow. (D) Expression of tdTomato in NK cells (TCRβ−CD19−DX5+NKp46+CD122+), liver-resident NK cells (TCRβ−CD19−DX5−NKp46+CD122+), ILC1 (TCRβ−CD19−DX5−NKp46+CD11b−CD27+CD122low), and NKp46+ ILC3 (TCRβ−CD19−DX5−NKp46+CD11b−CD27−CD122low that are potentially ILC3) in the liver. Data are representative of two to three experiments (n = 2 in each experiment). (E and F) NKp46-CreERT2 Tg mice with a heterozygous Rosa26-YFP allele (E and F) and homozygous Rosa26-YFP alleles (F) were treated with tamoxifen for 5 d, and then splenocytes were analyzed. (E) Expression of YFP in NK cells; T cells; B cells; and non-NK, non-T, and non-B cells (predominantly myeloid cells) in the spleen. (F) Expression of YFP by NK cells in the spleen. Data are representative of more than five experiments (n = 2–6 in each experiment). Bold and thin lines represent cells in mice treated with tamoxifen and cells in untreated mice, respectively.
MCMV infection induces differentiation of memory Ly49H+ NK cells and cytokine-activated Ly49H− NK cells
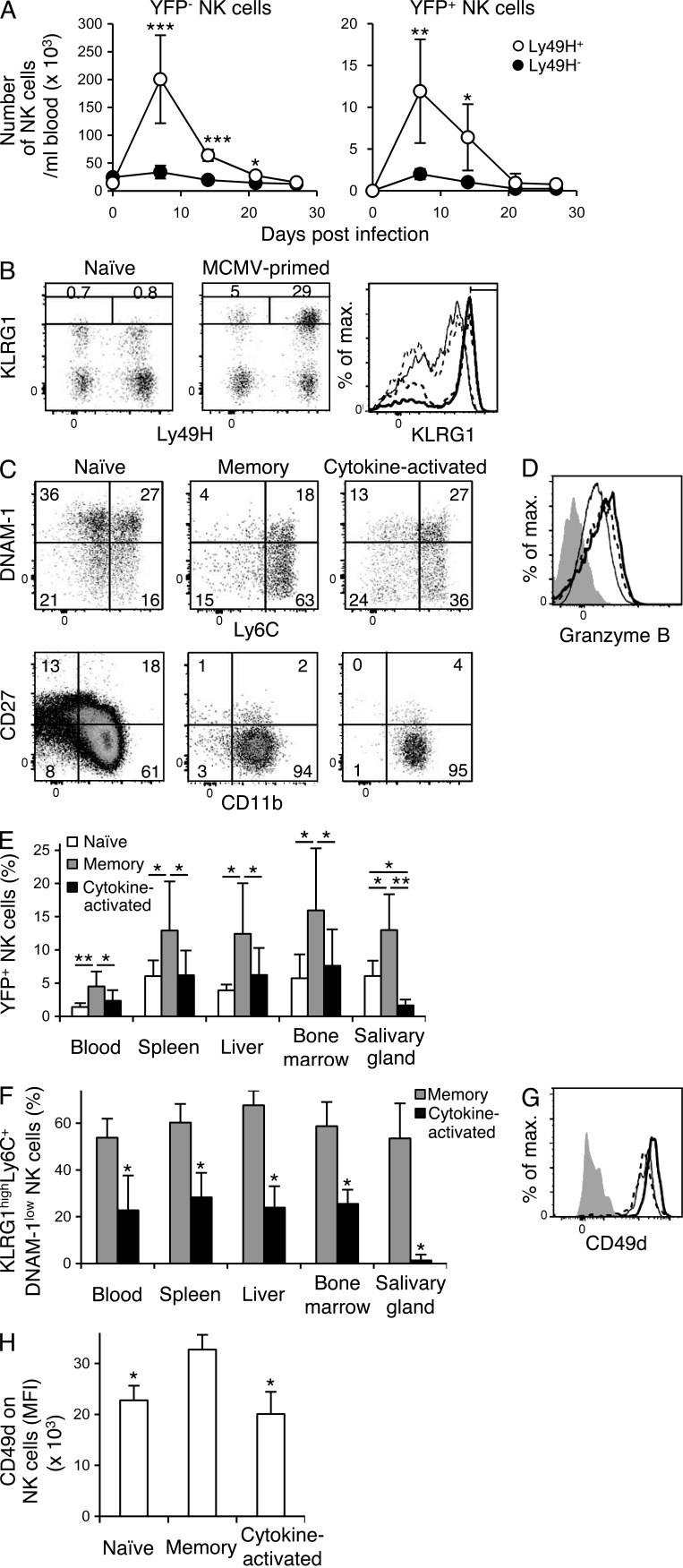
We investigated the fate of YFP+Ly49H+ NK cells in NKp46-CreERT2 x Rosa26-YFP Tg mice after tamoxifen administration during the course of MCMV infection. When NKp46-CreERT2 Tg mice carrying a heterozygous Rosa26-YFP allele were infected with MCMV on day 0 and treated with tamoxifen on days 0–4, YFP+ NK cells expressing Ly49H preferentially expanded and survived comparably to YFP−Ly49H+ NK cells (Fig. 2 A), allowing us to track the MCMV-primed long-lived NK cells as YFP+ NK cells in MCMV-infected mice. Consistent with our prior studies using Ly49H+ NK cells adoptively transferred into congenic recipients (Sun et al., 2009; Nabekura et al., 2014), these YFP+Ly49H+ NK cells persisted for >1 mo after MCMV infection and differentiated into MCMV-primed long-lived memory YFP+Ly49H+ NK cells expressing high levels of KLRG1 (Fig. 2 B). In contrast, <1% of YFP+ NK cells in naive uninfected NKp46-CreERT2 x Rosa26-YFP Tg mice expressed high levels of KLRG1 (Fig. 2 B). Unexpectedly, a subpopulation of YFP+Ly49H−KLRG1high NK cells in the MCMV-infected mice also survived for >1 mo after the infection (Fig. 2 B). These long-lived cytokine-activated YFP+Ly49H−KLRG1high NK cells that did not undergo clonal expansion possessed a lower frequency of cells expressing a memory cell phenotype (KLRG1highLy6C+DNAM-1low) compared with MCMV-specific YFP+Ly49H+KLRG1high memory NK cells (Fig. 2 C). When analyzed ex vivo 1 mo post infection (p.i.), both YFP+Ly49H+KLRG1high memory NK cells and the cytokine-activated YFP+Ly49H−KLRG1high NK cells showed a mature NK cell phenotype (CD11b+CD27−) and an increased expression of Granzyme B (Fig. 2, C and D). Compared with the frequencies of total YFP+ NK cells in naive uninfected NKp46-CreERT2 x Rosa26-YFP Tg mice at 1 mo after tamoxifen administration, a significantly higher percentage and absolute number of YFP+Ly49H+ memory NK cells were present in the lymphoid organs and, importantly, in the salivary glands, which is the site of persistent viral replication, in MCMV-infected mice (Fig. 2 E and not depicted). In contrast, cytokine-activatedYFP+Ly49H− NK cells did not increase in percentage or absolute number in the lymphoid organs during MCMV infection, and only a small number of YFP+Ly49H− NK cells were detected in the salivary glands (Fig. 2 E and not depicted). A significantly higher percentage of YFP+Ly49H+ memory NK cells displaying a memory immunophenotype (KLRG1highLy6C+DNAM-1low) were present in the lymphoid organs and salivary glands compared with cytokine-activated YFP+Ly49H− NK cells (Fig. 2 F). α4β1 integrin, which is composed of CD49d and CD29, is known to be an important adhesion molecule for localization of leukocytes in the salivary glands (Kunkel and Butcher, 2002, 2003). YFP+Ly49H+KLRG1high memory NK cells expressed higher amount of CD49d than cytokine-activated YFP+Ly49H−KLRG1high NK cells (Fig. 2, G and H). Both NK cell subsets equivalently expressed high amounts of CD29 on their surfaces (unpublished data). These results demonstrate that MCMV infection induces differentiation of two distinct long-lived NK cell subsets, the MCMV-specific memory Ly49H+ NK cells and cytokine-activated Ly49H− NK cells.
Figure 2.
NK cells differentiate into memory NK cells and cytokine-activated NK cells after MCMV infection. NKp46-CreERT2 Tg mice with a heterozygous Rosa26-YFP allele (A) or homozygous Rosa26-YFP alleles (B–H) were treated with tamoxifen on days 0–4 and infected or not with MCMV on day 0. (A) The number of YFP+ NK cells and YFP− NK cells in the blood over the course of infection. Data were pooled from two experiments (n = 7 in each group). *, P < 0.05; **, P < 0.01; ***, P < 0.005 versus Ly49H− cells. (B) Expression of Ly49H and KLRG1 on YFP+ NK cells in the spleens of uninfected and infected mice on day 27 p.i. The percentages of Ly49H+KLRG1high NK cells and Ly49H−KLRG1high NK cells are shown. Expression of KLRG1 on YFP+ NK cells in the spleens of naive uninfected mice and on YFP+Ly49H+ memory NK cells and cytokine-activated YFP+Ly49H− NK cells from MCMV-infected mice. Bold solid lines, bold dashed lines, thin solid lines, and thin dashed lines represent memory NK cells, cytokine-activated NK cells, naive Ly49H+ NK cells, and naive Ly49H− NK cells, respectively. (C) Phenotype of YFP+ NK cells in the spleens of naive uninfected mice, and YFP+Ly49H+KLRG1high memory NK cells and cytokine-activated YFP+Ly49H−KLRG1high NK cells in the spleens of infected mice on day 25–32 p.i. (D) Expression of Granzyme B in YFP+ NK cells in the spleens of naive uninfected mice, and in memory YFP+Ly49H+KLRG1high NK cells and cytokine-activated YFP+Ly49H−KLRG1high NK cells in the spleens of infected mice on day 32 p.i. Bold solid lines, bold dashed lines, and thin lines represent memory NK cells, cytokine-activated NK cells, and naive NK cells, respectively. Filled histograms represent staining with an isotype-matched control Ig. Data are representative of more than five experiments (n = 2–6 in each experiment). (E) The percentages of YFP+ NK cells in naive uninfected mice on days 25–27 after tamoxifen treatment and the percentages of YFP+Ly49H+KLRG1high memory NK cells and cytokine-activated YFP+Ly49H−KLRG1high NK cells in the organs of tamoxifen-treated infected mice on days 25–27 p.i. *, P < 0.05; **, P < 0.01. (F) The percentages of YFP+Ly49H+ NK cells and YFP+Ly49H− NK cells expressing a memory phenotype KLRG1highLy6C+DNAM-1low in the organs of tamoxifen-treated infected mice on days 25–27 p.i. Data are pooled from six experiments (n = 4–8 in each group). *, P < 0.01 versus memory cells. (G) Expression of CD49d on YFP+ NK cells in the blood of naive uninfected mice on day 25 after tamoxifen treatment and on YFP+Ly49H+KLRG1high memory NK cells and cytokine-activated YFP+Ly49H−KLRG1high NK cells in the blood of infected mice on day 25 p.i. Bold solid lines, bold dashed lines, and thin lines represent memory NK cells, cytokine-activated NK cells, and naive NK cells, respectively. A filled histogram represents staining with a control isotype-matched control Ig. Mean fluorescence intensity (MFI) of CD49d staining of NK cells are shown in H. Data are representative of two experiments (n = 4–6 in each group). *, P < 0.01 versus memory cells. P-values were calculated by a Student’s t test. Error bars show SEM.
IL-12 is required for the optimal differentiation of both memory Ly49H+ NK cells and cytokine-activated Ly49H− NK cells during MCMV infection
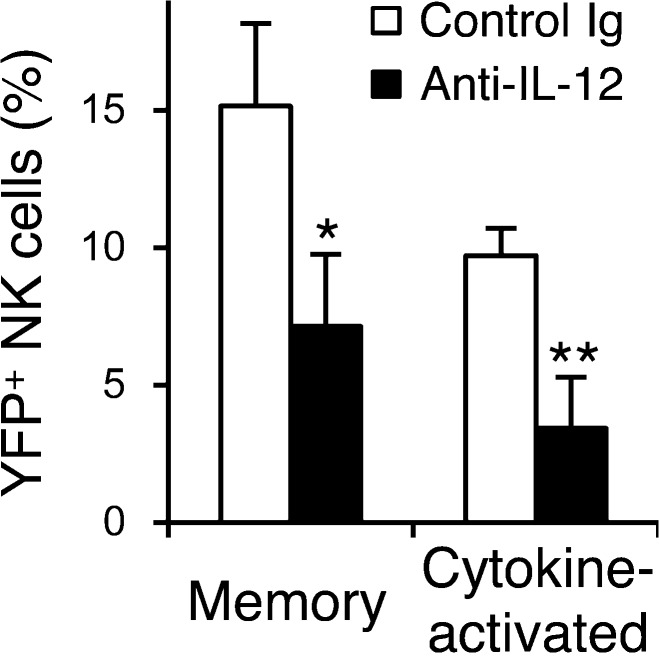
The differentiation of memory Ly49H+ NK cells during MCMV infection requires IL-12 (Sun et al., 2012). We examined the requirement of IL-12 for the generation of cytokine-activated Ly49H−KLRG1high NK cells in vivo during MCMV infection. When NKp46-CreERT2 x Rosa26-YFP Tg mice were infected with MCMV and treated with tamoxifen, neutralization of IL-12 on the day before MCMV infection and on days 3 and 6 p.i. significantly impaired the differentiation of both YFP+Ly49H+KLRG1high memory NK cells and cytokine-activated YFP+Ly49H−KLRG1high NK cells (Fig. 3). The neutralization of IL-12 did not affect the phenotype of Ly49H+KLRG1high memory NK cells or cytokine-activated Ly49H−KLRG1high NK cells (unpublished data), but did affect the magnitude of their response. These findings indicate that IL-12 is essential for the optimal differentiation of both Ly49H+ memory NK cells and cytokine-activated Ly49H− NK cells during MCMV infection.
Figure 3.
IL-12 is required for the optimal differentiation of memory NK cells and cytokine-activated NK cells. NKp46-CreERT2 Tg mice with homozygous Rosa26-YFP alleles were treated daily with tamoxifen on days 0–4 and infected with MCMV on day 0. Mice were injected with 200 µg control Ig or anti–IL-12 neutralizing mAb on the day before infection and on day 3 and 6 p.i. The percentages of YFP+Ly49H+KLRG1high memory NK cells and cytokine-activated YFP+Ly49H−KLRG1high NK cells in the spleens on day 26 p.i. are shown. Data are pooled from two experiments (n = 4–5 in each group). *, P < 0.01; **, P < 0.005 versus control Ig. P-values were calculated by a Student’s t test. Error bars show SEM.
Cytokine-activated NK cells persist longer than memory NK cells in an MCMV-free environment
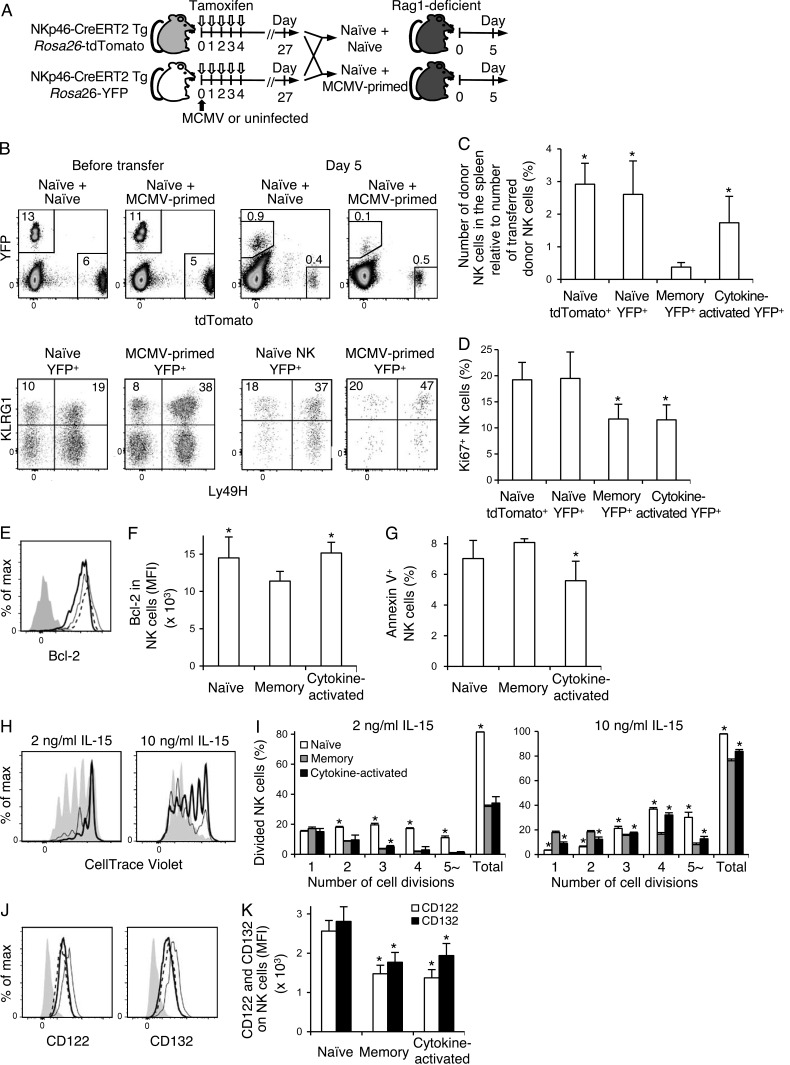
We investigated the persistence of MCMV-primed Ly49H+ memory NK cells and cytokine-activated Ly49H− NK cells in an MCMV-free environment. NKp46-CreERT2 x Rosa26-tdTomato Tg mice were treated with tamoxifen to label tdTomato+ NK cells in naive uninfected mice. NKp46-CerERT2 x Rosa26-YFP Tg were infected or not with MCMV and treated with tamoxifen. YFP+ NK cells from naive uninfected mice or YFP+ NK cells from MCMV-infected mice, which included both YFP+Ly49H+KLRG1high memory NK cells and cytokine-activated YFP+Ly49H−KLRG1high NK cells, were mixed with the tdTomato+ NK cells from naive uninfected mice as competitor NK cells, and then adoptively transferred into Rag1-deficient B6 mice (Fig. 4 A). A smaller number of MCMV-primed YFP+ NK cells, in particular YFP+Ly49H+KLRG1high memory NK cells, was detected in Rag1-deficient recipient mice on day 5 after transfer compared with YFP+ NK cells and tdTomato+ competitor NK cells from tamoxifen-treated naive uninfected mice (Fig. 4, B and C). YFP+Ly49H+KLRG1high memory NK cells and cytokine-activated YFP+Ly49H−KLRG1high NK cells were less proliferative in Rag1-deficient mice, as demonstrated by the smaller percentages of Ki67+ YFP+Ly49H+KLRG1high memory NK cells and cytokine-activated YFP+Ly49H−KLRG1high NK cells compared with YFP+ NK cells and tdTomato+ NK cells from tamoxifen-treated, naive uninfected mice on day 5 after transfer (Fig. 4 D). Consistent with the poor persistence of YFP+Ly49H+KLRG1high memory NK cells in MCMV-uninfected Rag1-deficient mice, YFP+Ly49H+KLRG1high memory NK cells expressed lower amounts of Bcl-2 as compared with cytokine-activated YFP+Ly49H−KLRG1high NK cells and YFP+ NK cells from naive uninfected mice on day 5 after transfer (Fig. 4, E and F). Furthermore, higher percentages of Annexin V+ NK cells were present in the YFP+Ly49H+KLRG1high memory NK cell subset than those in the cytokine-activated YFP+Ly49H−KLRG1high NK cell subset on day 5 after transfer (Fig. 4 G).
Figure 4.
Cytokine-activated NK cells preferentially persist in an MCMV-free environment. (A) NKp46-CreERT2 Tg mice with a heterozygous Rosa26-tdTomato allele or homozygous Rosa26-YFP alleles were treated with tamoxifen for 5 d and infected or not with MCMV on day 0. On day 27 after treatment with tamoxifen, tdTomato+ NK cells from naive uninfected mice were mixed with YFP+ NK cells from naive uninfected mice and with MCMV-primed YFP+ NK cells from infected mice, and then adoptively transferred into Rag1-deficient B6 mice. Donor NK cells were analyzed on day 5 after the transfer. (B) Expression of Ly49H and KLRG1 on donor tdTomato+ NK cells and YFP+ NK cells before transfer and in the spleens of Rag1-deficient recipient mice on day 5 after transfer. Splenocytes on day 5 after transfer were fixed and permeabilized for staining of Ki67. Thus, the fluorescence of tdTomato and YFP decreases as compared with before transfer. Data are representative of two experiments (n = 3–4 in each experiment). (C) The number of donor NK cells in the spleens of Rag1-deficient recipient mice on day 5 after transfer. The y axis represents the number of donor NK cells detected in the spleens of Rag1-deficient mice on day 5 compared with the number of donor NK cells adoptively transferred into Rag1-defiicent mice. *, P < 0.01 versus memory YFP+ cells. (D) The percentages of Ki67+ donor NK cells on day 5. Data were pooled from two experiments (n = 6–7 in each group). *, P < 0.05 versus naive tdTomato+ cells and naive YFP+ cells. (E–G) NKp46-CreERT2 Tg mice with homozygous Rosa26-YFP alleles were treated with tamoxifen for 5 d and infected or not with MCMV on day 0. On day 27 after tamoxifen treatment, YFP+ NK cells isolated from naive uninfected mice and MCMV-primed YFP+ NK cells were adoptively transferred into Rag1-deficient B6 mice. Donor NK cells were analyzed on day 5 after the transfer. (E) Expression of Bcl-2 in donor NK cells on day 5. Bold solid lines, bold dashed lines, and thin lines represent memory NK cells, cytokine-activated NK cells, and naive NK cells, respectively. A filled histogram represents staining with an isotype-matched control Ig. Data were representative of two experiments (n = 2–3 in each experiment). MFI of Bcl-2 staining in NK cells is shown in F. (G) The percentages of Annexin V+ donor NK cells on day 5. Data were pooled from two experiments (n = 5 in each group). *, P < 0.05 versus memory cells. (H–K) NKp46-CreERT2 Tg mice with homozygous Rosa26-YFP alleles were treated with tamoxifen for 5 d and infected or not with MCMV on day 0. (H and I) On day 25 after tamoxifen treatment, YFP+ NK cells from naive uninfected mice or MCMV-primed YFP+ NK cells were mixed with CD45.1+ WT B6 NK cells from naive uninfected mice, labeled with CellTrace Violet, and cultured in the presence of 2 or 10 ng/ml IL-15 for 4 d. (H) Cell divisions of NK cells on day 4. Filled histograms represent naive YFP+ NK cells. Bold and thin lines represent YFP+Ly49H+KLRG1high memory NK cells and cytokine-activated YFP+Ly49H−KLRG1high NK cells, respectively. The number of cell divisions of NK cells was quantified in I. Data are representative of two experiments (n = 3–6 in each experiment). *, P < 0.05 versus memory cells. (J) Expression of CD122 and CD132 on YFP+ NK cells in the spleens of naive uninfected mice and YFP+Ly49H+KLRG1high memory NK cells and cytokine-activated YFP+Ly49H−KLRG1high NK cells in the spleens of infected mice on days 25–27 p.i. Bold solid lines, bold dashed lines, and thin lines represent memory NK cells, cytokine-activated NK cells, and naive NK cells, respectively. Filled histograms represent staining with an isotype-matched control Ig. Data are representative of more than three experiments (n = 2–3 in each experiment). MFI of CD122 and CD132 staining of NK cells are shown in K. Data were pooled from three experiments (n = 6–7 in each group). *, P < 0.005 versus naive cells. P-values were calculated by a Student’s t test. Error bars show SEM.
To confirm the inefficient persistence of MCMV-primed NK cell subsets in MCMV-free hosts, YFP+ NK cells from naive uninfected mice and MCMV-primed YFP+ NK cells from infected mice were co-cultured with CD45.1+ WT B6 NK cells from naive uninfected mice in the presence of IL-15 for 4 d. YFP+Ly49H+KLRG1high memory NK cells and cytokine-activated YFP+Ly49H−KLRG1high NK cells responded to IL-15 in vitro less efficiently than YFP+ NK cells from naive uninfected mice (Fig. 4, H and I). Consistent with the poor persistence of YFP+Ly49H+KLRG1high memory NK cells in Rag1-deficient mice, YFP+Ly49H+KLRG1high memory NK cells showed an impaired response to IL-15 as compared with cytokine-activated YFP+Ly49H−KLRG1high NK cells (Fig. 4, H and I). YFP+Ly49H+KLRG1high memory NK cells and cytokine-activated YFP+Ly49H−KLRG1high NK cells expressed lower amounts of CD122 and CD132 on their cell surfaces than YFP+ NK cells from naive uninfected mice, which may partially explain their poor persistence and homeostatic proliferation in an MCMV-free environment in vivo and their impaired responses to IL-15 in vitro (Fig. 4, J and K). These results demonstrate that in vivo cytokine–activated Ly49H− NK cells persist longer than memory Ly49H+ NK cells in an MCMV-free environment, whereas memory Ly49H+ NK cells predominate over cytokine-activated NK cells in chronically MCMV-infected mice.
Memory Ly49H+ NK cells exert augmented antitumor activity
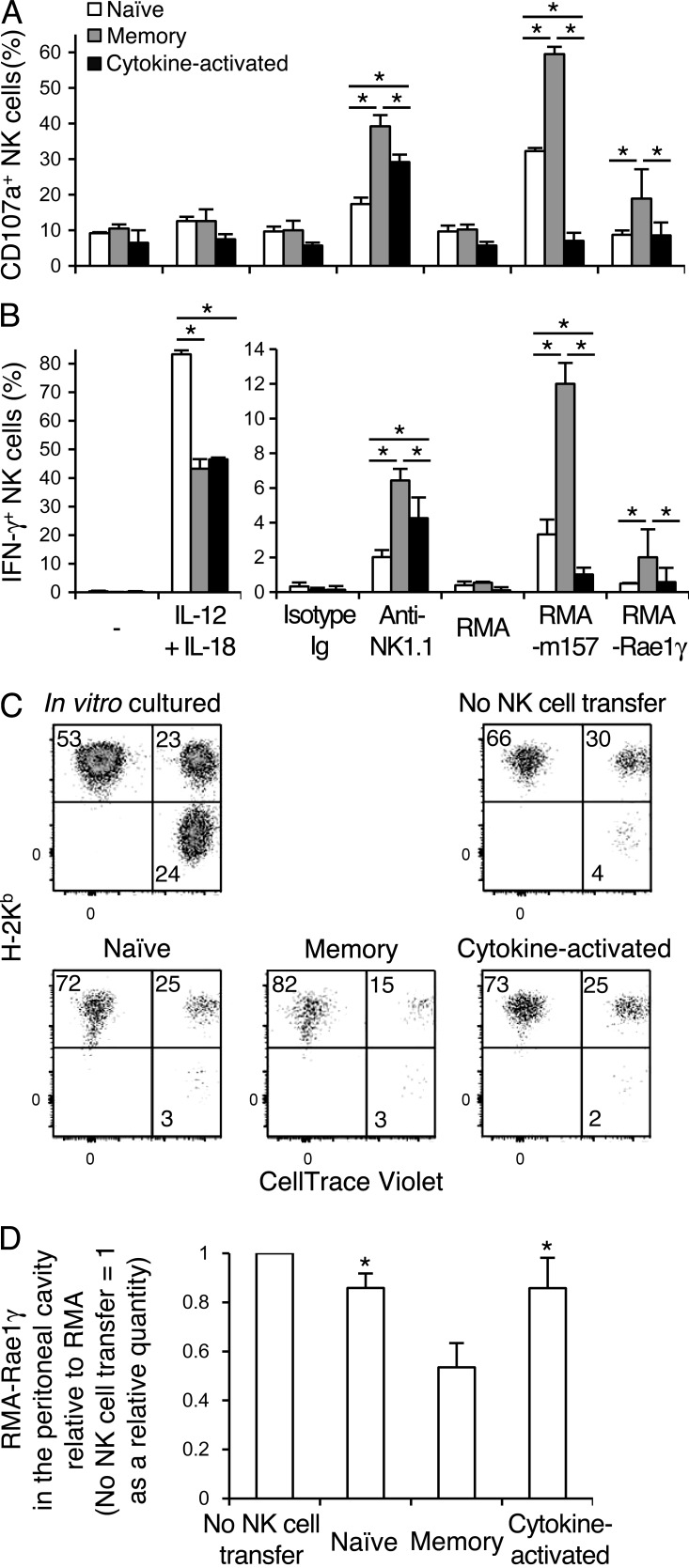
To evaluate effector functions of Ly49H+KLRG1high memory NK cells and cytokine-activated Ly49H−KLRG1high NK cells, YFP+ NK cells from naive uninfected NKp46-CreERT2 Tg x Rosa26-YFP mice treated with tamoxifen and MCMV-primed YFP+ NK cells from NKp46-CreERT2 Tg x Rosa26-YFP mice treated with tamoxifen and infected with MCMV were stimulated in vitro with IL-12 plus IL-18 by cross-linking NK1.1, or by co-culture with RMA transfectants expressing m157 or an NKG2D ligand, Rae1γ. A significantly lower frequency of YFP+Ly49H+KLRG1high memory NK cells and cytokine-activated YFP+Ly49H−KLRG1high NK cells produced IFN-γ compared with naive YFP+ NK cells after stimulation with IL-12 plus IL-18 (Fig. 5, A and B). These results are consistent with our prior study reporting reduced IFN-γ production from MCMV-primed memory Ly49H+ NK cells after IL-12 stimulation in the absence of Ly49H signaling (Min-Oo and Lanier, 2014) and a recent study in which human NK cells with memory features demonstrated a poor ability to produce IFN-γ after IL-12 plus IL-18 stimulation (Liu et al., 2016). Naive YFP+ NK cells, YFP+Ly49H+KLRG1high memory NK cells, and cytokine-activated YFP+Ly49H−KLRG1high NK cells expressed NK1.1, NKp46, 2B4, TIGIT, and Ly49C and/or Ly49I equivalently (unpublished data). YFP+Ly49H+KLRG1high memory NK cells and cytokine-activated YFP+Ly49H−KLRG1high NK cells equivalently expressed NKG2D, at levels slightly higher compared with naive YFP+ NK cells (unpublished data). However, a significantly higher frequency of YFP+Ly49H+KLRG1high memory NK cells degranulated and produced IFN-γ compared with both naive YFP+ NK cells and cytokine-activated YFP+Ly49H−KLRG1high NK cells after NK1.1 cross-linking or stimulation via NKG2D with RMA transfectants expressing Rae1γ (Fig. 5, A and B). A slightly larger fraction of cytokine-activated YFP+Ly49H−KLRG1high NK cells also showed degranulation and IFN-γ production compared with naive YFP+ NK cells after NK1.1 ligation (Fig. 5, A and B). To examine the effector functions of MCMV-primed NK cell subsets in vivo, YFP+ NK cells from naive uninfected mice and YFP+Ly49H+KLRG1high memory NK cells and cytokine-activated YFP+Ly49H−KLRG1high NK cells from MCMV-infected mice were purified and transferrred into T cell–depleted DAP10 and DAP12 double-deficient B6 mice, which lack expression of NKG2D. These recipient mice were then injected with a mixture of NK cell–resistant RMA cells, NK cell–sensitive Rae1γ-transfected RMA cells, and NK cell–sensitive RMA-S cells. Recipient mice receiving YFP+Ly49H+KLRG1high memory NK cells exhibited significantly improved clearance of Rae1γ−RMA cells, as compared with recipient mice that received naive YFP+ NK cells or cytokine-activated YFP+Ly49H−KLRG1high NK cells (Fig. 5, C and D). These results demonstrate that memory Ly49H+ NK cells exert enhanced effector functions in vitro and demonstrate augmented antitumor activity in vivo.
Figure 5.
Memory NK cells exert augmented antitumor activity. NKp46-CreERT2 Tg mice with homozygous Rosa26-YFP alleles were treated with tamoxifen for 5 d and infected or not with WT MCMV on day 0. (A and B) Degranulation (A) and IFN-γ production (B) of naive YFP+ NK cells from naive uninfected mice and YFP+Ly49H+KLRG1high memory NK cells and cytokine-activated YFP+Ly49H−KLRG1high NK cells from infected mice after culture in the presence of IL-12 and IL-18, stimulation with anti-NK1.1 mAb, and co-culture with RMA transfectants expressing m157 or Rae1γ. Data are representative of three experiments (n = 3–4 in each experiment). *, P < 0.05. (C) Splenic YFP+ NK cells from naive uninfected mice, and YFP+Ly49H+KLRG1high memory NK cells and cytokine-activated YFP+Ly49H−KLRG1high NK cells from infected mice were isolated on day 26 p.i. 15,000 YFP+ NK cells were transferred into T cell–depleted DAP10 and DAP12-double deficient recipient B6 mice. CellTrace Violet–labeled RMA-Rae1γ transfectants and RMA-S cells were mixed with RMA cells at a 1:1:1 (105 of each cell line) ratio, and injected intraperitoneally into these recipient mice on the day of NK cell transfer. An aliquot of the mixed tumor cells was cultured as a control. Proportions of these tumor cells (RMA, H-2Kb+ CellTrace Violet−; RMA-Rae1γ transfectants, H-2Kb+ CellTrace Violet+; and RMA-S cells, H-2Kb− CellTrace Violet+) in the peritoneal cavity were analyzed at 48 h after the injection. Data are representative of two experiments (n = 3 in each experiment). (D) NKG2D-mediated cytotoxicity against RMA-Rae1γ transfectants was quantified as the number of RMA-Rae1γ transfectants compared with the number of RMA cells detected in the peritoneal cavity at 48 h after injection. The y axis represents the clearance of RMA-Rae1γ transfectants in the peritoneal cavity of recipient mice receiving donor NK cells normalized to that of recipient mice not receiving donor NK cells expressed as a relative quantity. Data were pooled from two experiments (n = 6 in each group). *, P < 0.01 versus memory cells. P-values were calculated by a Student’s t test. Error bars show SEM.
Memory NK cells and cytokine-activated NK cells show impaired IFN-γ production after Listeria monocytogenes infection
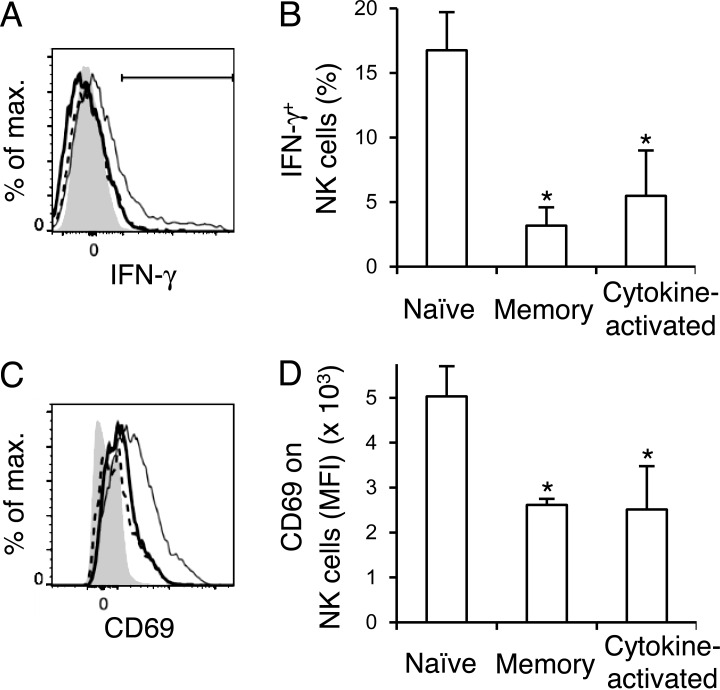
To assess functional responses of naive NK cells and MCMV-primed NK cells in response to infection with L. monocytogenes, we analyzed the activation and IFN-γ production in tamoxifen-treated uninfected and previously MCMV-infected NKp46-CerERT2 Tg x Rosa26-YFP mice on day 1.5 after L. monocytogenes infection. The percentages of YFP+ NK cells expressing IFN-γ after L. monocytogenes infection were significantly higher in the MCMV-uninfected mice than in the YFP+Ly49H+ memory NK cells and cytokine-activated YFP+Ly49H− NK cells in MCMV-infected mice (Fig. 6, A and B). YFP+Ly49H+ memory NK cells and cytokine-activated YFP+Ly49H− NK cells showed impaired activation after L. monocytogenes infection compared with naive YFP+ NK cells, as reflected by a lower frequency of NK cells up-regulating CD69 (Fig. 6, C and D). These findings demonstrate that MCMV-primed NK cell subsets are diminished in their responses to cytokine-driven bystander responses to L. monocytogenes infection.
Figure 6.
Memory NK cells and cytokine-activated NK cells show diminished responses to L. monocytogenes infection. NKp46-CreERT2 Tg mice with homozygous Rosa26-YFP alleles were treated with tamoxifen on days 0–4 and infected or not with MCMV on day 0. On day 25, these mice were infected intravenously with 5 × 104 CFU L. monocytogenes, and YFP+ NK cells in naive MCMV-uninfected mice and YFP+Ly49H+KLRG1high memory NK cells and cytokine-activated YFP+Ly49H−KLRG1high NK cells in MCMV-infected were analyzed on day 1.5 after infection with L. monocytogenes. (A) IFN-γ+ in naive NK cells, memory NK cells, and cytokine-activated NK cells in the spleens of L. monocytogenes–infected mice. Bold solid lines, bold dashed lines, and thin lines represent memory NK cells, cytokine-activated NK cells, and naive NK cells, respectively. A filled histogram represents NK cells in naive uninfected mice. The percentages of IFN-γ+ NK cells are quantified in B. (C) CD69 on naive NK cells, memory NK cells, and cytokine-activated NK cells in the spleens of L. monocytogenes–infected mice. Bold solid lines, bold dashed lines, and thin lines represent memory NK cells, cytokine-activated NK cells, and naive NK cells, respectively. A filled histogram represents staining with an isotype-matched control Ig. MFI of CD69 staining of NK cells is shown in D. Data were representative of two experiments (n = 4 in each experiment) in A and C. Data were pooled from two experiments (n = 4 in each group) in B and D. *, P < 0.05 versus naive cells. P-values were calculated by a Student’s t test. Error bars show SEM.
Ly49H signaling is required for differentiation of memory Ly49H+ NK cells during MCMV infection
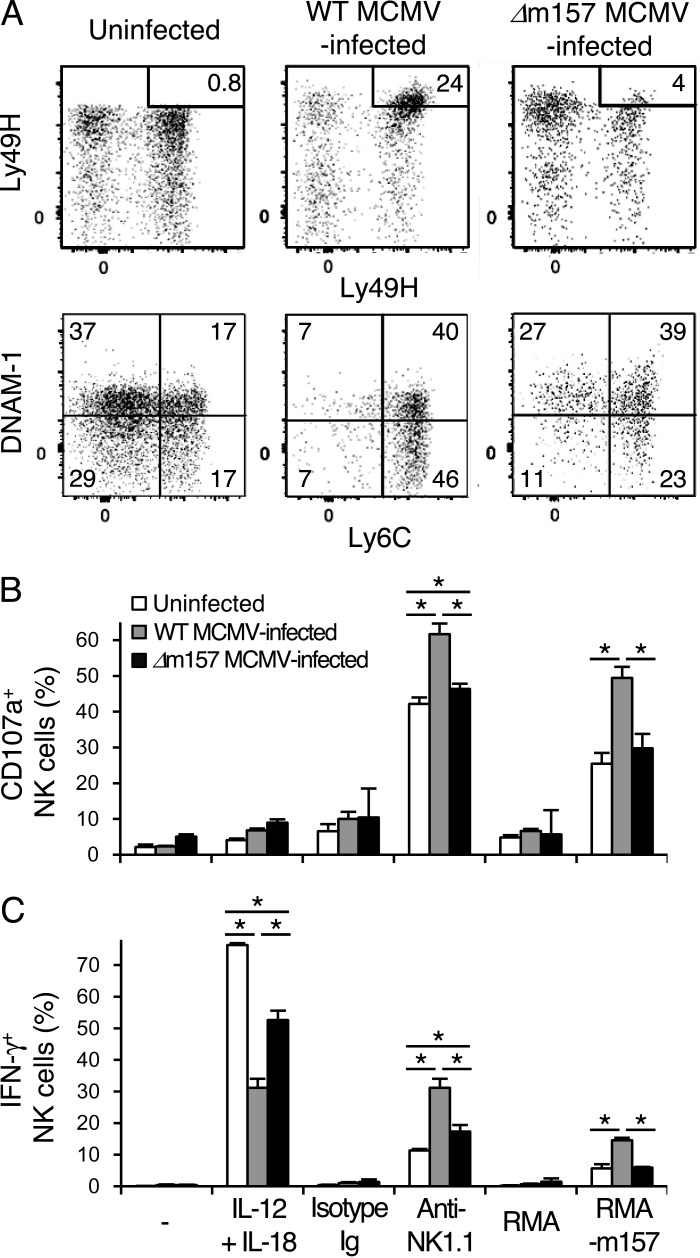
To determine whether an activating signal through Ly49H in addition to exposure to inflammatory cytokines during MCMV infection is essential for the generation of memory NK cells, NKp46-CreERT2 Tg x Rosa26-YFP mice were infected with WT MCMV or MCMV lacking the m157 gene (Δm157 MCMV) and treated with tamoxifen. The phenotype and effector functions of YFP+Ly49H+KLRG1high NK cells in WT MCMV-infected mice and Δm157 MCMV-infected mice were compared. Although infection with Δm157 MCMV induced the up-regulation of KLRG1 on YFP+ NK cells equivalent to WT MCMV, lower percentages of YFP+Ly49H+KLRG1high NK cells were present in mice infected with Δm157 MCMV compared with WT MCMV at 1 mo p.i. (Fig. 7 A). YFP+Ly49H+ NK cells in mice infected with WT and Δm157 MCMV showed a mature phenotype (CD11b+CD27−), up-regulated Granzyme B, and down-modulated CD122 and CD132 (unpublished data). However, significantly fewer of these YFP+Ly49H+ NK cells in Δm157 MCMV-infected mice displayed a memory immunophenotype (KLRG1highLy6C+DNAM-1low), but displayed a phenotype similar to the cytokine-activated YFP+Ly49H−KLRG1high NK cells in WT MCMV-infected mice (Fig. 2 C) and distinct from the YFP+Ly49H+KLRG1high memory NK cells in WT MCMV-infected mice (Fig. 7 A). To examine the effector functions of Δm157 MCMV-primed YFP+Ly49H+KLRG1high NK cells, YFP+ NK cells from naive uninfected mice, YFP+Ly49H+KLRG1high memory NK cells from WT MCMV-infected mice, and MCMV-primed YFP+Ly49H+KLRG1high NK cells from Δm157 MCMV-infected mice were stimulated in vitro with IL-12 plus IL-18 by cross-linking NK1.1 and by co-culture with RMA transfectants expressing m157. Similar to cytokine-activated YFP+Ly49H−KLRG1high NK cells in WT MCMV-infected mice (Fig. 5, A and B), Δm157 MCMV-primed YFP+Ly49H+KLRG1high NK cells secreted reduced amount of IFN-γ in response to IL-12 plus IL-18 and showed a slightly enhanced frequency of cells that degranulated and produced IFN-γ after cross-linking of NK1.1 compared with YFP+ NK cells from naive uninfected mice (Fig. 7, B and C). However, fewer of the YFP+Ly49H+KLRG1high NK cells from Δm157 MCMV-primed mice degranulated and produced IFN-γ compared with YFP+Ly49H+KLRG1high memory NK cells from WT MCMV-infected mice after NK1.1 ligation or Ly49H stimulation with m157 RMA transfectants (Fig. 7, B and C). These findings demonstrate that Ly49H signaling is required for the differentiation of bona fide MCMV-induced memory Ly49H+ NK cells with enhanced effector functions.
Figure 7.
Ly49H signaling is required for the differentiation of functional memory NK cells. NKp46-CreERT2 Tg mice with homozygous Rosa26-YFP alleles were treated with tamoxifen for 5 d and infected or uninfected with WT or Δm157 MCMV on day 0. (A) Phenotype of YFP+ NK cells in the spleens of naive uninfected mice, YFP+Ly49H+KLRG1high memory NK cells in the spleens of WT MCMV-infected mice, and MCMV-primed YFP+Ly49H+KLRG1high NK cells in the spleens of Δm157 MCMV-infected mice on days 25–27 p.i. The percentages of Ly49H+KLRG1high NK cells are shown. (B and C) Degranulation (B) and IFN-γ production (C) of YFP+ Ly49H+ NK cells from naive uninfected mice, YFP+Ly49H+KLRG1high memory NK cells from WT MCMV-infected mice, and MCMV-primed YFP+Ly49H+KLRG1high NK cells from Δm157 MCMV-infected mice after culture with IL-12 and IL-18, stimulation with anti-NK1.1 mAb, and co-culture with RMA cells expressing m157. Data are representative of two experiments (n = 3 in each experiment). *, P < 0.05. P-values were calculated by a Student’s t test. Error bars show SEM.
Discussion
Here, by using novel transgenic mice carrying inducible Cre under the control of expression by the Ncr1 gene, we found that two distinct NK cell subsets are generated by MCMV infection. One is an activating receptor ligand–driven memory NK cell and the other is an inflammatory cytokine-activated NK cell. Further, we directly compared the in vitro and in vivo functional properties and fate of activating receptor signal–driven Ly49H+KLRG1high memory NK cells versus cytokine-activated Ly49H−KLRG1high NK cells, both of which were generated in the same environment. Although prior studies have addressed the phenotypes, effector functions, persistence, and tissue distribution of mouse NK cells generated after MCMV infection in vivo or by in vitro activation with cytokines and then adoptive transfer into congenic mice (Cooper et al., 2009; Sun et al., 2009, 2010; Ni et al., 2012; van Helden et al., 2012), no studies have directly compared the functions and tracked the fate of activating receptor signal–driven memory NK cells versus cytokine-activated NK cells that are generated in vivo in a physiologically relevant viral infection. Our studies revealed that activating receptor signal–driven Ly49H+KLRG1high memory NK cells have augmented effector functions in vitro and in vivo, whereas cytokine-activated Ly49H−KLRG1high NK cells persist longer in an MCMV-free environment.
MCMV establishes latency after clearance of the primary infection and replicates chronically in the salivary glands (Vliegen et al., 2003). Prior studies have demonstrated that NK cells, T cells, and neutralizing antibodies against MCMV are critical for preventing recurrence and dissemination of MCMV from the salivary glands of latently infected mice (Jonjić et al., 1994; Polić et al., 1998). One of the notable differences between Ly49H+KLRG1high memory NK cells and cytokine-activated Ly49H−KLRG1high NK cells is their tissue distribution. Although both Ly49H+ and Ly49H− NK cells exist in the salivary glands of naive uninfected mice, Ly49H+KLRG1high memory NK cells, but not cytokine-activated Ly49H−KLRG1high NK cells, predominantly resided in the salivary glands of MCMV-infected mice. These findings suggest that the preferential accumulation of Ly49H+KLRG1high memory NK cells in the salivary glands might contribute to suppression of reactivation of MCMV in latently infected hosts. Despite our findings that Ly49H+KLRG1high memory NK cells predominate over cytokine-activated Ly49H−KLRG1high NK cells in the organs of MCMV-infected mice, cytokine-activated Ly49H−KLRG1high NK cells exhibited better persistence than memory Ly49H+KLRG1high NK cells in MCMV-free recipient mice. A possible explanation is that persistence of MCMV-specific Ly49H+KLRG1high memory NK cells is dependent on the equilibrium between viral replication and dissemination of MCMV and subsequent reactivation, causing proliferation of Ly49H+KLRG1high memory NK cells.
The previously described mouse cytokine-induced memory-like NK cells were generated by culturing NK cells in vitro with pharmacological amounts of a combination of cytokines (Cooper et al., 2009; Ni et al., 2012). However, there is no evidence that these cytokine-induced memory-like NK cells are generated in vivo in physiological situations. Here, we demonstrated that cytokine-activated Ly49H−KLRG1high NK cells are generated in vivo after MCMV infection. However, the functional properties of in vivo MCMV-induced cytokine-activated NK cells are different than the in vitro–generated cytokine-induced memory-like NK cells. The in vitro–derived cytokine-induced memory-like NK cells displayed enhanced IFN-γ production in response to ex vivo stimulation with cytokines, cross-linking of activating receptors, and tumor target cells, although they did not exert enhanced cytotoxicity (Cooper et al., 2009; Ni et al., 2012). However, we have demonstrated that in vivo MCMV-induced cytokine-activated NK cells display both enhanced degranulation and IFN-γ production in response to stimulation by cross-linking activating receptors and tumor target cells, but demonstrated poor IFN-γ production in response to IL-12 plus IL-18. Thus, the in vivo–generated cytokine-activated NK cells are not necessarily the counterparts of the cytokine-induced memory-like NK cells that were generated by in vitro culture.
Human CD94-NKG2Chigh NK cells preferentially expand during acute HCMV infection, and there is an elevated number and long-term persistence of mature CD94-NKG2ChighCD57+ NK cells in HCMV-seropositive individuals (Gumá et al., 2004; Lopez-Vergès et al., 2011; Foley et al., 2012a,b). These CD94-NKG2Chigh NK cells demonstrate enhanced cytokine production and degranulation when stimulated ex vivo, and there is a secondary expansion of CD94-NKG2ChighCD57+ NK cells in response to reactivation of HCMV in immunosuppressed transplant patients (Gumá et al., 2004; Lopez-Vergès et al., 2011; Foley et al., 2012a,b). Intriguingly, in some HCMV-seropositive individuals, there is an expansion of a unique subset of NK cells that lack expression of FcεRIγ. These NK cells predominantly express the CD94-NKG2C receptor and have enhanced antibody-dependent cellular cytotoxicity activity against antibody-coated HCMV-infected cells (Zhang et al., 2013). These findings support the hypothesis that activating signals through NKG2C drive the differentiation of human CD94-NKG2Chigh memory NK cells, similar to the m157-driven Ly49H+ memory NK cells in MCMV-infected mice (Gumá et al., 2004; Lopez-Vergès et al., 2011; Foley et al., 2012a,b). Additionally, these findings strongly suggest that an activating signal through the CD94-NKG2C-DAP12 receptor directly and quantitatively controls the differentiation of long-lived CD94-NKG2Chigh NK cells in the course of HCMV infection and that these cells may represent an activating receptor ligand–driven memory NK cell subset in humans.
HCMV-seropositive healthy individuals homozygously carrying the KLRC2 gene that encodes NKG2C show a significantly higher frequency and number of CD94-NKG2C+ NK cells than those carrying a heterozygous deletion of KLRC2 (Noyola et al., 2012; Muntasell et al., 2013). In KLRC2 hemizygous individuals, KLRC2+/− NK cells exhibit diminished calcium influx, degranulation, and proliferation upon NKG2C ligation compared with KLRC2+/+ NK cells (Muntasell et al., 2013). In response to acute HCMV infection, NK cells in patients transplanted with umbilical cord blood obtained from donors carrying a homozygous deletion of KLRC2 demonstrated an expansion of mature NK cells expressing an activating KIR, suggesting that receptors other than CD94-NKG2C may also drive NK cell differentiation (Della Chiesa et al., 2014). Similarly, in some HCMV-seropositive individuals carrying a heterozygous or homozygous deletion of KLRC2 there is also an expansion of the unique subset of NK cells lacking FcεRIγ (Liu et al., 2016; Muntasell et al., 2016). KLRC2−/− NK cells in HCMV-seropositive individuals display a mature phenotype (LILRB1highCD7lowCD161lowCD57highFcεR1γlowNKG2A−), which is similar but not identical to the CD94-NKG2Chigh NK cell subset in HCMV-seropositive KLRC2+/+ individuals (Liu et al., 2016). More importantly, these mature KLRC2−/− NK cells share enhanced TNF and IFN-γ production against antibody-coated target cells, impaired IFN-γ production after IL-12 plus IL-18 stimulation, and an epigenetic remodeling associated with demethylation of CpG motifs in the IFNG promoter region, similar to memory CD94-NKG2Chigh NK cells in HCMV-seropositive KLRC2+/+ individuals (Liu et al., 2016). The identification of an NK cell subset with memory features in HCMV-seropositive individuals lacking expression of NKG2C raises the possibility that inflammatory cytokines during HCMV infection may induce the differentiation of these mature NKG2C-deficient NK cells with augmented effector functions, which is reminiscent of the cytokine-activated Ly49H−KLRG1high NK cell subset in MCMV-infected mice.
Of note, both memory NK cells and cytokine-activated NK cells have several functional properties that may be beneficial for cancer immunotherapy, including augmented effector functions against tumors, persistence in vivo, and the capacity for secondary expansion. Further studies of the transcriptional signature and epigenetic modifications defining these distinct NK cell subsets are needed to better understand the critical regulatory factors for the divergence, differentiation, maintenance, and functional properties of these NK cells and to provide important insights into the development of effective NK cell–based vaccination strategies against infectious diseases and malignancies.
Materials and methods
Mice
C57BL/6 (B6) mice and congenic CD45.1+ B6 mice were purchased from Charles River. FVB/NJ mice were purchased from The Jackson Laboratory. Rosa26-YFP (Gt(ROSA)26Sortm1(EYFP)Cos; Srinivas et al., 2001), Rosa26-tdTomato (Gt(ROSA)26Sortm14(CAG-tdTomato)Hze; Madisen et al., 2010), Rag1-deficient (Rag1−/−; Mombaerts et al., 1992), and DAP10 (Hcst) and DAP12 (Tyrobp) double-deficient (Inui et al., 2009) mice on the C57BL/6 background (provided by T. Takai, Tohoku University, Japan) were maintained at the University of California (San Francisco, San Francisco, CA) in accordance with the guidelines of the Institutional Animal Care and Use Committee.
Generation of NKp46-CreERT2 Tg mice
The bacterial artificial chromosome (BAC) clone RP23-267N11 that encodes mouse chromosome 7 was modified to insert an internal ribosome entry site (IRES)-CreERT2 cassette (provided by M. Shlomchik, Yale University School of Medicine, New Haven, CT) into the 3′ untranslated region of exon 7 of the mouse Ncr1 gene by a BAC recombineering technology using reagents, vectors, and bacterial strains available from the National Cancer Institute (Bethesda, MD; https://ncifrederick.cancer.gov/research/brb/recombineeringInformation.aspx). The recombined BAC construct containing the IRES-CreERT2 element in the Ncr1 gene locus was microinjected into the pronucleus of single-cell fertilized zygotes of FVB/NJ mice. A founder mouse carrying the recombined BAC construct in its genome was intercrossed with Rosa26-YFP B6 mice or Rosa26-tdTomato B6 mice. The BAC transgenic mice (NKp46-CreERT2 Tg mice) carrying the Rosa26-YFP allele or the Rosa26-tdTomato allele, which express YFP or tdTomato, respectively, upon CreERT2-mediated excision of loxP-flanked stop codon were treated with tamoxifen (oral gavage of 200 µg/g body weight in corn oil) for 4 or 5 consecutive days.
MCMV and L. monocytogenes infection
A stock of Smith strain MCMV was prepared by homogenizing salivary glands harvested from infected BALB/c mice as described previously (Brune et al., 2001). Mice were infected by intraperitoneal injection of 5 × 103 PFU of salivary gland virus. In some experiments, Smith strain WT MCMV and Δm157 mutant MCMV (Bubić et al., 2004; provided by U. Koszinowski, Max von Pettenkofer-Institut, Munich, Germany) were prepared by infecting C57BL/6 3T3 cells in cell culture as described previously (Bubić et al., 2004). Mice were infected by intraperitoneal injection of 5 × 105 PFU WT MCMV or 2.5 × 105 PFU Δm157 MCMV. NKp46-CreERT2 Tg mice carrying the Rosa26-YFP allele were treated with tamoxifen for 4 or 5 d after MCMV infection on day 0. In some experiments, mice were inoculated intraperitoneally with 200 µg of a neutralizing mAb against mouse IL-12 p40 (clone C17.8) or an isotype-matched control rat IgG2a on the day before MCMV infection, and on days 3 and 6 p.i. L. monocytogenes 10403S strain was grown in brain–heart infusion broth to an OD600 of 0.2, and mice were infected intravenously with 5 × 104 CFU. Dose was determined by CFU assays for each infection.
Preparation of single-cell suspensions and NK cells
Leukocytes were isolated from spleen, bone marrow, blood, liver, and salivary glands. Livers were isolated after tissue perfusion with PBS and homogenized by using a Dounce homogenizer, and lymphocytes were prepared using centrifugation on a 40 and 60% Percoll gradient (GE Healthcare). Sublingual glands and submandibular glands were homogenized, and lymphocytes were prepared using centrifugation on a 40 and 70% Percoll gradient. NK cells were enriched by incubating splenocytes with purified rat mAbs against mouse CD4, CD5, CD8, CD19, Gr-1, and Ter119, followed by anti–rat IgG antibodies conjugated to magnetic beads (QIAGEN), as described previously (Nabekura et al., 2014).
Flow cytometry
Fc receptors (CD16 and CD32) were blocked with 2.4G2 mAb before surface or intracellular staining with the indicated fluorochrome-conjugated mAbs or isotype-matched control antibodies (BD, eBioscience, BioLegend, or TONBO Biosciences). In some experiments, cells were fixed with Cytofix (BD), followed by permeabilization with 0.1% Triton X-100 in PBS, and then stained with Alexa Flour 647–conjugated anti-Ki67 (BD). Samples were acquired on a LSRII or a FACSCalibur (BD) and data were analyzed with FlowJo software (FlowJo).
Adoptive transfer into Rag1-deficient mice
NK cells were enriched from spleens of uninfected NKp46-CreERT2 Tg x Rosa26-tdTomato mice, uninfected NKp46-CreERT2 Tg x Rosa26-YFP mice, and MCMV-infected NKp46-CreERT2 Tg x Rosa26-YFP mice on day 27 p.i. 30,000–50,000 tdTomato+ NK cells from naive uninfected NKp46-CreERT2 Tg x Rosa26-tdTomato mice were mixed with 40,000–60,000 YFP+ NK cells from naive uninfected NKp46-CreERT2 Tg x Rosa26-YFP mice or MCMV-primed YFP+ NK cells from MCMV-infected NKp46-CreERT2 Tg x Rosa26-YFP mice, and then injected intravenously into Rag1-deficient B6 mice. Donor NK cells in the spleens of Rag1-deficient recipient mice were analyzed on day 5 after the transfer.
In vitro culture
200,000 NK cells from uninfected and MCMV-infected NKp46-CreERT2 Tg x Rosa26-YFP mice were mixed with 5 × 104 CD45.1+ NK cells from uninfected WT B6 mice, labeled with 10 µM CellTrace Violet according to the manufacturer’s instructions (Invitrogen), and cultured in the presence of 2 or 10 ng/ml human IL-15 (R&D Systems) for 4 d at 37°C.
Ex vivo stimulation of NK cells
100,000 NK cells were incubated in 96-well tissue culture plates coated with anti-NK1.1 (clone PK136) or isotype-matched control mouse IgG2a as described previously (Nabekura et al., 2015); incubated with 2.5 ng/ml mouse IL-12 and 2.5 ng/ml mouse IL-18 (R&D Systems); or co-cultured with 105 RMA, RMA expressing m157, or RMA expressing Rae1γ for 5 h at 37°C in the presence of PE-conjugated anti-CD107a mAb and GolgiStop (BD), followed by staining for surface molecules and intracellular IFN-γ as previously described (Nabekura and Lanier, 2014).
In vivo cytotoxic assay
Enriched NK cells were stained with antibodies against TCRβ, B220, Ly49H, and KLRG1. NK cells in the spleens of naive uninfected NKp46-CreERT2 Tg x Rosa26-YFP mice were purified by sorting YFP+KLRG1+ cells gated on non–T cell and –B cell lymphocytes by using a FACSAria III (BD) on day 21–25 after the tamoxifen injection. Memory NK cells and cytokine-activated NK cells in the spleens of MCMV-infected NKp46-CreERT2 Tg x Rosa26-YFP mice were purified by sorting YFP+Ly49H+KLRG1high cells and YFP+Ly49H−KLRG1high cells gated on non–T cell and –B cell lymphocytes by using a FACSAria III on day 26 p.i. 15,000 NK cells were injected intraperitoneally into DAP10 and DAP12 double-deficient B6 mice. DAP10 and DAP12 double-deficient B6 mice were depleted of CD4+ and CD8+ T cells on the day before transfer of donor NK cells by intraperitoneal injection of 100 µg of anti-CD4 (clone GK1.5) and 100 µg of anti-CD8 (clone 2.43) mAbs. RMA transfectants expressing Rae1γ (RMA-Rae1γ) and RMA-S cells were labeled with CellTrace Violet, mixed with unlabeled RMA cells at a ratio of 1:1:1 (105 of each cell line; a total of 3 × 105 cells), and the mixed tumor cells were injected intraperitoneally into DAP10- and DAP12-deficient B6 mice on the day of donor NK cell transfer. An aliquot of the mixed tumor cells was cultured as a control. After 48 h, peritoneal cavity cells were collected and stained with anti-NK1.1, anti-Ly49A, and anti–H-2Kb mAbs. Tumor cells were gated on the basis of their characteristic forward and side light scatter properties, and then gated on NK1.1− and Ly49Ahigh cells. The different tumor cell types were discriminated by staining of H-2Kb and CellTrace Violet: RMA cells, H-2Kb+ CellTrace Violet−; RMA-Rae1γ cells, H-2Kb+ CellTrace Violet+; and RMA-S cells, H-2Kb− CellTrace Violet+. NKG2D-mediated cytotoxicity against RMA-Rae1γ was quantified as the proportion of viable recovered RMA-Rae1γ cells relative to that of RMA cells, and loss of RMA-S cells confirmed efficient NK cell cytolytic activity.
Statistical methods
The Student’s t test was used to compare results. P < 0.05 was considered statistically significant.
Acknowledgments
We thank the Lanier laboratory for comments and discussions. We are grateful to Helen Lu, Hong-Erh Liang, Richard M. Locksley, and Mark S. Anderson (University of California, San Francisco, San Francisco, CA) for helpful comments on BAC recombineering, and Mark J. Shlomchik (Yale University School of Medicine, New Haven, CT) for providing the IRES-CreERT2 cassette.
The work was supported by National Institutes of Health grant AI068129. L.L. Lanier is an American Cancer Society Professor. T. Nabekura is supported by the Friends of Leukemia Research Fund and the Nakajima Foundation.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- HCMV
- human cytomegalovirus
- ILC
- innate lymphoid cell
- IRES
- internal ribosome entry site
- ITAM
- immunoreceptor tyrosine-based activation motif
- MCMV
- mouse cytomegalovirus
- MFI
- mean fluorescence intensity
- p.i.
- post infection
- RSV
- respiratory syncytial virus
References
- Arase H., Mocarski E.S., Campbell A.E., Hill A.B., and Lanier L.L.. 2002. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 296:1323–1326. 10.1126/science.1070884 [DOI] [PubMed] [Google Scholar]
- Brune W., Hengel H., and Koszinowski U.H.. 2001. A mouse model for cytomegalovirus infection. Curr Protoc Immunol. Chapter 19:Unit 19.17. [DOI] [PubMed] [Google Scholar]
- Bubić I., Wagner M., Krmpotić A., Saulig T., Kim S., Yokoyama W.M., Jonjić S., and Koszinowski U.H.. 2004. Gain of virulence caused by loss of a gene in murine cytomegalovirus. J. Virol. 78:7536–7544. 10.1128/JVI.78.14.7536-7544.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper M.A., Elliott J.M., Keyel P.A., Yang L., Carrero J.A., and Yokoyama W.M.. 2009. Cytokine-induced memory-like natural killer cells. Proc. Natl. Acad. Sci. USA. 106:1915–1919. 10.1073/pnas.0813192106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Chiesa M., Falco M., Bertaina A., Muccio L., Alicata C., Frassoni F., Locatelli F., Moretta L., and Moretta A.. 2014. Human cytomegalovirus infection promotes rapid maturation of NK cells expressing activating killer Ig-like receptor in patients transplanted with NKG2C−/− umbilical cord blood. J. Immunol. 192:1471–1479. 10.4049/jimmunol.1302053 [DOI] [PubMed] [Google Scholar]
- Foley B., Cooley S., Verneris M.R., Curtsinger J., Luo X., Waller E.K., Anasetti C., Weisdorf D., and Miller J.S.. 2012a Human cytomegalovirus (CMV)-induced memory-like NKG2C+ NK cells are transplantable and expand in vivo in response to recipient CMV antigen. J. Immunol. 189:5082–5088. 10.4049/jimmunol.1201964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley B., Cooley S., Verneris M.R., Pitt M., Curtsinger J., Luo X., Lopez-Vergès S., Lanier L.L., Weisdorf D., and Miller J.S.. 2012b Cytomegalovirus reactivation after allogeneic transplantation promotes a lasting increase in educated NKG2C+ natural killer cells with potent function. Blood. 119:2665–2674. 10.1182/blood-2011-10-386995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumá M., Angulo A., Vilches C., Gómez-Lozano N., Malats N., and López-Botet M.. 2004. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood. 104:3664–3671. 10.1182/blood-2004-05-2058 [DOI] [PubMed] [Google Scholar]
- Inui M., Kikuchi Y., Aoki N., Endo S., Maeda T., Sugahara-Tobinai A., Fujimura S., Nakamura A., Kumanogoh A., Colonna M., and Takai T.. 2009. Signal adaptor DAP10 associates with MDL-1 and triggers osteoclastogenesis in cooperation with DAP12. Proc. Natl. Acad. Sci. USA. 106:4816–4821. 10.1073/pnas.0900463106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonjić S., Pavić I., Polić B., Crnković I., Lucin P., and Koszinowski U.H.. 1994. Antibodies are not essential for the resolution of primary cytomegalovirus infection but limit dissemination of recurrent virus. J. Exp. Med. 179:1713–1717. 10.1084/jem.179.5.1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel E.J., and Butcher E.C.. 2002. Chemokines and the tissue-specific migration of lymphocytes. Immunity. 16:1–4. 10.1016/S1074-7613(01)00261-8 [DOI] [PubMed] [Google Scholar]
- Kunkel E.J., and Butcher E.C.. 2003. Plasma-cell homing. Nat. Rev. Immunol. 3:822–829. 10.1038/nri1203 [DOI] [PubMed] [Google Scholar]
- Lanier L.L. 2005. NK cell recognition. Annu. Rev. Immunol. 23:225–274. 10.1146/annurev.immunol.23.021704.115526 [DOI] [PubMed] [Google Scholar]
- Leong J.W., Chase J.M., Romee R., Schneider S.E., Sullivan R.P., Cooper M.A., and Fehniger T.A.. 2014. Preactivation with IL-12, IL-15, and IL-18 induces CD25 and a functional high-affinity IL-2 receptor on human cytokine-induced memory-like natural killer cells. Biol. Blood Marrow Transplant. 20:463–473. 10.1016/j.bbmt.2014.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.L., Landskron J., Ask E.H., Enqvist M., Sohlberg E., Traherne J.A., Hammer Q., Goodridge J.P., Larsson S., Jayaraman J., et al. 2016. Critical role of CD2 co-stimulation in adaptive natural killer cell responses revealed in NKG2C-deficient humans. Cell Reports. 15:1088–1099. 10.1016/j.celrep.2016.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Vergès S., Milush J.M., Schwartz B.S., Pando M.J., Jarjoura J., York V.A., Houchins J.P., Miller S., Kang S.M., Norris P.J., et al. 2011. Expansion of a unique CD57+NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proc. Natl. Acad. Sci. USA. 108:14725–14732. 10.1073/pnas.1110900108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R., et al. 2010. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13:133–140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min-Oo G., and Lanier L.L.. 2014. Cytomegalovirus generates long-lived antigen-specific NK cells with diminished bystander activation to heterologous infection. J. Exp. Med. 211:2669–2680. 10.1084/jem.20141172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min-Oo G., Kamimura Y., Hendricks D.W., Nabekura T., and Lanier L.L.. 2013. Natural killer cells: walking three paths down memory lane. Trends Immunol. 34:251–258. 10.1016/j.it.2013.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P., Iacomini J., Johnson R.S., Herrup K., Tonegawa S., and Papaioannou V.E.. 1992. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 68:869–877. 10.1016/0092-8674(92)90030-G [DOI] [PubMed] [Google Scholar]
- Muntasell A., López-Montañés M., Vera A., Heredia G., Romo N., Peñafiel J., Moraru M., Vila J., Vilches C., and López-Botet M.. 2013. NKG2C zygosity influences CD94/NKG2C receptor function and the NK-cell compartment redistribution in response to human cytomegalovirus. Eur. J. Immunol. 43:3268–3278. 10.1002/eji.201343773 [DOI] [PubMed] [Google Scholar]
- Muntasell A., Pupuleku A., Cisneros E., Vera A., Moraru M., Vilches C., and López-Botet M.. 2016. Relationship of NKG2C copy number with the distribution of distinct cytomegalovirus-induced adaptive NK cell subsets. J. Immunol. 196:3818–3827. 10.4049/jimmunol.1502438 [DOI] [PubMed] [Google Scholar]
- Nabekura T., and Lanier L.L.. 2014. Antigen-specific expansion and differentiation of natural killer cells by alloantigen stimulation. J. Exp. Med. 211:2455–2465. 10.1084/jem.20140798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabekura T., Kanaya M., Shibuya A., Fu G., Gascoigne N.R., and Lanier L.L.. 2014. Costimulatory molecule DNAM-1 is essential for optimal differentiation of memory natural killer cells during mouse cytomegalovirus infection. Immunity. 40:225–234. 10.1016/j.immuni.2013.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabekura T., Girard J.P., and Lanier L.L.. 2015. IL-33 receptor ST2 amplifies the expansion of NK cells and enhances host defense during mouse cytomegalovirus infection. J. Immunol. 194:5948–5952. 10.4049/jimmunol.1500424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narni-Mancinelli E., Chaix J., Fenis A., Kerdiles Y.M., Yessaad N., Reynders A., Gregoire C., Luche H., Ugolini S., Tomasello E., et al. 2011. Fate mapping analysis of lymphoid cells expressing the NKp46 cell surface receptor. Proc. Natl. Acad. Sci. USA. 108:18324–18329. 10.1073/pnas.1112064108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J., Miller M., Stojanovic A., Garbi N., and Cerwenka A.. 2012. Sustained effector function of IL-12/15/18-preactivated NK cells against established tumors. J. Exp. Med. 209:2351–2365. 10.1084/jem.20120944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyola D.E., Fortuny C., Muntasell A., Noguera-Julian A., Muñoz-Almagro C., Alarcón A., Juncosa T., Moraru M., Vilches C., and López-Botet M.. 2012. Influence of congenital human cytomegalovirus infection and the NKG2C genotype on NK-cell subset distribution in children. Eur. J. Immunol. 42:3256–3266. 10.1002/eji.201242752 [DOI] [PubMed] [Google Scholar]
- O’Leary J.G., Goodarzi M., Drayton D.L., and von Andrian U.H.. 2006. T cell- and B cell-independent adaptive immunity mediated by natural killer cells. Nat. Immunol. 7:507–516. 10.1038/ni1332 [DOI] [PubMed] [Google Scholar]
- Peng H., Jiang X., Chen Y., Sojka D.K., Wei H., Gao X., Sun R., Yokoyama W.M., and Tian Z.. 2013. Liver-resident NK cells confer adaptive immunity in skin-contact inflammation. J. Clin. Invest. 123:1444–1456. 10.1172/JCI66381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polić B., Hengel H., Krmpotić A., Trgovcich J., Pavić I., Luccaronin P., Jonjić S., and Koszinowski U.H.. 1998. Hierarchical and redundant lymphocyte subset control precludes cytomegalovirus replication during latent infection. J. Exp. Med. 188:1047–1054. 10.1084/jem.188.6.1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romee R., Schneider S.E., Leong J.W., Chase J.M., Keppel C.R., Sullivan R.P., Cooper M.A., and Fehniger T.A.. 2012. Cytokine activation induces human memory-like NK cells. Blood. 120:4751–4760. 10.1182/blood-2012-04-419283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafini N., Vosshenrich C.A., and Di Santo J.P.. 2015. Transcriptional regulation of innate lymphoid cell fate. Nat. Rev. Immunol. 15:415–428. 10.1038/nri3855 [DOI] [PubMed] [Google Scholar]
- Smith H.R., Heusel J.W., Mehta I.K., Kim S., Dorner B.G., Naidenko O.V., Iizuka K., Furukawa H., Beckman D.L., Pingel J.T., et al. 2002. Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc. Natl. Acad. Sci. USA. 99:8826–8831. 10.1073/pnas.092258599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S., Watanabe T., Lin C.S., William C.M., Tanabe Y., Jessell T.M., and Costantini F.. 2001. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1:4 10.1186/1471-213X-1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J.C., Beilke J.N., and Lanier L.L.. 2009. Adaptive immune features of natural killer cells. Nature. 457:557–561. 10.1038/nature07665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J.C., Beilke J.N., and Lanier L.L.. 2010. Immune memory redefined: characterizing the longevity of natural killer cells. Immunol. Rev. 236:83–94. 10.1111/j.1600-065X.2010.00900.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J.C., Madera S., Bezman N.A., Beilke J.N., Kaplan M.H., and Lanier L.L.. 2012. Proinflammatory cytokine signaling required for the generation of natural killer cell memory. J. Exp. Med. 209:947–954. 10.1084/jem.20111760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Helden M.J., de Graaf N., Boog C.J., Topham D.J., Zaiss D.M., and Sijts A.J.. 2012. The bone marrow functions as the central site of proliferation for long-lived NK cells. J. Immunol. 189:2333–2337. 10.4049/jimmunol.1200008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vliegen I., Herngreen S., Grauls G., Bruggeman C., and Stassen F.. 2003. Improved detection and quantification of mouse cytomegalovirus by real-time PCR. Virus Res. 98:17–25. 10.1016/j.virusres.2003.08.009 [DOI] [PubMed] [Google Scholar]
- Zhang T., Scott J.M., Hwang I., and Kim S.. 2013. Cutting edge: antibody-dependent memory-like NK cells distinguished by FcRγ deficiency. J. Immunol. 190:1402–1406. 10.4049/jimmunol.1203034 [DOI] [PMC free article] [PubMed] [Google Scholar]